Abstract
The outer envelope of the extracellular form of vaccinia virus contains five virus-encoded proteins, F13, A33, A34, A56, and B5, that, with the exception of A56, are implicated in virus egress or infectivity. A34, a type II transmembrane glycoprotein, is involved in the induction of actin tails, the release of enveloped virus from the surfaces of infected cells, and the disruption of the virus envelope after ligand binding prior to virus entry. To investigate interactions between A34 and other envelope proteins, a recombinant vaccinia virus (vA34RHA) expressing an epitope-tagged version of A34 (A34HA) was constructed by appending an epitope from influenza virus hemagglutinin to the C terminus of A34. Complexes of A34HA with B5 and A36, but not with A33 or F13, were detected in vA34RHA-infected cells. A series of vaccinia viruses expressing mutated versions of the B5 protein was used to investigate the domain(s) of B5 required for interaction with A34. Both the cytoplasmic and the transmembrane domains of B5 were dispensable for binding to A34. Most of the extracellular domain of B5, which contains four short consensus repeats homologous to complement control proteins, was sufficient for A34 interaction, indicating that both proteins interact through their ectodomains. Immunofluorescence experiments on cells infected with A34-deficient virus indicated that A34 is required for efficient targeting of B5, A36, and A33 into wrapped virions. Consistent with this observation, the envelope of A34-deficient virus contained normal amounts of F13 but decreased amounts of A33 and B5 with respect to the parental WR virus. These results point to A34 as a major determinant in the protein composition of the vaccinia virus envelope.
Vaccinia virus, the most-studied poxvirus, replicates and assembles in the cytoplasm of the infected cell. Vaccinia virus assembly and release are complex processes involving several virus forms, including the noninfectious immature virus (IV), the intracellular mature virus (IMV), the intracellular enveloped virus (IEV), the cell-associated enveloped virus (CEV), and the extracellular enveloped virus (EEV) (25, 38, 39).
Fully infectious IMV particles are assembled in the cytoplasm and remain intracellular until cells are lysed. To accomplish cell-to-cell transmission, infectious virus particles must acquire an additional membrane. Thus, after IMV assembly, some IMV move from the assembly areas on microtubules and become wrapped by vesicles derived from the early endosomes (40, 42) or trans-Golgi network (36) to form IEV. Subsequently, IEV are transported to the cell surface, where the outer membrane fuses with the plasma membrane, thus generating virions which contain an extra membrane with respect to IMV. Virus particles that remain attached to the plasma membrane are termed CEV, while those released are called EEV (2). In cell culture, enveloped virions (CEV plus EEV) are responsible for virus transmission, since mutants that form normal amounts of IMV but are blocked in enveloped virus formation are impaired in transmission (1). Although both CEV and EEV can mediate virus transmission between cells, specific roles have been assigned to CEV and EEV in this process. CEV particles can induce the formation of actin tails that are important for efficient direct cell-to-cell spread, while EEV is responsible for long-range dissemination of virus (2, 5, 28).
Of the virus-encoded IEV envelope proteins that have been characterized, A33 (31), A34 (7), B5 (9, 16), and F13 (13) are present in IEV, CEV, and EEV. In addition, proteins K2 and A56 associate with and are present in EEV (6, 37, 41). In contrast to these EEV envelope proteins, A36 and F12 are present in IEV but absent from CEV and EEV envelopes (26, 42, 43, 50). The functions and immunological aspects of particular protein constituents of the IEV and EEV envelopes have been the focus of much attention. Studies of viruses carrying targeted mutations showed that proteins B5 and F13 are required for efficient and complete wrapping of IMV (1, 10, 47). Proteins A33, A34, A36, and B5 are required, either directly or indirectly, for the induction of actin tails (32, 34, 48, 49). Formation or release of EEV from the cell surface can be increased by mutations in the A33, A34, and B5 proteins (4, 12, 17, 18). Finally, IEV-specific proteins A36 and F12 are involved in the microtubule-mediated transport of IEV to the cell surface (30, 42, 45, 46).
We hypothesize that protein-protein interactions between IEV envelope proteins determine their localization in the wrapping membranes and might be needed not only for IMV wrapping but also for later events, such as IEV transport, egress, and induction of actin tails. To date, a number of protein interactions have been described, including A33-B5 (29), A33-A36 (33, 49), A34-B5 (33), A34-A36 (33), B5-F13 (27), A36-Nck (11), and A36-Grb2 (35), but in most cases no specific functions have been assigned to these interactions. In this work, we characterize the interactions established between A34 and other envelope proteins and demonstrate that the A34 protein is an important factor in determining the protein composition of the EEV envelope.
MATERIALS AND METHODS
Cells and viruses.
BSC-1 and CV-1 cells were grown in Eagle's minimal essential medium supplemented with 0.1 μg/ml penicillin, 0.1 μg/ml streptomycin, 2 mM l-glutamine (Bio Whittaker), and 5% fetal bovine serum (FBS; Bio Whittaker). BHK-21 cells (ATCC CCL10) were grown in BHK-21 Glasgow minimal essential medium (Gibco BRL) containing 5% FBS, 3 g/ml tryptose phosphate broth, 0.01 M HEPES, antibiotics, and glutamine. Virus infections were performed with 2% FBS for all cell types. Vaccinia virus vΔA34R (24) was provided by G. L. Smith.
Antibodies.
Mouse monoclonal antibody VMC-34 (anti-A33) was provided by G. H. Cohen. Anti-A36 mouse monoclonal antibody was made available by G. L. Smith. Rat monoclonal antibodies 19C2 (anti-B5) and 15B6 (anti-F13) were kindly made available by G. Hiller. Mouse monoclonal antibodies 4C5/CR3 (anti-porcine antigen) and 3B11/11 (anti-porcine sialoadhesin) were provided by C. Revilla and B. Álvarez and were used as control immunoglobulins G (IgGs) in coimmunoprecipitation experiments. Anti-A34 rabbit antiserum was obtained by immunization with a histidine-tagged soluble form of A34 expressed from a vaccinia virus recombinant (M. D. M. Lorenzo et al., unpublished data).
Plasmid construction.
