Abstract
The binding of herpes simplex virus type 1 ICP4, TATA-binding protein (TBP), and RNA polymerase II (polII) to the promoter regions of representative immediate-early (IE) (ICP0), early (E) (thymidine kinase [tk]), and late (L) (glycoprotein C [gC]) genes on the viral genome was examined as a function of time postinfection, viral DNA replication, cis-acting sites for TFIID in the tk and gC promoters, and genetic background of ICP4. The binding of TBP and polII to the IE ICP0 promoter was independent of the presence of ICP4, whereas the binding of TBP and polII to the tk and gC promoters occurred only when ICP4 also bound to the promoters, suggesting that the presence of ICP4 at the promoters of E and L genes in virus-infected cells is crucial for the formation of transcription complexes on these promoters. When the TATA box of the tk promoter or the initiator element (INR) of the gC promoter was mutated, a reduction in the amount of TBP and polII binding was observed. However, a reduction in the amount of ICP4 binding to the promoters was also observed, suggesting that the binding of TBP-containing complexes and ICP4 is cooperative. The binding of ICP4, TBP, and polII was also observed on the gC promoter at early times postinfection or when DNA synthesis was inhibited, suggesting that transcription complexes may be formed early on L promoters and that additional events or proteins are required for expression. The ability to form these early complexes on the gC promoter required the DNA-binding domain but in addition required the carboxyl-terminal 524 amino acids of ICP4, which is missing the virus n208. This region was not required to form TBP- and polII-containing complexes on the tk promoter. n208 activates E but not L genes during viral infection. These data suggest that a region of ICP4 may differentiate between forming TBP- and polII-containing complexes on E and L promoters.
The approximately 80 gene of herpes simplex virus (HSV) (34, 35) are transcribed by RNA polymerase II (polII) (1) in a sequential and coordinated manner (25, 26). The immediate-early (IE) (α) genes are expressed first, followed by the early (E) (β) and subsequently by the late (L) (γ) genes. The L genes are further classified as leaky L and true L genes, depending on their requirement for viral DNA replication. The IE proteins include infected-cell polypeptide 0 (ICP0), ICP4, ICP22, ICP27, and ICP47 (39). The expression of the IE proteins does not require prior viral protein synthesis (40). However, functional IE proteins are required for the optimal expression of both E and L gene products (25, 26).
Almost all of the HSV promoters possess a TATA box. By contrast only 10% of human polII genes possess TATA boxes (19). Members of each kinetic class of promoters share a general structural theme. IE promoters have upstream binding sites for cellular activators, most notably Sp1, and sites of action for the virion transactivator, VP16 (2, 3). The promoters for E genes possess upstream binding sites for cellular activators, again mostly Sp1. Upstream sites for transcription activators in true L promoters have not been described. These promoters appear to consist of simply a TATA box and an initiator element (described below). In addition, a downstream activator sequence has been described in some true L promoters (22, 23).
IE genes are the first to be expressed. Their promoters are activated by the binding of the virion transactivator, VP16, in a complex with cellular Oct1 and host cell factor to TAATGARAT elements, as well as by the action of Sp1 and other cellular activators. Recently it has also been shown that the VP16 activation domain results in the presence of acetylated histones on IE promoters, suggesting that it may recruit chromatin-remodeling factors or histone acetyltransferases (24). The protein products of the five IE genes are mainly regulatory proteins that affect different aspects of host cell metabolism. One of these, ICP4, is a transcription activator, which is required for the activated expression of HSV E and L genes. The protein products of E genes are mostly involved in viral DNA replication. With the expression of E proteins and the onset of viral DNA replication, both IE and E gene transcription are attenuated (25). In the absence of ICP4, IE transcription remains abundant (14, 16, 41, 48). With the expression of IE and E proteins and the onset of viral DNA replication, L gene transcription begins. L gene transcription is highly activated by ICP4, and L mRNAs and proteins are abundantly produced. This is interesting considering that L promoters contain binding sites only for TFIID (TATA box plus Inr). One reason for the abundant transcription of L genes is that ICP4 more efficiently activates transcription of promoters containing Inr elements (10, 21, 30).
ICP4 is a large and structurally complex molecule. Its mobility on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis corresponds to a molecular mass of 175 kDa (11), and it exists in cells as a ∼350-kDa dimer (36, 44). It has a Stokes radius of ∼90 Å, and its hydrodynamic properties suggest that it is a very elongated molecule (36, 44). This and the ability of ICP4 to bind to DNA (29) may provide the potential to function as an activator of transcription over long distances. Furthermore, ICP4 localizes in infected-cell nuclei in viral replication compartments (12), where it can be found with viral DNA and cellular RNA polII and ICP22 (32).
In reconstituted in vitro transcription reactions, ICP4 has been shown to activate transcription of a core RNA polII promoter with a relatively simple set of polII general transcription factors (4, 5, 21). ICP4 can form tripartite complexes on DNA with TFIID and TFIIB (46), and it can interact with TFIID in solution via its TBP-associated factor 1 (TAF1) subunit (5). It promotes the formation of transcription preinitiation complexes (20, 21) by enhancing TFIID binding to the promoter (20).
While the activities described above have been shown to be consistent with the transcriptional regulatory activities and genetics of ICP4 in virus-infected cells, they are mostly from in vitro reconstruction experiments. The present study examines the association of ICP4, the TATA-binding protein (TBP), and RNA polII with promoters representative of IE, E, and L genes as a function of time postinfection, DNA replication, cis-acting sites for TBP and hence any TBP-containing complex that might function on TATA box-containing promoters, and the genetic background of ICP4. All of the experiments were conducted in virus-infected cells by the use of specific viral mutants and the application of the technique of chromatin immunoprecipitation (ChIP).
MATERIALS AND METHODS
Cells and virus.
All experiments were performed using Vero cells (African green monkey kidney cells), which were obtained from the American Type Culture Collection. The viruses used in this study include the wild-type (wt) strain of HSV type 1 (HSV-1) (KOS), the tk TATA box mutant (LS-29-18) (7), the glycoprotein C (gC) initiator mutant (gCInr) (30), and the ICP4 mutants n12 (15), n208 (15), and i13 (42). The mutant LS-29-18 is severely impaired for tk transcription (7) and the binding of TBP to the mutant tk TATA box (9). gCInr contains base change mutations in three of the most important contacts in the initiator element of gC. The mutations render the promoter very inefficient for activation by ICP4 in vitro and in viral infection (30). n12 expresses an ICP4 molecule that is truncated at amino acid 251. It has no ICP4 activity (15). n208 expresses an ICP4 molecule that is truncated after amino acid 774. It can bind to DNA and activate many E genes but is deficient in L gene expression (15). Compared to wt ICP4, the n208 molecule cannot interact with TAF1 or TFIID in solution (5) and cannot multimerize on DNA (31).
