Abstract
Various domains of hepatitis B surface antigen (HBsAg) are essential for the assembly and secretion of hepatitis D virus (HDV). This study investigated the influences of the levels and sequences of HBsAg of naturally occurring HBV variants on the assembly and secretion of HDV. Six hepatitis B virus (HBV)-producing plasmids (three genotype B and three genotype C) and six HBsAg expression plasmids that expressed various HBsAg levels were constructed from the sera of HDV-infected patients. These plasmids were cotransfected with six expression plasmids of HDV of genotype 1, 2, or 4 into the Huh-7 hepatoma cell line. Serum HBsAg and HBV DNA levels were correlated with HDV RNA levels and outcomes of chronic hepatitis D (CHD) patients. The secretion of genotype 1, 2, or 4 HDV generally correlated with HBsAg levels but not with HBV genotypes or HBV DNA levels. Swapping and residue mutagenesis experiments of HBsAg-coding sequences revealed that the residue Pro-62 in the cytosolic domain-I affects the assembly and secretion of genotype 2 and 4 HDV and not those of genotype 1. The pre-S2 N-terminal deletion HBV mutant adversely affects secretion of the three HDV genotypes. In patients, serum HDV RNA levels correlated with HBsAg levels but not with HBV DNA levels. Viremia of HDV or HBV correlated with poor outcomes. In conclusion, the assembly and secretion of HDV were influenced by the amounts and sequences of HBsAg. For an effective treatment of CHD, reduction of HBsAg production in addition to the suppression of HBV and HDV replication might be crucial.
Hepatitis D virus (HDV) requires the help of hepatitis B virus (HBV) for viral assembly and transmission (43, 58). HDV is a 36-nm spherical virus particle consists of two major parts. The outer envelope is composed of hepatitis B surface antigen (HBsAg) (including large HBsAg [L-HBsAg], middle HBsAg [M-HBsAg], and small HBsAg [S-HBsAg], respectively) and lipids (3, 49, 50, 51). The inner viral particles are ribonucleoprotein (RNP) complexes, including a single-stranded circular 1.7-kb HDV RNA genome and the hepatitis delta antigen (HDAg) (11, 31, 54, 55). HDV synthesizes two forms of HDAg termed large (HDAg-L; 27 kDa) and small HDAg (HDAg-S; 24 kDa) from a single open reading frame on the antigenomic strand (54, 55). The HDAg-L is identical to the HDAg-S except for a 19-amino-acid (aa) extension at the C terminus by RNA editing during life cycle (7, 37). These two HDAgs have distinct functions in the HDV replication cycle. The HDAg-S plays an essential role in transactivating the replication of the HDV RNA (9, 30). For HDAg-L, two main functions were identified: (i) a key role interacting with HBsAg in the assembly of HDV (8, 10, 14) and (ii) a dominant-negative repressor of HDV RNA replication (9, 10, 15).
S-HBsAg is sufficient for HDV assembly but does not produce virions with infectivity (49, 52). Pre-S1 protein of the L-HBsAg has an HBV receptor-binding site, and the antigenic loop (AGL) of the major HBsAg also contains a determinant of infectivity. Both domains play important roles in the infectivity of HBV (5, 18, 25, 49), whereas M-HBsAg is dispensable for the morphogenesis of infectious HBV and HDV (4, 13, 48). Moreover, several studies suggest that mutations in HBsAg have detrimental effects on the assembly of HDV. For examples, deletion of amino acid residues 24 to 28 at cytosolic loop I (CYL-I) or substitutions of tryptophan at residues 196, 199, and 201 in cytosolic loop II (CYL-II) of S-HBsAg affected HDV assembly (1, 26, 27, 29).
The three forms of HBsAg have glycosylated and nonglycosylated isomers (32, 48, 53). N-linked glycosylation occurs at asparagine 4 (Asn4) of the M-HBsAg and at Asn146 of S-HBsAg, M-HBsAg, and L-HBsAg. However, about half of the HBsAg proteins remain unglycosylated at these sites. Removal of N-linked glycans on the HBsAg by tunicamycin treatment or N146T mutation is tolerated for the assembly of subviral HBV particles but is partially inhibitory for the assembly of HDV virions (48, 53).
HDV infection is an important etiology of fulminant hepatitis (16, 19, 56, 62). Since chronic HBV infection status could continuously provide HBsAg, more than 70 to 90% of chronic hepatitis B patients with HDV superinfection may progress to chronicity (59, 62). Furthermore, HDV superinfection may aggravate the underlying chronic hepatitis B to cirrhosis and hepatocellular carcinoma (HCC) (17, 44, 46, 59, 62).
Based on phylogenetic analysis, HDV was initially classified into three genotypes, increased to four genotypes later, and recently classified into at least eight major clades named genotypes 1 to 8 (6, 33, 60). Genotypes of HDV are closely related to the clinical course (6, 61). Genotype 3 HDV is prevalent in Southern America and is more frequently associated with fulminant hepatic failure (6). Our previous study has shown that both genotype 1 and 2 HDVs are prevalent in Taiwan and that genotype 2 HDV infection is less frequently associated with fulminant hepatitis at the acute stage and unfavorable long-term outcomes at the chronic stage than is genotype 1 (61). Furthermore, the efficiency of assembly of genotype 1 HDV is generally higher than that of genotype 2, which may at least partly explain the association of HDV genotypes with disease outcomes (20).
According to phylogenetic analysis of nucleotide sequences, HBV can also be classified into eight different genotypes A to H (12). In Taiwan, HBV genotypes B and C are the most prevalent (28), and genotype C HBV-infected patients more frequently develop cirrhosis or HCC than genotype B HBV-infected patients (28, 63). In addition, for patients with HBV and HDV dual infection, we recently observed that genotype 1 HDV, genotype C HBV, and persistent viremia were associated with poor prognosis in long-term follow-up (46). It is unclear whether genotype C HBV exerts its impact on worse outcomes directly or indirectly through a more efficient help in the assembly of HDV. Thus far, the interactions between hepatitis B and D viruses of different genotypes are still obscured. In the present study, we focused on the assembly and secretion efficiencies of HDV in the presence of various levels and sequences of HBsAg in a cell culture model. Plasmids producing genotype B and C HBVs or HBsAg constructed from natural HBV variants in patients were cotransfected with different genotypes of HDV (genotypes 1, 2, and 4, respectively) into a an HCC line, Huh-7 cells (40). We found that the assembly and secretion of HDV are more closely associated with the levels and sequences especially at residue 62 in the CYL-I region of S-HBsAg and the N-terminal region of middle-HBsAg than HBV genotypes per se. Serum HBsAg levels also correlated with HDV RNA levels in chronic hepatitis D (CHD) patients.
MATERIALS AND METHODS
Plasmids for HBV expression.
The sources of the different HBV and HDV plasmids in the present study were derived from CHD patients. An HBV partial genome sequence (approximately 3,056 bp) that contains complete HBV envelope open reading frame was amplified by using a pair of primers—B2600 NheI (5′-CCGCTAGCCTTACAGTAAATGAAAA-3′) and B25R (5′-TCCCACCTTATGTGTCCA-3′) Also, does the use of lowercase letters need to be explained?—via PCR and then cloned into the NheI/HindIII sites in the plasmid pcDNA3.1(−) (Invitrogen, San Diego, CA) vector. Another HBV partial genome sequence (approximately 1,859 bp) was amplified by the primer pair B980 NheI (5′-GGAAAGTATGCTAGCGAATTGTGG-3′) and B2839 BstEII (5′-CCAAGAATATGGTGACCC-3′) by PCR and then cloned into the NheI/BstEII sites in the plasmid containing the previously described 3-kb HBV partial sequence to become a replicative form of HBV construct. A total of six HBV-producing plasmids were constructed with three genotype B (numbered 1 to 3 [pHBV1, pHBV2, and pHBV3, respectively]) and three genotype C (numbered 4 to 6 [pHBV4, pHBV5, and pHBV6, respectively]) HBV. The accession numbers of GenBank for those six HBV-producing plasmids were EF494382, EF494380, EF494381, EF494376, EF494377, and EF494378, respectively. Also, the expression plasmids of the entire large, middle, and major HBsAgs (pHBVenv1, pHBVenv2, pHBVenv3, pHBVenv4, pHBVenv5, and pHBVenv6) were constructed from the previously described six HBV-producing plasmids. All of the plasmids expressing HBV genomes or HBsAg were driven under the control of cytomegalovirus (CMV) promoter.
Construction of plasmids expressing chimeric or mutated form of HBsAg.
Plasmids expressing chimeric forms of genotype C HBV were constructed by molecular cloning techniques. In summary, the pHBVenv4 and pHBVenv6 of HBV genotype C were digested with XbaI/XcmI and XbaI/BamHI, respectively. The digested products were swapped into the other XbaI/XcmI- and XbaI/BamHI-digested vector. For example, the chimeric plasmid, pHBVenv4-6-4 (aa 37 to 185) indicates that the pHBVenv4 contains a swapped insert coding aa 37 to 185 of HBsAg of pHBVenv6. Plasmids of pHBVenv6 for the expression of L61S, L62P, I110L, T126I, I131T, S140T, and K160R mutated or doubly mutated (L61S and L62P) HBsAg were constructed by using the PCR overlap extension method.
Construction of HDV expression plasmids.
HDV genotypes were determined by PCR-restriction fragment length polymorphism and verified by sequencing and phylogenetic analysis as described previously (61). To generate the replicative form of HDV cDNA, plasmids expressing HDV genotypes 1 (TWD2577-66), 2 (TWD2479-12), and 4 (TWD62) were constructed as described previously (20). The accession numbers of GenBank for these constructs were AF425644, AY261457, and AF018077, respectively. Upon transfection of these plasmids into Huh-7 cells, genomic-sense HDV RNA was produced.
Plasmids for L-HDAg expression.
A total of six plasmids expressing L-HDAg which were cloned into pCMV-EBNA (Clontech Laboratories, Palo Alto, CA) and constructed as previously described (20). TW1629 (16-L; AF352570), TW2577 (25-L), and TW2683 (83-L; AF352571) belong to genotype 1 HDV isolates, and TW842 (8-L; AF352568), TW2476 (24-L; AF104264), and TW3937 (39-L; AF352569) belong to genotype 2 HDV isolates.
