Abstract
A new Salmonella enterica phage, Det7, was isolated from sewage and shown by electron microscopy to belong to the Myoviridae morphogroup of bacteriophages. Det7 contains a 75-kDa protein with 50% overall sequence identity to the tail spike endorhamnosidase of podovirus P22. Adsorption of myoviruses to their bacterial hosts is normally mediated by long and short tail fibers attached to a contractile tail, whereas podoviruses do not contain fibers but attach to host cells through stubby tail spikes attached to a very short, noncontractile tail. The amino-terminal 150 residues of the Det7 protein lack homology to the P22 tail spike and are probably responsible for binding to the base plate of the myoviral tail. Det7 tail spike lacking this putative particle-binding domain was purified from Escherichia coli, and well-diffracting crystals of the protein were obtained. The structure, determined by molecular replacement and refined at a 1.6-Å resolution, is very similar to that of bacteriophage P22 tail spike. Fluorescence titrations with an octasaccharide suggest Det7 tail spike to bind its receptor lipopolysaccharide somewhat less tightly than the P22 tail spike. The Det7 tail spike is even more resistant to thermal unfolding than the already exceptionally stable homologue from P22. Folding and assembly of both trimeric proteins are equally temperature sensitive and equally slow. Despite the close structural, biochemical, and sequence similarities between both proteins, the Det7 tail spike lacks both carboxy-terminal cysteines previously proposed to form a transient disulfide during P22 tail spike assembly. Our data suggest receptor-binding module exchange between podoviruses and myoviruses in the course of bacteriophage evolution.
Double-stranded DNA bacteriophages can be grouped into several families, of which Myoviridae and Podoviridae are among the most common (1, 30). The two families have morphologically different tails. Podoviridae have a very short, noncontractile tail, normally carrying six stubby tail spikes. Tail spikes are responsible for cell attachment and frequently recognize and hydrolytically cleave bacterial cell surface polysaccharides. Myoviridae have a long, contractile tail with a base plate. Long and short tail fibers, normally attached to this base plate, bind to cell surface receptors but do not show enzymatic activity. Finally, Siphoviridae have a long, flexible, noncontractile tail.
The best-studied podoviral tail spike is that of bacteriophage P22, a well-characterized protein used as a model to study protein folding in vivo, as well as in vitro (5, 15, 39). It is a very stable homotrimer, and the structure of a tail spike subunit can be divided into three parts: an amino-terminal head-binding domain, a central parallel-β-helix domain containing 13 complete right-handed β-helical turns, and a carboxy-terminal, highly interdigitated part important for trimerization, thermostability, and the observed resistance of the trimer to dissociation by sodium dodecyl sulfate (SDS) (42, 45). The amino-terminal domain is thought to be flexibly attached to the other two parts, which together form a rigid unit; the flexibility of the short linker peptide may be important in the infection process (44). The central β-helix domain binds to the O-antigen region of the bacterial cell surface lipopolysaccharide. It exhibits an endorhamnosidase enzymatic activity cleaving the O-antigen polysaccharide.
The enzymatic activity is required for infection, but its function in the infection process is not well understood. It may contribute to the mobility of phage particles on the bacterial surface, and it is thought to be important for escaping bacterial debris after cell lysis, reflecting the role of the receptor-inactivating neuraminidase of influenza viruses (3, 27, 41).
Sequences coding for proteins with similarity to the P22 tail spike have been found in podo- and siphoviruses but not in myoviruses. Here we report the discovery of the new Salmonella bacteriophage Det7. Electron microscopy shows Det7 to be a myovirus, having a contractile tail. Several fragments of the Det7 genome were sequenced. Their predicted products either had no sequence homology to proteins from other bacteriophages in the database or showed low sequence identity with other myoviral proteins. The exception was one gene coding for a polypeptide with 50% identity to the tail spike from podovirus P22. Electron microscopy also showed that Det7 has a very large base plate harboring potential tail spikes.
We have produced an amino-terminally shortened Det7 tail spike protein by recombinant expression in Escherichia coli, purified the protein, and studied its stability, refolding, and receptor binding. We have also crystallized the protein and resolved its structure at 1.6-Å resolution and show that it is nearly identical to the P22 tail spike, despite missing cysteine residues presumed to be important for tail spike folding and assembly.
MATERIALS AND METHODS
Electron microscopy.
Virus samples were applied to carbon-coated, hydrophilized copper grids; negatively stained with 2% uranyl acetate (pH 4.5); and examined by using a CM12 transmission electron microscope (FEI Co., Eindhoven, The Netherlands) operated at 120 keV. Searching for suitable specimen areas was facilitated by a TV-rate charge-coupled device camera (model 673 III; Gatan GmbH, München, Germany). Electron micrographs were digitally recorded by means of a 1,000- by 1,000-pixel slow-scan charge-coupled device camera (TVIPS GmbH, Gauting, Germany).
Protein expression and purification.
A 1.7-kb fragment, encoding amino acids 152 to 708 of the Det7 tail spike protein preceded by a Met residue, was cloned into the expression vector pET21a (Novagen, Madison, WI). For protein expression, the resulting plasmid, pET21a-Det7tspΔ1-151 was transformed into E. coli HMS174 (DE3). The cultures were grown in Luria broth supplemented with 100 μg of ampicillin/ml at 30°C. Gene expression was induced by addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an optical density at 600 nm of 0.6 to 0.8. After overnight incubation under the same conditions, cells were harvested by centrifugation, resuspended in 50 mM Tris-HCl (pH 8.0)-2 mM EDTA, and disrupted by high-pressure lysis. The soluble protein was purified from the supernatant by using procedures similar to those described for the P22 tail spike protein lacking its head-binding domain (28). Briefly, the protein was precipitated with ammonium sulfate and purified by hydrophobic interaction chromatography, followed by anion-exchange chromatography. The purified protein was stored as a suspension in 40% saturated ammonium sulfate-10 mM Tris-HCl (pH 8.0). From here onward, the protein is referred to as Det7tspΔ1-151. The homologue from phage P22 (P22tspΔ1-108) was purified as described previously (28). A plasmid coding for P22tspΔ1-108 carrying the W365Y substitution was created by site-directed mutagenesis using pTSF1 (28) as a template and a QuikChange PCR mutagenesis kit (New England Biolabs) and according to the procedures suggested by the manufacturer. The oligonucleotides used (forward primer sequence 5′-GAGTGGCGTTAAAACTTACCAAGGTACTGTGGGCTCGACAAC-3′) introduce an additional StyI restriction site.
