Abstract
Expression of the budding yeast retrotransposon Ty3 results in production of viruslike particles (VLPs) and retrotransposition. The Ty3 major structural protein, Gag3, similar to retrovirus Gag, is processed into capsid, spacer, and nucleocapsid (NC) during VLP maturation. The 57-amino-acid Ty3 NC protein has 17 basic amino acids and contains one copy of the CX2CX4HX4C zinc-binding motif found in retrovirus NC proteins. Ty3 RNA, protein, and VLPs accumulate in clusters associated with RNA processing bodies (P bodies). This study investigated the role of the NC domain in Ty3-P body clustering and VLP assembly. Fifteen Ty3 NC Ala substitution and deletion mutants were examined using transposition, immunoblot, RNA protection, cDNA synthesis, and multimerization assays. Localization of Ty3 proteins and VLPs was characterized microscopically. Substitutions of each of the conserved residues of the zinc-binding motif resulted in the loss of Ty3 RNA packaging. Substitution of the first two of four conserved residues in this motif caused the loss of Ty3 RNA and protein clustering with P bodies and disrupted particle formation. NC was shown to be a mediator of formation of Ty3 RNA foci and association of Ty3 RNA and protein with P bodies. Mutations that disrupted these NC functions resulted in various degrees of Gag3 nuclear localization and a spectrum of different particle states. Our findings are consistent with the model that Ty3 assembly is associated with P-body components. We hypothesize that the NC domain acts as a molecular switch to control Gag3 conformational states that affect both assembly and localization.
Ty3 is a long-terminal-repeat (LTR) retrotransposon in Saccharomyces cerevisiae that encodes a Gag3 protein with domains similar to those of retrovirus capsid (CA) and nucleocapsid (NC) (51, 59). Our previous studies showed that Ty3 protein, RNA, and viruslike particles (VLPs) accumulate in association with RNA processing bodies (P bodies), and these are proposed to be the sites of assembly (4). The present study was undertaken to determine the role of the Ty3 NC domain in the association of Ty3 protein and RNA with P bodies and in the assembly of VLPs.
The similarity of retrovirus and Ty3 CA and NC domains suggests that they perform similar functions in their respective assembly contexts. The major structural proteins of retroviruses include matrix (MA), CA, and NC domains, as well as other less-conserved domains (12). The MA domain concentrates precursor polyproteins at the membrane (reviewed in references 1, 19, 46, 54, and 60) or the microtubule organizing center (65, 76) assembly sites. The Gag NC zinc-binding residues are directly implicated in the recognition of genomic RNA (17). In addition, the NC domain mediates less-specific interactions of Gag with RNA via patches of basic amino acids. Binding to RNA in turn facilitates Gag-Gag interactions within the CA domain that are required for assembly. Hence, these basic NC residues together with the spacer (SP) domain found between CA and NC domains of some retrovirus Gag proteins are designated the interaction or I domain (5, 6, 11, 13, 37, 42). Genetic studies with murine leukemia virus (MLV) (49) and Rous sarcoma virus (RSV) (37) show that mutations in NC are associated with a late phenotype, but the molecular basis of that phenotype is not known.
The NC domain also mediates interactions with several host factors, some of which are related to intracellular localization and assembly. Interactions have been documented with the structural protein actin (41, 47, 55, 73), elongation factor 1 alpha (10), ABCE1 (40), APOBEC3G (61), and nucleolin (2).
Despite the central role played by NC in RNA packaging and assembly, it is not completely clear when and how the first critical contacts occur between Gag and genomic RNA (8). There is evidence that Gag interactions with genomic RNA occur prior to arrival at the final assembly site. However, retroviruses differ in the mode of nuclear export of genomic RNA, the cis or trans nature of Gag association with template and/or genomic RNA, and localization of the assembly site. Human immunodeficiency virus type 1 (HIV-1) RNA and Gag concentrate early during expression at a pericentriolar site (38, 53). Delivery of RNA to the pericentriolar site is promoted by association with the A2 mRNA nuclear export protein (38). Interestingly, packaging efficiency is also partially dependent upon the HIV-1-encoded genomic RNA nuclear export factor, Rev (7, 68). These observations are consistent with the lack of bias for HIV-1 genomic RNA packaging by the proteins from which it is translated (50). Recent studies showed that mutations in the zinc-binding domain of HIV-1 NC disrupted trafficking of both Gag and genomic RNA to the plasma membrane assembly site (25). Association of Gag and genomic RNA prior to assembly occurs for other retroviruses as well. The MLV genomic RNA which originates from a nontranslated RNA pool (39) also associates intracellularly with Gag and Gag-Pol for trafficking to the plasma membrane assembly site (3). In cells infected by simian foamy virus (64) and RSV (9) Gag proteins traffic to the nucleus, and it has been proposed that they could capture genomic RNA for delivery to the assembly site. However, early nuclear localization of simian foamy virus Gag is not absolutely required for replication of virus in tissue culture cells. In the case of RSV, host factors have been identified as affecting nuclear export and packaging of genomic RNA (34). Thus, in both of these cases, the extent to which Gag or its NC domain is required for targeting the majority of genomic RNA to the assembly site remains unclear.
Despite the prevalence of LTR retrotransposons throughout eukaryotes, intracellular assembly of endogenous retroviruses and retrotransposons is relatively unexplored. Clusters of VLPs are observed by electron microscopy (EM) in cells expressing endogenous retroviruses, including murine MusD (56) and ovine enJSRV (48). Retrotransposons in Drosophila, such as copia (45), in S. cerevisiae, such as Ty1 (21) and Ty3 (26), and in Schizosaccharomyces pombe, such as Tf1 (70) are also observed in clusters. However, the localization of assembly and identity of determinants that target both RNA and protein to those assembly sites are relatively unexplored.
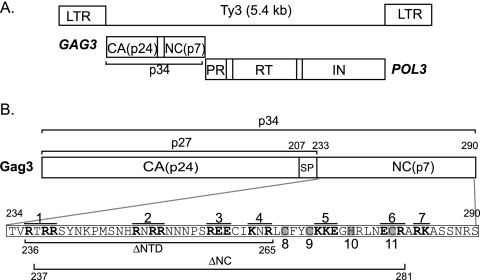
Ty3 is 5.4 kb in length and encodes Gag3 and Gag3-Pol3 precursor polyproteins (reviewed in reference 59) (Fig. 1A) which form VLPs of 40 to 50 nm in diameter. Gag3, the major structural protein, is processed by Ty3 protease (PR) into 24-kDa CA, p27, and 57-amino-acid (aa) NC. A 26-aa SP3 between CA and NC domains is inferred from positions of processing sites but has not been observed (31, 32). Processing of Gag3-Pol3, in addition to CA and SP3, produces a 76-aa NC, which spans the frameshift of Gag3-Pol3, protease (PR), 10-kDa junction fragment, reverse transcriptase (RT), and integrase (IN). Expression of Ty3 under a synthetic GAL1-10 promoter or the native pheromone-inducible promoter results in the formation of VLP clusters (4, 26). VLPs are associated with Ty3 RNA and, after longer times of induction, with cDNA. Ty3 protein, RNA, and VLPs are concentrated in association with P bodies, the proposed site of assembly (4). Two lines of evidence support a role for NC in the formation and function of VLPs. First, mutations in the conserved zinc-binding motif of the Gag3 NC domain were previously shown to cause decreased production and processing of VLPs (51). Second, in vitro data support a role for Ty3 NC in annealing of the primer tRNAMet to Ty3 at 5′ and 3′ sites (20) and in priming Ty3 RT (14).
FIG. 1.
Ty3 Gag3. (A) Ty3 genome and polyprotein organization. Protein domains are as marked; SP3, a predicted junction fragment of about 10 kDa between PR and RT, and the differentially processed ends of IN are indicated in that order as empty boxes. (B) Mutagenesis of Ty3 NC. Gag3 cleavage sites to produce CA, p27, SP3, and NC are indicated. The NC domain sequence (aa 234 to 290) is indicated in the lower panel. Ala-scanning mutations 1 to 7 are shown overlined and with substituted positions in boldface; the four conserved positions in the zinc-binding motif that were substituted with Ala are shown in gray and are numbered 8 to 11. The residues (aa 236 to 265) deleted in mutant (ΔNTD) and the residues (aa 237 to 281) deleted in mutant (ΔNC) are underlined.
The present study was undertaken to determine the role of Ty3 NC in the localization of Ty3 polyproteins and RNA with P-body components. The Ty3 NC domain was shown to be important for RNA cluster formation and for association of Ty3 proteins with P-body components. A subset of mutations disrupted particle formation and resulted in Gag3 nuclear localization. Our data support the model that NC mediates Ty3 RNA and protein association with P-body components and that this association occurs during assembly.
MATERIALS AND METHODS
Strains and culture conditions for S. cerevisiae and Escherichia coli.
The yeast strain yTM443 (MATa ura3-52 trp1-H3 his3-Δ200 ade2-101 lys2-1 leu1-12 can1-100 bar1::hisG Ty3 null) (43) was used in the present study except for cytological studies. For transmission EM, yeast strain BY4741 was used (MATa his3-Δleu2Δ met15Δ ura3Δ; Open Biosystems, Huntsville, AL) except in the case of the ΔNTD mutant, which was analyzed in strain yTM443. Direct fluorescence studies were performed in the BY4741 background using a strain in which the coding region for the P-body component Dhh1 was fused at the downstream end to the green fluorescent protein (GFP) coding sequence (Dhh1-GFP) (67). Immunofluorescence using anti-CA was performed in strain yTM443. Yeast transformations were performed by the lithium acetate procedure (63). The rich medium for S. cerevisiae cultures was YPD (1% yeast extract/2% peptone/2% dextrose). In order to select for cells with particular prototrophic markers, synthetic dextrose medium (0.67% yeast nitrogen base, 2% dextrose) or synthetic galactose (SG) medium (0.67% yeast nitrogen base, 2% galactose) or synthetic raffinose (SR) medium (0.67% yeast nitrogen base, 1% raffinose, 2% [vol/vol] glycerol, 2% [vol/vol] lactic acid) containing complete amino acids minus the appropriate amino acids was used (66).
