Abstract
Legionella pneumophila has been shown to utilize the icm/dot type IV secretion system for pathogenesis. This system was shown to be composed of icm/dot complex components and accessory proteins, as well as a large number of translocated substrates. Bioinformatic analysis of the regulatory regions of all the genes revealed that several icm/dot genes, as well as two genes encoding icm/dot translocated substrates, contain the conserved CpxR regulatory element, a regulator that has been shown previously to control the expression of the icmR gene. An experimental analysis, which included a comparison of gene expression in a L. pneumophila wild-type strain and gene expression in a cpxR deletion mutant, construction of mutants with mutations in the CpxR conserved regulatory elements, controlled expression studies, and mobility shift assays, demonstrated the direct relationship between the CpxR regulator and the expression of the genes. Furthermore, genomic analysis identified nine additional genes that contain a putative CpxR regulatory element; five of these genes (two legA genes and three ceg genes) were suggested previously to be putative icm/dot translocated substrates. The three ceg genes identified, which were shown previously to contain a putative PmrA regulatory element, were found here to be regulated by both CpxR and PmrA. The other six genes (two legA genes and four new genes products were found to be regulated by CpxR. Moreover, using the CyaA translocation assay, these nine gene products were found to be translocated into host cells in an Icm/Dot-dependent manner. Our results establish that the CpxR regulator is a fundamental regulator of the icm/dot type IV secretion system in L. pneumophila.
Legionella pneumophila, the causative agent of Legionnaires' disease, is an intracellular pathogen that multiplies in nature inside amoebae and in human macrophages during disease (24, 47). The icm/dot type IV secretion system is the major virulence system known in L. pneumophila; this system consists of 26 Icm/Dot proteins that probably constitute the secretion complex itself, as well as accessory proteins, such as chaperones (for a review, see reference 49). In the last 5 years, a growing number of protein substrates (RalF, LidA, Lep, Sid, Vip, Wip, Ylf, Leg, Vpd, Drr, and Ceg) that are translocated into host cells via the icm/dot secretion system were identified (for a review, see reference 42); in most cases the function of these proteins is not known, but two of them (SidF and SdhA) were shown to be required for inhibition of host cell death (5, 27) and four others (RalF, LidA, DrrA/SidM, and SidJ) were shown to be required for manipulation of host cell vesicular trafficking (28, 31, 37, 38), as well as other functions (11). Even though many icm/dot genes and many more icm/dot translocated substrates (IDTS), as well as potential IDTS, have been identified, there is only a limited amount of information about direct regulators that control the levels of expression of these genes.
A group of stationary-phase-related regulatory factors, including the stationary-phase sigma factor RpoS (2, 4, 20, 62), the two-component system LetAS (3, 19, 21, 30, 51), the ppGpp synthetase RelA (62), the translational repressor CsrA (16, 17, 36), the posttranscriptional regulator Hfq (32), and the response regulator LqsR (56), have been shown to be involved in L. pneumophila gene expression, activation of virulence traits, and physiological changes that occur in the stationary phase, but no direct target genes among the icm/dot genes themselves or among the IDTS-encoding genes (or any other genes) have been found for this regulatory network.
The only two regulators that were shown previously to directly regulate the expression of icm/dot genes or IDTS-encoding genes are PmrA and CpxR (18, 61). The PmrA response regulator, which is part of the PmrAB two-component system, was shown to directly regulate the expression of a large number of IDTS-encoding genes (61). The CpxR response regulator, which is part of the CpxRA two-component system, was shown to directly regulate the expression of icmR (18), as well as many functional homologues of icmR (encoding FIR proteins) in other Legionella species (15). In addition, after the discovery that CpxR is a direct regulator of icmR (18), this protein was identified in two additional screens related to pathogenesis. The CpxR regulator was found in a screen aimed at identification of suppressors of the IcmO/DotL lethality phenotype (57), as well as in a screen that was conducted to identify L. pneumophila mutants with defects in Rab1 recruitment (37). However, no additional genes directly regulated by CpxR were identified in these two screens.
The aims of the work described in this paper were to identify additional icm/dot-related genes (encoding complex components and accessory proteins, as well as IDTS) which are directly regulated by the CpxR response regulator and to characterize their roles in the icm/dot system. Our data clearly indicate that CpxR regulates the expression of different components of the icm/dot system, and its regulon includes at least 11 IDTS.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The L. pneumophila strains used in this study were L. pneumophila JR32, a streptomycin-resistant, restriction-negative mutant of L. pneumophila Philadelphia-1, which is a wild-type strain in terms of intracellular growth (48); OG2002, a cpxR mutant (18); HK-PQ1, a pmrA mutant (61); EA-CRPA, a cpxR pmrA double mutant (15); and GS3011, an icmT mutant (63). The Escherichia coli strains used were MC1022 containing the M15 deletion of the lacZ gene (9), MC1061 containing a deletion in the lacZ gene (10), BL21(DE3) (53), and cpxR deletion mutant PAD215, a kind gift from Thomas Silhavy. The bacterial media, plates, and antibiotic concentrations used have been described previously (50). The plasmids and primers used in this study are shown in Tables S1 and S2 in the supplemental material, respectively.
Construction of lacZ translational fusions.
To generate lacZ translational fusions, the regulatory regions of the genes of interest were amplified by PCR with the primers shown in Table S2 in the supplemental material. The PCR products were then digested with BamHI and EcoRI, cloned into pGS-lac-02, and sequenced to generate the plasmids shown in Table S1 in the supplemental material. The levels of expression from these plasmids were determined by the β-galactosidase assay as previously described (34, 61).
Generation of substitutions in the CpxR and PmrA binding sites.
To generate substitutions in the CpxR or PmrA binding sites in the regulatory regions of the genes examined, site-directed mutagenesis was performed with the consensus sequences by using the PCR overlap extension approach (22) and a method similar to the method described previously (61). In all the mutations generated the upstream part of the CpxR binding site was changed from GTAAA to TGAAA, and the upstream part of the PmrA binding site was changed from CTTAA to CTATA. The primers used for mutagenesis are shown in Table S2 in the supplemental material, and the plasmids resulting from site-directed mutagenesis are shown in Table S1 in the supplemental material.
Construction of isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible cpxR.
The L. pneumophila cpxR gene was amplified by PCR with primers CpxR-EcoRI and CpxR-pET-R (see Table S2 in the supplemental material). The PCR products were then digested with EcoRI and BamHI and cloned into pUC-18 to generate pEA-puc-CpxR; this plasmid was sequenced and then digested with the same enzymes and cloned into pMMB207 downstream from the Ptac promoter to generate pEA-pTac-CpxR. The resulting plasmid was then digested with XbaI and EheI, and the resulting fragment, containing Ptac-cpxR together with the lacI gene, was cloned into plasmids containing the regulatory region of the sidM and lvgA genes, as well as plasmids containing the mutations in the CpxR binding site of these two genes, resulting in plasmids shown in Table S1 in the supplemental material.
Construction of cyaA fusions.
