Abstract
The human papillomavirus (HPV) oncogene E6 has been shown to perform multiple functions (p53 degradation, telomerase activation, etc.) that play a role in oncogenic transformation. Beyond known E6 functions, an undefined mechanism that allows cellular invasion requires the E6 PDZ binding motif (PDZBM). Here, we show that HPV type 16 (HPV16) E6 interacts with and induces loss of a protein tyrosine phosphatase (PTPN13) in a PDZBM-dependent manner. PTPN13 loss induced either by the presence of E6 or by a short hairpin RNA strategy allows for anchorage-independent growth (AIG) and synergy with a known oncogene, Rasv12, resulting in invasive growth in vivo. Restoring PTPN13 expression reverses AIG in cells lacking PTPN13. A genomic analysis of colorectal carcinoma has identified an association between PTPN13 loss-of-function mutations and aberrant Ras signaling. Our findings support this correlation and provide methods for further evaluation of the mechanisms by which PTPN13 loss/Ras expression leads to invasive growth, the results of which will be important for treatment of HPV-related and non-HPV cancer.
Significant associative evidence supports a role for human papillomavirus type 16 (HPV16) in the carcinogenic progression of at least 25% of head and neck squamous cell cancer (HNSCC) cases (27, 38). In particular, the incidence of HNSCC of the tonsillar region is increasing (18, 20, 38). The epithelial surfaces surrounding the tonsil (oropharynx) are cancerous in a very high (60%) proportion of cases that are HPV positive, and these cases present with more metastatic and advanced disease states (34, 38) than HPV-negative cases. The incidence of HPV-related HNSCC is also increased in individuals with human immunodeficiency virus/AIDS disease (10). While the data presented in the following studies pertain to HPV-related HNSCC they may also be applicable to cases of HPV-related cervical cancer, which remains the second leading cause of cancer-related death in women. Therefore, understanding the viral mechanisms that lead to cancer will help meet a need for HPV-specific targeted therapy.
One of the major HPV viral oncogenes that allow progression to cancer is HPV16 E6. E6 has many potential carcinogenic effects, including telomerase activation and p53 degradation (26, 35). However, an understood mechanism that potentiates malignant progression has been associated with a PDZ (PSD-95, Discs Large, ZO-1) binding motif (PDZBM), a specific amino acid sequence that mediates protein-protein interactions with corresponding PDZ domain-containing proteins. High-risk HPV virus types 16, 18, 31, and 33 possess this binding motif at the E6 C terminus. In addition to this correlative evidence of increased malignant potential in humans, mouse studies demonstrated that PDZBM is required for HPV-related malignancy (37).
The studies described below explored the role of the E6 PDZBM in binding and inducing loss of a PDZ domain containing phosphatase PTPN13. PTPN13 is a member of the nonreceptor phosphatases and specifically falls into a class of phosphatases that contain FERM (four point one, ezrin, radixin, moesin) domains. Other members in this group are PTPD1 and PTPH1. PTPH1 has a PDZ binding domain and also been recently reported to bind E6 (22). PTPN13 has multiple names (PTPL1, PTP-Bas, PTP1e, FAP-1, and PTP-Bl [mouse]); for simplicity, we will use its genomic name, PTPN13. PTPN13 is a highly modular, large (270 kDa) protein with multiple interactive domains. The C terminus of PTPN13 has a kinase noncatalytic C-lobe (KIND) domain with a nondefined function. After the KIND domain, the next functional area is a FERM domain. This 300-amino-acid stretch has been shown to be critical for membrane targeting (15). PTPN13 also contains five different PDZ binding domains. A host of potential binding partners have been identified for these domains (7). Finally, PTPN13 has an N-terminal phosphatase domain; recent crystal structure analysis suggests that it has targets with multiple tyrosine-phosphorylated sites (42). PTPN13 has been implicated in several cellular pathways that play a role in cell survival. For instance, IκBA, EphrinB, B-catenin, and c-src have all been identified as potential targets (7). A mutational analysis of human colon cancers indicates that loss of function is critical for carcinogenesisis (43). The in vitro and in vivo findings in this paper supply further functional evidence of the important role PTPN13 plays as a tumor suppressor.
To further understand the functional significance of the HPV16 E6 PDZBM-induced loss of PTPN13 we have completed several assays that test invasiveness. The in vitro assay we use to assess invasive transformation is anchorage-independent growth (AIG), which can correlate with invasive growth in vivo (11). When normal, nontransformed epithelial cells are grown under AIG conditions, programmed cell death is induced (6). Our past work has demonstrated PDZBM-dependent induction by E6 of AIG in early-passage human tonsil epithelial cells (HTECs) (40). To further explore the relevance of these in vitro findings in a living animal we have also developed techniques to examine viral transformation of early-passage murine tonsil epithelial cells (MTECs). The following data show that both mouse and human tonsil keratinocytes require E6 PDZBM for AIG. We then use this similarity to identify a novel E6 PDZ interaction with a PDZ domain-containing protein whose loss has the physiologic consequence of allowing both AIG and invasive growth.
MATERIALS AND METHODS
Short hairpin RNA (shRNA) vectors.
Retroviral vectors targeting human PTPN13 were kindly provided by Tom Hei (21) (Columbia University, New York, NY). Nontargeting control vectors and vectors targeting mouse PTPN13 were obtained from Open Biosystems (source identification no. 64925). Virus was produced as previously described (8, 14).
Cell lysis immunoblot analysis.
