Abstract
The mechanisms involved in hepatitis C virus (HCV) RNA replication are unknown, and this aspect of the virus life cycle is not understood. It is thought that virus-encoded nonstructural proteins and RNA genomes interact on rearranged endoplasmic reticulum (ER) membranes to form replication complexes, which are believed to be sites of RNA synthesis. We report that, through the use of an antibody specific for double-stranded RNA (dsRNA), dsRNA is readily detectable in Huh-7 cells that contain replicating HCV JFH-1 genomes but is absent in control cells. Therefore, as that of other RNA virus genomes, the replication of the HCV genome may involve the generation of a dsRNA replicative intermediate. In Huh-7 cells supporting HCV RNA replication, dsRNA was observed as discrete foci, associated with virus-encoded NS5A and core proteins and identical in morphology and distribution to structures containing HCV RNA visualized by fluorescence-based hybridization methods. Three-dimensional reconstruction of deconvolved z-stack images of virus-infected cells provided detailed insight into the relationship among dsRNA foci, NS5A, the ER, and lipid droplets (LDs). This analysis revealed that dsRNA foci were located on the surface of the ER and often surrounded, partially or wholly, by a network of ER-bound NS5A protein. Additionally, virus-induced dsRNA foci were juxtaposed to LDs, attached to the ER. Thus, we report the visualization of HCV-induced dsRNA foci, the likely sites of virus RNA replication, and propose that HCV genome synthesis occurs at LD-associated sites attached to the ER in virus-infected cells.
For all positive-sense RNA [(+)RNA] viruses studied to date, the RNA synthesis machinery is associated with the cytoplasmic surfaces of intracellular membranes, and many of the proteins required for viral RNA synthesis contain membrane-targeting sequences. Virus-encoded proteins such as poliovirus 2BC/3A (46, 52), dengue virus NS4A (37, 38), and brome mosaic virus 1a (7, 48) have an intrinsic capability to promote intracellular membrane rearrangement, which serves to house viral replicase complexes, thereby creating membrane-wrapped factories for genome replication. The sequestration of viral RNA synthesis machinery into membrane-enclosed structures likely protects from host response proteins recognizing viral RNA genomes and also provides a stable and confined environment for replication. Consequently, a range of structures with altered intracellular membrane morphologies are frequently observed in (+)RNA virus-infected cells and include single-membrane vesicles or spherules, double-membrane vesicles, and convoluted membrane structures (2, 16, 25, 62).
Consistent with the strategy employed by other (+)RNA viruses, the replication of the genome of hepatitis C virus (HCV) is believed to occur in membrane-bound vesicles, probably derived from the endoplasmic reticulum (ER) (10, 15, 44, 51). Prior to the initiation of viral RNA synthesis, the translation of the HCV genome first yields a large polyprotein that is proteolytically processed into 10 individual proteins, of which nonstructural proteins NS3, NS4A, NS4B, NS5A, and NS5B are essential for HCV genome replication (3, 29). Membrane-targeting sequences have been identified in each of the nonstructural proteins, with the exception of NS3, which localizes to the ER membrane through interaction with NS4A (6, 21, 40, 63). It is thought that the nonstructural proteins, viral genomes, and certain host-encoded factors interact to form multiprotein assemblies termed replication complexes (RCs), which accumulate on rearranged ER membranes and are believed to be the sites of viral RNA synthesis (15, 17, 43, 59). Current evidence suggests that the oligomerization of NS4B is, at least in part, responsible for rearranging ER membranes to generate an intracellular platform for viral RNA replication, termed the membranous web (10, 21, 31, 64). Both nonstructural proteins and HCV RNA have been observed in close association with this structure (10, 15). Viewed under the electron microscope, the membranous web appears as a network of membrane-bound vesicles, which are believed to house HCV RCs (10, 15). In support of this hypothesis, purified membrane vesicles from Huh-7 cells that harbor HCV replicons contain virus-encoded nonstructural proteins and are able to synthesize HCV RNA in vitro (1, 18, 26, 44). While membrane association is critical for HCV RNA synthesis, the nature of this association and the organization of the RC are poorly understood.
Within the RC, the instigation of HCV RNA synthesis occurs by an unknown mechanism but is thought to involve the de novo initiation of genome replication by NS5B (32, 66), the virus-encoded RNA-dependent RNA polymerase, through the production of a negative-strand template (29, 42, 53). Once the negative strand is produced, it can remain as a free single-stranded RNA (ssRNA) molecule or be attached to the positive strand to form a double-stranded RNA (dsRNA) replicative intermediate (RI). It is not yet clear whether HCV RNA is replicated using the negative strand in a dsRNA RI form, as revealed for Kunjin virus (8), or in the form of a free ssRNA template, as demonstrated for the Qβ phage (11). However, dsRNA has been detected in cell-free HCV RNA synthesis assay mixtures (1, 26), suggesting that dsRNA RIs may be produced during HCV genome replication. dsRNA has been visualized in virus-infected cells, particularly those infected with (+)RNA viruses such as Sindbis virus (Togaviridae), encephalomyocarditis virus (Picornaviridae), severe acute respiratory syndrome coronavirus (Coronaviridae), and Kunjin virus and dengue virus (Flaviviridae), but is not seen in uninfected cells (37, 50, 60, 62). Furthermore, dsRNA has been used effectively as a marker to characterize both Kunjin and rubella virus RCs (27, 34, 61, 62). Hence, the detection of dsRNA in virus-infected cells provides a means to conveniently examine the formation and properties of viral RCs and to investigate the nature of interactions with both cell-derived factors and organelles and virus-encoded proteins.
In this study, we present the first report of the visualization of dsRNA in cells supporting HCV RNA replication. We have utilized immunodetection methods to identify and characterize sites of dsRNA accumulation in cells supporting the replication of subgenomic HCV RNA and in virus-infected cells. We demonstrate that the generation of dsRNA in cells is a result of HCV RNA replication and, using three-dimensional (3D) reconstruction of confocal laser-scanned images, provide a unique insight into the relationship between dsRNA-containing structures and viral and cellular components in HCV-infected cells.
MATERIALS AND METHODS
Cells.
Huh-7 cells were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum as described previously (54). The Huh-7-derived cell line termed 2/1 (55) supports the replication of subgenomic RNA from the JFH-1 strain of HCV (23) and was maintained in DMEM containing 10% fetal calf serum and G418 at 500 μg/ml.
Antibodies.
Antibodies used to detect HCV core protein (rabbit antiserum R308) and NS5A and human adipocyte differentiation-related protein (ADRP) have been described previously (19, 33, 54). Rabbit antiserum against calnexin was used according to the instructions of the manufacturer (Sigma). Mouse monoclonal antibody specific for dsRNA (J2) was purchased from Scicons (Hungary).
In vitro transcription of JFH-1 RNA and introduction by electroporation.