Plasmid constructions and mutagenesis.
The seven Ala-scanning mutants and six zinc-binding domain Ala replacement mutants were created by using site-directed mutagenesis of a subfragment of Ty3. The mutated fragment sequence was determined (CoGenics, Inc., Houston, TX), and the fragment was swapped into a full-length, galactose-inducible copy of Ty3 in the high-copy plasmid pDLC201, as previously described for mutations in the Ty3 CA-coding domain (33). The pDLC201 plasmid contains the Ty3-1 element (accession no. M34549). The two deletion mutants, ΔNTD and ΔNC, were created by site-directed mutagenesis of the subfragment but using overlap extension PCR (71). The PR− variants (PR−) of the deletion mutants were created by site-directed mutagenesis of the subfragment containing the deletion mutation in order to convert the active site Asp to Ile as previously described (31). The presence of the mutation in the final construct was confirmed by DNA sequence analysis. The mutagenic oligonucleotides are listed in Table 1.
TABLE 1.
Oligonucleotides used to produce Ty3 NC mutants
| Oligonucleotide set | Ty3 NC mutant | Forward sequencea (5′-3′) |
|---|---|---|
| 1 | R236A/R238A/R239A | GGATACGTCCATACCGTAGCCACAGCAGCATCTTACAATAAACC |
| 2 | R249A/R251A/R252A | CCAATGTCAAATCATGCAAACGCCGCAAATAACAACCCATCTAG |
| 3 | R258A/E259A/E260A | CAACCCATCTGCAGCAGCATGTATAAAAAATCGGCTATGCTTC |
| 4 | K263A/R265A | CCCATCTAGAGAAGAATGTATAGCAAATGCCCTATGCTTCTATTG |
| 5 | K271A/K272A/E273A | CGGCTATGCTTCTATTGTGCCGCAGCCGGACATCGCCTGAACG |
| 6 | E279A/R281A | GGGACATCGCCTGAACGCATGTGCAGCACGTAAGGCGAG |
| 7 | R283A/K284A | CGCCTGAACGAATGTAGAGCAGCTGCCGCGAGTTCTAACCGATC |
| 8 | C267A | GAATGTATAAAAAATCGGCTAGCCTTCTATTGTAAGAAAGAGGG |
| 9 | C270A | CGGCTATGCTTCTATGCTAAGAAAGAGGGACATCGCC |
| 10 | H275A | CGGCTATGCTTCTATTGTAAGAAAGAGGGAGCTCGCCTGAACG |
| 11 | C280A | GGGACATCGCCTGAACGAAGCTAGAGCACGTAAGGCGAG |
| 12 | C267A/C280A | GAATGTATAAAAAATCGGCTAGCCTTCTATTGTAAGAAAGAGGGACATCGCCTGAACGAAGCTAGAGCA |
| 13 | C270A/C280A | CGGCTATGCTTCTATGCTAAG-AAAGAGGGACATCGCCTGAACGAAGCTAGAGCACGTAAGGCGAG |
| 14 | ΔNTD (aa 236 to 265) | CTTTCTTACAATAGAAGCATAGTACGGTATGGACGTATCC |
| ΔNTD (aa 236 to 265) | GGATACGTCCATACCGTACTATGCTTCTATTGTAAGAAAG | |
| ΔNTD (aa 236 to 265) | TCGAATCACAGGATCCCCCTGAACTACCC | |
| ΔNTD (aa 236 to 265) | GTATCCTGTAGGTACCGTCTTTCTTCGGG | |
| 15 | ΔNC (aa 237 to 281) | CTCGCCTTACGTGCTCTTACGGTATGGACGTATCCATTC |
| ΔNC (aa 237 to 281) | GAATGGATACGTCCATACCGTAAGAGCACGTAAGGCGAG | |
| ΔNC (aa 237 to 281) | TCGAATCACAGGATCCCCCTGAACTACCC | |
| ΔNC (aa 237 to 281) | GTATCCTGTAGGTACCGTCTTTCTTCGGG | |
| 16 | PR D59I [ΔNTD(PR−)] | CACGAAAGTAAAAACCCTGTTTATCAGTGGATCACCCACGTC |
| 17 | PR D59I [ΔNC(PR−)] | CACGAAAGTAAAAACCCTGTTTATCAGTGGATCACCCACGTC |
Mutated bases are underlined.
In order to localize mutant Ty3 protein relative to P bodies, a plasmid carrying a galactose-inducible Ty3 fused to red fluorescent protein (Ty3-RFP) on a high-copy plasmid (pNB2266) was used (4). The RFP open reading frame (ORF) was fused in frame with the IN-coding region at the end of the Ty3 POL3 ORF. Ty3-RFP expression and VLP formation resulted in PR processing of Gag3 and Gag3-Pol3 and generation of an IN-RFP fusion protein. Mutations were introduced into the Ty3-RFP plasmid by swapping in the mutagenized fragment as described above for mutagenesis of wild-type (wt) Ty3. In order to localize Ty3 RNA, a plasmid carrying a galactose-inducible Ty3 marked with two copies of the MS2 coat protein binding site (Ty3-MS2) was used (4). Mutated fragments were swapped into the wt expression clone as described above for wt Ty3 and Ty3-RFP.
Transposition assays.
Transposition was measured by using a target plasmid assay described previously (30). Transcription of plasmid-borne Ty3 under control of the GAL1-10 upstream activating sequence was induced by growth on galactose-containing medium. Transposition of Ty3 into the low-copy target plasmid, pCH2bo19V, activates the expression of a suppressor tRNA gene. Transposition is scored by growth dependent upon the suppression of the ochre nonsense alleles, ade2-101 and lys2-1, in yeast strain yTM443 (30, 43). Transposition assays were performed at 24 and 30°C. Under each condition, two independent transformants for each mutant were patched onto synthetic dextrose-Ura-His medium. After 1 day, cells were replica plated onto SG-Ura-His to induce transposition. After 2 days, cells were replica plated onto minimal medium supplemented with Leu and Trp to select for Ade+ and Lys+ colonies.
Immunoblot analysis of Ty3 intermediates in extracts of cells expressing Ty3.
Two transformants containing either the high-copy expression plasmid, pDLC201, or mutant derivative plasmid, and target plasmid were grown in galactose-containing medium for 24 h at 24°C in order to allow Ty3 expression. Whole-cell extracts (WCEs) were prepared under denaturing conditions, and immunoblot analysis was performed with anti-CA and anti-IN immunoglobulin G (IgG) as described previously (33). Immunoblots were also performed with anti-NC peptide IgG (51), which was diluted 1:1,000.
Southern blot analysis of Ty3 cDNA in extracts of cells expressing Ty3.
Cells of yeast strain yTM443 transformed with either wt Ty3 high-copy expression plasmid pDLC201 or mutant derivative plasmids and the target plasmid pCH2bo19V were grown as described above for immunoblot analysis. RNA-free DNA (1.0 μg) was prepared and digested with EcoRI, fractionated by electrophoresis in a 0.8% agarose gel, and processed for Southern blot analysis as described previously (33). Hybridization was performed with a 32P-labeled, internal BglII fragment of Ty3, which hybridizes with the full-length 5.4-kbp Ty3 cDNA, as well as Ty3 donor plasmid and chromosomal Ty3 elements. Filters were washed and exposed to a Personal Molecular FX Phosphorimager screen, and the signals were quantified using Quantity One Software (Bio-Rad, Inc., Richmond, CA).
RNA protection assay.
Yeast strain yTM443 transformed with either wt Ty3 expression plasmid, pDLC201, or mutant derivative plasmids was grown as described for immunoblot analysis. RNA was extracted under nondenaturing conditions and subjected to benzonase digestion analysis as previously described (33). Ty3 control RNA was generated in vitro by T7 RNA polymerase transcription of a Ty3 template plasmid linearized at the downstream end of Ty3 by digestion with SnaBI. Extracted RNAs were denatured in 6.7% glyoxal for 1 h at 50°C, fractionated by gel electrophoresis in a 0.8% agarose gel, and processed for Northern blot analysis (57). The percentage of protection was determined based on protection in the plus benzonase condition relative to the amount of Ty3 RNA in the 4°C untreated sample. Labeled RNA was quantified as described for cDNA.
Particle analysis by velocity sedimentation.
In order to test for particle formation and multimerization, WCEs were subjected to velocity sedimentation. Cells of yeast strain yTM443 transformed with wt Ty3 expression plasmid pDLC201 or mutant derivative plasmids were grown in SG medium for 6 h at 24°C in order to allow Ty3 expression. Cells equivalent to 15 ml of an optical density at 600 nm of 0.7 were suspended in 200 μl of BEB native extraction buffer (100 mM KCl-10 mM Tris-HCl [pH 7.5]-10 mM EDTA with inhibitors [2.1 μg of aprotinin/ml, 6.6 μg of pepstatin/ml, 2 mM phenylmethylsulfonyl fluoride, 0.66 μg of leupeptin/ml, and 26 U of RNase inhibitor/ml]) and divided into two 100-μl portions; one was left untreated, and the other was adjusted to a final concentration of 1% NP-40 and supplemented with 2 μl of antifoam. Detergent-treated and untreated portions were vortex mixed vigorously with sterile glass beads. An additional 250 μl of extraction buffer was added to the detergent treated and untreated portions, and the cells were pipetted from the glass beads and subjected to centrifugation at 500 × g for 5 min to remove the cell debris. The supernatant fractions were removed. An aliquot of 40 μl was reserved for immunoblot analysis, and the remaining supernatants were subjected to centrifugation at 16,110 × g for 30 min. Pellets were resuspended directly into the original volume of supernatant with BEB buffer. For both detergent treated and untreated samples, equal amounts of the WCEs, pellet, and supernatant fractions were fractionated by electrophoresis in a sodium dodecyl sulfate (SDS)-4 to 20% polyacrylamide gel and analyzed by immunoblotting with anti-Ty3 CA as described previously with anti-Ty3 CA (33).
