Abstract
Noroviruses (family Caliciviridae) are the major cause of epidemic nonbacterial gastroenteritis in humans, but the mechanism of antibody neutralization is unknown and no structure of an infectious virion has been reported. Murine norovirus (MNV) is the only norovirus that can be grown in tissue culture, studied in an animal model, and reverse engineered via an infectious clone and to which neutralizing antibodies have been isolated. Presented here are the cryoelectron microscopy structures of an MNV virion and the virion in complex with neutralizing Fab fragments. The most striking differences between MNV and previous calicivirus structures are that the protruding domain is lifted off the shell domain by ∼16Å and rotated ∼40° in a clockwise fashion and forms new interactions at the P1 base that create a cagelike structure engulfing the shell domains. Neutralizing Fab fragments cover the outer surface of each copy of the capsid protein P2 domains without causing any apparent conformational changes. These unique features of MNV suggest that at least some caliciviruses undergo a capsid maturation process akin to that observed with other plant and bacterial viruses.
Human noroviruses are responsible for an estimated 23 million cases of and 50,000 hospitalizations due to nonbacterial acute gastroenteritis per year in the United States alone (24). Norovirus particles, as for the virions of other calicivirus genera (vesiviruses, sapoviruses, lagoviruses), are very stable in the environment and highly contagious. Disease symptoms include vomiting, diarrhea, low-grade fever, malaise, and abdominal cramping or pain. Symptoms typically resolve within 48 h, and the infection is usually mild and self-limiting (15).
The calicivirus virion is made up of a single major capsid protein, VP1, that ranges in molecular mass from 56 to 76 kDa (9, 20). A major advance in the study of calicivirus structure was the production of virus-like particles (VLPs) from insect cells infected with a recombinant baculovirus expressing the Norwalk virus (rNV) capsid protein (21). These rNV VLPs are antigenically similar to native virions (17, 21). VLP self-assembly is independent of minor capsid protein VP2, and both the full-length capsid protein of 58 kDa and a smaller 34-kDa cleavage product are detected in infected insect cells (21). The cryo-transmission electron microscopy (cryo-TEM) image reconstructions of the rNV VLP (28), primate calicivirus Pan-1 (27), Parksville VLP (7), Grimsby VLP (7), and the related vesivirus San Miguel virus (7) have been elucidated. Subsequently, the crystallographic structures of rNV VLPs (26) and the San Miguel vesivirus authentic virion (6) were determined. These structures exhibit T=3 icosahedral symmetry, with the particles being composed of a single capsid protein. With architectures very much like that of members of the plant Tombusviridae family (18), the capsid protein has three structural domains; the N terminus (N), shell (S), and “protruding” (P) domains. The N terminus of the rNV VLP capsid protein is buried in the particles, while the C-terminal amino acids are exposed on the VLP surface. The S domain is composed of an eight-stranded β sandwich, has a high degree of amino acid sequence homology among noroviruses, and forms a continuous 150-Å-diameter protein shell. This shell domain is connected by a flexible hinge to the P domain at the C-terminal half of the capsid protein. Similar to the Tombusviridae (18), dimers of the P domains are found at the icosahedral twofold axes (C-C subunit dimers) and encircling the fivefold axes (A-B subunit dimers). Unlike that of the family Tombusviridae, the P domain can be further divided into P1 and P2 subdomains. P1 forms a stem connecting the shell domain to a globular head region (P2). P1 shows moderate sequence diversity among noroviruses, while P2, located at the outermost extreme of the capsid, has a highly variable amino acid sequence. Since a monoclonal antibody (MAb) that binds to P2 was found to block attachment of rNV VLPs, it is thought that P2 is responsible for cellular recognition (34). In addition, the P2 domain also contains the determinants for antigenicity and host specificity.
Since murine norovirus (MNV) represents the first norovirus for which the immune response can be fully explored in an animal model, we have determined the cryo-TEM structures of the MNV T=3 authentic virion in the presence and absence of neutralizing Fab fragments. Unlike the previous structures of caliciviruses, the P domains of MNV twist and rise up off the surface of the shell domains by ∼16Å. In this new orientation, they form an outer shell via interactions among the P1 domains that could represent a form of maturation akin to that of other viral systems (e.g., see references 4, 16, and 35). Importantly, this structure is also observed in the structure of MNV bound with neutralizing antibodies, and no additional conformational changes in the virion are observed at this resolution. Together, these results suggest that MNV undergoes a maturation process that lifts the P domains up off the surface of the capsid shell and that antibody-mediated neutralization is independent of conformational changes and bivalent attachment.
MATERIALS AND METHODS
MNV propagation.