The in vitro transcription of RNA was accomplished using the T7 MEGAscript kit as directed by the manufacturer (Ambion). For transient replication assays, RNA was prepared from pSGR-Luc-JFH-1 or pSGR-Luc-JFH-1/GND. These plasmids carry bicistronic RNA containing the luciferase reporter gene in the first cistron and either the wild-type (wt) JFH-1 subgenomic replicon or a nonreplicative mutant form (encoding a GDD-to-GND mutation in NS5B) in the second cistron, respectively (55). Subgenomic JFH-1 RNA was introduced into Huh-7 cells by electroporation, and levels of RNA replication were monitored by luciferase assays as described previously (55). If appropriate, tissue culture medium was supplemented with ribavirin at 50 or 100 μg/ml and fresh ribavirin-containing medium was added daily to cells.
To generate infectious HCV, RNA generated from pJFH-1 was introduced into Huh-7 cells by electroporation as described previously (58, 65) and virus released into the growth medium was used to infect monolayers of naïve Huh-7 cells. Prior to infection, medium containing supernatant virus was subjected to low-speed centrifugation to remove cell debris and then diluted with fresh DMEM. Infected cells were fixed in methanol for indirect immunofluorescence, or cell extracts were prepared for Western blot analysis. HCV titers in tissue culture medium were determined by a 50% tissue culture-infective-dose assay (28).
Western blot analysis.
The preparation of cell extracts, polyacrylamide gel electrophoresis, and Western blot analysis were performed as described previously (54).
FISH.
Cells grown on glass coverslips were fixed for 20 min in methanol at −20°C. After a brief wash in phosphate-buffered saline (PBS), cells were prehybridized at 37°C for 30 min in hybridization buffer (50% formamide, 10% dextran sulfate, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]). Alexa Fluor 488-labeled DNA probes were generated using the fluorescent in situ hybridization (FISH) Tag DNA green kit and pJFH-1 (58) according to the instructions of the kit manufacturer (Invitrogen). This procedure generated Alexa Fluor 488-tagged dsDNA probes of approximately 300 bp that covered the entire JFH-1 genome. Following prehybridization, the probes were diluted in hybridization buffer to approximately 1 ng/μl and 20-μl aliquots were added to appropriate coverslips. Denaturation of both the sample and the probe was accomplished by incubation of the coverslips at 95°C for 2 min. The coverslips were inverted and placed onto glass slides, and the slides were incubated at 37°C overnight in humidity chambers. The following day, coverslips were washed once in 2× SSC at 60°C and once in 2× SSC at room temperature and either examined directly or processed for indirect immunofluorescence as described below.
Indirect immunofluorescence.
Cells, fixed for 20 min in methanol at −20°C, were washed with PBS and incubated with primary antibody (diluted in PBS) for 2 h at room temperature. After a second wash with PBS, cells were incubated with appropriate fluorophore-conjugated secondary antibodies (raised in donkey) for 1 h at room temperature. Following a final wash with PBS, cells were rinsed with H2O before being mounted onto slides with Citifluor (Citifluor Ltd.). For RNase A treatment, cells fixed in methanol were washed with PBS and incubated with 50 μg of RNase A/ml in either 0.1× SSC (low-salt conditions) or 2× SSC (high-salt conditions) for 2 h at 37°C. Cells were then quickly washed in PBS, and each coverslip was treated with 100 U of rRNasin (Promega), diluted in PBS, for 1 h at room temperature. Following a brief wash with PBS, cells were processed for indirect immunofluorescence as described above. Cell images were captured using a Zeiss LSM510 META inverted confocal microscope and associated software. For the 3D reconstructions, 60 to 80 z-stack images per sample were collected using a 63× lens objective with a numerical aperture of 1.4 and deconvolved by 3D blind deconvolution (10 iterations) using Autodeblur software (MediaCybernetics). 3D reconstructions of selected intracellular regions of interest were created with the 5D viewer extension in the isosurface mode by using a bin factor of 1.
RESULTS
Detection of intracellular HCV RNA by FISH.
The visualization of HCV RNA within cells supporting viral RNA synthesis provides insight into the localization and abundance of sites of replication. A limited analysis of cells containing HCV subgenomic replicons from genotype 1b using FISH has been performed previously and identified virus RNA as bright dots distributed throughout the cytoplasm, with some accumulation in the perinuclear region (15). Using the HCV JFH-1 isolate, we extended the analysis of intracellular viral RNA distribution. Sites of HCV RNA accumulation in an Huh-7-derived cell line, termed 2/1 (55), that supports the stable replication of JFH-1 subgenomic replicon RNA (23) were visualized by FISH (Fig. 1A). Cytoplasmic fluorescence, attributed to the binding of the FISH probe to JFH-1 RNA, was detected in >90% of 2/1 cells (Fig. 1A, panel i). In contrast, no signal was observed in control Huh-7 cells (Fig. 1A, compare panels i and ii). Closer inspection of 2/1 cells labeled with a JFH-1-specific FISH probe revealed that fluorescence was concentrated in a network of numerous small, discrete foci (Fig. 1A, panels iii and iv). These foci often accumulated in the perinuclear region in a manner that has been described previously (15). To gain insight into the relationship between JFH-1 RNA-containing foci and NS5A, which is essential for HCV genome replication and can bind HCV RNA (20), FISH-probed 2/1 cells were analyzed also by indirect immunofluorescence using an antiserum specific for NS5A (Fig. 1A, panels v to ix). The results revealed that the localization of JFH-1 RNA-containing foci closely mirrored that of NS5A (Fig. 1A, panels v and vi), and the examination of dually probed 2/1 cells at higher magnification verified that the distribution of JFH-1 RNA-containing foci and the network of seemingly ER-bound NS5A overlapped (Fig. 1A, panels vii to ix).
FIG. 1.
Localization of HCV RNA in JFH-1 replicon-containing and virus-infected Huh-7 cells. (A) 2/1 JFH-1 replicon-containing cells (panels i and iii to ix) and Huh-7 control cells (panel ii) were hybridized with a JFH-1-specific FISH probe and counterstained, where appropriate, with antiserum specific for NS5A (panels vi, viii, and ix). Boxed regions in panels iii and v are shown enlarged in panels iv and vii to ix, respectively. (B) Huh-7 cells, mock infected (panel ii) or infected with JFH-1 virus for 48 h (panels i and iii to ix), were hybridized with a FISH probe specific for JFH-1 and, in certain cases, counterstained for NS5A (panels vi, viii, and ix). The indicated regions of virus-infected cells in panels iii and v are shown enlarged in panels iv and vii to ix, respectively. Bars, 10 μm.