Fluorescence microscopy.
The ability of different NC mutants to localize to P bodies was determined by fluorescence microscopy of wt and mutant variants of Ty3-RFP and Ty3-MS2. In order to visualize the Ty3-MS2 RNA, phage RNA-binding protein MS2 was fused to RFP (MS2-RFP) and expressed in the same strain with Ty3-MS2 in which the P body was visualized by P-body marker protein Dhh1-GFP. Gag3 was also localized by immunofluorescence with anti-CA antibodies and anti-rabbit IgG conjugated to Alexa 488 (4). Induction of cells was as described above for immunoblot analysis except that the inductions were for 6 h. Control cells were uninduced cells containing the Ty3 expression plasmid. A total of 150 cells representing each of two independent transformants were observed for each mutant. The number of cells actually expressing detectable marker varied among the different transformants.
EM.
EM was performed as described previously (33, 70). Transformants were grown in noninducing SR-Ura medium until early log phase, induced by the addition of galactose to 2%, and grown for 6 h at 24°C. Cells were imaged with a transmission electron microscope (H7600; Hitachi, Tokyo, Japan). Approximately 100 cells were observed per wt or mutant Ty3 transformant. Cells used in this experiment were confirmed to have a pattern of protein expression consistent with previous immunoblot analysis (data not shown).
RESULTS
Ala-scanning mutagenesis strategy.
The Ty3 NC domain spans carboxyl-terminal residues 234 through 290 of Gag3 (Fig. 1B). (Residues are numbered according to the Gag3 precursor.) In order to characterize the contribution of the NC domain to the Ty3 life cycle, a set of 17 mutants was created (Table 2). The mutations included seven Ala-scanning mutations in which windows of five residues containing two or more charged residues were substituted at charged positions with Ala; four single and two double Ala substitutions of conserved positions within the zinc-binding domain; and two deletion mutants, one lacking all but two residues of the amino-terminal domain (NTD) of NC, including 10 of 17 basic residues (aa 236 to 265, ΔNTD), and the other lacking 45 of 57 residues in NC (aa 237 to 281, ΔNC) (Fig. 1B). Deletion mutants were designed to retain PR processing and Gag3-Pol3 frameshift sites. Mutants were characterized by using transposition, immunoblot, sedimentation, packaging, replication, and localization assays.
TABLE 2.
Summary of Ty3 NC mutant phenotypes
| Oligonucleotide set | Mutation(s) | CA processing | Integrase | cDNA/plasmid | Transposition score (30°C/24°C) | % RNA protection | P-body localization |
|---|---|---|---|---|---|---|---|
| wt | wt | + | 1.25 ± 0.31 | +++ | 33 ± 8 | + | |
| 1 | R236A/R238A/R239A | wt | + | 2.0 | +++ | NDa | + |
| 2 | R249A/R251A/R252A | wt | + | 1.9 | +++ | ND | + |
| 3 | R258A/E259A/E260A | wt | + | 1.4 | +++/++ | 28 | + |
| 4 | K263A/R265A | wt | + | 2.1 | +++ | ND | + |
| 5 | K271A/K272A/E273A | Reduced | + | 0.5 | +/++ | 43 | + |
| 6 | E279A/R281A | wt | + | 5.0 | +++ | 30 | + |
| 7 | R283A/K284A | wt | + | 3.4 | +++ | ND | + |
| 8 | C267A | Reduced | +/− | 0.0 | − | 11 | − |
| 9 | C270A | Reduced | + | 0.0 | − | 13 | − |
| 10 | H275A | Reduced | + | 0.0 | − | 13 | + |
| 11 | C280A | Reduced | + | 0.1 | ±/+ | 10 | + |
| 12 | C267A/C280A | Reduced | − | ND | ND | 2 | − |
| 13 | C270A/C280A | Reduced | +/− | ND | ND | 3 | + |
| 14 | ΔNTD | Reduced | +/− | ND | ND | 3 | +/− |
| 15 | ΔNC | Reduced | + | ND | ND | 1 | +/− |
| 16 | ΔNTD(PR−) | None | − | ND | ND | ND | ND |
| 17 | ΔNC(PR−) | None | − | ND | ND | ND | ND |
ND, not determined.
Retrotransposition of Ala-scanning mutants.
A qualitative genetic assay of integration into a plasmid containing a tRNA gene target was performed to determine the effects of NC mutations on retrotransposition frequency (see Materials and Methods). Two independent transformants were tested for wt Ty3 and each mutant at 24 and 30°C. The transposition properties of these mutants are summarized in Table 2.
Five of the seven Ala-scanning mutants (R236A/R238A/R239A, R249A/R251A/R252A, K263A/R265A, E279A/R281A, and R283A/K284A) had transposition frequencies similar to that of the wt at both temperatures (Table 2). Two mutants were affected by temperature; one was severely defective at 30°C (K271A/K272A/E273A), and one was modestly affected at 24°C (R258A/E259A/E260A). Single zinc-binding motif mutants (C267A, C270A, H275A, and C280A) were each severely blocked for transposition at both temperatures. Thus, transposition appeared relatively resistant to small changes in charged residues but was sensitive to any change in the conserved zinc-binding motif.
Precursor polyprotein maturation.
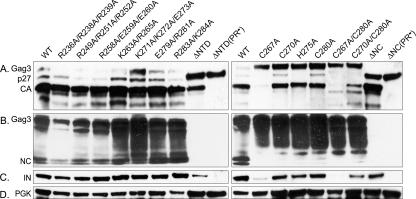
Ty3 PR processing was used as an indirect measure of proteins that were competent to assemble in a correctly folded state. YTM443 transformants carrying either pDLC201 with wt Ty3 or mutant derivatives were grown in SR-Ura medium and induced to express Ty3 for 24 h as described in Materials and Methods. WCEs were prepared, and the processing of wt and mutant Gag3 and Gag3-Pol3 was analyzed by immunoblot analysis with anti-CA, anti-NC, and anti-IN antibodies (Fig. 2 and Table 2).
FIG. 2.
Mutations in NC affect Ty3 Gag3 and Gag3-Pol3 protein processing. Cultures were induced for Ty3 expression for 24 h at 24°C, and WCEs were prepared under denaturing conditions and processed for immunoblot analysis as described in Materials and Methods. WCEs from cultures expressing the indicated mutants were fractionated by electrophoresis on a SDS-4 to 20% polyacrylamide gel and analyzed by immunoblotting with rabbit polyclonal IgG. (A) Immunoblot analysis of Ty3 CA domains. Immunoblot analysis used anti-CA antibodies. (B) Immunoblot analysis of Ty3 NC domains. IgG was raised against a peptide from the NTD of NC and used for immunoblot analysis (26). (C) Immunoblot analysis of Ty3 IN domains. The WCE prepared above was analyzed by immunoblotting with anti-IN. (D) Immunoblot analysis of PGK. As a loading control, the WCE was analyzed by immunoblotting with polyclonal mouse antibody against PGK. Separate gels were run for blots except for rows C and D, which were the same blot.
Extracts from cells expressing wt Ty3 analyzed with anti-CA showed Gag3, p27, and CA species (Fig. 2A). Analysis of extracts with anti-IN showed a predominant band consistent with the migration of the 61-kDa IN (Fig. 2C). The IN antibody also detected the slower mobility Gag3-Pol3 species (33; data not shown).
Extracts of cells expressing wt Ty3 and each of the five mutants with wt levels of transposition (R236A/R238A/R239A, R249/R251A/R252A, K263A/R265A, E279A/R281A, and R283A/K284A), as well as the mutant with modest temperature sensitivity (R258A/E259A/E260A), showed similar amounts of Gag3 and p27 and a significantly greater amount of CA than any other species (Fig. 2A). The amounts of NC and IN were comparable to that produced by wt Ty3 (Fig. 2B and C). The temperature-sensitive mutant K271A/K272A/E273A showed a modest reduction in Gag3 processing under permissive conditions (Fig. 2A,B) but the wt levels of IN (Fig. 2C).
All of the zinc-binding motif mutants, (C267A, C270A, H275A, C280A, C267A/C280A, and C270A/C280A) displayed reduced and aberrant processing of Gag3 (Fig. 2A and B). In immunoblot analyses with the anti-CA antibody, these mutants had high levels of precursor Gag3, and two showed one species migrating slightly more slowly than wt p27. Mutants C267A and C267A/C280A showed no detectable wt Gag3 processing products but did show an unidentified species migrating more slowly than Gag3 (data not shown) and some species migrating more slowly than the NC species. The zinc-binding motif mutants displayed no or barely detectable levels of either of the NC species which were most prominent in cells expressing wt Ty3 (Fig. 2B). However, they showed increased amounts of two slightly larger species, possibly arising from loss of SP-NC cleavage. Somewhat surprisingly, only one single zinc-binding motif mutant (C267A) displayed significantly reduced amounts of IN, but both double substitution mutants showed reduced amounts (Fig. 2C). This could have arisen from a reduction in processing or a reduction in the total amount of stable Gag3-Pol3 in cells, since the level of the higher-molecular-weight Gag3-Pol3 species was also reduced compared to that in cells expressing wt Ty3 (data not shown).
The wt or intermediate processing phenotypes of mutants were consistent with transposition results and confirmed that the basic residue replacements did not have significantly deleterious effects. In order to increase the probability that functions of the non-zinc-binding basic residues were disrupted, one additional mutation was created that was a deletion of residues 236 through 265, including a total of 10 of 17 basic amino acids (ΔNTD). Residues 237 to 281 of NC (45 of 57 residues), including the entire zinc binding motif (ΔNC) were deleted in another construct. Both of these mutants showed unprocessed, truncated Gag3, but also significant amounts of what appeared to be mature CA (Fig. 2A). As expected, because the anti-NC antibody was raised against a peptide representing the NTD, NC products were undetectable for the ΔNTD and the ΔNC mutants. In order to verify that the Gag3 processing pattern was attributable to Ty3 PR and therefore implied assembly, a second mutation was introduced into both deletion mutants to change the PR active site Asp to Ile. This mutation was previously shown to inactivate PR (31). In both cases, introduction of this mutation resulted in full-length Gag3, indicating that the mutants displayed bona fide PR processing activity.
