Abstract
Bovine viral diarrhea virus (BVDV) is a positive-strand RNA virus and a member of the genus Pestivirus in the family Flaviviridae. To identify and characterize essential factors required for BVDV replication, a library expressing random fragments of the BVDV genome was screened for sequences that act as transdominant inhibitors of viral replication by conferring resistance to cytopathic BVDV-induced cell death. We isolated a BVDV-nonpermissive MDBK cell clone that harbored a 1.2-kb insertion spanning the carboxy terminus of the envelope glycoprotein 1 (E1), the envelope glycoprotein E2, and the amino terminus of p7. Confirming the resistance phenotype conferred by this library clone, naïve MDBK cells expressing this fragment were found to be 100- to 1,000-fold less permissive to both cytopathic and noncytopathic BVDV infection compared to parental MDBK cells, although these cells remained fully permissive to vesicular stomatitis virus. This restriction could be overcome by electroporation of BVDV RNA, indicating a block at one or more steps in viral entry prior to translation of the viral RNA. We determined that the E2 ectodomain was responsible for the inhibition to BVDV entry and that this block occurred downstream from BVDV interaction with the cellular receptor CD46 and virus binding, suggesting interference with a yet-unidentified BVDV entry factor.
Bovine viral diarrhea virus (BVDV) is a member of the genus Pestivirus within the family Flaviviridae. Pestiviruses, including BVDV, border disease virus, and classical swine fever virus, are important animal pathogens. BVDV causes both acute and persistent infections in cattle, leading to substantial financial losses within the livestock industry each year (reviewed in reference 28). The pestiviruses contain a positive-sense RNA genome of approximately 12.5 kb (reviewed in reference 28). The viral genome harbors a large open reading frame flanked by 5′ and 3′ nontranslated regions. Within the 5′ nontranslated region is an internal ribosome entry site (IRES), which drives cap-independent translation of the viral open reading frame, generating a polyprotein of approximately 4,000 amino acids. The polyprotein is co- and posttranslationally processed by a combination of viral and host proteases to produce the mature viral proteins Npro-C-Erns-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B.
BVDV virions possess by a lipid envelope harboring three envelope glycoproteins, Erns, E1, and E2 (40, 44). E2 forms disulfide-linked homo- and heterodimers with E1 and has been implicated in receptor binding, as major neutralizing antibodies are directed against the E2 protein in an infected host (8, 36, 45). CD46 has been identified as a cellular receptor for BVDV (30), and viral entry occurs via clathrin-mediated endocytosis in a pH-dependent manner (23, 26). p7 is a small, hydrophobic protein that, although not found within BVDV particles, is essential for virion assembly (10, 13, 15). The remainder of the polyprotein is composed of nonstructural (NS) proteins which, likely partnered with cellular proteins, form complexes responsible for replication of the viral RNA genome.
Two BVDV biotypes, noncytopathic (ncp) and cytopathic (cp), defined by their respective effects on tissue culture cells, can be isolated from cows suffering from mucosal disease (31, 33). ncp BVDV NADLJiv90− and cp BVDV NADL are an isogenic pair of infectious cDNA clones. BVDV NADL harbors a cellular insertion, Jiv90, upstream of the cleavage site between the NS2 and NS3 proteins, which renders this virus cp (32). Jiv90 is a 90-amino-acid domain of Jiv (J-domain protein interacting with viral protein) which is a cellular cofactor for the NS2 autoprotease. cis or trans overexpression of Jiv during BVDV infection has been shown to increase cleavage at the NS2/3 site (24).
Viruses are obligate parasites and, therefore, in addition to interactions between viral proteins and RNA, host factor interactions are essential to the BVDV life cycle. Biochemical techniques that use a viral protein as bait to identify such interactions often give little hint as to the functional relevance of the association. Alternatively, functional genomics is a powerful approach to detect virus-virus and virus-cell factor interactions. One such technique is based on the isolation of genetic suppressor elements, sequences derived from a gene or genome of interest that can act as transdominant inhibitors of a particular biological function, for example, by binding to and blocking an essential interaction surface (9, 17). This type of approach allows selective inhibition of specific RNA or protein interactions and identifies a probe to further study the targeted process. Libraries of sequences derived from viral genomes have been used to study various phenomena, including the phage lambda life cycle (17) and human immunodeficiency virus type 1 (HIV-1) latency (9). Cellular processes have also been examined by screening for cellular cDNA fragments that inhibit mammalian cell growth or provide resistance to cytotoxic drugs (29, 37, 49).
In this study, using a random fragment BVDV cDNA library, we identified a fragment of the viral genome containing the carboxy terminus of E1, E2, and the amino terminus of p7 that acts as a transdominant inhibitor of BVDV replication. Madin-Darby bovine kidney (MDBK) cells expressing the fragment, called MDBK-E1*E2p7* cells, were defective at the level of BVDV entry, although CD46 receptor expression and binding of BVDV to the cells were normal. Analysis of the MDBK-E1*E2p7* cell population will provide insight into the molecules required for BVDV entry into MDBK cells.
MATERIALS AND METHODS
Cells, virus stocks, transcriptions, and transfections.
MDBK cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated horse serum. 293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% gamma-irradiated fetal calf serum. Media and reagents for cell culture were purchased from Gibco-BRL, Life Technologies Ltd.
Stocks of BVDV strains NADL, NADLJiv90−, and NADLJiv90− luc were generated as previously described (32) by electroporation of MDBK cells using a BTX ElectroSquarePorator with RNA transcribed in vitro from pBAC/NADL, pACNR/NADLJiv90−, and pACNR/NADLJiv90−/5′ubi-luc, respectively. For in vitro transcription, plasmids were linearized by digestion with SbfI (Promega) and transcribed using the T7 MEGAscript kit (Ambion) according to the manufacturer's instructions. RNA was purified using an RNeasy Mini column (Qiagen) or by precipitation with 7.5 M NH4Ac and analyzed by agarose gel electrophoresis.
Construction of random BVDV library.
To prepare BVDV random cDNA fragments, 10 to 20 μg of pBAC/NADL in TM buffer (50 mM Tris pH 8, 15 mM MgCl2) was nebulized in a chamber (Invitrogen) by application of 40 lb/in2 N2 gas for 3 min. The fragments were cleaned using a QIAquick PCR purification kit (Qiagen) and eluted in TM buffer. To create blunt ends, the fragments were incubated with a combination of T4 DNA polymerase and Klenow in the presence of deoxynucleoside triphosphates. After heat inactivation of the enzymes, the fragments were phosphorylated using T4 polynucleotide kinase. The fragments were run on a 0.7% agarose gel and 0.2 to 2-kb fragments were purified using the QIAquick gel purification kit (Qiagen).
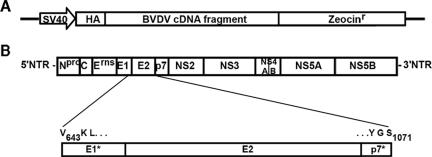
pBabeHAZSrfcI is a Moloney murine leukemia virus (MLV) expression vector which is derived from pBabeHAZ (12) and modified to contain a unique SrfI restriction site for cloning blunt-ended inserts. Inserts are expressed as fusion proteins with an amino-terminal influenza hemagglutinin (HA) epitope tag (YPYDVPDYA) and a carboxy-terminal zeocin resistance protein (Zeo) (Fig. 1A). In this vector, Zeo is out of frame with the start site of the HA tag and is not expressed unless the cloning site contains a stop codon-free insert to restore the open reading frame.
FIG. 1.