Plasmid pGem-A34RHAg, used for the construction of a recombinant vaccinia virus expressing the influenza virus hemagglutinin (HA) epitope fused to the C terminus of the A34 protein (vA34RHA), was obtained by the sequential cloning of four DNA fragments, containing the red-shifted green fluorescent protein (rsGFP) gene, HA epitope, A34R gene, and A34R recombination flanking sequences, into plasmid pGem-7Zf(−) (Promega). The rsGFP gene, under the control of the vaccinia virus synthetic early/late promoter, was amplified by PCR from plasmid pGem-pac-rsGFP (obtained by the cloning of rsGFP and puromycin acetyltransferase [pac] genes into pGem-7Zf) with oligonucleotides rsGFP-Nsi (5′-GCCTGACGCATGCATGAGAAAAATTG-3′ [NsiI site is underlined]) and Sp6 (5′-ATTTAGGTGACACTATAGAA-3′). The PCR product was cut with NsiI and inserted into NsiI-digested pGem-7Zf to generate pGem-rsGFP. The HA epitope was obtained by annealing oligonucleotide primers 34HA-X (5′-CTAGAGTACCCATATGATGTTCCAGATTATGCTTAAG-3′) and 34HA-E (5′-AATTCTTAAGCATAATCTGGAACATCATATGGGTACT-3′). The HA epitope was inserted into plasmid pGem-rsGFP, previously digested with XbaI and EcoRI, to generate pGem-HA-rsGFP. Primer 34HA-X inserted a stop codon at the end of the HA epitope. The left flank and the coding sequence of the A34R gene were amplified by PCR from the WR genome with oligonucleotides A34R-Aat II (5′-CAATAAATCCGACGTCTTGATTACC-3′ [AatII site is underlined]) and A34R-X (5′-CATTTTTTCTAGAGACTTGTAGAATT-3′ [XbaI site is underlined]). The PCR product was digested with AatII and XbaI and ligated to plasmid pGem-HA-rsGFP previously digested with AatII and XbaI to generate pGEM-fi-A34R-HA-rsGFP. Primer A34R-X eliminated the stop codon at the end of the A34R gene and provided an in-frame fusion with the HA epitope. Finally, the WR genome was used as the template to amplify the right flank of the A34R gene with oligonucleotides fd34-E (5′-AAAAGAATTCAAGTGACAACAAAAAATG-3′ [EcoRI site is underlined]) and A34Rfd-B (5′-GGATTGAGGATCCGATGATAATC-3′ [BamHI site is underlined]). The PCR product was cut with EcoRI and BamHI and inserted into EcoRI/BamHI-digested pGEM-fi-A34R-HA-rsGFP to generate pGEM-A34R-cHAg.
Plasmid pG-B5R-V5-Red2, used for the construction of the recombinant virus vA34RHAΔB5R, in which 94% of the B5R coding sequence has been replaced with a dsRed2 expression cassette, has been described previously (29).
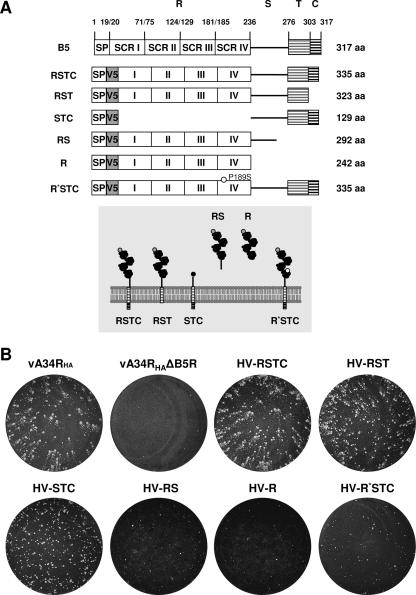
For the construction of recombinant viruses expressing A34HA and different mutated versions of V5-tagged B5 protein, six mutated versions of the B5R gene, depicted schematically in Fig. 3A, were inserted into the B5R locus of the virus mutant vA34RHAΔB5R. These mutated versions without a signal peptide sequence were amplified by PCR and inserted separately into XbaI/EcoRI-digested pG-B5R-V5-Red2 to generate plasmids pG-V5-B5Ra to pG-V5-B5Rf (29).
FIG. 3.
V5-tagged mutated versions of B5 protein. (A) (Top) Schematic representation of V5-tagged B5 versions. (Adapted from reference 29 with permission.) SP, signal peptide; V5, V5 epitope; R, four SCRs (I to IV); S, stalk; T, transmembrane domain; C, cytoplasmic tail. R* indicates an R region including a P189S mutation located in SCR IV. Versions were termed, as indicated on the left, according to the domains included in the construct. (Bottom) Predicted topology of the mutated B5 versions in the outer membrane of the EEV particle. Domains above the lipid bilayer are in the lumen of the wrapping membranes or the extracellular space, whereas domains below the membrane are cytosolic. (B) Plaque phenotypes of recombinant viruses expressing A34HA and V5-tagged mutated B5 versions. BSC-1 cell monolayers infected with vA34RHA, vA34RHAΔB5R, or the recombinant viruses indicated were incubated for 2 days, stained with crystal violet, and photographed.
Construction of recombinant viruses.
Recombinant viruses were isolated following infection/transfection experiments. Plasmids were transfected by use of the Fugene transfection reagent (Roche) following the manufacturer's recommendations. vA34RHA was constructed by transient dominant selection, using the rsGFP gene as the transiently selectable marker. CV-1 cells were infected with vΔA34R at 0.05 PFU per cell and transfected 1 h later with pGem-A34RHAg. vA34RHA was isolated from progeny virus by rounds of plaque purification on BSC-1 cells (8), during which plaques were screened for GFP fluorescence and plaque size (3). vA34RHAΔB5R was also constructed by transient dominant selection, using the rsGFP gene as the selectable marker. CV-1 cells were infected with vA34RHA at 0.05 PFU per cell and transfected 1 h later with pG-B5R-V5-Red2. vA34RHAΔB5R was isolated from progeny virus by rounds of plaque purification on CV-1 cells stably expressing B5 protein (Perdiguero and Blasco, unpublished), during which plaques were screened for GFP/dsRed2 fluorescence. Recombinant viruses expressing A34HA and different mutated versions of B5 protein tagged with the V5 epitope were constructed by transient dominant selection, using the rsGFP gene as the selectable marker. CV-1 cells were infected with vA34RHAΔB5R at 0.05 PFU per cell and transfected 1 h later with pG-V5-B5Ra to pG-V5-B5Rf. Recombinants were isolated from progeny virus by rounds of plaque purification on CV-1 cells stably expressing the B5 protein (B. Perdiguero and R. Blasco, unpublished data), during which plaques were screened for GFP/dsRed2 fluorescence.
Immunofluorescence microscopy.
BHK-21 cells grown to 70% confluence on round coverslips were infected with vaccinia virus at 5 PFU per cell. At 8 h postinfection, cells were washed with phosphate-buffered saline (PBS), fixed for 15 min at room temperature with cold 4% paraformaldehyde, washed with PBS, and permeabilized by incubation for 15 min with PBS-0.1% Triton X-100. After being washed with PBS, cells were incubated for 5 min with PBS containing 0.1 M glycine and then with primary antibody diluted in PBS-20% FBS (1:100 for anti-B5, 1:400 for anti-A33, 1:50 for anti-A36, 1:50 for anti-F13, and 1:20 for anti-HA-fluorescein [Roche]) for 30 min. After being washed with PBS, cells were incubated for 30 min with secondary antibody diluted in PBS-20% FBS (1:200 for rabbit anti-mouse IgG-tetramethyl rhodamine isocyanate [TRITC] [Dako]). Some preparations were also incubated with 2 mg/ml bisbenzimide (Hoechst dye) or TRITC-phalloidin (Sigma) for 30 min at room temperature. Finally, cells were washed extensively with PBS, mounted with FluorSave reagent (Calbiochem), and observed by fluorescence microscopy.
Western blotting.