Cell transfection.
The well-differentiated HCC cell line Huh-7 cells were used in the present study (40). Transfection of DNA was carried out by the calcium phosphate-DNA precipitation method as previously described (20, 21, 58). For production of HDV particles, 10 μg of HDV- and 10 μg of HBV-expressing plasmids were cotransfected into HuH-7 cells. The medium was changed after 6 to 8 h posttransfection, and subsequently medium was replaced and collected every 3 days for 9 days.
Western blot analysis.
For analysis of the expression of HDAgs and HBsAg, cell lysates and culture media from transfected Huh-7 cells were harvested on days 3, 6, and 9 posttransfection, respectively. The isolated proteins after sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis separation and blotting onto nitrocellulose membranes were stained for HDAgs with anti-HDV-positive human serum (1:5,000) (20, 21, 57) and for HBsAgs with monoclonal antibody A10F1 (1:2,000), respectively (36). The reference protein, heat shock cognate protein (Hsc70), was detected with monoclonal antibody HSC 70 (B-6; 1:5,000; Santa Cruz Biotechnology, Santa Cruz, CA). The antigen-bound antibodies on the membrane were detected by horseradish peroxidase-conjugated goat anti-human antibody (ab6858; Abcam Cambridge Science Park, Cambridge, United Kingdom) and horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG; ab6789; Abcam Cambridge Science Park) as previously described (20, 21). Finally, the signals were developed by Western blotting chemiluminescence reagent (MEN Life Science, Boston, MA).
Indirect double immunofluorescence staining and counting of transfected cells.
Immunofluorescence staining was modified and performed as previously described (57). Before DNA transfection, the Huh-7 cells were cultured overnight on coverslips. After days 3 and 6 posttransfection with different combinations of HBV- and HDV-producing plasmids, the cells were washed twice in ice phosphate-buffered saline (PBS [pH 7.4]) and then fixed and permeated with acetone at 4°C for 20 min. After multiple rinses with PBS, the cells were treated with PBS containing 1% bovine serum albumin at room temperature for 30 min. Double immunofluorescence staining of cells incubated with primary antibodies of polyclonal human anti-delta serum diluted 1:200 and monoclonal mouse antibody to HBsAg diluted 1:200 (m3506; DakoCytomation, Carpinteria, CA) was performed in the same buffer at room temperature for 30 min. After the cells were washed three times with PBS, secondary antibodies of rhodamine-conjugated rabbit antihuman IgG (ab6756; Abcam Cambridge Science Park) and fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG (ab6724; Abcam Cambridge Science Park, Cambridge, United Kingdom) were added, followed by incubation at room temperature for 30 min. Both conjugates were diluted 1:200 in the same buffer. Finally, the coverslips were washed thoroughly with PBS and then mounted with 50% glycerol. Photographs were taken by using a confocal fluorescence microscope (Axiovert 200M; Zeiss, Germany) with an oil ×63 objective lens (NA = 1.4; Zeiss). An argon 488-nm laser (45 mW; LASOS, Germany) and He-Ne 543-nm laser (2.5 mW; LASOS) were used to excite HDAg-rhodamine and HBsAg-FITC, respectively. To analyze the efficiency of transfection and the percentages of cells expressing HBsAgs, HDAgs, or both, we randomly selected 10 high-power fields to observe the intensity of fluorescence staining and count the number of cells with HBsAg and/or HDAg expression.
Analysis of HDV RNA.
Total cellular RNAs from harvested Huh-7 cells or viral particles from concentrated supernatant after high-speed centrifugation were extracted by TRIzol reagent (Life Technologies, Grand Island, NA). RNA purification was performed according to the manufacturer's instructions. A total of 20 μg of RNA was analyzed by Northern blotting as previously described (20, 21, 57, 58). After fixation by UV illumination, RNA was hybridized with digoxigenin (DIG)-labeled cDNA probes derived from different genotypes of HDV templates as previously described (20, 21). Hybridization was performed by incubation with a solution of DIG easy hyb granules (DIG labeling and detection kit; Roche Diagnostics System, Basel, Switzerland) at 55°C overnight. For controls, hybridization with DIG-labeled glyceraldehyde-3-phosphate dehydrogenase (G3PDH) gene was performed simultaneously.
Real-time PCR for HDV RNA quantitative assay.
Total RNA was extracted by using the acid guanidinium phenol-chloroform method and reverse transcription was carried out as previously described (59). A total of 20 μl of final cDNA solution was obtained from 50 μl of serum. To synthesize standard DNA, the HDV genome coding partial HDAg was amplified by PCR with primer 214 (5′-CACAAGAGCGGGTTCACCGACA-3′) and primer 120 (5′-ATGCCATGCCGACCCGAAGAGGAA-3′). The PCR products were inserted into the pCRII vector by TA cloning. The constructive plasmids were used as the standards for HDV cDNA quantification (plasmid 1078-5 is for HDV type 1, plasmid 110-A2 is for type 2, and plasmid D62-28 is for type 4). The purified plasmids were quantified by measurement of the optical density at a wavelength of 260 nm.
Real-time PCR was performed by using the BD QTaq DNA polymerase mix (Clontech Laboratories). Purified cDNA (5 μl) was added to a 20-μl PCR mixture containing 12.5 μl of BD QTaq DNA polymerase mix, 900 nM concentrations of each primer, and 250 nM fluorescent TaqMan MGB probe. The standard curve for this assay was calculated by using a series of 10-fold dilutions (from 5 to 5 × 106 copies/reaction) of previously titrated plasmids (p1078-5 is for HDV type 1, p110-A2 is for type 2, and pD62-28 is for type 4). The reaction consisted of one initiating step of 3 min at 95°C, followed by 40 cycles of amplification including 15 s at 95°C and 1 min at 60°C. The reactions, data acquisition, and analyses were performed with the ABI Prism 7700 sequence detection system (Applied Biosystems, Courtaboeuf, France).
The sensitivity and linearity of the real-time PCR method for the quantitative assay of HDV RNA were examined. The plasmids containing HDAg-coding sequences were used as standards to quantify HDV RNA in sera. Tenfold serial dilutions ranging from 5 to 5 × 106 copies were tested in triplicate, with mean cycle threshold (CT) values plotted against the copy number to establish a standard curve. The correlation coefficients were repeatedly >0.995, and the slopes were ranged from −3.1 to −3.4. Dilutions corresponding to inputs of five copies per reaction were repeatedly detected (results not shown). Thus, according to the dilution factors used during the RNA extraction and RT procedures, the sensitivity of the assay to detect HDV RNA in clinical samples was 400 copies/ml of serum, and the linearity of quantification ranged from 2 × 103 to 2 × 109 copies/ml.
Immunoprecipitation of intracellular nucleocapsids and extracellular virions of HBV.
The transfected cells and culture media were harvested on day 9 posttransfection. Cells were washed twice with ice-cold GKNP buffer (54 mM KCl, 4.4 mM KH2PO4, 136 mM NaCl, 0.34 mM Na2HPO4, 5.6 mM glucose, 0.001% phenol red [pH 7.4]) and then resuspended in 1 ml of NET buffer (50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 150 mM NaCl, 0.5% Nonidet P-40) containing 5 mM MgCl2 and kept on ice for 30 min. The cell lysate was centrifuged at 12,000 rpm for 10 min at 4°C to remove nuclei and cell debris. The remaining supernatant was treated with 10 μg of DNase I (Roche)/ml and 20 μg of RNase A (Roche)/ml at 37°C for 1 h. On the other hand, the culture media were clarified by low-speed centrifugation at 3,000 rpm at 4°C for 15 min to remove cell debris. The viral particles from 9 ml of clear culture supernatant were pelleted by high-speed centrifugation at 40,000 rpm at 4°C for 5 h in a SW41 rotor (Beckman) through a 20% sucrose cushion and then resuspended in 0.5 ml of 0.1× TE (0.1 mM EDTA, 1 mM Tris-HCl [pH 8.0]) buffer. The cell lysate and culture media sample were immunoprecipitated with an antibody to HBV core antigen (DakoCytomation) and protein A-Sepharose beads (Pharmacia, Germany) and an antibody to HBsAg (DakoCytomation) and protein A-Sepharose beads at 4°C for 4 h, respectively. By three washes with NET buffer, the sample was treated with NET buffer supplemented with 0.5% SDS, 20 μg of proteinase K (Roche)/ml, 20 μg of glycogen, and 5 N NaCl at 54°C for 30 min and then incubated at 37°C overnight. The DNA was extracted, precipitated, and finally dissolved in 0.1× TE for HBV DNA quantitative analysis.
Quantitative analysis of HBsAg and HBV DNA.
HBsAg in spent media or human serum was quantified by using a General Biological Surdine125 B kit (General Biologicals Corp., Taiwan). HBV DNA was measured by a Cobas Amplicor HBV monitor (Roche Diagnostic System, Basel, Switzerland). The detection limit of this assay is 300 copies/ml.
In order to show that the extracellular HBV DNA levels were not due to the contamination of residual plasmid DNA, PCR was carried out with three pairs of primers to amplify a product containing a segment of the HBV insert sequence and two products containing only segments of the vector sequences of pcDNA 3.1(−), respectively. The sensitivity of the PCR was determined by amplification of serial dilutions of the template plasmid (pHBV1). As few as 10 to 100 copies per reaction could be detected by using three sets of primers. After immunoprecipitation of HBV virions with antibody to the HBsAg envelope of HBV, the PCR experiments with primers based on the vector sequence all showed negative results, whereas the PCR experiments with primers based on the HBV insert sequence all showed positive results despite the fact that the primers based on vector sequences provided an equal or slightly higher sensitivity in PCR. These control experiments clearly excluded the contamination of residual plasmid DNA in immunoprecipitated HBV virions.
Long-term follow up of CHD patients.