A genetic screen identified the BVDV E1*E2p7* sequence as a transdominant inhibitor of BVDV replication. (A) Illustration of the library expression cassette within the pBabeHAZSrfcI retroviral vector. A simian virus 40 promoter drives expression of the BVDV cDNA library fragment fused to an HA tag at the amino terminus and a zeocin-selectable marker at the carboxy terminus. (B) Sequence alignment of the E1*E2p7* library insertion with the BVDV genome. The brackets descending from the pictured BVDV genome highlight the BVDV sequence present in the resistant clones. Amino acid positions relative to the BVDV NADL polyprotein are indicated.
For library construction, SrfI-digested pBabeHAZSrfcI vector was ligated with the BVDV cDNA fragments. Ligation products were purified using the Minelute QIAquick PCR purification kit (Qiagen) and added to DH10B ElectroMAX cells (Gibco BRL). Cells were electroporated using a Bio-Rad Gene Pulser 2 and plated on Luria-Bertani agar plates supplemented with 100 μg/ml ampicillin. Colonies were scraped into Luria-Bertani broth containing ampicillin and grown for 5 h at 37°C. Plasmid DNA was isolated using a Plasmid Maxi kit (Qiagen).
Packaging and screening of the random BVDV library.
For packaging of the BVDV library, 293T cells, grown in gamma-irradiated serum to eliminate contaminating BVDV, were seeded in 100-mm plates (2 × 106 cells/plate) and incubated overnight. A 1.5-μg aliquot of BVDV library in pBabeHAZSrfcI was mixed with 1 μg MLV-gagpol and 1 μg of vesicular stomatitis virus G envelope protein (VSV-G) expression plasmids (12) in a total of 15 μl TE (10 mM Tris, 1 mM EDTA). The mixture was added to a tube containing 18 μl Fugene 6 (Roche) in 200 μl OptiMEM (Gibco). Complexes were allowed to form for 15 min at room temperature and then added to 293T cells in fresh medium. At 48 h posttransfection, supernatants were harvested and filtered through a 0.45-μm syringe filter and Polybrene was added at 8 μg/ml. For transduction, medium from MDBK cells, seeded in a six-well plate (5 × 105/well), was replaced with the retrovirus-containing supernatant, and the cells were spun in a centrifuge (1,150 × g) for 30 min at room temperature. The plates were transferred to a 37°C incubator for an additional 1.5 h, after which the inoculum was removed and culture medium was added. At 48 h posttransduction, selective medium containing 400 μg/ml zeocin was added to each well. Cells were maintained in selective medium for the duration of the experiments. A control cell population was also created by transduction with the pBabeHAZ vector (12), where HA and Zeo were in frame, followed by zeocin selection. Zeocin-resistant cells expressing the BVDV library or the empty vector, pBabeHAZ, were expanded and infected with BVDV NADL at a multiplicity of infection (MOI) of 5. Colonies that developed after infection were expanded and reinfected with NADL. Colonies that survived secondary infection were further analyzed.
Analysis of resistant MDBK clones.
The genomic DNA from 2.5 × 106 cells of each resistant clone was harvested using the DNeasy tissue kit (Qiagen), based on the manufacturer's protocol. The viral insert was amplified with the primers 5′-GCTTATCCATATGATGTTCCAGATT-3′ and 5′-GCACCGGAACGGCACTGGTCAACTT-3′ using the Expand High-Fidelity PCR system (Roche) with 10 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 1.5 min and 20 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 1 min (adding 5 s/cycle). PCR products were gel purified and sequenced. To test if the library inserts could provide resistance to naïve cells, the PCR products were cloned into pBabeHAZSrfcI using EcoRI and NotI restriction sites. The pBabeHAZSrfcI clones were packaged and used to transduce MDBK cells as above.
Construction of pLenti6-EGFP, pLenti6-E1*E2p7*, pLenti6-Sig-E2, and pLenti6-Sig-E2*.
pLenti6-EGFP is a derivative of pLenti6/GW-V5/LacZ (Invitrogen). The enhanced green fluorescent protein (EGFP) sequence was amplified from pEGFP (Invitrogen) using the EGFP-specific primers 5′-CGGGATCCACCACCATGGGGCGCGCCGGCGGAAGCGGCGGAAGCATGGTGAGCAAGGGCGAGGAGCTGTTCAC-3′ and 5′-GGCTCGAGTTACTTGTACAGCTCGTCCATGCCGAGAGTGATC-3′, which added BamHI and XhoI restriction sites to the 5′ and 3′ ends of the EGFP sequence, respectively. The BamHI-EGFP-XhoI cassette was cloned into pLenti6/GW-V5/LacZ, which had been digested with BamHI and XhoI. To generate pLenti6-E1*E2p7*, nucleotides 2312 to 3598 from the BVDV NADL genome were PCR amplified. The forward primer added a BamHI restriction site, a Kozak consensus sequence, and an ATG nucleotide sequence (5′-GGGGATCCGCCACCATGGTGAAGTTAGTGTTGAGGGCAC-3′). The reverse primer added a stop codon followed by a XhoI restriction site (5′-GGGGCTCGAGTTATGATCCATACTGAATCCCTAA-3′). The BamHI- E1*E2p7*-XhoI cassette was inserted into the pLenti6 backbone that had been digested with BamHI and XhoI. To generate pLenti6-Sig-E2, the full-length E2 sequence (nucleotides 2462 to 3583) was amplified using the forward primer 5′-CGGGATCCGCCACCATGAAGACTATCATTGCTTTGAGCTACATTTTCTGTCTGGTTCTCG-3′ that added a BamHI restriction site, a Kozak consensus sequence, and ATG, followed by the HA signal sequence (MKTIIALSYIFCLVLG) and a short linker region containing unique NheI and AscI restriction sites. The reverse primer, 5′-CTCGAGTTACCCTAAGGCCTT-3′, added a stop codon and an XhoI restriction site. The BamHI-Sig-E2-XhoI cassette was inserted into the pLenti6 backbone that had been digested with BamHI and XhoI. For pLenti6-Sig-E2*, nucleotides 3148 to 3490 were PCR amplified using 5′-GAACGAGACTGGTTACAGGCTA-3′ and 5′-GGCTCGAGTTAGGACTCAGCGAAGTAATCCCG-3′. The reverse primer added a stop codon and XhoI restriction site. The PCR product was digested with BsrGI and XhoI and cloned into pLenti6-Sig-E2 that had been digested with BsrGI and XhoI.
For packaging, 1.5 μg lentiviral vector was cotransfected into 293T cells with 1 μg HIV-gagpol and 1 μg VSV-G expression constructs (Invitrogen) using Fugene 6. MDBK cells were transduced as described above for the BVDV library. Transduced MDBK cells were selected with blasticidin (4 μg/ml) for 3 days and infected with NADLJiv90− luc.
BVDV binding assay.
MDBK-E1*E2p7* and pBabe cells, seeded in 12-well plates (2.5 × 105/well), were cooled on ice and infected with BVDV NADLJiv90− (MOI of 10) for 1 h at 4°C. Cell monolayers were washed seven times with cold culture medium, and RNA was extracted using the TRIzol reagent (Invitrogen) or the RNeasy mini kit (Qiagen). BVDV RNA levels were quantified by real-time quantitative reverse transcription-PCR (RT-PCR). For antibody blocking experiments, cells were preincubated with hybridoma supernatant from a mixture of monoclonal antibodies (MAb; BVD/CA 17, 26, and 27; kindly provided by Till Rümenapf, Justus-Liebig University, Germany) for 2 h at 37°C. Cells were then chilled on ice and infected with BVDV NADLJiv90− in the presence of antibody at 4°C as above.
