Abstract
Replication of human cytomegalovirus (HCMV) produces large DNA concatemers of head-to-tail-linked viral genomes that upon packaging into capsids are cut into unit-length genomes. The mechanisms underlying cleavage-packaging and the subsequent steps prior to nuclear egress of DNA-filled capsids are incompletely understood. The hitherto uncharacterized product of the essential HCMV UL52 gene was proposed to participate in these processes. To investigate the function of pUL52, we constructed a ΔUL52 mutant as well as a complementing cell line. We found that replication of viral DNA was not impaired in noncomplementing cells infected with the ΔUL52 virus, but viral concatemers remained uncleaved. Since the subnuclear localization of the known cleavage-packaging proteins pUL56, pUL89, and pUL104 was unchanged in ΔUL52-infected fibroblasts, pUL52 does not seem to act via these proteins. Electron microscopy studies revealed only B capsids in the nuclei of ΔUL52-infected cells, indicating that the mutant virus has a defect in encapsidation of viral DNA. Generation of recombinant HCMV genomes encoding epitope-tagged pUL52 versions showed that only the N-terminally tagged pUL52 supported viral growth, suggesting that the C terminus is crucial for its function. pUL52 was expressed as a 75-kDa protein with true late kinetics. It localized preferentially to the nuclei of infected cells and was found to enclose the replication compartments. Taken together, our results demonstrate an essential role for pUL52 in cleavage-packaging of HCMV DNA. Given its unique subnuclear localization, the function of pUL52 might be distinct from that of other cleavage-packaging proteins.
The infection cycle of human cytomegalovirus (HCMV) comprises a phase in the cell nucleus where genome replication and assembly of new capsids take place (24). Replication of the 230-kbp viral DNA genome leads to the formation of concatemers of head-to-tail-linked viral genomes, which are believed to be highly branched. These concatemers are subsequently cleaved into unit-length genomes, which are packaged into preformed capsids. The DNA-filled capsids associate with some tegument proteins at the nuclear membrane and then are transferred into the cytoplasm, where they undergo further coating with tegument proteins. Final envelopment of the capsids most likely occurs in a cytoplasmic virus assembly compartment, which partly overlaps with and is possibly derived from the trans-Golgi network (30).
Cleavage-packaging of HCMV genomes and capsid maturation in the nucleus are not completely understood, yet several viral proteins have been implicated as involved in these processes. The HCMV terminase responsible for cleavage of concatemeric DNA was shown to consist of two essential proteins, pUL56 and pUL89 (31, 36). pUL56 binds to viral capsids as well as to the packaging signal located in the a-repeat of the HCMV genome and has been shown to possess ATPase activity. pUL89 directly interacts with pUL56 and seems to be required mainly for DNA cleavage (4, 19, 31). pUL104 is supposed to form the portal at one vertex of the capsid through which the viral genome is translocated (13).
Assembly of herpesvirus capsids commences with the arrangement of capsid and scaffold proteins, giving rise to spherical, unstable procapsids, which mature into stable, angular B capsids (for reviews, see references 17 and 29). Successful packaging of viral DNA results in sealed, DNA-filled C capsids, whereas A capsids, which contain neither DNA nor scaffold, are thought to arise from abortive DNA-packaging events. For nuclear egress, the nucleocapsids first have to overcome certain barriers in the nucleus such as the host cell chromatin and the lamin meshwork to reach the inner nuclear membrane. Budding at this membrane leads to enveloped particles in the perinuclear space. The following steps are discussed controversially (22, 29), with the currently most favored model proposing a de-envelopment/reenvelopment process, which occurs by fusion of the primary viral envelope with the outer nuclear membrane, releasing the capsids into the cytoplasm.
In addition to the proteins mentioned above, it is most likely that a number of additional viral factors play a role in cleavage-packaging of the virus genome and in the subsequent steps of maturation, tegumentation, and nuclear egress of the DNA-filled capsids. Among these are the HCMV proteins pUL51, pUL52, and pUL77, which are homologous to the herpes simplex virus type 1 (HSV-1) proteins pUL33, pUL32, and pUL25, respectively. To date, the knowledge about these HSV-1 proteins is limited, and no function has been assigned to their HCMV counterparts. HSV-1 pUL33 interacts with the HSV-1 terminase proteins pUL15 and pUL28 (2) as well as with the portal vertex pUL6, suggesting that pUL33 may help in stabilizing the interaction between the terminase subunits and/or in translocating viral DNA into the capsid (1, 41). HSV-1 pUL25 is not required for DNA cleavage but is required for efficient encapsidation (23, 34). There is now strong evidence that pUL25 forms a complex with pUL17 and binds to HSV-1 capsids, presumably stabilizing the capsid structure during DNA packaging or once the packaging process is completed (25, 37, 38). In pseudorabies virus, pUL25 was proposed to be required for nuclear egress of mature capsids (20). pUL32, the HSV-1 homolog of the HCMV UL52 protein, is an essential 67-kDa protein that exhibits a predominantly cytoplasmic localization and is able to bind zinc ions (10). In another study, an HSV-1 UL32 insertion mutant was shown to be defective for cleavage and encapsidation of HSV-1 DNA (21). When cells were transfected with an UL32 expression plasmid and superinfected with the UL32 mutant, the UL32 protein was found mainly in the cytoplasm but some of the protein could also be detected in nuclear replication compartments where UL32 colocalized with ICP8. Interestingly, in cells infected with the UL32 mutant, capsids accumulated at the nuclear periphery in a region which is devoid of ICP8 (21). It was therefore proposed that pUL32 may play a role in targeting capsids to replication compartments for packaging of viral DNA. HCMV UL52 was reported to be essential, as demonstrated by random and site-directed mutagenesis of the HCMV Towne and AD169 strains (14, 42). As for HSV-1 pUL32, HCMV pUL52 does not seem to be a virion constituent (39).
In this study, we asked which step of the HCMV infection cycle is blocked in the absence of UL52. To this end, a UL52 mutant was constructed and then its replication cycle was analyzed in noncomplementing cells. Failure of DNA cleavage and the absence of mature C capsids suggested that pUL52 plays a role in encapsidation and/or DNA cleavage. However, the UL52 protein did not colocalize with other cleavage-packaging proteins within the nucleus, suggesting that its role is distinct in these processes.
MATERIALS AND METHODS
Viruses and cells.
The recombinant viruses (RV) used in this study were derived from the bacterial artificial chromosome (BAC)-cloned genome (pHB5) of the HCMV laboratory strain AD169 (6). The HCMV BAC pHG contains an enhanced green fluorescent protein (EGFP) gene in the unique short region and lacks the open reading frames (ORFs) UL1 to UL10 (UL1-10), with only one Flp recognition target (FRT) site retained at this position (9). The BAC pHD corresponds to pHB5 with the UL1-10 locus deleted. RV reconstituted from these BACs (RV-HG and RV-HD, respectively) served as parental viruses for the mutants analyzed in this study. Culture of human foreskin fibroblasts (HFF), preparation of virus stocks, growth curves, and determination of virus titers were done essentially as described previously (6). The telomerase-immortalized human fibroblast line hTERT-BJ-1 (Clontech, Palo Alto, CA) was grown in a medium consisting of 4 parts of Dulbecco's modified Eagle's medium (DMEM) and 1 part of medium 199 (Sigma Aldrich, Munich, Germany) with 5% fetal calf serum (Biochrom AG, Berlin, Germany), 1 mM sodium pyruvate, 2 mM glutamine, 100 U/ml of penicillin, and 100 μg/ml of streptomycin sulfate. The retroviral packaging cell line RetroPack PT67 (Clontech) was propagated in DMEM containing 5% fetal calf serum and the same supplements. The telomerase-immortalized human retina pigment epithelial (RPE) cell line (Clontech) was cultured in DMEM-Nut-Mix F12 with 15 mM HEPES (Biochrom) with 0.35% (wt/vol) sodium bicarbonate, 2 mM glutamine, 100 U/ml of penicillin, and 100 μg/ml of streptomycin sulfate. For generation of UL52-expressing cells, the retroviral gene transfer and expression system Retro-X (Clontech) was applied following the instructions of the manufacturer. This system is based on a Moloney murine leukemia virus-derived vector and the NIH 3T3-derived packaging cell line RetroPack PT67. In brief, RetroPack PT67 cells were transfected with the retroviral vector pLXSN-UL52 expressing UL52 under control of the long terminal repeat (LTR) promoter. Stably transfected PT67 cells were selected in the presence of 200 μg/ml G418. The supernatant of the RetroPack PT67-UL52 cells containing the dualtropic recombinant retroviruses was filtered (0.45-μm pore size) and used to infect hTERT-BJ1 cells in the presence of 8 μg/ml Polybrene. Stably transduced BJ-UL52 cell clones were selected and further propagated in the presence of 200 μg/ml G418.
Plasmids.