In order to investigate whether serum HDV RNA levels also correlate with the amounts of serum HBsAg in CHD patients and clinical outcomes, 56 consecutive CHD patients who visited Taipei Veterans General Hospital and had sufficient stored serum samples were enrolled in a study to examine the clinical relevance of serum HBsAg, HBV DNA, and HDV RNA levels. All of the patients had underlying chronic hepatitis B based on a history of positive HBsAg for more than 6 months and being negative for IgM antibody to hepatitis B core antigen (IgM anti-HBc; Abbott Laboratories, North Chicago, IL). These patients were all positive for antibodies to HDV antigen (anti-HDV, anti-Delta; Abbott) and negative for both antibodies to hepatitis C virus and human immunodeficiency virus. Baseline serum HBsAg, HBV DNA, and HDV RNA levels were tested. The patients were monitored regularly at least every 3 to 6 months after enrollment, and outcomes such as cirrhosis, HCC, and clinical remission were recorded as previously described (46).
Statistical analysis.
All statistical analyses were performed by the Statistical Program for Social Sciences (SPSS 10.0 for Windows; SPSS., Inc., Chicago, IL). The correlation between serum HDV RNA, HBsAg, and HBV DNA levels was studied by Spearman rank correlation analysis. Clinical outcomes were compared by using the Fisher exact test. A two-tailed P value of <0.05 was considered significant in all tests.
RESULTS
Predicted amino acid sequences and levels of the HBsAg expressed by HBV-producing plasmids of different genotypes.
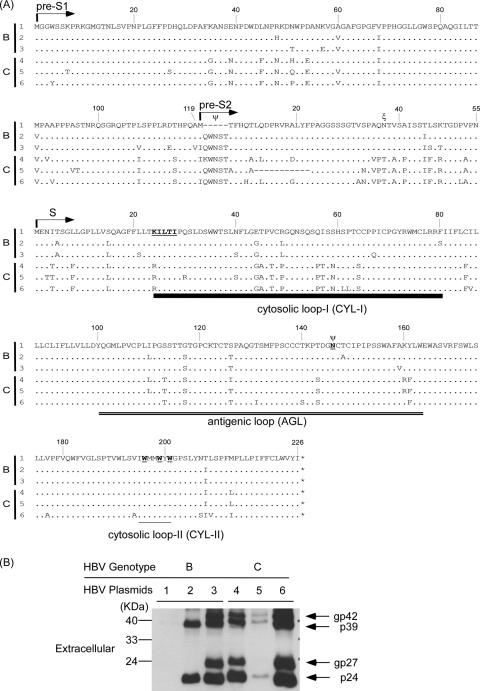
The predicted amino acid sequences of the L-, M-, and S-HBsAgs of the six HBV-producing plasmids are shown in Fig. 1A. The numbers of the amino acids start from the initiation codon of the L-, M-, and S-HBsAgs, respectively. There were minor variations in amino acid sequences within the same genotype. However, there were some genotype-specific changes in amino acids, e.g., residue 37 of the M-HBsAg associated with O glycosylation was threonine in genotype C but was Asn in genotype B HBV. Asn4 of M-HBsAg and residue 146 of S-HBsAg associated with N glycosylation were conserved in all of the six HBV isolates, except pHBV1 that has a small deletion at the coding sequence for the first 2 to 6 aa, including the Asn4 of the M-HBsAg. aa 12 to 22 of the M-HBsAg expressed by pHBV5 were also deleted. The HBsAgs expressed by these HBV-producing plasmids on day 9 posttransfection are shown in Fig. 1B. pHBV3, -4, and -6 did not express M-HBsAg due to the lack of the pre-S2 start codons (Fig. 1A), and pHBV1 and -5 had small deletions in the pre-S2 domains that might be associated with undetectable M-HBsAg. The secreted HBsAg levels varied among the cells transfected by the six HBV plasmids. As shown in Fig. 1B, the HBsAg level was the lowest and was hardly detectable by Western blot analysis in lane 1 and was highest in lane 6. There were no glycosylated forms of HBsAg in lane 2.
FIG. 1.
(A) Schematic representation of the predicted amino acid sequences of pre-S1, pre-S2, and S domains expressed by the six HBV-producing plasmids. The N termini of the pre-S1, pre-S2, and S proteins are indicated by arrows. The CYL-I is marked by a thick line. The AGL is marked by double underlines. The CYL-II is marked by a thin line. The N- and O-linked glycosylation sites are indicated by “Ψ” and “ξ,” respectively. The amino acids affected HDV assembly and secretion described in previously studies (1, 26, 27, 29) are marked by boldface underlined letters. (B) The HBsAg expressed and secreted into media by the six HBV-producing plasmids at day 9 posttransfection. The glycosylated and nonglycosylated isomers of L-HBsAg (p39 and gp42) and S-HBsAg (p24 and gp27) are indicated on the right site. The numbers 1 to 3 and the numbers 4 to 6 indicate the expression plasmids belonging to genotype B and C HBVs, respectively. pHBV3, -4, and -6 could not express M-HBsAg due to the lack of pre-S2 start cordons. The pHBV1 and -5 had small deletions in pre-S2 domains and also expressed barely detectable M-HBsAgs. The residues of N- and O-linked glycosylation sites in pHBV2 are conserved, whereas the glycosylated forms of L-, M-, and S-HBsAgs were not detected.
The influence of HBV genotypes, HBsAg expression, and HBV DNA levels on the assembly and secretion of HDV of different genotypes.
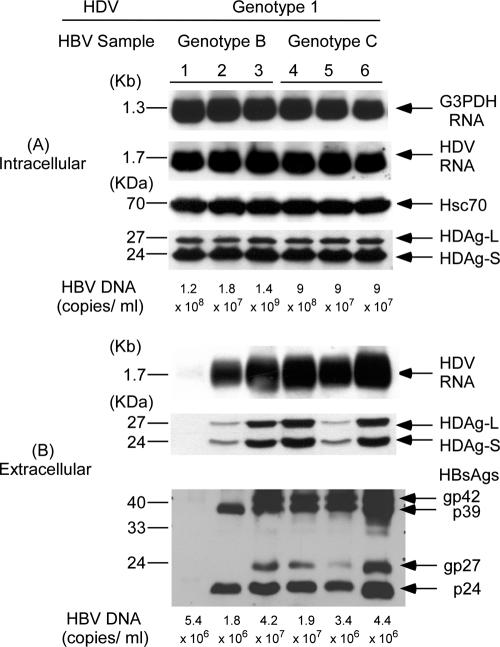
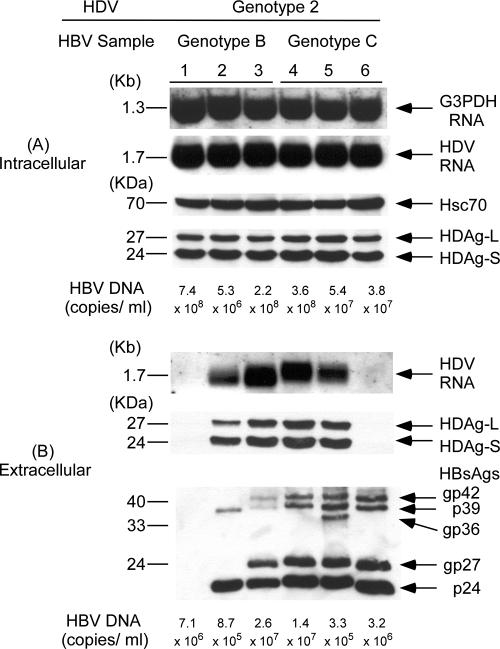
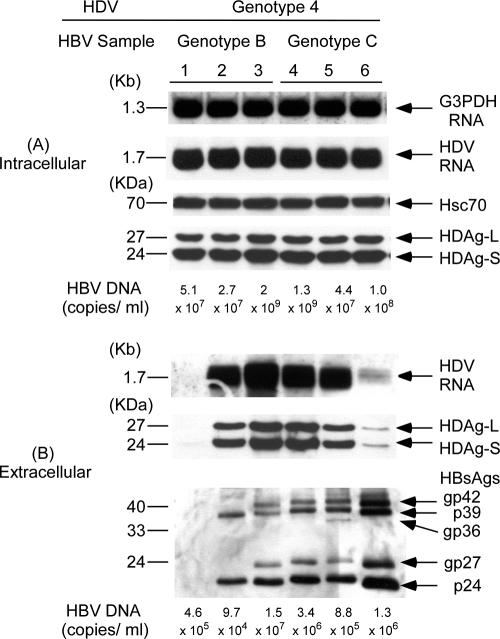
To investigate whether HBV genotypes affect HDV assembly, Huh-7 cells were cotransfected with an HDV genotype 1 plasmid and each one of the three HBV plasmids expressing genotype B or C HBV, respectively (Fig. 2). In Fig. 2A, the same amounts of HDV RNA and two forms of HDAgs were detected in cell lysates with any combination of HBV genotypes and genotype 1 HDV. The equal amounts of loaded samples were evidenced by similar amounts of G3PDH RNA and Hsc70 protein in each lane, respectively. In contrast, there were marked differences in HDV RNA and HDAg levels in culture media with different combinations of cotransfection (Fig. 2B). The difference in secreted HDV RNA shown by Northern blot analysis was supported by the results of real-time PCR. The hardly visible HDV RNA signal that could be seen only after prolonged exposure in lane 1 of Fig. 2B correlates with the lowest level of 2.36 × 105 copies/ml using a sensitive real-time PCR. The pHBV1 with a small deletion at the first 2 to 6 aa, including the N4 of the M-HBsAg, expressed and secreted the lowest amount of HBsAg and appeared to be the least competent helper for the assembly and secretion of HDV. The influence of the small deletion at the first 2 to 6 aa, including the N4 of the M-HBsAg, on HDV RNA assembly and secretion is described in greater detail below. The HDV RNA levels were 3.13 × 106, 1.70 × 107, 1.42 × 107, 8 × 106, and 1.43 × 107 copies/ml, respectively, in lanes 2 to 6, correlating with varied HDV RNA signals shown by Northern blot analysis. Experiments cotransfecting HDV genotype 2 or 4 plasmids with different genotypes of HBV plasmids showed similar results (Fig. 3 and 4, respectively). The secreted genotype 2 HDV RNA shown in lanes 1 to 6 of Fig. 3B correlated with 5.77 × 105, 2.22 × 106, 3.08 × 107, 1.02 × 107, 9.68 × 106, and 6.15 × 105 copies/ml using real-time PCR. The lowest HDV RNA level also correlated with the weakest signal in lane 1 of Fig. 3. In addition, a relatively lower level of secreted genotype 2 HDV RNA compared to those in lanes 2 to 5 was observed in lane 6 as either measured by Northern blot analysis or real-time PCR despite the highly secreted HBsAg levels. Similar correlation between the results of real-time PCR assay and those of Northern blot analysis was observed in the measurement of secreted genotype 4 HDV RNA in media (data not shown). Taken together, the results indicated that the assembly efficiency of HDV varied among different isolates within and between HBV genotypes. The influence of HBV genotypes was thus not obvious. Interestingly, larger amounts of HDV RNA and HDAg in medium appeared to correlate with the quantities of HBsAg in culture media, with the exception of lane 6 (these findings are described in greater detail below) (Fig. 2B, 3B, and 4B). Similar findings were observed in the cotransfections of HDV of various genotypes with HBsAg expression plasmids of different HBV genotypes (data not shown).