Immunoblotting.
To detect the BVDV E2 protein in MDBK cells, cells were lysed in sodium dodecyl sulfate (SDS) lysis buffer (0.5% SDS, 50 mM Tris-Cl, pH 7.5, 1 mM EDTA). Proteins were separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a nitrocellulose membrane, and detected using anti-E2 MAb 214 (Veterinary Laboratories Agency, Weybridge, United Kingdom) and a horseradish peroxidase-conjugated anti-mouse secondary antibody (Sigma). Detection was performed with the SuperSignal West Femto maximum sensitivity substrate (Pierce), according to the manufacturer's instructions.
To detect CD46 protein in MDBK cells, cells were lysed as above in SDS lysis buffer. Proteins were separated by 10% SDS-PAGE, transferred to a nitrocellulose membrane, and detected using CD46 polyclonal antiserum (kindly provided by Till Rümenapf, Justus-Liebig University), diluted 1:200 in Tris-buffered saline with 10% horse serum, and horseradish peroxidase-conjugated anti-rabbit secondary antibody (Sigma). Detection was performed with the SuperSignal West Pico chemiluminescent substrate (Pierce).
BVDV plaque- and focus-forming assays.
MDBK cells, seeded in six-well dishes, were infected with 10-fold dilutions of virus for 1 h at 37°C. The cells were overlaid with 1% SeaKem LE agarose in minimal essential medium containing 5% horse serum and penicillin-streptomycin and incubated at 37°C for 3 days. Monolayers were fixed with 7% formaldehyde, and the agarose overlays were removed. For plaque assays, the monolayer was stained with crystal violet as previously described (32). For focus-forming assays, the monolayer was permeabilized with 0.25% Triton X-100 in phosphate-buffered saline (PBS) and stained with anti-BVDV polyclonal antibody B224 (kindly provided by Kenny Brock, Auburn University), diluted 1:500 in PBS-0.25% Triton X-100 and peroxidase-conjugated anti-bovine immunoglobulin G (Sigma), and diluted 1:1,000 in PBS-0.25% Triton X-100 (38). BVDV-positive foci were visualized using the 3,3′-diaminobenzidine peroxidase substrate (Vector Laboratories).
Immunostaining and flow cytometry.
For cell surface staining of CD46, cells were detached with PBS-0.5 mM EDTA and resuspended in 5% goat serum, 0.1% sodium azide in Dulbecco's PBS. Cells were incubated with primary antibody, anti-CD46, diluted in 0.5% bovine serum albumin, 0.1% sodium azide (SB), washed with SB, and then incubated with Alexa Fluor 488 or 633 goat anti-mouse immunoglobulin G secondary antibody (Molecular Probes, Eugene, OR). Cells were washed with SB, fixed with 2% paraformaldehyde, and analyzed by flow cytometry on a FACSCalibur (Becton Dickinson), counting 104 cells.
Luciferase assays.
Cells were washed once with Dulbecco's PBS and lysed with cell culture lysis buffer (Promega). Lysates were harvested by scraping and mixed with luciferase assay substrate (Promega), as suggested by the manufacturer. Luciferase activity was measured using a Lumat LB9507 luminometer (EG&G Berthold).
Real-time quantitative RT-PCR.
Real-time quantitative RT-PCR was performed as described elsewhere (27). Briefly, total RNA was extracted from cells using either TRIzol reagent (Invitrogen) or the RNeasy Mini kit (Qiagen). RNA was mixed with primers and probe specific for the BVDV 5′ nontranslated region as well as for bovine β-actin RNA to normalize total RNA levels. BVDV-specific RNA and bovine β-actin RNA were amplified using the Platinum quantitative RT-PCR ThermoScript one-step system (Invitrogen) and detected using the ABI Prism 7700 sequence detection system (PE Applied Biosystems, Foster City, CA).
RESULTS
Functional genomics approach to identify essential interactions in the BVDV life cycle.
Throughout the life cycle of an RNA virus, viral RNAs and proteins are closely associated and presumably interact intimately with cellular factors. To identify interactions required for BVDV replication, a random cDNA library of the BVDV genome was constructed. A plasmid encoding the BVDV NADL genome was sheared into fragments, which were cloned into the retroviral vector pBabeHAZSrfcI. Inserts were flanked by the coding sequence for an HA epitope prefaced by a start codon and by a zeocin resistance gene (zeo), 5′ and 3′ to the fragments, respectively (Fig. 1A). The final library encoded approximately 40,000 BVDV clones, with inserts ranging in size from 0.2 to 2 kb, averaging approximately 0.7 kb.
To screen the library for transdominant inhibitors of BVDV replication, Zeocin-resistant MDBK cells expressing the BVDV library were infected with cp BVDV NADL to isolate cells able to survive BVDV-induced cytopathic effect (CPE). Of the 20 library-transduced resistant colonies that survived BVDV challenge, 11 survived expansion, and only 3 remained resistant after reinfection with BVDV NADL. To determine the identity of the library inserts contained in the resistant clones, we harvested genomic DNA from these cells and amplified the retroviral insertion by PCR. Two of the three clones contained an insertion of approximately 1.2 kb, which encoded 150 bases of BVDV E1, the entire E2 gene, and 15 bases of p7 and was named E1*E2p7* (Fig. 1B). In addition to E1*E2p7*, one of these two cell clones also contained a 0.7-kb library fragment derived from the BVDV NS5A gene cloned into pBabeHAZSrfcI in the reverse orientation with respect to the BVDV genome. The same 0.7-kb insertion was also found alone in the third resistant clone. This sequence, however, could not protect naïve cells from BVDV infection (data not shown) and was therefore eliminated from further analysis.
Naïve cells expressing E1*E2p7* are resistant to homologous BVDV infection.
To test if E1*E2p7* could provide resistance to naïve cells, the sequence was recloned into pBabeHAZSrfcI and packaged into pseudoparticles used to transduce naïve MDBK cells. After selection with zeocin, the resulting cell population was called MDBK-E1*E2p7*. As a control, an MDBK-pBabe population, transduced with the empty vector pBabeHAZ, was also generated.