Particle formation by Ty3 NC mutants.
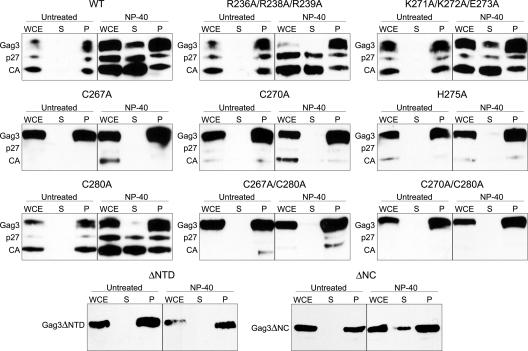
The processing pattern of many of the mutants indicated that protein multimerization or assembly was occurring in some, but possibly not all, mutants. In order to better understand the point at which transposition was disrupted, we next investigated the ability of individual mutants to form sedimentable species. A subset of mutants representing ones with wt transposition frequency (R236A/R238A/R239A) and those affected for transposition (K271A/K272A/E273A, C267A, C270A, H275A, and C280A), as well as double mutants (C267A/C280A and C270A/C280A), were tested. In order to maximize the possibility that mutant multimerization could be distinguished from wt, cells were induced for Ty3 expression for only 6 h at 24°C. Protein was extracted under nondenaturing conditions as described in Materials and Methods.
WCEs of cells expressing wt Ty3 had Gag3, p27, and CA in the pellet fraction in the absence or presence of detergent (Fig. 3). In the presence of detergent, some Gag3, but predominantly p27 and CA, were solubilized. This suggested that Gag3 and its processed species were found in different types of complexes, some of which either were associated with membranes or were more labile in the presence of detergent. For the NC mutants tested, all WCEs contained detergent-insoluble, pelletable material. This finding indicated that, overall, multimerization was robust (Fig. 3). Mutants that displayed substantial processing (R236A/R238A/R239A, K271A/K272A/E273A, and C280A) showed pelleting patterns similar to that of the wt. Consistent with the observation that predominantly processed Gag3 species were solubilized by detergent, the remaining mutants with little or no Gag3 processing did not release Gag3 species into the supernatant. These results indicated that mutations within hydrophilic patches or even within the zinc-binding motif in the NC domain did not significantly decrease the ability of Gag3 proteins to form complexes but did affect the nature of the Gag3-Gag3 interaction so that processing did not occur.
FIG. 3.
Sedimentation analysis of NC mutants. Cells were induced for 6 h at 24°C, and WCEs were prepared in native conditions and incubated with or without NP-40 as described in Materials and Methods. Samples were fractionated by electrophoresis on a SDS-4 to 20% polyacrylamide gel and analyzed by immunoblotting with polyclonal anti-CA antibody as described in the legend of Fig. 2.
Protection of genomic RNA.
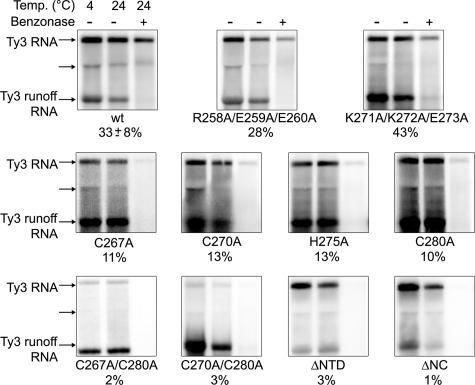
A subset of mutants displayed significant processing but were nonetheless profoundly defective in transposition. In order to better understand the basis of the transposition defect, the ability of particles to protect RNA was investigated (Fig. 4 and Table 2). wt Ty3 and Ty3 PR− (immature) VLPs have been previously shown to protect Ty3 RNA from benzonase digestion (33; unpublished data), so the inability to process alone does not cause a defect in RNA packaging.
FIG. 4.
NC mutations reduce packaging of genomic RNA. Cultures were induced for Ty3 expression for 24h at 24°C, and WCEs were prepared under native conditions as described in Materials and Methods. Positive control in vitro-transcribed Ty3 RNA (Ty3 runoff RNA) was added to each WCE. WCEs were left untreated at 4 and at 24°C for 6 min (first and second lanes, respectively) or treated with benzonase for 6 min at 24°C (third lanes). RNA was extracted and analyzed by Northern blotting with a 32P-labeled Ty3-specific probe. The Ty3 5.2-kb RNA and Ty3 runoff RNAs are indicated. Two or more independent experiments were performed for each mutant, and a representative example is shown. The percent protection of RNA is indicated under each set. A subgenomic 3.1-kb Ty3 RNA was observed to fractionate with a mobility similar to that of the 25S rRNA (unlabeled arrow).
Cells expressing wt Ty3, two temperature-sensitive phenotype mutants (R258A/E259A/E260A and K271A/K272A/E273A), the six mutants with zinc-binding domain substitutions (C267A, C270A, H275A, C280A, C267A/C280A, C270A/C280A), and the two deletion mutants (ΔNTD and ΔNC) were examined. Cells were induced to express Ty3 for 24 h at 24°C. WCEs were prepared under nondenaturing conditions and incubated in the presence or absence of benzonase as described in Materials and Methods. In vitro-generated, truncated Ty3 transcripts were added to all reactions to monitor completion of benzonase digestion. Nucleic acid in these extracts was analyzed by Northern blotting using a Ty3-specific probe. Under these conditions, 33% (±8%) of the wt Ty3 RNA was protected from benzonase digestion compared to control, untreated extracts incubated on ice. The in vitro Ty3 transcripts were completely digested by benzonase treatment (Fig. 4).
The Ala-scanning mutants that transposed at wt frequencies at the permissive temperature (R258A/E259A/E260A) showed protection of RNA similar to wt Ty3 (Fig. 4). Mutant K271A/K272A/E273A had decreased transposition and Gag3 processing. Surprisingly, it had 43% protection of RNA. Substitution of only one zinc-binding motif residue (C267A, C270A, H275A, and C280A) caused decreased protection, resulting in levels of RNA protection ranging from 10 to 13%. These results suggested that incomplete protection of RNA was achieved even in the presence of a compromised zinc-binding motif structure but clearly implicated the zinc-binding motif in the packaging of genomic RNA. Mutant C267A, which showed no Gag3 processing (Fig. 2), was not distinguished from other mutants by this criterion. Both double zinc-binding motif substitution mutants (C267A/C280A and C270A/C280A) and ΔNTD, which retained the zinc-binding motif, as well as ΔNC, showed lower levels of extractable RNA, suggesting that there was less protection of RNA.
Reverse transcription.
As discussed above, NC acts both in RNA genome packaging and as a chaperone in primer annealing. Analysis of RNA packaging indicated that at least one mutant (K271A/K272A/E273A) competent to protect RNA was deficient for transposition. Thus, we investigated the level of replicated extrachromosomal Ty3 cDNA in cells induced for Ty3 expression (Fig. 5 and Table 2).
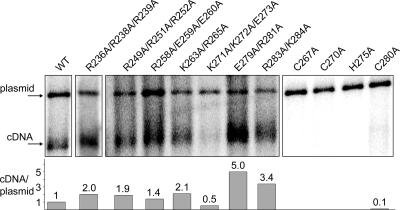
FIG. 5.
Mutations in basic residues and zinc-binding motif show different effects on cDNA production. Cultures were induced for Ty3 expression as described in the legend of Fig. 2 and processed for Southern blot analysis as described in Materials and Methods. DNA was purified from cells induced for Ty3 expression and cleaved with EcoRI to linearize the Ty3 expression plasmid. Southern blot analysis was performed with a 32P-labeled Ty3-specific probe, which hybridizes with the 5.4-kbp extrachromosomal Ty3 cDNA (cDNA), as well as the Ty3 donor plasmid (plasmid). The Ty3 cDNA/plasmid ratio in four independent wt cultures was 1.3 ± 0.3. Other ratios were normalized to the wt ratio. Two or more independent experiments were performed for each mutant, and a representative example is shown.
Coincident with wt processing phenotypes and near-wt transposition frequency, the basic mutants (R236A/R238A/R239A, R249A/R251A/R252A, R258A/E259A/E260A, K263A/R265A, E279A/R281A, and R283A/K284A) produced cDNA/plasmid ratios in the same range or higher than wt Ty3. Mutant K271A/K272A/E273A, which had temperature-sensitive transposition but only a modest decrease in processing and full RNA protection, had decreased levels of cDNA (Fig. 5). All single zinc-binding motif mutants (C267A, C270A, H275A, and C280A) that had reduced and aberrant Gag3 processing and did not produce mature NC had no detectable levels of cDNA (Fig. 5). Only mutants that showed transposition had detectable levels of cDNA.
Localization of mutant Ty3-RFP and P bodies.
Our earlier studies showed that Ty3 protein and RNA were both associated with P bodies, leading to the hypothesis that these were sites of assembly. In the present study, we probed the interdependence of Ty3 protein and RNA targeting.
We first tested whether the NC domain was important for Ty3 protein and RNA cluster formation and for colocalization with P bodies. In order to visualize Ty3 mutant proteins in living cells, the NC mutations were introduced into the Ty3-RFP expression vector in which POL3 is fused in frame to the RFP-coding region. wt and mutant Ty3-RFP elements were expressed in a strain that expresses the P-body marker Dhh1-GFP from its native chromosomal locus. In cells not expressing Ty3, there are multiple small Dhh1-GFP foci. However, in cells expressing Ty3 there is typically only one or two P bodies that are enlarged compared to those in wt cells (Fig. 6). Ty3-RFP and Dhh1-GFP, as well as other P-body markers, typically colocalize with these enlarged foci (4).