Plasmid pMMB-cyaA-C (61) was used to clone all of the cyaA fusions constructed. All genes examined were amplified by PCR using a pair of primers (see Table S2 in the supplemental material) containing suitable restriction sites at the 5′ end. The PCR products were subsequently digested with the relevant enzymes and cloned into the pMMB-cyaA-C vector to generate plasmids shown in Table S1 in the supplemental material; all the inserts were sequenced to verify that no mutations were incorporated during the PCR. To construct cyaA fusions which were expressed under regulation of the natural promoter of the gene, the regulatory regions of the relevant genes were amplified by PCR using primers shown in Table S2 in the supplemental material, digested, and cloned into plasmids containing the corresponding cyaA fusion instead of the Ptac promoter to generate plasmids shown in Table S1 in the supplemental material.
Protein purification and gel mobility shift assays.
L. pneumophila His6-CpxR was purified from E. coli BL21(DE3) containing plasmids pOG-ECP2 (18) and pRep4 using nickel bead columns (Qiagen) according to the manufacturer's instructions. After purification, the fractions containing the protein were dialyzed against a buffer containing 20 mM Tris-HCl (pH 7.9), 50 mM KCl, 0.2 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, and 20% glycerol for 2 h and with the same buffer containing 30% glycerol overnight. The purified protein was then stored at −20°C. Gel mobility shift assays were performed as previously described (61), with a few modifications. The regulatory regions of the sidM, lvgA, and cegC2 genes (∼200 bp) were amplified by PCR with primers shown in Table S2 in the supplemental material and 3′ end labeled with digoxigenin (DIG) by using DIG-11-ddUTP (Roche). Increasing amounts of the purified proteins were mixed with 136 pg of the sidM-labeled probe, 154 pg of the lvgA-labeled probe, or 123 pg of the cegC2-labeled probe in buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 0.1 mM dithiothreitol, 0.1 mg/ml bovine serum albumin, 1 μg/ml poly(dI-dC), 5% glycerol, and 10 ng/ml herring sperm DNA. For samples containing unlabeled probe, 50 ng (for sidM and lvgA) or 150 ng (for cegC2) of the probe was allowed to bind the CpxR protein for 15 min before addition of the DIG-labeled probe. A binding reaction was carried out for 30 min at room temperature, and samples were then loaded onto 6% polyacrylamide-0.25× Tris-acetate-EDTA gels in 0.5× Tris-acetate-EDTA running buffer. Following electrophoresis, the gels were transferred to nylon membranes and fixed by UV cross-linking. The DIG-labeled DNA fragments were detected by following the manufacturer's instructions.
Quantitative RT-PCR.
RNA was prepared as described elsewhere (18). Total RNA was digested with an RNase-free DNase reagent (Promega). The reverse transcription (RT)-PCR mixture contained the following components: 10 μg of total RNA, 20 pmol of reverse primer (see Table S2 in the supplemental material), 40 U of RNase inhibitor (RNasin; Promega), and 10 U of avian myeloblastosis virus reverse transcriptase (Promega). The reaction mixture was incubated for 5 min at 45°C, and cDNA was synthesized at 37°C for 1 h. The following cycle conditions were used for PCR: 30 s at 95°C, 30 s at 50°C, and 30 s at 72°C for extension. The number of cycles varied depending on the levels of expression of the various mRNAs to ensure that the comparison was performed in the linear range of amplification. 16S rRNA was used as the internal control to confirm that equal amounts of total RNA were used in the reactions.
CyaA translocation assay.
Differentiated HL-60-derived human macrophages plated in 24-well tissue culture dishes at a concentration of 2.5 × 106 cells/well were used for the assay. Bacteria were grown on ABCYE [N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered charcoal yeast extract] plates containing chloramphenicol for 48 h. The bacteria were scraped off the plates and suspended in AYE (ACES-buffered yeast extract) medium, the optical density at 600 nm (OD600) was adjusted to 0.1 in AYE containing chloramphenicol, and the resulting cultures were grown on a roller drum for 17 to 18 h until an OD600 of about 3 (stationary phase) was reached. The bacteria were then diluted in fresh AYE medium to obtain an OD600 of 0.2 and grown for 2 h. IPTG was added to final concentration of 1 mM, and the cultures were grown for an additional 2 h. Cells were infected with bacteria harboring the appropriate plasmids at a multiplicity of infection of 4, and the plates were centrifuged at 180 × g for 5 min, followed by incubation at 37°C under CO2 (5%) for 2 h. Cells were then washed twice with ice-cold phosphate-buffered saline buffer (1.4 M NaCl, 27 mM KCl, 100 mM Na2HPO4, 18 mM KH2PO4) and lysed with 200 μl of lysis buffer (50 mM HCl, 0.1% Triton X-100) at 4°C for 30 min. Lysed samples were boiled for 5 min and neutralized with NaOH. The levels of cyclic AMP (cAMP) were determined using the cAMP Biotrak enzyme immunoassay system (Amersham Biosciences) according to the manufacturer's instructions. The CyaA fusion proteins were detected by Western blotting, using monoclonal anti-CyaA 3D1 antibody (Santa Cruz Biotechnology, Inc.) diluted 1:500 and goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (Jackson Immunoresearch Laboratories, Inc.) diluted 1:10,000.
RESULTS
Deletion in CpxR reduces the level of expression of several Icm/Dot components.
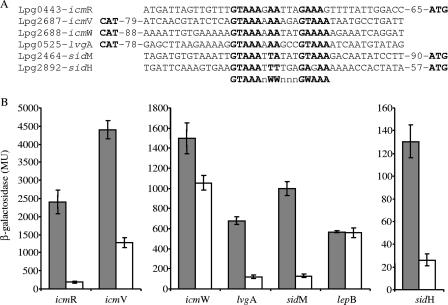
Previously, we demonstrated that the L. pneumophila CpxR regulator directly controls the expression of the icmR gene (18). In addition, the levels of expression of the icmV and icmW genes were also found to be reduced in the cpxR mutant strain (18, 57). Sequence analysis of the regulatory regions of all the icm/dot components, which include the icm/dot genes themselves, genes encoding accessory proteins (such as LvgA), and IDTS-encoding genes, indicated that a predicted CpxR binding site is present in the upstream regulatory region of several of these genes (Fig. 1A), including (i) the icmV and icmW genes, both of which are part of an operon along with one additional gene (icmV-dotA and icmW-icmX) and which were found to contain the CpxR regulatory element on the complementary strand (the CpxR regulatory element that was characterized previously in the icmR regulatory region was located on the direct strand [Fig. 1A]); (ii) the lvgA gene, whose product was shown previously to interact with the IcmS protein (58) and which was also found to contain the CpxR regulatory element on the complementary strand; and (iii) the sidH and sidM/drrA genes, whose products were shown previously to be translocated into host cells in an Icm/Dot-dependent manner (31, 37, 43) and which were found to contain the CpxR regulatory element on the direct strand.
FIG. 1.
Analysis of the expression of known pathogenesis-related genes containing the CpxR consensus sequence. (A) Sequence alignment of the regulatory regions of icm/dot genes (icmR, icmV, and icmW), the icm/dot accessory gene lvgA, and two IDTS-encoding genes (sidM and sidH). The putative consensus sequence is indicated by boldface type, and the distances to the first ATG codon are also indicated. The complementary strand is shown for genes in which the consensus regulatory element was identified on the reverse strand (icmV, icmW, and lvgA). (B) Expression of lacZ fusions in wild-type L. pneumophila and the cpxR deletion mutant. The expression of lacZ fusions of three icm/dot genes (icmR, icmV, and icmW), the lvgA gene, two IDTS-encoding genes (sidM and sidH) that contain the CpxR consensus regulatory element, and lepB that does not contain the consensus regulatory element in wild-type strain JR32 (shaded bars) and the cpxR deletion mutant OG2002 (open bars) was examined. β-Galactosidase activity was determined as described in Materials and Methods. The data (in Miller units [MU]) are the averages ± standard deviations (error bars) of at least three different experiments.