Whole-cell lysates were harvested at 4°C with protein lysis buffer as previously described (19). Expression of human p53 was measured by immunoblotting using 25 μg total cellular proteins with p53 antibody (OP-43; CalBiochem, San Diego, CA) at a 1:1,000 dilution and a standard Western blotting technique. The antibodies used were hDLG (BD catalog no. D67820-050), GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Ambion catalog no. 4300), PTPN13 (Santa Cruz Biotechnology catalog no. 15356), and actin (Santa Cruz catalog no. sc-1616). Blots were developed by ECL chemiluminescent detection (Pierce Biotechnology).
Soft agar assay.
AIG assays were performed following previously published methods, with triplicate experiments for each set of conditions (2). Briefly, a base layer of 1% noble agar was placed on a 12-mm-diameter 0.4-um-pore-size synthetic membrane insert (Costar). Cells (1 × 104) were suspended in 0.3% agar with medium over the base layer. Cells were fed every other day with their respective growth media introduced into the lower chamber. At 3 weeks, colonies greater than 100 μm in diameter were counted. The degree of anchorage-independent growth was calculated using the following equation for determining percent colony-forming efficiency: (no. of colonies counted at 3 weeks/no. of cells seeded at start of assay) × 100.
In vivo assay.
In vivo growth was assayed using previously described techniques (19). All experiments were performed in accordance with institutional and national guidelines and regulations, with the protocol reviewed and approved by the Animal Care Committee at the University of Iowa. Briefly, using an 18-gauge needle, C57BL/6 mice (six per group) were injected with 1 × 106 cells in the subcutaneous tissue of the flank. Animals were euthanized when the tumor size was >20 mm in its greatest dimension or when the animal was substantially emaciated. Mice were considered tumor free when they showed no evidence of tumor after 3 months. The data shown are from one representative experiment that was replicated two times.
Cell culture.
Normal HTECs and MTECs were isolated from the epithelium overlying C57BL/6 mouse and human tonsil tissue as previously described (19, 40). HEK293, UPCI SCC90 (provided by Sue Gorlin), and UMSCC84 human cell lines were maintained in 10% fetal bovine serum-Dulbecco modified Eagle medium with 1% penicillin/streptomycin.
RT-PCR of E6 and PTPN13.
The results of RNA isolation and reverse transcription-PCR (RT-PCR) quantitation of E6 oncogenes were analyzed as previously described (28, 29, 39). The message levels (see Fig. 2) were standardized to 18S rRNA levels by use of a primer probe kit from Applied Biosystems (Foster City, CA).
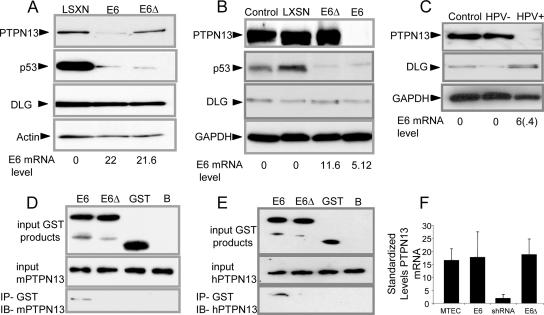
FIG. 2.
HPV16 E6 PDZBM is required for induction of loss and for physical association with PTPN13. The results of Western blot analyses of indicated proteins after retroviral transduction with the indicated vectors (LXSN, E6, E6Δ) are shown as follows. (A) HTECs with extended life in culture due to expression of hTert and shRNA in response to the presence of p16. (B) MTECs. (C) HNSCC lines with HPV16 (UPCI SCC90 [HPV+]) and without HPV16 (UMSCC84 [HPV−]), with E6 mRNA expression levels indicated as percents (with s.d. in parentheses); n = 3, with experiments repeated. (The E6 level shown for all cell lines refers to quantitative real-time RT-PCR of E6 mRNA standardized to an 18S rRNA signal.) (D and E) GST-E6 proteins (upper panel) bound directly to GST beads were incubated with lysates of cells of the 293 cell line (middle panel) expressing mouse or human PTPN13. Western blotting (IB) of mouse and human PTPN13 revealed coprecipitation (IP) of GST-E6 products with mouse and human PTPN13, respectively (lower panel). (F) PTPN13 quantitative real-time RT-PCR standardized to an 18S rRNA signal. Error bars represent s.d.; n = 3, with experiments repeated.
Coimmunoprecipitation of PTPN13 with E6-GST.
Fragments encoding HPV16 E6 and HPV16 E6Δ146-151 were cloned between BamHI and EcoRI sites into vector pGEX-2T (Amersham Biosciences) downstream of the glutathione S-transferase (GST) open reading frame. GST, GST-E6, and GST E6Δ146-151 were expressed and purified by following a protocol modified from reference 9 without dithiothreitol in the lysis buffer. 293T cells were transfected with pEGFPN3-PTPBL (4), and the cells were harvested 24 h after transfection and lysed in lysis buffer (50 mM TrisHCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 2 mM Na3VO4, 10 mM NaPPi, 100 mM NaF, 10% glycerol, 0.1% NP-40, protease inhibitors). Equal amounts of GST and the two fusion proteins (15 μg) and PTPBL-EGFP (50 μg total protein) were used for each precipitation. The GST, GST-E6, and GST E6Δ146-151 lysates were incubated directly with 10 μl GST beads (Amersham Biosciences) overnight at 4°C in TEN100 (20 mM Tris [pH 7.5], 0.1 mM EDTA, 100 mM NaCl, protease inhibitors). The beads were then washed three times with TEN 200 (20 mM Tris [pH 7.5], 0.1 mM EDTA, 200 mM NaCl, 0.5% NP-40, protease inhibitors). PTPN13 lysates were added to the beads, and the complexes were incubated at 4°C for 4 h in phosphate-buffered saline and then washed three times with TEN300 (20 mM Tris [pH 7.5], 0.1 mM EDTA, 300 mM NaCl, 1% Triton, protease inhibitors). The bound proteins were detected by Western blotting using anti-FAP-1 antibody (sc-15356).