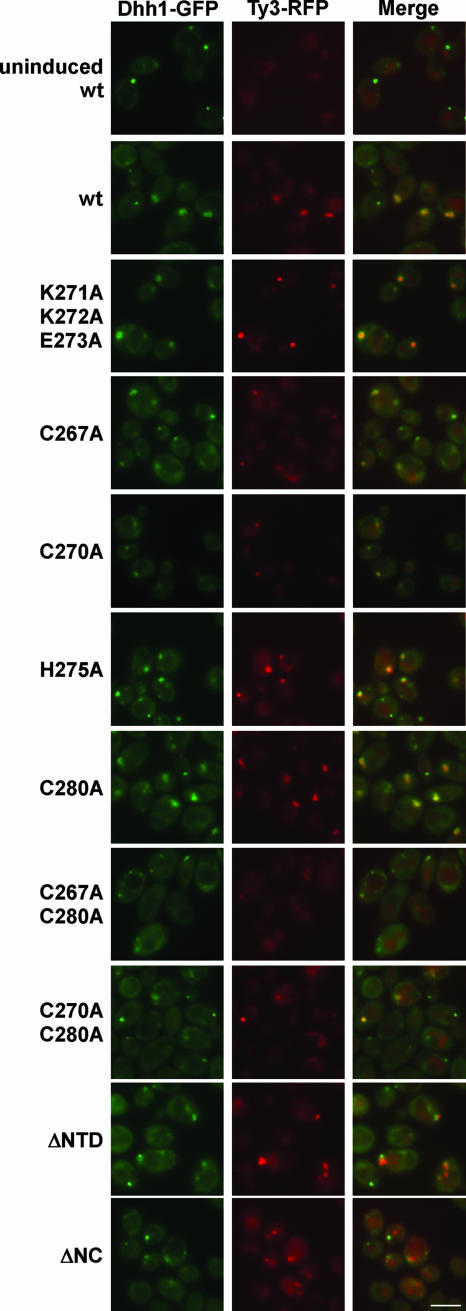
FIG. 6.
NC function is required for full association of Ty3-RFP and P-body clusters but not for Ty3-RFP cluster formation. wt and NC mutant Ty3-RFP was expressed in a strain with Dhh1-GFP under the native promoter. Mutants shown are representative of two independent transformants of each type. Cells were induced for 6 h as described in Materials and Methods (4). Control cells were uninduced cells containing the Ty3-RFP expression plasmid. Fluorescence imaging was performed with exposure times of 4,000 ms for TRITC (tetramethyl rhodamine isothiocyanate) and 2,500 ms for fluorescein isothiocyanate. (Vacuoles show low levels of red autofluorescence.) The scale bar represents 5 μm in this and subsequent figures.
Cells were induced for expression of wt Ty3-RFP and nine mutants for 6 h at 24°C prior to examination. Two independent transformants were tested for each mutant. For wt and mutant K271A/K272A/E273A, Ty3-RFP colocalized with Dhh1-GFP for two independent transformants (Fig. 6 and data not shown). Cells expressing the single zinc-binding motif substitution mutants, C267A and C270A and double substitution mutants, C267A/C280A and C270A/C280A, all of which showed reduced Gag3 processing, and RNA protection, generated notably smaller fluorescent clusters. However, Ty3-RFP and Dhh1-GFP fluorescent foci when detectable colocalized (Fig. 6 and data not shown). Cells expressing zinc-binding motif mutants H275A and C280A were similar to the wt in both size of Ty3-RFP clusters and colocalization with Dhh1-GFP.
Cells expressing the ΔNTD mutant had a significant fraction of Ty3-RFP foci with which the P-body marker did not colocalize, and for cells expressing the ΔNC mutant this was the case for the majority of Ty3-RFP foci (Table 3). These findings showed that NC is not essential for cluster formation but contributes to Ty3 protein association with P-body components.
TABLE 3.
Ty3-RFP and Dhh1-GFP localization in cells expressing NC deletion mutants
| wt or mutant | No. of foci
|
Foci not coincident (%) | |||||
|---|---|---|---|---|---|---|---|
| Total Ty3 foci | Coincident foci | Ty3 focia | Adjacent | Overlapping | Discrete | ||
| wt | 107 | 105 | 0 | 0 | 2 | 0 | 1.9 |
| ΔNTD | 121 | 81 | 5 | 3 | 9 | 23 | 33.1 |
| ΔNC | 108 | 45 | 3 | 6 | 4 | 50 | 58.3 |
These clusters were in cells that did not display accumulation of the P-body marker in fluorescent foci.
Localization of Ty3 RNA and P bodies.
We next determined the effect of mutations in the NC domain on localization of Ty3 RNA with P bodies. These experiments were designed similarly to those for localization of mutant Ty3 NC proteins except that Ty3 RNA was marked downstream of POL3 with two copies of the MS2 bacteriophage coat protein binding site (Ty3-MS2) and visualized by interaction with the MS2 phage capsid RNA-binding protein fused to RFP (MS2-RFP) (4). Cells were induced to express Ty3-MS2 for 6 h at 24°C and were examined by fluorescence microscopy.
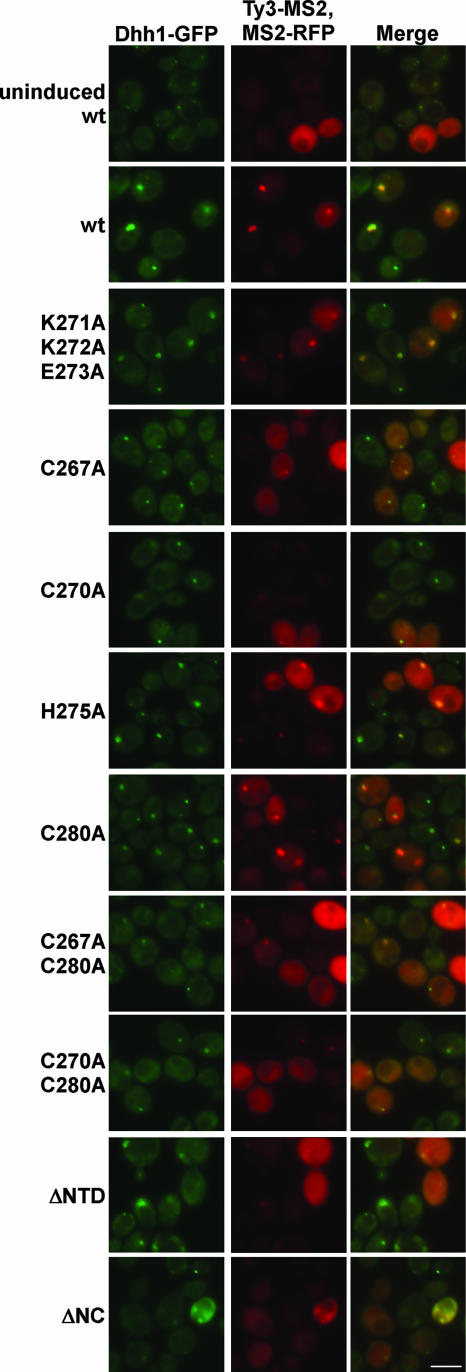
As previously reported, MS2-RFP fluorescence was diffuse, and Dhh1-GFP foci were small and scattered in cells not expressing Ty3-MS2 (4) (Fig. 7). Fluorescence imaging of cells expressing wt Ty3-MS2, MS2-RFP, and Dhh1-GFP showed one or two large, bright foci in which RFP and GFP fluorescence colocalized. The basic mutant K271A/K272A/E273A produced a similar pattern. Ty3-MS2 mutants containing substitutions C267A, C270A, and C267A/C280A produced significantly fewer and smaller Ty3-MS2 and Dhh1 foci. The C280A and C270A/C280A mutants also showed reduced numbers and size of foci, but they were more similar to wt and Ty3-MS2 and Dhh1 foci colocalized. Mutant H275A was most similar to the wt (Fig. 7). Cells expressing ΔNTD and ΔNC mutants displayed minimal Ty3-MS2 foci, and Dhh1-GFP foci were similar to those in cells not expressing Ty3. Thus, Ty3-MS2 RNA clusters, similar to Ty3-RFP protein clusters, were very sensitive to mutations at positions 267 and 270 of the zinc-binding motif. In cells expressing the ΔNTD and ΔNC mutants, Ty3-MS2 foci were also minimal (Fig. 7 and unpublished data).
FIG. 7.
NC function is required for Ty3 RNA cluster formation. NC mutations were introduced to pTy3-MS2 and yeast strain BY4741 expressing Dhh1-GFP was transformed with pTy3-MS2 and pMS2-RFP and induced for 6 h at 24°C. Control cells were not induced. The mutants shown are representative of two independent transformants of each type. Fluorescence imaging was performed with exposure times of 2,000 ms for TRITC and 2,500 ms for fluorescein isothiocyanate.
Nuclear localization of Gag3 in cells expressing specific NC mutants.
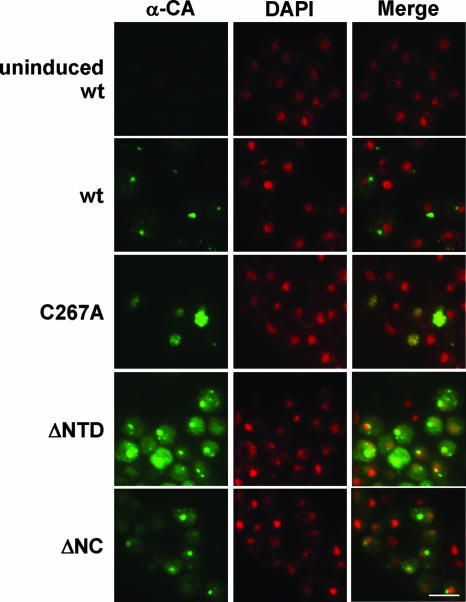
Because our RFP reporter was fused to Gag3-Pol3, we also used immunofluorescence with anti-CA antibodies to directly examine the localization of Gag3 in cells expressing the NC mutants (Fig. 8 and data not shown). Cells were induced to express wt and mutant Ty3 for 6 h at 24°C (4). As described previously, a majority of cells expressing wt Ty3 showed accumulation of CA in one or two large foci per cell, which were separate from the nuclear DAPI (4′,6′-diamidino-2-phenylindole) fluorescence. In contrast, cells expressing the C267A mutant showed diffuse anti-CA nuclear fluorescence and an irregular nuclear boundary. Cells expressing the other zinc-binding motif mutants, ΔNTD, and ΔNC mutants showed one to three Gag3 foci and some cytoplasmic Gag3. In about one-third of cells, one focus overlapped with DAPI nuclear staining (Fig. 8 and data not shown). In cells where fluorescence indicated the presence of the CA domain, the majority of the nuclei contained diffuse fluorescence. Because the Ty3-RFP reporter did not directly measure Ty3 Gag3-Pol3 species, we also used anti-IN in immunofluorescence experiments to monitor the localization of the IN domain. Although differential sensitivity of the antibodies used did not allow us to conclude that there is no Gag3-Pol3 in the nucleus, IN was detected in patterns similar to that of Ty3-RFP and was not detected in the nuclei of cells expressing the NC mutants (data not shown). Thus, mutations that disrupted Gag3 binding to Ty3 RNA resulted in the nuclear localization of Gag3 but not of Gag3-Pol3.