JFH-1 genomic RNA is infectious in Huh-7 cells and is capable of releasing infectious virus into the culture medium (58, 65). Therefore, JFH-1 allows the distribution of viral RNA in HCV-infected cells to be examined. FISH analysis of JFH-1-infected cells revealed foci containing JFH-1 RNA that were similar in appearance to those observed in 2/1 cells, although the number of foci in virus-infected cells was reduced compared to the number detected in 2/1 cells (Fig. 1B, panels i to iv). No signal was observed in mock-infected Huh-7 cells (Fig. 1B, compare panels i and ii). The RNA-containing foci were localized in the cytoplasm of virus-infected cells and, in the majority of cells, were detected in the perinuclear region (Fig. 1B, panels iii and iv). In a manner identical to that in 2/1 cells, the distribution of JFH-1 RNA-containing foci overlapped with that of NS5A protein in virus-infected cells (Fig. 1B, panels v to ix). Apart from NS5A, it was not possible to detect viral or cellular antigens in FISH-probed virus-infected cells reliably by indirect immunofluorescence, probably due to the destructive nature of the thermal denaturation step included in the FISH protocol (data not shown). Consequently, an alternative method to visualize sites containing HCV RNA under native conditions was sought in order to examine the interactions between viral RNA and both virus-encoded proteins and cell-derived components.
Visualization of dsRNA in cells constitutively replicating HCV subgenomic replicon RNA.
dsRNA has been used as a marker to identify Kunjin, rubella, and dengue virus RCs and has been observed in cells infected with a variety of (+)RNA viruses (27, 37, 60, 61). To determine whether dsRNA could be detected in Huh-7 cells supporting steady-state replication of JFH-1 subgenomic replicons, 2/1 cells were fixed and probed with monoclonal antibody J2 (30, 47, 60). J2 specifically recognizes dsRNA of more than 40 bp in length, and antibody binding is independent of the sequence and nucleotide composition of the antigen (47). J2 has recently been used to visualize dsRNA in cells infected with a number of (+)RNA viruses (37, 60). In >90% of 2/1 cells labeled with J2, distinct cytoplasmic structures that were not present in control Huh-7 cells were observed (Fig. 2A, compare panels i and ii). The dsRNA-containing structures observed in 2/1 cells were remarkably similar to JFH-1 RNA foci seen in the same cells by using FISH (compare Fig. 1A and 2A) and appeared as numerous discrete foci throughout the cytoplasm (Fig. 2A, panels iii and iv). Analogous to the distributions of foci and NS5A observed in 2/1 cells by FISH, the distributions of J2-labeled foci and NS5A coincided rather than exhibiting colocalization (Fig. 2A, panels v to ix). Specific colocalization of J2-labeled foci and NS5A was evident (Fig. 2A, panel ix) but not common. A low level of fluorescence was apparent in the nuclei of some 2/1 cells probed with J2, and we attributed this finding to nonspecific staining of this particular cell line since no signal was evident in the nuclei of Huh-7 cells supporting transient replication of JFH-1 replicons or those infected with JFH-1 virus (Fig. 3; also see Fig. 5).
FIG. 2.
Visualization of dsRNA in Huh-7 cells constitutively replicating subgenomic JFH-1 replicon RNA. (A) 2/1 JFH-1 replicon-containing cells (panels i and iii to ix) and Huh-7 control cells (panel ii) were probed for the presence of dsRNA using monoclonal antibody J2 and counterstained, where appropriate, for NS5A (panels vi to ix). Cellular regions boxed in panels iii and v are shown enlarged in panels iv and vii to ix, respectively. The arrow indicates the colocalization of dsRNA-containing foci and NS5A protein in 2/1 replicon-containing cells. (B) 2/1 cells were treated with RNase A under either low-salt (LS) or high-salt (HS) conditions and probed for dsRNA and NS5A. Bars, 10 μm.
FIG. 3.
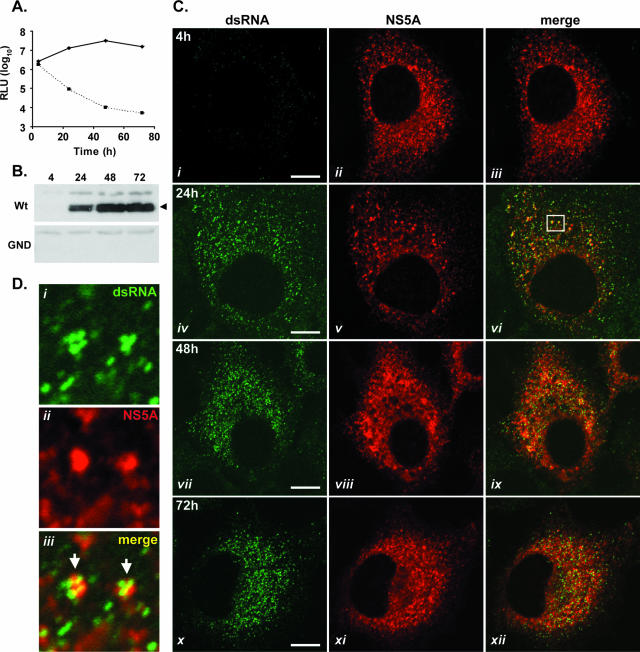
dsRNA-containing foci are formed during the transient replication of HCV replicons. JFH-1 wt and GND mutant subgenomic replicon RNAs were introduced into Huh-7 cells by electroporation, and cells were assessed at 4, 24, 48, and 72 h. (A) Cells were examined for luciferase activity (cells with wt replicon RNA, solid line; cells with GND mutant replicon RNA, dashed line). RLU, relative light units. (B) Cell extracts were probed with NS5A antiserum in a Western blot analysis. The arrowhead indicates NS5A protein. (C) Fixed cells were examined for the presence of dsRNA and NS5A by indirect immunofluorescence at 4, 24, 48, and 72 h postelectroporation. The boxed region in panel vi is shown enlarged in panel D. Arrows indicate the colocalization of dsRNA-containing foci and NS5A protein. Bars, 10 μm.
FIG. 5.
HCV-infected Huh-7 cells accumulate dsRNA foci that associate with NS5A and core proteins. (A) Huh-7 cells infected with JFH-1 virus were examined for the presence of dsRNA and NS5A at 24, 48, and 72 h postinfection. Selected regions of cells indicated in panels iii and vi are shown enlarged in panels B and C, respectively, and stained for dsRNA (panels i) and NS5A (panels ii). Arrows illustrate regions of colocalization of dsRNA-containing foci and NS5A protein in HCV-infected cells. (D) Huh-7 cells infected with JFH-1 virus for 48 h were labeled with antibodies specific for dsRNA and core protein. The boxed cellular region in panel iii is shown enlarged in panels iv to vi. The arrow indicates the colocalization of core protein-coated LDs and foci of dsRNA. Bars, 10 μm.
To establish whether foci detected by J2 were composed of dsRNA, 2/1 cells were fixed and treated with RNase A under either low- or high-salt conditions. Under low-salt conditions, RNase A digests both ssRRNA and dsRNA, while digestion under high-salt conditions degrades only ssRNA (61). In 2/1 cells treated with RNase A under low-salt conditions, J2 staining was completely lost while the detection of NS5A was unaffected (Fig. 2B, panels i and ii). By contrast, RNase A treatment of cells under high-salt conditions had no affect on the presence or distribution of J2-labeled foci. We did observe higher levels of background staining in cells treated with RNase A than in untreated cells (Fig. 2B, panel iii), probably due to the liberation of some dsRNA from within the cells. These data strongly suggested that the J2-labeled foci seen in 2/1 cells were composed of dsRNA. Taken together, these findings demonstrate the visualization of dsRNA in cells supporting steady-state replication of HCV subgenomic RNA.
dsRNA-containing foci are generated as a consequence of HCV RNA replication.
