Abstract
Jaagsiekte sheep retrovirus (JSRV) envelope (Env) is an active oncogene responsible for neoplastic transformation in animals and cultured cells. In this study, we used syncytium induction and fluorescence-based cell fusion assays to investigate JSRV Env fusion and its modulation by the cytoplasmic tail (CT). We found that JSRV Env induced syncytia in cells overexpressing the receptor for JSRV and that a low pH was required for this process to occur. Fusion kinetics studies revealed that cell-cell fusion by JSRV Env at neutral pH was poor, taking up to a day, in sharp contrast to fusion at low pH, which peaked within 2 min following a low-pH trigger. Deletion of the C-terminal 7 or 16 amino acids of the JSRV Env CT had no or little effect on fusion, yet additional truncation toward the membrane-spanning domain, resulting in mutants retaining as little as 1 amino acid of the CT, led to progressively increased syncytium formation at neutral pH that was further enhanced by low-pH treatment. Notably, the severely truncated mutants showed elevated levels of surface subunits in culture medium, suggesting that the CT truncations resulted in conformational changes in the ectodomain of Env that impaired surface subunit associations. Taken together, this study reveals for the first time that the fusion activity of the JSRV Env protein is dependent on a low pH and is modulated by the CT, whose truncation overcomes, at least partially, the low-pH requirement for fusion and enhances Env fusion activity and kinetics.
Fusion of enveloped viruses with host cells is a fundamental process essential for viral replication and productive infection. It occurs either at the plasma membrane at a neutral pH or in the intracellular compartments, where an acidic pH is required (17). Retroviral fusion is generally believed to be triggered by specific interactions between the viral envelope (Env) protein and its cognate cell entry receptor, as well as coreceptors in some cases. Upon receptor binding, the Env protein undergoes a series of conformational changes on the surface (SU) and transmembrane (TM) subunits, leading to insertion of the Env fusion peptide into the cell membrane and subsequent formation of trimers of hairpin structures (also referred to as six-helix bundles) that complete virion-cell fusion (18). Recent studies indicate that for five retroviruses, ecotropic murine leukemia virus (ecotropic MLV) (36, 42), avian sarcoma and leukosis virus subgroups A and B (15, 40), mouse mammary tumor virus (48, 51), foamy virus (44), and equine infectious anemia virus (5, 26), fusion occurs in endosomes or other intracellular compartments and requires an acidic pH. Moreover, depending on viral strains and cell types, some retroviruses may use both pH-dependent and -independent pathways for fusion and cell entry (27, 36, 42).
The fusion activity of retroviral Env is controlled by viral and host factors at multiple levels. In addition to the universal cleavage of a precursor Env protein into SU and TM subunits by cellular proteases in the Golgi complex, which converts the Env protein to a metastable state, a portion of the cytoplasmic tail (CT) of many retroviruses is removed by viral protease cleavage after virion assembly (10). For example, the Env proteins of Moloney MLV (MoMLV) (25, 37, 45, 49), gibbon ape leukemia virus (9), and several other simple retroviruses (7, 50) contain a ∼16-amino-acid (aa) segment at the C terminus of the CT, termed the R peptide, that modulates Env fusogenicity. The R peptide must be removed to fully activate the Env fusogenicity after new viral particles are released (21, 22, 25, 49, 62). This mode of regulation by the Env CT is believed to be important for viral fitness, because it potentially prevents syncytium formation in infected cells while activating Env on newly assembled viral particles so that they are competent to infect naive host cells.
Jaagsiekte sheep retrovirus (JSRV) is a simple betaretrovirus that causes a contagious pulmonary adenocarcinoma in sheep (19). Inoculation of concentrated JSRV virions into newborn lambs induces rapid lung tumor formation in as little as 2 weeks (57), showing JSRV is an acutely transforming retrovirus. It is now well established that Env of JSRV is an active oncogene that is responsible for JSRV oncogenesis (2, 8, 12, 13, 33, 35, 47, 60). Of particular interest, the CT of JSRV Env is essential for cell transformation, since replacement of this domain with those of other retroviruses completely abrogates cell transformation (32, 43). The JSRV Env CT is a 44-aa segment, relatively long compared to those of other simple retroviruses, and harbors several sequence motifs and residues known to be crucial for cell transformation and possibly for intracellular trafficking (34). Importantly, the N-terminal 18-aa segment of its CT is predicted to form an amphipathic alpha-helix structure, the hydrophobic side of which has a tendency to interact with the cellular membrane (23). Exactly how this structure might modulate cell transformation and other functions of JSRV Env, including fusion, is currently unknown.
The receptor for JSRV has been identified as hyaluronidase 2 (Hyal2), a glycosylphosphatidylinositol-anchored cell surface protein (47). Hyal2 belongs to the hyaluronidase gene family yet interestingly has a low hyaluronidase activity, which normally degrades the hyaluronic acid present in the extracellular matrix of cells (28, 29, 47, 59). A recent study suggests that the hyaluronidase activity of Hyal2 is not associated with the receptor activity (58). JSRV infects cells derived from many species, including human, ovine, bovine, and canine, but notably not from rodents, and this host range correlates with the receptor activities of Hyal2 isolated from these species (16, 38, 46, 47).
While the transforming properties of JSRV Env have been studied extensively, its fusion characteristics have not been investigated. In this study, we developed syncytium induction and cell-cell fusion assays to use with JSRV Env and performed a series of experiments to test the hypothesis that JSRV Env-mediated fusion may require an acidic pH and may be negatively regulated by its relatively long CT. Our results provide evidence that JSRV entry is pH dependent, and they indicate that JSRV Env employs multiple control mechanisms to regulate its fusion activity.
MATERIALS AND METHODS
Env constructs.
The parental JSRV Env gene derived from the JS7 strain (14) was tagged with a FLAG sequence at the N terminus of SU and was initially cloned into a MoMLV long-terminal-repeat-driven expression vector, pSX2neo (the resulting construct was referred to as pSX2neo-FLAG-Jenv [32]). In the present study, the Env-coding region was subcloned into pCIneo (Sigma, St. Louis, MO), a cytomegalovirus-driven expression vector, in order to achieve a higher level of expression in 293 and 293T cells (the resulting construct was referred to as pCIneo-FLAG-Jenv). To create the JSRV Env truncation mutants, PCR was carried out using a common upstream primer, 5′-GCCTGGTATGATGAAACTGC-3′, paired with the individual unique downstream primers listed below to amplify the TM subunits of truncation mutants: CT608, 5′-GCATAGATCTTCACCTCTCTTTATTTTTTAAAAGC-3′; CT599, 5′-GCATAGATCTCTAAAGATGTTGGTGCTGTA-3′; CT589, 5′-GCATAGATCTCTATTTCATATGCAGCATTTC-3′; CT574, 5′-GCATAGATCTTCACATGCCACGAACGAGGCAAGG-3′; CT572, 5′-GCATAGATCTTCAACGAACGAGGCAAGGAAA-3′; CT571, 5′-GCATAGATCTTCAAACGAGGCAAGGAAATATAAG-3′. The PCR products were either directly cloned into a pCIneo-FLAG-Jenv backbone or were shuttled first into the pSX2neo-FLAG-Jenv vector and subsequently into the pCIneo-FLAG-Jenv vector. The 10A1 amphotropic Env-expressing vector was generated by cloning the 10A1 Env-encoding sequence from pSX2 (39) into the pCIneo vector (referred to as pCIneo-10A1). The vesicular stomatitis virus glycoprotein (VSV-G) expression vector (pMD.G) has been previously described (41).
Retroviruses and cell lines.
The green fluorescent protein (GFP)-encoding retroviral pseudotypes bearing different Env proteins were produced by cotransfection of 293T cells with plasmids encoding individual Env or VSV-G proteins, a packaging plasmid encoding MoMLV Gag-Pol (pCMV-gag-pol-MLV), and a transfer vector encoding GFP (pCMV-GFP-MLV) (the two latter plasmids were kind gifts of François-Loïc Cosset, Lyon, France). The human immunodeficiency virus type 1 (HIV-1) lentiviral vector encoding GFP was generated by cotransfection of 293T cells with pMD.G (41), pCMV-HIVΔ 8.2 (41), and pHIV-eGFP (a gift from Eric Cohen, Institut de Recherches Cliniques de Montréal, Montréal, Canada). Viral pseudotypes were harvested 48 to 72 h posttransfection and were filtered through 0.45-μm-pore-size filters or centrifuged at 2,500 × g to remove the cell debris. The Hyal2-encoding retroviral vector was produced from a PT67/LH2SN producer cell line (47).