The retroviral expression vector pLXSN-UL52 was constructed by PCR amplification of the UL52 ORF by use of primers UL52.for (5′-CCCGTTAACCCTCAATCAGCGGCGCCGAT-3′) and UL52.rev (5′-GCGGGATCCCGCGGCGTGCGCACGCCGCT-3′) with BAC pHB5 as a template. The PCR product was cut with HpaI and BamHI and cloned into the respective sites of pLXSN (Clontech). The integrity of the resulting plasmid, pLXSN-UL52, was confirmed by sequencing. For ectopic expression of the hemagglutinin (HA)-tagged pUL52, the shuttle plasmid pOri6K-UL52-HA was generated, which encodes pUL52 with an HA epitope at the C terminus and contains 530 bp of the sequences upstream of the UL52 start codon, providing suitable promoter elements. Primers UL52-HA.for (5′-CGCGGTACCCGCACCGACGCCACCGCCGATT-3′), binding at the putative UL52 promoter region, and UL52-HA.rev (5′-CCCGTTAACCTAAGCGTAGTCTGGGACGTCGTATGGGTAGACATACTTGTCTATCACGTA-3′), located at the 3′ end of the UL52 ORF and encoding the HA tag (underlined), were used with pHB5 as the template. The resulting PCR product was treated with KpnI and HpaI and ligated to pOri6K-Kan1 (9) cut with KpnI and EcoRV. Plasmids derived from the pOri6K series contain a kanamycin resistance cassette plus one FRT site as well as the R6Kγ bacterial origin of replication (5) and thus cannot replicate in Escherichia coli strains that lack the phage lambda π protein. Plasmid pOri6K-UL52-NHA-P contains the UL52 ORF, the sequence encoding the N-terminal HA tag, and the UL52 promoter sequences. A derivative of pOri6K-Kan1 providing a modified polylinker with additional restriction sites (pOri6k-MfeI) was used as vector. The UL52 ORF was excised from pLXSN-UL52 via BamHI and HpaI and inserted into the respective sites of pOri6k-MfeI. The resulting plasmid, pOri6K-Mfe-UL52, was then cut with NheI followed by partial digestion with SacII to remove the 5′ end of the UL52 ORF. The UL52 5′ end was subsequently amplified by PCR with primers UL52-NHA.for (5′-CGCGGCTAGCCGCCATGTACCCATACGACGTCCCAGACTACGCTAATCCGAGTACCCACGTGAGC-3′; the sequence encoding the HA tag is underlined) and UL52-NHA.rev (5′-GGCGGGGTGTTGAGGATTTA-3′) by use of pLXSN-UL52 as a template. The PCR product was cut with NheI and SacII and inserted into pOri6K-Mfe-UL52, yielding pOri6K-UL52-NHA. To add promoter sequences to the N-terminally tagged UL52 ORF, primers UL52-P-NHA.for (5′-CGCCAATTGCGCACCGACGCCACCGCCGATT-3′) and UL52-P-NHA.rev (5′-CGCGGCTAGCATCGGCGCCGCTGATTGAGG-3′) were used to amplify the region comprising the 530 bp upstream of the UL52 start codon. The PCR product obtained was treated with MfeI and NheI and ligated to pOri6K-UL52-NHA cut with MfeI and NheI, giving rise to pOri6K-UL52-NHA-P. The integrity of the final plasmid was verified by sequencing. All plasmids harboring the R6Kγ origin of replication were propagated in the E. coli strain PIR1 (Invitrogen, Karlsruhe, Germany).
BAC mutagenesis and reconstitution of virus mutants.
The HCMV UL52 deletion BACs pHG-ΔUL52 and pHD-ΔUL52 (corresponding to pHG-ΔUL52 but lacking the EGFP gene) were constructed in E. coli DH10B by use of the ET mutagenesis procedure, which relies on homologous recombination between the BAC-cloned HCMV genome and linear DNA fragments (5). The linear fragment used encoded a kanamycin resistance marker (knR) flanked by FRT sites and was obtained by PCR using primers containing ∼40 bp of homology to the intended integration site in the BAC at the 5′ end and 20 bp specific for knR at the 3′ end. The recombination enzymes were expressed from plasmid pKD46, which carries the phage lambda recombination genes red α, β, and γ under control of an inducible promoter (12). Primers used to disrupt the UL52 gene in the HCMV BACs were UL52-ko.for (5′-GCGGCCGCCTCATACCAGGTAAATCCTCAACACCCCGCCAAGAAAAGTGCCACCTGCAGAT-3′) and UL52-ko.rev (5′-TTGGTGACGCGGATGTTGCCGGCGCACTGCGGGTCGCAGAACAGGAACACTTAACGGCTGA-3′), and plasmid pOri6K-F5 served as a template. pOri6K-F5 encodes a knR marker flanked by mutant FRT sites that do not interact with the wild-type FRT sites (9). Subsequent excision of the FRT-flanked KnR marker was done in E. coli via Flp recombinase expressed by plasmid pCP20 as described previously (5). Mutant BACs expressing HA-tagged versions of the UL52 protein were constructed by Flp recombinase-driven integration of plasmids pOri6K-UL52-HA and pOri6K-UL52-NHA-P into the wild-type FRT site located at the previous UL1-10 locus as reported elsewhere (9). pHG-52HA-1 and pHG-52HA-2 were generated by inserting pOri6K-UL52-HA into pHG-ΔUL52 and pHG, respectively, and pHG-52HA-3 and pHD-52HA were obtained by integration of pOri6K-UL52-NHA-P into pHG-ΔUL52 and pHD-ΔUL52, respectively. To reconstitute virus mutants, the recombinant BACs were transfected into HFF by use of an adenovirus-mediated gene delivery protocol as described previously (9).
Pulsed-field gel electrophoresis.
BJ-1 or BJ-UL52-4 cells (5 × 105) were infected with either RV-HG or RV-HG-ΔUL52 at a multiplicity of infection (MOI) of 1. Five days postinfection (p.i.), cells were harvested and resuspended in 40 μl of phosphate-buffered saline (PBS). After 40 μl of 2% low-melting-point agarose was added, cells were cast in block formers, followed by incubation of the agarose blocks in 10 mM Tris-HCl (pH 8.0)-100 mM EDTA (pH 8.0)-20 mM NaCl supplemented with 1% sarcosyl and 500 μg/ml proteinase K for 48 h at 45°C. Agarose blocks were washed with TE buffer (10 mM Tris-HCl [pH 8.0]-1 mM EDTA) five times for 15 min each at 45°C and equilibrated in 0.5× Tris-borate-EDTA buffer for 30 min on ice before being loaded into the wells of the gel. Conditions for gel electrophoresis were as reported earlier (8). Prior to Southern blotting, the gel was incubated in 0.25 M HCl for 45 min, followed by 30 min of incubation in 0.5 M NaOH-1.5 M NaCl. DNA fragments were transferred to nylon membranes by use of 0.4 M NaOH.
Analysis of viral and BAC DNA.
HCMV BAC DNA was isolated from 10-ml overnight E. coli cultures by use of an alkaline lysis protocol and characterized by restriction analysis and gel electrophoresis. Large-scale preparations of HCMV BAC DNA were obtained from 100-ml bacterial cultures by use of Nucleobond PC 100 columns (Macherey-Nagel, Düren, Germany) according to the instructions of the manufacturer. Total DNA was isolated from infected cells by lysing the cells in 50 mM Tris-HCl (pH 8.0)-10 mM EDTA-0.5% sodium dodecyl sulfate (SDS), followed by proteinase K digestion (500 μg/ml) for 3 h at 56°C. The samples were extracted twice with phenol-chloroform and the DNA was precipitated with isopropanol. HCMV DNA from virus stocks was prepared accordingly by mixing 1 volume of the virus stock (equivalent to approximately 4 × 106 PFU) with 1 volume of 100 mM Tris-HCl (pH 8.0)-20 mM EDTA-1% SDS. Proteinase K treatment and precipitation of DNA were then done as described above. To analyze the formation of genomic termini, BJ-1 cells were infected with the respective viruses at an MOI of 1.5 and total DNA was isolated on day 5 p.i. Five micrograms of each DNA sample was then subjected to restriction enzyme treatment followed by agarose gel electrophoresis and Southern blotting. The DNA was transferred to nylon membranes and analyzed by hybridization using probes specific for either the a- or the b-repeat sequence. The hybridization probe binding to the b-sequence was isolated as a 986-bp SalI fragment (corresponding to nucleotides [nt] 2973 to 3958 of the HCMV genome; GenBank accession number X17403 [11]) from a plasmid containing part of the HCMV terminal repeat region (E. M. Borst, unpublished data), and the probe specific for the a-sequence was excised as a 553-bp XhoI fragment (equivalent to nt 19 to 571 of the HCMV genome) from the HCMV amplicon vector (8). The probe specific for the 5′ end of the UL52 ORF was isolated as a 662-bp fragment by treatment of pLXSN-UL52 with HpaI and XhoI. Accumulation of replicated viral DNA was assessed by use of a slot blot assay. BJ-1 cells were infected at an MOI of 0.025 and total DNA was isolated on days 1 to 6 p.i. using the QiaAmp DNA blood mini kit (Qiagen, Hilden, Germany). One microgram of each DNA sample was transferred to a nylon membrane by use of a slot blot apparatus. Hybridization was done with a 32P-labeled DNA probe specific for the HCMV lytic origin region obtained as a 921-bp SacI fragment (nt 94645 to 95565 of the HCMV genome) from plasmid p6K-ori3.7-1 (9). The radioactive signals obtained were quantified with a phosphorimager.
Electron microscopy.
BJ-1 fibroblasts (1 × 105) were infected with RV-HG or with RV-HG-ΔUL52 at an MOI of 6. Likewise, the BJ-UL52-4 cells were infected with RV-HG-ΔUL52 with the same MOI. All cells were fixed at 6 days p.i. with 1% glutaraldehyde in 200 mM cacodylate buffer, pH 7.4, for 75 min at room temperature, stained with 1% (wt/vol) OsO4, 1.5% (wt/vol) K3Fe(CN)6 for 30 min followed by 0.5% (wt/vol) uranyl acetate in 50% ethanol overnight, dehydrated using a graded ethanol series and propylenoxid, pelleted, and embedded in Epon. Ultrathin sections of 50 to 100 nm in thickness were cut and further contrasted using lead citrate (33). Electron microscopy micrographs were recorded with a Zeiss EM10 microscope, and images printed on regular photo paper were scanned and further processed using Adobe Photoshop version 6.0.