The vast majority of HCV nonstructural proteins synthesized within Huh-7 cells supporting steady-state replication of HCV replicons are not involved in RNA replication (44). Therefore, the formation of dsRNA in cells following the onset of JFH-1 replication was monitored to examine levels of coincidence between dsRNA foci and NS5A at time points immediately following the initiation of replicon replication (Fig. 3). Additionally, this approach sought to determine whether the accumulation of dsRNA was a result of HCV RNA synthesis. To confirm the initiation of RNA replication, extracts prepared from Huh-7 cells electroporated with the wt replicon and a nonreplicative control replicon (encoding a GDD-to-GND mutation in NS5B) were monitored for luciferase activity at set intervals (Fig. 3A). Levels of luciferase activity from cell extracts containing the wt replicon rose sharply from 4 to 48 h postelectroporation, indicative of high levels of viral RNA replication (Fig. 3A) (55). In contrast, luciferase activities from cell extracts containing the nonreplicative GND mutant replicon diminished over the same time period (Fig. 3A). Western blot analysis of cells electroporated with the wt replicon revealed the accumulation of NS5A protein from 24 h postelectroporation onwards, which was further evidence for RNA replication (Fig. 3B) (55). By comparison, no NS5A protein was detected in extracts prepared from cells electroporated with GND mutant replicon RNA (Fig. 3B). In parallel with these experiments, examinations of Huh-7 cells containing wt or GND mutant replicon RNA by indirect immunofluorescence were performed at 4, 24, 48, and 72 h postelectroporation using antibodies specific for dsRNA and NS5A (Fig. 3C; also see Fig. S1 in the supplemental material). At 4 h postelectroporation, before the onset of viral RNA replication at levels detectable by luciferase assay (22), NS5A protein was present in approximately 60% of cells and was presumably derived from the translation of input replicon RNA, since cells containing wt or GND mutant versions of the replicon tested positive for NS5A (Fig. 3C, panel ii; also see Fig. S1 in the supplemental material). However, at this time point, dsRNA was not consistently detected in cells containing the wt replicon and no dsRNA was evident in cells containing the GND mutant replicon (Fig. 3C, panel i; also see Fig. S1 in the supplemental material). Furthermore, although NS5A in cells electroporated with the GND mutant replicon was still detectable up to 24 h postelectroporation, no dsRNA was observed in these cells at this time point or any later time point (see Fig. S1 in the supplemental material). At 24 h postelectroporation with the wt JFH-1 replicon, dsRNA-containing foci were present in the cytoplasm of cells and were indistinguishable from those present in 2/1 cells (compare Fig. 2A and 3C, panel iv). The appearance of dsRNA foci within cells containing the wt JFH-1 replicon correlated with rising levels of RNA synthesis, as judged by luciferase assay and the accumulation of NS5A protein (compare Fig. 3A and C, panels i and iv). In all cells, which were positively stained with J2, the distribution of dsRNA-containing foci closely mirrored that of NS5A protein (Fig. 3C, panels vi, ix, and xii) and previously unseen levels of colocalization of the two antigens could be detected (Fig. 3D). In such instances, it was common to encounter a cluster of up to four dsRNA-containing complexes associated with one larger focus of NS5A protein (Fig. 3D, panel iii).
To investigate further the relationship between the appearance of dsRNA foci and HCV RNA replication, the formation of foci containing dsRNA during the transient replication of the JFH-1 replicon in the presence of ribavirin, which is an inhibitor of HCV genome synthesis (24), was examined. wt JFH-1 replicon RNA was introduced by electroporation into Huh-7 cells, which were then incubated in standard tissue culture medium or medium supplemented with ribavirin (50 or 100 μg/ml). Cell extracts prepared at set intervals were assayed for luciferase activity (Fig. 4A). This assay revealed that ribavirin at 100 μg/ml was more effective at inhibiting HCV replication than ribavirin at 50 μg/ml; at 72 h postelectroporation, the level of luciferase activity in extracts prepared from cells treated with ribavirin at 100 μg/ml was 125-fold lower than that in extracts from untreated cells (Fig. 4A). In contrast, the level of luciferase activity in extracts from cells treated with ribavirin at 50 μg/ml was only eightfold lower than that in untreated samples at 72 h postelectroporation (Fig. 4A). Having established conditions under which the replication of the wt replicon was efficiently repressed by ribavirin, we examined the formation of dsRNA-containing foci in cells electroporated with wt JFH-1 replicon RNA and treated with ribavirin by indirect immunofluorescence (Fig. 4C). At 4 h postelectroporation, there was no difference between the untreated and ribavirin-treated cells; NS5A protein was often present, but dsRNA was absent (Fig. 4B and C, panels i and ii). At 24 h postelectroporation, dsRNA-containing foci were abundant in the cytoplasm of untreated cells but were not detected in the cytoplasm of ribavirin-treated cells, despite the presence of NS5A protein (Fig. 4B and C, panels iii and iv). A similar phenotype was evident at 48 h postelectroporation, although some ribavirin-treated cells containing NS5A demonstrated very weak staining for dsRNA (Fig. 4B and C, panels v and vi). At 72 h postelectroporation, levels of luciferase activity within cell extracts prepared from ribavirin-treated cells had risen threefold compared to values determined 48 h postelectroporation (Fig. 4A). This observation indicated either that the concentration of ribavirin used was not sufficient to entirely abolish the replication of the replicon or that ribavirin-resistant replicons had began to emerge. The rise in levels of replication in ribavirin-treated cells observed between 48 and 72 h postelectroporation was accompanied by the appearance of dsRNA foci in cells that also contained NS5A (Fig. 4C, panels vii and viii). Although levels of dsRNA in ribavirin-treated cells were diminished compared to those observed in untreated cells at 72 h postelectroporation (Fig. 4B and C, panels vii and viii), the appearance of dsRNA foci in cells treated with ribavirin directly correlated with the observed increase in replication as judged by levels of luciferase activity in cell extracts. Taken together, the results presented here demonstrate that dsRNA-containing foci are generated as a direct consequence of HCV RNA replication.
FIG. 4.
The generation of dsRNA-containing foci is a consequence of HCV RNA replication. wt JFH-1 subgenomic replicon RNA was introduced into Huh-7 cells by electroporation in the absence (untreated) or presence of ribavirin (50 or 100 μg/ml), and cells were assessed at 4, 24, 48, and 72 h. (A) Cells were examined for luciferase activity. RLU, relative light units. (B and C) Cells grown in the absence (B) or presence (C) of 100 μg of ribavirin/ml were examined for the appearance of dsRNA and NS5A by indirect immunofluorescence at 4, 24, 48, and 72 h postelectroporation. The arrow indicates dsRNA-containing foci in ribavirin-treated cells. Bars, 10 μm.