FIG. 2.
Correlation between secretion of genotype 1 HDV and different HBsAg expression provided by HBV-producing plasmids of various genotypes. (A) The cellular lysates were analyzed for intracellular HDV RNA and HDAg expression by Northern blotting with DIG-labeled cDNA probes and Western blotting with human anti-HDV, respectively. The equal loading of RNA and protein samples were assessed by hybridization with DIG-labeled DNA probe for G3PDH and monoclonal antibody specific for heat shock protein (Hsc70), respectively. (B) The HDV particles in 9 ml of culture medium were pelleted by high-speed centrifugation (40,000 rpm for 5 h at 4°C in a Beckman SW41 rotor) through a 20% sucrose cushion. The pellets were dissolved in the SDS sample buffer and analyzed for extracellular HDAg and HBsAg by Western blotting. The HDV RNA from pelleted HDV particles was analyzed by Northern blotting. After immunoprecipitation with anti-HBc and anti-HBs, respectively, the amounts of HBV DNA in cellular lysates and pelleted HBV virions were extracted, purified, and measured by using a Cobas Amplicor HBV monitor.
FIG. 3.
Correlation between secretion of genotype 2 HDV and different HBsAg expression provided by HBV-producing plasmids of various genotypes. (A) Analysis of HDV RNA and HDAgs in cellular lysates. (B) Analysis of HDV RNA, HDAg, HBsAg, and HBV DNA in culture media. Analyses of intracellular and extracellular HDV RNA, HDAgs, HBsAgs, and HBV DNA were performed as described in the legend of Fig. 2.
FIG. 4.
Correlation between secretion of genotype 4 HDV and different HBsAg expression provided by HBV-producing plasmids of various genotypes. (A) Analysis of HDV RNA and HDAgs in cellular lysates. (B) Analysis of HDV RNA, HDAgs, HBsAgs, and HBV DNA in culture media. Analyses of intracellular and extracellular HDV RNA, HDAgs, HBsAgs and HBV DNA were performed as described in the legend of Fig. 2.
Although pHBV2 provided similar amounts of unglycosylated S-HBsAg and L-HBsAg with those expressed by pHBV3, pHBV4, and pHBV5, the secreted HDV RNA were ca. 7.2 to 39.1% of those assembled by HBV-producing plasmids that expressed both glycosylated and unglycosylated HBsAg. The amounts of HDV RNA and HDAg in media did not necessarily correlate with HBV DNA levels in media or HBV DNA of nucleocapsids in transfected cells. The discrepant results were more obvious in lanes 1 of Fig. 2, 3, and 4 and also in the lanes 6 of Fig. 3 and 4.
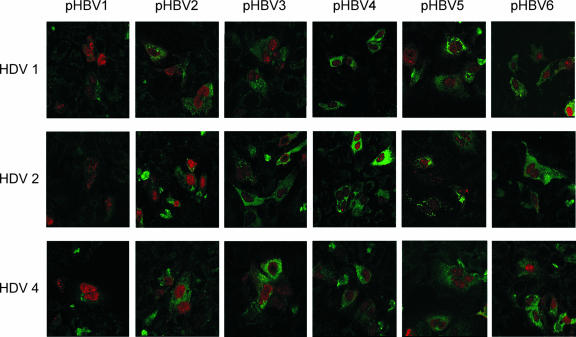
In order to further clarify whether HBsAg expression in cells transfected with various combinations of HBV- and HDV-producing plasmids correlated with secreted HBsAg and HDV virions, indirect double immunofluorescence staining of HBsAg and HDAg were performed at days 3 and 6 posttransfection. As shown in Fig. 5, HBsAg was shown in the cytoplasm with greenish FITC staining, and HDAg was shown in the nuclei with reddish rhodamine staining. About 15.6 to 18.1% of the cells were transfected in each combination of cotransfection, and ca. 77.9 to 86.5% of the transfected cells showed colocalization of HBsAg and HDAg on the same cells. No particularly higher efficiency of cotransfection or colocalizations of both antigens were observed in any of the 18 combinations of cotransfection in repeated experiments. The intensity of fluorescence staining peaked at day 3 (Fig. 5) and slightly decreased at day 6 (results not shown). In general, the intensities of HBsAg staining in the cytoplasm of cells transfected with different combinations of HBV and HDV-producing plasmids correlate with secreted HBsAg and HDV (Fig. 2, 3, and 4). For example, the cells transfected with pHBV1 showed the weakest HBsAg staining in the cytoplasm and secreted the lowest levels of HBsAg and HDV (Fig. 5; Fig. 2, 3, and 4, lanes 1), whereas cells transfected with pHBV3, -4, and -6 showed relatively stronger HBsAg staining and secreted higher levels of HBsAg and HDV RNA in media, with the exception of the lower secreted genotype 2 and 4 HDV RNA levels (Fig. 2B, 3B, and 4B, lanes 6). The intensities of HBsAg staining in cells transfected with pHBV2 and -5 were moderate, and the secreted HDV RNA levels were between the two above-mentioned groups.
FIG. 5.
Immunofluorescence staining of the expressed HBsAgs and HDAgs in transfected Huh-7 cells examined by using a confocal fluorescence microscope. HDV genotype 1, 2, and 4 expression plasmids (top, middle, and bottom panels, respectively) were cotransfected with six HBV-producing plasmids of genotype B or C. All cells were doubly stained with mouse anti-HBsAg and human anti-HDV antibodies after 3 days of transfection. The secondary antibodies of rhodamine-conjugated rabbit anti-human IgG and FITC-conjugated rabbit anti-mouse IgG were used to show the greenish HBsAg and reddish HDAg immunofluorescence stainings, respectively. Photographs (original magnification, ×630) were taken by using a confocal fluorescence microscope.
Assembly and secretion of viruslike particles (VLPs) in the presence of HBV plasmids expressing different levels of HBsAg.
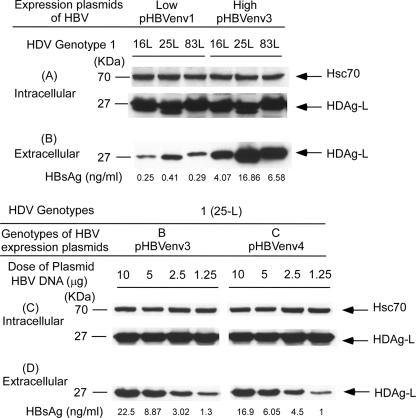
In order to further clarify whether different levels of HBsAg affect HDV assembly efficiency, a simple VLP model composed of HBsAg and HDAg-L was used. Plasmids encoding the HDAg-Ls of genotypes 1 (16L, 25L, and 83L) and 2 (8L, 24L, and 39L) were cotransfected with the plasmids expressing different levels of HBsAg (low expression, pHBVenv1; high expression, pHBVenv3) into Huh-7 cells. Transfected cells and culture media were collected on day 3 posttransfection to detect, respectively, the expression of HDAg-L and of the reference protein in cell lysates and secreted VLPs. As illustrated in Fig. 6A, the amounts of HDAg-L expressed in cell lysates were not significantly different. However, the secreted HDAg-L varied with the different levels of envelope proteins provided by the HBsAg-expression plasmids (Fig. 6B). The same findings were observed in the cotransfections of the HDAg-L expression plasmids of genotype 2 HDV with the HBsAg expression plasmids that provided different levels of envelope proteins (data not shown).
FIG. 6.
Assembly and secretion of VLPs in the presence of different levels of HBsAg. Intracellular (A) and extracellular (B) Huh-7 cells in 10-cm-diameter dishes (6 × 106 cells per dish) were cotransfected with 10 μg (each dish) of expression plasmids of HDAg-L (genotype 1: 16L, 25L, and 83L) and 10 μg of pHBVenv plasmids expressing different HBsAg levels. Low and high expression levels of HBsAg were from pHBVenv1 and -3, respectively. Intracellular (C) and extracellular (D) Huh-7 cells in 10-cm-diameter dishes were cotransfected with 10 μg (each dish) of expression plasmids of HDAg-L (25-L, HDAg-L expressed by a genotype 1 HDV) and serial dilutions of 10 μg of HBV genotype B (pHBVenv3) and C (pHBVenv4) plasmids. Both cells and spent media were collected on day 3 posttransfection. Analyses of intracellular and extracellular HDAg-L and equal loading of protein sample in each lane were performed as described in the legend of Fig. 2. HBsAg in spent media was quantified by using a General Biological Surdine125 B kit.
Assembly and secretion of VLPs in the presence of serial dilutions of HBsAg.
To determine whether HBsAg level was the main factor to affect the efficiency of HDV assembly, different concentrations of HBV plasmids were used. Fixed amounts of expression plasmids of the HDAg-L were cotransfected with HBsAg-expressing plasmids (pHBVenv3 and pHBVenv4) in serial dilutions of 10, 5, 2.5, and 1.25 μg into Huh-7 cells. As shown in Fig. 6C, the amounts of HDAg-L in the cell lysate of each lane were similar. However, regardless of the presence of genotype B or C HBsAg, the amounts of VLPs in culture media were notably reduced with serial dilutions of transfected HBsAg-expressing plasmids and decreasing HBsAg levels in culture media (Fig. 6D). The same results were observed in the cotransfection of HDV genotype 2 and 4 plasmids with serial dilutions of HBsAg-expressing plasmids (data not shown). It is apparent that HDV assembly efficiency is more closely associated with the levels of HBsAg and independent of HBV genotypes.