FIG. 8.
Loss of NC function results in nuclear localization of Gag3. NC mutations were introduced to Ty3 in pDLC201, and it was transformed into yeast strain yTM443. Cells were induced for 6 h at 24°C as described in Materials and Methods. Control cells were not induced. Ty3 Gag3 was visualized by immunofluorescence with rabbit polyclonal anti-CA and goat anti-rabbit IgG conjugated to Alexa 488 (green). DNA was stained with DAPI to visualize the nucleus (pseudo-colored red). Fluorescence imaging was performed with exposure times of 280 ms for Alexa 488 and 300 ms for DAPI.
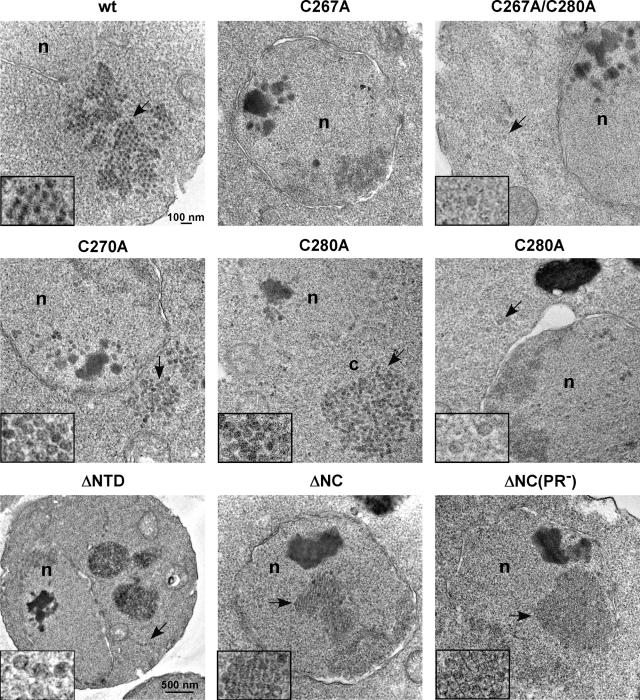
EM visualization of particle formation by NC mutants.
The analysis described above indicated that each of the eight mutants that was defective in transposition was competent to form some type of complex. Nonetheless, various degrees of difference from the wt were observed in the patterns of RNA and protein localization and RNA packaging. In addition, it appeared that in some mutants, localization of the Gag3-Pol3 RFP marker and Gag3 differed. In order to better understand the nature of the assembly defects, cells expressing a subset of these mutants were examined by EM. Cells expressing wt Ty3 and its mutant derivatives were induced for 6 h at 24°C and were processed as described previously (33). Ty3 wt, zinc-binding motif mutants C267A, C270A, C280A, C267A/C280A, and mutants ΔNTD, ΔNC, and ΔNC(PR−) were examined (Fig. 9). One hundred cells were examined for each mutant type. Cells expressing wt Ty3, as previously described (33), showed large multilobed clusters of particles averaging 40.9 ± 4.0 nm in diameter with electron-dense centers. These correlated well with the multilobed appearance of large fluorescent foci in cells expressing Ty3-RFP. Mutant C267A and mutant C267A/C280A were severely defective in both protein processing, RNA protection, and Ty3 clustering with P bodies. Expressing cells showed accumulation of electron-dense nuclear material. Multiple round patches with poorly defined edges that ranged from 37 to about 80 nm in diameter were also observed in these nuclei. Scattered, more discrete, particle-like forms were observed outside nuclei (Fig. 9 and data not shown). Measurement of seven of these particle-like forms in cells expressing mutant C267A showed that the diameter was close to that of wt Ty3 VLP at 40.2 ± 3.1 nm. However, these lacked the dense interior of wt particles. Despite the occurrence of unusual, apparently membrane-related structures in cells expressing wt Ty3, dense unstructured nuclear aggregates were not observed and nuclear particles similar to Ty3 particles were extremely rare. Cells expressing C270A showed types of molecular defects similar to those of cells expressing C267A and C267A/C280A but were less severely affected. The electron micrographs showed dense aggregates and associated round dense forms in nuclei similar to those in cells expressing C267A. However, cells expressing C270A differed in that the continuum of forms was shifted away from aggregates and unstructured, round patches toward particles that lacked dense centers but did have defined edges. The cytoplasm of a fraction of these cells showed distinct clusters of particles. Mutant C280A was the least severely affected of the zinc-binding motif mutants, and dense nuclear aggregates were clearly associated with particulate forms. This reinforced the previous impression that the dense nuclear aggregates formed as a result of Gag3 accumulation and could represent a precondensation stage of particle morphogenesis.
FIG. 9.
EM analysis reveals different Ty3 particle morphologies and localization controlled by NC function. BY4741 cells were transformed with the wt and Ty3 NC mutants, grown to log phase, and induced for Ty3 expression for 6 h at 24°C. Cultures were processed as described in Materials and Methods. Cells were imaged at 100 kV and ×10,000 direct magnification, except for the ΔNTD mutant, which was expressed in strain yTM443 and which was imaged at ×5,000 direct magnification. Abbreviations: n, nucleus; c, cytoplasm. All inset images are adjusted to ×24,000 magnification. Arrows indicate inset particles.
Cells expressing mutants ΔNTD and ΔNC were the only ones in which Ty3-RFP foci formed and were not consistently associated with P bodies. Ty3-MS2 foci were generally absent. Dense material and a few particles were observed in the nucleoplasm of these cells. A minority of cells showed small groups of cytoplasmic particles and individual particles lacking dense centers. However, a fraction of these cells also showed paracrystalline arrays. The diameter of 15 of the ΔNC particles was 40.7 ± 2.6 nm, indicating that they were also similar in diameter to wt VLPs. These also occurred in cells expressing the ΔNC(PR−) mutant. Taken together, these findings indicated that Ty3 can assemble into relatively stable, although not demonstrably separable, particles in the absence of the entire NC.
DISCUSSION
Cells expressing Ty3 form large clusters of Ty3 RNA, protein, and VLPs in association with P-body proteins. We have hypothesized that this effectively sequesters Ty3 components from the translation apparatus and concentrates them for assembly (4). We show here that the NC domain is a critical contributor to this localization of Ty3 proteins and RNA with P-body components (Table 2) and to the assembly process.
NC is required for wt Ty3 RNA cluster formation and association of Ty3 proteins with P bodies but not for protein cluster formation.
The basic residues and zinc-binding domain of NC mediate RNA interactions of the Gag proteins of most retroviruses (reviewed in references 12 and 17). Ty3 mutated at each of the zinc motif positions failed to protect genomic RNA, indicating that similar to retroviruses, NC, Ty3 NC plays a major role in genome packaging.
The NC domain of Ty3 plays an important role in the colocalization of Ty3 RNA and protein and P-body components. Ty3 lacking significant portions of NC (ΔNTD or ΔNC) did not form RNA clusters and formed protein clusters that failed to associate consistently with P-body components (Table 3). In addition, Ty3 elements with mutations at specific positions of NC (C267 or C270) failed to form RNA clusters and cytoplasmic Ty3-RFP clusters and instead localized Gag3 in the nucleus. A recent study of HIV-1 Gag localization similarly found that mutations in the zinc-binding motif or deletion of NC resulted in diffuse or mislocalized HIV RNA and Gag and even aberrant nuclear localization of Gag (25). These findings show that if the structural protein is not competent for interaction with genomic RNA, neither it nor the RNA are delivered to the assembly site efficiently. Thus, a ribonucleoprotein complex must form prior to arrival of protein and genomic RNA at the assembly site.
In the present study, we systematically disrupted short patches of charged residues within Ty3 NC but did not identify a mutant that failed to transpose. In contrast, basic residues in the I domain of retroviral NC contribute to nonspecific RNA binding which promotes assembly. Substitution of basic residues in MLV (27), RSV (6, 36), and HIV-1 (11, 58) NC disrupt Gag interactions and the formation of infectious particles. In RSV a leucine zipper dimerization interface can substitute for the NC domain in assembly, indicating that interaction of Gag with RNA facilitates multimerization (28). Therefore, if Ty3 has a basic domain that plays a role similar to that of the retrovirus I domain, multimerization must either involve redundant basic patches or be more extensive and resistant to disruption.
A set of mutations (K271A/K272A/E273A), located within the subdomain spanned by the zinc-binding motif, which did not decrease Ty3 RNA protection reduced the processing and cDNA levels (Table 2). Interestingly, Ty3 K271 is in the same position within the zinc-binding motif as the K30A substitution in MLV NC, which impaired cDNA production and RT processivity (22, 78). In vitro studies indicated that Ty3 NC anneals primer tRNA (20) and forms reverse transcription-competent complexes with Ty3 RT (14). Thus, it is possible that these residues in Ty3 also play a specific role in reverse transcription.