The viruses n208, n12, and i13 were grown on E5 cells as described previously (15). E5 cells are derived from Vero cells and express complementing levels of ICP4 upon HSV-1 infection (15). KOS, LS-29-18, and gCInr were propagated on Vero cells. For experiments, 5 × 106 Vero cells were seeded into 100-mm petri dishes 1 day prior to infection. The day after seeding, the monolayers were infected at a multiplicity of infection of 5 PFU/cell with the indicated viruses. The virus inoculum in 1.0 ml was adsorbed to the monolayer for 1 h at 4°C. After the 1-hour adsorption period, the inoculum was removed, 37°C medium was added, and incubation was continued at 37°C in a humidified 5% CO2 incubator. Zero hours postinfection is considered the time when the cultures were warmed to 37°C and put in the incubator. A 400-μg/ml concentration of phosphonoacetic acid (PAA) (Lancaster Synthesis, Eastgate, England) was added to the medium to inhibit viral DNA synthesis.
ChIP.
ChIP was carried out as described previously (28, 49) with a few modifications. At 4 and 8 h postinfection the cells were treated with 1% formaldehyde for 10 min, and washed three times with phosphate-buffered saline containing protease inhibitors (67 ng/ml aprotinin, 1 ng pepstatin, 0.16 mM TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone], 1 mM phenylmethylsulfonyl fluoride [PMSF]). The cells were pelleted at 3,000 rpm, resuspended in PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid] lysis buffer (100 μl per million cells) containing protease inhibitors (5 mM PIPES, 85 mM KCl, 0.5% NP-40, 4 μg/ml aprotinin, 2 μg/ml pepstatin, 0.15 mM TLCK, and 0.6 mM PMSF), and incubated on ice for 15 min. The cells were then pelleted at 5,000 rpm for 5 min in a refrigerated centrifuge, resuspended in SDS lysis buffer containing protease inhibitors (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 4 μg/ml aprotinin, 2 μg/ml pepstatin, 0.15 mM TLCK, and 0.6 mM PMSF), and incubated on ice for 15 min. The samples were then sonicated for a total of 50 seconds (five 10-second pulses) using a Cole Palmer ultrasonic processor model CP-130 with a microtip. The DNA was sheared to give fragment sizes of less than 400 bp as determined by agarose gel electrophoresis. The samples were then spun at 12,500 rpm for 10 minutes in a refrigerated centrifuge to remove the debris. The supernatant was diluted (10 times) in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 167 mM NaCl) and precleared twice for 2 h each with protein A and protein G beads. Antibodies N15 (3 μl) for ICP4, 8WG16 (1 μl) for polII, and sc273 (10 μl) for TBP were then added to the samples. 8WG16 (for polII) was obtained from Covance, sc273 (for TBP) was obtained from Santa Cruz, and N15 for ICP4 was purified in the laboratory. The samples were then kept overnight on a rotator at 4°C, and the next day 60 μl of the appropriate beads (protein A beads for N15 and protein G for sc273 and 8WG16) was added and kept on a rotator for 1 hour at 4°C. The beads containing the immune complexes were washed eight times for 4 min each with 1 ml of different buffers (two times with low-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 150 mM NaCl), two times with high-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 500 mM NaCl), once with LiCl immune complex wash buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl [pH 8.1]), and three times with TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The DNA-protein complexes were eluted from the beads by adding 250 μl of elution buffer (1% SDS, 0.1 M NaHCO3) and incubating on a rotator for 15 min. This was done twice, thereby giving a total volume of 500 μl of suspension containing the DNA-protein complexes. The cross-links were then reversed by adding 20 μl of 5 M NaCl and incubating at 65°C overnight. The next day, 1 ml of ethanol was added and the sample was put at −20°C overnight or at −80°C for 1 h. The DNA was then pelleted in a refrigerated centrifuge for 20 min at 12,500 rpm. The supernatant was discarded, and 70% cold ethanol (kept at −20°C) was added to the pellet. The sample was centrifuged at 12,500 rpm in a refrigerated centrifuge and the supernatant discarded. The DNA was resuspended in 489 μl of TE. To this 11 μl of 10× proteinase K buffer (2.5 ml 1 M Tris-HCl [7.5], 2.5 ml 0.5 M EDTA, 12.5 ml 10% SDS) and 1 μl of proteinase K (20 mg/ml) were added. The samples were then incubated at 55°C for 1 hour. The DNA from each sample was purified using a Qiaquick PCR purification kit (Qiagen). The DNA was dissolved in a total of 150 μl of distilled water. The samples were then used for real-time PCR. Each ChIP experiment was repeated a minimum of three times.
Real-time PCR.
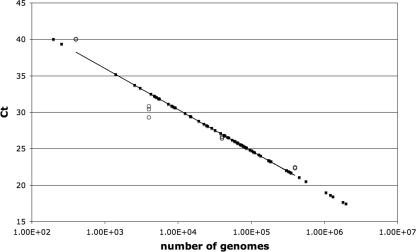
Reactions were set up in triplicate using 5 μl of DNA for each reaction. Before setting up the 96-well reaction plate, a master mix containing 0.625 μl of each primer (stock concentration, 1 mM), 12.5 μl iTaq SYBR green super mix with 1.0 μM 6-carboxy-X-rhodamine (Bio-Rad), and 6.25 μl of water for a total of 20 μl for each reaction was made. The final reaction volume was 25 μl, including the DNA. The primers used in these experiments and their locations relative to the transcription start site of the gene to be analyzed are given in Table 1. KOS DNA was also included in the plate, in the range of 400,000 to 40 copies of the genome, to provide a standard curve. This range was chosen because the threshold cycle values are in the range for the samples used for the ChIP experiment. After the plate was set up, it was run by using the ABI 7900HT Fast Real Time machine. The conditions for the run were as follows: stage 1. 50°C for 2 min; Stage 2, 95°C for 10 min; and stage 3, 40 cycle repeats of 95°C for 15 seconds and 60°C for 1 min. After the run was complete, the results were analyzed using the Applied Biosystems SDS 2.3 software, which generates a standard curve as shown in Fig. 1. Such curves were generated for each set of primers used in a given experiment. As Fig. 1 shows, most of the experimental samples are within the range of the viral DNA standards. Most of the triplicates for the standard curve coincided with each other (Fig. 1).
TABLE 1.