HCV-infected cells accumulate foci containing dsRNA that associate with virus-encoded proteins.
Utilizing JFH-1 virus, we next sought to investigate the formation of dsRNA-containing foci in HCV-infected cells. Cells were analyzed at 24, 48, and 72 h postinfection for the presence of dsRNA and NS5A (Fig. 5). dsRNA was detected in cells at all time points postinfection, and the distribution of dsRNA-containing foci changed from localization throughout the cytoplasm to mainly perinuclear localization as the time course progressed (Fig. 5A, panels i, iv, and vii). The observed pattern of dsRNA distribution was mirrored by that of NS5A (Fig. 5A, panels ii, v, and viii), and at the earliest time point, when the distribution of NS5A was more punctate than ER-like, high levels of colocalization of dsRNA-containing foci and NS5A protein were observed (Fig. 5B). At 48 h postinfection, absolute colocalization of dsRNA and NS5A was less evident (Fig. 5A, panels iv to vi). Nevertheless, in regions of JFH-1-infected cells that still exhibited punctate distribution of NS5A, several small dsRNA-containing structures were often found associated with a larger focus of NS5A in a manner similar to that observed during the transient replication of JFH-1 replicon RNA (Fig. 5C, panels i to iii). No dsRNA accumulated in mock-infected cells at any time point (data not shown).
Next, the relationship between dsRNA-containing foci and core protein, the major HCV capsid protein, in cells infected with JFH-1 virus was examined. Core protein is transferred to the surfaces of cytoplasmic lipid droplets (LDs), and we have previously established that complete transfer in virus-infected cells can take up to 48 h (5). Therefore, in order to examine the distribution of dsRNA foci in relation to core protein, cells were analyzed 48 h following infection with JFH-1 virus (Fig. 5D). dsRNA foci and core protein exhibited near-identical distributions in virus-infected cells (Fig. 5D, panels i to iii). At higher magnification, foci containing dsRNA were often observed juxtaposed to core protein-coated ring-like structures, presumed to be LDs (Fig. 5D, panels iv to vi). Thus, our data demonstrate the accumulation of dsRNA-containing foci within HCV-infected cells and highlight associations formed with virus-encoded structural (core) and nonstructural (NS5A) proteins.
Relationships between dsRNA-containing foci observed in HCV-infected cells and specific cellular factors.
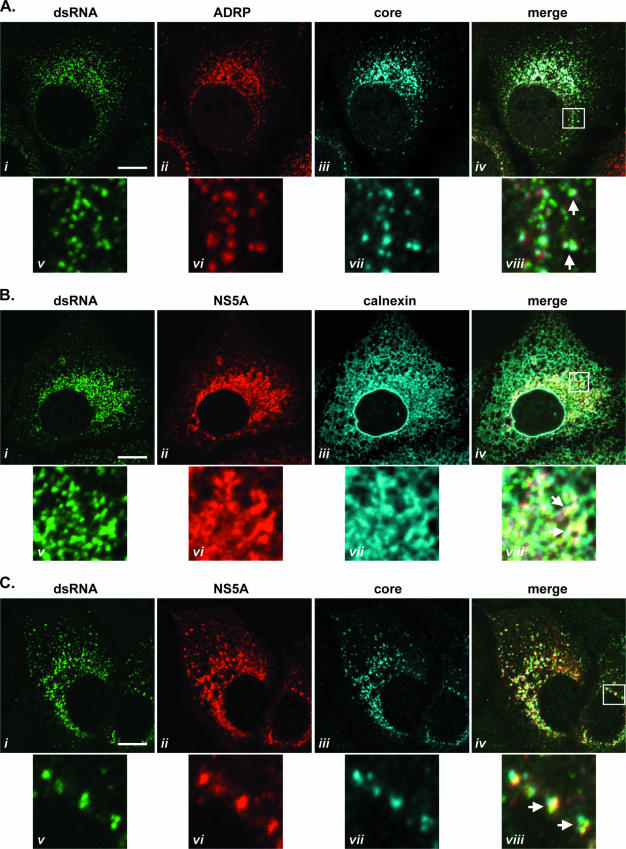
The above-described findings indicated that, in virus-infected cells, dsRNA-containing foci were associated with virus-encoded proteins known to attach to LDs (core protein) (4) and the ER (NS5A) (6). Therefore, the relationships between virus-induced dsRNA foci and these cellular organelles was examined in detail through the use of antibodies specific for ADRP and calnexin, markers for LDs and the ER membrane, respectively (54, 57). JFH-1-infected Huh-7 cells, probed with dsRNA-specific J2 antibody, were counterstained with antibodies specific for either ADRP and core protein (Fig. 6A) or NS5A and calnexin (Fig. 6B). In cells infected with HCV, core protein localized with ADRP on LDs, and dsRNA-containing foci were observed in close proximity and frequently found juxtaposed to core protein-coated LDs (Fig. 6A, panel viii). The calnexin-labeled ER appeared as a diffuse network distributed throughout the cytoplasm of virus-infected cells (Fig. 6B, panel iii). NS5A localized to this network, and the colocalization of NS5A and the ER was particularly evident in the perinuclear region (Fig. 6B, panels vi and vii). dsRNA-containing foci exhibited a pattern of localization similar to that of ER-bound NS5A, with analogous accumulation in perinuclear regions (Fig. 6B, panel i). When samples were viewed at greater magnification, a high level of colocalization of dsRNA-containing foci, NS5A, and calnexin was observed (Fig. 6B, panels v to viii). Finally, the distribution of dsRNA-containing foci in relation to the localization of core protein and NS5A in virus-infected cells was examined (Fig. 6C). This analysis revealed sites of colocalization of dsRNA, core protein, and NS5A (Fig. 6C, panels i to iv). Indeed, we found several smaller foci of dsRNA coincident with one larger focus of ER-bound NS5A protein at sites that included core protein-coated LDs (Fig. 6C, panels v to viii).
FIG. 6.
Distribution of dsRNA with NS5A, core protein, ADRP, and calnexin in HCV-infected cells. Huh-7 cells infected with JFH-1 virus for 48 h were analyzed for the presence of dsRNA together with ADRP and core protein (A), NS5A and calnexin (B), or NS5A and core protein (C) by indirect immunofluorescence. The selected cellular regions indicated in panels iv are shown enlarged in panels v to viii. Arrows indicate regions of close apposition of antigens. Bars, 10 μm.