MNV (strain MNV.CW1) was propagated in RAW 264.7 cells as previously described (36). In brief, RAW cells were plated in T175 flasks and allowed to attach overnight. Cells were infected with MNV at a multiplicity of infection of 0.05 for 42 to 48 h before freezing at −80°C. Virus particles were released by three freeze-thaw cycles. The virus lysate was then centrifuged at 10,000 rpm to remove cellular debris. The supernatant was then treated with DNase I (10 μg/ml) and MgCl2 (5 mM, final concentration) and allowed to incubate at room temperature for 1 h. After digestion, 10 mM EDTA and 1% lauryl sarcosine (final concentrations) were added to supernatant. This solution was then centrifuged at 48,000 rpm with a 70 Ti rotor (Beckmann) for 2.5 to 3.0 h. The resulting pellet was resuspended in phosphate-buffered saline (PBS) (∼250 to 300 μl), allowed to rest at 4°C for several hours, applied to a continuous sucrose gradient (7.5 to 45%), and centrifuged at 36,000 rpm for 2.5 to 3 h in a swinging-bucket rotor, SW41 Ti. The virus band was then collected with an ISCO gradient fractionator, and virus-containing fractions were pooled, diluted with PBS, pelleted at 48,000 rpm for 3 h, and resuspended in 250 to 300 μl PBS.
Fab preparation and neutralization assays.
MAb A6.2 was purified with the Affi Gel protein A MAPS II kit from Bio-Rad by using the protocol described in the kit. The purified antibody was stored in PBS containing 0.05% sodium azide, and the concentration was estimated spectrophotometrically by using an extinction coefficient of 1.4 ml/cm · mg. Fab fragments were generated with agarose-linked papain (Sigma catalogue no. P4406). The papain and MAb A6.2 were mixed in a 1:1 (wt/wt) ratio in the presence of 25 mM β-mercaptoethanol and incubated at 37°C for 8 to 10 h in a shaker water bath. After digestion, the papain was removed by centrifugation and the sample was dialyzed against PBS. The Fc fragments were removed by passing the dialyzed sample through a Vivapure protein A mini spin column. Plaque assays were performed as described previously (36).
Cryoelectron microscopy and three-dimensional (3D) image reconstruction.
Data were collected at the National Resource for Automated Molecular Microscopy (NRAMM) facility in San Diego. Small (∼3.5-μl) aliquots of purified MNV (∼1 mg/ml) and MNV-Fab (mixed at a ratio of 900 Fab fragments per MNV virion) samples were placed onto Quantifoil R2/4 grids (Canemco & Marivac, Lakefield, Quebec, Canada) and frozen with a Vitrobot (Fei Company, Hillsboro, OR) apparatus.
Micrographs were recorded on a Philips Tecnai F20 at 120 kV at a magnification of 50,000. Images were acquired digitally with a TVIPS 2k x 2k Peltier-cooled charge-coupled device camera. A total of 281 focal paired images (underfocus values differing by 1 μm) were used in the reconstruction for the MNV-apo particles that were collected with an electron dose of ∼12 e−/Å2 with a resulting pixel size of 1.63Å at the specimen. For the MNV-Fab image reconstruction, a total of 131 focal paired images were collected with an electron dose of ∼10 e−/Å2 with a resulting pixel size of 1.63Å at the specimen. The particles were selected from the micrographs with the program ×3d (11-13). The contrast transfer function was fitted to the power spectrum of each micrograph with the program Bshow (19). The orientation and center of each boxed particle were determined with the program PFT2, which is based on a model-based polar Fourier transform method (2) altered to use both phase and amplitude information in orientation selection (3). 3D reconstructions were computed with a Fourier-Bessel algorithm (14). For an initial model, a 25-Å Norwalk capsid was calculated with the program bsf (BSOFT package) (19) by using the atomic coordinates to calculate a 3.4-Å map (Protein Data Bank identification, 1IHM) (26).
The final 3D reconstructions for the MNV-apo and MNV-Fab particles were computed from 3,392 (selected out of 4,092) and 326 (selected out of 332) particles, respectively, to a resolution cutoff of 10 Å. The actual resolution of the density map was estimated by randomly splitting the image data into two roughly equal sets, computing independent 3D reconstructions, and comparing the structure factors calculated independently for each reconstruction. The coefficient of correlation between the two halves of the data sets was better than 0.5 to ∼12 Å and ∼22Å for the MNV-apo and MNV-Fab structures, respectively.
RESULTS
Structure of an authentic norovirus virion.
The mechanism of norovirus neutralization is unknown. However, the ability to propagate MNV in tissue culture and the availability of neutralizing MAbs enable us, for the first time, to address this topic by structural analysis. To ascertain whether this neutralizing antibody causes any changes in the virion, it was first necessary to determine the structure of MNV in the absence of bound antibody. The SMSV capsid protein is ∼5 kDa larger than that of NV, with most of the additional residues residing in the P2 domain at the connections between the various β strands. Since the MNV capsid protein is of the same length as that of NV and of the same virus genera, the amino acid sequence of the P domain aligns better with NV than with SMSV. Therefore, the atomic structure of rNV VLP was used in the following modeling experiments.