Assembly and secretion of HDVs of different genotypes in the presence of wild-type and chimeric HBV-producing plasmids.
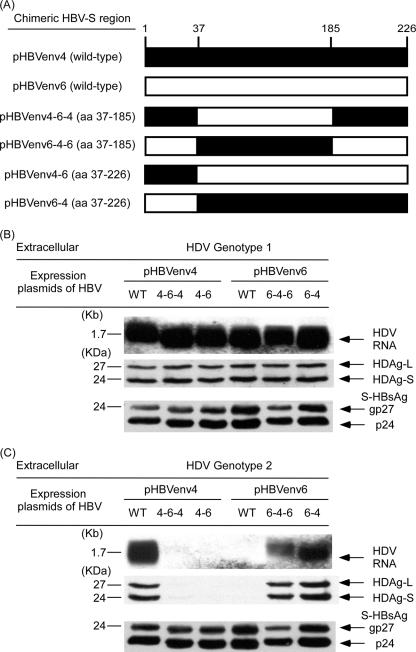
As indicated above, HBsAg levels played an important role in HDV assembly and secretion, except for a few discrepant results in the cotransfections of genotype 2 or 4 HDV expression plasmids with HBV plasmids shown in Fig. 3 and 4 (lanes 6). The pHBV6 of genotype C secreted the highest HBsAg level in medium among the six HBV-producing plasmids (Fig. 1) but assembled very low levels of HDV RNA in media when cotransfected with genotype 2 or 4 HDV (Fig. 3B and 4B). However, the amount of HDV RNA in the medium did not disproportionately diminish upon cotransfection of genotype 1 HDV-producing plasmid with pHBV6 (Fig. 2B). In order to study the mechanisms of the discrepant results, plasmids expressing chimeric forms of HBsAg were constructed by swapping experiments to determine which region of HBsAg sequence was crucial for HDV assembly. The amino acid sequences expressed by pHBVenv4 and pHBVenv6 were mostly the same, except for minor differences in the CYL-I, AGL, and CYL-II regions (Fig. 1A). A segment of sequence from aa 37 to 185 encompassing the CYL-I and AGL known to be critical for HDV assembly (1, 26, 27, 29, 50) was swapped between pHBVenv4 and pHBVenv6. The other segment of sequence from aa 37 to 226 encompassing the CYL-I, AGL, and CYL-II (1, 26, 27, 29), known to be critical for HDV assembly, was also swapped between pHBVenv4 and pHBVenv6. As shown in Fig. 7A, four plasmids expressing a chimeric form of HBV—pHBVenv4-6-4 (containing a swapped coding sequence of aa 37 to 185 of HBsAg from HBVenv expression plasmid 6), pHBVenv4-6 (containing a swapped coding sequence of aa 37 to 226 of HBsAg from HBVenv expression plasmid 6), pHBVenv6-4-6 (containing a swapped coding sequence of aa 37 to 185 of HBsAg from HBVenv-expression plasmid 4), and pHBVenv6-4 (containing a swapped coding sequence of aa 37 to 226 of HBsAg from HBVenv expression plasmid 4)—were constructed. Chimeric HBVenv plasmids and their parental wild-type HBVenv plasmids were cotransfected with HDV-producing plasmids. There were no significant differences in HDV assembly and secretion in the cotransfections of genotype I HDV with wild-type or chimeric HBVenv plasmids (Fig. 7B). However, the assembly efficiencies of genotype 2 and 4 HDV were markedly different between their cotransfections with wild-type and chimerical plasmids of HBVenv (Fig. 7C, genotype 2 HDV; HDV genotype 4 [data not shown]). In the cotransfection of genotype 2 HDV and wild-type pHBVenv6, the assembly and secretion of HDV was very low, as evidenced by hardly visible signals of HDV RNA and HDAgs in the medium (Fig. 7C). However, the assembly and secretion of HDV was markedly increased when the chimeric pHBVenv6-4 was used instead. On the contrary, the assembly and secretion of HDV was high when wild-type pHBVenv4 was cotransfected with genotype 2 HDV. It was strikingly decreased when the chimeric pHBVenv4-6 plasmid was used in the cotransfections (Fig. 7C). We then narrowed down the chimeric region by changing residues 37 to 185. In keeping with these findings, genotype C HBV with the residues 37 to 185 of HBsAg from pHBVenv6 appeared to be a poor helper HBV for the assembly of genotype 2 HDV (Fig. 7C) and genotype 4 HDV (data not shown).
FIG. 7.
Assembly and secretion of HDV of different genotypes with wild-type and chimeric HBsAg-expression plasmids. (A) Schematic diagram of wild-type and chimeric S-HBsAg expressed by pHBVenv4 (▪) and pHBVenv6 (□) of genotype C. A segment of the sequence from aa 37 to 185 encompassing the CYL-I and AGL known to be critical for HDV assembly was swapped between pHBVenv4 and pHBVenv6. The other segment of the sequence from aa 37 to 226 encompassing the CYL-I, AGL, and CYL-II known to be critical for HDV assembly was also swapped between pHBVenv4 and pHBVenv6. (B) Assembly and secretion of genotype 1 HDV by wild-type and chimeric HBsAgs. (C) Assembly and secretion of genotype 2 HDV by wild-type and chimeric HBsAgs. Both cells and spent media were collected on day 9 posttransfection. Analyses similar to those described in the legend of Fig. 2 of intracellular (data not shown) and extracellular HDV RNA, HDAg, and S-HBsAg were performed.
In addition to the amounts of HBsAg, the findings in the present study suggested that domains of the CYL-I and AGL of HBsAg (Fig. 1A) may affect the assembly efficiency of HDV when interacting with genotype 2 and 4 HDVs.
The influence of amino acid sequence variations within the CYL-I and AGL domains on HDV assembly and secretion.
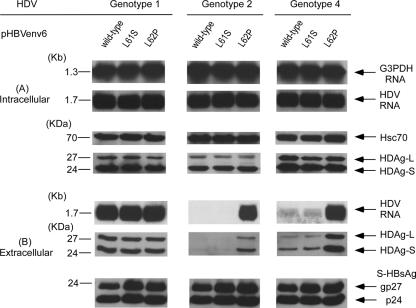
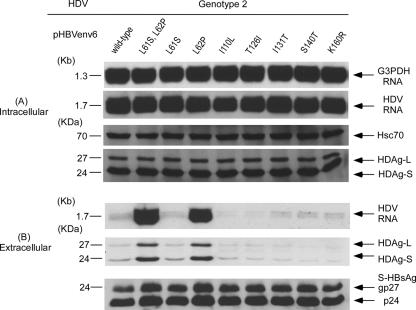
To investigate which residue in domains of the CYL-I and AGL spanning from aa 37 to 185 was important for the assembly and secretion of HDV of different genotypes. First, we compared the amino acid sequences of HBsAg between pHBVenv4 and pHBVenv6. There was a difference of nine residues (including two residues in the CYL-I region and five residues in AGL region) between them. Because previous studies demonstrated that the CYL-I of S-HBsAg was the candidate to interact with the HDV RNP during the assembly of HDV virions (47), wild-type pHBVenv6 and mutant plasmids of L61S and L62P in the CYL-I region were first constructed and cotransfected with HDV plasmids of three genotypes, respectively (Fig. 8A). As shown in the left panel of Fig. 8B, the assembly and secretion of genotype 1 HDV was not affected by the substitutions of lysine to serine or proline at residues 61 and 62 of S-HBsAg. Of note, the secretions of genotype 2 and 4 HDVs were markedly increased when pHBV mutants with L62P was used instead of the wild type in cotransfection (Fig. 8B, middle and right panels).
FIG. 8.
Assembly and secretion of HDV of different genotypes with help from the wild type (pHBVenv6) and from L61S and L62P mutations in the CYL-I region of HBsAg-expression plasmids. Both cells and spent media were collected on day 9 posttransfection. (A) Analysis of HDV RNA and HDAg in cellular lysates. (B) Analysis of HDV RNA, HDAg, HBsAg, and HBV DNA in culture media. Analyses of intracellular and extracellular HDV RNA, HDAg, and S-HBsAg were performed as described in the legend of Fig. 2.
To further clarify whether other residues in the AGL region that differ between pHBVenv4 and pHBVenv6 also affect the assembly or secretion of genotype 1, 2, and 4 HDVs, mutant expression plasmids of pHBVenv6 with I110L, T126I, I131T, S140T, and K160R substitutions were cotransfected with HDV plasmids of genotypes 1, 2, and 4. As shown in Fig. 9, the amounts of secreted HDV RNA and two forms of HDAgs of genotype 2 HDV via the help of HBsAg with the substitutions of amino acids at AGL regions according to those expressed by pHBVenv4 were not increased compared to those assembled by HBsAg provided by the wild-type pHBVenv6. Similar results were observed in the cotransfection of genotype 4 HDV plasmid with these mutant pHBVenv6 plasmids (data not shown). Again, the assembly and secretion of genotype 1 HDV were not significantly affected by the substitutions of these amino acids (including L62P) in CYL-1 and AGL domains that differ between pHBVenv4 and PHBVenv6 (data not shown). In summary, these results clearly indicate that residue Pro-62 in the CYL-I domain is the critical amino acid that increases the assembly and secretion of genotype 2 and 4 HDVs but does not affect the assembly and secretion of genotype 1 HDV.
FIG. 9.
Assembly and secretion of genotype 2 HDV with help from the wild type (pHBVenv6), double mutations (L61S and L62P), and single mutations (L61S, L62P, I110L, T126I, I131T, S140T, and K160R) in the CYL-I and AGL regions of HBsAg expression plasmids. Both cells and spent media were collected on day 9 posttransfection. (A) Analysis of HDV RNA and HDAg in cellular lysates. (B) Analysis of HDV RNA, HDAg, and S-HBsAg in culture media. Analyses of intracellular and extracellular HDV RNA, HDAg, and S-HBsAg were performed as described in the legend of Fig. 2.