Transfection.
HEK293 and UPCI SCC90 cell lines were transfected at 60% to 80% confluence by use of Polyfect reagent (Qiagen catalog no. 301105) following manufacturer protocols. Briefly, 8 μg of pEGFP-N1 with and without PTPN13 was transfected using the appropriate volumes of manufacturer reagents into cells on a 100-mm-diameter tissue culture dish. Addition of antibiotics (G418) to the cell culture media following transfection allowed for selection of stable cell lines expressing the plasmids of interest.
Immmunohistochemistry.
Formalin-fixed, paraffin-embedded tissues were deparaffinized in xylene and antigen unmasked in citrate buffer, and endogenous peroxidases were quenched with 2% H2O2. Tissues were blocked with 10% bovine serum albumin. PTPN13 was detected using either sc-15356 or sc-1138 (Santa Cruz Biotechnology, Santa Cruz, CA). Incubation with primary antibody (1:125) was performed overnight at 4°C. Secondary antibody incubation and detection were carried out with an EnVision+ system (for sc-15356) or LSAB+ system (for sc-1138) according to the instructions of the manufacturer (DakoCytomation). Slides were costained with hematoxylin. As a control to test specificity, either a fivefold excess of blocking peptide (sc-1138 P; Santa Cruz) prior to application or secondary antibody alone was used.
Statistics.
Standard errors and standard deviations (s.d.) as indicated were calculated using Microsoft Excel formulas.
RESULTS
E6 induces AIG in MTECs via a PDZ-dependent mechanism.
Our past work has used retroviral transduction of HPV16 viral oncogenes into early-passage tonsil keratinocytes to demonstrate the requirement of the E6 PDZBM in allowing AIG (40). Further investigation of invasive growth by human cells in vivo has obvious practical limitations in studies of humans. Therefore, to determine whether our AIG findings determined using human cells had implications for invasive growth in vivo we first explored whether MTECs demonstrated the same E6 PDZBM requirement for AIG and then determined whether the PDZBM was needed for invasive growth in vivo. MTECs expressing wild-type E6 displayed AIG, whereas E6 PDZBM-deleted MTECs (E6Δ146-151) and vector control cells did not display AIG (Fig. 1A). To assess whether the observed AIG in retroviral transduced cells also occurred in the available HPV16-positive head and neck cancer cell line we also examined growth of this cell line in soft agar. We observed AIG in an HPV16-positive cell line (UPCI SCC90) but not in an HPV-negative cell line (UMSCC84) (Fig. 1A). These data demonstrate that a mechanism associated with the E6 PDZBM plays a role in allowing AIG.
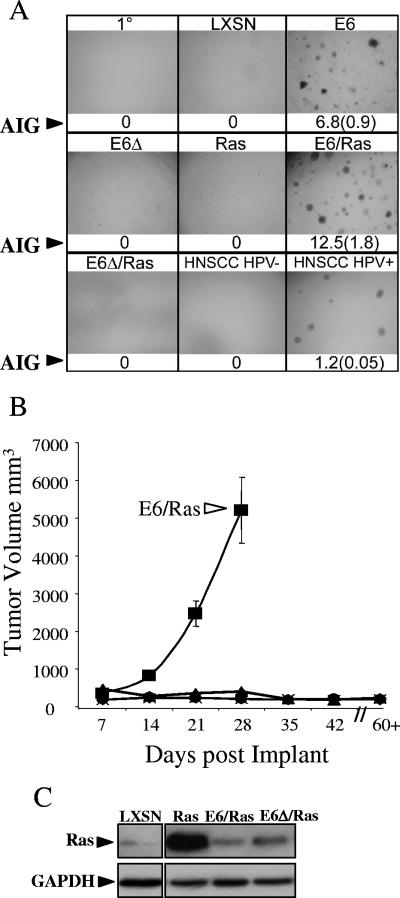
FIG. 1.
E6 induction of AIG and invasive growth is dependent on the PDZBM. (A) E6 induction of AIG. Single-cell suspensions of 104 primary (1°) MTECs and MTECs transduced with LXSN (vector control), E6, E6Δ (no PDZBM), H-Ras, E6/H-Ras, E6Δ/H-Ras, and HNSCC HPV− (UMSCC84) and HPV+ (UPCI SCC90) cell lines were seeded in triplicate into soft agar and photographed at a magnification of ×40 after 3 weeks. Colonies and corresponding quantifications of colony-forming efficiency (in percentages of AIG) along with standard errors (in parentheses) are shown; n = 3, with experiments repeated. (B) Average tumor growth curves for mice injected with the indicated cells subcutaneously in the flank. ▪, E6/Ras; ♦, E6; ▴, E6Δ/Ras; •, E6Δ; X, Ras. Error bars represent s.d.; n = 6. (C) Ras expression in MTECs after retroviral transduction with either vector expressing Ras. The upper panel shows total Western blotting results, comparing total Ras levels in the indicated cell lines. GAPDH levels are shown to compare protein loading results.
The PDZBM of E6 is required to synergize with Ras for invasive growth.