293/LH2SN and HTX/LH2SN cells were generated by infection of 293 or HTX cells with PT67/LH2SN viral stocks in the presence of 5 μg/ml polybrene (Sigma), similar to the generation of 3T3/LH2SN as previously described (47). The 293T/GFP cells were generated by infection of 293T cells with the GFP-encoding HIV-1 vectors bearing VSV-G, followed by ring cloning and flow cytometry to confirm GFP expression. All cells were grown in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 5 or 10% fetal bovine serum and were maintained at 37°C in a 10% CO2-air atmosphere at 100% relative humidity.
Antibodies and reagents.
The anti-FLAG monoclonal antibody, EZview Red anti-FLAG affinity gel, and the secondary anti-mouse immunoglobulin G (IgG) coupled to fluorescein isothiocyanate (FITC) or phycoerythrin were purchased from Sigma (St. Louis, MO). Anti-human IgG coupled to FITC was purchased from DAKO Cytometer (Glostrup, Denmark). The JSRV SU (JSU) fusion protein (JSU-hFc) for detection of cell surface receptor levels was produced and purified as described previously (31). The fluorescent dye 5-(and-6)-([{4-chloromethyl} benzoyl] amino)tetramethylrhodamine (CMTMR) and Lipofectamine 2000 were purchased from Invitrogen (Carlsbad, CA). FuGENE HD was purchased from Roche (Indianapolis, IN), and bafilomycin A1 (BafA1) was purchased from Sigma (St. Louis, MO). Pro-Mix 35S cell labeling mix was purchased from Amersham (Amersham, Buckinghamshire, England).
Syncytium induction assay.
Cells seeded in six-well plates were transfected with 2 μg of plasmid DNAs encoding each individual Env protein plus 0.5 μg of plasmid encoding GFP (to monitor transfection efficiency) using the calcium-phosphate method for 293, 293/LH2SN cells or FuGENE HD (Roche, Indianapolis, IN) for the other cell lines. The medium was changed 6 to 9 h following transfection, and syncytium formation was determined 12 to 24 h posttransfection under a light microscope. The numbers of syncytia formed per field and the numbers of nuclei per syncytium were quantified and were averaged from at least four different fields. Cells were subsequently treated with phosphate-buffered saline (PBS)-10 mM HEPES-10 mM morpholineethanesulfonic acid (Sigma) (pH 5.0) for 1 min; syncytium formation was evaluated under a microscope over a period of 5 min to 24 h.
Cell-cell fusion assay.
Effector 293T/GFP cells were transfected with 2 μg of DNA encoding individual Env using Lipofectamine 2000 (Invitrogen). Twelve to twenty-four hours posttransfection, cells were washed with Hanks balanced salt solution (5.4 mM KCl, 0.3 mM Na2HPO4, 0.4 mM KH2PO4, 4.2 mM NaHCO3, 5.6 mM glucose, 137 mM NaCl, pH 7.4) and then detached using PBS containing 5 mM EDTA. Target cells expressing endogenous Hyal2 or overexpressing human Hyal2 were labeled with 3.5 μM CMTMR (Invitrogen) in media without serum at 37°C for 30 min and then were incubated at 37°C for 30 min in fresh medium containing serum, followed by three washes with regular medium. Effector cells and target cells were then plated on 24-well plates at a ratio of 1:1 or 1:3, depending on experiments, and were cocultured at 4°C or 37°C for 1 h. Cells were then treated with PBS-10 mM HEPES and 10 mM morpholineethanesulfonic acid (pH 7.4 or pH 5) for 1 min and were incubated with normal growth medium at 37°C or different temperatures for various periods of time. Cell-cell fusion was examined under a fluorescence microscope (Carl Zeiss, Goettingen, Germany) and photographed. Alternatively, cells were washed with PBS, detached using 0.05% trypsin-0.02% EDTA (Invitrogen), and analyzed by flow cytometry using FACSCalibur (BD Bioscience, Mississauga, Ontario, Canada).
Determination of JSRV Env and Hyal2 surface expression by flow cytometry.
Cells were washed with Hanks balanced salt solution, detached using PBS plus 5 mM EDTA, and resuspended in wash buffer (PBS plus 2% fetal bovine serum). Cells (5 × 105) were incubated on ice with anti-FLAG antibody for 1 h, washed twice with wash buffer, and incubated with anti-mouse IgG coupled to FITC for 293 cells or phycoerythrin for 293T-GFP cells for 1 h. Cells were then washed and analyzed by flow cytometry. Hyal2 expression on the cell surface was analyzed similarly except that cells were incubated with the purified JSU-hFc fusion protein for 4 h on ice, followed by incubation with anti-human IgG Fc antibody coupled to FITC (31).
Retroviral vector transduction and BafA1 treatment.
Cells (105) seeded in 12-well plates on day 0 were pretreated with 10 or 25 nM BafA1 on day 1 for 2 h, followed by transduction with JSRV/MoMLV pseudotypes in the presence of 5 μg/ml polybrene plus 10 or 25 nM BafA1 at 37°C for 6 h. Noninternalized viral particles were inactivated for 1 min with citrate buffer (40 mM sodium citrate, 10 mM KCl, 135 mM NaCl, pH 3.15). Cells were washed three times with PBS and then grown for an additional 2 days before GFP titers were determined by flow cytometry.
Metabolic labeling.
293T cells were transiently transfected with plasmids encoding JSRV Env using the calcium-phosphate coprecipitation method. Twenty-four hours posttransfection, cells were starved in Dulbecco's modified Eagle medium without cysteine and methionine (Invitrogen) for 30 min and pulse-labeled with a 62.5-μCi mixture of cysteine plus methionine (Amersham) for 1 h, followed by chase-labeling for 4 h in the presence of complete growth medium. Cells were then washed with cold PBS once and lysed in lysis buffer (PBS containing 50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) in the presence of 10 μg/ml aprotinin (Sigma), 10 μg/ml leupeptin (Sigma), and 1 mM phenylmethylsulfonyl fluoride (Sigma). Env proteins in cell lysates and in the culture medium were immunoprecipitated using anti-FLAG beads, and the proteins in the immune complexes were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, the dried gels were submitted to autoradiography, and band intensities were quantitated using Quantity One (Bio-Rad, Hercules, CA) image analysis software.
RESULTS
JSRV Env induces syncytium formation at low pH.
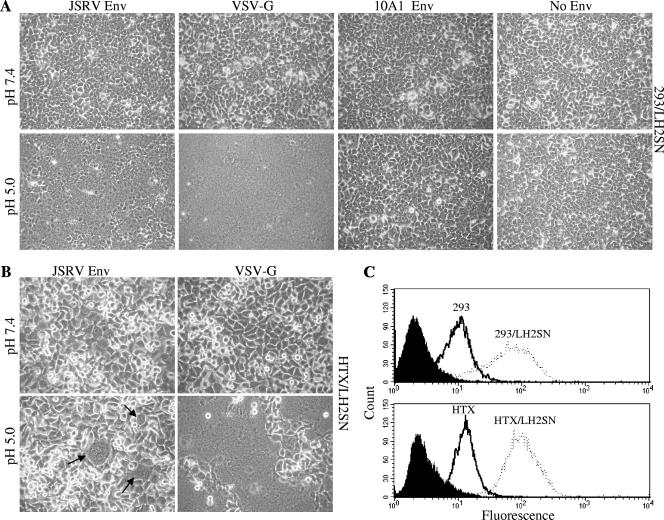
We first sought to determine if JSRV Env induces syncytium formation in cells overexpressing human Hyal2, a standard fusion assay not yet applied to JSRV Env. 293/LH2SN cells were transfected with plasmids encoding JSRV Env, 10A1 amphotropic MLV Env, or VSV-G or left untransfected, along with a plasmid encoding GFP as a transfection control. Twelve to twenty-four hours posttransfection, cells were examined for possible syncytium formation under microscopy and then exposed to either pH 7.4 or pH 5 solution and again examined for syncytia. VSV-G-mediated fusion has been shown to be pH dependent, while negative control amphotropic 10A1 MLV Env does not induce cell fusion at acidic or neutral pH (20, 36). At neutral pH, no syncytium formation was observed in any of the transfected cells (Fig. 1A, upper panels). By contrast, a 1-min incubation at pH 5 resulted in substantial levels of syncytium formation for JSRV Env (Fig. 1A, lower panels). The JSRV Env-induced syncytium formation appeared at approximately 10 min after the low-pH trigger, while that of VSV-G induced massive syncytia within 5 min of low-pH treatment. The difference between results for JSRV Env and for VSV-G was not distinguishable after 30 min of low-pH treatment, since virtually all cells fused for both proteins. These data clearly demonstrate that JSRV Env induces syncytium formation in 293 cells overexpressing human Hyal2 and that a low-pH treatment is required for this process to occur.