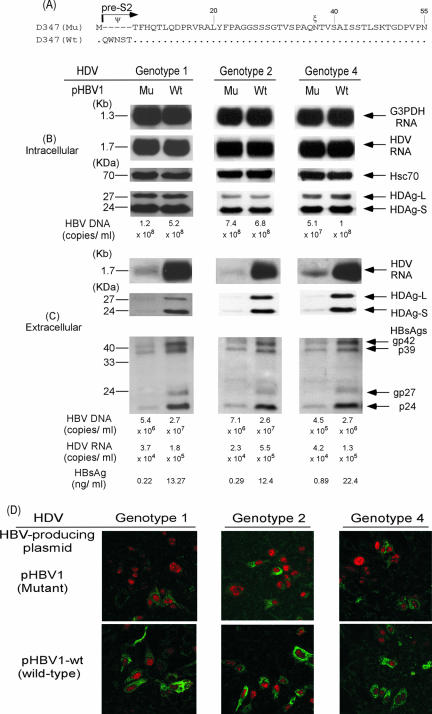
Assembly and secretion of HDV of different genotypes in the presence of wild-type (pHBV1-wt) and pHBV1 with an N-terminal deletion of pre-S2.
According to previous studies (2, 34, 38), the N-terminal 5 aa, including the glycosylation site at Asn4 are crucial for the secretion of HBV. To examine whether the low amounts of extracellular HBsAg, HDV RNA, and HDAg in lanes 1 of Fig. 2B, 3B, and 4B resulted from a naturally occurring deletion at this region, we isolated and constructed the wild-type partner of pHBV1(pHBV1-wt, Fig. 10A) from the viral quasispecies of the same HDV-infected patient. The pHBV1-wt has a sequence identical to that of pHBV1 except that the former has an intact pre-S2 domain. Both pHBV1 and pHBV1-wt were cotransfected with the HDV-producing plasmids of the genotypes 1, 2, and 4, respectively. To ascertain whether pHBV1 with an N-terminal deletion of the pre-S2 domain indeed produced trace but detectable HBsAg, HDV RNA, and HDAg in culture media, we increased the exposure time for Northern and Western blotting analysis to 2 h and 20 min, respectively. As shown in Fig. 10C, the amounts of secreted HBsAg as measured by immunoassay were markedly increased to 25.2- to 60-fold in the culture medium of cells transfected by pHBV1-wt compared to those transfected by pHBV1, a finding consistent with the results shown by Western blotting. The differences in the levels of secreted HBV DNA were 3.7- to 6-fold between pHBV1-wt and pHBV1, while the levels of secreted HDV RNA in the presence of HBsAg provided by pHBV1-wt were 3.1- to 23.9-fold higher than those in the presence of HBsAg provided by pHBV1. The indirect double immunofluorescence staining of HBsAg and HDAg was performed at days 3 (data not shown) and 9 posttransfection. As shown in Fig. 10D, the intensities of HBsAg staining in the cytoplasm of cells transfected by pHBV1-wt were stronger than those of cells transfected by pHBV1 and accounted for larger amounts of secreted HBsAg, HBV DNA, and HDV RNA (Fig. 10C).
FIG. 10.
Effects of wild-type (pHBV1-wt) and pre-S2 deletion mutant (pHBV1) of the HBV-producing plasmids on HBsAg secretion and HDV assembly. Equal amounts of three different genotypes of HDV- and HBV- expressing plasmids were cotransfected into Huh-7 cells. Both cells and spent media were collected on day 9 posttransfection. (A) Schematic representation of the predicted amino acid sequences of the pre-S2 domains expressed by the wild type (pHBV1-wt) and a mutant (pHBV1) of the HBV-producing plasmids. (B) Analysis of HDV RNA, HDAg, and HBV DNA in cellular lysates. (C) Analysis of HDV RNA, HDAg, HBV DNA, and HBsAg in culture media. Analyses of intracellular and extracellular HDV RNA, HDAg, HBV DNA and HBsAgs were performed as described in the legend of Fig. 2. (D) Immunofluorescence staining of HBsAg and HDAg in transfected Huh-7 cells. All cells were doubly stained with mouse anti-HBsAg and human anti-HDV antibodies after 9 days of transfection.
Correlation of serum HDV RNA levels with HBsAg and HBV DNA levels and the implications for patients with chronic HDV infection.
In order to determine whether the correlation between HBsAg, HBV DNA, and HDV RNA levels observed in transfected human hepatoma cell lines also existed in CHD patients, stored serum samples of 56 patients with chronic HDV infection were analyzed (Fig. 11).
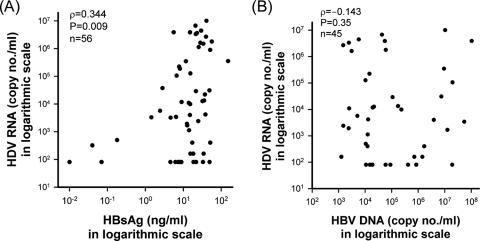
FIG. 11.
(A) Spearman rank correlation analysis of the association of serum HDV RNA levels with serum HBsAg levels in CHD patients. (B) Spearman rank correlation analysis of the association of serum HDV RNA levels with serum HBV DNA RNA levels in CHD patients with positive serum HBV DNA.
At the time of enrollment, 45 of 56 patients with CHD had detectable HBV DNA. The correlation between serum HDV RNA and HBsAg levels was significant as determined by Spearman rank correlation analysis (ρ = 0.344, P = 0.009, Fig. 11A). This correlation mainly existed in patients with undetectable HBV DNA in sera (ρ = 0.618, P = 0.043) but not in patients with positive serum HBV DNA (ρ = 0.239, P = 0.114). Moreover, there was no correlation between HDV RNA and HBV DNA in patients with positive serum HBV DNA (ρ = −0.143, P = 0.35, Fig. 11B).
After a median follow-up of 66 months, 18 (31.6%) patients were in biochemical remission with persistently normal alanine aminotransferase levels, 18 (31.6%) had chronic hepatitis, 9 (15.8%) progressed to cirrhosis, and 12 (21.1%) had HCC. Of the 21 patients with poor prognosis, including cirrhosis and HCC, 14 patients had serum HBV DNA levels more than 104 copies/ml (group 1), and the remaining 7 had serum HBV DNA levels below this level (group 2). The median level of HBsAg of these 21 patients was 15.36 ng/ml. There was no significant difference in the percentages of patients with HBsAg levels greater than this value between group 1 and group 2 patients (6 of 14 versus 4 of 7). Also, there was no significant difference in the percentages of patients with serum HDV RNA levels of greater than 104 copies/ml between group 1 and 2 patients. Therefore, the expression of HBsAg and HDV RNA levels appear to be independent of the presence of HBV DNA. Of note, all of the subjects had at least one kind of viral genome in their sera, and 16 (76%) had both circulating HBV and HDV.
Of the 51 patients with HDV viremia, 15 (29.4%) patients became in biochemical remission, 15 (29.4%) had chronic hepatitis, 9 (17.6%) had cirrhosis, and 12 (23.5%) developed HCC. Of the six patients negative for HDV RNA, three (50%) became in biochemical remission, another three (50%) had chronic hepatitis, and none progressed to cirrhosis or HCC during the follow-up period. Of note, four of the six patients without detectable serum HDV RNA had serum HBsAg levels below the median value of HBsAg of the 56 CHD patients. There was a trend of correlation between HDV viremia and poor prognosis, including cirrhosis and HCC (21 of 51 versus 0 of 6; P = 0.075 [Fisher exact test]).
DISCUSSION
In a recent report, genotype C HBV, in addition to genotype 1 HDV, was shown to be more commonly linked to adverse outcomes than genotype B HBV in patients with dual HBV-HDV infections similar to its closer association with unfavorable outcomes in patients with HBV monoinfections (12, 28, 46, 63). The present study took an original approach to comparing the interactions of HDV and HBV of different genotypes in a cell culture model. As shown in Fig. 2, 3, and 4, the assembly efficiency of HDV varied among different isolates within and between HBV genotypes. The role of HBV genotypes in HDV assembly and secretion was not apparent. The findings presented here indicate that the association of genotype C HBV with worse outcomes in patients with chronic HDV infection is not simply due to providing larger amounts of HBsAg for HDV assembly.
The major HBsAg is sufficient for HDV assembly (52). However, there has been no report of the relationship between the amount of HBsAg and the efficiency of HDV assembly and secretion. Although HBsAg is usually expressed in greater amounts than the amount needed for HBV envelopment, there are four lines of evidence in the present study to support that secreted HDV is dependent of HBsAg levels: first, a larger amount of secreted HDV was detected in the presence of helper HBV providing larger amounts of HBsAg; second, this trend still exists in a simple VLP model when different clones of HDAg-L expression plasmids were cotransfected with HBsAg-expressing plasmids that provided different levels of HBsAg; third, the secreted VLP levels were decreased with the reductions in HBsAg due to serial dilutions of cotransfected HBV plasmids; and finally, clinical observation revealed that serum HBsAg levels correlated with HDV RNA levels in CHD patients, particularly in patients with undetectable HBV viremia.
In addition to the finding of the influence of HBsAg levels on the assembly and secretion of HDV, another important finding of the present study is the discovery that amino acid sequence variations in HBsAg expressed by naturally occurring HBV variants affect the assembly efficiency of genotype 2 and 4 HDVs but not genotype 1. In the reports of Sureau and coworkers, the deletion of residues 24 to 28 at the CYL-I, the N146T mutation, or substitutions of tryptophan residues at 196, 199, and 201 in the CYL-II of S-HBsAg affected HDV assembly (1, 26, 27, 29). There were no deletions or substitutions on coding sequences for these amino acid residues of the HBV expression plasmids used in the present study. Interestingly, swapping and residue mutagenesis experiments of HBsAg-coding sequences revealed that P62L substitution in the CYL-I adversely affects the assembly and secretion of genotype 2 and 4 HDV but not those of genotype 1. The HDV package signal at the 19 C-terminal amino acid sequences of the HDAg-L is markedly divergent (73.7%) between genotypes 1 and 2, whereas genotype 4 is closer to genotype 2 in this domain (78.9% homology). According to previous reports, the assembly efficiency of HDV genotype 1 was reduced or defective in the presence of P201A, P204A, P205A, and P213A mutations in the nuclear export signal region of HDAg-L (35, 42). The prolines at residues 205 and 208 of HDAg-L of the genotype 1 HDV were substituted by arginine and lysine, respectively, in naturally occurring genotype 2 and 4 HDV isolates in the present study. Our preliminary data indicated that the substitution of arginine at residue 205 of HDAg-L of genotypes 2 and 4 by proline increased the assembly of these two genotypes even in the presence of pHBV6 with P62L substitution in the CYL-I (unpublished data). Whether the change P62L results in a conformation change of HBsAg and a poor protein-protein interaction between S-HBsAg and HDAg-L of genotype 2 and 4 HDVs remains to be determined.