Western blots of infected cell lysates were performed using BSC-1 cells grown in six-well plates and harvested at 24 h postinfection in denaturant buffer (80 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate [SDS], 10% glycerol, and 0.01% bromophenol blue solution, with or without 0.71 M 2-mercaptoethanol). Proteins were resolved by electrophoresis in 12% SDS-polyacrylamide gels. After SDS-polyacrylamide gel electrophoresis (PAGE), separated proteins were transferred to nitrocellulose. Membranes were incubated overnight at 4°C with primary antibody (diluted in PBS containing 0.05% Tween 20 and 1% nonfat dry milk) conjugated with horseradish peroxidase (1:500 for anti-HA-horseradish peroxidase [anti-HA-HRP] [Roche] and 1:5,000 for anti-V5-HRP [Invitrogen]). After being washed extensively with PBS-0.05% Tween 20, membranes were incubated for 1 min with a 1:1 mix of solution A (2.5 mM luminol [Sigma], 0.4 mM ρ-coumaric acid [Sigma], 100 mM Tris-HCl, pH 8.5) and solution B (0.018% H2O2, 100 mM Tris-HCl, pH 8.5) and exposed to X-ray film.
For Western blot analysis of proteins present in extracellular virus, supernatants of BHK-21-infected cells were clarified, and extracellular virus was pelleted by centrifugation in an SW-41 rotor at 18,000 rpm for 2 h. The pellet was resuspended in 300 μl of denaturant buffer (80 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, and 0.01% bromophenol blue solution, with or without 0.71 M 2-mercaptoethanol). Western blotting was carried out as described above.
Immunoprecipitation.
BSC-1 cells (7.5 × 106) were infected with vaccinia virus at 5 PFU per cell. At 24 h postinfection, cells were lysed with digitonin lysis buffer (1% digitonin [Sigma], 10 mM triethanolamine, pH 7.8 [Merck], 150 mM NaCl, 1 mM EDTA), with 10 mM phenylmethylsulfonyl fluoride (Boehringer Mannheim) as a protease inhibitor, for 30 min at 4°C. The lysed cells were scraped and spun down for 20 min at 13,000 rpm and 4°C. The supernatant was harvested and precleared by incubation with protein G-Sepharose beads (Amersham) for 1 h at 4°C in a tube rotator. After centrifugation for 1 min at 13,000 rpm and 4°C, the precleared supernatant was incubated with protein G-Sepharose beads and control IgG (300 μl) or specific antibodies (5 μl for anti-A33, 100 μl for anti-A36, anti-B5, and anti-F13, and 1.5 μg for anti-V5 [Invitrogen] and anti-HA [Sigma-Aldrich]) overnight for 1 h at 4°C while being mixed end over end in a tube rotator. After centrifugation for 1 min at 13,000 rpm and 4°C, the supernatant was removed and immunoprecipitates bound to protein G-Sepharose beads were washed extensively with lysis buffer. Finally, immunoprecipitates were resuspended in denaturant buffer, boiled for 5 min at 95°C, and resolved by SDS-PAGE.
RESULTS
Construction of a recombinant vaccinia virus expressing HA-tagged A34 protein.
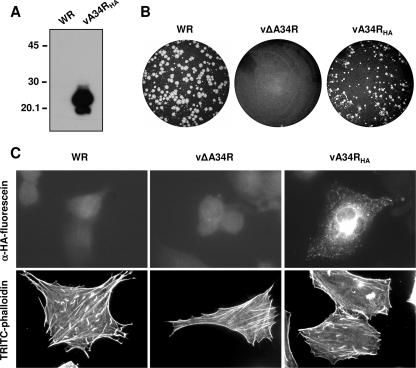
To facilitate experiments on interactions involving glycoprotein A34, a modified version of the protein (A34HA) was obtained by fusing an epitope tag derived from influenza virus HA at the C terminus of the A34 protein. The modified A34RHA gene was inserted into the virus genome by recombination in the A34R deletion mutant. Virus recombinant vA34RHA is expected to produce a protein of 25 to 29 kDa, resulting from the fusion of A34 with the HA epitope. The correct expression of the tagged A34HA fusion protein was confirmed by Western blot analysis of extracts of vA34RHA-infected cells, which revealed the presence of a protein with the expected electrophoretic mobility that reacted with anti-HA antibody (Fig. 1A).
FIG. 1.
Characterization of a recombinant vaccinia virus expressing an HA-tagged version of the A34 protein. (A) A34HA expression by Western blot analysis. Extracts of BSC-1 cells infected for 24 h with a recombinant vaccinia virus expressing the fusion protein A34HA (vA34RHA) or with control virus (WR) were probed with anti-HA-HRP antibody. The positions of protein molecular mass markers (in kDa) are indicated. (B) Plaque phenotype of vA34RHA. BSC-1 cell monolayers infected with WR, vΔA34R, or vA34RHA were incubated for 2 days, stained with crystal violet, and photographed. (C) Subcellular distribution of A34HA and induction of actin tails by vA34RHA. BHK-21 cells infected for 7 h were fixed, permeabilized, and incubated with anti-HA-fluorescein (top panels) or TRITC-phalloidin (bottom panels). Note the presence of actin tails in vA34RHA-infected cells.
To evaluate the functionality of A34HA, the phenotype of vA34RHA regarding virus transmission was compared with those of the parental WR virus strain and the A34-deficient virus (vΔA34R). Reportedly, vΔA34R produces elevated amounts of enveloped virions, albeit with reduced infectivity, but is unable to induce actin tails and has a tiny plaque phenotype (7, 24, 48). Thus, deletion of A34R does not block virus egress but results in severe defects in virus cell-to-cell transmission. In a standard plaque assay, vA34RHA plaques were significantly larger than those of vΔA34R, indicating that A34HA provides A34 function. However, vA34RHA plaques were smaller than plaques formed by the WR virus strain, indicating some effect of introducing the HA epitope tag (Fig. 1B).
Immunofluorescence of vA34RHA-infected cells with anti- HA antibody revealed a distribution of A34HA that was similar to that of normal A34 in WR-infected cells, with a strong juxtanuclear staining and a punctate staining revealing peripheral virions (Fig. 1C).
Since deletion of A34R completely abrogates the ability of vaccinia virus to induce actin tails (48), we also tested the function of vA34RHA in promoting actin tail formation. vA34RHA-infected cells were stained with TRITC-phalloidin and examined by fluorescence microscopy (Fig. 1C). In contrast with vΔA34R, vA34RHA was able to induce actin tails in infected cells, although the number of tails was smaller than that found in WR-infected cells. The above results indicate that the epitope-tagged A34HA fusion protein can functionally replace the normal A34 protein, albeit with reduced efficiency.
Coimmunoprecipitation of A34HA with IEV envelope proteins.