FIG. 1.
JSRV Env protein induces syncytium formation at low pH in cells overexpressing human Hyal2. 293/LH2SN (A) or HTX/LH2SN (B) cells were transfected with plasmid DNA encoding JSRV Env, VSV-G, 10A1 MLV Env, or none of these (No Env); 12 to 24 h posttransfection, cells were examined for syncytium induction at neutral pH and phase contrast micrographs of representative fields captured (pH 7.4, top panels). Cells were then treated with a pH 5.0 buffer for 1 min, after which the pH was brought back to neutral and cells were examined for syncytium induction under a light microscope over a period of 5 min to 24 h after the low-pH pulse. Images shown are representative phase-contrast micrographs captured at the 1-h time point (pH 5.0, bottom panels). Arrows indicate syncytia induced by JSRV Env in HTX/LH2SN cells. (C) Hyal2 surface expression in cells used in syncytium induction assays. 293 (solid line), 293/LH2SN (broken line), HTX (solid line), or HTX/LH2SN cells (broken line) were incubated with purified JSRV SU-human IgG Fc fusion protein, followed by an incubation with FITC-labeled anti-human Fc antibody, and were analyzed by flow cytometry. Filled areas represent 293/LH2SN or HTX/LH2SN cells incubated with secondary antibody alone. Representative results of three independent experiments are shown.
We next examined syncytium induction in 293T and 293 cells that express an endogenous level of human Hyal2. These two cell lines are highly susceptible to JSRV vector transduction, although the titers of JSRV in these cells were approximately fivefold lower than that in 293/LH2SN cells (data not shown). While syncytium formation by JSRV Env was occasionally observed in these two cell lines at pH 5 but not at pH 7.4, the efficiency was extremely low, as reflected by the very low numbers of syncytium formation (∼2 per dish) and the small sizes of syncytia (∼4 nuclei per syncytium) (data not shown). To determine if this was a cell-type-specific effect, we next performed syncytium formation assays with human HTX cells and HTX cells overexpressing human Hyal2 (referred to as HTX/LH2SN cells hereafter). Even more surprisingly, only two to three syncytia per field were observed in HTX/LH2SN cells at pH 5 (Fig. 1B, lower panels), and none was detected in HTX cells (data not shown) even though they are as susceptible as 293 cells to JSRV vector transduction (46). In vitro binding assays showed the Hyal2 expression level on the surface of HTX/LH2SN cells was equivalent to that in 293/LH2SN cells (Fig. 1C), suggesting that the low efficiency of syncytium formation by JSRV Env in HTX/LH2SN cells was not due to a difference in Hyal2 expression. While the transfection efficiency of HTX/LH2SN cells (∼60%; Roche FuGene HD method) was not as high as that of 293/LH2SN cells (∼100%), this resulted in a small reduction in JSRV Env expression in HTX/LH2SN cells relative to that in 293/LH2SN cells (data not shown). Furthermore, VSV-G invariably induced large and comparable numbers of syncytia in both HTX/LH2SN (Fig. 1B) and HTX cells (data not show), with efficiencies similar to that in 293/LH2SN cells, suggesting that the low syncytium formation efficiency for JSRV Env in HTX/LH2SN cells was not solely due to its relative low transfection efficiency. Therefore, syncytium formation by JSRV Env not only is determined by the levels of Hyal2 expression in the indicator cells but appears to be affected by the cell lines used in the experiments.
Development of cell-cell fusion assay for measurement of JSRV Env fusogenicity.
To better understand JSRV Env fusogenicity and kinetics, we next developed a cell-cell fusion assay that could measure the redistributions of cytoplasmic contents once cells became fused. To facilitate this assay, we first used lentiviral vector transduction to establish a 293T effector cell line that stably expressed a GFP. One clone that expressed an intermediate level of GFP (referred to as 293T/GFP) was chosen as the effector cell line for the cell-cell fusion assay described below.
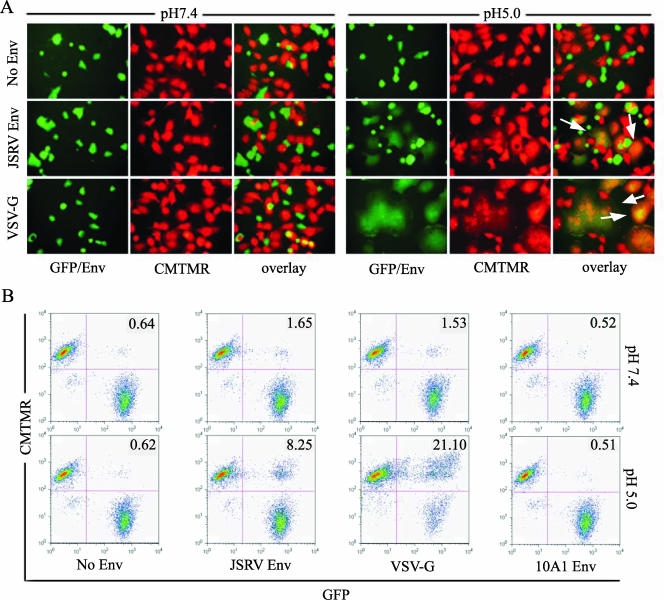
293T/GFP cells were transfected with plasmids encoding individual Env proteins; 24 h posttransfection, cells were cocultured with target HTX/LH2SN cells preloaded with CMTMR, an orange fluorescent dye that is activated by cellular esterases and retained in the cytoplasm. The conditions for CMTMR labeling were optimized so that the orange fluorescent cytoplasmic content signals in the target cells were comparable to that of GFP in the Env-expressing effector cells in order to facilitate fluorescence-activated cell sorting (FACS) analysis. Cocultured cells were subjected to either neutral pH or pH 5.0 for 1 min, rapidly brought to neutral pH, and further incubated at 37°C for 1 h, after which cell-cell fusion was analyzed by fluorescence microscopy and flow cytometry. Similar to the case with syncytium formation (Fig. 1A), no obvious cell-cell fusion was observed at neutral pH for JSRV Env or for VSV-G and 10A1 Env (Fig. 2A, left panels). Interestingly, FACS analysis reproducibly showed low levels of cell-cell fusion at neutral pH for VSV-G and JSRV Env (two- to threefold above that for the no-Env control) (Fig. 2B). Coculturing of effector and target cells at 4°C for 1 or 2 h did not reduce the low levels of fusion activities at neutral pH but rather yielded similar results (data not shown), indicating that the observed fusion at neutral pH was not due to coculturing at 37°C.
FIG. 2.
The JSRV Env protein induces cell-cell fusion at low pH. Effector 293T/GFP (green) cells were transfected with plasmid DNA encoding JSRV Env, VSV-G, or none (No Env); 16 to 24 h posttransfection, cells were counted and cocultured at 37°C for 1 h with HTX/LH2SN target cells prelabeled with CMTMR (red). Cells were then treated with pH 7.4 or pH 5.0 buffers for 1 min and cultured in regular medium for an additional 1 h, and cell-cell fusion was analyzed by fluorescence microscopy (A) or flow cytometry (B). Arrows indicate fused cells (orange-yellow in the overlay images) induced by JSRV Env or VSV-G. Numbers shown in the upper-right quadrant of cytometry profiles represent percentages of fused cells.
Importantly, a pH 5.0 treatment markedly enhanced cell-cell fusion by both JSRV Env and VSV-G but not by 10A1 MLV Env, as evidenced by the appearance of giant cells fluorescing both green and orange that appear yellow in the merged fluorescent images (Fig. 2A, right panels) and by significantly increased percentages of GFP-positive (GFP+)/CMTMR-positive cells examined by FACS (Fig. 2B, bottom panels). These results indicated that JSRV Env fusion is greatly enhanced by low pH.
Cell-cell fusion induced by JSRV Env is Hyal2 specific.
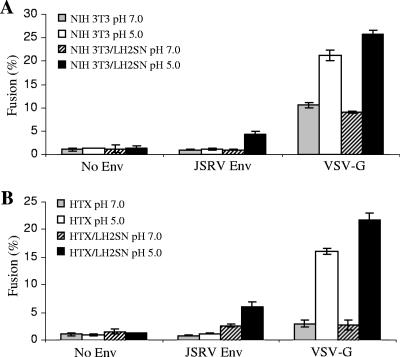
Given the low yet detectable level of cell-cell fusion activity induced by JSRV Env at neutral pH, we next determined if the fusion by JSRV Env was Hyal2 specific. NIH 3T3 (TK−) cells that do not express functional Hyal2 and NIH 3T3/LH2SN (TK−) cells that were engineered to express human Hyal2 (47) were used as target cells. At neutral pH, no apparent cell-cell fusion (<1%) was observed for either cell line (Fig. 3A). A brief exposure to pH 5 resulted in approximately 5% of NIH 3T3/LH2SN cells fusing but did not give 3T3 cell fusion (Fig. 3A). The relatively low percentages of fusion induced by JSRV Env in NIH 3T3/LH2SN cells were in a sharp contrast to that for VSV-G, which induced 20 to 25% cell-cell fusion with both the NIH 3T3 and NIH 3T3/LH2SN cell lines (Fig. 3A).