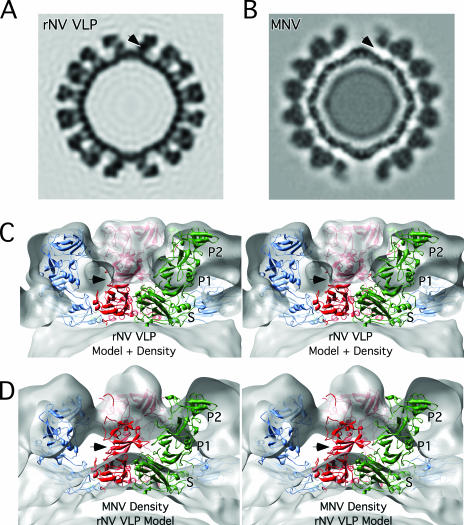
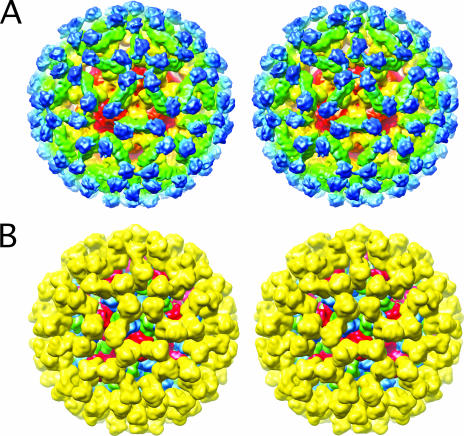
Even from the initial examination of the MNV electron density, it was clear that the structure of MNV was significantly different than that of rNV VLP (Fig. 1). The P domains of rNV VLP rest upon the shell domain and form dimeric protrusions at the icosahedral twofold axes (C-C dimers) and around the icosahedral fivefold axes (A-B dimers). In stark contrast, there is a large gap in electron density between the shell and protruding domains of MNV. When the rNV VLP structure was overlaid onto the MNV density, it was clear that the entire P1 domain lies in the gap between the P and S domains and that the electron density for the MNV protruding domains extended far beyond the outer extents of the P2 domains of the rNV VLP structure. At first, it seemed possible that this difference might be due to an error in the magnification or contrast transfer function corrections. However, this was immediately deemed unlikely since the inner and outer extents of the apparent shell domains in MNV matched that of the rNV VLP model extremely well. In addition, separate data sets confirmed the MNV virion structure (see below). Therefore, it appears that the MNV P domains lift up off the shell (S domains) to form a second proteinaceous layer.
FIG. 1.
Cross sections of rNV VLP and MNV electron density maps. (A) Cross section of the rNV VLP electron density based on the known crystal structure (26) and calculated to a resolution of 12 Å. (B) Cryo-TEM electron density of MNV. (C) Stereo image of the magnified view of the calculated rNV VLP density superimposed with the structures of subunits A, B, and C colored blue, green, and red, respectively. (D) Stereo image of the cryo-TEM electron density of MNV in gray with the superimposed atomic model of rNV VLP for comparison. The black arrow in each panel shows the approximate location of the P1 domain in the rNV VLP structure. Note that this corresponds to a region void of density in MNV.
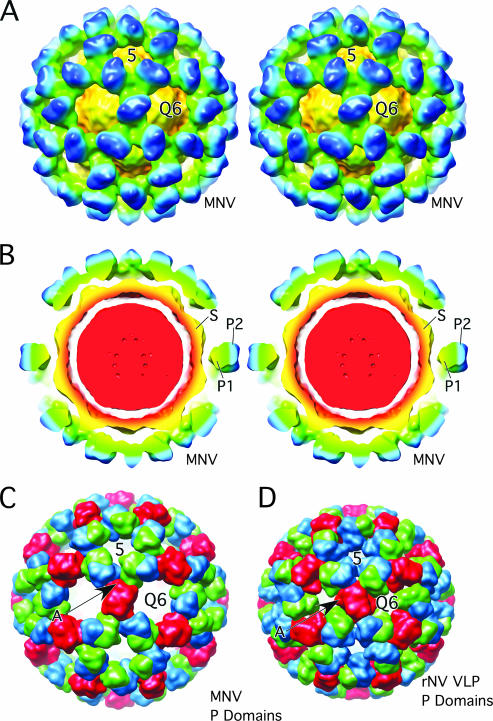
To examine this further, a model for MNV was constructed by modifying the structure of rNV VLP (Fig. 2 and 3). During the fitting process, both possible hands of the cyro-TEM image reconstruction were considered since there is a handedness to the protruding domains themselves. As indicated by arrow A in Fig. 2, the P1 domain sticks out at an angle compared to the top P2 domain. The hand that best accommodated this “twist” was used for modeling. The rNV VLP P domains were raised by ∼16 Å off the surface of the shell domains via extension of the flexible connection between the S and P1 domains. This moved the P1 domain out of the void immediately above the shell surface of MNV (arrow in Fig. 1). The A-B and C-C protruding domain dimers were then rotated by ∼40° in a clockwise fashion in order to fit into the “floating” shell of protruding domains. This translation and rotation increased the outer radius by ∼10% and significantly opened the holes at the quasi-sixfold axes.
FIG. 2.
Comparison of MNV and rNV VLP models. (A) Stereo, depth-cued rendering of MNV electron density going from blue (outside) to yellow (inside). Approximately, the P2 domains are blue, the P1 domains are green/cyan, and the shell domains are yellow/orange. (B) Stereo representation of a cross section of MNV with the same color radial cueing as in panel A. The approximate locations of the S, P1, and P2 domains are noted. The floating nature of the P domains is most notable in this view. (C) Calculated electron density of the P domains of the MNV model made by modifying the structure of rNV VLP. The A, B, and C subunits are blue, green, and red, respectively. To emphasize the differences in the orientation and positions, only the P domains were used in this calculation. (D) Representation of the calculated electron density of the rNV VLP P domains colored in the same manner as in panel C.