Interestingly, the assembly and secretion of genotype 1 HDV was not obviously affected in cotransfections with either the wild type, a chimeric mutant, or the L62P mutant of pHBVenv6. The assembly efficiency of HDV genotype 1 is more related to the amounts of HBsAg (Fig. 2B). In our previous study, genotype 1 HDV has been reported to have greater assembly efficiency than genotype 2 (20). We further demonstrated here that higher assembly efficiency of genotype 1 HDV generally exists irrespective of HBV genotypes or variants, while the assembly and secretion of genotype 2 or 4 HDV may be further compromised in the presence of some HBV variants with L62 in the CYL-I of HBsAg. Genotype 1 HDV has been reported to be widely distributed worldwide, whereas genotype 2 and 4 HDV are limited to Taiwan, Japan, and Yakutia of far eastern Asia (6, 23, 24, 45, 60, 61). In addition to possible historical reasons, the finding of more efficient interactions between genotype 1 HDV and HBVs of different genotypes in the present study suggested that viral factors may also play important roles for the wide distribution of genotype 1 HDV in the world. Greater assembly and secretion efficiency of HDV may facilitate the spread of HDV to more hepatocytes, which may subsequently influence clinical courses and outcomes.
The M-HBsAg was not expressed in all of the six HBV-producing plasmids examined here due to either mutation of the start codon of M-HBsAg or deletion of the pre-S2 domains. Although the start codon and M-HBsAg coding sequence of pHBV2 appeared conserved, M-HBsAg was still undetectable. The level of M-HBsAg may be too low to be detected by Western blot analysis. Despite of the absence of M-HBsAg due to various causes, HDV was assembled and secreted into the medium. This finding is consistent with previous reports that M-HBsAg is dispensable for the morphogenesis of infectious HBV and HDV (4, 13, 47). Interestingly, the small deletion of aa 2 to 6 of the N-terminal region of the pre-S2 domain of pHBV1 resulted in markedly reduced intracellular HBsAg expression and subsequently reduced HBsAg, HBV DNA, and HDV RNA in the media despite the fact that the HBV DNA in nucleocapsids produced by this HBV plasmid is at a moderate level compared to the remaining five HBV plasmids. It has been reported that the large envelope protein lacking the first 5 aa of pre-S2 was unable to support HBV secretion (34). In contrast to the negative impact of L62 of S-HBsAg on the assembly and secretion of genotype 2 and 4 HDV, the pre-S2 N-terminal deletion HBV mutant adversely affected the expression and secretion of HBsAg and subsequently markedly reduced the assembly and secretion of all of the three HDV genotypes investigated here.
Glycosylation at Asn4 of M-HBsAg and Asn146 of S-HBsAg has been reported to affect HDV assembly (2, 38, 48, 53). Recently, additional glycosylated isoforms of L-HBsAg were found by using an improved detection system, but the posttranslational N glycosylation of L-HBsAg seemed to be dispensable for HBV morphogenesis (32). The role of the glycosylation of HBsAg on the secretion of HBV appears to be somewhat controversial (32, 39). The N146 was conserved in all of the six HBV-expression plasmids tested in the present study. However, Asn4 was deleted in pHBV1. Because the expressed HBsAg was low in pHBV1, it is unclear whether the deleted Asn4 had further impact on HDV assembly and secretion. Interestingly, pHBV2 had conserved Asn4 and Asn146 but did not express glycosylated HBsAg. It has been reported that only half of the glycosylation sites are really glycosylated. The absence of glycosylated HBsAg in pHBV2 appeared to have some negative impact on HDV assembly compared to other isolates expressing both glycosylated and unglycosylated HBsAg but seemed to have no obvious adverse influence on the secretion of HBV. However, more HBV isolates expressing only unglycosylated HBsAg are needed to support this observation. Of note, all three of the genotype B HBV expression plasmids in the present study did not have threonine at the O glycosylation site at residue 37 of M-HBsAg, whereas all three of the genotype C HBV plasmids were conserved. The virological and clinical implications remain to be elucidated.
In addition to the close association of HDV assembly and secretion with HBsAg expression in the cell culture system, serum HDV RNA levels also correlated with HBsAg expression in CHD patients but not with HBV DNA levels. Active HBV replication may compete with HDV for HBsAg envelopes in the assembly of virions. These findings are consistent with previous reports that the replication of HDV is independent of HBV, but the package needs the supply of HBsAg, mainly major HBsAg, from HBV (52, 58). Recently, HBV DNA levels of more than 104 copies/ml have been reported to correlate with an increased risk of cirrhosis and HCC (22, 63). Not surprisingly, two-thirds of the 21 HDV patients who progressed to cirrhosis or HCC had circulating HBV DNA levels higher than this cutoff value. However, group 2 patients who had sufficient HBsAg for continuous HDV assembly also progressed to grave outcomes despite having lower or undetectable HBV DNA levels in sera. All of the 21 patients with adverse outcomes had either HDV and/or HBV viremia consistent with our previous reports in a larger number of patients (46, 59). The treatment of CHD by inhibition of HBV replication with nucleoside analogues is apparently insufficient and not satisfactory (41).
In conclusion, the assembly and secretion of HDV are more closely associated with the amounts of HBsAg than with the HBV genotypes per se. In addition to the amount of HBsAg, the residue 62 position in the CYL-I domain of S-HBsAg sequences plays an important role in determining the assembly and secretion efficiency of HDV genotypes 2 and 4 not genotype 1. The pre-S2 N-terminal deletion HBV mutant adversely affects secretion of the three HDV genotypes. Cases of CHD patients with undetectable HBV viremia, but with high serum HBsAg levels and HDV viremia, may lead to dismal outcomes. For an effective treatment of CHD, reduction of HBsAg production in addition to the suppression of HBV and HDV replication may be necessary.
Acknowledgments
We thank Sheng-Chung Lee for the kind provision of the monoclonal antibody (A10F1) to HBsAg and Pui-Ching Lee for statistical analysis.
This study was supported by grants from the National Science Council (NSC91-2314-B-010-079, NSC92-2314-B-010-015, and NSC93-2314-B-010-005) and partly by a grant (grant V96S4-023) from the Taipei Veterans General Hospital, Taipei, Taiwan.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Blanchet, M., and C. Sureau. 2006. Analysis of the cytosolic domains of the hepatitis B virus envelope proteins for their function in viral particle assembly and infectivity. J. Virol. 8011935-11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Block, T. M., X. Lu, F. M. Platt, G. R. Foster, W. H. Gerlich, B. S. Blumberg, and R. A. Dwek. 1994. Secretion of human hepatitis B virus is inhibited by the imino N-butyldeoxynojirimycin. Proc. Natl. Acad. Sci. USA 912235-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonino, F., K. H. Heermann, M. Rizzetto, and W. H. Gerlich. 1986. Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J. Virol. 58945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 881059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruss, V., J. Hagelstein, E. Gerhardt, and P. R. Galle. 1996. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology 218396-399. [DOI] [PubMed] [Google Scholar]
- 6.Casey, J. L., T. L. Brown, E. J. Colan, F. S. Wignall, and J. L. Gerin. 1993. A genotype of hepatitis D virus that occurs in northern South America. Proc. Natl. Acad. Sci. USA 909016-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casey, J. L., and J. L. Gerin. 1995. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J. Virol. 697593-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, F. L., P. J. Chen, S. J. Tu, C. J. Wang, and D. S. Chen. 1991. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 888490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao, M., S. Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 645066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, P. J., F. L. Chang, C. J. Wang, C. J. Lin, S. Y. Sung, and D. S. Chen. 1992. Functional study of hepatitis delta virus large antigen in packaging and replication inhibition: role of the amino-terminal leucine zipper. J. Virol. 662853-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, P. J., G. Kalpana, J. Goldberg, W. Mason, B. Werner, J. Gerin, and J. Taylor. 1986. Structure and replication of the genome of the hepatitis delta virus. Proc. Natl. Acad. Sci. USA 838774-8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu, C. J., and A. S. Lok. 2002. Clinical significance of hepatitis B virus genotypes. Hepatology 351274-1276. [DOI] [PubMed] [Google Scholar]
- 13.Fernholz, D., P. R. Galle, M. Stemler, M. Brunetto, F. Bonino, and H. Will. 1993. Infectious hepatitis B virus variant defective in pre-S2 protein expression in a chronic carrier. Virology 194137-148. [DOI] [PubMed] [Google Scholar]
- 14.Glenn, J. S., J. A. Watson, C. M. Havel, and J. M. White. 1992. Identification of a prenylation site in delta virus large antigen. Science 2561331-1333. [DOI] [PubMed] [Google Scholar]
- 15.Glenn, J. S., and J. M. White. 1991. trans-Dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 652357-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govindarajan, S., K. P. Chin, A. G. Redeker, and R. L. Peters. 1984. Fulminant B viral hepatitis: role of delta agent. Gastroenterology 861417-1420. [PubMed] [Google Scholar]
- 17.Govindarajan, S., K. M. De Cock, and A. G. Redeker. 1986. Natural course of delta superinfection in chronic hepatitis B virus-infected patients: histopathologic study with multiple liver biopsies. Hepatology 6640-644. [DOI] [PubMed] [Google Scholar]