To determine whether the E6 PDZBM requirements for AIG also played a role in invasive growth in vivo we also examined an in vivo growth assay that implants the various transduced MTECs back into immune-competent syngeneic C57BL/6 mice. Our past work has found that E6/E7 was sufficient to immortalize but not allow invasive growth of MTECs after reimplantation (19). The addition of Rasv12 to cells expressing E6/E7 allowed invasive tumorigenic growth. The results showed not only that the mice developed subcutaneous nodules but also that the cells comprising these nodules directly invaded surrounding muscle, bone, and nerve, resulting in paralysis (19). We also show that in MTECs, E6 alone is sufficient to induce this same invasive growth in combination with Rasv12. In this report we expand on these previous studies by showing that expression of E6 alone with or without the PDZBM was not sufficient to allow invasive tumorigenic growth, suggesting that the requirements for AIG and invasive growth in vivo are different (Fig. 1B) (1, 19). We hypothesized that other cellular changes are required to support invasive growth in vivo. The idea that cells would require another mutation or oncogene for neoplastic growth is supported by the observation that Ras signaling is frequently aberrant in HNSCC; furthermore, in past studies Ras has been shown to act synergistically with E6 in mouse skin cancer models (23, 24, 32, 36). Therefore, we tested whether Rasv12 could synergize with E6 for enhanced AIG and in vivo growth of MTECs. Addition of Rasv12 to E6-expressing cells substantially increased AIG, whereas the presence of Rasv12 alone or E6Δ146-151/Rasv12 failed to result in growth (Fig. 1A), suggesting that a mechanism involving the E6 PDZBM is required to synergize with Rasv12 to increase AIG. When the cells were injected into syngeneic mice, E6/Rasv12 cells displayed growth whereas E6Δ146-151/Rasv12 cells failed to grow (Fig. 1B). To determine whether the E6 PDZBM enhanced levels of Rasv12, thus allowing invasion, we examined the levels of Rasv12 in the cell lines. E6/Rasv12 cells showed less expression, suggesting that E6 does not enhance invasion by increasing Rasv12 expression (Fig. 1C), although it is possible that E6 alters the downstream signaling of Ras pathways. These data show that anchorage-independent growth in vitro and invasive growth in vivo are dependent on a mechanism associated with the E6 PDZBM.
Loss of PTPN13 requires expression of E6 with a PDZBM.
This E6 PDZBM-dependent induction of AIG in both MTECs and HTECs led us to look for relevant PDZ domain-containing proteins interacting with E6 to exert this effect. Based on protein sequencing results, E6's PDZBM is predicted to interact with class 1 PDZ domains. Yeast two-hybrid and coprecipitation studies with E6 fusion proteins have generated a list of candidate proteins that interact with the E6 PDZBM (12, 13, 22, 25, 30, 33, 41), but whether these proteins play a role in malignant transformation has not been addressed. E6 has been shown to target interacting proteins for degradation in vitro (35). To determine whether loss of candidate PDZ-interacting proteins correlated with the presence of the E6 PDZBM, whole-cell lysates from HTECs and MTECs transduced with either E6 or E6Δ146-151 were examined by Western blot analysis. As shown for the DLG antibody (Fig. 2A and C), none of the previous reported candidates (results for Scribble, PTPH1, and Magi1-3 are not shown) demonstrated PDZBM-dependent loss. These results are consistent with findings from Simonson et al. showing that neither DLG and Scribble was degraded in transgenic mice expressing E6 or in human HPV-positive (HPV+) cancer cell lines (37). Since our data suggested that a mechanism of the E6 PDZBM synergized with Ras, we investigated PTPN13, a PDZ domain-containing phosphatase whose loss has been associated with aberrant Ras activity in colon cancer (43). PTPN13 levels were decreased in HTECs and MTECs expressing E6 but not E6Δ146-151 in vitro (Fig. 2A and B). Both wild-type E6- and E6Δ146-151-expressing cells exhibited p53 degradation, as has been previously shown (5), demonstrating that the E6Δ146-151 mutant still degrades p53 (Fig. 2B). We also found that the HPV+ HNSCC cell line had associated loss of PTPN13, whereas an HPV-negative (HPV−) cell line did not (Fig. 2C). Taken together, these findings show a consistent association between E6 expression and loss of PTPN13.
Interaction between E6 and PTPN13 requires the PDZBM.
The observed loss of PTPN13 could be due to direct physical association, transcriptional regulation, or targeted degradation through other intermediate proteins. To determine whether E6 interacts with PTPN13 via its PDZBM, we incubated GST-E6 or GST-E6Δ146-151 fusion proteins with cell lysates, coprecipitated with GST beads, and blotted for PTPN13. We were unable to detect association in MTECs or untransfected 293 cells, likely because of the relatively small amount of this protein present. We were able to coprecipitate human as well as mouse PTPN13 with full-length E6 but not with the E6Δ146-151 mutant (Fig. 2D and E) when we used lysates from transiently transfected 293 cells that overexpressed the protein. To determine whether E6 altered transcriptional regulation, we examined PTPN13 mRNA levels by quantitative RT-PCR using primary, E6, E6Δ146-151, and shPTPN13 cells. Similar levels of PTPN13 mRNA were detected in MTEC controls, E6, and E6Δ146-151 cells, and as expected, PTPN13 levels were diminished in shPTPN13 cells (Fig. 2F). These data indicate that downregulation of PTPN13 protein expression is modulated posttranscriptionally by a PDZ-mediated physical interaction between E6 and PTPN13.
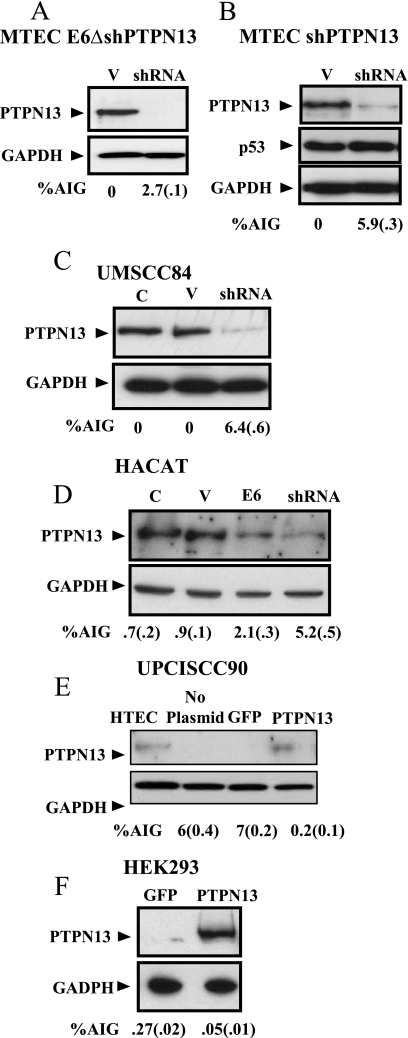
Loss of PTPN13 induces AIG in mouse and human epithelial cells.