LacZ fusions were constructed for all the genes described above and examined to determine their levels of expression in the L. pneumophila wild-type strain and the cpxR deletion mutant, and they were all found to be expressed at lower levels in the cpxR mutant (Fig. 1B). As a negative control, an IDTS-encoding gene (lepB) that was shown previously to be regulated by the PmrA regulator (61), which belongs to the same family of regulators as CpxR (1), was not affected by deletion of the CpxR regulator (Fig. 1B). To further support the lacZ fusion results, the levels of expression of the icmV and sidM genes were also examined at the mRNA level using quantitative RT-PCR and were found to be reduced in the cpxR deletion mutant compared to the wild-type strain (Fig. 2). These results demonstrate that the CpxR regulator participates in the regulation of the pathogenesis-related genes examined and that it can also regulate the expression of genes when its regulatory element is located on the complementary strand (as it is in the case of icmV, icmW, and lvgA) (Fig. 1A).
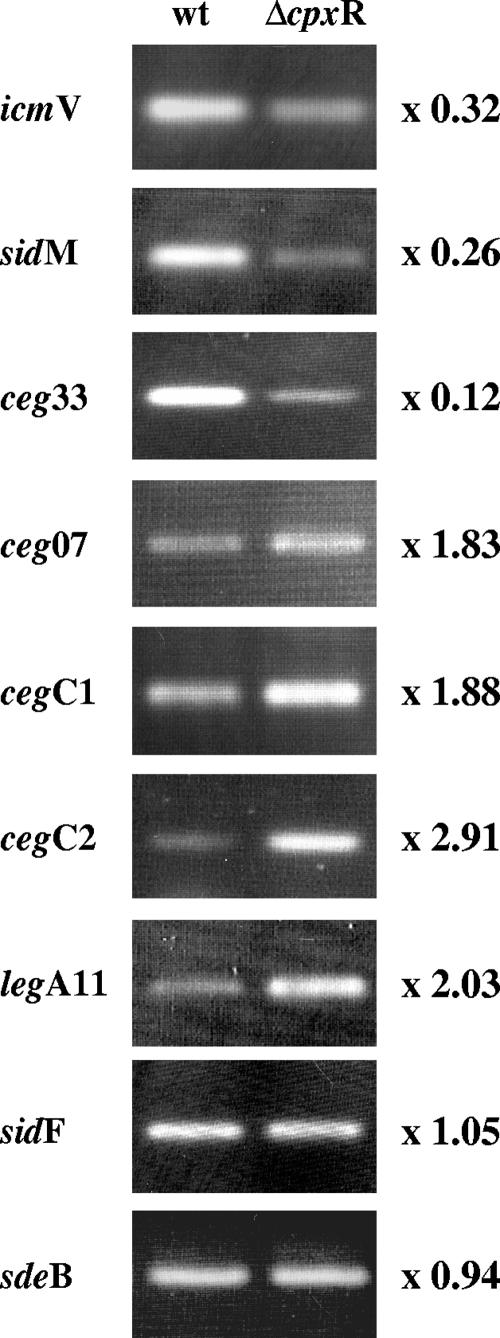
FIG. 2.
Analysis of mRNA levels for genes containing the CpxR consensus sequence in the wild type and a cpxR deletion mutant. Total RNA derived from the wild type (wt) and the cpxR deletion mutant (ΔcpxR) was used in RT-PCR to assess the mRNA levels for seven genes containing the CpxR consensus sequence (icmV, sidM, ceg33, ceg07, cegC1, cegC2, and legA11) and two genes that do not contain the CpxR consensus sequence (sidF and sdeB). The PCR was carried out for 19 cycles for icmV, legA11, and sidM, for 21 cycles for ceg07, ceg33, cegC1, sidF, and sdeB, and for 23 cycles for cegC2. Quantification (values are indicated on the right) was performed using the TINA image analysis software, and the values were obtained by dividing the mRNA level in the cpxR mutant by the mRNA level in the wild-type strain.
Consensus sequence identified is the target sequence of the CpxR response regulator.
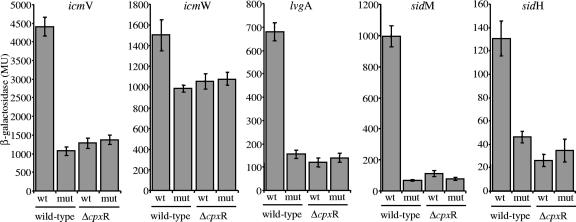
To examine the relationship between the consensus sequence identified (Fig. 1A) and the CpxR regulator, the consensus sequence of the five genes identified (icmV, icmW, lvgA, sidM, and sidH) was mutagenized by changing the upstream part of the consensus sequence from GTAAA to TGAAA, and the results of this analysis are shown in Fig. 3. The results obtained clearly demonstrate that the mutations in the regulatory element had the same effect as deletion of the CpxR regulator. Moreover, when the fusions containing the mutated consensus sequence were introduced into the cpxR deletion mutant, no additional reduction in the level of expression was observed (Fig. 3), supporting the conclusion that the conserved regulatory element identified is recognized by the CpxR response regulator itself.
FIG. 3.
Analysis of the relationship between the cpxR consensus and the CpxR regulator. Expression of icmV, icmW, lvgA, sidM, and sidH lacZ fusions in L. pneumophila was determined. For each of the fusions, data are shown for the wild-type regulatory region (wt) and the mutated regulatory region (mut) in the JR32 wild-type strain (wild-type) and cpxR mutant strain OG2002 (ΔcpxR). β-Galactosidase activity was determined as described in Materials and Methods. The data are the averages ± standard deviations (error bars) of at least three different experiments. MU, Miller units.
CpxR activates the expression of the genes containing the conserved regulatory element.
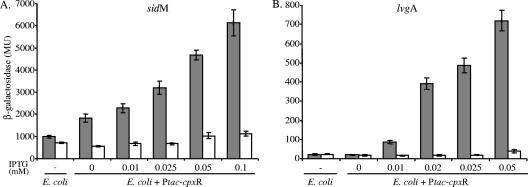
In order to define the role of CpxR in the regulation of the genes that contain the conserved regulatory element, the effect of CpxR in a surrogate host, E. coli, was examined. In this analysis the L. pneumophila cpxR gene was cloned under control of the Ptac promoter (induced by IPTG) and introduced into E. coli together with the sidM fusion (containing the CpxR binding site in the direct orientation) or the lvgA lacZ fusion (containing the CpxR binding site on the complementary strand). The results obtained in this analysis clearly demonstrated that the concentration of IPTG used (which controls the level of expression of the CpxR regulator) determined the levels of expression of the sidM and lvgA lacZ fusions in E. coli (Fig. 4), proving that CpxR has a role in their regulation. Moreover, the sidM and lvgA lacZ fusions that contained substitutions in the CpxR consensus sequence were not influenced by the addition of IPTG (Fig. 4). The results described here show that the CpxR response regulator is strongly related to the consensus sequence identified and show that it positively regulates the expression of the genes examined.