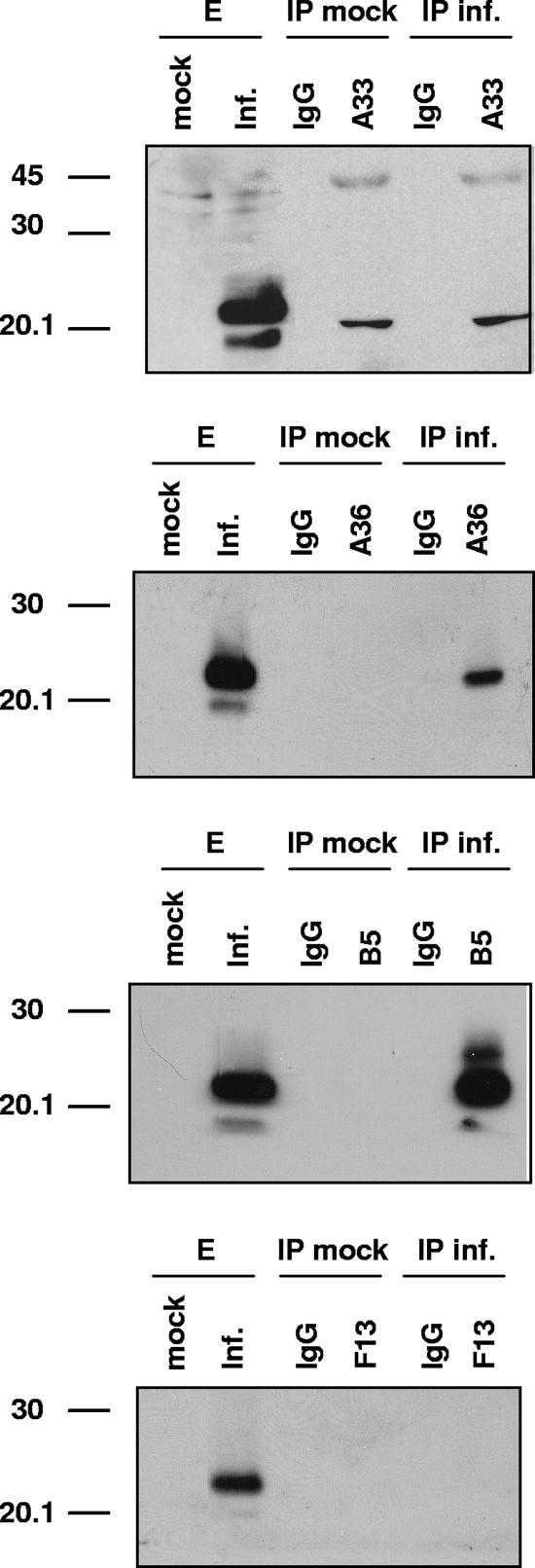
We next searched for interactions between A34 and other IEV envelope proteins. Extracts of cells infected with vA34RHA were immunoprecipitated with antibodies to A33, A36, B5, and F13 and then subjected to Western blot analysis with anti-HA antibody (Fig. 2). Both anti-A36 and anti-B5 antibodies immunoprecipitated the A34HA protein, whereas anti-A33 and anti-F13 failed to immunoprecipitate A34HA. This experiment confirmed a previous study (33) showing that A34 is associated with A36 and B5.
FIG. 2.
Coimmunoprecipitation of A34 from infected cells. BSC-1 cells were mock infected or infected with 5 PFU per cell of vA34RHA and then lysed at 24 h postinfection. Immunoprecipitations were performed with either control IgG or anti-A33, anti-A36, anti-B5, or anti-F13 antibody (indicated at the top of each panel). Western blots were probed with anti-HA-HRP antibody. Lanes E, infected cell extracts prior to immunoprecipitation; IP mock, immunoprecipitation of mock-infected cell extracts; IP inf., immunoprecipitation of vA34RHA-infected cell extracts. Molecular mass markers (in kDa) are indicated on the left.
Construction of vaccinia virus recombinants expressing A34HA and mutated versions of B5.
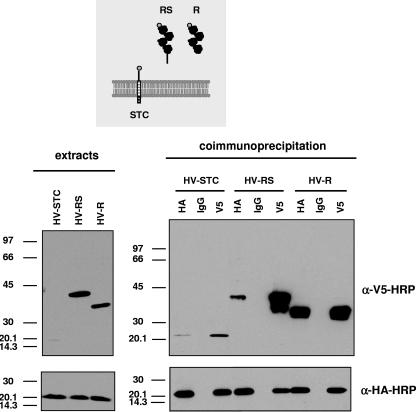
To facilitate the study of the A34-B5 interaction, a series of recombinant viruses harboring both A34HA and mutated versions of B5 was obtained. A similar set of B5 mutations in the WR context was used in a previous study (29). To isolate these recombinants, a B5R deletion mutant was first constructed from vA34RHA, and subsequently, mutated versions of B5 were introduced into the B5R locus. In the mutated versions, a V5 epitope was included at the N terminus of the mature B5 protein (i.e., after signal peptide removal) to assist in detection. Viruses were termed according to the domains of B5 included (see the scheme in Fig. 3A), with R being the short consensus repeats (SCRs), homologous to complement control proteins (amino acids 20 to 236), S being the stalk region (amino acids 237 to 276), T being the transmembrane domain (amino acids 277 to 303), and C being the cytoplasmic tail (amino acids 304 to 317). Thus, virus HV-RSTC is a recombinant virus expressing HA-tagged A34 and the complete V5-tagged B5 protein. Viruses lacking the cytoplasmic domain (HV-RST), the SCR domains (HV-STC), the transmembrane and cytoplasmic domains (HV-RS), or the stalk, transmembrane, and cytoplasmic domains (HV-R) were isolated. Finally, a recombinant virus (termed HV-R*STC) containing a B5 version with a proline-to-serine mutation at position 189 (P189S) was constructed. The P189S mutation has been shown to enhance the release of enveloped virus into the extracellular medium and to produce a comet-shaped plaque phenotype (17).
The plaque phenotypes of these recombinant viruses are shown in Fig. 3B. Regarding plaque size, the results are congruent with data for the same B5 mutations in the WR background (29), considering the smaller plaque size of the parental virus (vA34RHA). As expected, inclusion of the V5 epitope or deletion of the cytoplasmic domain of B5 had little or no effect on plaque size. Recombinant viruses HV-RS and HV-R produced small plaques, suggesting that membrane anchoring is essential for B5 function in virus transmission. Deletion of the SCR domains (HV-STC) or the P189S mutation (HV-R*STC), which confer a comet-forming phenotype to WR virus, produced small plaques with no apparent comet tail when introduced into the vA34RHA virus. At present, we do not have an explanation for this discrepancy.
Mapping the A34-B5 interaction by coimmunoprecipitation experiments.
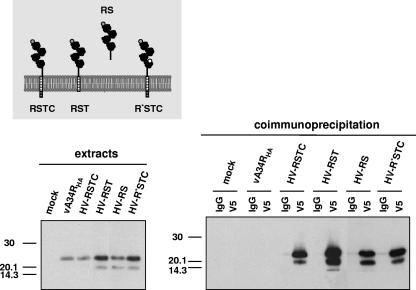
Taking advantage of the observation that the A34HA-B5 complex can be immunoprecipitated, we sought to identify the B5 domain involved in A34 binding. Thus, extracts prepared from cells infected with recombinant viruses expressing both A34HA and mutated forms of V5-tagged B5 protein were subjected to immunoprecipitation with anti-V5 antibody, followed by immunoblot detection with anti-HA antibody (Fig. 4). A34HA was immunoprecipitated from cells infected with viruses expressing the complete B5 protein (HV-RSTC) or the B5 version lacking the cytoplasmic domain (HV-RST), indicating that the cytoplasmic tail of B5 was not required for A34 binding. The soluble form of B5 (HV-RS), lacking the transmembrane and cytoplasmic domains, was also efficiently coimmunoprecipitated with A34HA. These results indicate that the ectodomain of B5 is sufficient to mediate binding to A34, while the transmembrane and cytoplasmic domains of B5 are not necessary for the interaction.