The Ty3 NC domain was required for wt VLP formation but, in the absence of NC, paracrystalline particle arrays formed and Gag3 and Gag3-Pol3 processing were observed. Dependence upon NC for assembly appears to differ among retroviruses. In the case of MLV, deletion of the NC domain blocked significant particle formation (49). In contrast, HIV-1 lacking the NC domain does form particles, but they are unstable in the presence of wt PR (52, 72). Not only were Ty3 ΔNTD and ΔNC mutant proteins produced at significant levels and processed, but mutating the Ty3 PR catalytic site in ΔNTD and ΔNC mutants did not significantly increase protein levels or, in the case of ΔNC, dramatically change the frequency of particles observed by EM (data not shown). These observations are consistent with a greater role for the Ty3 CA domain in particle stability than is played by the retroviral CA domain and also with our inability to identify a conventional I domain.
Substitution of zinc-binding residues causes pleiotropic particle assembly defects.
Each substitution in the Ty3 NC zinc-binding motif caused the loss of genomic RNA protection, a finding consistent with a role for this motif in packaging, as has been described for retroviruses and as was predicted in our previous study (51). However, there was also evidence for a distinct function of the zinc-binding motif. Substitution of Cys with Ala at position 267 and, to a lesser extent, at position 270, showed effects that were different from those resulting from substitutions at the two other positions of the zinc-binding motif (Table 2). These included the loss of Ty3 RNA and protein and P-body clustering and the failure to assemble VLPs and process proteins. These additional defects are therefore presumed to arise from changes distinct from loss of zinc binding and genomic RNA packaging. Despite the accumulation of dense nuclear material, C267A and C270A substitutions seem unlikely to have caused immediate global misfolding of Gag3. Improperly folded protein would be expected to be degraded, and C267A and C270A Gag3 were even competent for nuclear entry. Because we observed assembly of the ΔNC mutant into paracrystalline arrays, NC is not required for protein association per se. In cells expressing the C267A and, to a lesser extent the C270A mutant, Gag3 was concentrated in nuclei and electron-dense spheres, slightly larger than VLPs, were observed (Fig. 8 and 9). Thus, these mutations affected P-body association and particle condensation. We propose that NC could function as a conformational switch and that the state of conformation controls aspects of Gag3 cellular localization and assembly.
Ty3 assembly has some parallels with retrovirus assembly. First, similar to retroviruses, the Ty3 zinc-binding motif is not essential for particle formation. Substitution of two of the four zinc-binding residues did not disrupt particle formation or completely eliminate processing, which is consistent with the effects of analogous mutations in the single zinc motif in MLV (23, 24, 49) and substitutions or specific deletions of the two motifs in other retroviruses (see, for example, references 16, 18, 44, and 69). Second, similar to HIV-1 NC, Ty3 NC contains a domain that controls particle condensation. An extensive mutagenesis of the basic residues in the HIV-1 NC domain implicated nonbasic residues in a function required for wt particle density (11). It is possible that the condensation function lacking in mutant C267A corresponds to this function. Third, in retroviruses and in Ty3, there is evidence for multiple functions of residues in the zinc-binding domain. Substitutions of zinc-binding motif residues in RSV NC produced more severe effects than did deletion of the entire motif and substitutions at different positions of the second motif differed with respect to the severity of effect (35). Fourth, in both systems, zinc-binding motif substitutions can disrupt assembly at late stages (37, 49).
Mutations in NC cause accumulation of nuclear Gag3.
Cells expressing each of the zinc-binding motif substitution mutants and other mutants affected in Ty3 RNA packaging localized Gag3 in the nucleus. There are several possible explanations of this phenotype. The first possibility is that Gag3 nuclear accumulation in cells expressing RNA-binding mutants reflects only aberrant nuclear localization activity. For example, Gag3 could be normally retained by association with P bodies, Ty3 RNA binding, and Gag3 multimerization, and in the absence of this, diffuse into the nucleus and become trapped by multimerization and aggregation. It is interesting that, similar to Ty3, RSV and simian foamy virus both lack N-terminal myristylation (12, 75), which targets newly made Gag to membranes, and both are found in nuclei soon after synthesis (62, 64). Furthermore, in cells expressing HIV-1 psi− mutants, some nuclear localization of Gag is also observed that does not occur in cells expressing psi+ RNA (38). In a recent HIV-1 study, the wt patterns of both Gag and genomic RNA trafficking was dependent upon the zinc-binding motif, and the loss of normal trafficking was associated with some nuclear localization of Gag (25). Together, these observations suggest that reduced cytoplasmic retention contributes to nuclear localization of Gag in both natural and nonphysiologic contexts.
The second possibility is that Gag3 normally traffics to the nucleus and binds genomic RNA prior to export. If Gag3 normally localizes to the nucleus to bind genomic RNA, this localization is transient, since it is not observed even at early times in cells expressing wt Ty3 (unpublished data). Alternatively, it is possible that Gag3 binds the RNA in cis early during expression, but at later times, as Gag3 accumulates, the equilibrium shifts toward nuclear localization. This would be consistent with the combination of cis- and trans-packaging of RNA we observed in genetic assays (59; unpublished data). As mentioned above, both RSV and simian foamy virus Gag proteins are found in the nucleus, and nuclear localization signals were identified in NC (9) and the GRII carboxyl-terminal domain of Gag (77), respectively. However, in neither case has disruption of nuclear localization been shown to completely block infection of cells in culture (62, 64). Therefore, although it is attractive to speculate that this contributes to RNA packaging, other mechanisms must operate as well.
The third possibility is that Gag3 normally mediates docking of Ty3 VLPs on the nuclear pore. Since neither VLPs nor high levels of Gag3 are typically observed within nuclei, either very few particles transit the pore or uncoating of mature particles occurs during transit. In the case of mutants that fail to bind RNA, free Gag3, possibly in a novel folding state, could associate with the pore, allowing premature nuclear entry. Once inside, mutant Gag3 could aggregate or multimerize and become trapped. A nuclear localization signal has been identified in Ty3 IN. However, although it is required for transposition, it has not been shown specifically to be required for nuclear entry of the preintegration complex. The potential for retroviral Gag subdomains to affect nuclear entry is still being explored. CA has been implicated in the ability of HIV-1 to infect nondividing cells (74).
There are precedents for observations of nuclear VLPs among retrotransposons. VLPs formed by wt copia in Drosophila are found in large clusters in the nuclei of expressing cells (45), although the site of assembly has not been reported. Our results bear striking resemblance to the results of mutagenesis of the S. pombe Tf1 element. Tf1 is expressed from a single ORF, and clusters of cytoplasmic particles are observed in a small percentage of cells. Introduction of a premature frameshift to allow production of a discrete Gag species resulted in accumulation of nuclear particles (70). A short basic NLS has been mapped to the amino-terminal region of Tf1 Gag (15). Nup124 is required for wt transposition and appearance of nuclear Gag in stationary cells associated with transposition, but this interaction is dispensable for nuclear entry of some assembly incompetent mutants (29), suggesting that normally particles, rather than free Gag, are pore associated. Although these Gag mutations also affect genomic RNA packaging (70), Tf1 does not contain a zinc-binding motif, complicating more extensive comparisons.
Our findings indicate that the Ty3 NC domain mediates clustering of Ty3 RNA and interactions of Ty3 with P bodies. The central role of NC supports the model that association of Ty3 with P-body components occurs during assembly. Mutations in the NC domain of Gag3 were associated with a complete spectrum of Gag3 condensation states, ranging from dense nuclear aggregates to fuzzy balls larger than VLPs, to paracrystalline arrays. Collectively, these properties argue that the NC domain acts as a molecular switch controlling conformational states of Gag3 that affect assembly and nuclear entry.
Acknowledgments
This research was supported by Public Health Service grants GM33281 to S.B.S. and GM68903 to G.W.H. This study was funded in part by funds to K.N. from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. N.B.-B. was supported by National Institutes of Health grant T32 AI07319.