FIG. 4.
CpxR directly regulates the genes containing the consensus sequence. The expression of wild-type fusions (grey bars) of sidM::lacZ (A) and lvgA::lacZ (B), as well as the same fusions containing a mutation in the CpxR regulatory element (open bars), was examined in E. coli having a deletion in the cpxR gene (PAD215). The bacteria examined contained a plasmid with the L. pneumophila cpxR gene cloned under control of the Ptac promoter (activated by IPTG), and they were grown in media containing different concentrations of IPTG (indicated below the bars). Bacteria containing the same lacZ fusions without the cpxR gene were used as controls (left columns in both panels). β-Galactosidase activity was determined as described in Materials and Methods. The data are the averages ± standard deviations (error bars) of at least three different experiments. MU, Miller units.
L. pneumophila His6-CpxR protein directly binds to the regulatory regions of the genes examined.
To further support the direct relationship between the CpxR regulator and its binding site, the L. pneumophila CpxR protein was His tagged, overexpressed, purified, and used for gel mobility shift assays with ∼200-bp fragments that cover the sidM and lvgA regulatory regions. The L. pneumophila His6-CpxR was found to bind to the regulatory regions of both genes, as shown by a shift in the migration of the DNA probe (Fig. 5). The degree of the band shift and the amount of the shifted probe were correlated with increased amounts of the His6-CpxR protein (Fig. 5A and B). In addition, competition with an unlabeled probe reduced the band shift (Fig. 5A and B, compare the second and sixth lanes), and no effect was observed when the same amount of herring sperm DNA was added (Fig. 5A and B, compare the second and seventh lanes).
FIG. 5.
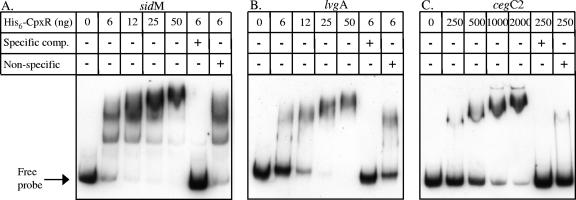
L. pneumophila His6-CpxR protein binds to the sidM, lvgA, and cegC2 regulatory regions. Mobility shift assays were performed with pure His6-CpxR protein and DIG-labeled sidM (136 pg) (A), lvgA (154 pg) (B), and cegC2 (123 pg) (C) regulatory regions in the presence (plus sign) or absence (minus sign) of unlabeled probe (Specific comp.) (50 ng in panels A and B and 150 ng in panel C) or the same amount of herring sperm DNA (Non-specific). The amount of His6-CpxR is indicated above each lane.
The mobility shift assay, together with the examination of gene expression in the cpxR null mutant, the controlled expression studies, and the analysis of mutations in the consensus sequence, strongly demonstrated the direct relationship between the CpxR regulator and its binding site, as well as its key role in the regulation of different components of the Icm/Dot secretion system in L. pneumophila.
Identification of new L. pneumophila CpxR-regulated genes.
The relative conservation of the CpxR regulatory element identified and the involvement of the CpxR-regulated genes in pathogenesis led us to search for additional genes that contain this conserved regulatory element in the L. pneumophila genome. This analysis was performed using whole-genome DNA pattern analysis (from the Regulatory Sequence Analysis Tools), which allowed one base mismatch from the consensus described in Fig. 1 only in the AAA part of the consensus sequence. The analysis which we performed resulted in identification of nine L. pneumophila genes that contain a putative CpxR consensus sequence (Table 1 and Fig. 6A and 7A). These genes include (i) two legA (Legionella eukaryotic gene) genes (lpg0038 [legA10] and lpg0436 [legA11]) that have been suggested previously to be putative IDTS-encoding genes due to the presence of an ankyrin domain (6, 12); (ii) three ceg (coregulated with effector-encoding genes) genes (lpg0227 [ceg7], lpg0898 [ceg18], and lpg2591 [ceg33]) that have been suggested previously to be putative IDTS-encoding genes due to the presence of a PmrA-encoding consensus sequence in their regulatory region (Fig. 7B) (61); and (iii) four cegC (coregulated with effector-encoding genes by CpxR) genes (lpg0012 [cegC1], lpg0126 [cegC2], lpg1144 [cegC3], and lpg2200 [cegC4]) that are unique genes with unknown functions that have not been described previously. A previous analysis of large groups of predicted L. pneumophila IDTS indicated that even though the genes encoding many of these proteins exhibit no homology to any entry in the GenBank database, several of them were found to have paralogues in the L. pneumophila genome (29), and it was also shown that the genes coding for some of these IDTS are occasionally located next to one another (28, 29, 61). Analysis of the genes identified (Table 1) revealed that three of these genes are located next to previously identified IDTS-encoding genes and three other genes have one or more paralogues in the L. pneumophila genome (Table 1).
TABLE 1.
Properties of proteins encoded by new genes containing the CpxR consensus sequence
| Genea | lpg no. | Nearby gene | Paralog(s) | Motif or domain |
|---|---|---|---|---|
| legA10 | lpg0038 | ceg1 (lpg0035) | Ankyrin repeat | |
| legA11 | lpg0436 | ceg14 (lpg0437) | Ankyrin repeat | |
| ceg7 | lpg0227 | lpg2248 | ||
| ceg18 | lpg0898 | |||
| ceg33 | lpg2591 | legS1 (lpg2588) | ||
| cegC1 | lpg0012 | lpg0502, lpg1455 | ||
| cegC2 | lpg0126 | lpg2433 (ceg30), lpg1120, lpg1484 (ceg22) | ||
| cegC3 | lpg1144 | Coiled coil | ||
| cegC4 | lpg2200 |
All the genes are also present in the genomes of L. pneumophila strains Lens, Paris, and Corby.
FIG. 6.
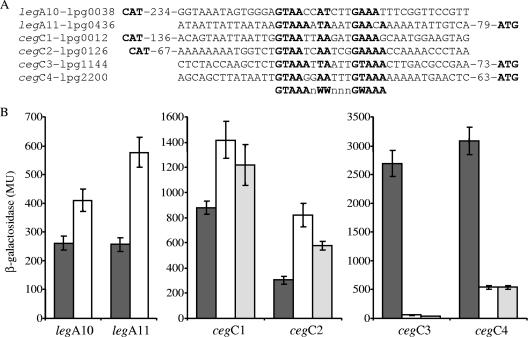
legA and cegC genes are regulated by the CpxR regulator. (A) Sequence alignment of the regulatory regions of the legA and cegC genes (legA10, legA11, cegC1, cegC2, cegC3, and cegC4). The putative consensus sequence is indicated by boldface type, and the distances to the first ATG codon are also indicated. The complementary strand is shown for genes (legA10, cegC1, and cegC2) in which the consensus regulatory element was identified on the reverse strand. (B) Expression of the four cegC genes and the two legA genes in L. pneumophila wild-type strain (dark grey bars) and cpxR mutant strain OG2002 (open bars). For the cegC genes lacZ fusions containing a mutation in the CpxR consensus sequence were also examined in the L. pneumophila wild-type strain (light grey bars). β-Galactosidase activity was determined as described in Materials and Methods. The data are the averages ± standard deviations (error bars) of at least three different experiments. MU, Miller units.