Fluorescence assay of carbohydrate binding.
Binding of Salmonella enterica serovar Typhimurium octasaccharide corresponding to two O-antigen repeats was measured essentially as described previously (3). A solution of Det7tspΔ1-151 (19.7 μg in 1 ml, equivalent to 330 nM subunits) in 20 mM sodium phosphate buffer (pH 7) was titrated with 2- to 5-μl aliquots of solutions of octasaccharide (5.4 mM) at 10 or 25°C in a stirred quartz cell. After each addition, the sample was equilibrated for 4 min, before the fluorescence emission at 340 nm with excitation at 295 nm was recorded in a Spex Fluorolog spectrofluorometer. The data were corrected for the fluorescence of trace impurities in the oligosaccharide preparation and for the volume change, which did not exceed 5%.
Thermal unfolding in the presence of SDS.
Dissociation and unfolding of detergent-resistant tail spike trimers was analyzed by SDS-gel electrophoresis essentially as described for the P22 protein (28). Samples contained Det7tspΔ1-151 (30 μg/ml) or P22tspΔ1-108 (32 μg/ml) in 20 mM sodium phosphate buffer-2% SDS-150 mM mercaptoethanol (pH 7). Densitometric analysis of trimer bands on grayscale images created with the aid of a transparency scanner (UMAX Astra 1220S) after electrophoresis gels were stained with Coomassie blue R was done with Gelscan (BioSciTec, Frankfurt/Main, Germany).
Refolding and reassembly.
Det7tspΔ1-151 (2 mg/ml) was denatured in the presence of 5 M guanidinium chloride for at least 1 h to achieve complete dissociation and unfolding, as determined by circular dichroism and tryptophan fluorescence spectroscopy. To initiate refolding and reassembly, a large excess of buffer (1 ml of 20 mM sodium phosphate, 2 mM EDTA, and 20 mM dithiothreitol) was rapidly added to a small volume (2.5 to 5 μl) of denatured protein, resulting in a final denaturant concentration of 25 mM or less. Higher denaturant concentrations may lead to the precipitation of guanidinium dodecyl sulfate during the subsequent assay for SDS-resistant trimers. Both the reaction vessel containing the denatured protein and the dilution buffer had been pre-equilibrated at the desired temperature. Reconstitution was quenched by the addition of SDS, and samples were analyzed by SDS-gel electrophoresis and silver staining as described for the P22 tail spike protein (8). To analyze reconstitution kinetics, refolding of individual samples was initiated by dilution at different time points, before all samples were quenched and assayed for SDS-resistant trimers.
Crystallization, crystallographic data collection, and data processing.
Ammonium sulfate precipitated Det7tspΔ1-151 was redissolved in 10 mM Tris-HCl (pH 8.5)-1 mM EDTA to a final concentration of between 20 and 30 mg/ml. Protein concentrations were determined by using UV absorption measurements at 280 nm; the absorbance of a 1-mg/ml protein solution was assumed to be 1.0 at a path length of 1 cm. Crystals were obtained by vapor diffusion in sitting drop CompactClover plates (Jena Biosciences, Jena, Germany), with 0.1- to 0.15-ml reservoirs and drops of 2 to 5 μl of protein solution mixed with 2 to 5 μl of reservoir solution. Reservoirs contained 10 to 30% (wt/vol) PEG4000, 0.1 M sodium citrate (pH 4.5), and 0 to 15% (vol/vol) glycerol. Hexagonal prisms of up to 1.2 by 1.2 by 0.25 mm were obtained, although for data collection a single crystal of approximately 0.3 by 0.3 by 0.1 mm was used. This crystal was sequentially transferred to 25% (vol/vol) glycerol in crystallization buffer, using 5% increments, and subsequently flash-cooled and stored in liquid nitrogen. The data were collected at 100 K; the total rotation angle was 90° (180 images of 0.5°). Reflections were integrated with the program MOSFLM (26, 34) and scaled by using SCALA (7). For data statistics and parameters, see Table 1.
TABLE 1.
Crystallographic data and refinement statistics
| Data and refinement parameters | Value(s) |
|---|---|
| Beam-line | ESRF ID23-1 |
| Wavelength (Å) | 0.95375 |
| Detector | Mosaic 225 mm (MAR Research) |
| Space group | R32 |
| Cell parameters (hexagonal setting) (Å) | a = b = 100.4; c = 330.0 |
| Wilson temp factor (Å2) | 15 |
| Resolution range (Å) | 15.0-1.6 (1.69-1.60)a |
| Multiplicity | 5.2 (4.8) |
| Completeness | 0.946 (0.859) |
| Rsymb | 0.081 (0.245) |
| Refinement | |
| Resolution range (Å) | 15.0-1.6 (1.66-1.60) |
| No. of reflections used in refinement | 77,590 (5,136) |
| No. of reflections used for R-free analyses | 1598 (96) |
| R-factorc (%) | 0.141 (0.19) |
| R-free (%) | 0.170 (0.24) |
| No. of atoms (protein/water/Na+) | 4,201/831/1 |
| Avg B-value (protein/solvent/Na+) (Å2) | 14.3/33.3/11.0 |
| Ramachandran statisticsd (%) | 88.7/10.7/0.7/0.0 |
| RMS deviationse (bonds/angles) (Å/°) | 0.018/1.7 |
Values in parentheses are for the highest-resolution bin, where applicable.
Rsym = Σh Σi Ihi − 〈Ih〉/Σh Σi Ihi, where Ihi is the intensity of the ith measurement of the same reflection and 〈Ih〉 is the mean observed intensity for that reflection.