FIG. 3.
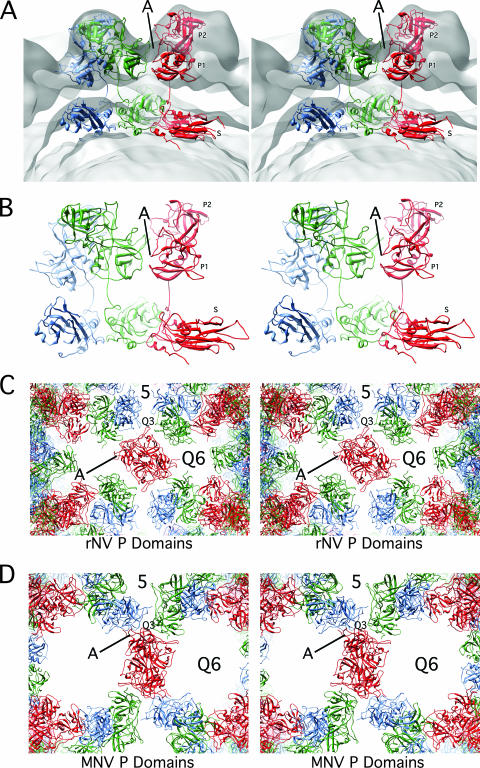
Detailed views of the modified rNV VLP atomic model. (A) Stereo representation of the cross section of the rNV VLP structure as modified to fit into the MNV structure. The orientation shown here differs from that in Fig. 1 in order to demonstrate the new and extensive interactions (denoted by the line marked A) between the P1 domains (red and green P1 interactions). (B) For clarity, this is a slightly expanded view of panel A with only the ribbon diagram displayed. In this view, the new and extensive interactions between the B and C subunits (green and red, respectively) are clearly visible. (C) Stereo view of the ribbon diagrams of the rNV P domains as assembled in the rNV particle. The view here is into the icosahedral twofold axes. Note that there are relatively few interactions among the P domains and that the side protrusion of the P1 domain (denoted by A) points into the space between the P domains. Also note that the spaces at the fivefold and quasi-sixfold (Q6) axes are significantly smaller than MNV in the panel below. (D) Stereo view of MNV in the same orientation. Note that the P1 domain protrusions (denoted by the letter A) now interact at the quasi-threefold axes as also shown in panel B. The lifting and rotation of the P domains open the gaps at the fivefold and quasi-sixfold (icosahedral threefold) axes.
This rotation also established new contacts that are likely required to create an ordered outer shell (area marked A in Fig. 3). This is particularly noticeable in the atomic architecture of the P domains of rNV compared to those of MNV. Panels C and D of Fig. 3 show the arrangement of the rNV and MNV P domains in the T=3 icosahedral lattice. In rNV, there is a loop at the top of the P2 domain that points toward the quasi-threefold axis (Q3), and there are relatively few interactions between the P domain dimers in the icosahedral lattice. In contrast, the relatively flat face of the P1 domains meet at the quasi-threefold axes in the MNV structure and form extensive interactions. It seems logical that without such extensive interactions among the P domains that they would collapse back onto the shell domains or at least be somewhat disordered in this cryo-TEM structure.
Antibody neutralization of MNV.
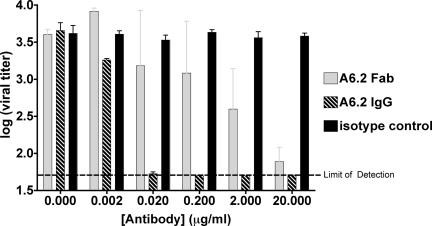
We previously demonstrated that MAb A6.2 neutralizes MNV in plaque-based assays (36). A6.2 does not react with capsid protein in Western assays, suggesting that this particular MAb binds to a 3D epitope rather than a linear peptide sequence. Fab fragments rather than intact antibodies were used in these studies since they are more soluble than immunoglobulins G (IgGs) and do not cause immunoprecipitation via interparticle cross-linking. However, it was uncertain whether intact antibodies are necessary for binding and/or neutralization. To this end, MNV neutralization assays were performed with Fab fragments of A6.2 (Fig. 4). The Fab fragment of A6.2 can neutralize MNV with ∼100 times lower efficacy than the intact antibody. This lower neutralization efficacy of the Fab fragment is consistent with a number of other antibody neutralization studies and can be due to loss of bivalent binding (e.g., see reference 10) or due to loss of intrinsic binding during the papain digestion and purification process. Nevertheless, these results demonstrate that neutralization does not require bivalent binding and that the structure of the Fab/MNV complex therefore represents a neutralized norovirus particle.
FIG. 4.
Neutralization of MNV by MAb and Fab A6.2. Equal amounts of MNV were incubated with MAb 6.2, its Fab fragment, or an isotype control antibody and analyzed in a plaque neutralization assay. The assay was repeated twice, and plaque assays were performed in duplicate to calculate standard deviations.
Structure of MNV complexed with neutralizing Fab fragments.