Immunofluorescence microscopy.
To test for expression of the viral DNA processivity factor (pUL44), BJ-1 cells were seeded on coverslips and infected at an MOI of 0.5. On day 3 p.i. cells were probed with a mouse monoclonal antibody directed against the pUL44 protein (a kind gift of B. Plachter, University of Mainz, Germany). An Alexa Fluor 568 goat anti-mouse antibody (catalog number [cat. no.] A-11031; Molecular Probes/Invitrogen, Karlsruhe, Germany) was used as the secondary antibody. To analyze the intracellular distribution of the HA-tagged pUL52, fibroblasts were infected at an MOI of 0.5 and incubated for the times indicated. A rat monoclonal anti-HA antibody (cat. no. 11867423001; Roche, Mannheim, Germany) was applied, followed by incubation of the coverslips with an Alexa Fluor 488 or an Alexa Fluor 568 goat anti-rat antibody (dilution, 1:500; cat. no. A-11006 or A11077, respectively; Molecular Probes). Antibodies directed against pUL56, pUL89, and pUL104 affinity purified from high-titer human serum were a kind gift of E. Bogner (Charite, Berlin, Germany). The secondary antibody was an Alexa Fluor 568 goat anti-human immunoglobulin G (cat. no. A-21090; Molecular Probes) applied at a 1:500 dilution. In brief, labeling of cells was done as follows. Cells were fixed with 3% (wt/vol) paraformaldehyde in PBS for 15 min, followed by an incubation in 20 mM glycine-50 mM NH4Cl for 10 min. Cells were permeabilized by addition of 0.2% (vol/vol) Triton X-100-PBS for 10 min, and unspecific binding sites were blocked with 0.2% gelatin in PBS for 10 min. All steps were done at room temperature. Incubation with the first and second antibodies, which were diluted in 0.2% (wt/vol) gelatin in PBS, was done for 45 min each at room temperature. Cells were then analyzed using a Zeiss LSM 510 Meta confocal laser scanning microscope. Digitalized images were further processed using the Zeiss LSM image browser and Adobe Photoshop version 6.0.
Immunoblotting.
To test the expression kinetics of pUL52-HA and pUL44, HFF were infected at an MOI of 3 with RV-HG-52HA-3 and incubated for the indicated time periods. Cells were lysed in reducing buffer Roti-Load1 (Roth, Karlsruhe, Germany) and an equivalent of 1 × 105 cells was loaded into each lane of a 10% polyacrylamide-SDS gel. After transfer of the proteins to a nitrocellulose membrane, pUL52-HA was detected with a polyclonal rabbit anti-HA antibody (cat. no. H6908; Sigma), followed by incubation of the blot with a horseradish peroxidase-linked anti-rabbit antibody (NA934; Amersham Biosciences, Freiburg, Germany). To visualize pUL44, the mouse monoclonal anti-UL44 was applied and detected with a horseradish peroxidase-linked anti-mouse antibody (NIF 825; Amersham). Infections in the presence of inhibitors of immediate-early or early gene expression were done as follows. To inhibit immediate-early gene expression, infected HFF were incubated for 30 min in the presence of 100 μg/ml cycloheximide (Sigma Aldrich, Munich, Germany) before infection at an MOI of 3. After incubation of the cells in the presence of cycloheximide for another 3 h at 37°C, actinomycin D (Sigma) was added to a final concentration of 5 μg/ml and incubation was continued for 30 min. HFF were then washed with PBS containing actinomycin D three times for 5 min, and then complete growth medium containing actinomycin D was added. Three hours later, cells were harvested and proteins were analyzed by Western blotting using the anti-HA and the anti-UL44 antibody as described above. The HCMV immediate-early 1 (IE1) protein was detected with a mouse monoclonal anti-IE1 antibody (cat. no. NEA-9221; Perkin Elmer Life Sciences Inc., Boston, MA). For the inhibition of early gene expression, cells were infected in the presence of 250 μg/ml phosphonoacetic acid (Sigma) and incubated for 72 h at 37°C with daily changes of phosphonoacetic acid-containing medium. Late gene expression was analyzed by incubating infected fibroblasts for 72 h at 37°C in the absence of any inhibitors.
RESULTS
Generation of an HCMV UL52 mutant and a complementing cell line.
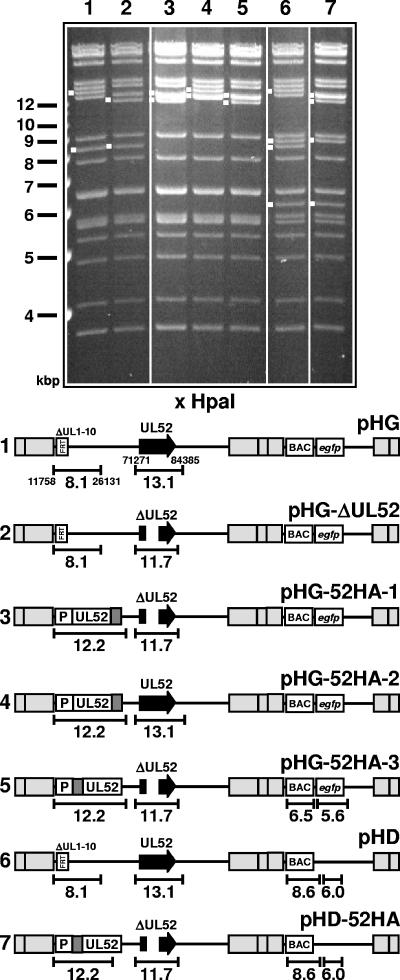
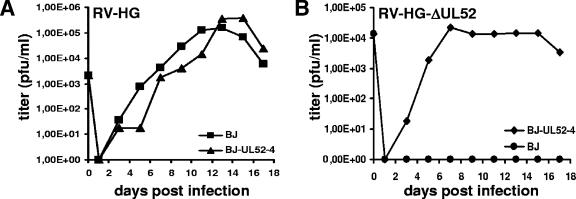
To investigate the function of the UL52 protein, we constructed an HCMV UL52 knockout mutant and a complementing cell line providing the UL52 protein in trans. Since the UL52 ORF is directly adjacent to the UL51 ORF and overlaps with the UL53 ORF, both of which are essential for growth of HCMV, we deleted only the middle part of the UL52 ORF (nt 74100 to 75500) to preserve the putative UL51 and UL53 promoter regions located in the 5′ and 3′ regions of the UL52 ORF. The mutation was introduced into the HCMV BAC pHG, which contains an EGFP gene in the unique short region and lacks the nonessential genes UL1 to UL10 (9), giving rise to pHG-ΔUL52. As a consequence, a 13.1-kbp HpaI fragment present in the parental BAC pHG is shifted to 11.7 kbp in the UL52-deleted BAC (Fig. 1, compare lanes 1 and 2). This genome was not infectious when transfected into permissive HFF, confirming that UL52 is an essential viral gene. To reconstitute a viral mutant from the pHG-ΔUL52 genome, we generated complementing cell lines by transducing the human fibroblast line hTERT-BJ-1 (3) with a recombinant retrovirus carrying the UL52 ORF under control of the retroviral LTR promoter. Twenty of the BJ-1 cell clones obtained were transfected with pHG-ΔUL52. In eight of the transfected cell clones, plaque formation was observed, and cell clone BJ-UL52-4 was chosen for further analysis because it supported growth of the UL52 deletion mutant most efficiently. Since HCMV gene expression occurs in a highly regulated temporal manner and since UL52 was not expected to be expressed throughout the entire replication cycle, we wondered whether the continuous expression of UL52 in the complementing cell line might interfere with virus growth. In order to test this, BJ-UL52-4 and BJ-1 cells were infected with the parental virus RV-HG, and the amounts of virus released in the supernatants of both cell types were determined by plaque assay on BJ-1 cells. The growth of RV-HG was at most slightly delayed on the complementing BJ-UL52-4 cells, and the final titers obtained from BJ-1 and BJ-UL52-4 cells were comparable (Fig. 2A). We concluded that the permanent presence of the UL52 protein throughout the viral replication cycle did not severely interfere with the growth of HCMV and that the cell line was appropriate for propagation of the UL52 mutant.
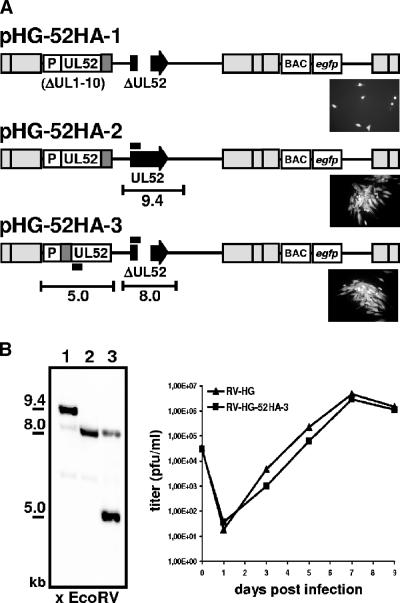
FIG. 1.
HCMV BAC genomes constructed in this study. The gel shows the HpaI restriction patterns of the recombinant BAC genomes. Relevant fragments are marked with white dots, and size markers are indicated to the left. The numbers of the lanes correspond to the genome structures shown below. (Bottom) Schematic drawing of the structures of the respective genomes. The sizes of fragments characteristic of each mutant are indicated. The nucleotide positions of HpaI restriction sites located around the UL1-10 and the UL52 locus are given in the first line (pHG). Gray boxes indicate repeat regions flanking the unique long and unique short genomic regions. The illustration is not drawn to scale.