FIG. 3.
Cell-cell fusion induced by the JSRV Env protein is receptor dependent and requires an overexpression of Hyal2. Cell-cell fusion was performed as described in the legend to Fig. 2, except that NIH 3T3 and NIH 3T3/LH2SN (A) or HTX and HTX/LH2SN (B) cells were used as target cells, respectively. Percentages of CMTMR-GFP double-positive cells, indicative of fusion, were determined by flow cytometry using the FlowJo program. Values are the means of results from representative experiments performed in duplicate. These experiments were repeated three or four times, with similar results.
We next performed the assay using HTX and HTX/LH2SN cells as target cells, in parallel with that using NIH 3T3 and NIH 3T3/LH2SN cells. To ensure comparability, exactly the same Env-transfected 293T/GFP dishes were split and used for coculturing with these four different target cell lines. Cell-cell fusion was detected for JSRV Env in HTX/LH2SN cells, but not in HTX cells, at pH 5 (Fig. 3B), consistent with the results of the syncytium formation assay (Fig. 1B). In contrast, VSV-G induced substantial and comparable levels of cell-cell fusion in both cell lines at low pH but not at neutral pH (Fig. 3B). These results for VSV-G were apparently different from what was observed in NIH 3T3 and NIH 3T3/LH2SN cells (Fig. 3A), suggesting that the VSV-G-induced fusion at neutral pH in NIH 3T3 cells was cell type specific. Importantly, these data confirmed that an overexpression of Hyal2 was required for JSRV Env fusion in HTX cells.
Progressive truncation of JSRV Env CT renders syncytium formation at neutral pH, which is further enhanced by low-pH treatment.
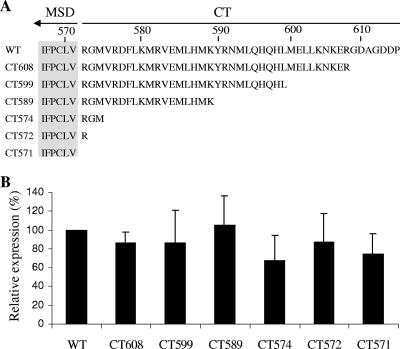
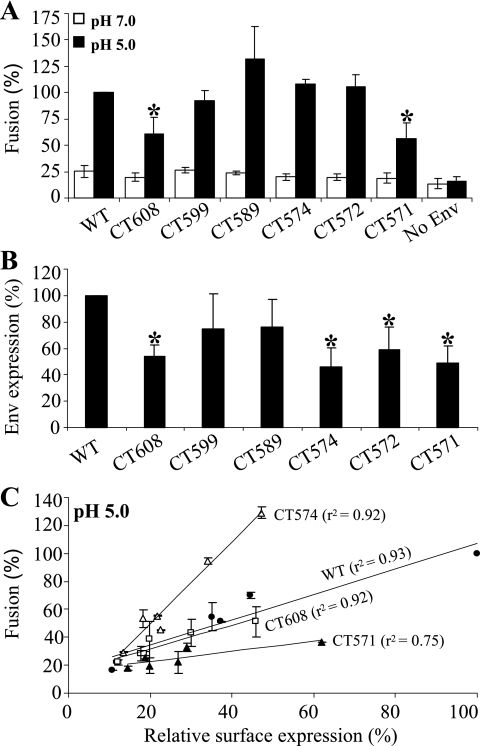
We next investigated a possible role of the CT of JSRV Env in fusion using the newly developed syncytium formation assay. A series of mutants truncated at the C terminus of JSRV Env was created, namely, CT608, CT599, CT589, CT574, CT572, and CT571, according to the position of the terminal amino acid (Fig. 4A). All mutants were tagged with a FLAG sequence at the N terminus of SU (31), and their surface expression levels were determined by an anti-FLAG antibody using flow cytometry. As shown in Fig. 4B, all of these mutants were efficiently expressed on the surfaces of 293/LH2SN cells, the cell line used for syncytium formation, and expression was roughly comparable among the mutants and the wild type.
FIG. 4.
CT sequences and surface expressions of JSRV Env wild type and truncation mutants. (A) CT sequences of JSRV Env wild type (WT) and truncation mutants. Amino acid numbering of the JSRV Env WT is according to that of the JS7 strain (14), and truncation mutants were named according to the positions of their terminal amino acids. Note that all constructs were tagged with a FLAG tag sequence at the N terminus of SU. (B) Env surface expression in 293/LH2SN cells. Cells transfected with plasmids encoding the JSRV Env WT or truncation mutants were incubated with an anti-FLAG antibody and examined for Env expression by flow cytometry. The fluorescence intensities (geometric means) of truncation mutants were normalized relative to that of the WT (%). Values are the means of results from four independent experiments ± standard deviations.
The syncytium-forming activities of these truncation mutants were then assessed in 293/LH2SN cells. At neutral pH, CT608 and CT599 showed no apparent syncytium formation, similar to that of the wild type (Table 1; also data not shown). Intriguingly, mutants with further truncation toward the membrane spanning domain (MSD) of JSRV Env exhibited progressively increased syncytium formation at neutral pH (Table 1; also data not shown). Yet the complete truncation of the CT in mutant CT571 resulted in a background level of fusion similar to that of the wild type (Table 1). Mutants CT574 and CT572, in particular, induced approximately five- to eightfold more syncytia (Table 1), of larger size, than wild-type Env (data not shown). A pH 5 treatment substantially enhanced the syncytium formation of all Env constructs, including CT571, whose fusion activity at low pH was apparently lower than that of wild-type Env and other truncation mutants (Table 1). Again, the two severe truncation mutants, CT574 and CT572, exhibited faster kinetics within the first 10 min of pH 5 treatment than wild-type Env and other mutants (Table 1). Taken together, these results indicate that progressive truncation of JSRV Env renders syncytium formation at neutral pH, which is further enhanced by a low-pH treatment.
TABLE 1.
Syncytium induction by JSRV Env and truncation mutantsa
| Env protein | Syncytium formation (per field)
|
||
|---|---|---|---|
| pH 7.4b | pH 5.0c
|
||
| 10 min | 15 min | ||
| None | 0.5 ± 0.6 | ||
| WT | 1.5 ± 0.6 | + | +++ |
| CT608 | 1.3 ± 0.5 | + | +++ |
| CT599 | 2.8 ± 1.2 | ++ | ++++ |
| CT589 | 7.3 ± 1.0 | +++ | +++++ |
| CT574 | 11.5 ± 1.9 | ++++ | +++++ |
| CT572 | 13.8 ± 3.1 | ++++ | +++++ |
| CT571 | 1.5 ± 0.6 | ± | ++ |
| VSV-G | 3.0 ± 1.1 | +++++ | +++++ |
293/LH2SN cells transfected with plasmids encoding the JSRV Env wild type (WT), truncation mutants, or VSV-G were evaluated for syncytium induction at pHs 7.4 and 5. Transfection efficiencies were similar for all constructs as determined by the GFP signals encoded by a cotransfected plasmid.
Number of syncytia per field at pH 7.4; averaged results from at least four fields; syncytia with four nuclei or greater were counted.
Percentage of syncytium formation per field 10 or 15 min after a pH-5.0 treatment. +, 10 to 20%; ++, 20 to 40%; +++, 40 to 60%; ++++, 60 to 90%; +++++, >90%. Results are from a representative experiment. Experiments were repeated at least four times, with similar results.
Cell-cell fusion activities of JSRV Env truncation mutants.
The cell-cell fusion assay allowed us to quantitatively measure the fusion activities of JSRV Env truncation mutants. At neutral pH, low levels of cell-cell fusion activities were detected for all mutants, with no significant differences observed among these mutants and the wild type (Fig. 5A). A pH 5 treatment resulted in substantially increased cell-cell fusion activities for all those mutants, including CT608 and CT571, whose fusion activities were low compared to those of other mutants (Fig. 5A). Under fluorescence microscopy, significant numbers of yellow GFP and CMTMR double-positive cells were detected at pH 5 (data not shown). While it was difficult to accurately quantify fused cells using fluorescence microscopy, it was noticeable that the sizes of cell-cell fusion induced by the severe truncation mutants, in particular CT574 and CT572, were consistently greater than those of the wild type and CT571 (data not shown). These results were in agreement with those of syncytium formation assays (Table 1).