To gain more insight into the distribution pattern of dsRNA-containing foci in relation to viral and cellular factors in HCV-infected cells, we analyzed confocal laser-scanned images of virus-infected cells with software designed to generate high-resolution 3D reconstructions. This approach allowed us to gain a unique perspective regarding the relationship of dsRNA to LDs, the ER, and NS5A in virus-infected cells. Raw z-stack confocal images of appropriately labeled HCV-infected cells were subjected to deconvolution, a computational 3D-image restoration technique, which serves to remove the unavoidable blurring effect of the point spread function in stacks of optical confocal sections (36). Z-stacks of images of HCV-infected cells labeled with antibodies specific for dsRNA and ADRP, dsRNA and calnexin, ADRP and calnexin, dsRNA and NS5A, and dsRNA, ADRP, and calnexin (see Fig. S2 in the supplemental material) were deconvolved. Regions of interest from the deconvolved z-stacks were reconstructed as 3D images to achieve a more in-depth analysis of the relationships between selected antigens than that offered by 2D confocal microscopy (Fig. 7). Consistent with information gained from previous examinations of HCV-infected cells (Fig. 5D and 6A), dsRNA-containing foci were clearly detected in close association with LDs and foci of dsRNA were often directly visualized either adjacent to single LDs or coating clusters of several LDs (Fig. 7A; also see Fig. S3 in the supplemental material). A similar level of detail was revealed for the localization of dsRNA and calnexin in cells infected with HCV and analyzed in an identical manner (Fig. 7B). The calnexin-labeled ER appeared as an interconnected network, with the majority of dsRNA-containing structures located on the surface of the ER (Fig. 7B). A smaller portion of the dsRNA foci were also seen to be surrounded, either partially or wholly, by the ER (Fig. 7B). Consistent with findings from previous confocal analyses of cells infected with HCV (Fig. 6B), very few dsRNA foci were found to be unconnected to the ER. The reconstruction of z-stacks from ADRP- and calnexin-labeled HCV-infected cells was performed to highlight the association of LDs with the ER (Fig. 7C). The results demonstrated that, in agreement with previous findings (54), LDs appeared to be tethered to the ER. The intimate association that exists between LDs and the ER provides a likely rationale for the close proximity of dsRNA foci to both organelles.
FIG. 7.
3D reconstruction of deconvolved z-stack images generated from HCV-infected cells. Selected regions from deconvolved z-stack sections of images of cells infected with JFH-1 virus for 48 h were reconstructed as 3D images. (A) Arrowheads highlight dsRNA-containing foci juxtaposed to LDs. (B) Large and small arrowheads indicate dsRNA-containing foci located on the surface of, and those surrounded by, the calnexin-labeled ER, respectively. (C) Arrowheads indicate close connections between ADRP-coated LDs and the calnexin-stained ER. (D) Large and small arrowheads indicate foci of dsRNA situated on the surface of, and those partially bounded by, ER-associated NS5A, respectively. (E and F) Arrowheads in panel E highlight close relationships among the calnexin-labeled ER, ADRP-coated LDs, and dsRNA foci. Numbered arrowheads in panel F correspond to the range of distributions observed in virus-infected cells labeled for calnexin, dsRNA, and ADRP, as follows: arrowhead 1, a single dsRNA focus on the ER surface; arrowhead 2, a single ER-attached LD; arrowhead 3, a complex composed of a single dsRNA focus juxtaposed to an LD, situated adjacent to the ER; and arrowhead 4, an ER-associated complex composed of a single LD and multiple foci of dsRNA. Bars, 1 μm.
Since NS5A is an ER-bound protein, the connection formed between dsRNA and NS5A was similar to that between dsRNA and calnexin; foci of dsRNA were located either on the surface of or surrounded by an ER-bound network of NS5A protein (Fig. 7D). Furthermore, very few dsRNA complexes were not connected with NS5A on the ER (Fig. 7D). To gain a definitive view of the relationships among dsRNA, LDs, and the ER, a z-stack of images of a virus-infected cell labeled for all three antigens was deconvolved and reconstructed as 3D images (Fig. 7E and F). The results revealed the extent of the interactions between dsRNA foci and the network of the ER and LDs, with all three antigens located in close proximity to one another (Fig. 7E). The full scope of interactions among dsRNA, the ER, and LDs is revealed in Fig. 7F. In this 3D image, four levels of distribution are evident: (i) a single dsRNA focus on the ER surface; (ii) a single ER-attached LD; (iii) a complex composed of a single dsRNA focus juxtaposed to an LD, situated adjacent to the ER; and (iv) an ER-associated complex composed of a single LD and multiple foci of dsRNA. Taken together, data generated from 3D reconstructions of virus-infected cells suggest that the ER network and associated LDs are the sites where dsRNA-containing foci generated in cells replicating HCV genomes are localized.
DISCUSSION
This study provides evidence for the formation of dsRNA-containing foci in cells supporting HCV RNA replication. Immunodetection methods allowed us to visualize intracellular structures containing dsRNA formed de novo after the initiation of HCV genome synthesis in transient replication assays and during virus infection. We examined the localization of dsRNA foci with respect to both virus-encoded and cell-derived factors by using conventional 2D confocal microscopy and 3D reconstruction techniques. The latter revealed novel interactions of dsRNA-containing foci with both LDs and the ER membrane in HCV-infected cells.
Taking advantage of the high replicative ability and infectious capacity of the HCV JFH-1 isolate, we have extended the analysis of intracellular viral RNA distribution by FISH to include an examination of the relationship of the RNA with virus-encoded NS5A in replicon-containing and virus-infected cells. Distinct structures containing viral RNA were visualized under both conditions and appeared as a network of foci whose distribution overlapped with that of NS5A protein. NS5A is an HCV-encoded nonstructural protein that localizes to the ER membrane and is essential for replication (3, 6, 29). The precise role of NS5A in HCV genome synthesis has yet to be determined, although it is known that the protein can bind viral RNA and may act to regulate the switch between RNA replication and virion assembly (12, 20, 41, 56). The RNA foci associated with NS5A on the ER membrane presumably reflected sites of viral RNA replication and, possibly, genome translation. To complement and expand the study of HCV RNA distribution instigated by using FISH, we examined cells supporting HCV RNA replication for the presence of dsRNA by immunodetection using monoclonal antibody J2. dsRNA is generated during the replication of (+)RNA viruses but is not found in uninfected cells (27, 34, 60-62). dsRNA was readily detected in both replicon-containing and virus-infected cells and formed foci whose distribution, abundance, and morphology were indistinguishable from those of foci observed by FISH. Additionally, foci of dsRNA associated with NS5A on the ER. Thus, we believe that the same RNA foci are identified by FISH and J2. The dsRNA foci generated in cells supporting the replication of HCV RNA likely represent sites at which HCV genome synthesis occurs, and the intimate link between RNA replication and the accumulation of dsRNA foci is highlighted by several lines of evidence. Firstly, no dsRNA foci were detected in cells electroporated with nonreplicative GND mutant replicon RNA, mock-infected cells, or control Huh-7 cells. Secondly, the appearance of dsRNA foci correlated directly with the initiation of JFH-1 RNA replication in transient replication assays. Lastly, the appearance of dsRNA foci in ribavirin-treated cells containing the wt JFH-1 replicon was delayed until there was evidence of RNA replication.