FIG. 4.
Mapping of A34-B5 interaction site by coimmunoprecipitation. BSC-1 cells were mock infected or infected with the viruses indicated at the top of each panel at 5 PFU per cell and then harvested at 24 h postinfection. The panel on the left shows the infected cell extracts prior to immunoprecipitation, probed with anti-HA antibody conjugated with HRP. Immunoprecipitation was performed with either control IgG (lanes IgG) or anti-V5 antibody (lanes V5), as indicated at the top of the right panel. Cell extracts (left) and immunoprecipitated material (right) were resolved by SDS-PAGE and subjected to Western blot analysis with anti-HA-HRP antibody. Molecular mass markers are indicated (in kDa).
It has been reported that specific mutations in both A34 and B5 result in enhanced release of virus into the extracellular medium and produce a comet-shaped plaque phenotype (4, 12, 17, 18, 21). Among these, a point mutation (P189S) in the fourth SCR domain of B5 has been shown to increase EEV release. To determine if the phenotypic effect of the P189S mutation was related to a loss of the A34-B5 interaction, we performed immunoprecipitation experiments on extracts of cells infected with HV-R*STC, expressing a V5-tagged B5 protein containing the P189S mutation. From these extracts, A34HA was efficiently coimmunoprecipitated with B5 by use of an anti-V5 antibody, indicating that the P189S mutation did not affect the A34-B5 interaction.
The ectodomain of B5 comprises four SCR domains and a stalk region. Since the soluble version of B5 (version RS) was able to form a complex with A34, we tested which portion of the ectodomain was involved in the interaction. With this aim, three mutant viruses, expressing the C terminus of B5, including the stalk portion (HV-STC), the complete soluble ectodomain of B5 (HV-RS), or the four SCR domains (HV-R), were used to infect cells and perform immunoprecipitation experiments (Fig. 5). As in previous experiments, version RS, comprising the complete B5 ectodomain, was coimmunoprecipitated with A34. Interestingly, the SCR domain region alone (version R) was also complexed with A34. These results suggest that the main interaction site between A34 and B5 lies within the SCR domain of B5. Version STC was immunoprecipitated with anti-HA antibody, albeit in smaller amounts. Also, A34HA coimmunoprecipitated with B5 version STC by use of anti-V5 antibody. These results indicate that in addition to the interaction site in the SCR domains, the C-terminal one-third of the protein includes a second interaction site.
FIG. 5.
Interaction of SCR domains of B5 with A34. BSC-1 cells were infected with the viruses indicated at the top of the panels at 5 PFU per cell and then harvested at 24 h postinfection. The panels on the left show Western blots of infected cell extracts prior to immunoprecipitation, probed with anti-V5-HRP antibody (top) or anti-HA-HRP antibody (bottom). Immunoprecipitation was performed with either control IgG (lanes IgG), anti-HA antibody (lanes HA), or anti-V5 antibody (lanes V5), as indicated at the top of the right panels. Immunoprecipitated material was resolved by SDS-PAGE and subjected to Western blot analysis with anti-V5-HRP antibody (top) or anti-HA-HRP antibody (bottom). Molecular mass markers are indicated (in kDa).
To further confirm that A34 and B5 interact through their extracellular portions, we coexpressed the ectodomains of both proteins. This was accomplished by coinfection with a recombinant virus expressing a soluble form of A34 tagged with a c-myc epitope and virus HV-RS or HV-R. In both cases, a complex formed between the A34 ectodomain and the B5 extracellular portion was detected by immunoprecipitation (not shown), reinforcing the idea that the SCR domains of B5 are sufficient to mediate the interaction with the extracellular portion of A34.
Effect of A34R deletion on targeting of IEV envelope proteins.
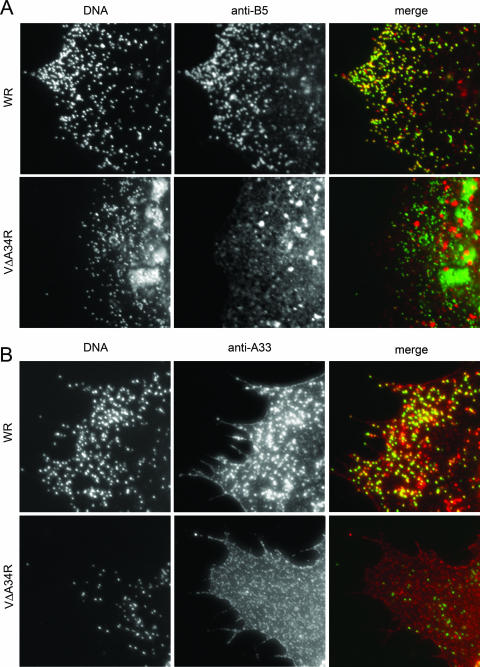
Vaccinia virus envelope proteins are targeted to trans-Golgi network vesicles or late endosomes that wrap IMV particles. The IEV thus formed are then transported from the wrapping areas to the cell periphery and subsequently released into the extracellular medium. Considering the known interactions between envelope proteins, we investigated the potential role of A34 in the incorporation of other envelope proteins into IEV particles. With that aim, we performed immunofluorescence experiments designed to detect the colocalization of envelope proteins with virions in the periphery of the cell (IEV and CEV). First, we compared the distribution of B5 in cells infected with either WR or A34-deficient viruses (Fig. 6A). To distinguish between vesicles and viral particles, Hoechst staining was performed on the same samples. Most of the staining with anti-B5 antibody in the periphery of WR-infected cells was coincident with the DNA-containing virus particles (IEV and CEV). In contrast, anti-B5 staining in vΔA34R-infected cells revealed an altered distribution of B5, with a more diffuse B5 signal, together with structures larger than virions that may represent vesicles. The poor colocalization of B5 with peripheral virions in vΔA34R-infected cells indicates that A34 is required for efficient targeting of B5 to enveloped virions.
FIG. 6.
Distribution of B5 and A33 envelope proteins in A34-deficient virus. BHK-21 cells were infected for 7 h, fixed, permeabilized, and stained with antibodies against envelope protein B5 or A33. DNA was visualized by Hoechst staining. (A) Colocalization of B5 and virus particles. Cells infected with WR or A34-deficient virus (vΔA34R) were stained with anti-B5 antibody. Merged images resulted from the combination of monochrome images pseudocolored green (Hoechst) and red (anti-B5). Note the colocalization of B5 protein with viral DNA in cells infected with WR but not in vΔA34R-infected cells. (B) Colocalization of A33 and virus particles. Cells infected with WR or vΔA34R were stained with anti-A33 antibody. Merged images resulted from combining monochrome images pseudocolored green (Hoechst) and red (anti-A33). Note that the colocalization of A33 protein with viral DNA in cells infected with WR is affected in vΔA34R-infected cells.