Primers used
| Primer | Sequence | Position |
|---|---|---|
| tkprom | CAGCTGCTTCATCCCCGTGG | −200 to +56 |
| AGATCTGCGGCACGCTGTTG | ||
| tkDS1 | GCAGCGACCCGCTTAACA | +15 to +84 |
| GAAGAGGTGCGGGAGTTTCAC | ||
| tkDS2 | TAGATGTTCGCGATTGTCT | +169 to +410 |
| GGCCATAACCGACGTAC | ||
| tkDS3 | GCGTCGGTCACGGCATAAG | +432 to +516 |
| GGGTGAGATATCGGCCGGG | ||
| tkDS4 | CCATCTCCCGGGCAAACGTG | +1134 to +1225 |
| ACTTACCTCCGGGATGGTCCAGACC | ||
| ICP8 | TGTACATATACCAACCGCATATCAGA | −32 to 40 |
| CGAGAGGCACAGATGCTTACG | ||
| gC | CGCCGGTGTGTGATGATTT | −90 to +80 |
| TTTATACCCGGGCCCCAT | ||
| ICP0 | ATAAGTTAGCCCTGGCCCGA | −24 to +36 |
| GCTGCGTCTCGCTCCG |
FIG. 1.
Standard real-time PCR curve generated with each primer pair in each experiment. The results are plotted on a semilogarithmic graph. Ct, threshold cycle. The KOS DNA standard described in Materials and Methods are shown as open circle. The software package SDS 2.3 from Applied BioSystems generates the curves and places on it the values for the experimental unknowns (closed circles).
RESULTS
Requirement of ICP4 for binding of polII to the tk and ICP0 promoters.
It has long been known that in the absence of ICP4, transcription from the viral genome is largely limited to that of the IE genes, and in the presence of ICP4, E and L genes are transcribed. Therefore, we might expect to observe the association of polII with a representative IE promoter, such as ICP0, to be independent of ICP4. Likewise, the association of polII with a representative E promoter, such as the tk promoter, might require ICP4. Therefore, testing these hypotheses represents a good way to partially validate the ChIP approach to observe ICP4 and key transcriptional proteins on HSV promoters in the context of viral infection.
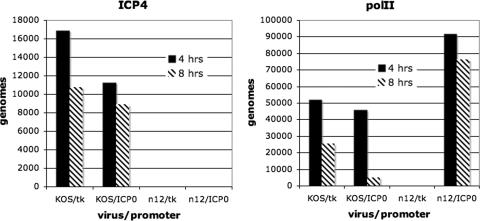
Vero cells were infected with KOS (wt) and n12, and ChIP analysis was performed on infected cells at 4 and 8 h postinfection as described in Materials and Methods. Figure 2 show the association of ICP4 and polII with the tk and ICP0 promoters at 4 and 8 h postinfection in KOS- and n12-infected cells. The ICP4 in KOS-infected cells associated with both the ICP0 and tk promoters at 4 and 8 h postinfection. n12 synthesizes a peptide of ICP4 that does not bind to DNA and has no regulatory activity, but still reacts with the N15 antibody. There was no ICP4 reactivity found on either the ICP0 or tk promoter in i12-infected cells. However, the association of polII with the ICP0 promoter was independent of ICP4 and remained higher in the absence of ICP4. Importantly, no polII reactivity was found on the tk promoter in the absence of ICP4 at the later time postinfection (8 h). Therefore, the interactions observed by the in situ ChIP approach reflect what we might expect from patterns of transcription from wt and ICP4 mutant viruses.
FIG. 2.
Association of ICP4 and polII with the ICP0 and tk promoters in n12- and KOS-infected cells. ChIP experiments were performed as described in Materials and Methods. The graphs give the number of genomes bound by the tk and ICP0 promoters on the n12 and KOS genomes to ICP4 and polII. In this experiment we compare wt (KOS) to the ICP4 mutant n12 at 4 and 8 h postinfection.
Binding of ICP4, TBP, and polII to the tk and gC promoters as a function of DNA replication and core promoter cis-acting sites.
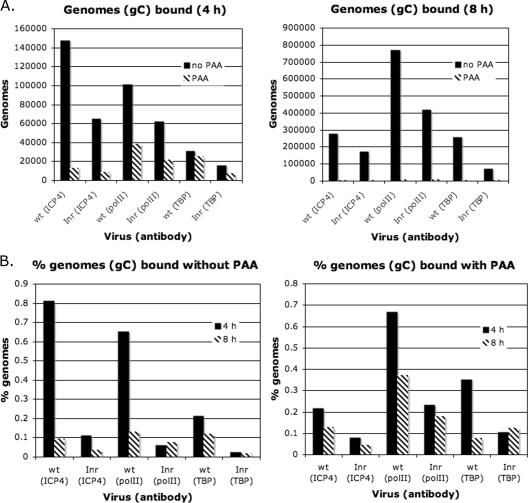
The tk gene is expressed as an E gene. Its transcription peaks at between 4 and 6 h postinfection and is subsequently shut off. tk mRNA synthesis is not sensitive to inhibition of viral DNA synthesis. The gC gene is a true L gene. It is not transcribed prior to viral DNA synthesis and is not detected when viral DNA synthesis is inhibited. There are also defined cis-acting sites for cellular factors, such as TFIID, in both the tk and gC core promoters that are important for the expression of these genes. The linker-scanning mutant LS-29-18 contains base changes in the TATA box of the tk gene such that tk transcription is markedly reduced (7), as is the affinity for TBP (9). The mutant gCInr contains base changes at key positions within the initiator element (INR) spanning the start site of gC transcription. Expression or the induction of the gC promoter by ICP4 is markedly reduced in the context of viral infection and in reconstituted in vitro transcription reactions (30). These mutants, along with wt virus (KOS), were used to examine how these mutations and DNA synthesis affect the formation of ICP4-, TBP-, and polII-containing complexes on the gC and tk promoters on the viral genome during viral infection.
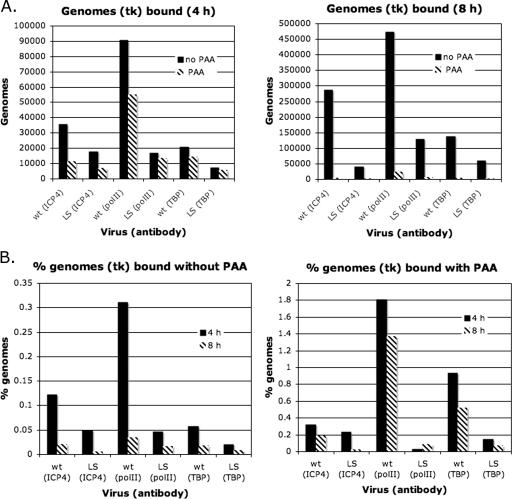
Vero cells were infected with KOS and LS-29-18 in the presence and absence of PAA to inhibit viral DNA synthesis. At 4 and 8 h postinfection, ChIP analysis was performed on the cultures. The data in Fig. 3 show the real-time PCR quantification using the primers that would amplify the tk promoter (Table 1). The data in Fig. 3 are represented in two different ways for clarity and also to establish different points. Whether the data are represented as the absolute number of genomes (Fig. 3A) or as the percentage of amplified immunoprecipitated genomes relative to the total number of genomes (Fig. 3B), it is clear that mutation of the TATA box resulted in reduced binding of TBP to the tk promoter on the viral genome. As might be expected, mutation of the TATA box also reduced binding of polII to the tk promoter. Interestingly, mutation of the TATA box also reduced binding of ICP4 to the tk promoter. This suggests that the presence of ICP4 may be affected not only by its intrinsic DNA-binding properties but also by the ability of cellular factors that ICP4 interacts with to bind to their DNA-binding sites. This and other possibilities are further discussed below.