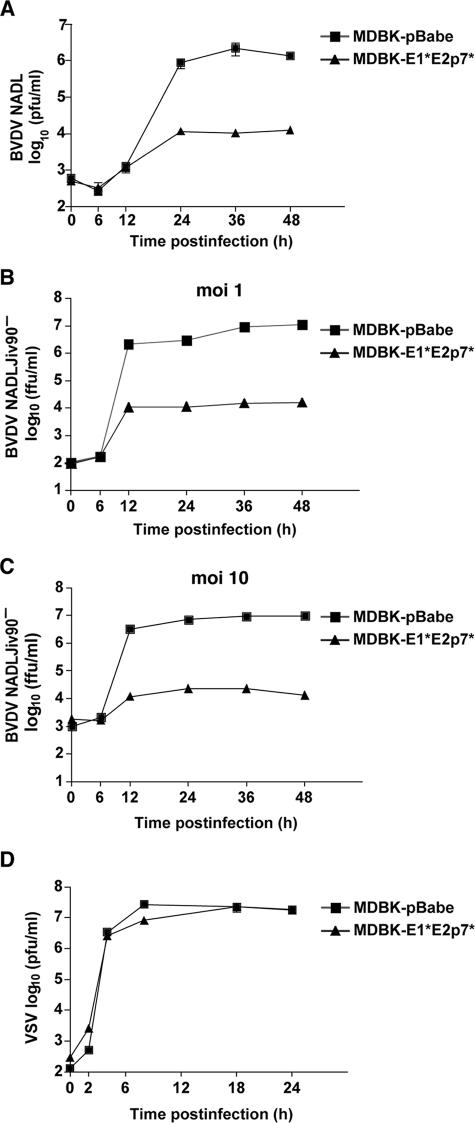
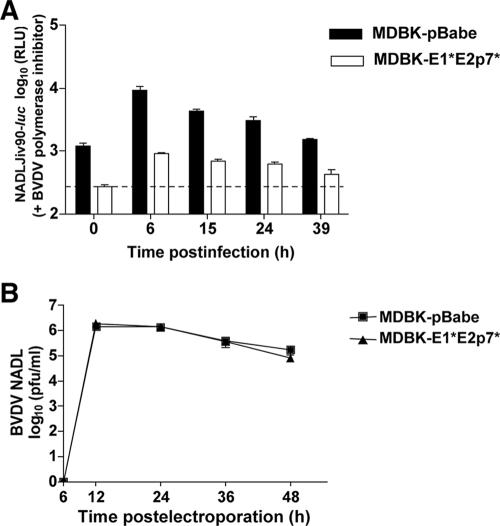
MDBK-E1*E2p7* and pBabe cells were infected with BVDV NADL, and cell supernatants were harvested at various time points to determine the viral titer. Over time, the BVDV titers obtained from MDBK-E1*E2p7* cells were 100- to 1,000-fold lower than from pBabe control cells (Fig. 2A). In addition, while total CPE was observed in the pBabe cells, there was minimal CPE in the E1*E2p7* cells (data not shown). A similar 1,000-fold difference in susceptibility to ncp BVDV NADLJiv90− between the control and E1*E2p7* cells was observed (Fig. 2B). Even a high-MOI infection (MOI of 10) with BVDV NADLJiv90− could not overcome the inhibition to BVDV replication in the E1*E2p7* cells (Fig. 2C). These data demonstrate that the E1*E2p7* insertion was responsible for the resistance observed in the original MDBK clones.
FIG. 2.
Cells expressing BVDV E1*E2p7* are resistant to homologous BVDV infection. (A) Cells transduced with a retroviral vector expressing the E1*E2p7* sequence, MDBK-E1*E2p7* cells (triangles), and MDBK-pBabe control cells (squares) were infected with BVDV NADL. Each point represents the mean titer of virus present in the supernatants of duplicate wells at the indicated time point postinfection. This graph shows representative data from at least two independent experiments. (B and C) MDBK-E1*E2p7* (triangles) and pBabe (squares) cells were infected with ncp BVDV NADLJiv90− at an MOI of 1 (B) or 10 (C), and supernatants were harvested as described above. Titers were determined by focus-forming assays performed in duplicate. (D) MDBK-E1*E2p7* cells are permissive for heterologous VSV infection. MDBK-E1*E2p7* (triangles) and pBabe (squares) cells were infected with VSV. Supernatants were harvested as described above, and titers were determined in duplicate by plaque assay. For all panels, error bars show the standard errors of the means.
To determine if the phenotype observed in the MDBK-E1*E2p7* cells was a BVDV-specific effect or a general virus inhibition, the E1*E2p7* and pBabe cells were infected with the interferon-sensitive rhabdovirus VSV. VSV was equally able to grow and form plaques on the E1*E2p7* and pBabe cells (Fig. 2D and data not shown). Therefore, the resistance in the MDBK-E1*E2p7* cells was not due to a general antiviral mechanism.
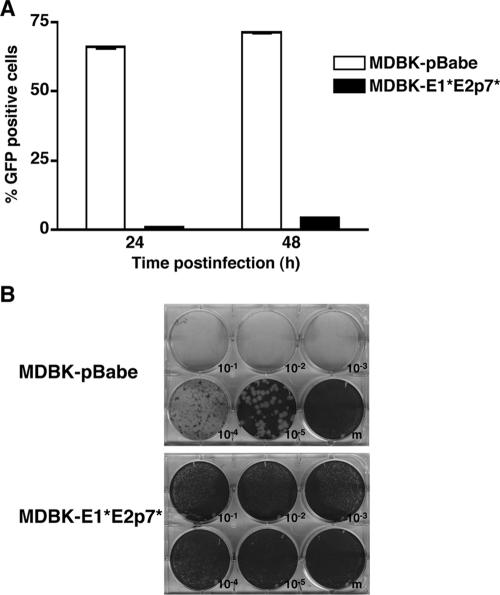
A small amount of virus could be detected in the supernatants of MDBK-E1*E2p7* cells after infection with either cp or ncp BVDV (Fig. 2A, B, and C). This could be due to a low level of virus infection in all of the E1*E2p7* cells or to infection of a subset of permissive cells in the population. To examine this further, we infected the E1*E2p7* and pBabe cells with NADLJiv90− GFP, an ncp BVDV that expresses GFP, and harvested the cells for flow cytometry. At 48 h postinfection (p.i.), only 4 to 5% of E1*E2p7* cells were GFP positive compared to more than 70% of pBabe cells (Fig. 3A). Since it is likely that even very weak replication would result in some detectable GFP signal, this result indicates that the majority of cells in the MDBK-E1*E2p7* cell population cannot support BVDV replication. Moreover, BVDV NADL could not form plaques on E1*E2p7* cell monolayers (Fig. 3B), suggesting that although some cells were infected, virus spread was inefficient due to a high proportion of nonpermissive cells.
FIG. 3.
The MDBK-E1*E2p7* cell population contains a small subset of permissive cells. (A) MDBK-E1*E2p7* and pBabe cells were infected with NADLJiv90− GFP. At 24 and 48 h p.i., cells were harvested for fluorescence-activated cell sorter analysis to determine the number of GFP-positive cells. Values are the averages of duplicate samples; error bars show the standard errors of the means. (B) Serial dilutions of BVDV NADL were used to infect monolayers of MDBK-E1*E2p7* or pBabe cells. The cells were then overlaid with agarose, incubated for 3 days, and stained with crystal violet.
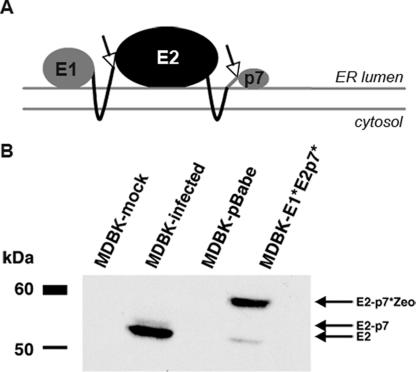
BVDV E2 is a heavily glycosylated, endoplasmic reticulum (ER)-resident, integral membrane protein which is cleaved from the BVDV polyprotein by host signal peptidase (15, 47) (Fig. 4A). To further characterize the MDBK-E1*E2p7* cells, we compared E2 protein expression in MDBK-E1*E2p7* cells to that from BVDV-infected MDBK cells by immunoblotting cell lysates with an anti-E2 antibody. BVDV-infected cell lysates displayed a dominant band between 50 and 60 kDa, corresponding to the molecular mass of the mature E2 protein, and a minor species migrating slightly slower, most likely due to the stable E2-p7 product in BVDV-infected cells (15) (Fig. 4B). Two bands were also detected from MDBK-E1*E2p7* cell lysates. The major band detected in these cells was of a higher molecular mass than in BVDV-infected cell lysates (Fig. 4B), likely representing the E2p7*-Zeo fusion protein. The presence of a minor species in MDBK-E1*E2p7* cells, migrating similarly to the molecular mass of the dominant band within the infected cell lysates, suggests that there is some processing of E2 at its carboxy terminus from the E1*E2p7* protein (Fig. 4B). In addition, the localization of E2 protein in E1*E2p7* cells was analogous to E2 expressed after BVDV infection, as judged by indirect immunofluoresence using an anti-E2 antibody and sensitivity of immunoprecipitated E2 to endoglycosidase H or PNGase F (data not shown).