FIG. 2.
Characterization of the UL52 complementing cell line and growth analysis of the ΔUL52 mutant. (A) Growth of the parental virus RV-HG on the UL52-expressing cell line BJ-UL52-4 and on normal BJ-1 cells. Cells were infected at an MOI of 0.1 and supernatants were analyzed for the presence of infectious virus by plaque assay on BJ-1 cells. (B) Growth properties of the UL52-knockout virus RV-HG-ΔUL52 on the complementing cell line BJ-UL52-4 and on normal BJ-1 cells. Cells were infected at an MOI of 0.1, and viral progeny produced (virus in the supernatant plus cell-associated virus) was determined by plaque assay on BJ-UL52-4 cells.
To generate the mutant lacking the UL52 gene, the BJ-UL52-4 cell clone was transfected with pHG-ΔUL52. After complete cytopathic effect had developed, virions were purified from both the supernatant and the infected cells and concentrated by centrifugation. The titer of the resulting stock of RV-HG-ΔUL52 was determined by plaque assay on the complementing cell line BJ-UL52-4 as 1.3 × 107 PFU/ml.
Since only part of the UL52 ORF was deleted, it was theoretically possible that the residual UL52 sequences in the pHG-ΔUL52 BAC may recombine with the full-length UL52 ORF present in the genome of the complementing cell line, thus resulting in the generation of replication-competent revertants. To test if this had occurred, DNA was extracted from BJ-UL52-4 cells infected with RV-HG-ΔUL52 and analyzed by Southern blotting using a UL52-specific probe. Only DNA fragments corresponding to RV-HG-ΔUL52 and not to viruses with the parental genome structure were detected, indicating that no revertants were produced upon reconstitution of mutant virus (data not shown). Moreover, the growth of RV-HG-ΔUL52 on normal BJ-1 cells and on the BJ-UL52-4 cell clone was examined (Fig. 2B). Cells were infected at an MOI of 0.1 and harvested together with the supernatants on days 1 to 17 p.i. Intracellular virions were released by three freeze-thaw cycles, combined with the supernatants, and tested for the presence of infectious virus particles by plaque assay on the complementing cell line BJ-UL52-4. No plaque formation was observed following inoculation of BJ-UL52-4 cells with the BJ-1-derived samples, whereas virus titers of up to 5 × 104 PFU/ml were obtained on the BJ-UL52-4 cells. Thus, only the UL52 deletion mutant and no replication-competent virus was produced by the complementing cell line.
pUL52 is not involved in viral DNA replication.
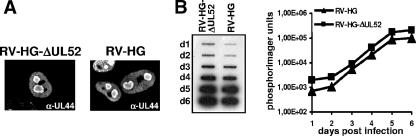
In order to figure out in which phase of the HCMV life cycle the UL52 gene product is required, we first analyzed whether the formation of replication compartments was affected in normal cells infected with the ΔUL52 mutant. BJ-1 cells were infected with either RV-HG-ΔUL52 or the parental virus RV-HG and on day 3 p.i. were analyzed by immunofluorescence microscopy with an antibody directed against the accessory factor of the viral DNA polymerase, pUL44 (Fig. 3A), which is localized in HCMV replication compartments (26). We did not observe any obvious differences in the appearance of the subnuclear structures containing pUL44, irrespective of the presence or absence of pUL52. Thus, the formation of replication compartments was not dependent on pUL52.
FIG. 3.
Genome replication of the ΔUL52 virus and the parental virus. (A) Formation of replication compartments in the absence of UL52. BJ-1 cells were infected with either RV-HG or RV-HG-ΔUL52 at an MOI of 0.5. Three days p.i. the cells were labeled with an antibody directed against the viral DNA polymerase processivity factor pUL44 and analyzed by confocal laser scanning microscopy. (B) Normal BJ-1 cells were infected with the UL52 knockout mutant RV-HG-ΔUL52 or the parental virus RV-HG at an MOI of 0.025. On days 1 to 6 p.i. (d1 to d6), total DNA was isolated from the infected cells and analyzed by slot blot hybridization using a probe specific for the HCMV lytic origin region. The signals obtained were quantified using a phosphorimager.
Next, we tested whether UL52 was needed for replication of viral DNA. Normal BJ-1 cells were infected with either the mutant RV-HG-ΔUL52 or the parental virus RV-HG. On days 1 to 6 p.i., total DNA was extracted from the infected cells and analyzed by slot blot hybridization using a probe specific for the HCMV oriLyt region. The signals were also quantified using a phosphorimager. Viral DNA accumulated with comparable kinetics in BJ-1 cells infected with either the parental virus or the deletion mutant (Fig. 3B). This result showed that the UL52 protein does not play a role in HCMV DNA replication.
The UL52 protein is required for cleavage of concatemeric HCMV DNA.
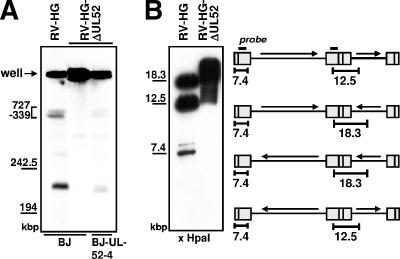
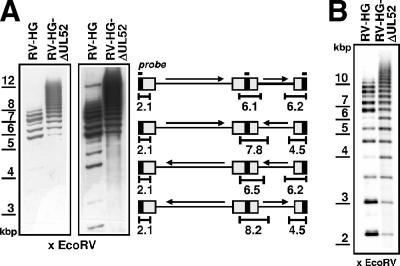
Due to its homology to the HSV-1 UL32 protein, which was proposed to be involved in cleavage-packaging of HSV-1 DNA, we wondered whether the UL52 protein might possess a similar function. Therefore, we asked whether cleavage of HCMV concatemeric DNA took place in the absence of the UL52 gene product. Normal BJ-1 cells were infected with either RV-HG or RV-HG-ΔUL52 and the complementing BJ-UL52-4 cells were infected with RV-HG-ΔUL52. On day 5 p.i., cells were embedded in agarose blocks and lysed, and the DNA was subjected to pulsed-field gel electrophoresis and Southern blotting using an HCMV-specific probe to examine the formation of viral unit-length genomes. In BJ-1 cells infected with the UL52 knockout mutant, unit-length genomes were not detected (Fig. 4A, second lane), whereas in cells infected with the parental virus RV-HG, unit-length genomes with a size of approximately 230 kbp were readily observed (Fig. 4A, first lane). Likewise, in complementing cells infected with the UL52 deletion mutant, a faint signal corresponding to unit-length HCMV genomes could be found (Fig. 4A, third lane). The lower abundance of cleaved genomes in the BJ-UL52-4 cells is consistent with the finding that these cells do not support growth of the UL52 knockout virus to titers that can be achieved with the parental virus (Fig. 2B). In all three lanes, the majority of the hybridization signal was found at the positions of the wells, representing viral concatemers which are too large to migrate into the gel. Taken together, these data indicated that viral concatemers were not cleaved in the absence of pUL52. In order to confirm this, total DNA was extracted from BJ-1 cells infected with either RV-HG or RV-HG-ΔUL52 and analyzed for the appearance of genomic termini by treatment with HpaI followed by Southern hybridization using a probe specific for the b-repeat sequences, which allowed the detection of fragments corresponding to the internal repeat as well as to free genomic termini derived from the left end of the viral genome (Fig. 4B). In Fig. 4B, the first lane shows the result of this experiment with DNA extracted from cells infected with RV-HG. Depending on the four possible isoforms of the HCMV genome, fragments of 12.5 and 18.3 kbp were generated from the internal repeat region. In addition, there was a distinct signal at a size of 7.4 kbp, which matched the size of the fragment comprising the left genomic terminus (the weak signal at a size of approximately 8 kbp was probably caused by the terminal fragment containing an additional copy of the a-sequence). The 7.4-kbp terminal fragment originated from cleavage of the viral concatemers and was clearly absent in DNA isolated from cells infected with the UL52 deletion mutant (Fig. 4B, second lane). In summary, these results demonstrated that the HCMV DNA was not cleaved in the absence of pUL52.
FIG. 4.
Viral concatemeric DNA is not cleaved in the absence of pUL52. (A) Normal BJ-1 cells were infected with the parental virus RV-HG or the UL52 deletion virus RV-HG-ΔUL52, and complementing BJ-UL52-4 cells were infected with RV-HG-ΔUL52 at an MOI of 1. Cells were harvested on day 5 p.i., embedded in agarose, and treated with proteinase K. Then, the DNA was resolved by pulsed-field gel electrophoresis and analyzed by Southern blotting using a probe specific for the a-repeat region. The positions of size markers derived from λ DNA are indicated to the left. (B) Normal BJ-1 cells were infected with the parental virus RV-HG or with the RV-HG-ΔUL52 mutant at an MOI of 1.5 and total DNA was isolated 5 days p.i. The DNA was cut with HpaI, followed by gel electrophoresis and Southern blotting using a probe derived from the b-sequence. The schematic drawing depicts the localization of the hybridization probe (black bar) as well as the DNA fragments corresponding to the internal repeat (12.5 and 18.3 kbp) and the left genomic terminus (7.4 kbp), depending on the four isoforms of the HCMV genome. The illustration is not drawn to scale.
