Abstract
Gag-FP (fluorescent protein) fusion constructs are commonly used to study human immunodeficiency virus type 1 assembly, yielding diffuse signals throughout the cytoplasm along with punctate signals routinely described as virus-like particles (VLPs) representing assembled but unprocessed Gag. However, these particles cannot be accurately described as VLPs, since fluorescence microscopy cannot provide structural resolution. We demonstrate here that the inability of a monoclonal p24 antibody to bind its cognate epitope when unprocessed Gag is assembled distinguishes VLPs from unassembled, monomeric Gag. Furthermore, we show that assembled and unassembled Gag punctate signals travel along microtubules. These monoclonal antibody studies provide a new tool for examining retroviral assembly.
Human immunodeficiency virus (HIV) is a complex retrovirus that assembles on lipid membranes within infected cells (13). However, the precise location of this site remains unclear. Historically, HIV assembly was believed to occur at the plasma membrane of the cell, an idea supported by several facts. Gag contains a myristoylation site that targets the protein to membranes (12, 16); the Env protein, which decorates the outer membrane of viral particles, is readily expressed on the surface of infected cells (17); and budding structures at the plasma membrane have been revealed by the use of electron microscopy (7).
Alternatively, HIV type 1 (HIV-1) utilizes multivesicular bodies (MVBs) to productively bud virion particles in primary macrophages and T cells (8, 10, 11). Recently, several published reports suggested that intracellular vesicles are capable of providing budding sites for HIV-1 in other cell types as well, including the HOS and HeLa cell types. These studies included the utilization of the ESCRT trafficking pathway, as evidenced by the roles played by several factors in the pathway, including the Chmp proteins (18) and AP-3 (1).
The uses of and requirement for MVBs have been contested in the past two years. Specifically, the Bieniasz laboratory has shown that blocking the MVB pathway does not abrogate viral particle production in macrophages (4). Moreover, they have also shown that targeting Gag to the MVB pathway rather than to the plasma membrane results in the loss of viral budding. They conclude that productive viral budding occurs only at the plasma membrane (4) and that mature virions found within cells have been reinternalized by endocytosis (6).
In many of these studies, Gag-FP (Gag-fluorescent protein) fusions were widely used to study the intracellular trafficking and assembly of HIV-1. These constructs typically yield diffuse reticular staining throughout the cytoplasm along with additional punctate signals of much brighter fluorescence intensity. These brighter signals are routinely assumed to represent virus-like particles (VLPs) in the form of assembled but immature virions. However, the actual structural status of these signals has not been directly examined. While they may be VLPs, they may also be localized concentrations of Gag that are precursors to viral assembly. Using monoclonal antibodies to HIV-1 Gag, we report a simple method of immunostaining that makes use of epitope masking to identify which punctate signals are actually VLPs. We also analyzed the intracellular movement of these signals on microtubules. Both VLPs and unassembled concentrations of Gag move along these cellular highways, further emphasizing the importance of this new technique as it applies to live-cell studies. We therefore present a new method to researchers in the field of HIV-1 biology for studying viral assembly.
A monoclonal p24 antibody cannot recognize all punctate Gag-FP signals within a cell.