A previous report implicated B5 in the incorporation of A33 into the virus envelope (29). Consequently, given the effect of A34 deficiency on the targeting of B5, one could expect an indirect effect of the A34 deficiency on the distribution of A33. Thus, we performed immunofluorescence with anti-A33 antibody and vΔA34R-infected cells and compared the distribution of A33 with that found in WR-infected cells (Fig. 6B). A33 staining was prominent in the periphery of WR-infected cells, both in DNA-containing particles and in the plasma membrane. In contrast, anti-A33 antibody did not label DNA-containing virus particles in vΔA34R-infected cells, where most of the staining was found in the plasma membrane and was not coincident with DNA labeling. Redistribution of A33 as a consequence of A34R deletion has been reported before (33). These results indicate that A34 deficiency has a dramatic effect on the incorporation of B5 and A33 into IEVs.
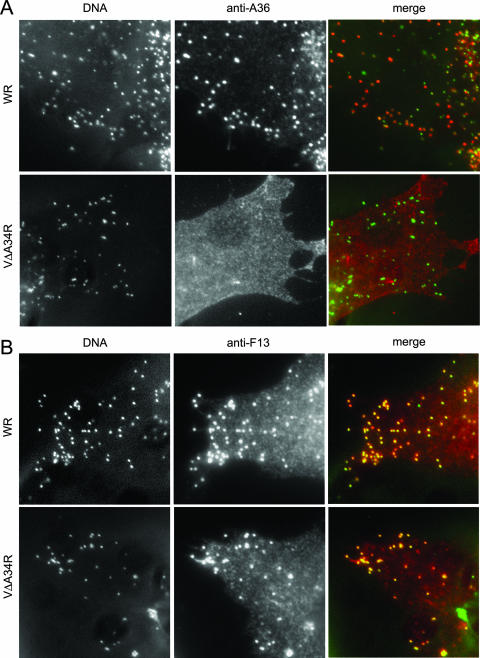
We also investigated the effect of A34 deficiency on the distribution of other constituents of IEV. We performed immunofluorescence microscopy with anti-A36 or anti-F13 antibody and vΔA34R-infected cells, together with DNA staining, to visualize virus particles (Fig. 7). In control WR-infected cells, A36 was present in a portion of the peripheral virus particles, consistent with the previous observation that A36 is present in IEV but absent from CEV. In contrast, anti-A36 antibody produced diffuse staining in vΔA34R-infected cells and did not label DNA-containing particles in the periphery of the cell, indicating a defect in the incorporation of A36 into IEV due to the absence of A34.
FIG. 7.
Distribution of A36 and F13 envelope proteins in A34-deficient virus. BHK-21 cells were infected for 7 h, fixed, permeabilized, and stained with anti-A36 or anti-F13 antibodies. Virion DNA was visualized by Hoechst staining. (A) Colocalization of A36 and virus particles. Cells infected with WR or A34-deficient virus (vΔA34R) were stained with anti-A36 antibody. Merged images resulted from the combination of monochrome images pseudocolored green (Hoechst) and red (anti-A36). Note the coincidence of the A36 protein with some virus particles (presumably IEV and maybe some CEV) in cells infected with WR but not in vΔA34R-infected cells. (B) Colocalization of F13 and virus particles. Cells infected with WR or vΔA34R were stained with anti-F13 antibody. Merged images resulted from combining monochrome images pseudocolored green (Hoechst) and red (anti-A33). Note the colocalization of the F13 protein with viral DNA in cells infected with both WR and ΔA34R.
The presence of F13 in enveloped, cell-associated virus (IEV plus CEV) was also revealed by immunofluorescence. In contrast to the results obtained with B5, A33, and A36, protein F13 staining clearly labeled peripheral virions both in WR- and in vΔA34R-infected cells, indicating that F13 was incorporated into enveloped virus in the absence of A34. These results indicate that targeting of F13 to IEV is not dependent on A34. In contrast, targeting of proteins A33, B5, and A36 to IEV is dependent on the presence of A34.
Envelope proteins in vΔA34R extracellular virus.
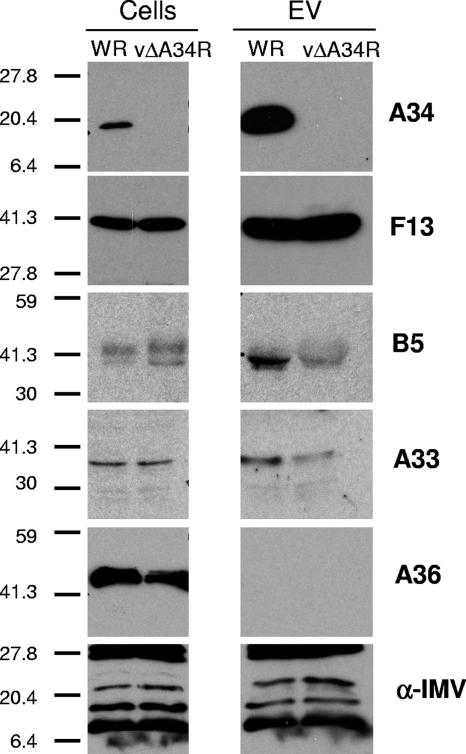
The above results showed an abnormal distribution of envelope proteins in cells infected with the A34-deficient virus. To test whether this resulted in an enveloped virus with altered amounts of proteins in the envelopes of extracellular viruses, extracts of extracellular virus in the medium of WR- or vΔA34R-infected cells were analyzed by Western blotting, using antibodies to different envelope components (Fig. 8). Similar amounts of IMV proteins were present in WR and vΔA34R extracellular virions, as revealed by a polyclonal antiserum to IMV proteins. Also, the lack of contamination from cellular membranes was consistent with the absence of A36 in extracellular virus for both WR and vΔA34R. Interestingly, the amounts of B5 and A33 present in vΔA34R extracellular virus were diminished with respect to those found in WR extracellular virus. In contrast, the amounts of F13 present in extracellular virus were similar for WR and vΔA34R. A previous study (24) reported that EEV envelope proteins were present in both WR and vΔA34R. Careful inspection shows that our analysis is in agreement with the amounts of F13 and B5 detected in vΔA34R extracellular virus in that report (see Fig. 5 in reference 24). Also, the relative amounts of the proteins present in extracellular virus are consistent with the immunofluorescence results obtained and confirm the idea that F13, but not A33 or B5, is incorporated in normal amounts into enveloped virus in the absence of A34.
FIG. 8.
Protein composition of extracellular virus envelope of A34-deficient virus. Extracts from BHK-21 cells infected with WR or vΔA34R virus (cells) or from the respective extracellular virus (EV) were subjected to SDS-PAGE and analyzed by Western blotting with antibodies to envelope proteins or with polyclonal antiserum to IMV proteins (α-IMV). Molecular weight markers are indicated on the left. Note the diminished amounts of glycoproteins A33 and B5 in vΔA34R extracellular virus with respect to those in WR extracellular virus.