The cryo-TEM structure of the Fab A6.2/MNV complex is shown in Fig. 5 and 6. It is evident that the P domains in this complex are in the same conformation as that observed in the apo virus structure (Fig. 1 to 3). When this electron density was overlaid with that of the unbound virion, the only significant difference at this resolution was the additional density belonging to the bound Fab (data not shown), demonstrating that neutralization does not induce major conformational changes in the virion. In addition, this is important validation of the apo structure since these data were collected with a different virus preparation on a different microscope at NRAMM. This strongly argues that the gap between the P domains and the S domains is real and that this conformation is immunologically relevant. Finally, since the density of the bound Fab fragment is as strong as the protruding domains, it is also apparent that a Fab is bound to each of the 180 P2 domains.
FIG. 5.
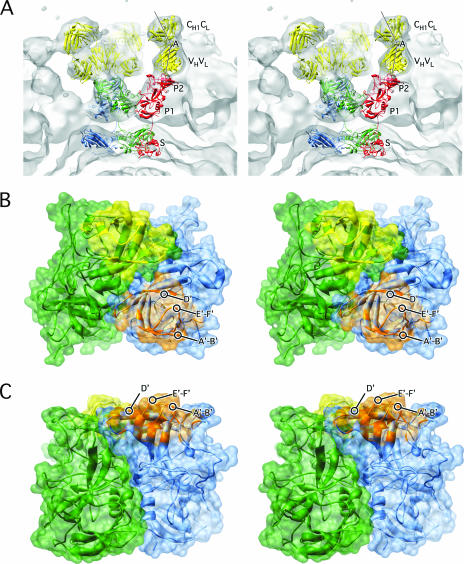
Structure of MNV complexed with neutralizing Fab A6.2. (A) Stereo representation of a depth-cued image reconstruction of MNV complexed with Fab fragments from MAb A6.2. Approximately, the constant domains (CH1·CL) of the bound Fab are blue, the variable domains (VH·VL) are green, the protruding domains are green/yellow, and the shell domains are red/orange. (B) Stereo image of the calculated electron density using the modified rNV VLP model and the structure of Fab1 (5), where Fab1 and subunits A through C are yellow, blue, green, and red, respectively.
FIG. 6.
Details of the Fab/MNV model. (A) Stereo image of the fit of the modified rNV VLP structure and Fab1 into the cryo-TEM density of the MNV/Fab complex. The colors used for the atomic models are the same as in Fig. 5. The lines show the approximate locations of the pseudo-twofold axes in the variable and constant domains. The angle between these axes defines the “elbow angle.” (B) Stereo image showing the areas of the P2 domains that are in contact with the bound antibody. Here, ribbon diagrams are shown in translucent surface renderings of the P2 domains within A-B dimers in green and blue. The areas in contact with the two different Fab fragments are yellow and orange. The viewing angle is from the top of the P domain dimers, and panel C is a side view of the same structure.
To facilitate interpretation of this cryo-TEM structure, the modified NV structure from above and that of a known Fab (Fab1 to HRV14) (5) was placed into the electron density. As shown in Fig. 6, the models fit extremely well. Indeed, the elbow angle (the angle between the variable and constant domains) was clearly visible (labeled A in Fig. 6). The two Fab fragments bound to each P2 domain dimer at a glancing angle and are so close to each other that the densities from the two sets of variable domains tend to merge. The Fab arms of an IgG are remarkably flexible and can bind bivalently to virions (e.g., see reference 30). The angles of the bound Fab fragments shown here suggest that the IgGs could bind with both arms within the A-B or C-C dimer, but with some difficulty. It is still possible that some IgGs could bridge between the two types of dimers, but this would preclude having all of the antibodies bound in a bivalent manner, as was observed in HRV14 (30). There, the Fab fragments bound in such an orientation that little to no conformational change was required to model the intact IgG. In contrast, the Fab fragments of MAb1 to the same epitope on HRV14 bound with a slight twist that prevented bivalent binding of the intact antibody (5). Therefore, while the flexibility of antibodies can theoretically afford bivalent binding in many cases, the orientation of the bound Fab arms determines the likelihood of such binding.
By using the model of the modified rNV VLP structure and the bound Fab, it is possible to map out the approximate epitope regions on the virus (Fig. 6). The bound antibody mainly contacts the protrusion formed by the A′-B′ and the E′-F′ loops that are at the extreme tips of the outer P2 domain. If the antibodies bound anywhere else on the top surface of the P2 domains, it seems unlikely that each copy of capsid protein would be bound with a Fab. Recently, an escape mutation in MNV to this antibody has been identified as L386F (23). The equivalent position of this mutation in NV is just to the carboxyl side of the E′-F′ loop. Since it is a hydrophobic residue and unlikely to be exposed, it seems likely that the mutation affects the structure of the E′-F′ loop indirectly.