FIG. 3.
Binding of ICP4, polII, and TBP to the tk promoters of KOS and LS-29-18. (A) ChIP experiments were done to determine the numbers of genomes bound to ICP4, polII, and TBP at 4 and 8 h postinfection in the presence and absence of PAA. (B) Representation of results in panel A as a percentage of genomes bound.
The data in Fig. 3A also indicate that the number of tk promoters bound by ICP4 increased from 4 to 8 h postinfection and was also greater in the absence of PAA than in its presence. This observation by itself is not entirely consistent with the known kinetics of tk transcription. However, it is interesting to note that the percentage of tk promoters bound by ICP4, TBP, and polII following DNA replication is markedly less than that in the absence of DNA replication, possibly suggesting the existence of a pool of genomes during DNA replication that are not expressing tk mRNA.
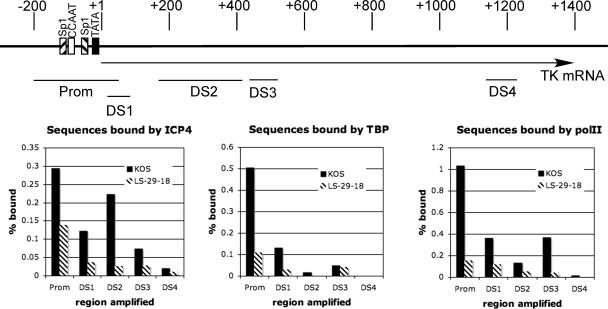
Figure 3 examines the association of ICP4, TBP, and polII with the tk promoter region on the viral genome. Mutation of the TATA box clearly affected the binding of all three to the promoter region. To observe the association of these proteins with nonpromoter regions of the tk gene and to determine how the TATA box mutation may affect this binding, ChIP analysis was performed on KOS- and LS-29-18-infected cells at 4 h postinfection using the PCR primers indicated in Fig. 4 and listed in Table 1. The data (Fig. 4) are represented as percent bound, as in Fig. 3B. The reason for this is that the PCR amplification efficiency under the conditions used to simultaneously amplify all the samples is not the same for all five primer pairs. As in Fig. 3, mutation of the TATA box markedly reduced binding of ICP4, TBP, and polII to the promoter region. The binding of TBP to the DS1 region was also detected, but it was approximately 20% of that detected at the promoter. It was also affected by the TATA box mutation. This most likely represents TBP binding to the tk promoter region. The proximal extent of this fragment is +15 relative to the tk mRNA start site, and the DNA in the ChIP procedure is sheared to several hundred base pairs. Moving further downstream (DS2, -3, and -4), the maximum binding of TBP was less than 10% of that of the promoter region, and the LS-29-18 mutation had no effect. These results are consistent with the lack of TATA boxes in these regions and the fact that TFIID does not participate in transcription elongation. Unlike TBP, polII was detected in significant amounts downstream of the promoter. The amount of polII observed downstream was still less than that observed at the promoter, possibly reflecting that the association of polII with any given sequence in the process of elongation is more transient than in initiation of transcription at the promoter. The reduced presence of polII observed downstream of the promoter in LS-29-18-infected cells is consistent with the reduced initiation of transcription at the promoter. ICP4 was also found downstream of the promoter. However, again the greatest amount of binding was observed at the promoter region, and mutation of the TATA box affected binding further downstream. It also appears that as one moves farther downstream of the promoter, there is a tendency to observe less ICP4 binding. This further supports the possibility that the presence of ICP4 may be affected not only by its intrinsic DNA-binding properties but also by the ability of cellular factors that ICP4 interacts with to bind to their DNA-binding sites.
FIG. 4.
Binding of ICP4, polII, and TBP to different regions of the tk gene. The top panel shows a schematic of the tk gene and promoter and the regions which are amplified by the designated primers, which are listed in Table 1. The bottom panels give the percentages of genomes bound by ICP4, TBP, and polII at the promoter and different regions of the tk gene in KOS- and LS-29-18-infected cells. The results are for 4 h postinfection.
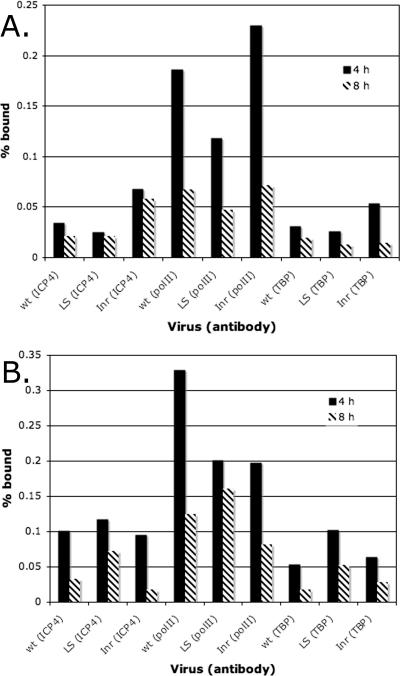
Figure 3 examined the binding of ICP4, TBP, and polII to the tk promoter. We next applied the same approach to the analysis of the gC promoter. Therefore, Vero cells were infected with KOS and gCInr in the presence and absence of PAA to inhibit viral DNA synthesis. At 4 and 8 h postinfection, ChIP analysis was performed on the cultures. As in the case of the tk promoter, alteration of a cis-acting site for TFIID (INR in this case) not only affected the binding of TBP and polII but also affected the binding of ICP4 (Fig. 5). In addition, many more gC promoter sequences were bound by all three proteins in the absence of PAA than in its presence (Fig. 5A), and the percentage of promoter sequences bound decreased following DNA replication (Fig. 5B). However, ICP4, TBP, and polII were observed on the gC promoter region at 4 h postinfection and in the presence of PAA. Therefore, while the association of these proteins with the gC promoter as a function of the Inr sequence is consistent with the activity of the promoter, the temporal association of TBP and polII it is not entirely consistent with the observed kinetics of expression.