DISCUSSION
Vaccinia virus assembly and transmission depend on several virus-encoded proteins. To date, a number of studies have focused on the role of particular proteins or their domains in specific functions carried out by the virus envelope. Of the protein constituents of the virus envelope, proteins F13 and B5 are required for IMV wrapping (1, 47), while proteins A34 and B5 are involved in the retention of CEV on the plasma membrane (4, 12, 21), the induction of actin tails (12, 21, 23, 48), and the disruption of the viral membrane prior to virus entry (15, 19).
We have characterized here the interaction between the A34 and B5 proteins and have shown that the extracellular portions of both proteins are sufficient to mediate their interaction. We have considered that in establishing the interaction, both proteins could be placed in the same or opposite membranes. Significantly, mutations in the extracellular domains of A34 and B5 produce similar comet-shaped plaque phenotypes as a result of increased release of enveloped virus from infected cells (4, 12, 21). We have tested if these mutations prevent the formation of the A34-B5 complex. Deletion of the SCR domains in B5 (12), which is known to increase virus release from the cell, affects the interaction, which is in agreement with the notion that CEV could be retained at the cell surface by A34-B5 interaction between the CEV membrane and the plasma membrane. However, we did not detect differences in the A34-B5 interaction when using an A34 protein containing a K151E mutation (4) that increases CEV release (not shown). Similarly, a P189S mutation in the extracellular domain of B5 that causes enhanced EEV formation (17) did not have a noticeable effect on the A34-B5 interaction. Therefore, we have to conclude that the interaction between A34 and B5 is not the only factor determining CEV retention at the plasma membrane.
Our results indicate that the interaction between A34 and B5 can be mediated not only by the SCR domains but also by the stalk region. It is noteworthy that both portions of the protein contain negatively charged domains (with isoelectric points of 4.49 and 4.41 for SCR domains 2 and 3, respectively, and 3.88 for the stalk region) that could interact with the positively charged ectodomain of A34 (isoelectric point of 9.34). Therefore, it seems likely that electrostatic interactions between the ectodomains of the two proteins contribute to their binding.
We have studied here the consequences of A34 deficiency in the distribution of other envelope proteins and, more specifically, in their presence in enveloped virus forms. The subcellular localization of different envelope proteins in infected or noninfected cells suggests that several proteins cooperate to achieve their destinations in the wrapping compartment during infection (20). Our results reinforce this notion by showing that in the absence of A34, the B5 protein is not targeted properly to enveloped viruses. In addition, it has been shown that the distribution of B5 is modified by coexpression of F13 (14). Interestingly, the B5 protein is known to contain signals for ER export to the Golgi complex and for plasma membrane retrieval (44) and is able to reach the Golgi complex in the absence of other virus proteins (20). Therefore, it is likely that the roles of F13 and A34 in the localization of B5 occur at a Golgi or post-Golgi step. In particular, the function of the A34-B5 interaction might be necessary for the incorporation of B5 into the IEV membranes during virus wrapping rather than for the targeting of B5 to the wrapping compartment.
A34 deficiency also results in the anomalous distribution of A33, although no direct interaction between the two proteins has been demonstrated. Recently, we reported that the incorporation of A33 into IEV is dependent on the expression of B5 (29). Also, Rottger et al. (33) described the redistribution of A33 as a consequence of A34R or B5R deletion. Given the effect of A34 on B5 targeting and that of B5 on A33 targeting, the effect of A34R deletion on the distribution of A33 could be mediated by B5. One possibility to explain the data available is that the three proteins form an A34-B5-A33 complex. However, the inability of us and others (33) (Fig. 2) to coimmunoprecipitate A33 and A34 argues against this hypothesis. One appealing hypothesis that needs further investigation is that B5 forms alternative complexes with either A33 or A34.
One intriguing question is how A34-deficient viruses, which seem to be affected in the distribution of multiple envelope proteins, are able to exit cells efficiently. The fact that virus wrapping, transport, and exit can occur with an altered distribution of B5 is difficult to reconcile with the fact that B5 is required for EEV formation. One possible explanation is that B5 is required in only small amounts for virus wrapping or that it can carry out its function without being massively incorporated into IEV. The possibility that decreased amounts of B5 protein may be sufficient for normal virus wrapping has been proposed before (22). The observation that F13 is efficiently targeted to IEV and EEV in the absence of A34, leading to increased EEV release, even when B5 and A33 are recruited to EEV in smaller amounts, reinforces the idea that F13 is the main requirement for virus wrapping and egress. Our data support the idea that A34, which is not needed for IEV formation or transport, is a key factor for determining the protein composition of the virus envelope, leading to the incorporation of proteins B5 and A33.
At least part of the phenotype of the A34-deficient virus can be explained by reduced amounts of different proteins in the virus envelope. For instance, the decrease of infectivity of EEV and their inability to rupture the envelope before entry can be the direct result of a small amount of B5 in the envelope, since B5 is required for this process. Likewise, the lack of induction of actin tails caused by A34 deficiency could be the result of the altered incorporation of B5, A33, and A36 into vΔA34R IEV.
Acknowledgments
We thank G. L. Smith, G. H. Cohen, R. Eisenberg, and G. Hiller for the generous gift of antibodies and G. L. Smith for vaccinia virus mutant vΔA34R.
This work was supported by contracts QLK2-CT2002-01867 and CT2006-037536 from the European Commission and by grant BFU2005-05124 from Dirección General de Investigación Científica y Técnica, Spain.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Blasco, R., and B. Moss. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J. Virol. 655910-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasco, R., and B. Moss. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell virus spread. J. Virol. 664170-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco, R., and B. Moss. 1995. Selection of recombinant vaccinia viruses on the basis of plaque formation. Gene 158157-162. [DOI] [PubMed] [Google Scholar]
- 4.Blasco, R., J. R. Sisler, and B. Moss. 1993. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J. Virol. 673319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulter, E. A., and G. Appleyard. 1973. Differences between extracellular and intracellular forms of poxvirus and their implications. Prog. Med. Virol. 1686-108. [PubMed] [Google Scholar]
- 6.Brum, L. M., P. C. Turner, H. Devick, M. T. Baquero, and R. W. Moyer. 2003. Plasma membrane localization and fusion inhibitory activity of the cowpox virus serpin SPI-3 require a functional signal sequence and the virus encoded hemagglutinin. Virology 306289-302. [DOI] [PubMed] [Google Scholar]
- 7.Duncan, S. A., and G. L. Smith. 1992. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J. Virol. 661610-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Earl, P., and B. Moss. 1991. Expression of proteins in mammalian cells using vaccinia viral vectors, p. 16.15.1-16.18.10. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Wiley-Interscience, New York, NY.