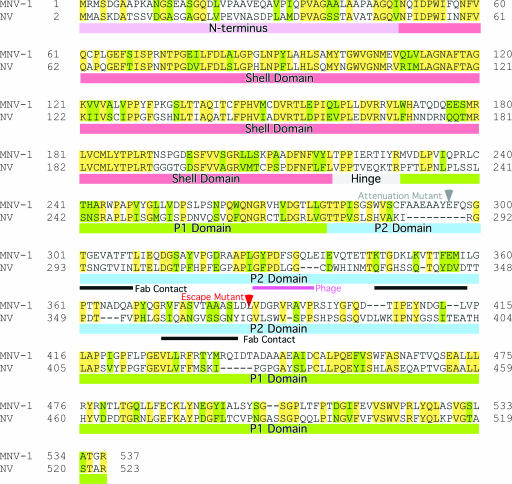
The approximate contact regions are summarized in Fig. 7. For this figure, the antibody contact regions were identified by using the rNV structure fitted into the MNV electron density and BLAST (1) was used for the sequence alignment. As noted in the introduction, the hinge region connects the shell domain with the P domains. Since the MNV P domains are floating above the shell domains while the rNV P domains are not, the conformation of the hinge regions must also differ between these two noroviruses. There is not an obvious reason for this difference based on the sequence alignment in this region. This figure also shows the relatively low homology between NV and MNV in the outermost, P2, domain. Not surprisingly, some of the regions of lowest homology within the P2 domain are around the antibody contact regions. Immediately adjacent to the third antibody contact region is the escape mutation L386F (23). Since this residue is adjacent to another hydrophobic residue, V387, it seems likely that neither side chain is exposed to the antibody and therefore the escape mutation may indirectly affect the conformation of the contact loop immediately preceding this escape mutation. Previous studies of this same antibody used phage display to identify a possible epitope (23). The antibody used in this study was found to bind to a peptide with the sequence GWWEDHGQL but only when constricted to a loop conformation via a disulfide bond. From this sequence, they identified a GXXXXXGQL region in MNV that was suggested to be at least part of the epitope (purple bar in Fig. 7). The modeling studies discussed here suggest that this region is not in contact with the antibody. From the location of this region in rNV and the low homology of MNV to this peptide, it seems possible that this peptide is acting as an epitope mimic. Similar functional mimicry has been observed in other antibodies. For example, a peptide has been found to be an effective functional mimic of carbohydrate antigens (33). Nevertheless, the location of the escape mutation is in good agreement with the observed antibody contacts.
FIG. 7.
NV-versus-MNV-1 sequence alignment. Colored bars beneath the sequences represent the N terminus and the S, hinge, P1, and P2 domains in mauve, red, gray, green, and blue, respectively. The red arrow indicates the location of the known escape mutation (L386F) (23). The black bars indicate the approximate contacts with the antibody, and the purple bar denotes the peptide identified by the phage display that binds to MAb A6.2 (23). The gray arrow denotes the site of the attenuation mutation (36).
DISCUSSION
Structure of an authentic norovirus virion.
The novel finding from this study is that the structure of MNV shows a surprising departure from previously defined calicivirus structures. Two crystal structures (6, 26) and several cryo-TEM structures (7, 32) of caliciviruses have been determined to date. In all of these cases, the P domains lie directly upon the S domains. Notably, the atomic structures all have a long flexible loop that connects the P domain to its S domain. In the case of MNV, the P domains were found to rise up off the S domain sphere to form a second shell around the virion. This major structural difference is unlikely to be due to an artifact of image reconstruction since it was observed from different virus preparations collected at different times and on different instruments. Further, the density is well accounted for by moving the P domains of the NV structure at an obvious flex point. The size, shape, and location of the bound Fab fragment clearly define the outer extent of the viral envelope, further substantiating the validity of the image reconstruction. Furthermore, it is unlikely that this altered structure of MNV is due to damage incurred during purification since these samples are highly infectious and none of the micrographs had notable amounts of damaged or empty particles. Perhaps even more important is the fact that neutralizing Fab fragments are fully capable of binding to the capsid in both enzyme-linked immunosorbent assay and cryo-TEM studies and therefore this structure represents at least the immunologically relevant structure. Finally, the new conformation of the P domain, with its new and extensive interactions among the P1 domains, is a means by which to form a stable secondary shell at the larger radius. Otherwise, it is not clear what would keep the P domains from collapsing back upon the shell domains.
One possible origin for the unique conformation of MNV compared to prior structures is that all of the other norovirus structures to date have been of recombinant VLPs since there has not been a tissue culture system available. It may be that these VLPs do not undergo the same assembly and maturation process as authentic virions. The crystal structure of authentic SMSV has been determined (8) and, like that of the noroviruses, does not have this “floating P domain” shell. However, the P domains of this Vesivirus are in a conformation that is quite distinct from that observed in the crystal structure of rNV VLP and therefore the structure observed here with MNV may be Norovirus genus specific. It is also possible that the crystallization conditions used for SMSV and rNV VLP favored the more compressed state of the viral shell. However, the fact that the cryo-TEM structure of SMSV agrees well with its crystal structure seems to negate this possibility (7).
Since the flexible connecting loop between the P1 and S domains is common to all caliciviruses, it seems possible that this conformational transition is also shared among these viruses. This proposed conformation change may be similar to the structural changes that reoviruses undergo during the various stages of the infection process (31), perhaps priming the virus for receptor interactions and genome release. Alternatively, perhaps the structure of MNV represents a mature state of the virion that is inherently more stable than the compressed form. Bacteriophage HK97 is an excellent example of such a process (11, 35).