FIG. 5.
Cell-cell fusion activities of JSRV Env CT truncation mutants. Cell-cell fusion assays were performed as described in the legend to Fig. 2, except that different JSRV Env CT truncation mutants were used for transfection of 293T/GFP cells. (A) Relative fusion efficiency measured by flow cytometry. Cell-cell fusion was performed as described for Fig. 2. The fusion activities of CT truncation mutants were compared to that of wild-type (WT) Env at pH 5 (which was set to 100%). Results are averages from three independent experiments performed in duplicate ± standard deviations. An asterisk represents a P value of <0.05, indicating that the difference between the truncation of interest and the WT was statistically significant. (B) Relative Env surface expression of JSRV Env CT truncation mutants in 293T/GFP cells. A portion of transfected 293T/GFP cells was incubated with an anti-FLAG antibody to determine the Env surface expression by flow cytometry. The average fluorescence intensities per cell (geometric means) of truncation mutants were normalized to that of the WT (%); values are averages for four independent experiments ± standard deviations. An asterisk indicates a P value of <0.05. (C) Correlations between Env surface expression and cell-cell fusion activity at low pH. 293T/GFP cells were transfected with various amounts of plasmid DNA encoding JSRV Env WT, CT608, CT574, or CT571, ranging from 0.5 to 4 μg. Env surface expression and cell-cell fusion activity at pH 5 were determined by flow cytometry. For analysis, the highest surface expression of the JSRV Env WT and its corresponding fusion activity were set to 100% in the x axis and the y axis, respectively, and were used to normalize the other data. Values are averages for two independent experiments ± standard deviations; the r2 value was calculated for each experiment and was averaged.
The surface expression of JSRV Env truncation mutants on 293T/GFP cells ranged from 50 to 75% of the level seen for wild-type Env, as judged in terms of mean fluorescence intensity per cell (Fig. 5B). The low level of surface expression detected was particularly apparent for CT608, CT574, CT572, and CT571. To address this issue, we transfected 293T/GFP cells with different amounts of plasmid DNA encoding the JSRV Env wild type or some of these mutants, ranging from 0.5 to 4 μg, and found that the more the Env protein was expressed, the higher the level of fusion activity detected (Fig. 5C). Importantly, despite similarly low levels of surface expression, mutant CT574 exhibited an overall two- to threefold-higher level of fusion activity than wild-type Env, while the CT608 fusion activity was similar to that of the wild type and CT571 had an approximately twofold-reduced fusion activity compared to that of the wild type (Fig. 5C). A graph of the relative surface expression versus relative fusion activity appeared to be linear, with a squared correlation coefficient (r2) up to >0.9 for most of the constructs tested (Fig. 5C). The correlation clearly demonstrated that the fusion activity of JSRV Env was linearly dependent on the surface expression and indicated that severe truncation of JSRV Env CT did enhance its cell-cell fusion activity at low pH.
Fusion kinetics of JSRV Env at neutral pH is slow and is enhanced by CT truncations.
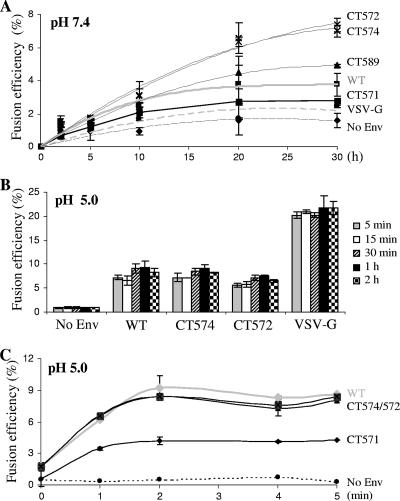
Noting that some of the JSRV Env CT truncations, in particular those proximal to the MSD, resulted in apparent syncytium formation at neutral pH (Table 1) but failed to show enhanced cytoplasmic content transfer relative to that of the wild type (Fig. 5A), we investigated whether this apparent discordance was due to experimental conditions used in our cell-cell fusion assays. The period of coculture was expanded from the original 1-h period to 2, 5, 10, 20, or 30 h. While an extension of coculture to 2 or 5 h had no significant effect on the fusion activities of constructs tested at neutral pH (Fig. 6A; also data not shown), prolonged coculture revealed that after the initial few hours, the truncation mutants CT589, CT574, and CT572 induced more rapid and extensive fusion that did not peak even by 30 h. This was in spite of the fact the surface expression levels of these constructs were 25 to 50% lower than that of the wild type (Fig. 5B). By contrast, fusion by VSV-G and wild-type JSRV Env peaked by 20 h at levels that were about half that of the two most active truncation mutants. These results were confirmed by fluorescence microscopy, where a substantial proportion of GFP+/CMTMR-positive double-positive cells were observed, with sizes of fused cells increasing over time (data not shown). Collectively, these results demonstrate that progressive truncation of the JSRV Env CT enhanced the cell-cell fusion activity of JSRV Env at neutral pH, although a prolonged coculture was required.
FIG. 6.
Fusion kinetics of JSRV Env and truncation mutants. Cell-cell fusion assays were performed as described in the legend to Fig. 2, except that different JSRV Env truncation mutants were used for transfection of 293T/GFP cells and that different periods of recovery time were applied to cocultured cells after a pH 7.4 or pH 5 pulse. (A) Fusion kinetics at neutral pH. Following 1-min treatment with a pH 7.4 solution, cocultured cells were incubated at 37°C for 2, 5, 10, 20, or 30 h, and cell-cell fusion activity was measured for each construct. (B and C) Fusion kinetics at low pH. Following a 1-min treatment with a pH 5 solution, cocultured cells were incubated with regular medium at 37°C for the indicated time periods, and fusion activity was determined by flow cytometry. Results are from a representative experiment performed in duplicate; experiments were repeated at least three times with similar results. Note that the Env surface expression of CT574, CT572, and CT571 in these experiments was approximately 25 to 50% lower than that of the WT (similar to the results shown in Fig. 5B) and that fusion activities presented here were not normalized by their expression levels.
Kinetics of JSRV Env-mediated fusion at low pH is rapid.
The fusion kinetics of JSRV wild-type Env and representative truncation mutants were also determined at low pH. In this case, the effector and target cells were cocultured at 37°C for 1 h and were subjected to pH 5 for 1 min as in the standard assay described above, and cell-cell fusion was analyzed following various periods of neutralization or recovery time, i.e., 5 min, 15 min, 30 min, 1 h, and 2 h. Surprisingly, even within 5 min of pH 5 treatment, fusion reached peak levels, with only small changes afterwards for all constructs (Fig. 6B). These results suggested that fusion by JSRV Env and truncation mutants at pH 5 occurred rapidly and was almost complete within 5 min of the pH 5 trigger.
We next shortened the recovery period to 0, 1, 3, or 4 min and determined the effects on the fusion kinetics of JSRV Env and truncation mutants. Surprisingly, again, a pH 5 treatment alone, even in the absence of a recovery time (time point 1 in Fig. 6C), resulted in approximately 80% of the maximal fusion activities for all of the JSRV Env proteins tested, including CT571, which showed the lowest fusion activity at all time points examined (Fig. 6C). An additional 1-min recovery incubation at 37°C (time point 2 in Fig. 6C) substantially increased their fusion activities to peak levels, yet to comparable extents for all constructs except for CT571, which exhibited a much smaller increase than the others (Fig. 6C). Of note, here also, despite their overall low levels of surface expression compared to that of wild-type Env, the fusion activities and kinetics of CT574 and CT572 were comparable to that of JSRV wild-type Env (Fig. 6C). These results, together with those shown in Fig. 6B, strongly suggested that cell-cell fusion induced by JSRV Env and truncation mutants at low pH occurred very rapidly, almost within the first 2 min after a pH 5 trigger. For comparison, fusion by VSV-G peaked at 1 min (∼37% in fusion activity), followed by a decline at 2 min (∼35%) and 4 min (∼24%), respectively, and with a plateau at 5 min (23%) (data not shown).
BafA1 inhibits transduction by JSRV Env-coated pseudovirions.