R = Σ‖Fobs(hkl)| − |Fcalc(hkl)‖/ΣFobs(hkl).
Calculated according to the program PROCHECK (24). The percentages are indicated of residues in the “most favored/additionally allowed/generously allowed/disallowed” regions of the Ramachandran plot, respectively.
Estimates were provided by the program REFMAC (31).
Structure solution and refinement.
A clear molecular replacement solution was identified by the AMORE program (32), using data between 15.0 and 3.0 Å and the structure of the equivalent fragment of the bacteriophage P22 tail spike as a model (PDB-code 1TSP [40]). The solution was input into ARP-WARP (33) using data to 1.6-Å resolution, and a model resulted containing 547 residues in three separate chains, but with a large fraction traced out-of-phase. Rebuilding of the model and addition of extra amino acids was done with O (18) and yielded a model containing 553 amino acids, Ala156 to Val708. Refinement was done by using the REFMAC program (31); water molecules were built by using ARP (23).
Coordinate and sequence deposition.
Coordinates and structure factors have been deposited in the Protein Data Bank (accession codes 2V5I and R2V5ISF, respectively). The nucleotide sequence of the Det7 tail spike gene (ORF708) has been deposited in GenBank (accession number AM765843).
RESULTS
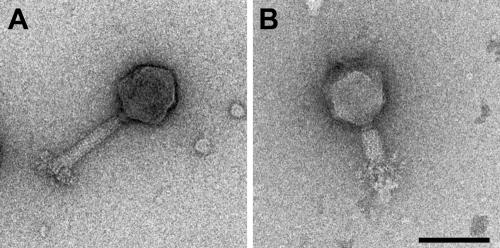
A new bacteriophage, Det7, was isolated from a local sewage sample plated on S. enterica serovar Typhimurium as a host. Electron micrographs of Det7 particles negatively stained with uranyl acetate showed a myovirus having an icosahedral head with a diameter of ∼90 nm (Fig. 1A). The contractile tail (Fig. 1B) has a total length of ∼120 nm. It consists of a small collar (10 nm), a shaft (95 by 20 nm), and a large base plate of approximately 15 by 70 nm.
FIG. 1.
Electron microscopy of bacteriophage Det7. Transmission electron micrographs of Det7 phage particles were negatively stained with uranyl acetate. (A) Intact phage, with extended tail; (B) phage with contracted tail. Bar, 0.1 μm.
Identification of a “podoviral” tail spike in Det7.
DNA sequence analysis of several genome fragments of the Det7 phage revealed only very low sequence similarities to other phages (and, if any, then with other myoviruses). The exception was a single gene which was found to be homologous to gene 9 of phage P22 coding for the prototype podoviral tail spike endoglycosidase. The protein encoded by the new tail spike gene has 50% overall sequence identity with the phage P22 tail spike and is 42 residues longer (Fig. 2). Using an antiserum to phage P22 tail spike in Western blots (data not shown), we found the Det7 protein to be present in the phage particle.
FIG. 2.
Amino acid sequences of the Det7 and P22 tail spikes. The alignment of the bacteriophage P22 tail spike sequence with the bacteriophage Det7 tail spike sequence was prepared by using CLUSTAL W (17). The first residues present in the amino terminally shortened, crystallized tail spike proteins are in boldface.
The sequence identity between the Det7 and P22 tail spikes varies according to the domains of the P22 protein. The amino-terminal domain, which is responsible for the interaction with the phage head in P22, has 13% sequence identity and is 45 residues longer in Det7, whereas the more carboxy-terminal parts responsible for O-antigen-binding and trimer stability in P22 have 60% sequence identity and are of almost exactly identical length in the two proteins. The homologous part of 557 residues (Det7tspΔ1-151) was produced as a soluble protein in E. coli and purified to homogeneity (see Materials and Methods). Like P22tspΔ1-108, the homologous receptor-binding part of P22 tail spike, Det7tspΔ1-151 forms trimers that are resistant to denaturation by SDS at ambient temperature (compare to Fig. 6, below).
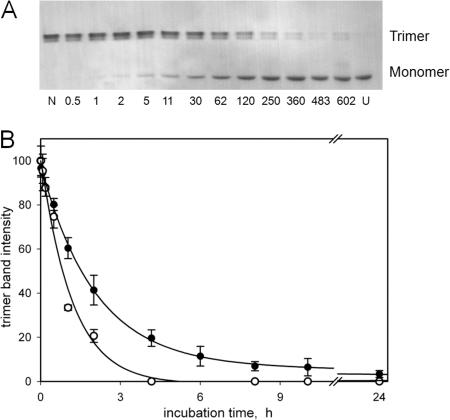
FIG. 6.
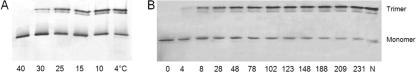
Thermal stability of Det7 tail spike protein. A: Purified Det7tspΔ1-151 was incubated in the presence of 2% SDS at 78.5°C for the time (in minutes) indicated and rapidly cooled on ice, and samples were analyzed by SDS-gel electrophoresis at room temperature. The Det7 tail spike trimer appears as a doublet band in the upper part, whereas the SDS complex of the unfolded polypeptide is the single band in the lower part of the figure. Lanes N and U indicate a native control left in SDS at room temperature and an unfolded (boiled) control, respectively. (B) Kinetics of thermal unfolding of Det7tspΔ1-151 (•) and of the corresponding tail spike fragment TSPΔN from phage P22 (○), as determined by densitometry of trimer bands. Conditions were identical to those in panel A; data and error bars correspond to averages and standard deviations from four Coomassie blue-stained gels.
Structure determination.