FIG. 7.
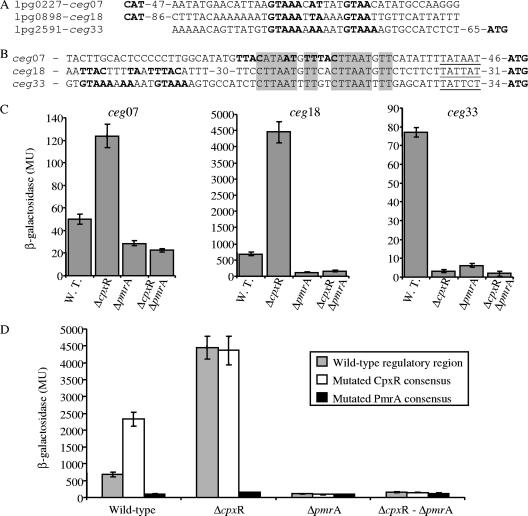
ceg genes are regulated by the CpxR and PmrA regulators. (A and B) Sequence alignment of the regulatory regions of the three ceg genes according to the CpxR consensus sequence (A) or the PmrA consensus sequence (B). The putative PmrA consensus sequence is indicated by shading, the putative CpxR consensus sequence is indicated by boldface type, the putative −10 promoter elements are underlined, and the distances to the first ATG codon are also indicated. (C) Expression of the three lacZ fusions in wild-type L. pneumophila (W.T.), cpxR deletion mutant OG2002 (ΔcpxR), pmrA deletion mutant HK-PQ1 (ΔpmrA), and cpxR pmrA double-deletion mutant EA-CRPA (ΔcpxR ΔpmrA). (D) Expression of the wild-type ceg18::lacZ fusion (grey bars), the ceg18::lacZ fusion containing a mutation in the CpxR consensus sequence (open bars), and the ceg18::lacZ fusion containing a mutation in the PmrA consensus sequence (black bars) in wild-type L. pneumophila (Wild-type), cpxR deletion mutant OG2002 (ΔcpxR), pmrA deletion mutant HK-PQ1 (ΔpmrA), and cpxR pmrA double-deletion mutant EA-CRPA (ΔcpxR-ΔpmrA). β-Galactosidase activity was determined as described in the Materials and Methods. The data are the averages ± standard deviations (error bars) of at least three different experiments. MU, Miller units.
Four cegC genes and two legA genes are regulated by CpxR.
To examine if the four cegC genes and two legA genes identified are regulated by CpxR, they were all fused to the lacZ reporter. The six resulting fusions were examined to determine their levels of expression in the L. pneumophila wild-type strain compared to their levels of expression in a cpxR deletion mutant. Similar to the results obtained for the CpxR-regulated genes described above, the levels of expression of all the genes examined were found to be affected in the cpxR mutant compared to the wild-type strain (Fig. 6B). However, the levels of expression of some of the genes examined were found to be higher in the cpxR mutant strain than in the wild-type strain, indicating that CpxR acts as a repressor for these genes, a result similar to the results described previously for the CpxR regulator in other systems (44). In addition, the CpxR consensus sequence of the four cegC genes was mutated, and when the mutated fusions were examined in the L. pneumophila wild-type strain, effects (increases in the levels of expression of cegC1 and cegC2 and decreases in the levels of expression of cegC3 and cegC4) similar to the effects obtained when the corresponding wild-type fusions were examined in the cpxR deletion mutant were obtained (Fig. 6B). To confirm the CpxR repression observed, the cegC1, cegC2, and legA11 genes were also examined at the mRNA level using quantitative RT-PCR, and the levels of their mRNAs were found to be higher in the cpxR deletion mutant (Fig. 2), as expected based on the lacZ fusions results. Furthermore, since the two genes (sidM and lvgA) that were examined in the gel mobility shift assay were activated by CpxR, we examined the regulatory region of the cegC2 gene (repressed by CpxR) using this assay as well. L. pneumophila His6-CpxR was found to bind to the regulatory regions of the cegC2 gene, as shown by a shift in the migration of the DNA probe (Fig. 5C). Increases in the size of the band shift and in the amount of the shifted probe were correlated with increased amounts of the His6-CpxR protein (Fig. 5C). It is interesting that no band shift was observed when the assay was performed with amounts of the CpxR protein that were sufficient to obtain a clear band shift with sidM and lvgA (between 6 and 50 ng) and that larger amounts of CpxR (250 ng) were required to obtain a clear band shift. These results might indicate that the CpxR protein has higher affinity for the sites on which it acts as an activator than for the sites on which it acts as a repressor, or they might be related to the degree of identity of each site to the CpxR consensus sequence.
Three ceg genes are regulated by both PmrA and CpxR.
In a previous study we found that PmrA regulates the expression of many L. pneumophila IDTS-encoding genes; in addition, the PmrA consensus sequence was found in the regulatory region of 35 uncharacterized genes (61). As described above, three of these genes (ceg7, ceg18, and ceg33) were also found to contain a putative CpxR binding site (Fig. 7A). The organizations of the CpxR and PmrA putative regulatory elements in relation to one another were found to be different in these three genes. In ceg7 the two sites overlap, and the CpxR site was found to be located on the complementary strand; in ceg18 the two sites were found to be located about 35 bp apart, and the CpxR site was located on the complementary strand; and in ceg33 the two sites were found to be located about 10 bp apart, and they both were located on the direct strand (Fig. 7B) (this type of organization was observed previously for the fir genes in different Legionella species [15]). To investigate the effect of the CpxR and PmrA regulators on the levels of expression of these genes, three lacZ fusions were constructed and examined in the L. pneumophila wild-type strain, the cpxR deletion mutant (OG2002), the pmrA deletion mutant (HK-PQ1), and the cpxR pmrA double mutant (EF-CRPA), and the results are shown in Fig. 7C. The results obtained clearly show that both the CpxR and PmrA regulators affect the expression of the three genes examined. PmrA always serves as an activator, and CpxR serves as a repressor for two of the genes (ceg7 and ceg18) and as an activator for ceg33. The effect of the cpxR deletion mutation on the expression of the ceg7 and ceg33 genes was also examined at the mRNA level, and the results were similar to the results obtained for the lacZ fusions (Fig. 2). The data indicated very clearly that the results for the cpxR pmrA double mutant were always the same as the results for the pmrA single mutant, indicating that the relief of repression that was observed in the cpxR deletion mutant with ceg7 and ceg18 requires activation of the PmrA regulator. The effects of both regulators on the level of expression of the ceg18::lacZ fusion were the most pronounced effects and opposite each other (a 6.5-fold increase in the cpxR mutant and 6-fold decrease in the pmrA mutant). Therefore, this gene was analyzed further by constructing mutants with mutations in both the pmrA and cpxR putative regulatory elements, and the resulting fusions were analyzed in the four strains indicated above; the results are shown in Fig. 7D. The results clearly demonstrated that the expression of the ceg18 gene is completely dependent on the presence of the PmrA regulatory element and the PmrA regulator, since no expression was obtained when either of these two factors was missing. Mutations in the cpxR binding site, in the CpxR regulator, or in both resulted in an increase in the level of expression of the gene, but this effect was observed only if the pmrA regulatory element and the PmrA regulator were intact, indicating that CpxR represses the PmrA activation of this gene.