The combined action of E6 and PTPN13 loss resulting in induction of AIG prompted the question of whether PTPN13 loss alone was sufficient to induce AIG or whether there were other important functions of E6 contributing to cellular transformation. To determine the physiologic consequence of PTPN13 loss, we used an shRNA approach. As with E6-associated PTPN13 loss, the presence of shPTPN13 induced loss in early-passage MTECs and MTECs expressing E6Δ146-151 and was sufficient to induce AIG (Fig. 3A and B). Expression of p53 protein remained constant (Fig. 3B). These data and the fact that E6Δ146-151 degrades p53 but not PTPN13 suggest that the mechanism of E6 PDZBM-induced loss is a separate function from E6's ability to degrade p53. Interestingly, shPTPN13 induced more AIG for the MTECs than for the E6Δ146-151 cells. This finding may be related to small differences in induced loss of PTPN13 between these cells. As seen with MTECs, shRNA-mediated loss of PTPN13 was sufficient to induce AIG in the previously mentioned HPV− UMSCC84 HNSCC cell line (Fig. 3C). To test whether depletion of this protein would also alter the AIG of a different epithelial cell we examined AIG in HaCaT cells, a well-characterized immortal skin keratinocyte cell line (Fig. 3D). When one compares the loss of PTPN13 induced by E6 to that induced by shPTPN13 in these cells, it is clear that neither method results in a total loss of protein; shPTPN13 shows a slightly increased level of loss compared to E6 results. The association of PTPN13 loss with AIG is further corroborated by the reexpression of PTPN13 in epithelial cell lines that lack PTPN13 due to E6 degradation (UPCI SCC90) or cell lines that display AIG and spontaneously lack PTPN13 (Fig. 3E and F). We were unable to establish primary cells that overexpressed this protein due to cell death induced by overexpression. However, in the established cell lines we were able to find clones that grew at a normal rate, expressed full-length PTPN13, and were viable over the long term in culture. Interestingly, even in these cell lines, clones that overexpressed the protein did not survive (data not shown). Therefore, it became apparent that even transformed cells could tolerate only so much PTPN13. Upon restoration of PTPN13 protein expression to near-physiologic (compared to HTECs) amounts (Fig. 3E), AIG was substantially reduced or even blocked in these cells (Fig. 3E and F), demonstrating a critical role of PTPN13 in determining the cell's ability to grow in an anchorage-independent manner.
FIG. 3.
Modulation of PTPN13 alters AIG. (A) Levels of AIG with loss of PTPN13 protein in MTECs expressing E6Δ in isolated clonal populations of cells after transduction with shRNA constructs targeting PTPN13 compared to the results obtained with control (V) nontargeting shRNA vector. In all panels, standard errors are shown in parentheses; n = 3, with experiments repeated. (B) Levels of AIG with loss of shPTPN13 in early-passage MTECs cells transduced with shRNA targeting PTPN13 (clonal population) compared to the results obtained with nonsense shRNA vector (V). (C) PTPN13 in an HPV− HNSCC cancer line (UMSCC84). C, no treatment; V, nonsense shRNA vector; shRNA, vector targeting PTPN13. (D) Loss of PTPN13 in HaCaT cells, representing a skin keratinocyte, induced by the presence of either E6 or shPTPN13 increased AIG. Lysates of cells transduced with the indicated constructs were examined for AIG and loss of PTPN13. (E and F) UPCI SCC90 (HPV+) and HEK293 cell lines were transfected and selected with G418 to obtain stable lines expressing either GFP or PTPN13. AIG assay results and PTPN13 protein levels after transfection are shown. HTEC PTPN13 protein levels are shown in panel E for comparison to the amount of PNPN13 in transfected cells.
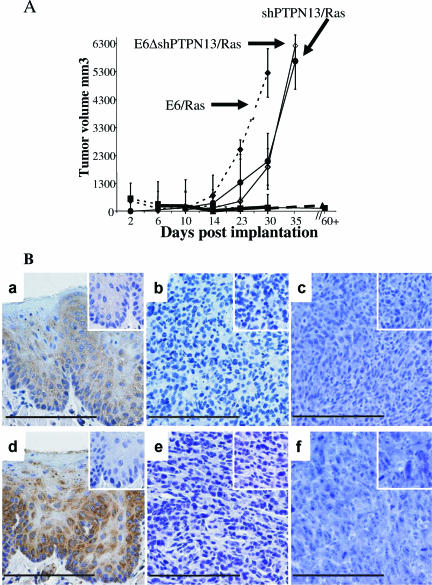
Loss of PTPN13 synergizes with Ras to allow invasive growth.