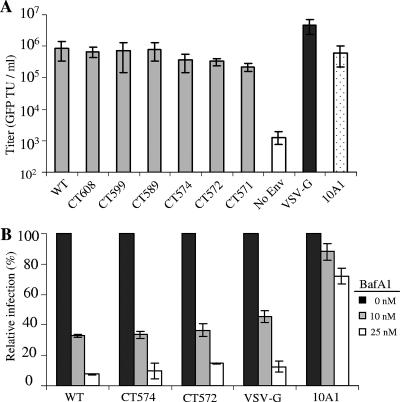
The ability of JSRV Env truncation mutants to induce syncytium formation and to enhance cell-cell fusion at neutral pH prompted us to examine whether infection was affected by the endosomal proton pump inhibitor BafA1, an assay commonly used to determine the pH dependence of virus entry. MoMLV pseudovirions bearing wild-type JSRV Env or individual truncation mutants were titered on 3T3/LH2SN cells by quantifying the GFP-transducing units per ml (Fig. 7A). The infectivities of truncation pseudotypes and the wild type were comparable (Fig. 7A).
FIG. 7.
MoMLV pseudovirions bearing the JSRV Env wild type (WT) and CT truncations are sensitive to BafA1 treatment. (A) NIH 3T3/LH2SN cells were infected with GFP-encoding MoMLV pseudovirions bearing JSRV Env WT or truncation mutants, and percentages of GFP+ cells were determined by flow cytometry 48 h postinfection. Titers are expressed as GFP transducing units (TU) per ml. (B) NIH 3T3/LH2SN cells were pretreated with indicated concentrations of BafA1 for 2 h and infected by MoMLV pseudovirions bearing different Env (multiplicity of infection = 0.1 to 0.5) in the presence of the drug for 6 h. Noninternalized viral particles were inactivated by citrate buffer at pH 3.15 for 1 min, and percentages of GFP positive cells were determined 48 h postinfection by flow cytometry. Values are percentages of transduction relative to that of untreated cells and are the averages of two independent experiments ± standard deviations.
BafA1 strongly inhibited transduction by the wild-type JSRV pseudotypes (Fig. 7B), consistent with the pH-dependent fusion by JSRV Env observed above and with the results we report in the accompanying article (4). Interestingly, transduction by the CT574 and CT572 pseudotypes was also strongly, and almost equally, inhibited by the BafA1 treatments, either at a 10 nM or 25 nM concentration of BafA1 (Fig. 7B) and at different multiplicities of infection (0.1 to 0.5) (data not shown). Similar results were also obtained using pseudovirions transducing alkaline phosphatase (data not shown). To explore if BafA1 might have any effect on the entry kinetics of JSRV pseudovirions bearing the wild-type or mutant Env protein, NIH 3T3/LH2SN cells were prebound by pseudovirions at 4°C for 2 h, followed by an incubation at 37°C for 4 to 6 h in the presence of 25 nM BafA1 added at 0, 1, 2, and 3 h following the temperature switch. Again, the extents of inhibition by BafA1 for the wild type and these truncation mutants at every time point were similar (data not shown). Taken together, we concluded that while progressive truncation of JSRV Env enhanced cell-cell fusions at neutral pH, the cell entry by pseudovirions bearing these truncations was still predominantly pH dependent.
Secretion of JSRV SU into culture medium is enhanced by CT truncations.
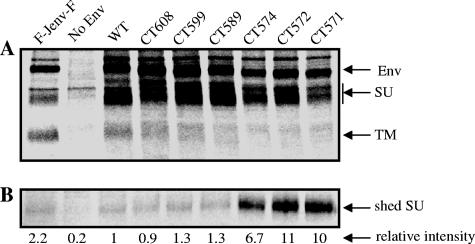
As an initial effort to understand possible mechanisms by which truncation of JSRV Env CT enhances its fusogenicity, we examined the synthesis, processing, and release of SU into the culture medium in metabolic labeling experiments. 293T cells transiently transfected with plasmids encoding individual Env proteins were pulse-labeled with [35S]Met/Cys for 1 h, and then the label was chased for 4 h. Cell lysates and culture medium were separately subjected to immunoprecipitation with an anti-FLAG antibody that recognizes the FLAG sequence tagged at the N terminus of SU, followed by SDS-polyacrylamide gel electrophoresis and autoradiography. All of the JSRV Env truncation mutants were efficiently synthesized and processed, with efficiencies approximately similar to that of the wild type (Fig. 8A). Strikingly, release of SU into culture medium was markedly greater for CT574, CT572, and CT571, by almost 10-fold compared to that of the wild type and other mutants (Fig. 8B). Noticeably, the associations between the SU and TM subunits of these three truncation mutants decreased relative to those of the wild type and other mutants, although it was difficult to quantify these differences because of overall low intensities of TM (Fig. 8A). Nonetheless, these results demonstrated that secretion of JSRV SU was enhanced by the CT truncations, possibly resulting from conformational changes of the ectodomain of TM.
FIG. 8.
Severe truncation of the JSRV Env CT results in an increased release of SU into the culture medium. 293T cells transfected with plasmid DNA encoding the JSRV Env wild type (WT) or CT truncation mutants were pulse-labeled with [35S]Met/Cys for 1 h and chased for 4 h. Cell lysates and culture medium were harvested and immunoprecipitated using anti-FLAG beads. The immunoprecipitated products were resolved by SDS-PAGE, and analyzed by autoradiography. (A) Synthesis and processing of JSRV Env proteins in cell lysates. Positions of radiolabeled JSRV Env precursor (Env), SU, and TM that were recovered in the immune complexes precipitated by antibody against the FLAG tag on SU are indicated by arrows. An unrelated JSRV Env mutant having a FLAG sequence at both the N terminus of SU and the C terminus of TM (first lane, F-Jenv-F) was also included in these experiments in order to indicate the positions of TM. (B) Release of JSRV SU in the wild type and truncation mutants into cell culture medium. The radiolabeled SU recovered in the immune complexes from each culture medium sample was quantified from densitometric scans of the autoradiographs using Quantity One software. The band intensity of SU recovered from the WT sample was set to 1, and the relative intensity of each CT mutant was calculated as the ratio of the intensity of the mutant SU band to that of the WT sample. Results shown are the averages for two independent experiments. No Env, samples from parental, nontransfected 293T cells.
DISCUSSION
We and others have previously attempted to investigate JSRV Env-mediated fusion with little success, largely because of insufficient levels of Hyal2 expression in the target cells and low transfection efficiencies of effector cells used in these assays. In the present study, we overcame these problems by generating the 293/LH2SN and HTX/LH2SN cell lines that overexpress human Hyal2. Using these cell lines, we developed an effective syncytium formation assay and a fluorescence-based cell-cell fusion assay that quantifies cytoplasmic content transfer, and we employed them to investigate the fusogenicity of JSRV Env. We demonstrated that rapid activation of JSRV Env fusion activity requires a low-pH treatment and is Hyal2 specific (Fig. 1, 2, and 3; Table 1).
In this study, we discovered that the CT of JSRV Env modulates its fusion activity and that the fusion-regulating region within its CT appears to differ in location from the fusion-modulating R peptide found in many other retroviral Env CTs (7, 9, 25, 37, 45, 49, 50). Our results strongly argue that the N-terminal and central regions of the CT (aa 572 to 599) of JSRV Env, rather than its C terminus (aa 600 to 615), are critical for fusion modulation. Truncation of the last 16 aa of JSRV Env (CT599) had no significant effect on syncytium formation and cytoplasmic transfer activity (Table 1; Fig. 4 and 5). Further truncations to within one residue of the MSD, e.g., CT589, CT574, and CT572, progressively enhanced fusion (Table 1; Fig. 4, 5, and 6). In contrast, others have shown that truncation of the CT of ecotropic MoMLV (25, 37, 56) and Mason-Pfizer monkey virus (6, 54) to within one residue of the MSD does not increase fusion, and fusion is not enhanced by progressively truncating its CT to lengths shorter than the natural R peptide cleavage site. We noticed also that the tailless CT571 exhibited a reduced fusion activity, which was possibly due to a disruption of the Env membrane structure resulting from the loss of an arginine (R572) residue adjacent to the MSD. It has been previously shown that the positively charged amino acids located either in the membrane-proximal CT or within the MSD of some enveloped viruses can influence Env fusion activities (3, 30, 52).
More intriguingly, several JSRV Env CT truncations, in particular those proximal to the MSD (e.g., CT574 and CT572), induced syncytia (Table 1; Fig. 5C) and enhanced cytoplasmic transfer at neutral pH (Fig. 6A), although the fusion activity at neutral pH required a much longer incubation period than did fusion at an acidic pH (Fig. 6B and C). Hence, progressive truncation of the JSRV Env CT not only enhanced Env-mediated cell-cell fusion but also appeared to render it less dependent on low pH. This unexpected finding raised the possibility that low pH might be the trigger that relieves JSRV Env from the fusion modulation imposed by this CT region. That is, exposure to low pH might be providing the same function for JSRV Env as viral protease cleavage of the C-terminal R peptide does for many other retroviral Env proteins.