- 9.Engelstad, M., S. T. Howard, and G. L. Smith. 1992. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology 188801-810. [DOI] [PubMed] [Google Scholar]
- 10.Engelstad, M., and G. L. Smith. 1993. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology 194627-637. [DOI] [PubMed] [Google Scholar]
- 11.Frischknecht, F., V. Moreau, S. Rottger, S. Gonfloni, I. Reckmann, G. Superti-Furga, and M. Way. 1999. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401926-929. [DOI] [PubMed] [Google Scholar]
- 12.Herrera, E., M. M. Lorenzo, R. Blasco, and S. N. Isaacs. 1998. Functional analysis of vaccinia virus B5R protein: essential role in virus envelopment is independent of a large portion of the extracellular domain. J. Virol. 72294-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirt, P., G. Hiller, and R. Wittek. 1986. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J. Virol. 58757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Husain, M., and B. Moss. 2001. Vaccinia virus F13L protein with a conserved phospholipase catalytic motif induces colocalization of the B5R envelope glycoprotein in post-Golgi vesicles. J. Virol. 757528-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain, M., A. S. Weisberg, and B. Moss. 2007. Resistance of a vaccinia virus A34R deletion mutant to spontaneous rupture of the outer membrane of progeny virions on the surface of infected cells. Virology 366424-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaacs, S. N., E. J. Wolffe, L. G. Payne, and B. Moss. 1992. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 667217-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katz, E., B. M. Ward, A. S. Weisberg, and B. Moss. 2003. Mutations in the vaccinia virus A33R and B5R envelope proteins that enhance release of extracellular virions and eliminate formation of actin-containing microvilli without preventing tyrosine phosphorylation of the A36R protein. J. Virol. 7712266-12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katz, E., E. Wolffe, and B. Moss. 2002. Identification of second-site mutations that enhance release and spread of vaccinia virus. J. Virol. 7611637-11644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law, M., G. C. Carter, K. L. Roberts, M. Hollinshead, and G. L. Smith. 2006. Ligand-induced and nonfusogenic dissolution of a viral membrane. Proc. Natl. Acad. Sci. USA 1035989-5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenzo, M. M., I. Galindo, G. Griffiths, and R. Blasco. 2000. Intracellular localization of vaccinia virus extracellular enveloped virus envelope proteins individually expressed using a Semliki Forest virus replicon. J. Virol. 7410535-10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathew, E., C. M. Sanderson, M. Hollinshead, and G. L. Smith. 1998. The extracellular domain of vaccinia virus protein B5R affects plaque phenotype, extracellular enveloped virus release, and intracellular actin tail formation. J. Virol. 722429-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathew, E. C., C. M. Sanderson, R. Hollinshead, M. Hollinshead, R. Grimley, and G. L. Smith. 1999. The effects of targeting the vaccinia virus B5R protein to the endoplasmic reticulum on virus morphogenesis and dissemination. Virology 265131-146. [DOI] [PubMed] [Google Scholar]
- 23.Mathew, E. C., C. M. Sanderson, R. Hollinshead, and G. L. Smith. 2001. A mutational analysis of the vaccinia virus B5R protein. J. Gen. Virol. 821199-1213. [DOI] [PubMed] [Google Scholar]
- 24.McIntosh, A. A., and G. L. Smith. 1996. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J. Virol. 70272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moss, B. 1996. Poxviridae: the viruses and their replication, p. 2637-2671. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 26.Parkinson, J. E., and G. L. Smith. 1994. Vaccinia virus gene A36R encodes a M(r) 43-50 K protein on the surface of extracellular enveloped virus. Virology 204376-390. [DOI] [PubMed] [Google Scholar]
- 27.Payne, L. G. 1992. Characterization of vaccinia virus glycoproteins by monoclonal antibody precipitation. Virology 187251-260. [DOI] [PubMed] [Google Scholar]
- 28.Payne, L. G. 1980. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J. Gen. Virol. 5089-100. [DOI] [PubMed] [Google Scholar]
- 29.Perdiguero, B., and R. Blasco. 2006. Interaction between vaccinia virus extracellular virus envelope A33 and B5 glycoproteins. J. Virol. 808763-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmstrom, F. Frischknecht, M. Zettl, T. Zimmermann, and M. Way. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3992-1000. [DOI] [PubMed] [Google Scholar]
- 31.Roper, R. L., L. G. Payne, and B. Moss. 1996. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J. Virol. 703753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roper, R. L., E. J. Wolffe, A. Weisberg, and B. Moss. 1998. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol. 724192-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rottger, S., F. Frischknecht, I. Reckmann, G. L. Smith, and M. Way. 1999. Interactions between vaccinia virus IEV membrane proteins and their roles in IEV assembly and actin tail formation. J. Virol. 732863-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanderson, C. M., F. Frischknecht, M. Way, M. Hollinshead, and G. L. Smith. 1998. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J. Gen. Virol. 791415-1425. [DOI] [PubMed] [Google Scholar]
- 35.Scaplehorn, N., A. Holmstrom, V. Moreau, F. Frischknecht, I. Reckmann, and M. Way. 2002. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr. Biol. 12740-745. [DOI] [PubMed] [Google Scholar]
- 36.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans-Golgi network. J. Virol. 68130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shida, H. 1986. Variants of vaccinia virus hemagglutinin altered in intracellular transport. Mol. Cell. Biol. 63734-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, G. L., and A. Vanderplasschen. 1998. Extracellular enveloped vaccinia virus. Entry, egress, and evasion. Adv. Exp. Med. Biol. 440395-414. [PubMed] [Google Scholar]
- 39.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 832915-2931. [DOI] [PubMed] [Google Scholar]
- 40.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60163-178. [PubMed] [Google Scholar]
- 41.Turner, P. C., and R. W. Moyer. 2006. The cowpox virus fusion regulator proteins SPI-3 and hemagglutinin interact in infected and uninfected cells. Virology 34788-99. [DOI] [PubMed] [Google Scholar]
- 42.van Eijl, H., M. Hollinshead, G. Rodger, W. H. Zhang, and G. L. Smith. 2002. The vaccinia virus F12L protein is associated with intracellular enveloped virus particles and is required for their egress to the cell surface. J. Gen. Virol. 83195-207. [DOI] [PubMed] [Google Scholar]
- 43.van Eijl, H., M. Hollinshead, and G. L. Smith. 2000. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped virus particles. Virology 27126-36. [DOI] [PubMed] [Google Scholar]
- 44.Ward, B. M., and B. Moss. 2000. Golgi network targeting and plasma membrane internalization signals in vaccinia virus B5R envelope protein. J. Virol. 743771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward, B. M., and B. Moss. 2004. Vaccinia virus A36R membrane protein provides a direct link between intracellular enveloped virions and the microtubule motor kinesin. J. Virol. 782486-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 7511651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolffe, E. J., S. N. Isaacs, and B. Moss. 1993. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J. Virol. 674732-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolffe, E. J., E. Katz, A. Weisberg, and B. Moss. 1997. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J. Virol. 713904-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolffe, E. J., A. S. Weisberg, and B. Moss. 1998. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology 24420-26. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, W. H., D. Wilcock, and G. L. Smith. 2000. Vaccinia virus F12L protein is required for actin tail formation, normal plaque size, and virulence. J. Virol. 7411654-11662. [DOI] [PMC free article] [PubMed] [Google Scholar]