FIG. 5.
Binding of ICP4, polII, and TBP to the gC promoters of KOS and gCInr. (A) ChIP experiments were done to determine the numbers of genomes bound to ICP4, polII, and TBP at 4 and 8 h postinfection in the presence and absence of PAA. (B) Representation of results in panel as a percentage of genomes bound.
The results in Fig. 3 and 5 show that when core promoter cis-acting sites for TFIID in the tk and gC promoters were mutated, there was is a reduction in the binding of TBP, polII, and ICP4. In addition, the results in Fig. 4 demonstrate that when the tk TATA box was mutated, then the binding of ICP4 downstream of the tk promoter was also affected. Given these results, we sought to examine the binding of ICP4, TBP, and polII to a promoter in LS-29-18 and gCInr that does not contain mutations and is relatively distant from the tk and gC promoters. Therefore, KOS, LS-29-18 and gCInr were used to infect Vero cells in the presence and absence of PAA. At 4 and 8 h postinfection, the cultures were processed and analyzed by ChIP analysis using a primer pair that amplifies a region that spans the core ICP8 promoter (Table 1). The results of the analysis are shown in Fig. 6, where Fig. 6A shows results in the absence of PAA and Fig. 6B shows results in the presence of PAA. There was no consistent effect of the mutations in the tk and gC promoters on the binding of TBP and ICP4, in either the presence or absence of PAA or at 4 and 8 h postinfection. In this experiment it appears that the mutations in the tk and gC promoters affected the binding of polII to the ICP8 promoter at 4 h postinfection in the presence of PAA (Fig. 6B). However, this was not seen at 8 h postinfection in the presence of PAA (Fig. 6B) or in the absence of PAA (Fig. 6A). Therefore, we conclude that the binding of ICP4, tk, and polII to the ICP8 promoter is not affected by mutations in the tk and gC core promoters.
FIG. 6.
Similar binding pattern of ICP4, polII, and TBP to the ICP8 promoter in KOS-, LS-29-18-, and gCInr-infected cells. ChIP analysis was conducted on KOS-, LS-19-18-, and gCInr-infected cells at 4 and 8 h postinfection to examine ICP4, TBP, and polII binding to the ICP8 promoter. (A) Percentage of genomes bound at 4 and 8 h postinfection in the absence of PAA. (B) Percentage of genomes bound at 4 and 8 h postinfection in the presence of PAA.
Binding of ICP4, TBP, and polII to promoters as a function of ICP4 functional domains.
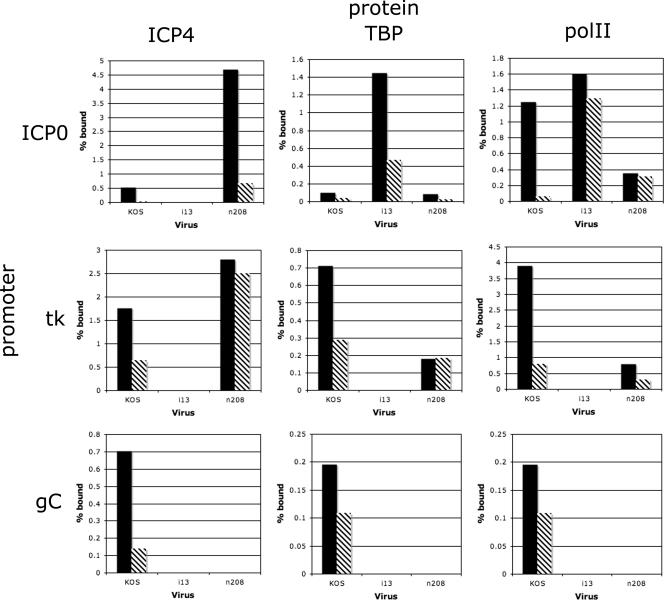
The experiments thus far examined the binding of ICP4, TBP, and polII to promoters as a function of cis-acting sites within the promoters. They also established that ICP4 is required for the binding of polII to the tk promoter. ICP4 is a large and complex protein, with multiple regions of the protein contributing to its regulatory activities. From the study of many temperature-sensitive, deletion, truncation, amino acid insertion, and missense mutants, it appears that there are two general phenotypes of ICP4 mutants (8, 13-15, 42, 43). One of these phenotypes is characterized by the overexpression of IE genes and very low or undetectable levels of E and L gene expression. Examples of such mutations are those that delete most of the gene, inactivate the DNA-binding properties of the protein, or that retain DNA-binding properties but delete the amino- and carboxyl-terminal activation regions. Another phenotype is characterized by a mutant (n208) that is truncated at amino acid 774, thereby deleting the carboxyl-terminal 524 amino acids. E genes are activated, IE genes are repressed, but true L genes are not expressed. Many temperature-sensitive mutants with mutations in the region deleted in n208 share this phenotype. Given these two phenotypes, we sought to determine how the binding of ICP4, TBP, and polII to IE, E, and L promoters on the viral genome during infection is influenced by the composition of ICP4 and how this may contribute to the observed phenotype in infected cells. Two ICP4 mutants were compared to wt virus. i13 contains a 2-amino-acid insertion at residue 338. The expressed protein exists as a dimer with the same hydrodynamic properties as the wt, but it does not bind to DNA, overexpresses IE genes, and is completely defective in activation of E and L genes (5, 42, 43). The other mutant is n208 (described above).
Vero cells were infected with KOS, i13, and n208. At 4 and 8 h postinfection, the cultures were processed and analyzed by ChIP for the presence of ICP4 (wt and mutant forms), TBP, and polII on the ICP0, tk, and gC promoters. The antibody used to precipitate ICP4 (N15) reacts with both the i13 and n208 forms of ICP4. The results are shown in Fig. 7. Both n208 and KOS ICP4 bound to the ICP0 promoter, while i13 did not. Consistent with Fig. 2, the binding of TBP and polII to the ICP0 promoter was independent of ICP4 binding. The relatively high abundance of TBP and polII on the ICP0 promoter on the i13 genome is consistent with the expression phenotype of i13 (42). Both KOS and n208 ICP4 also bound to the tk promoter, as did TBP and polII. i13 did not bind to the tk promoter, nor did TBP or polII. These observations are consistent with the phenotypes of i13 and n208. In the case of gC, the presence of ICP4, TBP, and polII at 4 h postinfection was again observed. gC was not expressed at this time. Therefore, the presence of these proteins on the gC promoter may be necessary but not sufficient for gC transcription. Interestingly, n208 did not bind to the gC promoter. TBP and polII were also not observed on the gC promoter. While n208 ICP4 has the same affinity for strong ICP4 binding sites as KOS ICP4, it is somehow differentially binding to the tk and gC promoters regions, and this appears to affect the binding of TBP and polII to the promoters. Possible explanations for this differentiation are discussed below.