This possibility predicted that the CT truncations should render virus infection less dependent on low pH. To test this concept, we examined the effect of the proton pump inhibitor BafA1 on infection of MoMLV pseudovirions bearing these truncated JSRV Env proteins. Interestingly, these viruses were still as sensitive to BafA1 treatments as virus containing wild-type Env (Fig. 7B). Two interpretations occur to us: the first is that the modulation of fusion by the CT is not relieved by an acidic pH; the second is that a low pH relieves the modulation of fusion by the CT but an acidic pH is also required to trigger the initial conformation changes that expose the fusion peptide. We favor the latter case as the reason because this possibility seems most in line with the observation that a low pH rapidly triggers syncytium formation and cell-cell fusion by JSRV Env (Table 1; Fig. 1, 2, and 6B and C) and that the syncytium formation and cytoplasmic transfer induced by the JSRV truncation mutants at neutral pH were still strongly enhanced by low-pH treatment (Table 1; Fig. 5).
One demonstrable effect of the CT truncations was altered conformations in the ectodomain of TM that may directly promote fusion. Release of SU from CT574, CT572, and CT571 into culture medium was markedly greater than that for wild-type Env and other mutants (Fig. 8B). Others have shown that deletion of the R peptide of MoMLV Env or truncation of the HIV-1 or simian immunodeficiency virus Env CT induces conformational changes in the ectodomain of TM and can result in increased SU shedding and cell fusion (1, 55, 61). It is possible that the secreted SU may function as a soluble receptor-binding domain that promotes Env fusion at neutral pH, and in the case of JSRV, this may partially relieve the low-pH requirement. However, conditioned medium of cells expressing truncation mutants that fuse at neutral pH did not show an enhancement in the fusion activity of wild-type JSRV Env at neutral pH, and a purified soluble SU did not render the JSRV entry less sensitive to BafA1 treatment (data not shown). Alternatively, the unconstrained TM on the surface of cells may directly enhance Env fusion activity, as has been demonstrated for HIV-1 and other retroviruses (18). Characterization of the conformational changes induced by the CT truncations should provide important clues for a better understanding of the mechanisms of JSRV Env-mediated fusion and its regulation.
We propose two mechanisms that may explain an inhibitory effect of the JSRV Env CT on fusion. First, similar to the case with several other retroviruses (53, 54, 56), the N terminus of the JSRV Env CT preferentially folds into an alpha-helical structure (23) that facilitates coiled-coil formation (Y.-M. Zheng and S.-L. Liu, unpublished results) and as such may greatly stabilize the multimeric structure of the prefusion Env trimer. Whether or not the truncations presented here affect JSRV Env multimerization remains to be investigated. Second, any cellular factor(s) that binds to the Env CT to trigger intracellular signaling and thereby induce oncogenic transformation (34) may also be involved in regulation of fusion, directly or indirectly. In fact, the severely truncated JSRV Env mutants that exhibited enhanced fusogenicities, i.e., CT589, CT574, and CT572, were incapable of inducing cell transformation in vitro (Y.-M. Zheng and S.-L. Liu, unpublished results). Alternatively, an independent cellular factor(s) unrelated to transformation might be responsible for fusion regulation by the Env CT. Work is ongoing to identify cellular binding partners of the JSRV Env CT, and these molecules, once identified, will be tested for their roles in cell fusion.
Members of the betaretrovirus family, to which JSRV belongs, differ in their pH dependence and in the presence or absence of a cleavable R peptide in the CT of their TM. Mouse mammary tumor virus entry is pH dependent, and the CT of its TM is not cleaved by the viral protease (54), whereas MPMV entry is pH independent and its Env has a C-terminal 16-residue R peptide that is cleaved off by the viral protease after budding (6, 7). While our current data showing the pH dependence of JSRV Env-mediated fusion and its negative regulation by the CT do not imply a role or a presence of the R peptide in the JSRV Env CT, they do not rule out this possibility either, and it will have to be critically addressed in future studies. One essential question that remains to be addressed is the evolutional and functional role of the JSRV Env CT in vivo, particularly from the viral replication and oncogenesis perspectives. In this respect, it is of interest to note that the pH of human and likely other mammals’ lung fluids is relatively acidic (24) and that JSRV MoMLV pseudovirions are quite resistant to the denaturing environment of lung fluids (11) and to low-pH inactivation (4). Whether or not the pH-dependent entry of JSRV reported here is also important for viral replication in vivo remains to be investigated.
Acknowledgments
We thank A. D. Miller for reagents and for continuous support and encouragement. We also thank Eric Cohen for helpful discussions.
This work was supported by funding from the Canadian Institute of Health Research (CIHR) to S.-L.L. and a grant from the U.S. National Institutes of Health to L.M.A. (AI 33410). M.C. was supported by scholarships from the Natural Sciences and Engineering Research Council of Canada (NSERC) and Fonds de la Recherche en Santé du Québec (FRSQ). S.-L.L. is a Canada Research Chair in Virology and Gene Therapy.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Aguilar, H. C., W. F. Anderson, and P. M. Cannon. 2003. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R Peptide. J. Virol. 771281-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. E., K. J. Sherrill, S. M. Crispell, M. R. Perrott, J. O. Carlson, and J. C. DeMartini. 2002. The jaagsiekte sheep retrovirus envelope gene induces transformation of the avian fibroblast cell line DF-1 but does not require a conserved SH2 binding domain. J. Gen. Virol. 832733-2742. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong, R. T., A. S. Kushnir, and J. M. White. 2000. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 151425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertrand, P., M. Côté, Y.-M. Zheng, L. M. Albritton, and S.-L. Liu. 2008. Jaagsiekte sheep retrovirus utilizes a pH-dependent endocytosis pathway for entry. J. Virol. 822555-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brindley, M. A., and W. Maury. 2005. Endocytosis and a low-pH step are required for productive entry of equine infectious anemia virus. J. Virol. 7914482-14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brody, B. A., S. S. Rhee, and E. Hunter. 1994. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J. Virol. 684620-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brody, B. A., S. S. Rhee, M. A. Sommerfelt, and E. Hunter. 1992. A viral protease-mediated cleavage of the transmembrane glycoprotein of Mason-Pfizer monkey virus can be suppressed by mutations within the matrix protein. Proc. Natl. Acad. Sci. USA 893443-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caporale, M., C. Cousens, P. Centorame, C. Pinoni, M. De las Heras, and M. Palmarini. 2006. Expression of the jaagsiekte sheep retrovirus envelope glycoprotein is sufficient to induce lung tumors in sheep. J. Virol. 808030-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christodoulopoulos, I., and P. M. Cannon. 2001. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J. Virol. 754129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin, J. M., S. H. Hughes, and H. E. Varmus (ed.). 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 11.Coil, D. A., J. H. Strickler, S. K. Rai, and A. D. Miller. 2001. Jaagsiekte sheep retrovirus Env protein stabilizes retrovirus vectors against inactivation by lung surfactant, centrifugation, and freeze-thaw cycling. J. Virol. 758864-8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dakessian, R., Y. Inoshima, and H. Fan. 2007. Tumors in mice transgenic for the envelope protein of Jaagsiekte sheep retrovirus. Virus Gene 3573-80. [DOI] [PubMed] [Google Scholar]
- 13.Danilkovitch-Miagkova, A., F. M. Duh, I. Kuzmin, D. Angeloni, S.-L. Liu, A. D. Miller, and M. I. Lerman. 2003. Hyaluronidase 2 negatively regulates RON receptor tyrosine kinase and mediates transformation of epithelial cells by jaagsiekte sheep retrovirus. Proc. Natl. Acad. Sci. USA 1004580-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeMartini, J. C., J. V. Bishop, T. E. Allen, F. A. Jassim, J. M. Sharp, M. de las Heras, D. R. Voelker, and J. O. Carlson. 2001. Jaagsiekte sheep retrovirus proviral clone JSRV(JS7), derived from the JS7 lung tumor cell line, induces ovine pulmonary carcinoma and is integrated into the surfactant protein A gene. J. Virol. 754239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz-Griffero, F., S. A. Hoschander, and J. Brojatsch. 2002. Endocytosis is a critical step in entry of subgroup B avian leukosis viruses. J. Virol. 7612866-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dirks, C., F. M. Duh, S. K. Rai, M. I. Lerman, and A. D. Miller. 2002. Mechanism of cell entry and transformation by enzootic nasal tumor virus. J. Virol. 762141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 28525-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70777-810. [DOI] [PubMed] [Google Scholar]