An alternative explanation for the difference between the MNV structure and prior calicivirus structures is that this conformation is only observed in MNV. This may be akin to the differences among the rhinovirus serotypes where capsid transitions are easier, for example, in HRV14 than in HRV16 (22, 25). While HRV14 rapidly uncoats in the presence of its receptor (25) and rapidly extrudes the normally buried VP1 and VP4 N termini at room temperature (22), HRV16 only exhibits such dynamics at significantly higher temperatures (22, 25). Similarly, perhaps we are seeing this unique structure in MNV because it is more facile than the other caliciviruses or VLPs. Clearly, structural studies of MNV VLPs are necessary to determine which of these possibilities is correct. Whatever the role of this “enlarged” state of MNV, the availability of an animal model, cell culture system, and infectious clone makes its elucidation a tractable problem for further study.
Neutralization of a norovirus.
The structure of the Fab/MNV complex is another example of antibody-mediated neutralization that does not involve induced conformational changes. The binding pattern is similar to that found with IgG binding to the rabbit hemorrhagic disease virus, which had an apparent binding occupancy of 50% (32). It is possible that an intact A6.2 MAb would be able to bind bivalently to each P2 dimer. However, this would require a great deal of movement at the elbow and hinge regions of the antibody, and hence, interparticle cross-linking may occur before the virion can be substantially saturated with bivalently bound IgGs. More importantly, it is clear that the virus is neutralized by Fab fragments and therefore neutralization is not dependent upon bivalent binding, although bivalent binding would likely increase the apparent affinity (avidity) of the MAb. Furthermore, the lack of antibody-induced conformational changes is also consistent with nearly all of the antibody-virus complex structures studied to date (29) and supports the contention that antibodies mainly tag the virions for recognition by the immune system rather than neutralize via imposition of conformational changes. This leaves the abrogation of cellular attachment as the most likely in vitro mechanism of neutralization for MNV.
Acknowledgments
This work was supported by startup funds from the Donald Danforth Plant Science Center to T.J.S. K.D. was supported by NIH (GM-066087). A portion of the work presented here was conducted at NRAMM, which is supported by NIH through the National Center for Research Resources P41 program (RR17573). C.E.W. was supported by training grant T32-CA09547 and NIH grant U54 AI057160 to the Midwest Center of Excellence for Biodefense and Emerging Infectious Diseases Research. H.W.V. was supported by RO1 AI054483.
We are sincerely grateful to all of the staff of NRAMM for all of their help and support in preparing the samples and collecting the images. Finally, we thank David Belnap for help in installing and executing the various image analysis programs on the Macintosh.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116120-130. [DOI] [PubMed] [Google Scholar]
- 3.Bubeck, D., D. J. Filman, N. Cheng, A. C. Steven, J. M. Hogle, and D. M. Belnap. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-Angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 797745-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canady, M. A., M. Tihova, T. N. Hanzlik, J. E. Johnson, and M. Yeager. 2000. Large conformational changes in the maturation of a simple RNA virus, nudaurelia capensis omega virus (NomegaV). J. Mol. Biol. 299573-584. [DOI] [PubMed] [Google Scholar]
- 5.Che, Z., N. H. Olson, D. Leippe, W.-M. Lee, A. Mosser, R. R. Rueckert, T. S. Baker, and T. J. Smith. 1998. Antibody-mediated neutralization of human rhinovirus 14 explored by means of cryo-electron microscopy and X-ray crystallography of virus-Fab complexes. J. Virol. 724610-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, R., J. D. Neill, M. K. Estes, and B. V. V. Prasad. 2006. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. USA 1038048-8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, R., J. D. Neill, J. S. Noel, A. M. Hutson, R. I. Glass, M. K. Estes, and B. V. V. Prasad. 2004. Inter- and intragenus structural variations in caliciviruses and their functional implications. J. Virol. 786469-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, R., J. D. Neill, and B. V. V. Prasad. 2003. Crystallization and preliminary crystallographic analysis of San Miguel sea lion virus: an animal calicivirus. J. Struct. Biol. 141143-148. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, I. N., and P. R. Lambden. 1997. The molecular biology of caliciviruses. J. Gen. Virol. 78291-301. [DOI] [PubMed] [Google Scholar]
- 10.Colonno, R. J., P. L. Callahan, D. M. Leippe, and R. R. Rueckert. 1989. Inhibition of rhinovirus attachment by neutralizing monoclonal antibodies and their Fab fragments. J. Virol. 6336-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conway, J. F., R. L. Duda, N. Cheng, R. W. Hendrix, and A. C. Steven. 1995. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J. Mol. Biol. 25386-99. [DOI] [PubMed] [Google Scholar]