- 18.Gripon, P., J. Le Seyec, S. Rumin, and C. Guguen-Guillouzo. 1995. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology 213292-299. [DOI] [PubMed] [Google Scholar]
- 19.Hadler, S. C., M. De Monzon, A. Ponzetto, E. Anzola, D. Rivero, A. Mondolfi, A. Bracho, D. P. Francis, M. A. Gerber, and S. Thung. 1984. Delta virus infection and severe hepatitis: an epidemic in the Yucpa Indians of Venezuela. Ann. Intern. Med. 100339-344. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, S. C., W. J. Syu, I. J. Sheen, H. T. Liu, K. S. Jeng, and J. C. Wu. 2002. Varied assembly and RNA editing efficiencies between genotypes I and II hepatitis D virus and their implications. Hepatology 35665-672. [DOI] [PubMed] [Google Scholar]
- 21.Hsu, S. C., W. J. Syu, L. T. Ting, and J. C. Wu. 2000. Immunohistochemical differentiation of hepatitis D virus genotypes. Hepatology 321111-1116. [DOI] [PubMed] [Google Scholar]
- 22.Iloeje, U. H., H. I. Yang, J. Su, C. L. Jen, S. L. You, and C. J. Chen. 2006. Predicting liver cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology 130678-686. [DOI] [PubMed] [Google Scholar]
- 23.Imazeki, F., M. Omata, and M. Ohto. 1990. Heterogeneity and evolution rates of delta virus RNA sequences. J. Virol. 645594-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivaniushina, V., N. Radjef, M. Alexeeva, E. Gault, S. Semenov, M. Salhi, O. Kiseiev, and P. Deny. 2001. Hepatitis delta virus genotype I and II cocirculate in an endemic area of Yakutia, Russia. J. Gen. Virol. 822709-2718. [DOI] [PubMed] [Google Scholar]
- 25.Jaoude, G. A., and C. Sureau. 2005. Role of the antigenic loop of the hepatitis B virus envelope proteins in infectivity of hepatitis delta virus. J. Virol. 7910460-10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenna, S., and C. Sureau. 1998. Effect of mutations in the small envelope protein of hepatitis B virus on assembly and secretion of hepatitis delta virus. Virology 251176-186. [DOI] [PubMed] [Google Scholar]
- 27.Jenna, S., and C. Sureau. 1999. Mutations in the carboxyl-terminal domain of the small hepatitis B virus envelope protein impair the assembly of hepatitis delta virus particles. J. Virol. 733351-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2000. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 118554-559. [DOI] [PubMed] [Google Scholar]
- 29.Komla-Soukha, I., and C. Sureau. 2006. A tryptophan-rich motif in the carboxyl terminus of the small envelope protein of hepatitis B virus is central to the assembly of hepatitis delta virus particles. J. Virol. 804648-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo, M. Y., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 631945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai, M. M. 1995. The molecular biology of hepatitis delta virus. Annu. Rev. Biochem. 64259-286. [DOI] [PubMed] [Google Scholar]
- 32.Lambert, C., and R. Prange. 2007. Posttranslational N glycosylation of the hepatitis B virus large envelope protein. Virol. J. 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Gal, F., E. Gault, M. P. Ripault, J. Serpaggi, J. C. Trinchet, E. Gordien, and P. Deny. 2006. Eighth major clade for hepatitis delta virus. Emerg. Infect. Dis. 121447-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1998. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J. Virol. 725573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, C. H., S. C. Chang, C. H. Wu, and M. F. Chang. 2001. A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J. Biol. Chem. 2768142-8148. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S. C., J. Y. Shew, and A. C. Chang. 1984. Monoclonal antibodies to hepatitis B surface antigen (HBsAg) produced by somatic cell hybrids. J. Formosan Med. Assoc. 83142-148. [PubMed] [Google Scholar]
- 37.Luo, G. X., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 641021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, X., A. Mehta, R. Dwek, T. Butters, and T. Block. 1995. Evidence that N-linked glycosylation is necessary for hepatitis B virus secretion. Virology 213660-665. [DOI] [PubMed] [Google Scholar]
- 39.Mehta, A., X. Lu, T. M. Block, B. S. Blumberg, and R. A. Dwek. 1997. Hepatitis B virus (HBV) envelope glycoproteins vary drastically in their sensitivity to glycan processing: evidence that alteration of a single N-linked glycosylation site can regulate HBV secretion. Proc. Natl. Acad. Sci. USA 941822-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakabayashi, H., K. Teketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 423858-3863. [PubMed] [Google Scholar]
- 41.Niro, G. A., F. Rosina, and M. Rizzetto. 2005. Treatment of hepatitis D. J. Viral Hepatitis 122-9. [DOI] [PubMed] [Google Scholar]
- 42.O'Malley, B., and D. W. Lazinski. 2005. Roles of carboxyl-terminal and farnesylated residues in the functions of the large hepatitis delta antigen. J. Virol. 791142-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizzetto, M., M. G. Canese, J. L. Gerin, W. T. London, D. L. Sly, and R. H. Purcell. 1980. Transmission of the hepatitis B virus-associated delta antigen to chimpanzees. J. Infect. Dis. 141590-602. [DOI] [PubMed] [Google Scholar]
- 44.Rizzetto, M., G. Verme, S. Recchia, F. Bonino, P. Farci, S. Arico, R. Calzia, A. Picciotto, M. Colombo, and H. Popper. 1983. Chronic hepatitis in carriers of hepatitis B surface antigen, with intrahepatic expression of the delta antigen: an active and progressive disease unresponsive to immunosuppressive treatment. Ann. Intern. Med. 98437-441. [DOI] [PubMed] [Google Scholar]
- 45.Sakugawa, H., H. Nakasone, T. Nakayoshi, Y. Kawakami, S. Miyazato, F. Kinjo, A. Saito, S. P. Ma, H. Hotta, and M. Kinoshita. 1999. Hepatitis delta virus genotype IIb predominates in an endemic area, Okinawa, Japan. J. Med. Virol. 58366-372. [DOI] [PubMed] [Google Scholar]
- 46.Su, C. W., Y. H. Huang, T. I. Huo, H. H. Shih, I. J. Sheen, S. W. Chen, P. C. Lee, S. D. Lee, and J. C. Wu. 2006. Genotypes and viremia of hepatitis B and D viruses are associated with outcomes of chronic hepatitis D patients. Gastroenterology 1301625-1635. [DOI] [PubMed] [Google Scholar]
- 47.Sureau, C. 2006. The role of the HBV envelope proteins in the HDV replication cycle. Curr. Top. Microbiol. Immunol. 307113-131. [DOI] [PubMed] [Google Scholar]
- 48.Sureau, C., C. Fournier-Wirth, and P. Maurel. 2003. Role of N glycosylation of hepatitis B virus envelope proteins in morphogenesis and infectivity of hepatitis delta virus. J. Virol. 775519-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sureau, C., B. Guerra, and R. E. Lanford. 1993. Role of the large hepatitis B virus envelope protein in infectivity of the hepatitis delta virion. J. Virol. 67366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sureau, C., B. Guerra, and H. Lee. 1994. The middle hepatitis B virus envelope protein is not necessary for infectivity of hepatitis delta virus. J. Virol. 684063-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor, J. M. 1999. Hepatitis delta virus. Intervirology 42173-178. [DOI] [PubMed] [Google Scholar]
- 52.Wang, C. J., P. J. Chen, J. C. Wu, D. Patel, and D. S. Chen. 1991. Small-form hepatitis B surface antigen is sufficient to help in the assembly of hepatitis delta virus-like particles. J. Virol. 656630-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, C. J., S. Y. Sung, D. S. Chen, and P. J. Chen. 1996. N-linked glycosylation of hepatitis B surface antigens is involved but not essential in the assembly of hepatitis delta virus. Virology 22028-36. [DOI] [PubMed] [Google Scholar]
- 54.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence, and expression of the hepatitis delta (delta) viral genome. Nature 323508-514. [DOI] [PubMed] [Google Scholar]
- 55.Weiner, A. J., Q. L. Choo, K. S. Wang, S. Govindarajan, A.′G. Redeker, J. L. Gerin, and M. Houghton. 1998. A single antigenomic opening reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta. J. Virol. 62594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, J. C., C. L. Chen, M. C. Hou, T. Z. Chen, S. D. Lee, and K. J. Lo. 1994. Multiple viral infection as the most common cause of fulminant and subfulminant viral hepatitis in an area endemic for hepatitis B: application and limitations of the polymerase chain reaction. Hepatology 19836-840. [PubMed] [Google Scholar]
- 57.Wu, J. C., C. L. Chen, S. D. Lee, I. J. Sheen, and L. P. Ting. 1992. Expression and localization of the small and large delta antigens during the replication cycle of hepatitis D virus. Hepatology 161120-1127. [PubMed] [Google Scholar]
- 58.Wu, J. C., P. J. Chen, M. Y. Kuo, S. D. Lee, D. S. Chen, and L. P. Ting. 1991. Production of hepatitis delta virus and suppression of helper hepatitis B virus in a human hepatoma cell line. J. Virol. 651099-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, J. C., T. Z. Chen, Y. S. Huang, F. S. Yen, L. T. Ting, W. Y. Sheng, S. H. Tsay, and S. D. Lee. 1995. Natural history of hepatitis D viral superinfection: significance of viremia detected by polymerase chain reaction. Gastroenterology 108796-802. [DOI] [PubMed] [Google Scholar]
- 60.Wu, J. C., T. Y. Chiang, and I. J. Sheen. 1998. Characterization and phylogenetic analysis of a novel hepatitis D virus strain discovered by restriction fragment length polymorphism analysis. J. Gen. Virol. 791105-1113. [DOI] [PubMed] [Google Scholar]
- 61.Wu, J. C., K. B. Choo, C. M. Chen, T. Z. Chen, T. I. Huo, and S. D. Lee. 1995. Genotyping of hepatitis D virus by restriction-fragment length polymorphism and relation to outcome of hepatitis D. Lancet 346939-941. [DOI] [PubMed] [Google Scholar]
- 62.Wu, J. C., S. D. Lee, S. Govindarajan, T. W. Kung, Y. T. Tsai, K. J. Lo, and L. P. Ting. 1990. Correlation of serum delta RNA with clinical course of acute hepatitis delta virus superinfection in Taiwan: a longitudinal study. J. Infect. Dis. 1611116-1120. [DOI] [PubMed] [Google Scholar]
- 63.Yu, M. W., S. H. Yeh, P. J. Chen, Y. F. Liaw, C. L. Lin, C. J. Liu, W. L. Shih, J. H. Kao, D. S. Chen, and C. J. Chen. 2005. Hepatitis B virus genotype and DNA level and HCC: a prospective study in men. J. Natl. Cancer Inst. 97265-272. [DOI] [PubMed] [Google Scholar]