FIG. 7.
Binding of ICP4, TBP, and polII to the ICP0, tk, and gC promoters in KOS-, i13-, and n208-infected cells. ChIP analysis was conducted on KOS-, i13-, and n208-infected cells to examine the binding of ICP4, TBP, and polII to the ICP0, tk, and gC promoters on the viral genomes. The graphs give percentages of genomes bound at 4 h postinfection (solid bars) and 8 h postinfection (hatched bars).
DISCUSSION
In this study ChIP was used to examine the binding of HSV-1 ICP4, TBP, and polII to the promoter regions of representative IE (ICP0), E (tk), and L (gC) genes on the viral genome as a function of time postinfection, viral DNA replication, cis-acting sites for TFIID in the tk and gC promoters, and genetic background of ICP4. This was done in an effort to further our understanding of the mechanism by which ICP4 activates transcription and to examine some of the events that are relevant to the ordered expression of HSV IE, E, and L genes. The information gained using the ChIP assay with virus-infected cells reflects events that occur on viral promoters on the genome during infection. Therefore, the resulting observations reflect physiologically relevant events.
We infer that the presence of TBP and polII on HSV promoters reflects the formation of transcription initiation complexes. All HSV genes are transcribed by polII (1), and most contain TATA boxes, which are binding sites for TBP. However, TBP exists in cells as a part of multisubunit complexes. With respect to polII transcription, TFIID is the most studied TBP-containing complex. It consists of TBP plus approximately 14 TAFs (52). However, there may be other TBP-containing core promoter recognition complexes in cells (37). Therefore, in this study, by examining TBP binding to HSV promoters in infected cells, we are examining the binding of TBP-containing complexes that may be TFIID as previously defined (52) but that, given the possible variability of TBP-containing complexes and the possible effects of viral infection, may also be complexes that differ from the classic TFIID composition.
The presence of TBP and polII on the tk and gC promoters correlates with the presence of ICP4.
It has previously been shown that ICP4 will help form transcription preinitiation complexes including TFIID and polII on a DNA fragment containing the gC promoter immobilized on magnetic beads and that the ability to ultimately recruit polII follows from the ability to recruit purified TFIID (20). ICP4 has also been shown to interact with TFIID, with one of the interactions with TFIID being via the TAF1 component (5). Therefore, this study confirms and extends the in vitro studies described above. When ICP4 was not present on the tk or gC promoter, TBP and polII were also not present (Fig. 2 and 7), suggesting that the presence of ICP4 at the promoters of E and L genes in virus-infected cells is crucial for the formation of transcription complexes on these promoters. The formation of TBP- and polII-containing complexes on the ICP0 promoter was independent of the presence of ICP4 (Fig. 2 and 7). This is consistent with its expression as an IE gene.
The presence of ICP4 near the promoters of genes it activates is consistent with its participation with transcription initiation complexes via protein-protein interactions. The inability of i13 to promote the formation of these complexes suggests that binding to DNA is also required (Fig. 7). From work with ICP4 mutants, it has been previously shown that its DNA-binding properties are important for activation (38, 43). ICP4 has been shown to bind to specific sites, which are present in the HSV genome (18). However, no such sites have been associated with induction of genes (7, 45). More recent studies have suggested that ICP4 is abundantly deposited on the HSV genome shortly after infection (17). Therefore, it is reasonable to propose that DNA binding, while not necessarily to specific sites, is important for its role in activation. This binding may be to DNA proximal to the promoter or may also involve ICP4 bound at sites distal to the promoter.
Cooperativity of TBP and ICP4 binding to tk and gC promoters.
The TATA box in the tk promoter and the initiator element (INR) in the gC promoter are both binding sites for TFIID. TBP binds to the TATA box, and TBP, TAF1, and TAF2 are required to bind to the INR (47). As previously mentioned, ICP4 interacts with TAF1 (5), and the INR is important for activation of the gC promoter by ICP4 (21, 30). When the mutants LS-29-18 (Fig. 3) and gCInr (Fig. 5) were compared to KOS for ICP4, TBP, and polII binding to the promoters, not only were TBP and polII present in reduced abundance but so was ICP4. These data suggest two conclusions. (i) TFIID is probably a TBP-containing complex associated with the gC promoter. The effect of the mutant INR on TBP binding suggests a requirement for TAF1 and -2. Further ChIP studies are needed to support this. (ii) The binding of TFIID and ICP4 may be cooperative. These studies not only show that ICP4 binding near the promoter is crucial for TBP-complex binding but also show that if TFIID binding to the promoter is reduced by mutating its binding sites, then ICP4 binding is reduced as well.
In the case of the tk promoter, not only was ICP4 binding to the promoter fragment reduced in LS-29-18-infected cells, but so was binding of ICP4 to regions downstream of the promoter (Fig. 4). Binding of ICP4 to the ICP8 promoter was not affected by the mutation in LS-29-18 (Fig. 6). This suggests that the association of ICP4 with the viral genome can be affected over some distance by the presence of binding sites for other complexes it interacts with, such as TFIID. Factors that are likely to influence TFIID binding on distal ICP4 binding include the previously mentioned ability of ICP4 to interact with TFIID and the ability of ICP4 to multimerize on DNA. We have recently shown that ICP4 forms higher-order multimers on DNA, and we hypothesized that this and its large size and elongated shape may allow ICP4 to act at a distance. This property may also transmit the effect of reduced interactions at the promoter over distances. This may have implications for the expression of nearby genes.
Role of DNA replication in the formation of ICP4-, TBP-, and polII-containing complexes.
Because viral DNA replication is a determinant in the distinction between E and L genes, one of the objectives of this study was to determine the effect of viral DNA replication on the formation of ICP4-, TBP-, and polII-containing complexes on the tk (E) and gC (L) promoters. For both the tk and gC promoters, the number of ICP4-, TBP-, and polII-bound promoters increased with time and DNA replication; however, the percentage of promoters bound following DNA replication was less than that prior to DNA replication (Fig. 3 and 5). Therefore, while there were more gC promoters bound late than tk promoters and fewer in the presence of PAA, these results cannot explain the different kinetics of expression of these genes. This may reflect that transcription initiation complexes are forming on L promoters prior to when they are expressed and that this occurs in the absence of viral DNA replication. This may reflect a requirement for additional factors not present in the infected cells prior to DNA replication for transcription of L genes. The transcriptional environment may change to be more conducive to late transcription by the dynamics of DNA replication, altered intranuclear localization (12), or changes resulting from DNA replication-induced damage responses (27, 33).
ICP4 domain requirements for the formation of ICP4-, TBP-, and polII-containing complexes.