During these experiments, we noticed that in the DNA samples derived from RV-HG-ΔUL52-infected cells, the fragments corresponding to the internal repeat region migrated more slowly than in the control DNA isolated from RV-HG-infected BJ-1 fibroblasts and that instead of distinct bands there was rather a smear of the hybridization signal (Fig. 4B, second lane). Such hybridization patterns are reminiscent of HCMV genomes containing a variable number of copies of the a-sequence. To test this, the DNA samples were treated with EcoRV followed by gel electrophoresis and Southern blotting with a probe comprising the a-sequence (Fig. 5A). Several bands were detected in the lane containing the DNA isolated from RV-HG-infected cells (Fig. 5A, first lane), corresponding to fragments derived from the genomic termini as well as from the internal repeat sequences. Please note that the sizes of the fragments vary depending on the presence of one or more a-repeats. In the lane containing the DNA obtained from RV-HG-ΔUL52-infected cells (Fig. 5A, second lane), a ladder of hybridization signals was detected, reflecting the presence of fragments with many more copies of the a-sequence. As expected, fragments corresponding to free genomic ends were missing from RV-HG-ΔUL52-infected fibroblasts (Fig. 5A, second panel [long exposure]). These experiments suggested that in the absence of pUL52 (i.e., in the absence of DNA cleavage), the a-sequences were highly amplified within the viral concatemers. However, since amplification of a-sequences is believed to occur concomitantly with cleavage-packaging, the possibility remained that viral genomes containing amplified a-sequences were not generated in the fibroblasts but were already present in the genomes of the inoculated virus. We therefore isolated viral DNA from virus stocks of either the parental virus or the UL52 deletion mutant and analyzed the samples by Southern hybridization using the probe specific for the a-sequence (Fig. 5B). As is obvious from the figure, RV-HG-ΔUL52 genomes harbored a higher number of a-sequences than RV-HG genomes. Thus, the amplified a-sequences were acquired by RV-HG-ΔUL52 during its replication on the complementing cell line.
FIG. 5.
Increased copy number of a-sequences in the RV-HG-ΔUL52 genomes. (A) DNA samples derived from normal BJ-1 cells infected with either the parental virus RV-HG or the UL52 knockout mutant RV-HG-ΔUL52 (obtained as described for Fig. 4B) were cut with EcoRV and subjected to gel electrophoresis and Southern blotting using a probe specific for the HCMV a-sequence. The two panels show different exposure times of the blot. (Right) Schematic drawing of the four isomeric forms of the HCMV genome and localization of the hybridization probe (black bar) to the a-sequence (black rectangles). The sizes of the DNA fragments expected to hybridize to the a-specific probe are indicated (calculation of the fragment sizes was done by considering one a-sequence only and the presence of a 217-bp heterogeneity in the a-sequence [35], which was found before in the HCMV BAC used in this study). (B) Analysis of viral DNA isolated from virions of the parental virus RV-HG or the deletion mutant RV-HG-ΔUL52. The viral DNA was cut with EcoRV, followed by gel electrophoresis and Southern blotting using the probe binding to the a-sequence.
Intranuclear distribution of the HCMV cleavage-packaging proteins pUL56, pUL89, and pUL104 in the absence of pUL52.
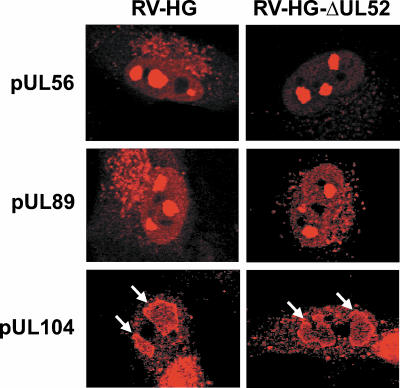
Three viral proteins participating in cleavage and packaging of HCMV DNA have been identified and characterized so far. The products encoded by the ORFs UL56 and UL89 constitute the viral terminase, and pUL104 represents the portal protein of the capsid (13, 31). The lack of cleavage of viral concatemers in cells infected with the UL52 deletion mutant would be explainable if pUL52 would regulate the expression or localization of one or more of these viral proteins. In order to examine this, normal BJ-1 fibroblasts were infected with RV-HG or RV-HG-ΔUL52 and analyzed by immunofluorescence microscopy on day 3 p.i. using human antibodies directed against pUL56, pUL89, and pUL104 (Fig. 6). In HCMV-infected cells, these proteins typically localize to nuclear patches representing the sites of cleavage-packaging of viral DNA (18). The subnuclear distributions of all three viral proteins were very similar if not identical in the presence or absence of pUL52 (Fig. 6). In uninfected fibroblasts, no signal was obtained with any of the antibodies (data not shown), proving their specificity for the respective HCMV proteins. Thus, pUL52 did not seem to influence the expression or localization of other proteins known to be required for cleavage-packaging of HCMV DNA.
FIG. 6.
Subnuclear localization of the HCMV cleavage-packaging proteins pUL56, pUL89, and pUL104 in the absence of the UL52 protein. BJ-1 cells were infected at an MOI of 0.5 with RV-HG or RV-HG-ΔUL52. On day 3 p.i., cells were probed with antibodies specific for the pUL56, pUL89, and pUL104 proteins and analyzed by confocal laser scanning microscopy. Please note that the labeling of the structures in the cytoplasm probably results from the binding of the human antibodies to the viral Fc receptors. Therefore, only the labeling of the proteins in the nucleus is considered to be specific. Arrows indicate the specific staining pattern of pUL104 within the nuclei.
Formation of capsids in the absence of pUL52.
Cleavage and encapsidation of herpesvirus genomes are tightly linked processes (24). Viral DNA is spooled into preformed capsids following the head-full mechanism, with cleavage occurring after packaging of one unit-length genome. The interaction of procapsids with viral concatemeric genomes leads to either DNA-filled C capsids or—if packaging is not completed successfully—empty A capsids that contain neither DNA nor scaffold proteins (29). A deficiency in capsid formation in cells infected with the UL52 deletion mutant would therefore result in concatemers remaining uncut and not being encapsidated.
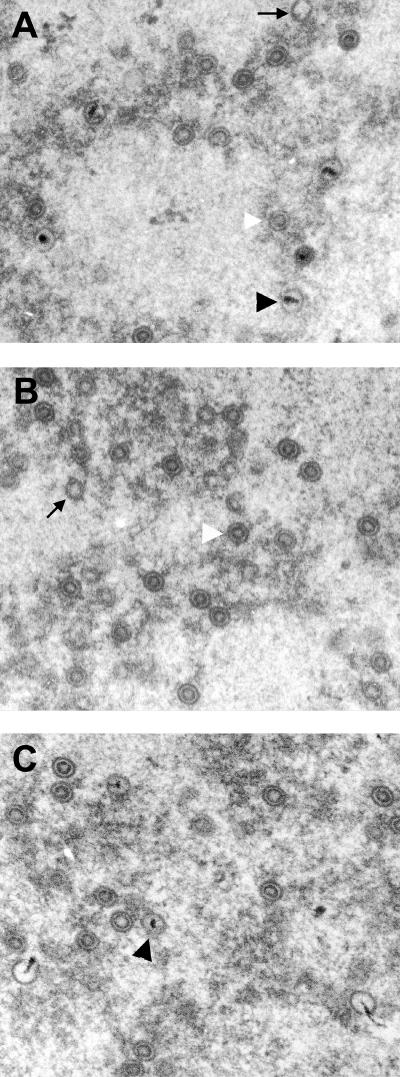
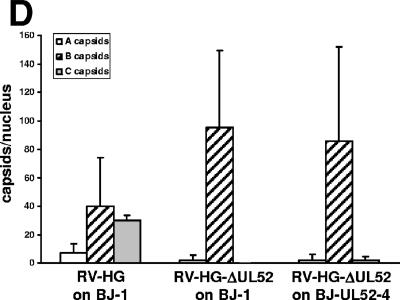
To test whether pUL52 was required for capsid formation, we analyzed fibroblasts infected for 6 days by electron microscopy. In normal fibroblasts infected with the parental virus, all three capsid types were readily detected in most of the nuclei (Fig. 7A). Many capsids contained an electron-dense material, indicative for viral DNA packaging, and were therefore classified as C capsids. Moreover, there were many B capsids, which were characterized by the internal spherical scaffold, and some cross sections of the infected nuclei even contained only B capsids. A quantification of the capsids in 20 nuclei showed that approximately 40% of all were C capsids, 50% were B capsids, and 10% were A capsids (Fig. 7D). A capsids contain neither a scaffold nor genomic DNA (29) and thus appeared empty.
FIG. 7.
Analysis of HCMV capsid formation and DNA packaging in the absence of pUL52. Electron micrographs of infected fibroblasts. (A) After 6 days of infection, the nuclei of fibroblasts infected with the parental RV-HG contained many C capsids (black arrowhead), many B capsids (white arrowhead), and very few A capsids (arrows). (B) In cells infected with RV-HG-ΔUL52, the nuclei contained mainly B capsids (white arrowhead), very rarely A capsids (arrow), and no C capsids. (C) In the complementing BJ-UL52-4 cells infected with RV-HG-ΔUL52, a few C capsids (black arrowhead) and mostly B capsids were found in the nuclei. (D) The amounts of the different capsid types in 20 nuclei were quantified for each condition depicted in panels A to C. Shown are the average values per nucleus and the standard deviations.
The nuclei of fibroblasts infected with RV-HG-ΔUL52 contained mainly B capsids (Fig. 7B), and in only very few nuclei did we detect some A capsids. However, we never found any C capsids in these cells. Thus, capsid formation was not impaired in the absence of pUL52, but genome packaging did not occur. In contrast, when the complementing BJ-UL52-4 cells were infected with RV-HG-ΔUL52, some C capsids could be detected (Fig. 7C). Again, we quantified the capsid types in 20 nuclei and detected more than 90% B capsids, approximately 2.5% C capsids, and 2.5% A capsids (Fig. 7D). Thus, C capsid formation and DNA packaging were restored in the pUL52-expressing fibroblasts, albeit with lower efficiency. This was consistent with the fact that the BJ-UL52-4 cells were not able to support growth of RV-HG-ΔUL52 to titers the same as those for the parental virus (Fig. 2B).