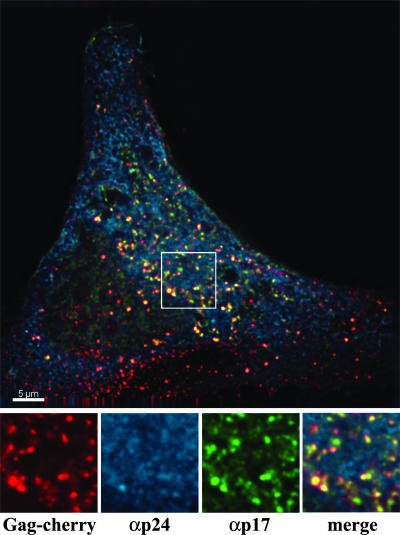
Cherry-C1 vector (a gift from R. Tsien [14]) was used to generate Gag-cherry. Briefly, the cherry coding sequence replaced the enhanced green FP (GFP) reading frame of pEGFP (Clontech) by use of AgeI and NotI restriction sites. The codon-optimized Gag reading frame of Gag-GFP (a gift from M. Resh), which contains only Pr55gag (nucleotides 790 to 2287, where +1 represents the first nucleotide of the 5′ proviral U3) and lacks the viral protease, was inserted in frame and upstream of the cherry sequence. This construct was transfected into HeLa cells on glass coverslips by use of Effectene (Qiagen) following the manufacturer's protocol. At 18 h posttransfection, the cells were fixed with 3.7% formaldehyde in 0.1 M PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer (pH 6.7) for 3 to 4 min. Cells were washed twice with 1× phosphate-buffered saline (PBS) and concurrently permeabilized and blocked using 10% normal donkey serum, 0.1% Triton X-100, 0.01% sodium azide, and PBS for 10 min. Cells were washed twice with PBS and stained with a 1:250 dilution of a mouse α-p24 antibody (Ag3; AIDS Reagent Repository) (15) in the above-described blocking solution lacking Triton X-100. Samples were incubated at room temperature for 1 h and washed twice with PBS. Samples were then treated with a Cy5-labeled donkey α-mouse antibody (Jackson ImmunoResearch) in blocking solution lacking Triton X-100 for 1 h at room temperature. Samples were washed three times with PBS and then stained with a second Gag p17 antibody (Capricorn) that was fluorescein isothiocyanate labeled by use of a Zenon antibody labeling kit (Invitrogen) in blocking solution lacking Triton X-100. This commercially available antibody targets the N terminus of the matrix. Samples were washed three times with PBS, mounted, and dried. Images were acquired using a DeltaVision deconvolution microscope and SoftWoRx software (Fig. 1). By using the “Data Inspector” function, the average fluorescent intensity within the cell from the diffuse signal was measured for each color channel. These values were then doubled, and all signals below this value were subtracted from the image. The remaining signals were considered for colocalization analysis.
FIG. 1.
A monoclonal p24 antibody does not recognize all Gag within transfected cells. HeLa cells were transiently transfected with a Gag-cherry plasmid. At 18 to 21 h posttransfection, cells were fixed and stained for p24 by use of a monoclonal antibody to p24 (Ag3; αp24) and p17 (Capricorn). An individual Z stack of a representative cell is shown. The smaller boxes along the bottom are blowups of the white-bordered box in the larger image. From left to right, they show Gag-cherry expression, Ag3 αp24 staining, Capricorn αp17 staining, and the merged image. Scale bar, 5 μm.
The p17 antibody readily detected both the diffuse Gag signal seen throughout the cytoplasm and the punctate signals (proportion of punctate signals detected, 95.91% ± 2.46%; n = 15). The Ag3 antibody also detected the diffuse signals within the transfected cells. However, this antibody was only able to detect a minor fraction of the punctate signals (5.97% ± 1.87%; n = 20). In a very few cases, the Ag3 antibody was able to detect up to 30% of the punctate signals that were Ag3 positive. Although investigation of this point is beyond the scope of this report, we reasonably speculate that this minor population of cells (<10%) with increased Ag3-positive punctate signals represents the early stages of expression of Pr55gag in which Gag monomers are being concentrated just prior to the initiation of assembly. Similar results were observed with HOS cells. Additionally, the results were not dependent on the nature of the attached fluorophore, as a Gag-GFP construct yielded similar results (data not shown).
The inability of a p24 antibody to recognize VLPs is due to epitope masking.