The availability of mutants with mutations in different domains of ICP4 having different phenotypes affords the opportunity to examine the ability of ICP4 to promote TBP- and polII-containing complexes on promoters in infected cells and correlate the observations with the phenotypes of the mutants. The studies described above show that the association of ICP4 with the tk and gC promoters is crucial for the binding of TBP-containing complexes and polII to the promoters. The results with the mutant i13 (Fig. 7) as described above further underscore the importance of the DNA-binding activity to the recruitment of TBP and polII. In the absence of the DNA-binding activity, the recruitment of TBP and polII to the tk and gC promoters did not occur. However, it did occur on the IE promoter for ICP0. Presumably this is due to the action of VP16. Moreover, the abundance of TBP- and polII containing complexes on the ICP0 promoter on the i13 genome remains relatively high late after infection. These observations are consistent with the phenotype of i13, which expresses very low levels of E genes and very high levels of IE genes (42).
The virus n208 expresses an ICP4 protein that lacks the carboxyl-terminal 524 amino acids. The remaining 774 amino acids are sufficient for DNA binding, repression, and activation of E genes in virus infection. True L genes are not expressed. Figure 5 shows that the ability of ICP4 to promote TBP- and polII-containing complexes on the gC promoter was similar in both the presence and absence of PAA, showing that the formation of these complexes is not a differentiating factor for E and L gene expression. However the formation of TBP- and polII-containing complexes on the tk and gC promoters differed in KOS- and n208-infected cells (Fig. 7). As expected from the results in Fig. 5, ICP4, TBP, and polII bound to the gC promoter at both early and late times postinfection in KOS-infected cells. However, while ICP4, TBP, and polII bound to the tk promoter in n208-infected cells, ICP4-, TBP-, and polII-containing complexes were not observed on the gC promoter at any time in n208-infected cells. The correlation between the phenotype of n208 and its inability to form these early ICP4-, TBP-, and polII-containing complexes suggests that these complexes are relevant to the expression of L genes. Furthermore, the data suggest that a region of ICP4 may differentiate between formation of TBP- and polII-containing complexes on E and L promoters.
The basis for the difference between the abilities of wt and n208 ICP4 to recruit TBP-containing complexes and polII to E and L promoters can be considered by contrasting the phenotypes and biochemical properties of the n208 virus and protein with those of the wt virus and protein. The n208 protein was not found to interact with TAF1 or TFIID in vitro (5). This may help explain why n208 is not found on the gC promoter, since it cannot cooperatively bind with TFIID. However, it still recruits TBP- and polII-containing complexes to the tk promoter. Thus, the processes of recruiting the transcription machinery to E and L promoters may be different. The mechanisms of activation of E and L promoters by ICP4 are most probably different. It has been shown that while TFIIA is required for ICP4 to activate the tk promoter, it is not required to activate the gC promoter. The INR in the gC promoter is required for the TFIIA-independent activation of the gC promoter by ICP4 (50). Another activity that n208 lacks relative to wt ICP4 is the ability to multimerize on DNA. This has the effect of increasing the affinity of wt ICP4 for weak DNA-binding sites. Thus, while wt ICP4 and n208 can bind equally well to strong ICP4 binding sites, the observed affinity of n208 for the viral genome in the vicinity of weak sites will be much lower than that of wt virus. It may be that the intrinsic affinity of ICP4 for DNA in the vicinity of L genes is less than that in the vicinity of E genes, increasing the importance of the multimerization property inherent in the carboxyl-terminal 524 amino acids. The last 524 amino acids of ICP4 exhibit a large degree of conservation between the ICP4 analogs of other alphaherpes viruses. Given this and the size of this region, there are probably other properties specified by this region in addition to those described above. At the very least, the regions responsible for interacting with TAF1, multimerizing on DNA, and activating transcription need to be further dissected by genetic analysis.
The studies described here begin to examine the contribution of ICP4 to the formation of transcription complexes on HSV promoters representative of different kinetic classes. A remaining issue with ChIP experiments and viral infection that needs to be addressed is the relatively low values for percent input bound in all such experiments. This is probably due to many factors, such as the efficiency by which an antibody precipitates its antigen, the efficiency of cross-link reversal, the amount of viral DNA in the stocks used to infect the cells, the probability that an infecting genome enters the productive cycle, and most likely other factors as well. However, considering the positive ChIP results and the fact that polII is needed to transcribe HSV genes, it is clear from these studies that ICP4 recruits the cellular transcription machinery to activate promoters in the context of viral infection. These studies confirm in cells some of what has been shown in in vitro reconstruction experiments. They also expand on previous studies by showing that domains of ICP4 may possibly differentiate between the recruitment of TBP- and polII-containing complexes to E and L promoters on the genome during infection. These studies do not rigorously elucidate the nature of the recruited TBP-containing complexes, although they support the idea that TFIID is probably one such complex. In addition, they do not address the important possibility that other viral proteins, such as ICP27 and ICP8 (6, 51), may be involved in early and late transcription. These are also areas for further study.