Purified Det7tspΔ1-151 was subjected to crystallization trials, and hexagonal plates or prisms grew at several different conditions, one of which was optimized (see Materials and Methods). Crystals belonged to the rhombohedral space group R32, contained one molecule in the asymmetric unit (i.e., one third of a trimer), and had a solvent content of 54% (Matthews coefficient = 2.7). We solved the structure by molecular replacement using the known structure of P22tspΔ1-108 (pdb-code 1TSP [40]), followed by automatic tracing of 547 amino acids and partial docking of their side chains. The resulting map showed good electron density for residues 156 to 708 (residues 6 to 558 of Det7tspΔ1-151); the amino-terminal five amino acids could not be modeled. The final model has very good correlation with the crystallographic data and good stereochemistry (Table 1).
Overview of the structure.
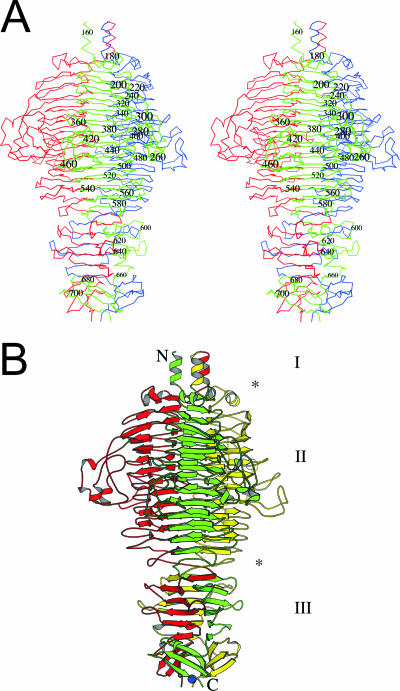
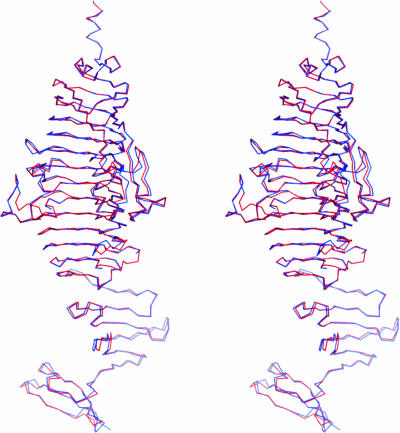
The bacteriophage Det7 tail spike is a parallel homotrimer, virtually identical to the bacteriophage P22 tail spike (40). The structure can be divided into several parts (Fig. 3): an amino-terminal three-helix bundle, which connects to the absent base-plate binding domain (42, 44); a central β-helical domain (amino acids 168 to 586); and a carboxy-terminal intertwined domain (amino acids 587 to 708). The surface area of the monomer is 25.5 × 103 Å2, of which 8.7 × 103 Å2 (34%) is buried in the trimer. This rather large amount of buried surface explains the extraordinary stability and SDS resistance of the trimer. Not surprisingly, considering the high sequence similarity, the Det7 tail spike structure is very similar to the P22 tail spike structure (Fig. 4), and aligned residues can be superimposed with a root mean square (RMS) differences between C-alphas of only just over 0.8 Å when monomers are superposed and under 0.9 Å when trimers are superposed. If we consider the β-helix domain (amino acids 168 to 586) alone, it can be superimposed onto the P22 tail spike β-helix domain with RMS differences between C-alpha atoms of only 0.5 Å.
FIG. 3.
Overall crystal structure of the bacteriophage Det7 tail spike lacking amino acids 1 to 151. (A) Wall-eyed stereo view of C-alphas connected by lines. The three monomers are colored red, green, and blue, and every twentieth residue of the green monomer is numbered. (B) Ribbon diagram in the same orientation as in panel A. Here the monomers are colored red, green, and yellow. A sodium ion observed in the crystal structure is shown as a blue ball. The amino and carboxy termini of the green monomer are indicated with an N and a C, respectively. The different domains are also shown, with asterisks indicating the approximate borders. “I” indicates the connector to the phage-binding domain, “II” indicates the O-antigen binding and hydrolysis domain, and “III” indicates the intertwined carboxy-terminal domain. The figure was prepared using Bobscript (12), a modified version of Molscript (22). Secondary structure elements were identified by using an algorithm developed by Kabsch and Sander (21), which is based on hydrogen bonding observed in the structure.
FIG. 4.
Comparison of Det7 and P22 tail spike structures. The tail spikes of bacteriophage Det7 (red) and P22 (blue) (42) are superimposed and shown as backbone traces. Wall-eyed stereo images for Fig. 4 and 5B were prepared by using Pymol (10).
The receptor-binding and hydrolysis domain.
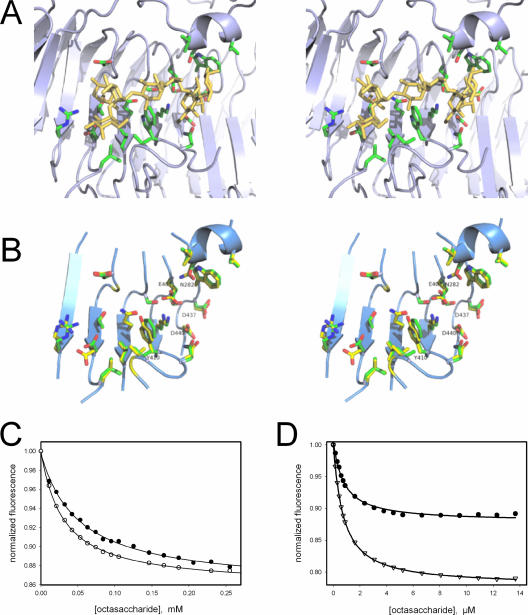
The binding site for the lipopolysaccharide O-antigen repeat region is known for the P22 tail spike (41) and involves exclusively the central β-helix domain (Fig. 5A). Residues reported to contact the O antigen are well conserved between the two tail spikes: the three carboxylate residues involved in the catalytic mechanism of the P22 tail spike endorhamnosidase are all preserved in Det7 (Glu404, Asp437, and Asp440). Among the other 18 residues significantly interacting with the ligand in the crystal structure of the complex of P22 tail spike with an octasaccharide hydrolysis product, there are only two differences: Trp365 of P22 is Tyr410 in Det7 and Ser237 of P22 is Asn282 in the Det7 tail spike sequence (Fig. 5B). As observed for P22 tail spike (3), oligosaccharide binding to Det7 tail spike protein in solution results in a decrease of the protein's tryptophan fluorescence. Binding titrations with an octasaccharide from S. enterica serovar Typhimurium at 10°C and 25°C yielded dissociation equilibrium constants of close to 30 and 50 μM, respectively (Fig. 5C). Dissociation constants measured with P22 tail spike under identical conditions are more than 10-fold smaller (compare Fig. 5D). The maximum fluorescence change at saturation with the octasaccharide was 14% for Det7 tail spike compared to 20% observed for P22 tail spike. To assess whether the substitution of Trp365 in P22 tail spike by a tyrosine is responsible for the differences, we created a W365Y mutant of P22tspΔ1-108 and analyzed its oligosaccharide binding. When titrated with the octasaccharide at 10°C, the tryptophan fluorescence decreased by 12%, and the dissociation equilibrium constant was 0.7 μM, virtually identical to the dissociation constant measured for the wild-type under these conditions (Fig. 5D). Thus, the higher fluorescence change observed with the P22 protein can be attributed to Trp365, whereas the difference in the binding affinity appears to be unrelated to the substitution of this tryptophan by Tyr410 in the Det7 tail spike.