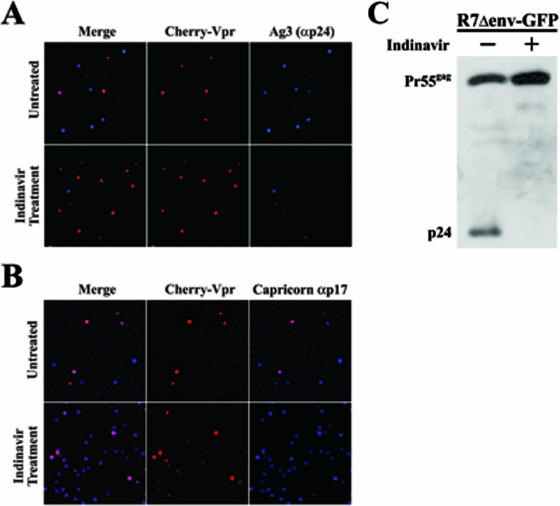
The Ag3 antibody could detect the diffuse, monomeric Gag-cherry signal but not the majority of punctate signals, suggesting that epitope masking may be involved. When assembled into VLPs, the p24 domain of Gag is buried within the VLP and is potentially inaccessible to the antibody. To test this hypothesis, two plates of HeLa cells were transfected with the proviral HIV-1 clone R7Δenv and a cherry-Vpr expression plasmid. Since HIV efficiently packages Vpr, progeny virions are labeled red. One sample was also treated with 5 μM indinavir (AIDS Reagent Repository) to prevent Gag processing. At 48 h later, viruses were harvested and purified by use of 0.45-μm-pore-size filters. Viruses were then spun onto glass coverslips in a 24-well plate at room temperature, fixed, and stained with either the Ag3 p24 antibody or the Capricorn p17 antibody (Fig. 2A and B).
FIG. 2.
The inability of the Ag3 antibody to recognize all Gag is due to epitope masking. HeLa cells were transfected with an R7Δenv proviral clone and the expression plasmid cherry-Vpr in the presence and absence of indinavir. At 48 h posttransfection, virus was purified from the supernatant, put on glass, fixed, and stained with the Ag3 αp24 antibody (A) or αp17 (Capricorn) (B). (C) HeLa cells were transiently transfected with R7Δenv-GFP in the absence (−) and presence (+) of indinavir. At 48 h posttransfection, the cells were lysed in protein sample buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting using the Ag3 antibody.
The majority (74.35%; n = 113) of red-labeled viral particles grown in the absence of indinavir were detected by the Ag3 antibody (Fig. 2A, top row). Conversely, only a small percentage (1.37%; n = 73) of viral particles generated in the presence of indinavir were detected by Ag3 (Fig. 2A, bottom row). In turn, the Capricorn antibody readily detected the red-labeled particles in the absence (95.7%; n = 161) (Fig. 2B, top row) or presence (94.4%; n = 177) (Fig. 2B, bottom row) of indinavir. This further suggests that the Ag3 antibody cannot detect VLPs due to epitope masking.
To confirm and extend this hypothesis, two 60-mm-diameter plates of HeLa cells were transfected using the proviral HIV construct R7Δenv-GFP and Effectene. One plate was also treated with 5 μM indinavir to inhibit viral protease activity. After 48 h, cells were scraped, lysed in protein sample buffer, and analyzed by Western blotting with the Ag3 antibody (sodium dodecyl sulfate-polyacrylamide gel electrophoresis; 5% stacking gel [pH 6.8] and 10% resolving gel [pH 8.8]). The proteins were visualized using donkey α-mouse-conjugated horseradish peroxidase (Pierce) and enhanced chemiluminescence (Pierce) (Fig. 2B). The antibody readily detected both the Pr55gag polyprotein and the cleaved p24 subunit in cells without indinavir treatment. Expectedly, the antibody only detected the polyprotein in cells treated with the inhibitor. These results verify the ability of the antibody to detect both processed p24 and unprocessed Gag when the corresponding epitope is accessible. Taken together, the results of Fig. 2 verify that the Ag3 antibody can identify Gag but cannot do so when assembled into a VLP.