Nine new genes identified code for new L. pneumophila IDTS.
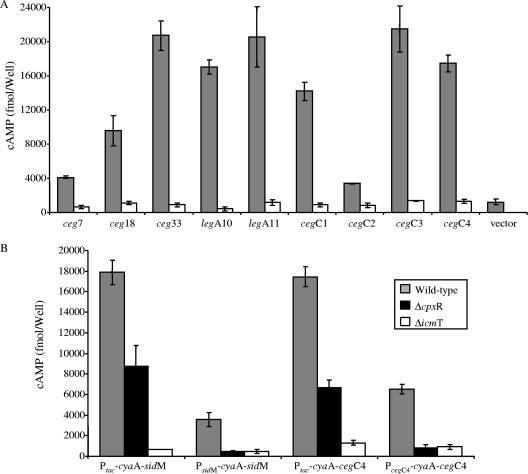
To determine whether the proteins encoded by the nine new CpxR-regulated genes identified (two legA genes, three ceg genes, and four cegC genes) are translocated into host cells by the L. pneumophila icm/dot system, the Bordetella pertussis CyaA reporter gene fusion that has been used previously to study the translocation of many bacterial effectors into eukaryotic cells (52) was utilized. The legA10, legA11, ceg7, ceg18, ceg33, cegC1, cegC2, cegC3, and cegC4 genes were fused to the CyaA reporter at their N-terminal ends (leaving their C-terminal ends exposed) and transformed into wild-type strain JR32 and an icmT mutant in which the icm/dot system is inactive. HL-60-derived human macrophages were infected with L. pneumophila expressing the CyaA hybrid proteins or unfused CyaA, and translocation was examined (Fig. 8A). Infection with bacteria containing all the genes examined resulted in very large differences in the cAMP levels produced when the wild-type and icmT mutant strains were used (although the levels of the CyaA fusion proteins were similar, as deduced by Western blot analysis using a CyaA antibody [data not shown]), indicating that a functional Icm/Dot secretion system was required for translocation (Fig. 8A). A construct expressing only CyaA or several unrelated proteins fused to the CyaA reporter resulted in very low levels of cAMP (Fig. 8A and data not shown), indicating that increased cAMP levels required the presence of the proteins examined. We concluded that the nine proteins examined (five proteins that were suggested previously to be putative IDTS, as well as the four newly identified proteins) that contain the CpxR consensus sequence in their regulatory regions are new IDTS.
FIG. 8.
Icm/Dot-dependent translocation of CpxR-regulated genes. (A) Wild-type strain JR32 (grey bars) and icmT mutant GS3011 (open bars) harboring the CyaA fusion proteins (indicated below the bars) were used to infect HL-60-derived human macrophages, and the cAMP levels of the infected cells were determined as described in Materials and Methods. (B) Effect of the cpxR deletion mutant on the translocation of sidM and cegC4 cyaA fusions in wild-type strain JR32 (grey bars), cpxR mutant OG2002 (black bars), and icmT mutant GS3011 (open bars). The fusions were cloned under control of Ptac or under control of their natural promoter. The data are the means for the amount of cAMP per well obtained in three independent experiments, and the error bars indicate standard deviations.
Deletion in cpxR affects IDTS translocation into host cells.
To examine the effect of the cpxR deletion on IDTS translocation, the translocation of two cyaA fusions (sidM and cegC4) was examined. These two cyaA fusions were examined when they were expressed from the Ptac promoter (induced by IPTG), as well as when each of them was expressed from its natural promoter (which should reflect the overall effect of the cpxR deletion on the translocation of the genes). When the genes were under Ptac control, high levels of translocation were observed in the wild-type strain for both fusions, and a clear reduction in translocation was seen in the cpxR deletion mutant (Fig. 8B). The reduction in translocation was probably due to the effect of the cpxR regulator on the expression of icm/dot components (see above). When the same cyaA fusions were expressed from their natural promoters (PsidM-cyaA-sidM and PcegC4-cyaA-cegC4), moderate levels of translocation were observed in the wild-type strain and a dramatic reduction in translocation was observed in the cpxR deletion mutant (Fig. 8B). The reduction in translocation most likely resulted from the combined effects of the cpxR deletion mutation on the level of expression of icm/dot components and the level of expression of the IDTS themselves. These results established that the CpxR regulator is a fundamental regulator of the icm/dot type IV secretion system in L. pneumophila.
DISCUSSION
The L. pneumophila Icm/Dot type IV secretion system is composed of about 23 complex components, three accessory proteins, and a large number of IDTS (for reviews, see references 42 and 49), and many additional proteins have been suggested to be putative IDTS (6, 12, 29, 61). This very large number of components must be coordinated at the level of gene expression in order to result in a successful infection and intracellular multiplication.
In a previous study, we identified the PmrA response regulator as a direct regulator of 16 IDTS, and 32 potential IDTS were found to contain a putative PmrA site. In addition, the PmrA regulator was found to be required for intracellular growth in the protozoan host Acanthamoeba castellanii and to be partially required for intracellular growth in HL-60-derived human macrophages (61). Several years ago, we identified the L. pneumophila CpxR response regulator as a direct regulator of one of the icm/dot genes (icmR), and it was also shown to affect the level of expression of several other icm/dot genes. However, the CpxR regulator was found to be dispensable for intracellular growth in the two hosts mentioned above (18). In this study, we expanded the analysis of icm/dot components that are regulated by CpxR and found that the icmR and lvgA genes, the icmW-icmX and icmV-dotA transcriptional units, and 11 IDTS, 9 of which are new IDTS that were validated in this study, are regulated by CpxR (Fig. 9). The identification of CpxR as an activator or repressor of IDTS (which might indicate that different IDTS are required at different times during infection), as well as an activator of icm/dot genes, might seem to conflict with the lack of an intracellular growth phenotype. One explanation for these results might be related to the very large number of IDTS in L. pneumophila, many of which have redundant functions (42). It is possible that some of the IDTS regulated by CpxR have functionally redundant IDTS which are not regulated by CpxR and that these proteins are expressed and perform their functions also in the cpxR deletion mutant. A second explanation might be related to the very large number of different protozoans that were shown to serve as hosts for L. pneumophila. It might be that replication in different hosts requires different sets of IDTS (overlapping or nonoverlapping), and it is possible that the set of IDTS which is regulated by CpxR is less critical for intracellular growth in the two hosts examined; however, the cpxR mutant might have an intracellular growth phenotype in a different host similar to the phenotype observed for the pmrA mutant in A. castellanii. Even though no intracellular growth defect was observed for the cpxR deletion mutant, the CpxR regulator was found to affect several pathogenesis-related phenotypes: (i) the CpxR regulator was found in a screen aimed at identification of suppressors of the IcmO/DotL lethality phenotype (57); (ii) the CpxR regulator was identified in a screen that was conducted to identify L. pneumophila mutants with defects in Rab1 recruitment (37); and (iii) the cpxR deletion mutation was found to strongly affect IDTS translocation (this study). All these results indicate that CpxR is a central regulator of L. pneumophila pathogenesis.