Construction of HCMV genomes expressing HA-tagged versions of pUL52.
To characterize the protein encoded by the UL52 ORF, we generated an HCMV mutant expressing an HA-tagged UL52 protein. We first constructed an HCMV BAC genome in which the DNA sequence encoding the HA epitope derived from the influenza virus HA (amino acid sequence, YPYDVPDYA) was inserted between the last codon and the stop codon of UL52, leading to the fusion of the epitope to the C terminus of pUL52. Because of the overlap of the UL52 ORF with the essential viral gene UL53, this mutation could not be introduced at the original position of the UL52 gene without interrupting ORF UL53. Instead, the tagged UL52 ORF together with 530 bp of upstream sequences comprising the putative UL52 promoter region was inserted at an ectopic position of the BAC pHG-ΔUL52 by making use of Flp recombinase-mediated recombination between a shuttle plasmid providing UL52 and an FRT site that had been previously inserted into the BAC genome (Fig. 1, bottom, lines 1 and 2) (9). As a consequence, the 8.1-kbp HpaI fragment of pHG-ΔUL52 comprising the FRT site shifted to 12.2 kbp in pHG-52HA-1 (Fig. 1, compare lanes 2 and 3; please note that the 12.2-kbp fragment in lane 3 is a double band). The resulting BAC, pHG-52HA-1, was transfected into HFF but did not lead to plaque formation (Fig. 8A, micrograph in the first line). Only single green fluorescent cells were detected even at 2 weeks after transfection, indicating that this BAC genome was not infectious. This suggested that the C terminus of pUL52 is essential for its function. We next tested whether this mutation exerted a dominant-negative effect on the function of the authentic pUL52. To do this, the shuttle vector containing the C-terminally tagged UL52 ORF was recombined with the parental BAC pHG, which contains the intact UL52 gene at its original genomic position. The HpaI restriction pattern of the resulting BAC genome, pHG-52HA-2, showed a 12.2-kbp double band, reflecting the successful insertion of the shuttle plasmid, as well as a 13.1-kbp fragment corresponding to the intact UL52 locus (Fig. 1, lane 4). pHG-52HA-2 readily produced plaques when transfected into permissive cells (Fig. 8A, micrograph in the second line), demonstrating that the C-terminal mutation did not confer a strict dominant-negative effect. We also examined the growth kinetics of RV-HG-52HA-2 and did not find a difference between these growth kinetics and those of the parental virus (data not shown). Therefore, a partial dominant-negative effect of the C-terminally tagged pUL52 was also excluded.
FIG. 8.
Properties of HCMV BAC genomes expressing HA-tagged variants of the UL52 protein. (A) First line: a modified UL52 gene encoding pUL52 tagged with the HA epitope (dark gray box) at the C terminus was inserted at an ectopic position (the previous UL1-10 locus) into the UL52-deleted genome, resulting in the BAC pHG-52HA-1. P, promoter; 530 bp of upstream sequences of the UL52 start codon. Second line: BAC pHG-52HA-2 encodes a C-terminally tagged pUL52 (at the ectopic position) plus the authentic pUL52 (from the normal UL52 ORF; black arrow). Third line: A UL52 gene encoding an N-terminally tagged version was inserted into the ΔUL52 BAC, giving rise to the BAC pHG-52HA-3. The micrographs show HFF transfected with the respective HCMV BAC constructs 2 weeks posttransfection. Pictures were taken using UV light to visualize GFP fluorescence. (B) (Left) Analysis of viral DNA isolated from virions of RV-HG-52HA-3 (lane 3). DNA was cut with EcoRV and subjected to gel electrophoresis and Southern blotting using a probe specific for the 5′ end of the UL52 ORF (indicated in panel A by the black bars). DNAs of the BACs pHG (lane 1) and pHG-ΔUL52 (lane 2) were used as controls. The sizes of the expected EcoRV fragments (not drawn to scale) originating from the authentic and ectopic UL52 ORF are indicated in panel A. Please note that the DNA of RV-HG-52HA-3 produced a weaker hybridization signal at 8 kbp because only 304 bp of the 5′ portion of the UL52 ORF was available for hybridization to the 660-bp probe. (Right) Growth analysis of the parental virus RV-HG and the mutant virus reconstituted from pHG-52HA-3 on HFF. Cells were infected at an MOI of 0.1, supernatants were harvested at the time points indicated, and viral titers were measured by plaque assay on HFF.
Next, we constructed a viral genome in which the DNA sequence encoding the HA epitope was located between the start codon and the second codon of the ectopically inserted UL52 ORF, leading to a viable virus expressing an N-terminally tagged UL52 protein. The resulting BAC, pHG-52HA-3, was characterized by an HpaI 12.2-kbp double band and a 11.7-kbp fragment containing the UL52 deletion (Fig. 1, lane 5). Following transfection, infectious virus was recovered from pHG-52HA-3 (Fig. 8A, micrograph in the third line). This virus carried the UL52 gene at an ectopic position, and therefore recombination between these sequences and the remaining UL52 sequences at the original position could potentially occur, leading to a repaired UL52 gene. Viral DNA was isolated from RV-HG-52HA-3 virions and compared to the parental genomes pHG and pHG-ΔUL52 (Fig. 8B, left) by Southern hybridization. Only the signals characteristic of the UL52 locus with the deletion (8.0 kbp) (Fig. 8B, lanes 2 and 3) and the UL52 gene present at the ectopic position (5.0 kbp) (Fig. 8B, lane 3) were detected, thus showing that the RV-HG-52HA-3 genome remained stable during generation of the mutant. Next, the growth kinetics of the mutant was investigated. HFF were infected with either the parental virus RV-HG or the mutant RV-HG-52HA-3, and infectious virus particles released in the supernatant of the infected cells were analyzed at several time points p.i. by plaque assay (Fig. 8B). The mutant expressing the N-terminally tagged UL52 protein grew with kinetics comparable to those of the parental virus and was therefore used in further experiments to examine the UL52 protein in infected cells.
pUL52 is expressed with true late kinetics.
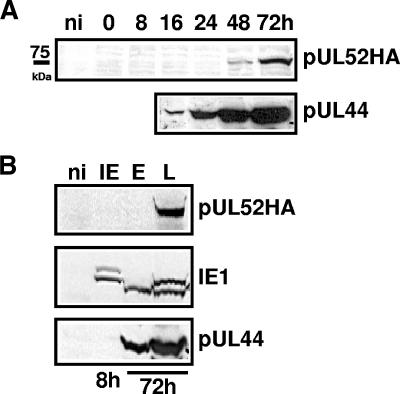
To characterize the UL52 protein and its expression kinetics, HFF were infected with pHG-52HA-3 and cell lysates were prepared at different time points p.i. (0 to 72 h), followed by Western blotting using antibodies directed against the HA tag or the early viral protein pUL44 (Fig. 9A). A band of approximately 75 kDa was detected with the anti-HA antibody at 48 and 72 h after infection. This matches very well the predicted molecular mass of 74 kDa of the UL52 protein (28). Compared to pUL44, pUL52 was expressed at late time points in infection. This suggested that pUL52 was possibly expressed with late kinetics. In order to verify this, viral gene expression in infected cells was restricted to the immediate-early, early, or late phase by use of appropriate infection conditions and inhibitors (for details, see Materials and Methods) and samples of cell lysates were subjected to Western blotting with antibodies against the HA tag, the IE1 protein, and the early protein pUL44 (Fig. 9B). pUL52 was detected only under late conditions, and the other viral control proteins were expressed with their expected specific kinetics. These data demonstrated that pUL52 is a true late protein.
FIG. 9.
Expression kinetics of the UL52 protein. (A) HFF were infected with RV-HG-52HA-3 at an MOI of 3. Cells were harvested at the time points indicated, and proteins were analyzed by immunoblotting using an antibody directed against the HA tag or against the viral early protein pUL44. ni, noninfected HFF. (B) To analyze specifically viral protein expression of the immediate-early (IE), early (E), or late (L) phase of the HCMV infection cycle, HFF were infected as described for panel A and either incubated in the presence of cycloheximide for 3 h followed by incubation in medium containing actinomycin D for another 5 h (IE), incubated for 72 h in the presence of phosphonoacetic acid (E), or left without inhibitors and harvested at 72 h p.i. (L). Cell lysates were analyzed by immunoblotting with antibodies directed against the HA tag, the immediate-early protein IE1, or the early protein pUL44.
Subcellular localization of pUL52.