FIG. 5.
O-antigen oligosaccharide binding. (A) Oligosaccharide complex of P22 tail spike protein (pdb entry 1tyx [41]). The octasaccharide, corresponding to two O-antigen repeats from S. enterica serovar Typhimurium, and side chains of amino-acid residues in contact with the ligand are shown as sticks. (B) Binding site residues of P22 (green carbons) and Det7 tail spikes (yellow carbons) aligned by their C-alpha coordinates depicted together with fragments of the Det7 backbone (ribbon diagram). Putative active-site carboxylate residues of Det7 tail spike and the two residues not conserved between both proteins are labeled. (C) Fluorescence titration of Det7tspΔ1-151 with the octasaccharide at 25°C (•) or 10°C (○). The solid lines are nonlinear fits to a binding isotherm for a single class of binding sites resulting in dissociation equilibrium constants of KD, 25°C = 50.7 μM and KD 10°C = 29.9 μM and a 14% fluorescence decrease at saturation. (D) Titration of the P22tspΔ1-108 mutant W365Y (•) and the corresponding wild type (▿) with the octasaccharide at 10°C.
Carboxy-terminal domain.
The carboxy-terminal domain contains three parts connected by loops (42): an intertwined region with extensive intersubunit hydrogen bonding (amino acids 585 to 596), followed by a five-stranded β-sheet (residues 601 to 664) and a three-stranded β-sheet (residues 679 to 707). These segments enclose a central hydrophobic core. Although the sequence identity of the last 53 residues of Det7 tail spike with the corresponding part of P22 tail spike protein is only 25%, the three-dimensional structure is well conserved between these parts of both proteins (Fig. 2 and 4). At the very carboxy terminus of the Det7 protein, a polar contact patch involving the main-chain oxygen of Leu706, a sodium ion, and water molecules is present. The sodium ion (Fig. 3) may be a consequence of the crystallization conditions. Therefore, its biological relevance is unknown.
Tail spike stability and folding.
The bacteriophage Det7 tail spike is even more thermostable than its P22 equivalent, if both proteins are compared without their phage-particle binding domains (Fig. 6). The half-life of the Det7tspΔ1-151 trimer in 2% SDS at 78.5° (∼1.5 h) was twice that of the equivalent P22 tail spike fragment (∼50 min). Upon unfolding of Det7tspΔ1-151 in guanidinium chloride solutions, a 70% decrease in the tryptophan fluorescence emission measured at 336 nm was observed. This fluorescence change was used to measure unfolding kinetics at 10°C, which were well described by a single exponential decay. Plots of the logarithm of the rate constant of unfolding over the denaturant concentration were linear, with the rate constant decreasing from 0.12 s−1 at 5.75 M to 3.5 × 10−4 s−1 at 4.1 M denaturant. If the data are extrapolated to buffer without denaturant, the resulting half-life would be 66 years, which is similar to that determined for the complete P22 tail spike and ∼10-fold longer than the extrapolated half-life of the P22tspΔ1-108 trimer (9).
Upon dilution from guanidinium chloride solutions at low temperature, Det7tspΔ1-151 refolded and reassembled efficiently to form SDS-resistant trimers (Fig. 7). Above ambient temperature, however, Det7 tail spike refolding yields decreased, until no reconstitution of SDS-resistant trimers was observed at 40°C (Fig. 7A). A very similar temperature sensitivity of folding has been observed for the P22 protein and has been attributed to the marginal thermal stability of essential subunit folding intermediates (8, 9). Although efficient at low temperature, the reconstitution of SDS-resistant Det7 tail spike trimers was very slow, occurring with a half time of several hours at 4 to 10°C (Fig. 7B) and again reflecting observations with the P22 protein (9, 28). Thus, Det7 tail spike shares characteristic folding and unfolding properties of the P22 tail spike protein.
FIG. 7.
Renaturation of Det7 tail spike protein. (A) Temperature-sensitive folding. Unfolded Det7tspΔ1-151 protein was diluted 1:400 from 5 M guanidinium chloride at the temperature indicated and allowed to refold and reassemble for 4 days before the formation of detergent-resistant trimers was probed by SDS gel electrophoresis and silver staining. (B) Time course of SDS-resistant trimer formation. Reconstitution of Det7tspΔ1-151 was initiated by a 1:200 dilution from 5 M guanidinium chloride at 4°C. After the times indicated (in hours), trimer reconstitution was quenched by the addition of SDS, followed by SDS-gel electrophoresis.