FIG. 9.
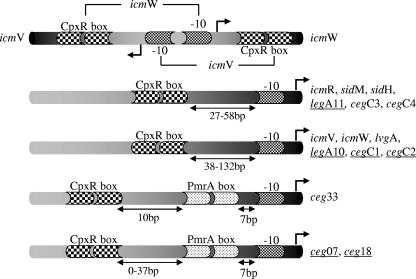
Schematic diagrams of CpxR-regulated promoters. The CpxR boxes, PmrA boxes, −10 promoter elements, and transcription start sites (arrow) are indicated along with their orientations with respect to the first ATG codon of the gene (black region). For icmV and icmW (upper bar) the relevant regulatory elements for each of the two genes are indicated. The genes that were found to have a certain type of regulation are indicated on the right; the genes that were found to be repressed by CpxR are underlined.
Although the PmrA and CpxR regulators belong to the same group of DNA binding proteins containing a winged helix-turn-helix motif (1) and are both involved in the regulation of L. pneumophila pathogenesis-related genes, their effects on their target genes and the locations of their regulatory elements were found to be different. The PmrA binding sites were found to be located at a precise distance from the −10 promoter, and they were always found to be located on the direct strand (61). In contrast, the regulatory elements of the CpxR regulator were found to be located at different distances from the predicted −10 promoter element and on both the direct and complementary strands (Fig. 9). A situation similar to the situation described here has been observed for the CpxR regulators in other bacteria (14). Besides the differences in the locations of the two regulators and in the orientations of their regulatory elements, the PmrA regulator was found to function as an activator of its target genes in all the genes examined thus far, whereas the CpxR regulator was found to function as a repressor or as an activator of its target genes (Fig. 9). The CpxR regulator was also shown to function as an activator as well as a repressor in other bacteria (44).
The CpxRA and PmrAB two-component systems have both been found to be involved in the pathogenesis of bacteria other than L. pneumophila. The CpxR response regulator has been shown to be involved in the pathogenesis of Salmonella enterica, Shigella sonnei, enteropathogenic E. coli, and Yersinia sp., and in most cases its involvement was related to contact with eukaryotic cells (7, 8, 25, 40, 41). The PmrA response regulator has been shown to be involved in the virulence of S. enterica, Pseudomonas aeruginosa, and Francisella sp., and its involvement was mainly related to its relationship with the PhoP response regulator and resistance to antimicrobial peptides (13, 33, 35, 55). The CpxR and PmrA regulators are assumed to function together with their cognate sensor histidine kinases (CpxA and PmrB, respectively) (Fig. 10), which are located immediately downstream from them. Since these two response regulators are key regulators of the L. pneumophila icm/dot pathogenesis system, it is important to determine the environmental signals that control PmrB and CpxA phosphorylation, as these stimuli might provide clues about how the intracellular environment influences the expression of the different IDTS. Currently, the environmental signals that control the phosphorylation of L. pneumophila CpxA and PmrB are not known, but analysis of the activation of these systems in other bacteria produced interesting and relevant findings. The CpxRA system in E. coli was shown to be activated in response to periplasmic stress and accumulation of unfolded proteins in the periplasm, and this activation was shown to involve the CpxP protein (26, 46); however, no CpxP homolog was found to be present in L. pneumophila. In S. enterica, the CpxA protein was shown to be involved in sensing low pH (39). The S. enterica PmrAB system was shown to be activated by low pH, as well as in response to high levels of Fe(III) (45, 60). Since L. pneumophila resides in a phagosome that does not fuse with lysosomes early during infection (23, 59) but it was shown to acquire lysosomal markers late during infection (54), it is tempting to believe that one or both of these sensor histidine kinases respond to changes in pH. However, since we found three ceg genes which were shown here to be regulated by both CpxR and PmrA and since for two of these genes CpxR was found to act as a repressor and PmrA was found to act as an activator, it is unlikely that both CpxA and PmrB sense pH and that they control the levels of expression of the same target genes in opposite ways. The environmental stimuli that activate the PmrAB and CpxRA two-component systems in L. pneumophila and the time during infection when these two-component systems are activated remain to be elucidated.
FIG. 10.
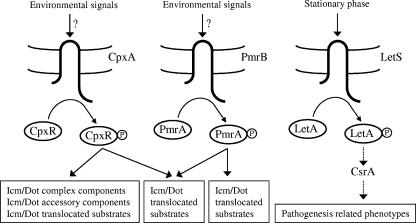
Model of regulation of the L. pneumophila Icm/Dot pathogenesis system. The three two-component systems (CpxRA, PmrAB, and LetAS) which were found to be involved in the regulation of pathogenesis-related genes and/or phenotypes are shown. The environmental signals sensed by CpxA and PmrB are currently not known, and the phosphorylation of these components is expected to activate by transfer of the phosphate group to the cognate response regulators CpxR and PmrA, respectively, which then activate or repress the transcription of their target genes. The target genes of CpxR and PmrA overlap in some cases, but only CpxR was found to control the expression of icm/dot genes; both regulators control the expression of IDTS-encoding genes. No target genes are currently known for the LetA-CsrA regulatory cascade. The dotted line indicates connections that might be indirect.
Supplementary Material
Acknowledgments
We thank Thomas J. Silhavy for the E. coli cpxR deletion mutant. We thank Michal Feldman for her help with the mobility shift assays and Elena Degtyar for her help with the CyaA translocation system.
This work was supported by grant 393/03 from the Israeli Science Foundation to G.S.