FIG. 4.
E2 expression in MDBK-E1*E2p7* cells. (A) Putative membrane topology of BVDV E1*E2p7* (adapted from reference 41). The E2 glycoprotein is an ER-resident protein with a carboxy-terminal transmembrane anchor (right-hand black line) and a short carboxy-terminal tail (gray line). Arrows indicate sites of cleavage by host signal peptidase. (B) Lysates from MDBK-E1*E2p7*- and BVDV NADL-infected MDBK cells were assayed by Western blotting using an antibody against BVDV E2 (MAb 214). Lysates from mock-infected MDBK cells and pBabe cells were used as negative controls. Arrows indicate the expected identity of the protein in each band based on molecular mass.
Expression of the BVDV E2 ectodomain is sufficient for BVDV inhibition.
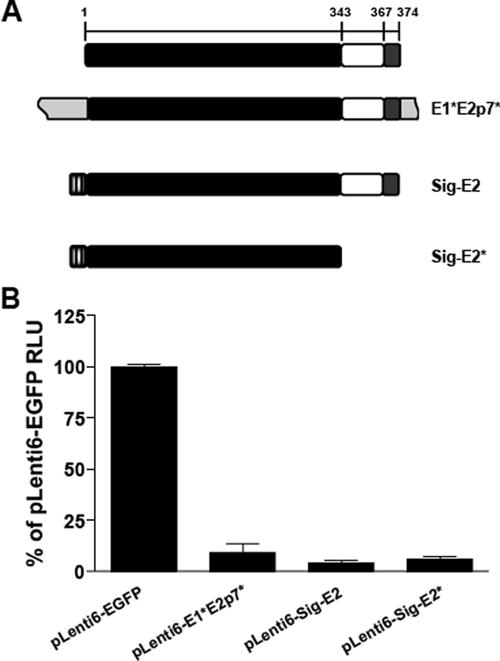
We sought to determine if E2 expression alone, in the absence of the E1* and p7* flanking sequences present in the MDBK-E1*E2p7* cells, was sufficient for BVDV inhibition. MDBK cells were infected with lentiviral pseudoparticles expressing the original construct, E1*E2p7* (pLenti6-E1*E2p7*), the E2 coding region alone, preceded by the signal sequence from influenza virus HA to direct E2 into the ER lumen and followed by a stop codon (pLenti6-Sig-E2) or EGFP (pLenti6-EGFP) (Fig. 5A).
FIG. 5.
The E2 ectodomain is sufficient for BVDV inhibition. (A) Organization of the E2 protein and E2 expression constructs in the pLenti6 lentiviral vector. The predicted ectodomain (black bar), transmembrane anchor (white bar), and carboxy-terminal domain (gray bar) are indicated. The numbers on the top bar show the amino acid number within E2 corresponding to the start of a particular domain (21). The light gray boxes (E1*E2p7*) correspond to the E1* and p7* sequences of E1*E2p7*. The textured gray boxes (Sig-E2 and Sig-E2*) correspond to the HA signal sequence inserted upstream of the first amino acid of the E2 protein. (B) MDBK cells were transduced with packaged lentiviral vectors expressing EGFP, E1*E2p7*, Sig-E2, or Sig-E2*. After blasticidin selection, each cell population was infected with NADLJiv90− luc. At 48 h p.i., cells from triplicate wells were harvested for luciferase assays. The graph shows the average of two independent experiments done in triplicate; error bars show the standard errors of the means.
Transduced MDBK cells were infected with NADLJiv90− luc (27), a bicistronic ncp BVDV that expresses a luciferase reporter gene driven by the BVDV IRES and the BVDV polyprotein via the EMCV IRES. After infection with NADLJiv90− luc, cells expressing E1*E2p7* or E2 alone had a similar reduction in luciferase activity compared to the EGFP control cells (Fig. 5B). These results indicate that the flanking E1* and p7* sequences did not contribute to the BVDV inhibition observed using the original E1*E2p7* construct.
To determine if the E2 ectodomain alone was responsible for BVDV inhibition, pLenti6-Sig-E2*, expressing the E2 ectodomain with no transmembrane or carboxy-terminal domain, was constructed (Fig. 5A). MDBK cells were infected with pLenti6-Sig-E2* pseudoparticles and challenged with NADLJiv90− luc. Following challenge, luciferase activity from the Sig-E2* cells was similar to that from E1*E2p7* and Sig-E2 cells and more than 10-fold lower than from the negative control EGFP-expressing cells (Fig. 5B). Immunofluorescence staining for E2 within the Sig-E2* cells showed a significant amount of E2 within the cell, possibly in the ER and secretory pathway (data not shown). These data demonstrate that the E2 ectodomain is sufficient to confer the BVDV inhibition observed from E1*E2p7*.
BVDV is blocked prior to translation in MDBK-E1*E2p7* cells.
To examine the block to BVDV infection in the original MDBK-E1*E2p7* cells, we again used NADLJiv90− luc, which provided a sensitive assay to monitor BVDV translation and replication. Similar to the above experiments, a 100- to 1,000-fold decrease in luciferase activity was observed after NADLJiv90− luc infection of MDBK-E1*E2p7* cells compared to pBabe cells (data not shown).
We examined translation of the incoming NADLJiv90− luc genome in the absence of viral RNA replication using a BVDV-specific polymerase inhibitor (4). We previously showed, based on experiments comparing luciferase activity after BVDV infection of untreated and polymerase inhibitor-treated MDBK cells, as well as after transfection of replication-competent and polymerase-defective BVDV replicons, that appreciable BVDV replication does not occur before 6 h (27). After infection with NADLJiv90− luc, the luciferase activity within the pBabe cells peaked at 6 h and steadily decreased as expected, likely due to RNA degradation without replication (Fig. 6A). At the 6-h time point, the luciferase activity on the E1*E2p7* cells was approximately 10-fold less than on pBabe cells (Fig. 6A).
FIG. 6.
MDBK-E1*E2p7* cells have a defect at the level of BVDV entry. (A) MDBK-E1*E2p7* and pBabe cells were infected with NADLJiv90− luc in the presence of a BVDV polymerase inhibitor (4). At each time point, cells were harvested for luciferase assays. Each bar represents the mean value of duplicate wells; error bars show the standard errors of the means. The dashed line indicates the background level of the assay from naïve MDBK cells. (B) MDBK-E1*E2p7* and pBabe cells were electroporated with in vitro-transcribed BVDV NADLJiv90− luc RNA. At each time point, supernatants from electroporated cells were harvested and titers were determined on MDBK cells. Each point represents the mean titer of duplicate wells; error bars show the standard errors of the means.
This reduction could be due to a defect in translation or in the ability of BVDV to enter the cells and deliver its genome to the cytoplasm. To distinguish between these options, we tested whether bypassing the virus entry steps by transfection could overcome the block to BVDV replication in E1*E2p7* cells. Virus titer in the supernatants of MDBK-E1*E2p7* and pBabe cells was examined after electroporation of in vitro-transcribed BVDV NADL RNA. Over a 48-h period there was no difference in the titer of BVDV from MDBK-E1*E2p7* compared to the pBabe control cells (Fig. 6B). Because BVDV could be efficiently translated and replicated if allowed to bypass the entry steps into MDBK-E1*E2p7* cells, the block to BVDV replication is presumably at the level of virus entry, defined here as a step that occurs prior to translation.