The observation of AIG associated with decreased PTPN13 levels alone led us to further assess the in vivo physiologic significance of the loss of PTPN13 by injecting the various cells into mice and quantifying growth. As with E6-expressing cells, the MTECs with shPTPN13 alone were not able to grow in vivo; however, shPTPN13 cells with Ras grew invasively (Fig. 4A). Likewise, shRNA-induced loss of PTPN13 in the E6Δ146-151/Ras cells also allowed invasive growth. The cells with E6 did exhibit a higher growth rate than the cells with shPTP-BL, possibly suggesting that E6 PDZBM has other modifying functions that help with invasive growth. Histologically, all tumors were squamous cell cancers. As is customary for squamous cancers, intratumoral variation with regard to differentiation did occur. All tumors showed muscle and capillary lymphatic space and nerve invasion. No metastasis was observed in autopsies performed on these mice. To determine whether the E6- and shPTPN13-associated loss of PTPN13 observed in culture persists in vivo, we compared PTPN13 immunostaining results for normal tonsil epithelium with those seen with murine E6/Ras or Ras/shPTPN13 tumors. PTPN13 immunostaining was observed in the suprabasal cells in the normal epithelium. This location corresponds to the localization of E6 oncogene expression in HPV lesions (16, 17). None of the mouse E6/Ras or shPTPN13/Ras tumors showed detectable PTPN13 immunostaining (Fig. 4B). Taken together, these data demonstrate that loss of PTPN13 either with or without the intervention of a mechanism associated the E6 PDZBM is required for AIG and that this loss, in conjunction with expression of an oncogene (Rasv12), will permit invasive growth of MTECs in vivo.
FIG. 4.
Loss of PTPN13 synergizes with H-Ras for invasive growth in immune-competent mice. (A) Average growth curves for mice injected with the indicated cells subcutaneously in the flank. ♦, E6/Ras; •, shPTPN13/Ras; ⋄, E6Δ/shPTPN13/Ras; ▪, shPTPN13; ▴, E6Δ/shPTPN13. Error bars represent s.d.; n = 6. (B) Immunohistochemistry of PTPN13 protein in normal mouse tonsil epithelium (panels a and d), E6/Ras tumors (panels b and e), and shPTPN13/Ras tumors (panels c and f) in vivo. Panels a to c and panels d to f present the results of the use of antibodies against epitopes for the C and N termini of the protein, respectively. Insets show the results obtained with either blocking peptide (panels a to c) or secondary controls (panels d to f) alone. Bars, 130 um.
DISCUSSION
The propensity of high-risk HPV16 E6 to participate in the transformation of an epithelial cell correlates with the ability of the viral oncogene of high-risk HPVs to perform multiple mechanisms that are necessary but not sufficient to allow invasion. While other reported non-PDZBM-dependent functions (e.g., degradation of p53, activation of telomerase, etc.) are also likely required, the mechanism associated with the PDZBM plays a critical role in allowing invasive growth. As discussed above, multiple PDZ domain-containing proteins can interact with PDZBM of E6 (12, 13, 22, 25, 30, 33, 41). It is likely that E6 can interact with multiple PDZ domain-containing proteins and that each interaction may have a different effect on transformation. Our data show an E6 PDZBM interaction, E6 PDZBM-induced loss, and the physiologic consequence of that loss with regard to PTPN13 for AIG of human and mouse keratinocytes. The data also show the significance of this interaction in synergy with a second oncogene-activated Ras in vivo for invasive growth of MTECs. The discovery of this E6-PTPN13 link will allow the exploration of several pertinent questions. (i) Is the binding direct, and, if so, which domain(s) of PTPN13 does E6 bind? (ii) How does the interaction result in degradation? (iii) What is the physiologic relevance of the E6/Ras synergy for human HPV-related HNSCC? Our data suggest that the addition of another intracellular “hit” such as a modification of Ras or of upstream pathways that lead to increased Ras signaling (by, e.g., Erb receptor tyrosine kinases) may explain the progression from HPV infection to carcinogenic events. (iv) What are the transformation roles of the other cellular proteins that have been reported to interact with the E6 PDZBM? The transformation methods utilized in this study now provide an avenue for further exploration these questions with respect to mouse and human tonsillar keratinocytes.
Finally, these findings suggest that PTPN13 and its downstream signaling events play a role in preventing AIG. PTPN13 is a nonreceptor protein tyrosine phosphatase with multiple functional domains (7). The mouse and human homologs have many components that are nearly identical (e.g., PDZ domain 2 is 96% identical between mice and humans) and share an overall homology of 80% (3, 15), suggesting similar functions in human and mouse cell lines. Multiple potential PTPN13-interacting proteins and phosphatase targets point to a role for PTPN13 in cell growth, motility, and apoptosis (7). However, the physiological relevance of the reported interactions has thus far not been proven in vivo. An engineered mouse with a mutation that deletes PTPN13's catalytic phosphatase domain has been generated, and subtle defects in neuron regrowth following injury and in retinal ganglion cell survival were noted (31, 44). Our data suggest that the other domains in PTPN13 may play a more critical role, since we observed a change in the ability of cells to grow in an anchorage-independent manner, both in vitro and in vivo, upon loss of the entire protein. Importantly, the gene encoding PTPN13 is among the six tyrosine-specific phosphatase genes that are frequently mutated in conjunction with aberrant Ras signaling in human colon cancer specimens (43). In line with our findings, these data from human cancers point to a suppressive role for PTPN13 in invasive growth. The growth assays exploited in this study now provide a method to further explore the relevance of the various functional domains of PTPN13 and to examine the downstream cellular pathways affected by PTPN13 that result in AIG suppression. In summary, these findings show that HPV16 E6 PDZBM mediates the loss of PTPN13 and that loss of PTPN13 plays a critical role in allowing invasive growth for epithelial cells.
Acknowledgments
This work is supported by merit funding through the Veterans Affairs Medical Center.
We thank Lieke van den Berk, Jan Schepens, and Willem Melchers for help in plasmid constructions and Michael Welsh, Joseph Zabner, and Zoya Kurago for critical insight and review of our work.