We thank M. Oakes for assistance with fluorescence microscopy. We thank Vuk Kovacevic and Becky Irwin for technical assistance.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Adamson, C. S., and I. M. Jones. 2004. The molecular basis of HIV capsid assembly: five years of progress. Rev. Med. Virol. 14107-121. [DOI] [PubMed] [Google Scholar]
- 2.Bacharach, E., J. Gonsky, K. Alin, M. Orlova, and S. P. Goff. 2000. The carboxy-terminal fragment of nucleolin interacts with the nucleocapsid domain of retroviral Gag proteins and inhibits virion assembly. J. Virol. 7411027-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basyuk, E., T. Galli, M. Mougel, J. M. Blanchard, M. Sitbon, and E. Bertrand. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 5161-174. [DOI] [PubMed] [Google Scholar]
- 4.Beliakova-Bethell, N., C. Beckham, T. Giddings, Jr., M. Winey, R. Parker, and S. Sandmeyer. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 1294-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett, R. P., T. D. Nelle, and J. W. Wills. 1993. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus gag proteins. J. Virol. 676487-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 729034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandt, S., M. Blissenbach, B. Grewe, R. Konietzny, T. Grunwald, and K. Uberla. 2007. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog. 3518-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butsch, M., and K. Boris-Lawrie. 2002. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 763089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butterfield-Gerson, K. L., L. Z. Scheifele, E. P. Ryan, A. K. Hopper, and L. J. Parent. 2006. Importin-beta family members mediate alpharetrovirus gag nuclear entry via interactions with matrix and nucleocapsid. J. Virol. 801798-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cimarelli, A., and J. Luban. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 735388-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 743046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffin, J. M., S. H. Hughes, and H. E. Varmus (ed.). 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- 13.Craven, R. C., A. E. Leure-DuPree, C. R. Erdie, C. B. Wilson, and J. W. Wills. 1993. Necessity of the spacer peptide between CA and NC in the Rous sarcoma virus Gag protein. J. Virol. 676246-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cristofari, G., C. Gabus, D. Ficheux, M. Bona, S. F. Le Grice, and J. L. Darlix. 1999. Characterization of active reverse transcriptase and nucleoprotein complexes of the yeast retrotransposon Ty3 in vitro. J. Biol. Chem. 27436643-36648. [DOI] [PubMed] [Google Scholar]
- 15.Dang, V. D., and H. L. Levin. 2000. Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p. Mol. Cell. Biol. 207798-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorfman, T., J. Luban, S. P. Goff, W. A. Haseltine, and H. G. Gottlinger. 1993. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 676159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Souza, V., and M. F. Summers. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3643-655. [DOI] [PubMed] [Google Scholar]
- 18.Dupraz, P., S. Oertle, C. Meric, P. Damay, and P. F. Spahr. 1990. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J. Virol. 644978-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed, E. O. 2002. Viral late domains. J. Virol. 764679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabus, C., D. Ficheux, M. Rau, G. Keith, S. Sandmeyer, and J.-L. Darlix. 1998. The yeast Ty3 retrotransposon contains a 5′/3′ bipartite primer binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO. J. 174873-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garfinkel, D. J., J. D. Boeke, and G. R. Fink. 1985. Ty element transposition: reverse transcriptase and virus-like particles. Cell 42507-517. [DOI] [PubMed] [Google Scholar]
- 22.Gonsky, J., E. Bacharach, and S. P. Goff. 2001. Identification of residues of the Moloney murine leukemia virus nucleocapsid critical for viral DNA synthesis in vivo. J. Virol. 752616-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorelick, R. J., D. J. Chabot, D. E. Ott, T. D. Gagliardi, A. Rein, L. E. Henderson, and L. O. Arthur. 1996. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J. Virol. 702593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA 858420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grigorov, B., D. Decimo, F. Smagulova, C. Pechoux, M. Mougel, D. Muriaux, and J. L. Darlix. 2007. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen, L. J., D. L. Chalker, K. J. Orlinsky, and S. B. Sandmeyer. 1992. Ty3 GAG3 and POL3 genes encode the components of intracellular particles. J. Virol. 661414-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Housset, V., H. De Rocquigny, B. P. Roques, and J. L. Darlix. 1993. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J. Virol. 672537-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, M. C., H. M. Scobie, Y. M. Ma, and V. M. Vogt. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 7611177-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, M. K., K. C. Claiborn, and H. L. Levin. 2005. The long terminal repeat-containing retrotransposon Tf1 possesses amino acids in gag that regulate nuclear localization and particle formation. J. Virol. 799540-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinsey, P., and S. Sandmeyer. 1995. Ty3 transposes in mating populations of yeast: a novel transposition assay for Ty3. Genetics 13981-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirchner, J., and S. B. Sandmeyer. 1993. Proteolytic processing of Ty3 proteins is required for transposition. J. Virol. 6719-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuznetsov, Y. G., M. Zhang, T. M. Menees, A. McPherson, and S. Sandmeyer. 2005. Investigation by atomic force microscopy of the structure of Ty3 retrotransposon particles. J. Virol. 798032-8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen, L. S., M. Zhang, N. Beliakova-Bethell, V. Bilanchone, A. Lamsa, K. Nagashima, R. Najdi, K. Kosaka, V. Kovacevic, J. Cheng, P. Baldi, G. W. Hatfield, and S. Sandmeyer. 2007. Ty3 capsid mutations reveal early and late functions of the amino-terminal domain. J. Virol. 816957-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeBlanc, J. J., S. Uddowla, B. Abraham, S. Clatterbuck, and K. L. Beemon. 2007. Tap and Dbp5, but not Gag, are involved in DR-mediated nuclear export of unspliced Rous sarcoma virus RNA. Virol. 363376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, E. G., A. Alidina, C. May, and M. L. Linial. 2003. Importance of basic residues in binding of Rous sarcoma virus nucleocapsid to the RNA packaging signal. J. Virol. 772010-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, E. G., and M. L. Linial. 2004. Basic residues of the retroviral nucleocapsid play different roles in gag-gag and Gag-Psi RNA interactions. J. Virol. 788486-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, E. G., and M. L. Linial. 2006. Deletion of a Cys-His motif from the alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects. Virol. 347226-233. [DOI] [PubMed] [Google Scholar]
- 38.Levesque, K., M. Halvorsen, L. Abrahamyan, L. Chatel-Chaix, V. Poupon, H. Gordon, L. DesGroseillers, A. Gatignol, and A. J. Mouland. 2006. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic 71177-1193. [DOI] [PubMed] [Google Scholar]
- 39.Levin, J. G., and M. J. Rosenak. 1976. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. USA 731154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lingappa, J. R., J. E. Dooher, M. A. Newman, P. K. Kiser, and K. C. Klein. 2006. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J. Biol. Chem. 2813773-3784. [DOI] [PubMed] [Google Scholar]
- 41.Liu, B., R. Dai, C. J. Tian, L. Dawson, R. Gorelick, and X. F. Yu. 1999. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J. Virol. 732901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma, Y. M., and V. M. Vogt. 2004. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 7852-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menees, T. M., and S. B. Sandmeyer. 1994. Transposition of the yeast retroviruslike element Ty3 is dependent on the cell cycle. Mol. Cell. Biol. 148229-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meric, C., E. Gouilloud, and P. Spahr. 1988. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His boxes. J. Virol. 623228-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyake, T., N. Mae, T. Shiba, and S. Kondo. 1987. Production of virus-like particles by the transposable genetic element, copia, of Drosophila melanogaster. Mol. Gen. Genet. 20729-37. [DOI] [PubMed] [Google Scholar]
- 46.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20395-425. [DOI] [PubMed] [Google Scholar]
- 47.Mortara, R. A., and G. L. Koch. 1989. An association between actin and nucleocapsid polypeptides in isolated murine retroviral particles. J. Submicrosc. Cytol. Pathol. 21295-306. [PubMed] [Google Scholar]
- 48.Murcia, P. R., F. Arnaud, and M. Palmarini. 2007. The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of Jaagsiekte sheep retrovirus Gag. J. Virol. 811762-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muriaux, D., S. Costes, K. Nagashima, J. Mirro, E. Cho, S. Lockett, and A. Rein. 2004. Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 7812378-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nikolaitchik, O., T. D. Rhodes, D. Ott, and W. S. Hu. 2006. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J. Virol. 804691-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orlinsky, K. J., and S. B. Sandmeyer. 1994. The Cys-His motif of Ty3 NC can be contributed by Gag3 or Gag3-Pol3 polyproteins. J. Virol. 684152-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, K. Nagashima, R. C. Sowder, D. T. Poon, and R. J. Gorelick. 2003. Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J. Virol. 775547-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perlman, M., and M. D. Resh. 2006. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic 7731-745. [DOI] [PubMed] [Google Scholar]
- 54.Resh, M. D. 2005. Intracellular trafficking of HIV-1 Gag: how Gag interacts with cell membranes and makes viral particles. AIDS Rev. 784-91. [PubMed] [Google Scholar]
- 55.Rey, O., J. Canon, and P. Krogstad. 1996. HIV-1 Gag protein associates with F-actin present in microfilaments. Virol. 220530-534. [DOI] [PubMed] [Google Scholar]
- 56.Ribet, D., F. Harper, M. Dewannieux, G. Pierron, and T. Heidmann. 2007. Murine MusD retrotransposon: structure and molecular evolution of an “intracellularized” retrovirus. J. Virol. 811888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 58.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55(Gag). J. Virol. 747238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sandmeyer, S. B., M. Aye, and T. M. Menees. 2002. Ty3: a position-specific, gypsylike element in Saccharomyces cerevisiae, p. 663-682. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 60.Scarlata, S., and C. Carter. 2003. Role of HIV-1 Gag domains in viral assembly. Biochim. Biophys. Acta Biomembr. 161462-72. [DOI] [PubMed] [Google Scholar]
- 61.Schafer, A., H. P. Bogerd, and B. R. Cullen. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328163-168. [DOI] [PubMed] [Google Scholar]
- 62.Scheifele, L. Z., R. A. Garbitt, J. D. Rhoads, and L. J. Parent. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 993944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schiestl, R. H., and D. R. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16339-346. [DOI] [PubMed] [Google Scholar]
- 64.Schliephake, A. W., and A. Rethwilm. 1994. Nuclear-localization of foamy virus Gag precursor protein. J. Virol. 684946-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sfakianos, J. N., R. A. LaCasse, and E. Hunter. 2003. The M-PMV cytoplasmic targeting-retention signal directs nascent Gag polypeptides to a pericentriolar region of the cell. Traffic 4660-670. [DOI] [PubMed] [Google Scholar]
- 66.Sherman, F. 1998. Getting started with yeast, p. 3-21. In C. Guthrie and G. R. Fink (ed.), Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, CA.
- 67.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swanson, C. M., B. A. Puffer, K. M. Ahmad, R. W. Doms, and M. H. Malim. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 232632-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanchou, V., D. Decimo, C. Pechoux, D. Lener, V. Rogemond, L. Berthoux, M. Ottmann, and J. L. Darlix. 1998. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J. Virol. 724442-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teysset, L., V. D. Dang, M. K. Kim, and H. L. Levin. 2003. A long terminal repeat-containing retrotransposon of Schizosaccharomyces pombe expresses a Gag-like protein that assembles into virus-like particles which mediate reverse transcription. J. Virol. 775451-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Urban, A., S. Neukirchen, and K. E. Jaeger. 1997. A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res. 252227-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, S. W., K. Noonan, and A. Aldovini. 2004. Nucleocapsid-RNA interactions are essential to structural stability but not to assembly of retroviruses. J. Virol. 78716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilk, T., B. Gowen, and S. D. Fuller. 1999. Actin associates with the nucleocapsid domain of the human immunodeficiency virus Gag polyprotein. J. Virol. 731931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 785670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu, S.F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 2711579-1582. [DOI] [PubMed] [Google Scholar]
- 76.Yu, S. F., S. W. Eastman, and M. L. Linial. 2006. Foamy virus capsid assembly occurs at a pericentriolar region through a cytoplasmic targeting/retention signal in Gag. Traffic 7966-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu, S. F., K. Edelmann, R. K. Strong, A. Moebes, A. Rethwilm, and M. L. Linial. 1996. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J. Virol. 708255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang, W. H., C. K. Hwang, W. S. Hu, R. J. Gorelick, and V. K. Pathak. 2002. Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo. J. Virol. 767473-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]