CD46 expression levels and BVDV binding on MDBK-E1*E2p7* cells are similar to control cells.
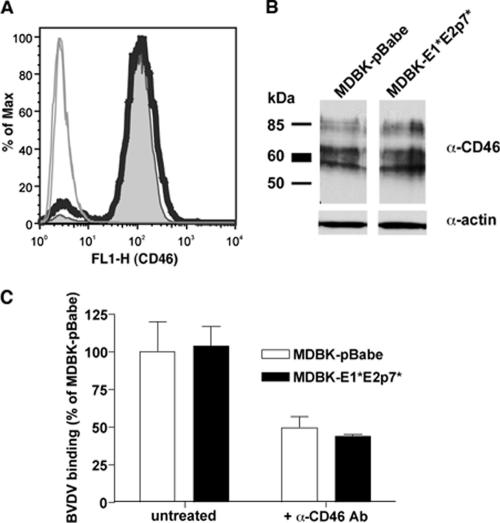
We hypothesized that the resistance to BVDV entry in MDBK-E1*E2p7* cells may be at the level of the viral receptor, CD46 (30). MDBK-E1*E2p7* and pBabe cells were analyzed for CD46 expression using flow cytometry and found to have similar levels of this protein on their surfaces (Fig. 7A). Western blotting of total cell lysates also revealed comparable levels of total CD46 in the two cell types (Fig. 7B). Taken together, these results suggest that the BVDV inhibition observed in cells expressing the E1*E2p7* insertion is not due to CD46 downregulation.
FIG. 7.
CD46 expression on MDBK-E1*E2p7* cells is similar to that in control pBabe cells. (A) To analyze CD46 expression on the cell surface, cells were detached and stained with a polyclonal antibody against bovine CD46. Cells were processed in the absence of detergent to selectively stain only the surface of the cells. CD46 expression on MDBK-E1*E2p7* (thick black line) and pBabe cells (tinted gray curve) was analyzed by fluorescence-activated cell sorting after gating on live cells. Results with cells stained with preimmune serum are indicated by thin solid lines. (B) To examine total CD46 expression, lysates from MDBK-E1*E2p7* and pBabe cells were analyzed by Western blotting with an antibody against bovine CD46 (α-CD46) or actin (α-actin). The presence of multiple bands in the anti-CD46 blot is due to multiple spliced isoforms of CD46 in the cell. (C) BVDV binding to MDBK-E1*E2p7* cells is similar to control pBabe cells. Cells were incubated for 1 h at 4°C with BVDV NADLJiv90− at an MOI of 10. Cells were washed extensively, and RNA was harvested for quantitative RT-PCR to determine cell-associated BVDV RNA levels. For bars marked +α-CD46 Ab, cells were pretreated with a mixture of anti-CD46 MAbs and then infected with BVDV as described above. Values are expressed as the percentage of untreated MDBK-pBabe BVDV binding. The data shown are the averages of at least duplicate samples; error bars show the standard deviations. Values shown for the untreated samples are representative of at least three independent experiments.
Although surface levels of CD46 were apparently similar between MDBK-pBabe and E1*E2p7* cells, it remained possible that E1*E2p7* cells could have a functional defect in cell surface binding of BVDV. To investigate this possibility, cells were chilled to 4°C and infected with ncp BVDV NADLJiv90− at an MOI of 10. Cell-associated BVDV RNA was quantified by real-time quantitative RT-PCR analysis. The assay revealed similar levels of BVDV binding on the surface of both MDBK-pBabe and E1*E2p7* cells (Fig. 7C). Preincubation with an anti-CD46 antibody reduced cell-associated BVDV RNA levels in both cell types by approximately 50% (Fig. 7C), indicating that much of the detected binding was dependent on CD46. These results indicate that the block to BVDV infection in MDBK-E1*E2p7* cells lies downstream of BVDV binding to the cell surface.
DISCUSSION
To identify essential interactions in the BVDV life cycle, we took a functional genomics approach by screening a random BVDV cDNA library for genome fragments that could act as transdominant inhibitors of viral replication. Of the two unique BVDV sequences that were isolated from resistant colonies after cp BVDV infection, one aligning with the E2 protein (E1*E2p7*) and the other with the NS5A protein, only the former insertion could provide resistance to naïve cells. Further experiments demonstrated that BVDV was blocked at the level of entry in the MDBK-E1*E2p7* cells in a CD46-independent manner. However, if allowed to bypass the entry step, BVDV replication was unaffected by expression of the E1*E2p7* insertion.
Further mapping of the E1*E2p7* insertion revealed that the E2 ectodomain was required for the inhibitory activity. We observed that Sig-E2*-expressing cells, encoding the signal sequence of the HA protein, and the E2 ectodomain followed by a stop codon were as resistant to BVDV entry as those expressing the original E1*E2p7* construct. Therefore, the E1 and p7 flanking sequences, as well as the E2 transmembrane and carboxy-terminal domains, are not required for BVDV inhibition. Moreover, the signal sequence for E2, normally provided by E1, acting to direct the E2 ectodomain into the ER lumen, could be replaced by a heterologous signal sequence to provide similar inhibition as the original insertion. Hence, the ectodomain of E2, independent from the transmembrane and carboxy-terminal domain, can act as a transdominant inhibitor of BVDV entry. The observation that the BVDV structural proteins can prevent BVDV infection is supported by previously published data. A cell line expressing the BVDV C through E2 proteins was shown to be nonpermissive for BVDV replication (39). Additionally, Harada et al. described an MDBK cell line expressing BVDV E2 and p7 separated by an IRES that was similarly inhibited for BVDV infection (15).
BVDV inhibition provided by the E1*E2p7* insertion may be due to a number of possible mechanisms. BVDV entry could be inhibited as a result of E2 interfering directly with the activity of a particular BVDV entry factor or indirectly by antagonizing a host signaling pathway required for virus uptake. Expression of BVDV E2 may prevent BVDV from entering the cell by a similar mechanism as superinfection exclusion observed in acutely infected cells (27). Superinfection exclusion, the ability of an established virus infection to prevent or interfere with a homologous secondary infection, can occur by a number of different mechanisms (1, 20, 42, 48). We and others have examined the phenomenon of BVDV superinfection exclusion in MDBK cells and showed that superinfecting BVDV encounters blocks at both the levels of entry and replication (27, 34). For the entry block, it was found that cells harboring a BVDV genome lacking the E2 gene failed to exclude superinfecting BVDV at the level of entry. In light of these data, it is possible that expression of E2 within MDBK cells may mimic the phenomenon of superinfection exclusion at the level of viral entry.
Envelope protein-mediated receptor downregulation and interference with the activity of a cellular receptor are well-documented mechanisms of viral superinfection exclusion. For example, the downregulation of CD4 from the surface of HIV infected cells by the viral envelope precursor gp160 (7, 19, 43) or the removal of sialic acid by influenza neuraminidase from the glycoproteins within the secretory pathway and the surface of infected cells (3, 25, 35) prevents superinfection of these viruses. Additionally, the Env protein of the retrovirus foamy virus (FV) was shown to be sufficient for mediating superinfection exclusion in FV-infected cells (6). Interference with a cellular receptor has also been observed for mouse gammaretroviruses. Mouse cells harboring certain integrated proviruses, ecotropic Fv4 or polytropic Rmcf or Rmcf2, which express MLV envelope genes, are resistant to exogenous ecotropic or polytropic MLV infection, respectively (5, 16, 18, 50). Although the entry pathway for BVDV is CD46-dependent (30), we found that CD46 expression in MDBK-E1*E2p7* cells did not differ from pBabe control cells. Consistent with this result, BVDV binding, which is mediated by CD46 (23, 30), was also equivalent between MDBK-E1*E2p7* and the control cells. It remains possible that E2 could be interfering with a specific molecular form or localization of CD46 that is expressed on the cell surface and required for BVDV to be internalized into MDBK cells, although our experiments suggest that the block to BVDV entry lies downstream of the CD46-dependent step.