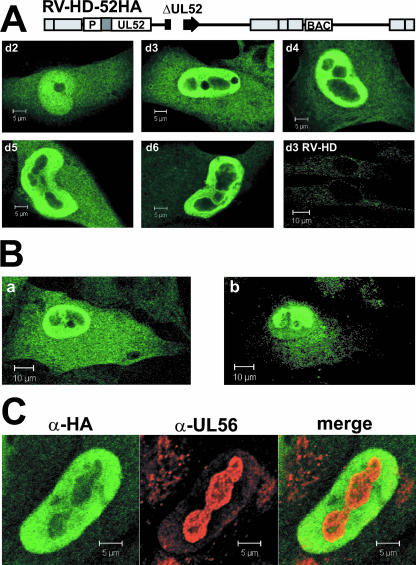
The HCMV proteins known to be involved in cleavage-packaging of viral DNA pUL56, pUL89, and pUL104 are localized in distinct patches in the nuclei of infected cells, which overlap with replication compartments (18) (Fig. 6). To investigate the subcellular localization of pUL52 by immunofluorescence microscopy, an additional virus mutant was constructed that expressed the N-terminally tagged UL52 protein but lacked the EGFP. The resulting HCMV BAC, pHD-52HA, and the parental BAC pHD were characterized by HpaI restriction analysis. Compared to the BAC genomes containing the EGFP gene in the unique short region, fragments of 6.5 and 5.6 kbp were replaced by fragments of 8.6 and 6.0 kbp in pHD and pHD-52HA (Fig. 1, lanes 6 and 7). Please note that the 6.5- and 5.6-kbp fragments present in the EGFP gene-containing BACs comigrate with other fragments of 6.4 and 6.3 kbp and of 5.7 and 5.5 kbp, respectively (e.g., lane 5). The deletion of UL52 and ectopic insertion of the HA-tagged UL52 gene was reflected by the lack of the 13.1-kbp fragment and the appearance of the 11.7- and 12.2-kbp bands (Fig. 1, compare lanes 6 and 7). HFF were infected with RV-HD-52HA, labeled on days 2 to 6 p.i. with an antibody directed against the HA tag, and analyzed by confocal laser scanning microscopy (Fig. 10A). HFF infected with the parental virus RV-HD served as a negative control (Fig. 10A, bottom right). On day 2 after infection, some of the UL52HA protein was localized in the cytoplasm, but a substantial portion was evenly distributed in the nucleus, sparing only the nucleoli. On day 3 and later, the majority of pUL52 was present in the nucleus, but large areas in the center of the nuclei and the nucleoli remained unlabeled. This pattern did not change significantly during the infection cycle. When confocal sections from the top or the bottom of the cell were examined, the labeling for pUL52 looked different (Fig. 10B). Areas that appeared dark in a central section became brightly fluorescent (Fig. 10B, compare panels a and b), implying that nuclear compartments were surrounded by pUL52. In summary, the localization of pUL52 was predominantly nuclear, and its subnuclear distribution was different from that of other HCMV proteins participating in cleavage-packaging of the viral genome.
FIG. 10.
Subcellular localization of the UL52 protein. (A) Cells were infected with RV-HD-52HA at an MOI of 0.5. On days 2 to 6 p.i. (d2 to d6), cells were labeled with an antibody specific for the HA tag and analyzed by confocal laser scanning microscopy. The images show the central section of the cells. HFF infected with the parental virus RV-HD were used as a negative control. (B) Different sections of HFF infected with RV-HD-52HA for 3 days were examined. Shown are central (a) and top (b) sections through the same cell. (C) HFF were infected with RV-HD-52HA at an MOI of 0.5 and labeled on day 3 p.i. with antibodies (α) directed against the HA tag and the pUL56 terminase protein.
In order to find out which nuclear compartment was enclosed by pUL52, HFF infected with RV-HD-52HA were labeled on day 3 p.i. with antibodies directed against the HA tag or the UL56 protein. pUL52 was localized around the compartments containing pUL56, with some colocalization being apparent at the boundaries (Fig. 10C). pUL56 is known to be targeted to replication compartments late in the HCMV infection cycle (18). Thus, in infected cells pUL52 encloses the nuclear compartments in which replication and packaging of HCMV genomes take place.
The BAC genome pHG-52HA-1, encoding the C-terminally tagged UL52 protein, was not infectious when transfected into fibroblasts (Fig. 8A), indicating that the pUL52 C terminus is crucial for its function. To investigate the subcellular localization of the C-terminally tagged pUL52, pHG-52HA-1 was transfected into RPE cells and analyzed 3 days posttransfection by immunofluorescence by use of the anti-HA antibody. As a control, RPE cells were transfected with pHG-52HA-3 encoding the N-terminally tagged UL52. The RPE cell line was chosen because these cells are permissive to HCMV infection and can be transfected more efficiently with large BACs than primary fibroblasts (7). pHG-52HA-3-transfected cells showed the expected nuclear localization of pUL52 (Fig. 11A). In cells transfected with pHG-52HA-1, pUL52 was distributed throughout the cells with a significant portion being present in the cytoplasm (Fig. 11B), but an accumulation in the nucleus was not observed. We concluded from these data that the C terminus of pUL52 was crucial for its correct subnuclear localization, which in turn seemed to be required for the essential role of the UL52 protein in the HCMV replication cycle.
FIG. 11.
Subcellular localization of the C-terminally tagged pUL52 variant. RPE cells were transfected with the EGFP-expressing BAC pHG-52HA-3 encoding pUL52 with an N-terminally fused HA tag (A) or with pHG-52HA-1, which encodes the C-terminally tagged UL52 ORF (B). Three days posttransfection, cells were labeled with an anti-HA antibody (α-HA) and analyzed by confocal laser scanning microscopy.
DISCUSSION
In this report, we provide the first characterization of the protein encoded by the essential UL52 gene of HCMV. pUL52 was found to be a true late protein with an apparent molecular mass of approximately 75 kDa that localized mainly to the nucleus, surrounding the viral replication and packaging compartments. In the absence of pUL52, viral DNA was replicated to normal levels, but cleavage of genome concatemers did not occur. Assembly of capsids was not impaired in the absence of the UL52 protein, but only B capsids were detected, demonstrating that genome encapsidation did not take place. pUL52 does not seem to affect the subnuclear localization of other viral proteins known to mediate cleavage-packaging of HCMV genomes. Since its nuclear localization was different from that of the known cleavage-packaging proteins, pUL52 might have a distinct role in this process.
Since we expected that pUL52 acts in concert with other viral proteins, it appeared reasonable to generate and study a UL52 mutant in order to learn which step of the HCMV infection cycle was blocked in the absence of pUL52. Disruption of the UL52 ORF was easily achieved by applying established recombination techniques to the BAC-cloned cytomegalovirus (CMV) genome in E. coli. However, the complete UL52 ORF could not be deleted without affecting the promoters or ORFs of the neighboring essential viral genes. The remaining UL52 sequences bore the risk that upon propagation in the complementing cell line, the mutant could regain the missing sequences. Reconstitution of replication-competent viruses was not observed, suggesting that such recombination events are rather unlikely. The unaltered growth kinetics of the recombinant RV-HG-52HA-3, which carried the UL52 gene at an ectopic position, indicated that the expression of UL51 and UL53 was not impaired by the mutation. Thus, the observed phenotype of RV-HG-ΔUL52 was due solely to the disruption of UL52.
For complementation, we generated stably transduced cell lines by retroviral transfer of UL52. Expression of UL52 driven by the retroviral LTR promoter was constitutive and probably below the physiological level obtained during normal viral replication, since the complementing cell line did not support production of the mutant to yields that can be reached with the parental CMV strain. The marginal delay in the growth kinetics of the parental virus on the complementing cell line may represent a specific property of the chosen cell clone or could be due to the continuous presence of the UL52 protein. Retroviral gene transfer for complementation of CMV mutants has been employed by others with similar results (27, 32). There is clearly a need for the establishment of improved complementing systems for CMV mutants, perhaps by using authentic viral promoters to guarantee the correct temporal expression of the respective viral gene. Even so, we could produce amounts of the UL52 mutant on the complementing cell line that were sufficient to perform the study.
The infection experiments indicated that replication of the viral genome was not affected in the absence of pUL52. This result is consistent with the observation that formation of replication compartments was unchanged in cells infected with the mutant. Also, in hindsight of the late expression kinetics of pUL52, one would not have expected an impact on viral DNA synthesis. Similarly, it is unlikely that pUL52 plays a role in capsid assembly, since the morphology of the B capsids in ΔUL52- and RV-HG-infected cells was indistinguishable in ultrathin sections analyzed by electron microscopy.
In the absence of pUL52, neither unit-length genomes nor DNA-filled C capsids were detected in infected cells, indicating that there was a block in genome packaging and cleavage. Herpesvirus genome cleavage and encapsidation are tightly linked processes, and at this stage we cannot resolve whether one or both of these steps were blocked. The low frequency of A capsids, which are believed to arise from loss of DNA due to an abortive encapsidation (29), may suggest that packaging had not even been attempted in these cells. A similar phenotype was reported for an HSV-1 UL32 mutant (21). These authors showed that the HSV-1 UL32 mutant synthesized viral DNA in normal amounts but was defective for cleavage of concatemeric DNA and encapsidation. Moreover, the expression of other HSV-1 cleavage-packaging proteins such as the terminase subunits and the portal protein was not disturbed in the absence of UL32. Similarly, we found in this study that the HCMV terminase pUL56/pUL89 and the putative portal protein pUL104 were expressed and localized correctly in normal cells infected with the UL52 knockout virus, implying that pUL52 did not act via these proteins.
An interesting observation for ΔUL52-infected cells was the high level of amplification of the a-repeat in the uncut concatemers. We could show that the amplification occurred already during propagation of the mutant in the complementing cell line, and we assume therefore that this phenomenon is not specifically due to the lack of the UL52 protein. Since packaging of the mutant genome in the complementing cell line is probably not as efficient as packaging of the wild-type virus genomes, there might be more time for amplification of the a-sequence in the concatemers before encapsidation occurs.