Acknowledgments
This work was supported by NIH grant AI30612.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Alwine, J. C., W. L. Steinhart, and C. W. Hill. 1974. Transcription of herpes simplex type 1 DNA in nuclei isolated from infected HEp-2 and KB cells. Virology 60302-307. [DOI] [PubMed] [Google Scholar]
- 2.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 1801-19. [DOI] [PubMed] [Google Scholar]
- 4.Carrozza, M. J., and N. DeLuca. 1998. The high-mobility-group protein 1 is a coactivator of herpes simplex virus ICP4 in vitro. J. Virol. 726752-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrozza, M. J., and N. A. DeLuca. 1996. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol. Cell. Biol. 163085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, I. H., L. Li, L. Silva, and R. M. Sandri-Goldin. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 793949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coen, D. M., S. P. Weinheimer, and S. L. McKnight. 1986. A genetic approach to promoter recognition during trans induction of viral gene expression. Science 23453-59. [DOI] [PubMed] [Google Scholar]
- 8.Compel, P., and N. A. DeLuca. 2003. Temperature-dependent conformational changes in herpes simplex virus ICP4 that affect transcription activation. J. Virol. 773257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook, W. J., B. Gu, N. A. DeLuca, E. B. Moynihan, and D. M. Coen. 1995. Induction of transcription by a viral regulatory protein depends on the relative strengths of functional TATA boxes. Mol. Cell. Biol. 154998-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook, W. J., S. M. Lin, N. A. DeLuca, and D. M. Coen. 1995. Initiator elements and regulated expression of the herpes simplex virus thymidine kinase gene. J. Virol. 697291-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courtney, R. J., and M. Benyesh-Melnick. 1974. Isolation and characterization of a large molecular-weight polypeptide of herpes simplex virus type 1. Virology 62539-551. [DOI] [PubMed] [Google Scholar]
- 12.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55857-868. [DOI] [PubMed] [Google Scholar]
- 13.DeLuca, N. A., M. A. Courtney, and P. A. Schaffer. 1984. Temperature-sensitive mutants in herpes simplex virus type 1 ICP4 permissive for early gene expression. J. Virol. 52767-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLuca, N. A., and P. A. Schaffer. 1988. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 62732-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon, R. A., and P. A. Schaffer. 1980. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein VP175. J. Virol. 36189-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., G. Sourvinos, and A. Orr. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 773680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faber, S. W., and K. W. Wilcox. 1986. Association of the herpes simplex virus regulatory protein ICP4 with specific nucleotide sequences in DNA. Nucleic Acids Res. 146067-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.FitzGerald, P. C., A. Shlyakhtenko, A. A. Mir, and C. Vinson. 2004. Clustering of DNA sequences in human promoters. Genome Res. 141562-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grondin, B., and N. DeLuca. 2000. Herpes simplex virus type 1 ICP4 promotes transcription preinitiation complex formation by enhancing the binding of TFIID to DNA. J. Virol. 7411504-11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu, B., and N. DeLuca. 1994. Requirements for activation of the herpes simplex virus glycoprotein C promoter in vitro by the viral regulatory protein ICP4. J. Virol. 687953-7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzowski, J. F., J. Singh, and E. K. Wagner. 1994. Transcriptional activation of the herpes simplex virus type 1 UL38 promoter conferred by the cis-acting downstream activation sequence is mediated by a cellular transcription factor. J. Virol. 687774-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzowski, J. F., and E. K. Wagner. 1993. Mutational analysis of the herpes simplex virus type 1 strict late UL38 promoter/leader reveals two regions critical in transcriptional regulation. J. Virol. 675098-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrera, F. J., and S. J. Triezenberg. 2004. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 789689-9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 148-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honess, R. W., and B. Roizman. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 721276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwahori, S., N. Shirata, Y. Kawaguchi, S. K. Weller, Y. Sato, A. Kudoh, S. Nakayama, H. Isomura, and T. Tsurumi. 2007. Enhanced phosphorylation of transcription factor sp1 in response to herpes simplex virus type 1 infection is dependent on the ataxia telangiectasia-mutated protein. J. Virol. 819653-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, K. D., and E. H. Bresnick. 2002. Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods 2627-36. [DOI] [PubMed] [Google Scholar]
- 29.Kattar-Cooley, P., and K. W. Wilcox. 1989. Characterization of the DNA-binding properties of herpes simplex virus regulatory protein ICP4. J. Virol. 63696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, D. B., S. Zabierowski, and N. A. DeLuca. 2002. The initiator element in a herpes simplex virus type 1 late-gene promoter enhances activation by ICP4, resulting in abundant late-gene expression. J. Virol. 761548-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuddus, R. H., and N. A. DeLuca. 2007. DNA-dependent oligomerization of herpes simplex virus type 1 regulatory protein ICP4. J. Virol. 819230-9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leopardi, R., P. L. Ward, W. O. Ogle, and B. Roizman. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the U(L)13 proteinkinase. J. Virol. 711133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lilley, C. E., C. T. Carson, A. R. Muotri, F. H. Gage, and M. D. Weitzman. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 1025844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 691531-1574. [DOI] [PubMed] [Google Scholar]
- 35.McGeoch, D. J., A. Dolan, S. Donald, and D. H. Brauer. 1986. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 141727-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metzler, D. W., and K. W. Wilcox. 1985. Isolation of herpes simplex virus regulatory protein ICP4 as a homodimeric complex. J. Virol. 55329-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller, F., M. A. Demeny, and L. Tora. 2007. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J. Biol. Chem. 28214685-14689. [DOI] [PubMed] [Google Scholar]
- 38.Paterson, T., and R. D. Everett. 1988. The regions of the herpes simplex virus type 1 immediate early protein Vmw175 required for site specific DNA binding closely correspond to those involved in transcriptional regulation. Nucleic Acids Res. 1611005-11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira, L., M. H. Wolff, M. Fenwick, and B. Roizman. 1977. Regulation of herpesvirus macromolecular synthesis. V. Properties of alpha polypeptides made in HSV-1 and HSV-2 infected cells. Virology 77733-749. [DOI] [PubMed] [Google Scholar]
- 40.Post, L. E., S. Mackem, and B. Roizman. 1981. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell 24555-565. [DOI] [PubMed] [Google Scholar]
- 41.Preston, C. M. 1979. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J. Virol. 29275-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shepard, A. A., and N. A. DeLuca. 1991. Activities of heterodimers composed of DNA-binding- and transactivation-deficient subunits of the herpes simplex virus regulatory protein ICP4. J. Virol. 65299-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shepard, A. A., A. N. Imbalzano, and N. A. DeLuca. 1989. Separation of primary structural components conferring autoregulation, transactivation, and DNA-binding properties to the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 633714-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shepard, A. A., P. Tolentino, and N. A. DeLuca. 1990. trans-dominant inhibition of herpes simplex virus transcriptional regulatory protein ICP4 by heterodimer formation. J. Virol. 643916-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smiley, J. R., D. C. Johnson, L. I. Pizer, and R. D. Everett. 1992. The ICP4 binding sites in the herpes simplex virus type 1 glycoprotein D (gD) promoter are not essential for efficient gD transcription during virus infection. J. Virol. 66623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, C. A., P. Bates, R. Rivera-Gonzalez, B. Gu, and N. A. DeLuca. 1993. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J. Virol. 674676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verrijzer, C. P., J. L. Chen, K. Yokomori, and R. Tjian. 1995. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 811115-1125. [DOI] [PubMed] [Google Scholar]
- 48.Watson, R. J., and J. B. Clements. 1980. A herpes simplex virus type 1 function continuously required for early and late virus RNA synthesis. Nature 285329-330. [DOI] [PubMed] [Google Scholar]
- 49.Wells, J., and P. J. Farnham. 2002. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods 2648-56. [DOI] [PubMed] [Google Scholar]
- 50.Zabierowski, S., and N. A. DeLuca. 2004. Differential cellular requirements for activation of herpes simplex virus type 1 early (tk) and late (gC) promoters by ICP4. J. Virol. 786162-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, C., and D. M. Knipe. 2002. Association of herpes simplex virus type 1 ICP8 and ICP27 proteins with cellular RNA polymerase II holoenzyme. J. Virol. 765893-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, Q., P. M. Lieberman, T. G. Boyer, and A. J. Berk. 1992. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 61964-1974. [DOI] [PubMed] [Google Scholar]