DISCUSSION
Despite their occurrence in structurally dissimilar phage particles, the structures and the folding and ligand binding properties of Det7 and P22 tail spike proteins are very similar. Both proteins bind octasaccharide fragments from Salmonella lipopolysaccharide with micromolar affinity, although the binding affinity of the Det7 protein is somewhat lower. In addition to most of the residues contacting the ligand in the P22 tail spike endorhamosidase, all three active-site carboxylates are conserved in the Det7 protein. Since the substitution of any of the three carboxylate residues by the corresponding carboxamides inactivates the enzyme but does not affect ligand binding (3), their presence in Det7 tail spike suggests that the receptor hydrolyzing activity is required in the life cycle of the myovirus Det7 like it is for the podovirus P22.
Temperature-sensitive folding.
The P22 tail spike has been used as a model protein to study the kinetic competition between protein folding and inclusion body formation in vivo and in vitro (5, 15, 39). Many single amino acid substitutions in the β-helix and the inserted “dorsal fin” domain cause a temperature-sensitive folding (tsf) phenotype (16), because they destabilize an essential intermediate in the folding of the β-helix domain (8). Most residues at the corresponding sites in Det7 tail spike are identical to the respective wild-type residues of P22 (16 of 23 listed by Haase-Pettingell and King (16), a small polar residue is replaced by another small polar residue at four of the remaining sites. Only three substitutions may be considered nonconservative: Pro250, Glu344, and Gly435 in P22 are Gln, Ser, and Asn in Det7, respectively. With the exception of Ser389, which is at the subunit interface and packed against a tryptophan of a neighboring subunit, the side chains of these residues are solvent exposed in the Det7 tail spike structure. None of the replacements in Det7 relative to P22 are identical to residues present in P22 tail spike tsf mutant proteins.
Two sets of intragenic global suppressors for tsf mutations of the P22 tail spike have been described (13, 25). Mutating Val331 of the P22 tail spike to Ala or Gly and Ala334 to Val or Ile increase the folding efficiency of the P22 tail spike. In the phage Det7 tail spike protein, the position equivalent to 331 is occupied by a glycine (Gly376), and the 334 equivalent is a valine. Thus, both positions are occupied by tsf suppressor residues in the Det7 protein. Taken together, the data suggest that the Det7 tail spike is optimized for folding efficiency. In this context, it is interesting that the oligosaccharide dissociation equilibrium constant for the V331G tsf suppressor mutant P22 tail spike is virtually identical to that of the Det7 protein (4).
Tail spike assembly pathway.
In the case of the P22 protein, folding of the large β-helix precedes subunit association and the very slow folding reaction reflected by the acquisition of SDS resistance is attributed to the slow intertwining of the carboxy-terminal parts occurring after subunit association.
In a recent site-directed mutagenesis study, King and coworkers (45) have targeted the part of the carboxy-terminal region of P22 tail spike, where the chains begin to wrap around each other. These researchers found the first residues of this segment (540 to 544 in the P22 tail spike) to be very tolerant to amino acid substitutions, whereas substitutions beyond residue 545 prevent trimer maturation. As may be expected, if the folding pathways are preserved, there is no sequence conservation between P22 and Det7 tail spikes in the tolerant region (LAEEG in P22 is LLDPV in Det7). In contrast, the residues immediately following the tolerant region (LGN in P22 and LGQ in Det7) are well preserved, as is the sequence of the beta-prism between residues 560 and 610 in P22 (residues 601 and 655 in Det7) with 75% sequence identity and only five nonconservative substitutions, all at solvent-exposed sites. Beyond the beta-prism, sequence identity decreases sharply to 25% for residues 656 to 708 (residues 611 to 666 in P22). This region constitutes the only major part of the crystallized protein with low sequence identity and the only domain where the structures do not align perfectly and superpose less well (see Fig. 4); furthermore, a two-amino-acid deletion is present in the loop between the third-to-last and second-to-last β-strand of the Det7 tail spike compared to the P22 tail spike. Thus, although even for this part the structural architecture is well conserved between the two proteins, it may not play a crucial role in a conserved assembly pathway of the tail spike proteins, as has been suggested (45). The P22 tail spike residues Cys613 and Cys635 have been proposed to form transient inter-subunit disulfide bridges in the incompletely folded protrimer (35), and a double mutant with both cysteines replaced by serines does not assemble. The tail spikes of phages Det7 and SP6 (37, 38) do not contain the two cysteines (Cys613 of the P22 tail spike is Tyr in Det7 and Phe in SP6 and Cys635 of the P22 tail spike is Thr in Det7 and Ser in SP6). Together with our observations on Det7 tail spike refolding, this finding strongly suggests that disulfide formation is not an essential feature of the folding and assembly of this class of proteins which, in vivo, occurs in the reducing cytoplasm of the enterobacterial cell.
Evolution of bacteriophage tail spikes.
Although the sequences of the major parts of the P22 and Det7 tail spike polypeptides are 60% identical, there is no significant sequence homology between their amino-terminal parts. This difference in sequence homology reflects function, since the amino-terminal domains have to bind different phage particles, whereas the carboxy-terminal parts preserve the same receptor-binding and hydrolysis specificities. A BLAST search (2) with residues 1 to 160 of the Det7 tail spike polypeptide turned up segments of several bacteriophage endosialidases, such as amino acids 179 to 215 of the phage K1F tail protein (29, 36), which are 81% identical to residues 123 to 159 of the Det7 tail spike sequence. K1F is a T7-like phage with the ability to replicate on encapsulated E. coli K1. The capsule-degrading endosialidase portion of its tail protein (residues 246 to 911) has been structurally characterized (43). It may be connected to a head-binding domain via the segment exhibiting sequence similarity to Det7 tail spike. We did not find a sequence match to any known protein for Det7 tail spike residues 1 to 120, which might thus represent the base-plate binding domain.