The observation that CD46 downregulation from the surface of MDBK-E1*E2p7* cells is not the mechanism of BVDV inhibition suggests that CD46 may not be the only molecule required for BVDV entry. This hypothesis has been proposed previously by Maurer et al. (30). In their study, nonpermissive cells transduced to express CD46 remained nonpermissive for BVDV infection. Moreover, BVDV enters cells by clathrin-mediated endocytosis and CD46 is excluded from endosomes due to a sorting signal (14, 23, 30), suggesting that an additional factor may act downstream from CD46 to mediate subsequent internalization steps. Therefore, although CD46 expression and binding of BVDV to the MDBK-E1*E2p7* cells is unaffected, the possibility of E2-mediated downregulation of an unidentified, essential coreceptor required for BVDV internalization and fusion still exists. The requirement for multiple cellular entry factors is certainly true for hepatitis C virus and is likely for BVDV as well, considering that both of these viruses undergo an activation step which occurs after primary binding (23, 46, 46a). MDBK-E1*E2p7* cells may then be resistant to BVDV entry due to the ability of E2 to sequester this additional entry factor away from the cell surface or from other BVDV entry factors. Furthermore, the observed BVDV entry defect in MDBK-E1*E2p7* cells is analogous to that of CRIB cells (cells resistant to infection with BVDV) (11). CRIB cells are also resistant to BVDV entry and harbor functional CD46 on their surfaces. It was originally thought that these cells were deficient in the low-density lipoprotein receptor (2), a proposed BVDV entry factor, although a recent study suggested that these cells do in fact express the low-density lipoprotein receptor and furthermore that this molecule is not required for BVDV infection (22). Even so, CRIB cells can be used in conjunction with MDBK-E1*E2p7* cells to further define a complete set of BVDV entry factors, possibly by cataloging differences in cell surface molecules between these BVDV-resistant cell types and wild-type MDBK cells.
Functional genomics is a powerful approach to examine essential interactions between individual viral components, as well as between viral and cellular factors. Viral proteins or RNA associate with and recruit cellular cofactors for various aspects of their life cycle. There are numerous advantages of employing a strategy such as that described here to identify transdominant inhibitors of viral replication. First, the BVDV inhibitor was identified in a functional context where it prevented viral infection, directly demonstrating the relevance of an E2-mediated interaction in BVDV replication. Notably, only one cDNA, spanning the full-length E2 gene, out the 40,000 cDNA clones included in the random BVDV library, was pulled out of our screen and conferred resistance to naïve cells. This suggests that expression of the E1*E2p7* insertion in MDBK cells provides a strong, inhibitory mechanism against BVDV. Although the E2 ectodomain was sufficient for BVDV inhibition, the larger insertion could have provided some advantage in the stability or expression level during screening in MDBK cells. Another advantage of the approach used here is that a random genomic library is inherently unbiased and thus allows the identification of functional domains or regions that may not be tested by a rationally designed method. In addition, simply based on the number of different sequences being screened, the use of a library increases the likelihood of identifying functional interactions required for viral replication. In one study, screening of a random fragment library derived from the HIV-1 genome identified inhibitors of productive HIV-1 infection and latency which aligned with seven regions of the virus genome (9).
Future studies to analyze BVDV replication could utilize less-stringent screening methods compared to cell death, which could allow selection for cDNA insertions offering intermediate levels of BVDV resistance to MDBK cells. This method has the potential to further identify essential interactions that occur with components of the full-length BVDV genome. Moreover, the MDBK- E1*E2p7* cells characterized above will serve as a useful tool for further study of the mechanism of BVDV superinfection exclusion at the level of entry and the molecules required for BVDV to productively enter a cell.
Acknowledgments
We are grateful to many colleagues for helpful discussions during the course of this work and especially to Catherine Murray for critical reading of the manuscript.
This work was supported by PHS grants (CA57973 and AI072613), the Ellison Medical Foundation, the Starr Foundation, the Greenberg Medical Research Institute, and by the Northeast Biodefense Center U54-AI057158-Lipkin. C.M.R. is an Ellison Medical Foundation Scholar in Global Infectious Diseases. D.M.T. was supported in part through a Women and Science Fellowship. Additional support was provided by NIH grant AI058976 (M.R.M.).
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Adams, R. H., and D. T. Brown. 1985. BHK cells expressing Sindbis virus-induced homologous interference allow the translation of nonstructural genes of superinfecting virus. J. Virol. 54351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 9612766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Air, G. M., and W. G. Laver. 1989. The neuraminidase of influenza virus. Proteins 6341-356. [DOI] [PubMed] [Google Scholar]
- 4.Baginski, S. G., D. C. Pevear, M. Seipel, S. C. Sun, C. A. Benetatos, S. K. Chunduru, C. M. Rice, and M. S. Collett. 2000. Mechanism of action of a pestivirus antiviral compound. Proc. Natl. Acad. Sci. USA 977981-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassin, R. H., S. Ruscetti, I. Ali, D. K. Haapala, and A. Rein. 1982. Normal DBA/2 mouse cells synthesize a glycoprotein which interferes with MCF virus infection. Virology 123139-151. [DOI] [PubMed] [Google Scholar]
- 6.Berg, A., T. Pietschmann, A. Rethwilm, and D. Lindemann. 2003. Determinants of foamy virus envelope glycoprotein mediated resistance to superinfection. Virology 314243-252. [DOI] [PubMed] [Google Scholar]
- 7.Crise, B., L. Buonocore, and J. K. Rose. 1990. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J. Virol. 645585-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deregt, D., S. R. Bolin, J. van den Hurk, J. F. Ridpath, and S. A. Gilbert. 1998. Mapping of a type 1-specific and a type-common epitope on the E2 (gp53) protein of bovine viral diarrhea virus with neutralization escape mutants. Virus Res. 5381-90. [DOI] [PubMed] [Google Scholar]
- 9.Dunn, S. J., S. W. Park, V. Sharma, G. Raghu, J. M. Simone, R. Tavassoli, L. M. Young, M. A. Ortega, C. H. Pan, G. J. Alegre, I. B. Roninson, G. Lipkina, A. Dayn, and T. A. Holzmayer. 1999. Isolation of efficient antivirals: genetic suppressor elements against HIV-1. Gene Ther. 6130-137. [DOI] [PubMed] [Google Scholar]
- 10.Elbers, K., N. Tautz, P. Becher, D. Stoll, T. Rumenapf, and H. J. Thiel. 1996. Processing in the pestivirus E2-NS2 region: identification of proteins p7 and E2p7. J. Virol. 704131-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flores, E. F., and R. O. Donis. 1995. Isolation of a mutant MDBK cell line resistant to bovine viral diarrhea virus infection due to a block in viral entry. Virology 208565-575. [DOI] [PubMed] [Google Scholar]
- 12.Gao, G., X. Guo, and S. P. Goff. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 2971703-1706. [DOI] [PubMed] [Google Scholar]
- 13.Griffin, S. D., L. P. Beales, D. S. Clarke, O. Worsfold, S. D. Evans, J. Jaeger, M. P. Harris, and D. J. Rowlands. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 53534-38. [DOI] [PubMed] [Google Scholar]