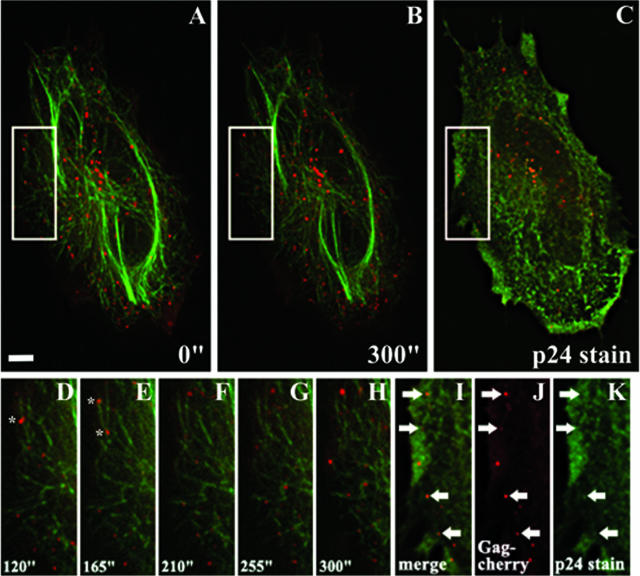
Having noticed two classes (assembled and unassembled) of punctate signal seen within cells and being able to distinguish between the two, we wished to examine how these signals move within living cells. For this purpose, HeLa cells in Delta T dishes (Fisher) were transfected with 0.3 μg of the Gag-cherry construct and 0.01 μg of enhanced yellow FP-tubulin (Clontech). At 18 to 21 h posttransfection, the cells were supplemented with 1 μM taxol for 10 min at 37°C. The cells were transferred to a preheated environmental chamber (37°C) on the microscope stage and imaged every 15 s for 5 min. After the last image was acquired, the growth medium was immediately removed and the cells were fixed on the stage. The cells were then subsequently stained and imaged as previously described.
A representative result is shown in Fig. 3 (for the complete movie of the cell and particle movement, see supplemental movie S1). The results clearly demonstrate the movement of punctate Gag-cherry signals along the microtubules of transfected cells. Furthermore, both Ag3-negative and Ag3-positive signals were shown to move along microtubules. Concurrently, a small group of unassembled localized Gag (as evidenced by their Ag3-positive staining) is not initially attached to microtubules but quickly attaches and begins moving along them. This evidence directly demonstrates that both assembled Gag and unassembled Gag utilize microtubules for movement. Further, because of Gag association with membranes, microtubules, and actin (3, 5, 9; this report) as well as with CD63 (2) and the ESCRT pathways (1, 18), we hypothesize that the Ag3-positive signals are endosomal vesicles. Future work will address this hypothesis.
FIG. 3.
Assembled and unassembled Gag traffics within cells by use of the microtubule network. HeLa cells were transiently transfected with Gag-cherry and yellow FP-tubulin. At 18 to 21 h posttransfection, complete Z-stack images were acquired every 15 s for 5 min. Immediately following the last image acquisition, the cells were fixed and stained for p24 by use of the Ag3 antibody. (A to C) Cell images at the start (A) and end (B) of acquisition and after immunostaining (C). In panels A and B, Gag is shown in red and tubulin is shown in green. In panel C, Gag is shown in red and the antibody staining is shown in green. (D to H) Enlargements of the boxes shown in panels A and B, detailing Gag movement in 45-s intervals starting at 120 s. Gag is shown in red, and tubulin is shown in green. The asterisks identify fluorescent signals that split into two viral particles. (I to K) Enlargements of the white box shown in panel C. Arrows pointing right indicate Ag3-negative (assembled) punctate signals; arrows pointing left indicate Ag3-positive (unassembled) punctate signals. All images shown represent volume projections of four Z-stack images of the cell. Scale bar, 5 μm.