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Alonio, L. V., M. A. Picconi, D. Dalbert, J. Mural, O. Bartt, G. Bazan, M. Dominguez, and A. R. Teyssie. 2003. Ha-ras oncogene mutation associated to progression of papillomavirus induced lesions of uterine cervix. J. Clin. Virol. 27263-269. [DOI] [PubMed] [Google Scholar]
- 2.Berger, K. L., F. Barriga, M. J. Lace, L. P. Turek, G. J. Zamba, F. E. Domann, J. H. Lee, and A. J. Klingelhutz. 2006. Cervical keratinocytes containing stably replicating extrachromosomal HPV-16 are refractory to transformation by oncogenic H-Ras. Virology 35668-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuppen, E., S. Nagata, B. Wieringa, and W. Hendriks. 1997. No evidence for involvement of mouse protein-tyrosine phosphatase-BAS-like Fas-associated phosphatase-1 in Fas-mediated apoptosis. J. Biol. Chem. 27230215-30220. [DOI] [PubMed] [Google Scholar]
- 4.Cuppen, E., M. Wijers, J. Schepens, J. Fransen, B. Wieringa, and W. Hendriks. 1999. A FERM domain governs apical confinement of PTP-BL in epithelial cells. J. Cell Sci. 112(Pt. 19)3299-3308. [DOI] [PubMed] [Google Scholar]
- 5.Dalal, S., Q. Gao, E. J. Androphy, and V. Band. 1996. Mutational analysis of human papillomavirus type 16 E6 demonstrates that p53 degradation is necessary for immortalization of mammary epithelial cells. J. Virol. 70683-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douma, S., T. Van Laar, J. Zevenhoven, R. Meuwissen, E. Van Garderen, and D. S. Peeper. 2004. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature 4301034-1039. [DOI] [PubMed] [Google Scholar]
- 7.Erdmann, K. S. 2003. The protein tyrosine phosphatase PTP-Basophil/ Basophil-like. Interacting proteins and molecular functions. Eur. J. Biochem. 2704789-4798. [DOI] [PubMed] [Google Scholar]
- 8.Foster, S. A., G. W. Demers, B. G. Etscheid, and D. A. Galloway. 1994. The ability of human papillomavirus E6 proteins to target p53 for degradation in vivo correlates with their ability to abrogate actinomycin D-induced growth arrest. J. Virol. 685698-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frangioni, J. V., and B. G. Neel. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210179-187. [DOI] [PubMed] [Google Scholar]
- 10.Frisch, M., R. J. Biggar, and J. J. Goedert. 2000. Human papillomavirus-associated cancers in patients with human immunodeficiency virus infection and acquired immunodeficiency syndrome. J. Natl. Cancer Inst. 921500-1510. [DOI] [PubMed] [Google Scholar]
- 11.Frisch, S. M., and R. A. Screaton. 2001. Anoikis mechanisms. Curr. Opin. Cell Biol. 13555-562. [DOI] [PubMed] [Google Scholar]
- 12.Gardiol, D., C. Kuhne, B. Glaunsinger, S. S. Lee, R. Javier, and L. Banks. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 185487-5496. [DOI] [PubMed] [Google Scholar]
- 13.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 195270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1991. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 65473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendriks, W., J. Schepens, D. Bachner, J. Rijss, P. Zeeuwen, U. Zechner, H. Hameister, and B. Wieringa. 1995. Molecular cloning of a mouse epithelial protein-tyrosine phosphatase with similarities to submembranous proteins. J. Cell. Biochem. 59418-430. [DOI] [PubMed] [Google Scholar]
- 16.Higgins, G. D., D. M. Uzelin, G. E. Phillips, and C. J. Burrell. 1991. Presence and distribution of human papillomavirus sense and antisense RNA transcripts in genital cancers. J. Gen. Virol. 72(Pt. 4)885-895. [DOI] [PubMed] [Google Scholar]
- 17.Higgins, G. D., D. M. Uzelin, G. E. Phillips, P. McEvoy, R. Marin, and C. J. Burrell. 1992. Transcription patterns of human papillomavirus type 16 in genital intraepithelial neoplasia: evidence for promoter usage within the E7 open reading frame during epithelial differentiation. J. Gen. Virol. 73(Pt. 8)2047-2057. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman, H. T., L. H. Karnell, G. F. Funk, R. A. Robinson, and H. R. Menck. 1998. The National Cancer Data Base report on cancer of the head and neck. Arch. Otolaryngol. Head Neck Surg. 124951-962. [DOI] [PubMed] [Google Scholar]
- 19.Hoover, A. C., W. C. Spanos, G. F. Harris, M. E. Anderson, A. J. Klingelhutz, and J. H. Lee. 2007. The role of human papillomavirus 16 E6 in anchorage-independent and invasive growth of mouse tonsil epithelium. Arch. Otolaryngol. Head Neck Surg. 133495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ioka, A., H. Tsukuma, W. Ajiki, and A. Oshima. 2005. Trends in head and neck cancer incidence in Japan during 1965-1999. Jpn. J. Clin. Oncol. 3545-47. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov, V. N., Z. Ronai, and T. K. Hei. 2006. Opposite roles of FAP-1 and dynamin in the regulation of Fas (CD95) translocation to the cell surface and susceptibility to Fas ligand-mediated apoptosis. J. Biol. Chem. 2811840-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing, M., J. Bohl, N. Brimer, M. Kinter, and S. B. Vande Pol. 2007. Degradation of tyrosine phosphatase PTPN3 (PTPH1) by association with oncogenic human papillomavirus E6 proteins. J. Virol. 812231-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalyankrishna, S., and J. R. Grandis. 2006. Epidermal growth factor receptor biology in head and neck cancer. J. Clin. Oncol. 242666-2672. [DOI] [PubMed] [Google Scholar]