- 14.Grummer, B., S. Grotha, and I. Greiser-Wilke. 2004. Bovine viral diarrhoea virus is internalized by clathrin-dependent receptor-mediated endocytosis. J. Vet. Med. B 51427-432. [DOI] [PubMed] [Google Scholar]
- 15.Harada, T., N. Tautz, and H. J. Thiel. 2000. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J. Virol. 749498-9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartley, J. W., R. A. Yetter, and H. C. Morse III. 1983. A mouse gene on chromosome 5 that restricts infectivity of mink cell focus-forming recombinant murine leukemia viruses. J. Exp. Med. 15816-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holzmayer, T. A., D. G. Pestov, and I. B. Roninson. 1992. Isolation of dominant negative mutants and inhibitory antisense RNA sequences by expression selection of random DNA fragments. Nucleic Acids Res. 20711-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda, H., F. Laigret, M. A. Martin, and R. Repaske. 1985. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J. Virol. 55768-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jabbar, M. A., and D. P. Nayak. 1990. Intracellular interaction of human immunodeficiency virus type 1 (ARV-2) envelope glycoprotein gp160 with CD4 blocks the movement and maturation of CD4 to the plasma membrane. J. Virol. 646297-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston, R. E., K. Wan, and H. R. Bose. 1974. Homologous interference induced by Sindbis virus. J. Virol. 141076-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohl, W., A. Grone, V. Moennig, and G. Herrler. 2007. Expression of the surface glycoprotein E2 of bovine viral diarrhea virus by recombinant vesicular stomatitis virus. J. Gen. Virol. 88157-165. [DOI] [PubMed] [Google Scholar]
- 22.Krey, T., E. Moussay, H. J. Thiel, and T. Rumenapf. 2006. Role of the low-density lipoprotein receptor in entry of bovine viral diarrhea virus. J. Virol. 8010862-10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krey, T., H. J. Thiel, and T. Rumenapf. 2005. Acid-resistant bovine pestivirus requires activation for pH-triggered fusion during entry. J. Virol. 794191-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lackner, T., A. Muller, A. Pankraz, P. Becher, H. J. Thiel, A. E. Gorbalenya, and N. Tautz. 2004. Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus. J. Virol. 7810765-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamb, R. A., and P. W. Choppin. 1983. The gene structure and replication of influenza virus. Annu. Rev. Biochem. 52467-506. [DOI] [PubMed] [Google Scholar]
- 26.Lecot, S., S. Belouzard, J. Dubuisson, and Y. Rouille. 2005. Bovine viral diarrhea virus entry is dependent on clathrin-mediated endocytosis. J. Virol. 7910826-10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, Y. M., D. M. Tscherne, S. I. Yun, I. Frolov, and C. M. Rice. 2005. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J. Virol. 793231-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 29.Mahon, G. M., and I. P. Whitehead. 2001. Retrovirus cDNA expression library screening for oncogenes. Methods Enzymol. 332211-221. [DOI] [PubMed] [Google Scholar]
- 30.Maurer, K., T. Krey, V. Moennig, H. J. Thiel, and T. Rumenapf. 2004. CD46 is a cellular receptor for bovine viral diarrhea virus. J. Virol. 781792-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClurkin, A. W., S. R. Bolin, and M. F. Coria. 1985. Isolation of cytopathic and noncytopathic bovine viral diarrhea virus from the spleen of cattle acutely and chronically affected with bovine viral diarrhea. J. Am. Vet. Med. Assoc. 186568-569. [PubMed] [Google Scholar]
- 32.Mendez, E., N. Ruggli, M. S. Collett, and C. M. Rice. 1998. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J. Virol. 724737-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyers, G., N. Tautz, P. Becher, H. J. Thiel, and B. M. Kummerer. 1996. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J. Virol. 708606-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittelholzer, C., C. Moser, J. D. Tratschin, and M. A. Hofmann. 1998. Porcine cells persistently infected with classical swine fever virus protected from pestivirus-induced cytopathic effect. J. Gen. Virol. 792981-2987. [DOI] [PubMed] [Google Scholar]
- 35.Palese, P., K. Tobita, M. Ueda, and R. W. Compans. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61397-410. [DOI] [PubMed] [Google Scholar]
- 36.Paton, D. J., J. P. Lowings, and A. D. Barrett. 1992. Epitope mapping of the gp53 envelope protein of bovine viral diarrhea virus. Virology 190763-772. [DOI] [PubMed] [Google Scholar]
- 37.Pestov, D. G., and L. F. Lau. 1994. Genetic selection of growth-inhibitory sequences in mammalian cells. Proc. Natl. Acad. Sci. USA 9112549-12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qu, L., L. K. McMullan, and C. M. Rice. 2001. Isolation and characterization of noncytopathic pestivirus mutants reveals a role for nonstructural protein NS4B in viral cytopathogenicity. J. Virol. 7510651-10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reimann, I., G. Meyers, and M. Beer. 2003. Trans-complementation of autonomously replicating bovine viral diarrhea virus replicons with deletions in the E2 coding region. Virology 307213-227. [DOI] [PubMed] [Google Scholar]
- 40.Rumenapf, T., G. Meyers, R. Stark, and H. J. Thiel. 1991. Molecular characterization of hog cholera virus. Arch. Virol. Suppl. 37-18. [DOI] [PubMed] [Google Scholar]
- 41.Rumenapf, T., G. Unger, J. H. Strauss, and H. J. Thiel. 1993. Processing of the envelope glycoproteins of pestiviruses. J. Virol. 673288-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon, K. O., J. J. Cardamone, Jr., P. A. Whitaker-Dowling, J. S. Youngner, and C. C. Widnell. 1990. Cellular mechanisms in the superinfection exclusion of vesicular stomatitis virus. Virology 177375-379. [DOI] [PubMed] [Google Scholar]
- 43.Stevenson, M., C. Meier, A. M. Mann, N. Chapman, and A. Wasiak. 1988. Envelope glycoprotein of HIV induces interference and cytolysis resistance in CD4+ cells: mechanism for persistence in AIDS. Cell 53483-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiel, H. J., R. Stark, E. Weiland, T. Rumenapf, and G. Meyers. 1991. Hog cholera virus: molecular composition of virions from a pestivirus. J. Virol. 654705-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toth, R. L., P. F. Nettleton, and M. A. McCrae. 1999. Expression of the E2 envelope glycoprotein of bovine viral diarrhoea virus (BVDV) elicits virus-type specific neutralising antibodies. Vet. Microbiol. 6587-101. [DOI] [PubMed] [Google Scholar]
- 46.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 801734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.von Hahn, T., and C. M. Rice. Hepatitis C virus entry. J. Biol. Chem. Epub 19 Sept. 2007. doi: 10.1074/jbc.R700024200. [DOI] [PubMed]
- 47.Weiland, E., R. Stark, B. Haas, T. Rumenapf, G. Meyers, and H. J. Thiel. 1990. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J. Virol. 643563-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitaker-Dowling, P., J. S. Youngner, C. C. Widnell, and D. K. Wilcox. 1983. Superinfection exclusion by vesicular stomatitis virus. Virology 131137-143. [DOI] [PubMed] [Google Scholar]
- 49.Whitehead, I., H. Kirk, and R. Kay. 1995. Expression cloning of oncogenes by retroviral transfer of cDNA libraries. Mol. Cell. Biol. 15704-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, T., Y. Yan, and C. A. Kozak. 2005. Rmcf2, a xenotropic provirus in the Asian mouse species Mus castaneus, blocks infection by polytropic mouse gammaretroviruses. J. Virol. 799677-9684. [DOI] [PMC free article] [PubMed] [Google Scholar]