Given this paper's scope, we cannot address the question of the destinations of these signals. It could be that the unassembled accumulations are moving to the plasma membrane for assembly. Since they are on microtubules and since MVBs also travel on microtubules, they may be in the process of assembling. Alternatively, it could be that they been assembled at the start of the image acquisition but were being recycled during the time course (as suggested by the Bieniasz laboratory [6]), which would lead to disassembly and restored epitope accessibility for the Ag3 antibody. Similarly, Ag3-negative signals may have begun the time course unassembled and may have become assembled at the conclusion. Addressing these issues would require tests that are more rigorous and elaborate; such tests lie outside the scope of this brief communication. However, this experiment underscores the importance and impact of this technique for future HIV-1 assembly studies.
Implications and future directions for the study of assembly.
The findings of this report present a simple and accurate method of determining the assembly status of Gag accumulations within a cell. As previously stated, there exists a debate regarding where these viral proteins assemble within the cell. The Bieniasz group asserts that assembly occurs only at the plasma membrane (4). Conversely, the Spearman group and others state that intracellular assembly within MVBs is part of the natural and productive viral assembly pathway (1, 10, 18). Using the structural determinations that use of this antibody can provide, we have uncovered a powerful tool that could potentially resolve these differences.
It will be of great interest to screen other monoclonal antibodies that possess unique recognition qualities that can further characterize the assembly of Gag proteins. The discovery of these properties will allow new studies to be performed using wild-type virus for infections that will allow for detailed in vitro studies that more accurately portray what biological and assembly events occur in vivo.
Supplementary Material
Acknowledgments
Monoclonal antibody to HIV-1 p24 (AG3.0) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Jonathan Allan.
T.J.H. is an Elizabeth Glaser Scientist.
This work was supported by the Elizabeth Glaser Pediatric AIDS Foundation (35-04T) and NIH grant 1P50GM082545 (W. Sundquist, principal investigator).
Footnotes
Published ahead of print on 19 December 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Dong, X., H. Li, A. Derdowski, L. Ding, A. Burnett, X. Chen, T. R. Peters, T. S. Dermody, E. Woodruff, J.-J. Wang, and P. Spearman. 2005. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 120663-674. [DOI] [PubMed] [Google Scholar]
- 2.Gomez, C. Y., and T. J. Hope. 2006. Mobility of human immunodeficiency virus type 1 Pr55Gag in living cells. J. Virol. 808796-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jolly, C. T., I. Mitar, and Q. J. Sattentau. 2007. Requirement for an intact T cell actin and tubulin cytoskeleton for efficient HIV-1 assembly and spread. J. Virol. 815547-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jouvenet, N., S. J. D. Neil, C. Bess, M. C. Johnson, C. A. Virgen, S. M. Simon, and P. D. Bieniasz. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 4e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neil, S. J. D., S. W. Eastman, N. Jouvenet, and P. D. Bieniasz. 2006. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ono, A., and E. O. Freed. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 781552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orenstein, J. M., M. S. Meltzer, T. Phipps, and H. E. Gendelman. 1988. Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: an ultrastructural study. J. Virol. 622578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. Sowder II, Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 26642-51. [DOI] [PubMed] [Google Scholar]
- 10.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3718-729. [DOI] [PubMed] [Google Scholar]
- 12.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 837246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Resh, M. D. 2005. Intracellular trafficking of HIV-1 Gag: how Gag interacts with cell membranes and makes viral particles. AIDS Rev. 784-91. [PubMed] [Google Scholar]
- 14.Shu, X., N. C. Shaner, C. A. Yarbrough, R. Y. Tsien, and S. J. Remington. 2006. Novel chromophores and buried charges control color in mFruits. Biochemistry 459639-9647. [DOI] [PubMed] [Google Scholar]
- 15.Simm, M., M. Shahubuddin, W. Chao, J. S. Allan, and D. J. Volsky. 1995. Aberrant Gag protein composition of a human immunodeficiency virus type 1 vif mutant produced in primary lymphocytes. J. Virol. 694582-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spearman, P., J. J. Wang, N. Vander Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag domains essential to membrane binding and particle assembly. J. Virol. 683232-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Woodbury, NY. [PubMed]
- 18.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114701-713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.