We considered two potential scenarios that may account for the origin of the dsRNA foci detected by monoclonal antibody J2 in cells supporting HCV RNA replication. Firstly, consistent with the strategy adopted by (+)RNA viruses other than HCV (8, 9, 14, 45), dsRNA foci detected in cells replicating HCV RNA may represent dsRNA RIs generated during genome synthesis (27, 61). RIs are formed following the synthesis of complementary negative-sense RNA, which forms a dsRNA molecule with the plus strand, to act as a recycling template for the synthesis of nascent plus-strand RNA. Nascent strands are synthesized from the RI by strand displacement, and the number of nascent strands in the RI varies from one (Kunjin virus) (8) to between four and eight (polio-, dengue, and bovine viral diarrhea viruses) (9, 14, 45). Alternatively, the visualization of dsRNA may be a consequence of J2 antibody binding to intramolecular secondary structures within HCV genomes. Although we do not absolutely exclude the possibility that the J2 antibody also reacts with double-stranded regions of the HCV genome, we favor the former scenario for the following reasons: (i) dsRNA-containing foci have previously proven to be accurate markers for sites of viral genome replication in cells infected with other (+)RNA viruses (Kunjin, rubella, and Semliki Forest viruses) (27, 34, 61, 62); (ii) RIs have been detected in cell-free HCV RNA synthesis systems, suggesting that RIs are a strategy employed by HCV for genome replication (1, 26); and (iii) monoclonal antibody J2 is not able to bind RNA-RNA helices of less than 40 bp (47), and no predicted RNA secondary structures within the HCV genome contain more than 40 bp of unbroken helix. Thus, our findings suggest that dsRNA foci likely represent RIs formed during HCV RNA synthesis.
In this study, we describe the use of 3D reconstruction techniques applied to deconvolved z-stack images to examine the associations between dsRNA and cellular organelles. This analysis revealed that foci containing dsRNA were localized on the surface of, or surrounded by, the ER membrane, as judged by association with ER-retained calnexin and NS5A proteins. This finding is consistent with the current model for HCV RNA synthesis, in which genome replication occurs within ER-derived vesicles of the membranous web whose formation is believed to be driven, at least in part, by the oligomerization of NS4B (10, 15, 64). Additionally, we found foci of dsRNA in close proximity with, and often juxtaposed to, LDs, which are attached to the ER (54). LDs are storage organelles for intracellular lipid reserves and are actively involved in cellular lipid homeostasis (13, 35, 54). HCV core protein accumulates on the surfaces of LDs, and this attachment is linked to the production of infectious virus in cultured cells (4, 5, 39). Additionally, HCV RNA has been shown to accumulate around the surfaces of LDs in cells containing JFH-1 genomes, and LD-enriched fractions isolated from the same cells exhibit the ability to synthesize HCV RNA (39). These observations suggest that in cells transfected with JFH-1 genomes, LDs represent sites at which viral RNA replication can occur. Furthermore, the same study reported that core protein was responsible for the recruitment of HCV RCs to the LD surface (39). Our findings are consistent with this paradigm and further highlight the significance of LDs during HCV infection. However, we also observed dsRNA foci juxtaposed to LDs in replicon-containing cells (see Fig. S4 in the supplemental material), albeit to a far lesser extent than that in virus-infected cells (Fig. 6A). This observation suggested that the association between dsRNA and LDs in virus-infected cells was not dependent solely on the presence of core protein. In addition to core protein, NS5A has been found on the surfaces of LDs (49) and may be involved in the localization of HCV RNA to LDs, perhaps as a consequence of the RNA binding properties of the protein (20). In virus-infected cells, therefore, RNA replication may be directed to LD-connected regions of the ER through the actions of both core protein and NS5A, thereby facilitating the transfer of replicated genomes to sites of virus assembly.
To conclude, the findings presented in this report broaden our knowledge of HCV RNA distribution in virus-infected cells. The discovery of the close apposition of LDs to sites of HCV-induced foci containing dsRNA complements a growing appreciation of the involvement of LDs in the virus life cycle (5, 39). Furthermore, the combined use of immunological reagents directed at dsRNA and 3D image reconstruction procedures expands the number of approaches available to investigate the HCV life cycle.
Supplementary Material
Acknowledgments
We are grateful to Takaji Wakita for supplying JFH-1 plasmid DNA and to Mark Harris and Steve Griffin for providing the NS5A antiserum.
This work was supported by the Medical Research Council (United Kingdom) and by a Marie Curie intra-European fellowship to S.B. (contract number 025198).
Footnotes
Published ahead of print on 19 December 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ali, N., K. D. Tardif, and A. Siddiqui. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 7612001-12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1983. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology 13139-48. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 2901972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Boulant, S., R. Montserret, R. G. Hope, M. Ratinier, P. Targett-Adams, J. P. Lavergne, F. Penin, and J. McLauchlan. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 28122236-22247. [DOI] [PubMed] [Google Scholar]
- 5.Boulant, S., P. Targett-Adams, and J. McLauchlan. 2007. Disrupting the ability of HCV core protein to associate with lipid droplets abolishes production of infectious virus. J. Gen. Virol. 882204-2213. [DOI] [PubMed] [Google Scholar]
- 6.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 2778130-8139. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 744310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu, P. W., and E. G. Westaway. 1985. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology 14068-79. [DOI] [PubMed] [Google Scholar]
- 9.Cleaves, G. R., T. E. Ryan, and R. W. Schlesinger. 1981. Identification and characterization of type 2 dengue virus replicative intermediate and replicative form RNAs. Virology 11173-83. [DOI] [PubMed] [Google Scholar]
- 10.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 765974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eigen, M., C. K. Biebricher, M. Gebinoga, and W. C. Gardiner. 1991. The hypercycle. Coupling of RNA and protein biosynthesis in the infection cycle of an RNA bacteriophage. Biochemistry 3011005-11018. [DOI] [PubMed] [Google Scholar]
- 12.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. USA 10113038-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimoto, T., and Y. Ohsaki. 2006. Cytoplasmic lipid droplets: rediscovery of an old structure as a unique platform. Ann. N. Y. Acad. Sci. 1086104-115. [DOI] [PubMed] [Google Scholar]
- 14.Gong, Y., A. Shannon, E. G. Westaway, and E. J. Gowans. 1998. The replicative intermediate molecule of bovine viral diarrhoea virus contains multiple nascent strands. Arch. Virol. 143399-404. [DOI] [PubMed] [Google Scholar]
- 15.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 775487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 763697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamamoto, I., Y. Nishimura, T. Okamoto, H. Aizaki, M. Liu, Y. Mori, T. Abe, T. Suzuki, M. M. Lai, T. Miyamura, K. Moriishi, and Y. Matsuura. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 7913473-13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, R. W., J. Marcotrigiano, K. J. Blight, J. E. Majors, and C. M. Rice. 2003. Hepatitis C virus RNA synthesis in a cell-free system isolated from replicon-containing hepatoma cells. J. Virol. 772029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hope, R. G., and J. McLauchlan. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 811913-1925. [DOI] [PubMed] [Google Scholar]