A central question is how pUL52 contributes to cleavage and/or packaging of the viral genomes. We expected that the characterization of the protein and its subcellular localization might give us some hints. Since the RV that expressed the N-terminally tagged pUL52 replicated as the parental virus, we assumed that the modified and the authentic pUL52 possess identical properties. The apparent molecular weight was virtually identical to the calculated one (28), indicating that there were no major posttranslational modifications such as glycosylation. This is of significance since pUL52 displays sequence homology to the equine herpesvirus type 1 major envelope glycoprotein gp300, and therefore pUL52 and its homologs in other herpesviruses were grouped into a protein family termed herpesvirus major envelope proteins (28). We did not detect any potential signal peptide or transmembrane regions in the pUL52 amino acid sequence and never observed any pUL52 labeling at the plasma membrane. Therefore, we consider it very unlikely that pUL52 represents a transmembrane glycoprotein. Since for at least one other protein of this family, pUL32 of HSV-1, glycosylation and membrane association were excluded (10), the designation of this protein family is probably misleading. Although pUL52 was also localized in the cytoplasm, the majority accumulated in the nucleus already early in and throughout the infection cycle. As early as 3 days p.i., pUL52 was found to enclose the replication compartments. The presence of a putative nuclear localization signal at position 315 (PYNKPRR) suggests that pUL52 may be actively imported into the nucleus. The HSV-1 homolog pUL32 may differ from pUL52 with respect to its subcellular distribution. It was not concentrated in the nucleus but was localized predominantly in the cytoplasm, and only at later time points of infection did it colocalize with sites of viral DNA replication in the nucleus (10, 21). In contrast, HCMV pUL52 was excluded from viral replication compartments. In fact, it enclosed the nuclear compartments where viral genome replication and packaging proceeded.
The C-terminally tagged pUL52, which could not support viral replication, was distributed throughout the cell and did not exhibit a specific subnuclear localization. One may therefore conclude that the C terminus is important for the correct subcellular distribution of pUL52 and hence for its function. Perhaps the C terminus of pUL52 interacts with other cellular or viral proteins, which retain pUL52 in specific areas of the nucleus where it fulfills its role during the viral infection cycle. Interestingly, the C-terminally tagged version did not exert a dominant-negative effect when tested in the context of the wild-type genome. A dominant-negative effect of a mutant can occur when an essential interaction partner of the protein becomes sequestered and thus is no longer available for interaction with the authentic protein. Since the C-terminally tagged pUL52 did not act in a dominant-negative manner, one could hypothesize that pUL52 may interact only transiently with its yet unknown partner(s), that the interaction partners are available in high abundance, or, as suggested above, that the tagged protein does not reach the appropriate location. Further experiments will help to discriminate between these possibilities.
In this study, we could not reveal the operating mode of pUL52, but based on our data, several hypotheses are conceivable. (i) pUL52 might be directly involved in the cleavage of the genome concatemers. This seems unlikely, because the terminase subunits responsible for the actual cleavage reaction were located in a different subnuclear compartment. However, association of the terminase subunits with the viral DNA and the final cleavage reaction may occur at different places. (ii) pUL52 could contribute to the association of capsids and genomes. At least for HSV-1, different compartments for replication of the viral DNA and for assembly of the capsids (assemblons) were defined (40). Genome cleavage and encapsidation can occur only if the concatemers and capsids become physically linked. In fact, it was suggested that HSV-1 UL32 directs procapsids to the sites of DNA packaging (21). Our observation that the replication compartments and the compartment defined by the pUL52 labeling abut or even slightly overlap at their boundaries would be compatible with such a function. Further studies are needed to define the replication compartments and assemblons of HCMV and to learn whether they are different entities. (iii) Maturation of the capsids could be affected in the absence of pUL52. Although pUL52 is not a component of the mature virion (39), it may transiently associate with capsids. For instance, in pseudorabies virus there is evidence that components of the primary tegument and envelope acquired in the nucleus are lost after transfer of the particles through the nuclear membrane (16). The phenotype observed here for the UL52 mutant is somehow reminiscent of that of HSV-1 UL17 mutants that also fail to perform DNA packaging. A recent study indicated that UL17 together with its binding partner, UL25, constitute a minor component of the HSV capsids and upon genome encapsidation stabilize the mature C capsids (38). Although UL52 is not a homolog of UL17 or UL25, it may provide a similar function in the maturation process of HCMV capsids. (iv) pUL52 might also play a role in the transport of the capsids to the inner nuclear membrane. The localization of pUL52 between the nuclear envelope and the compartments where replication and packaging take place would be compatible with the idea that pUL52 supports the transfer of capsids to the sites where nuclear egress occurs. Indeed, active transport of herpesvirus capsids in the nucleus has recently been shown, and it was proposed that herpesviruses may synthesize a novel intranuclear motor system (15). Perhaps UL52 constitutes a part of such a transport system.
We favor hypotheses (ii) and (iii), but in order to decide which of these scenarios does apply, the cellular and viral interaction partners of pUL52 have to be identified in further studies. This will also enable us to elucidate the molecular working mechanism of this essential HCMV protein. The detailed knowledge of the processes of genome encapsidation and capsid maturation prior to nuclear egress opens the perspective necessary to interfere with these events and to develop more-effective antiviral compounds against HCMV or herpesviruses in general.
Acknowledgments
We thank Elke Bogner (Charite, Berlin) and Bodo Plachter (University of Mainz) for providing antibodies. We are grateful to Rudi Bauerfeind (Confocal Microscopy Facility, Hannover Medical School) for invaluable support with the confocal laser scanning microscopy.
The work was in part supported by DFG grant So403/3 to B.S.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Beard, P. M., and J. D. Baines. 2004. The DNA cleavage and packaging protein encoded by the UL33 gene of herpes simplex virus 1 associates with capsids. Virology 324475-482. [DOI] [PubMed] [Google Scholar]
- 2.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 764785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279349-352. [DOI] [PubMed] [Google Scholar]
- 4.Bogner, E., K. Radsak, and M. F. Stinski. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 722259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borst, E. M., C. Benkartek, and M. Messerle. 2007. Use of bacterial artificial chromosomes in generating targeted mutations in human and mouse cytomegaloviruses, p. 10.32.1-10.32.30. In J. E. Coligan, B. Bierer, D. H. Margulies, E. M. Shevach, W. Strober, and R. Coico (ed.), Current protocols in immunology. John Wiley & Sons, New York, NY. [DOI] [PubMed]
- 6.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 738320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borst, E. M., S. Mathys, M. Wagner, W. Muranyi, and M. Messerle. 2001. Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J. Virol. 751450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borst, E. M., and M. Messerle. 2003. Construction of a cytomegalovirus-based amplicon: a vector with a unique transfer capacity. Hum. Gene Ther. 14959-970. [DOI] [PubMed] [Google Scholar]
- 9.Borst, E. M., and M. Messerle. 2005. Analysis of human cytomegalovirus oriLyt sequence requirements in the context of the viral genome. J. Virol. 793615-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. E., A. P. Poon, and B. Roizman. 1996. Properties of the protein encoded by the UL32 open reading frame of herpes simplex virus 1. J. Virol. 703938-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154125-169. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dittmer, A., J. C. Drach, L. B. Townsend, A. Fischer, and E. Bogner. 2005. Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-d-ribonucleosides. J. Virol. 7914660-14667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 10014223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forest, T., S. Barnard, and J. D. Baines. 2005. Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7429-431. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39389-400. [DOI] [PubMed] [Google Scholar]
- 18.Giesen, K., K. Radsak, and E. Bogner. 2000. The potential terminase subunit of human cytomegalovirus, pUL56, is translocated into the nucleus by its own nuclear localization signal and interacts with importin alpha. J. Gen. Virol. 812231-2244. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, J. S., and E. Bogner. 2002. ATPase activity of the terminase subunit pUL56 of human cytomegalovirus. J. Biol. Chem. 2776943-6948. [DOI] [PubMed] [Google Scholar]
- 20.Klupp, B. G., H. Granzow, G. M. Keil, and T. C. Mettenleiter. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 806235-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 722463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leuzinger, H., U. Ziegler, E. M. Schraner, C. Fraefel, D. L. Glauser, I. Heid, M. Ackermann, M. Mueller, and P. Wild. 2005. Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 7913047-13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 721060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2674. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 25.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 806286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penfold, M. E., and E. S. Mocarski. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 23946-61. [DOI] [PubMed] [Google Scholar]
- 27.Prichard, M. N., N. Gao, S. Jairath, G. Mulamba, P. Krosky, D. M. Coen, B. O. Parker, and G. S. Pari. 1999. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J. Virol. 735663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rigoutsos, I., J. Novotny, T. Huynh, S. T. Chin-Bow, L. Parida, D. Platt, D. Coleman, and T. Shenk. 2003. In silico pattern-based analysis of the human cytomegalovirus genome. J. Virol. 774326-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 30.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheffczik, H., C. G. Savva, A. Holzenburg, L. Kolesnikova, and E. Bogner. 2002. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res. 301695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 7710594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1361007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 7510755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamashiro, J. C., D. Filpula, T. Friedmann, and D. H. Spector. 1984. Structure of the heterogeneous L-S junction region of human cytomegalovirus strain AD169 DNA. J. Virol. 52541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thoma, C., E. Borst, M. Messerle, M. Rieger, J. S. Hwang, and E. Bogner. 2006. Identification of the interaction domain of the small terminase subunit pUL89 with the large subunit pUL56 of human cytomegalovirus. Biochemistry 458855-8863. [DOI] [PubMed] [Google Scholar]
- 37.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 802118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trus, B. L., W. W. Newcomb, N. Cheng, G. Cardone, L. Marekov, F. L. Homa, J. C. Brown, and A. C. Steven. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 7810960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward, P. L., W. O. Ogle, and B. Roizman. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J. Virol. 704623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, K., F. Homa, and J. D. Baines. 2007. Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J. Virol. 816419-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 10012396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]