Until very recently (19), podoviral tail spike proteins, as represented by the prototype P22 tail spike, were the only oligomeric right-handed β-helix structures known, although the fold is widespread among monomeric microbial endoglycosidases and polysaccharide lyases and appears to dominate the autotransporter family of secreted bacterial proteins (20). BLAST searches have revealed two major classes of sequence homologues of the P22 tail spike protein. Homologues with very high sequence identity (ca. 98%) throughout the protein are found in the Salmonella phages A1, A18a, ST104, and ST64T and in putative prophages contained within Salmonella genomes. In a second class of homologues, high sequence identity to the P22 tail spike (≥60%) is limited to its amino-terminal 115 residues, which form the head-binding domain. These proteins are encoded by Shigella phage Sf6, E. coli H phage HK620, and podoviruses of other enterobacteria. No sequence homology is detectable in the more carboxy-terminal parts of these proteins, although biochemical data suggest that the overall structures of the Sf6 and HK620 tail spikes may be similar to that of the functional homologue from P22 (14). The data suggest a modular structure of the tail spike genes of P22-like bacteriophages, in which the part coding for the head-binding domain is combined with different gene fragments coding for endoglycosidase modules with different receptor-binding specificities (6).
On the other hand, Salmonella phage SP6, which is more similar to coliphage T7 than to P22 in appearance, carries a tail spike protein of 550 residues. SP6 tail spike exhibits close to 60% sequence identity to the receptor binding segment of P22 tail spike but lacks the head-binding domain (11, 37, 38). It may be anchored to the phage particle through noncovalent interaction with an adaptor protein (gp37) that exhibits sequence similarity in its amino-terminal part of 160 residues with the putative particle binding domain of the T7 tail fiber (11). In our newly discovered Det7 tail spike, the receptor-binding segment now appears to be covalently linked to a myoviral particle binding module. Among the P22-like tail spike proteins, SP6 is the one most similar to the Det7 protein (76% identical of 542 aligned residues), and the sequence identity extends into the extreme carboxy-terminal parts of the two proteins (58% identity for residues 656 to 708), in contrast to the lower similarity to the P22 tail spike. This suggests that the two tail spike proteins lacking the P22-like head-binding module have a common precursor. Interestingly, the recently sequenced Salmonella bacteriophage KS7 (GenBank accession number NC_006940) appears to contain a P22-like tail spike with sequence similarity to the Det7 tail spike. The similarity extends amino terminally beyond residue 153, i.e., the amino terminus of the shortened protein studied here, to residue 120, thus covering the region with sequence similarity to endosialidases of T7-like bacteriophages mentioned above. Again, this might indicate that the Det7 tail spike, with the exception of its extreme amino-terminal part of approximately 100 residues, originates from a T7-like phage.
In conclusion, we provide here evidence for receptor-binding domain transfer between podoviridae and myoviridae. The high sequence and structural homology, combined with the short generation times and high mutation rate of bacteriophages, suggests that in this case transfer was relatively recent. It also appears not the whole protein was transferred but that a recombination of a P22-like receptor-binding domain with a myovirus-binding amino-terminal domain has taken place. Our evidence for recent exchange of adhesion modules also suggests that the cell attachment and DNA injection mechanisms of podoviruses and myoviruses are more closely related than may be expected on the basis of their distinct tail morphologies.
Acknowledgments
We are grateful to Gavin Fox (Spanish CRG BM16; ESRF, Grenoble, France) and Dave Hall of the EMBL-ESRF Joint Structural Biology Group for providing crystallographic data collection facilities and help therewith. We thank Adriane Lochner for excellent technical assistance with Electron Microscopy.
M.J.V.R. was supported by a Ramón y Cajal contract of the Spanish Ministry of Education and Science. This study was supported by the grants BFU2005-02974 (Spanish Ministry of Education and Science) and PGIDIT-03-PXIC-20307-PN (Xunta de Galicia) to M.J.V.R. Both grants were cofinanced by the European Union. Work by the Potsdam group was supported by the German Research Foundation through grants Se517/16-2 and Se517/16-3. C.F. is the recipient of a fellowship by the State of Brandenburg in the framework of the International Max Planck Research School on Biomimetic Systems.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Ackermann, H. W. 2005. Bacteriophage classification, p. 67-89. In E. Kutter and A. Sulakvelidze (ed.) Bacteriophages: biology and applications. CRC Press, Boca Raton, FL.
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. W. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxa, U., S. Steinbacher, S. Miller, A. Weintraub, R. Huber, and R. Seckler. 1996. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys. J. 712040-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baxa, U., S. Steinbacher, A. Weintraub, R. Huber, and R. Seckler. 1999. Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity. J. Mol. Biol. 293693-701. [DOI] [PubMed] [Google Scholar]
- 5.Betts, S., and J. King. 1999. There's a right way and a wrong way: in vivo and in vitro folding, misfolding and subunit assembly of the P22 tailspike. Structure Fold. Des. 7R131-139. [DOI] [PubMed] [Google Scholar]
- 6.Casjens, S. R. 2005. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8451-458. [DOI] [PubMed] [Google Scholar]
- 7.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D 50760-763. [DOI] [PubMed] [Google Scholar]
- 8.Danner, M., and R. Seckler. 1993. Mechanism of phage P22 tailspike protein folding mutations. Protein Sci. 111869-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danner, M., A. Fuchs, S. Miller, and R. Seckler. 1993. Folding and assembly of phage P22 tailspike endorhamnosidase lacking the N-terminal, head-binding domain. Eur. J. Biochem. 215653-661. [DOI] [PubMed] [Google Scholar]
- 10.DeLano, W. L. 2002. The PyMOL molecular graphics system. [Online.] http://www.pymol.org.