- 24.Khan, A. J., B. L. King, B. D. Smith, G. L. Smith, M. P. DiGiovanna, D. Carter, and B. G. Haffty. 2002. Characterization of the HER-2/neu oncogene by immunohistochemical and fluorescence in situ hybridization analysis in oral and oropharyngeal squamous cell carcinoma. Clin. Cancer Res. 8540-548. [PubMed] [Google Scholar]
- 25.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 9411612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 38079-82. [DOI] [PubMed] [Google Scholar]
- 27.Kreimer, A. R., G. M. Clifford, P. Boyle, and S. Franceschi. 2005. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol. Biomarkers Prev. 14467-475. [DOI] [PubMed] [Google Scholar]
- 28.Lanham, S., A. Herbert, and P. Watt. 2001. HPV detection and measurement of HPV-16, telomerase, and survivin transcripts in colposcopy clinic patients. J. Clin. Pathol. 54304-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, J. H., S. M. Yi, M. E. Anderson, K. L. Berger, M. J. Welsh, A. J. Klingelhutz, and M. A. Ozbun. 2004. Propagation of infectious human papillomavirus type 16 by using an adenovirus and Cre/LoxP mechanism. Proc. Natl. Acad. Sci. USA 1012094-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 749680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorber, B., W. J. Hendriks, C. E. Van der Zee, M. Berry, and A. Logan. 2005. Effects of LAR and PTP-BL phosphatase deficiency on adult mouse retinal cells activated by lens injury. Eur. J. Neurosci. 212375-2383. [DOI] [PubMed] [Google Scholar]
- 32.Lu, S. L., H. Herrington, D. Reh, S. Weber, S. Bornstein, D. Wang, A. G. Li, C. F. Tang, Y. Siddiqui, J. Nord, P. Andersen, C. L. Corless, and X. J. Wang. 2006. Loss of transforming growth factor-beta type II receptor promotes metastatic head-and-neck squamous cell carcinoma. Genes Dev. 201331-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakagawa, S., and J. M. Huibregtse. 2000. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 208244-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritchie, J. M., E. M. Smith, K. F. Summersgill, H. T. Hoffman, D. Wang, J. P. Klussmann, L. P. Turek, and T. H. Haugen. 2003. Human papillomavirus infection as a prognostic factor in carcinomas of the oral cavity and oropharynx. Int. J. Cancer 104336-344. [DOI] [PubMed] [Google Scholar]
- 35.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75495-505. [DOI] [PubMed] [Google Scholar]
- 36.Schreiber, K., R. E. Cannon, T. Karrison, G. Beck-Engeser, D. Huo, R. W. Tennant, H. Jensen, W. M. Kast, T. Krausz, S. C. Meredith, L. Chen, and H. Schreiber. 2004. Strong synergy between mutant ras and HPV16 E6/E7 in the development of primary tumors. Oncogene 233972-3979. [DOI] [PubMed] [Google Scholar]
- 37.Simonson, S. J., M. J. Difilippantonio, and P. F. Lambert. 2005. Two distinct activities contribute to human papillomavirus 16 E6's oncogenic potential. Cancer Res. 658266-8273. [DOI] [PubMed] [Google Scholar]
- 38.Smith, E. M., J. M. Ritchie, K. F. Summersgill, J. P. Klussmann, J. H. Lee, D. Wang, T. H. Haugen, and L. P. Turek. 2004. Age, sexual behavior and human papillomavirus infection in oral cavity and oropharyngeal cancers. Int. J. Cancer 108766-772. [DOI] [PubMed] [Google Scholar]
- 39.Spanos, W. C., M. El-Deiry, and J. H. Lee. 2005. Cidofovir incorporation into human keratinocytes with episomal HPV 16 results in nonselective cytotoxicity. Ann. Otol. Rhinol. Laryngol. 114840-846. [DOI] [PubMed] [Google Scholar]
- 40.Spanos, W. C., J. Gieger, G. F. Harris, A. Bossler, R. B. Smith, A. Klingelhutz, and J. H. Lee. 2007. Deletion of the PDZ motif of HPV16 E6 preventing immortalization and anchorage-independent growth in human tonsil epithelial cells. Head Neck [Epub ahead of print.] doi: 10.1002/hed.20673. [DOI] [PMC free article] [PubMed]
- 41.Thomas, M., R. Laura, K. Hepner, E. Guccione, C. Sawyers, L. Lasky, and L. Banks. 2002. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene 215088-5096. [DOI] [PubMed] [Google Scholar]
- 42.Villa, F., M. Deak, G. B. Bloomberg, D. R. Alessi, and D. M. van Aalten. 2005. Crystal structure of the PTPL1/FAP-1 human tyrosine phosphatase mutated in colorectal cancer: evidence for a second phosphotyrosine substrate recognition pocket. J. Biol. Chem. 2808180-8187. [DOI] [PubMed] [Google Scholar]
- 43.Wang, Z., D. Shen, D. W. Parsons, A. Bardelli, J. Sager, S. Szabo, J. Ptak, N. Silliman, B. A. Peters, M. S. van der Heijden, G. Parmigiani, H. Yan, T. L. Wang, G. Riggins, S. M. Powell, J. K. Willson, S. Markowitz, K. W. Kinzler, B. Vogelstein, and V. E. Velculescu. 2004. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 3041164-1166. [DOI] [PubMed] [Google Scholar]
- 44.Wansink, D. G., W. Peters, I. Schaafsma, R. P. Sutmuller, F. Oerlemans, G. J. Adema, B. Wieringa, C. E. van der Zee, and W. Hendriks. 2004. Mild impairment of motor nerve repair in mice lacking PTP-BL tyrosine phosphatase activity. Physiol. Genomics 1950-60. [DOI] [PubMed] [Google Scholar]