- 19.Fan, H. (ed.). 2003. Jaagsiekte sheep retrovirus and lung cancer. Current topics in microbiology and immunology, no. 275. Springer, Berlin, Germany. [DOI] [PubMed]
- 20.Florkiewicz, R. Z., and J. K. Rose. 1984. A cell line expressing vesicular stomatitis virus glycoprotein fuses at low pH. Science 225721-723. [DOI] [PubMed] [Google Scholar]
- 21.Green, N., T. M. Shinnick, O. Witte, A. Ponticelli, J. G. Sutcliffe, and R. A. Lerner. 1981. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc. Natl. Acad. Sci. USA 786023-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson, L. E., R. Sowder, T. D. Copeland, G. Smythers, and S. Oroszlan. 1984. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described Gag and Env cleavage products. J. Virol. 52492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hull, S., and H. Fan. 2006. Mutational analysis of the cytoplasmic tail of jaagsiekte sheep retrovirus envelope protein. J. Virol. 808069-8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inglis, S. K., S. M. Wilson, and R. E. Olver. 2003. Secretion of acid and base equivalents by intact distal airways. Am. J. Physiol. Lung Cell Mol. Physiol. 284L855-L862. [DOI] [PubMed] [Google Scholar]
- 25.Januszeski, M. M., P. M. Cannon, D. Chen, Y. Rozenberg, and W. F. Anderson. 1997. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J. Virol. 713613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, S., B. Zhang, O. A. Weisz, and R. C. Montelaro. 2005. Receptor-mediated entry by equine infectious anemia virus utilizes a pH-dependent endocytic pathway. J. Virol. 7914489-14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katen, L. J., M. M. Januszeski, W. F. Anderson, K. J. Hasenkrug, and L. H. Evans. 2001. Infectious entry by amphotropic as well as ecotropic murine leukemia viruses occurs through an endocytic pathway. J. Virol. 755018-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lepperdinger, G., J. Mullegger, and G. Kreil. 2001. Hyal2—less active, but more versatile? Matrix Biol. 20509-514. [DOI] [PubMed] [Google Scholar]
- 29.Lepperdinger, G., B. Strobl, and G. Kreil. 1998. HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J. Biol. Chem. 27322466-22470. [DOI] [PubMed] [Google Scholar]
- 30.Lin, X., C. A. Derdeyn, R. Blumenthal, J. West, and E. Hunter. 2003. Progressive truncations C terminal to the membrane-spanning domain of simian immunodeficiency virus Env reduce fusogenicity and increase concentration dependence of Env for fusion. J. Virol. 777067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, S.-L., F. M. Duh, M. I. Lerman, and A. D. Miller. 2003. Role of virus receptor Hyal2 in oncogenic transformation of rodent fibroblasts by sheep betaretrovirus Env proteins. J. Virol. 772850-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, S.-L., M. I. Lerman, and A. D. Miller. 2003. Putative phosphatidylinositol 3-kinase (PI3K) binding motifs in ovine betaretrovirus Env proteins are not essential for rodent fibroblast transformation and PI3K/Akt activation. J. Virol. 777924-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, S.-L., and A. D. Miller. 2005. Transformation of Madin-Darby canine kidney epithelial cells by sheep retrovirus envelope proteins. J. Virol. 79927-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, S.-L., and A. D. Miller. 2007. Oncogenic transformation by the jaagsiekte sheep retrovirus envelope protein. Oncogene 26789-801. [DOI] [PubMed] [Google Scholar]
- 35.Maeda, N., M. Palmarini, C. Murgia, and H. Fan. 2001. Direct transformation of rodent fibroblasts by jaagsiekte sheep retrovirus DNA. Proc. Natl. Acad. Sci. USA 984449-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClure, M. O., M. A. Sommerfelt, M. Marsh, and R. A. Weiss. 1990. The pH independence of mammalian retrovirus infection. J. Gen. Virol. 71767-773. [DOI] [PubMed] [Google Scholar]
- 37.Melikyan, G. B., R. M. Markosyan, S. A. Brener, Y. Rozenberg, and F. S. Cohen. 2000. Role of the cytoplasmic tail of ecotropic Moloney murine leukemia virus Env protein in fusion pore formation. J. Virol. 74447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, A. D. 2003. Identification of Hyal2 as the cell-surface receptor for jaagsiekte sheep retrovirus and ovine nasal adenocarcinoma virus. Curr. Top. Microbiol. Immunol. 275179-199. [DOI] [PubMed] [Google Scholar]
- 39.Miller, A. D., and F. Chen. 1996. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J. Virol. 705564-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103679-689. [DOI] [PubMed] [Google Scholar]
- 41.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272263-267. [DOI] [PubMed] [Google Scholar]
- 42.Nussbaum, O., A. Roop, and W. F. Anderson. 1993. Sequences determining the pH dependence of viral entry are distinct from the host range-determining region of the murine ecotropic and amphotropic retrovirus envelope proteins. J. Virol. 677402-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmarini, M., N. Maeda, C. Murgia, C. De-Fraja, A. Hofacre, and H. Fan. 2001. A phosphatidylinositol 3-kinase docking site in the cytoplasmic tail of the Jaagsiekte sheep retrovirus transmembrane protein is essential for envelope-induced transformation of NIH 3T3 cells. J. Virol. 7511002-11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Picard-Maureau, M., G. Jarmy, A. Berg, A. Rethwilm, and D. Lindemann. 2003. Foamy virus envelope glycoprotein-mediated entry involves a pH-dependent fusion process. J. Virol. 774722-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 683220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rai, S. K., J. C. DeMartini, and A. D. Miller. 2000. Retrovirus vectors bearing jaagsiekte sheep retrovirus Env transduce human cells by using a new receptor localized to chromosome 3p21.3. J. Virol. 744698-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rai, S. K., F. M. Duh, V. Vigdorovich, A. Danilkovitch-Miagkova, M. I. Lerman, and A. D. Miller. 2001. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. USA 984443-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Redmond, S., G. Peters, and C. Dickson. 1984. Mouse mammary tumor virus can mediate cell fusion at reduced pH. Virology 133393-402. [DOI] [PubMed] [Google Scholar]
- 49.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 681773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice, N. R., L. E. Henderson, R. C. Sowder, T. D. Copeland, S. Oroszlan, and J. F. Edwards. 1990. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J. Virol. 643770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ross, S. R., J. J. Schofield, C. J. Farr, and M. Bucan. 2002. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad. Sci. USA 9912386-12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shmulevitz, M., J. Salsman, and R. Duncan. 2003. Palmitoylation, membrane-proximal basic residues, and transmembrane glycine residues in the reovirus p10 protein are essential for syncytium formation. J. Virol. 779769-9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song, C., S. R. Dubay, and E. Hunter. 2003. A tyrosine motif in the cytoplasmic domain of Mason-Pfizer monkey virus is essential for the incorporation of glycoprotein into virions. J. Virol. 775192-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song, C., K. Micoli, and E. Hunter. 2005. Activity of the Mason-Pfizer monkey virus fusion protein is modulated by single amino acids in the cytoplasmic tail. J. Virol. 7911569-11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spies, C. P., G. D. Ritter, Jr., M. J. Mulligan, and R. W. Compans. 1994. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J. Virol. 68585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor, G. M., and D. A. Sanders. 2003. Structural criteria for regulation of membrane fusion and virion incorporation by the murine leukemia virus TM cytoplasmic domain. Virology 312295-305. [DOI] [PubMed] [Google Scholar]
- 57.Verwoerd, D. W., E. M. De Villiers, and R. C. Tustin. 1980. Aetiology of jaagsiekte: experimental transmission to lambs by means of cultured cells and cell homogenates. Onderstepoort J. Vet. Res. 4713-18. [PubMed] [Google Scholar]
- 58.Vigdorovich, V., A. D. Miller, and R. K. Strong. 2007. Ability of hyaluronidase 2 to degrade extracellular hyaluronan is not required for its function as a receptor for jaagsiekte sheep retrovirus. J. Virol. 813124-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vigdorovich, V., R. K. Strong, and A. D. Miller. 2005. Expression and characterization of a soluble, active form of the jaagsiekte sheep retrovirus receptor, Hyal2. J. Virol. 7979-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wootton, S. K., C. L. Halbert, and A. D. Miller. 2005. Sheep retrovirus structural protein induces lung tumours. Nature 434904-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyss, S., A. S. Dimitrov, F. Baribaud, T. G. Edwards, R. Blumenthal, and J. A. Hoxie. 2005. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J. Virol. 7912231-12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao, Y., L. Zhu, C. A. Benedict, D. Chen, W. F. Anderson, and P. M. Cannon. 1998. Functional domains in the retroviral transmembrane protein. J. Virol. 725392-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]