Footnotes
Published ahead of print on 11 January 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aravind, L., V. Anantharaman, S. Balaji, M. M. Babu, and L. M. Iyer. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29231-262. [DOI] [PubMed] [Google Scholar]
- 2.Bachman, M. A., and M. S. Swanson. 2004. Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect. Immun. 722468-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachman, M. A., and M. S. Swanson. 2004. The LetE protein enhances expression of multiple LetA/LetS-dependent transmission traits by Legionella pneumophila. Infect. Immun. 723284-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 401201-1214. [DOI] [PubMed] [Google Scholar]
- 5.Banga, S., P. Gao, X. Shen, V. Fiscus, W. X. Zong, L. Chen, and Z. Q. Luo. 2007. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc. Natl. Acad. Sci. USA 1045121-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruggemann, H., C. Cazalet, and C. Buchrieser. 2006. Adaptation of Legionella pneumophila to the host environment: role of protein secretion, effectors and eukaryotic-like proteins. Curr. Opin. Microbiol. 986-94. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson, K. E., J. Liu, P. J. Edqvist, and M. S. Francis. 2007. Extracytoplasmic-stress-responsive pathways modulate type III secretion in Yersinia pseudotuberculosis. Infect. Immun. 753913-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlsson, K. E., J. Liu, P. J. Edqvist, and M. S. Francis. 2007. Influence of the Cpx extracytoplasmic-stress-responsive pathway on Yersinia sp. -eukaryotic cell contact. Infect. Immun. 754386-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadaban, M. J., J. Chou, and S. N. Cohen. 1980. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J. Bacteriol. 143971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138179-207. [DOI] [PubMed] [Google Scholar]
- 11.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 3031358-1361. [DOI] [PubMed] [Google Scholar]
- 12.de Felipe, K. S., S. Pampou, O. S. Jovanovic, C. D. Pericone, S. F. Ye, S. Kalachikov, and H. A. Shuman. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 1877716-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delgado, M. A., C. Mouslim, and E. A. Groisman. 2006. The PmrA/PmrB and RcsC/YojN/RcsB systems control expression of the Salmonella O-antigen chain length determinant. Mol. Microbiol. 6039-50. [DOI] [PubMed] [Google Scholar]
- 14.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 27726652-26661. [DOI] [PubMed] [Google Scholar]
- 15.Feldman, M., and G. Segal. 2007. A pair of highly conserved two-component systems participates in the regulation of the hypervariable FIR proteins in different Legionella species. J. Bacteriol. 1893382-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fettes, P. S., V. Forsbach-Birk, D. Lynch, and R. Marre. 2001. Overexpression of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int. J. Med. Microbiol. 291353-360. [DOI] [PubMed] [Google Scholar]
- 17.Forsbach-Birk, V., T. McNealy, C. Shi, D. Lynch, and R. Marre. 2004. Reduced expression of the global regulator protein CsrA in Legionella pneumophila affects virulence-associated regulators and growth in Acanthamoeba castellanii. Int. J. Med. Microbiol. 29415-25. [DOI] [PubMed] [Google Scholar]
- 18.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 1854908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gal-Mor, O., and G. Segal. 2003. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb. Pathog. 34187-194. [DOI] [PubMed] [Google Scholar]
- 20.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 1814879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44107-118. [DOI] [PubMed] [Google Scholar]
- 22.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 23.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 1582108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Investig. 60441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humphreys, S., G. Rowley, A. Stevenson, M. F. Anjum, M. J. Woodward, S. Gilbert, J. Kormanec, and M. Roberts. 2004. Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect. Immun. 724654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isaac, D. D., J. S. Pinkner, S. J. Hultgren, and T. J. Silhavy. 2005. The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. Proc. Natl. Acad. Sci. USA 10217775-17779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laguna, R. K., E. A. Creasey, Z. Li, N. Valtz, and R. R. Isberg. 2006. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. USA 10318745-18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, Y., and Z. Q. Luo. 2007. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect. Immun. 75592-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo, Z. Q., and R. R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch, D., N. Fieser, K. Gloggler, V. Forsbach-Birk, and R. Marre. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol. Lett. 219241-248. [DOI] [PubMed] [Google Scholar]
- 31.Machner, M. P., and R. R. Isberg. 2006. Targeting of host rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 1147-56. [DOI] [PubMed] [Google Scholar]
- 32.McNealy, T. L., V. Forsbach-Birk, C. Shi, and R. Marre. 2005. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J. Bacteriol. 1871527-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPhee, J. B., S. Lewenza, and R. E. Hancock. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50205-217. [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1972. Experiments in molecular biology. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Mohapatra, N. P., S. Soni, B. L. Bell, R. Warren, R. K. Ernst, A. Muszynski, R. W. Carlson, and J. S. Gunn. 2007. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect. Immun. 753305-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molofsky, A. B., and M. S. Swanson. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50445-461. [DOI] [PubMed] [Google Scholar]
- 37.Murata, T., A. Delprato, A. Ingmundson, D. K. Toomre, D. G. Lambright, and C. R. Roy. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8971-977. [DOI] [PubMed] [Google Scholar]
- 38.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295679-682. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama, S., A. Kushiro, T. Asahara, R. Tanaka, L. Hu, D. J. Kopecko, and H. Watanabe. 2003. Activation of hilA expression at low pH requires the signal sensor CpxA, but not the cognate response regulator CpxR, in Salmonella enterica serovar Typhimurium. Microbiology 1492809-2817. [DOI] [PubMed] [Google Scholar]
- 40.Nakayama, S., and H. Watanabe. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 1803522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nevesinjac, A. Z., and T. L. Raivio. 2005. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J. Bacteriol. 187672-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ninio, S., and C. R. Roy. 2007. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15372-380. [DOI] [PubMed] [Google Scholar]
- 43.Ninio, S., D. M. Zuckman-Cholon, E. D. Cambronne, and C. R. Roy. 2005. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 55912-926. [DOI] [PubMed] [Google Scholar]
- 44.Ogasawara, H., J. Teramoto, K. Hirao, K. Yamamoto, A. Ishihama, and R. Utsumi. 2004. Negative regulation of DNA repair gene (ung) expression by the CpxR/CpxA two-component system in Escherichia coli K-12 and induction of mutations by increased expression of CpxR. J. Bacteriol. 1868317-8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez, J. C., and E. A. Groisman. 2007. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol. Microbiol. 63283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raivio, T. L., and T. J. Silhavy. 1999. The σE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2159-165. [DOI] [PubMed] [Google Scholar]
- 47.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 331179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 615361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segal, G., M. Feldman, and T. Zusman. 2005. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 2965-81. [DOI] [PubMed] [Google Scholar]
- 50.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 655057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi, C., V. Forsbach-Birk, R. Marre, and T. L. McNealy. 2006. The Legionella pneumophila global regulatory protein LetA affects DotA and Mip. Int. J. Med. Microbiol. 29615-24. [DOI] [PubMed] [Google Scholar]
- 52.Sory, M. P., A. Boland, I. Lambermont, and G. R. Cornelis. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. USA 9211998-112002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 54.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 1921261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamayo, R., S. S. Ryan, A. J. McCoy, and J. S. Gunn. 2002. Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar Typhimurium. Infect. Immun. 706770-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tiaden, A., T. Spirig, S. S. Weber, H. Bruggemann, R. Bosshard, C. Buchrieser, and H. Hilbi. 2007. The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell. Microbiol. 92903-2920. [DOI] [PubMed] [Google Scholar]
- 57.Vincent, C. D., B. A. Buscher, J. R. Friedman, L. A. Williams, P. Bardill, and J. P. Vogel. 2006. Identification of non-dot/icm suppressors of the Legionella pneumophila ΔdotL lethality phenotype. J. Bacteriol. 1888231-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vincent, C. D., and J. P. Vogel. 2006. The Legionella pneumophila IcmS-LvgA protein complex is important for Dot/Icm-dependent intracellular growth. Mol. Microbiol. 61596-613. [DOI] [PubMed] [Google Scholar]
- 59.Wiater, L. A., K. Dunn, F. R. Maxfield, and H. A. Shuman. 1998. Early events in phagosome establishment are required for intracellular survival of Legionella pneumophila. Infect. Immun. 664450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wosten, M. M., L. F. Kox, S. Chamnongpol, F. C. Soncini, and E. A. Groisman. 2000. A signal transduction system that responds to extracellular iron. Cell 103113-125. [DOI] [PubMed] [Google Scholar]
- 61.Zusman, T., G. Aloni, E. Halperin, H. Kotzer, E. Degtyar, M. Feldman, and G. Segal. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 631508-1523. [DOI] [PubMed] [Google Scholar]
- 62.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and the role of RelA and RpoS in virulence gene expression. J. Bacteriol. 18467-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zusman, T., G. Yerushalmi, and G. Segal. 2003. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect. Immun. 713714-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.