- 11.Dobbins, A. T., M. George, Jr., D. A. Basham, M. E. Ford, J. M. Houtz, M. L. Pedulla, J. G. Lawrence, G. F. Hatfull, and R. W. Hendrix. 2004. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J. Bacteriol. 1861933-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esnouf, R. M. 1999. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. Sect. D 55938-940. [DOI] [PubMed] [Google Scholar]
- 13.Fane, B., R. Villafane, A. Mitraki, and J. King. 1991. Identification of global suppressors for temperature-sensitive folding mutations of the P22 tailspike protein. J. Biol. Chem. 26111640-11648. [PubMed] [Google Scholar]
- 14.Freiberg, A., R. Morona, L. van den Bosch, C. Jung, J. Behlke, N. Carlin, R. Seckler, and U. Baxa. 2003. The tailspike protein of Shigella phage Sf6. A structural homolog of Salmonella phage P22 tailspike protein without sequence similarity in the beta-helix domain. J. Biol. Chem. 2781542-1548. [DOI] [PubMed] [Google Scholar]
- 15.Goldenberg, D. P., and T. E. Creighton. 1994. Phage tailspike protein: a fishy tale of protein folding. Curr. Biol. 41026-1029. [DOI] [PubMed] [Google Scholar]
- 16.Haase-Pettingell, C., and J. King. 1997. Prevalence of temperature-sensitive folding mutations in the parallel beta coil domain of the phage P22 tailspike endorhamnosidase. J. Mol. Biol. 26788-102. [DOI] [PubMed] [Google Scholar]
- 17.Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and location of errors in these models. Acta Crystallogr. Sect. A 47110-119. [DOI] [PubMed] [Google Scholar]
- 19.Jung, W.-S., C.-K. Hong, S. Lee, C.-S. Kim, S.-J. Kim, S.-I. Kim, and S. Rhee. 2007. Structural and functional insights into intramolecular fructosyl transfer by inulin fructotransferase. J. Biol. Chem. 2828413-8423. [DOI] [PubMed] [Google Scholar]
- 20.Junker, M., C. C. Schuster, A. V. McDonnell, K. A. Sorg, M. C. Finn, B. Berger, and P. L. Clark. 2006. Pertactin beta-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc. Natl. Acad. Sci. USA 1034918-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 222577-2637. [DOI] [PubMed] [Google Scholar]
- 22.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24946-950. [Google Scholar]
- 23.Lamzin, V. S., and K. S. Wilson. 1997. Automated refinement for protein crystallography. Methods Enzymol. 277269-305. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26283-291. [Google Scholar]
- 25.Lee, S. C., H. Koh, and M. H. Yu. 1991. Molecular properties of global suppressors of temperature-sensitive folding mutations in P22 tailspike endorhamnosidase. J. Biol. Chem. 26623191-23196. [PubMed] [Google Scholar]
- 26.Leslie, A. G. 1999. Integration of macromolecular diffraction data. Acta Crystallogr. Sect. D 551696-1702. [DOI] [PubMed] [Google Scholar]
- 27.Liu, C., M. C. Eichelberger, R. W. Compans, and G. M. Air. 1995. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J. Virol. 691099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, S., B. Schuler, and R. Seckler. 1998. Phage P22 tailspike protein: removal of head-binding domain unmasks effects of folding mutations on native-state thermal stability. Protein Sci. 72223-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mühlenhoff, M., K. Stummeyer, M. Grove, M. Sauerborn, and R. Gerardy-Schahn. 2003. Proteolytic processing and oligomerization of bacteriophage-derived endosialidases. J. Biol. Chem. 27812634-12644. [DOI] [PubMed] [Google Scholar]
- 30.Murphy, F. A. 1995. Virus taxonomy, p. 15-57. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott/The Williams & Wilkins Co., Philadelphia, PA.
- 31.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 32.Navaza, J. 2001. Implementation of molecular replacement in AMoRe. Acta Crystallogr. Sect. D 571367-1372. [DOI] [PubMed] [Google Scholar]
- 33.Perrakis, A., R. Morris, and V. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6458-463. [DOI] [PubMed] [Google Scholar]
- 34.Powell, H. R. 1999. The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr. Sect. D 551690-1695. [DOI] [PubMed] [Google Scholar]
- 35.Robinson, A. S., and J. King. 1997. Disulphide-bonded intermediate on the folding and assembly pathway of a non-disulphide bonded protein. Nat. Struct. Biol. 4450-455. [DOI] [PubMed] [Google Scholar]
- 36.Scholl, D., and C. Merril. 2005. The genome of bacteriophage K1F, a T7-like phage that has acquired the ability to replicate on K1 strains of Escherichia coli. J. Bacteriol. 1878499-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scholl, D., S. Adhya, and C. R. Merril. 2002. Bacteriophage SP6 is closely related to phages K1-5, K5, and K1E but encodes a tail protein very similar to that of the distantly related P22. J. Bacteriol. 1842833-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scholl, D. J. Kieleczawa, P. Kemp, J. Rush, C. C. Richardson, C. Merril, S. Adhya, and I. J. Molineux. 2004. Genomic analysis of bacteriophages SP6 and K1-5, an estranged subgroup of the T7 supergroup. J. Mol. Biol. 3351151-1171. [DOI] [PubMed] [Google Scholar]
- 39.Seckler, R. 1998. Folding and function of repetitive structure in the homotrimeric phage P22 tailspike protein. J. Struct. Biol. 122216-222. [DOI] [PubMed] [Google Scholar]
- 40.Steinbacher, S., R. Seckler, S. Miller, B. Steipe, R. Huber, and P. Reinemer. 1994. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265383-386. [DOI] [PubMed] [Google Scholar]
- 41.Steinbacher, S., U. Baxa, S. Miller, A. Weintraub, R. Seckler, and R. Huber. 1996. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc. Natl. Acad. Sci. USA 9310584-10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinbacher, S., S. Miller, U. Baxa, N. Budisa, A. Weintraub, R. Seckler, and R. Huber. 1997. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 Å, fully refined structure of the endorhamnosidase at 1.56 Å resolution, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 267865-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stummeyer, K., A. Dickmanns, M. Mühlenhoff, R. Gerardy-Schahn, and R. Ficner. 2005. Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat. Struct. Mol. Biol. 1290-96. [DOI] [PubMed] [Google Scholar]
- 44.Tang, L., W. R. Marion, G. Cingolani, P. E. Prevelige, Jr., and J. E. Johnson. 2005. Three-dimensional structure of the bacteriophage P22 tail machine. EMBO J. 242087-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weigele, P. R., C. Haase-Pettingell, P. G. Campbell, D. C. Gossard, and J. King. 2005. Stalled folding mutants in the triple beta-helix domain of the phage P22 tailspike adhesin. J. Mol. Biol. 3541103-1117. [DOI] [PubMed] [Google Scholar]