- 12.Conway, J. F., and J. M. Steven. 1999. Methods for reconstructing density maps of ‘single’ particles from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol. 128106-118. [DOI] [PubMed] [Google Scholar]
- 13.Conway, J. F., B. L. Trus, F. P. Booy, W. W. Newcomb, J. C. Brown, and A. C. Steven. 1993. The effects of radiation damage on the structure of frozen hydrated HSV1 capsids. J. Struct. Biol. 111222-233. [DOI] [PubMed] [Google Scholar]
- 14.Crowther, R. A., L. A. Amos, J. T. Finch, D. J. De Rosier, and A. Klug. 1970. Three dimensional reconstructions of spherical viruses by Fourier synthesis from electron micrographs. Nature 226421-425. [DOI] [PubMed] [Google Scholar]
- 15.Dolin, R., N. R. Blacklow, H. DuPont, S. Formal, R. F. Buscho, J. A. Kasel, R. P. Chames, R. Hornick, and R. M. Chanock. 1971. Transmission of acute infectious nonbacterial gastroenteritis to volunteers by oral administration of stool filtrates. J. Infect. Dis. 123307-312. [DOI] [PubMed] [Google Scholar]
- 16.Dryden, K. A., G. Wang, M. Yeager, M. L. Nibert, K. M. Coombs, D. B. Furlong, B. N. Fields, and T. S. Baker. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 1221023-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green, K. Y., J. F. Lew, X. Jiang, A. Z. Kapikian, and M. K. Estes. 1993. Comparison of the reactivities of baculovirus-expressed recombinant Norwalk virus capsid antigen with those of the native Norwalk virus antigen in serologic assays and some epidemiologic observations. J. Clin. Microbiol. 312185-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison, S. C., A. J. Olson, C. E. Schutt, F. K. Winkler, and G. Bricogne. 1978. Tomato bushy stunt virus at 2.9Å resolution. Nature (London) 276368-373. [DOI] [PubMed] [Google Scholar]
- 19.Heymann, J. B. 2001. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 133156-169. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 19551-61. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, X., W. M. Zhong, T. Farkas, P. W. Huang, N. Wilton, E. Barrett, D. Fulton, R. Morrow, and D. O. Matson. 2002. Baculovirus expression and antigenic characterization of the capsid proteins of three Norwalk-like viruses. Arch. Virol. 147119-130. [DOI] [PubMed] [Google Scholar]
- 22.Katpally, U., and T. J. Smith. 2007. Pocket factors unlikely play a major role in the life cycle of human rhinovirus. J. Virol. 816307-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lochridge, V. P., and M. E. Hardy. 2007. A single amino acid substitution in the P2 domain of VP1 of murine norovirus is sufficient for escape from antibody neutralization. J. Virol. 8112316-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson, N. H., P. R. Kolatkar, M. A. Oliveira, R. H. Cheng, J. M. Greve, A. McClelland, T. S. Baker, and M. G. Rossmann. 1993. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. 90507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasad, B. V. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286287-290. [DOI] [PubMed] [Google Scholar]
- 27.Prasad, B. V., D. O. Matson, and A. W. Smith. 1994. Three-dimensional structure of calicivirus. J. Mol. Biol. 240256-264. [DOI] [PubMed] [Google Scholar]
- 28.Prasad, B. V. V., M. E. Hardy, and M. K. Estes. 2000. Structural studies of recombinant Norwalk capsids. J. Infect. Dis. 181S317-S321. [DOI] [PubMed] [Google Scholar]
- 29.Smith, T. J. 2003. Structural studies on antibody-virus complexes. Adv. Protein Chem. 64409-443. [DOI] [PubMed] [Google Scholar]
- 30.Smith, T. J., N. H. Olson, R. H. Cheng, E. S. Chase, and T. S. Baker. 1993. Structure of a human rhinovirus-bivalently bound antibody complex: implications for virus neutralization and antibody flexibility. Proc. Natl. Acad. Sci. USA 907015-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturzenbecker, L. J., M. Nibert, D. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 612351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thouvenin, E., S. Laurent, M.-F. Madelaine, D. Rasschaert, J.-F. Vautherot, and E. A. Hewat. 1997. Bivalent binding of a neutralizing antibody to a calicivirus involves the torsional flexibility of the antibody hinge. J. Mol. Biol. 270238-246. [DOI] [PubMed] [Google Scholar]
- 33.Vyas, N. K., M. N. Vyas, M. C. Chervenak, D. R. Bundle, B. M. Pinto, and F. A. Quiocho. 2003. Structural basis of peptide-carbohydrate mimicry in an antibody-combining site. Proc. Natl. Acad. Sci. USA 10015023-15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White, L. J., J. M. Ball, M. E. Hardy, T. N. Tanaka, N. Kitamoto, and M. K. Estes. 1996. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J. Virol. 706589-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wikoff, W. R., J. F. Conway, J. Tang, K. K. Lee, L. Gan, N. Cheng, R. L. Duda, R. W. Hendrix, A. C. Steven, and J. E. Johnson. 2006. Time-resolved molecular dynamics of bacteriophage HK97 capsid maturation interpreted by electron cryo-microscopy and X-ray crystallography. J. Struct. Biol. 153300-306. [DOI] [PubMed] [Google Scholar]
- 36.Wobus, C. E., S. M. Karst, L. B. Thackray, K.-O. Chang, S. V. Sosnovtsev, G. Belliot, A. Krug, J. M. Mackensie, K. Y. Green, and H. W. Virgin IV. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2e432. [DOI] [PMC free article] [PubMed] [Google Scholar]