- 20.Huang, L., J. Hwang, S. D. Sharma, M. R. Hargittai, Y. Chen, J. J. Arnold, K. D. Raney, and C. E. Cameron. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 28036417-36428. [DOI] [PubMed] [Google Scholar]
- 21.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 28470-81. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. M., S. N. Gretton, J. McLauchlan, and P. Targett-Adams. 2007. Mobility analysis of an NS5A-GFP fusion protein in cells actively replicating hepatitis C virus subgenomic RNA. J. Gen. Virol. 88470-475. [DOI] [PubMed] [Google Scholar]
- 23.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 1251808-1817. [DOI] [PubMed] [Google Scholar]
- 24.Kato, T., T. Date, M. Miyamoto, M. Sugiyama, Y. Tanaka, E. Orito, T. Ohno, K. Sugihara, I. Hasegawa, K. Fujiwara, K. Ito, A. Ozasa, M. Mizokami, and T. Wakita. 2005. Detection of anti-hepatitis C virus effects of interferon and ribavirin by a sensitive replicon system. J. Clin. Microbiol. 435679-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 753873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai, V. C., S. Dempsey, J. Y. Lau, Z. Hong, and W. Zhong. 2003. In vitro RNA replication directed by replicase complexes isolated from the subgenomic replicon cells of hepatitis C virus. J. Virol. 772295-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, J. Y., J. A. Marshall, and D. S. Bowden. 1994. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology 200307-312. [DOI] [PubMed] [Google Scholar]
- 28.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 29.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 30.Lukacs, N. 1994. Detection of virus infection in plants and differentiation between coexisting viruses by monoclonal antibodies to double-stranded RNA. J. Virol. Methods 47255-272. [DOI] [PubMed] [Google Scholar]
- 31.Lundin, M., M. Monne, A. Widell, G. Von Heijne, and M. A. Persson. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 775428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macdonald, A., K. Crowder, A. Street, C. McCormick, K. Saksela, and M. Harris. 2003. The hepatitis C virus non-structural NS5A protein inhibits activating protein-1 function by perturbing ras-ERK pathway signaling. J. Biol. Chem. 27817775-17784. [DOI] [PubMed] [Google Scholar]
- 34.Magliano, D., J. A. Marshall, D. S. Bowden, N. Vardaxis, J. Meanger, and J. Y. Lee. 1998. Rubella virus replication complexes are virus-modified lysosomes. Virology 24057-63. [DOI] [PubMed] [Google Scholar]
- 35.Martin, S., and R. G. Parton. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7373-378. [DOI] [PubMed] [Google Scholar]
- 36.McNally, J. G., T. Karpova, J. Cooper, and J. A. Conchello. 1999. Three-dimensional imaging by deconvolution microscopy. Methods 19373-385. [DOI] [PubMed] [Google Scholar]
- 37.Miller, S., S. Kastner, J. Krijnse-Locker, S. Buhler, and R. Bartenschlager. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 2828873-8882. [DOI] [PubMed] [Google Scholar]
- 38.Miller, S., S. Sparacio, and R. Bartenschlager. 2006. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 2818854-8863. [DOI] [PubMed] [Google Scholar]
- 39.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 91089-1097. [DOI] [PubMed] [Google Scholar]
- 40.Moradpour, D., V. Brass, E. Bieck, P. Friebe, R. Gosert, H. E. Blum, R. Bartenschlager, F. Penin, and V. Lohmann. 2004. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J. Virol. 7813278-13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moradpour, D., V. Brass, and F. Penin. 2005. Function follows form: the structure of the N-terminal domain of HCV NS5A. Hepatology 42732-735. [DOI] [PubMed] [Google Scholar]
- 42.Negro, F., D. Pacchioni, Y. Shimizu, R. H. Miller, G. Bussolati, R. H. Purcell, and F. Bonino. 1992. Detection of intrahepatic replication of hepatitis C virus RNA by in situ hybridization and comparison with histopathology. Proc. Natl. Acad. Sci. USA 892247-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okamoto, T., Y. Nishimura, T. Ichimura, K. Suzuki, T. Miyamura, T. Suzuki, K. Moriishi, and Y. Matsuura. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 255015-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinkert, D., R. Bartenschlager, and V. Lohmann. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 7913594-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richards, O. C., and E. Ehrenfeld. 1990. Poliovirus RNA replication. Curr. Top. Microbiol. Immunol. 16189-119. [DOI] [PubMed] [Google Scholar]
- 46.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 759808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schonborn, J., J. Oberstrass, E. Breyel, J. Tittgen, J. Schumacher, and N. Lukacs. 1991. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 192993-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9505-514. [DOI] [PubMed] [Google Scholar]
- 49.Shi, S. T., S. J. Polyak, H. Tu, D. R. Taylor, D. R. Gretch, and M. M. Lai. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292198-210. [DOI] [PubMed] [Google Scholar]
- 50.Stollar, B. D., and V. Stollar. 1970. Immunofluorescent demonstration of double-stranded RNA in the cytoplasm of Sindbis virus-infected cells. Virology 42276-280. [DOI] [PubMed] [Google Scholar]
- 51.Stone, M., S. Jia, W. D. Heo, T. Meyer, and K. V. Konan. 2007. Participation of Rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J. Virol. 814551-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 748953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takehara, T., N. Hayashi, E. Mita, H. Hagiwara, K. Ueda, K. Katayama, A. Kasahara, H. Fusamoto, and T. Kamada. 1992. Detection of the minus strand of hepatitis C virus RNA by reverse transcription and polymerase chain reaction: implications for hepatitis C virus replication in infected tissue. Hepatology 15387-390. [DOI] [PubMed] [Google Scholar]
- 54.Targett-Adams, P., D. Chambers, S. Gledhill, R. G. Hope, J. F. Coy, A. Girod, and J. McLauchlan. 2003. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J. Biol. Chem. 27815998-16007. [DOI] [PubMed] [Google Scholar]
- 55.Targett-Adams, P., and J. McLauchlan. 2005. Development and characterization of a transient-replication assay for the genotype 2a hepatitis C virus subgenomic replicon. J. Gen. Virol. 863075-3080. [DOI] [PubMed] [Google Scholar]
- 56.Tellinghuisen, T. L., J. Marcotrigiano, and C. M. Rice. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wada, I., D. Rindress, P. H. Cameron, W. J. Ou, J. J. Doherty II, D. Louvard, A. W. Bell, D. Dignard, D. Y. Thomas, and J. J. Bergeron. 1991. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J. Biol. Chem. 26619599-19610. [PubMed] [Google Scholar]
- 58.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, C., M. Gale, Jr., B. C. Keller, H. Huang, M. S. Brown, J. L. Goldstein, and J. Ye. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18425-434. [DOI] [PubMed] [Google Scholar]
- 60.Weber, F., V. Wagner, S. B. Rasmussen, R. Hartmann, and S. R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 805059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westaway, E. G., A. A. Khromykh, and J. M. Mackenzie. 1999. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology 258108-117. [DOI] [PubMed] [Google Scholar]
- 62.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 716650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolk, B., D. Sansonno, H. G. Krausslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 742293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, G. Y., K. J. Lee, L. Gao, and M. M. Lai. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 806013-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 1029294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 742017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.