Abstract
Herpesvirus DNA replication proceeds via concatemeric replicative intermediates that are comprised of head-to-tail-linked genomes. Genome maturation is carried out by the terminase, a protein complex that mediates both insertion of concatemer DNA into capsids and its subsequent cleavage to release genomes within these capsids. This cleavage is sequence specific, but the governing cis-acting DNA sequences are only partially characterized. Two highly conserved motifs called pac1 and pac2 lie near the ends of herpesvirus genomes and are known to be critical for genome maturation. However, the potential importance of other sequences has not been fully investigated. We have undertaken to define all of the sequences necessary for efficient genome maturation for a herpesvirus by inserting ectopic cleavage sites into the murine cytomegalovirus genome and assessing their ability to mediate genome maturation. A combination of deletion and substitution mutations revealed that the minimal cleavage site is large (∼180 bp) and complex. Sequences distal of pac1 (relative to the point of cleavage) were dispensable, suggesting that pac1 may be the sole cis-acting element on this side of the cleavage site. In contrast, a region distal to pac2 up to 150 bp from the point of cleavage was essential. Scanning substitutions revealed that the pac2 side of the cleavage site is complex and may contain multiple cis-acting sequence elements in addition to pac2. These results should facilitate the identification of trans-acting factors that bind to these elements and the elucidation of their functions. Such information will be critical for understanding the molecular basis of this complex process.
Herpesviruses have large (130- to 235-kb) linear double-stranded DNA genomes that replicate via concatemeric intermediates consisting of head-to-tail-linked genomes (1, 2, 14, 17, 18, 25, 27, 32). The concatemers are packaged into preformed capsids and cleaved at precise locations to release unit-length genomes within the capsids (3). The cis-acting sequences that are recognized by the cleavage/packaging machinery have not been fully defined, but two motifs denoted pac1 and pac2 were initially identified by their strong conservation near the ends of herpesvirus genomes (7). Typically, pac1 motifs consist of 3- to 7-bp A- or T-rich regions flanked on each side by short poly(C) tracts (7, 29), whereas pac2 motifs consist of 5- to 10-bp A-rich regions (7). In some viruses a consensus CGCGGCG motif is located just distal of pac2 (relative to the terminus) (7).
In order to mutagenically define the cis cleavage/packaging sequences of a herpesvirus, we developed an assay based on cleavage at a second or “ectopic” cleavage site that was inserted into the murine cytomegalovirus (MCMV) genome. In 1984, Marks and Spector (15) showed that the two ends of the MCMV genome (designated X and C, respectively) (Fig. 1) fuse shortly after infection to form “junctions.” Upon concatemer synthesis, such junctions presumably serve as natural cleavage/packaging sites. We predicted that an ectopically inserted copy of a junction would function as a cleavage/packaging site if it contained all the necessary cis-acting sequences. This hypothesis was tested by construction of virus RM4072, in which a 1.9-kb fusion of X and C sequences was inserted 45 kb from the right end of the MCMV genome in an orientation that created direct repeats of X and C sequences at the natural termini (Fig. 1) (21).
FIG. 1.
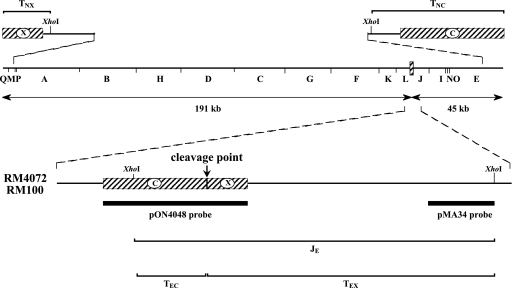
The MCMV ectopic cleavage site system. A HindIII restriction map of the MCMV genome is shown with terminal sequences, designated X and C (hatched boxes), expanded above. The region between the HindIII L and J fragments is expanded below to illustrate ectopic cleavage sites in viruses RM4072 or RM100 that are comprised of a 1.9-kb duplication/fusion of C and X terminal sequences (hatched boxes) in direct orientation relative to terminal sequences. A vertical arrow indicates the point where ectopic cleavage is predicted to occur. Brackets below represent XhoI fragments that result if ectopic sites are cleaved (TEC and TEX) or uncleaved (JE), while brackets above represent natural terminal fragments (TNX and TNC). Note: the XhoI site within the ectopic copy of C was artificially engineered and is not present at the natural C terminus. Thick bars represent sequences contained in probes used for Southern hybridizations.
The ectopic cleavage site in RM4072 was judged to be functionally equivalent to natural cleavage sites by the following reasoning. Within concatemers produced by RM4072, ectopic and natural cleavage sites should alternate and should be separated by 191 and 45 kb of intervening sequences (Fig. 1). Because herpesvirus DNA packaging is governed by a head-full constraint that requires entry of near-genome lengths of DNA into the capsid before cleavage can occur (8) and because neither 45 nor 191 kb was deemed likely to satisfy this requirement, we predicted that cleavage of RM4072 concatemers would occur in two frames: a natural frame in which cleavage occurs only at natural cleavage sites and skips over the intervening ectopic sites, and an ectopic frame in which cleavage occurs only at ectopic cleavage sites and skips over the intervening natural sites. Consequently, RM4072 virions should contain a mixture of two genome types: those cleaved at natural sites, having natural termini and uncleaved internal ectopic junctions, and those cleaved at ectopic sites, having ectopic termini and uncleaved internal natural junctions. Functional equivalence of ectopic and natural cleavage sites should give rise to an equal ratio of the two genome types.
To confirm these predictions, Southern hybridization was used to detect the restriction fragments that are illustrated in Fig. 1. RM4072 virion DNA contained restriction fragments deriving from natural termini (TNC and TNX) but also contained restriction fragments deriving from cleaved ectopic termini (TEC and TEX). Moreover, uncleaved natural junctions (JN) were present in RM4072 virion DNA but not in DNA from a control virus, and a proportion of ectopic junctions (JE) in RM4072 DNA remained uncleaved. Indeed, the roughly equimolar amounts of JE and TEX fragments established that the RM4072 ectopic cleavage sites are functionally equivalent to natural sites (21).
This result set the stage to evaluate the effects of mutations on ectopic cleavage site function. We reasoned that mutations that impaired cleavage/packaging at ectopic sites would reduce the proportion of genomes with ectopic termini. Hence, we used the ratio of JE to TEX fragment intensities to calculate efficiencies for mutant ectopic cleavage sites. By constructing recombinant viruses containing ectopic sites with mutations targeting conserved sequence motifs, we showed that the A-rich region of pac2, a CGCGGCG motif distal of pac2, and the left poly(C) tract of pac1 each are highly important for cleavage/packaging efficiency (21). These studies, however, were incomplete. The importance of additional conserved motifs was not addressed and, as the mutations were constructed in the context of the 1.9-kb ectopic site, the existence of cis elements outside of the core pac1-pac2 region was not determined.
In the present study we report results in which this original system was used to evaluate two additional site-directed mutations within or near pac1. We then describe the use of Tn7-mediated transposition to introduce ectopic sites into a bacterial artificial chromosome (BAC) clone of the MCMV genome to facilitate rapid virus construction. The new approach was used to further define the minimal sequences necessary for efficient cleavage site function. The results suggest that in addition to pac1 and pac2, sequences distal of pac2 are needed for the efficient maturation of MCMV genomes.
MATERIALS AND METHODS
Recombinant BAC construction.
BAC pSM3 is an infectious clone of the MCMV genome constructed and kindly provided by Martin Messerle (23). In previous work we modified pSM3 by insertion of a lacZα-mini-attTn7 site to generate BAC pSM3-117K (11). Ectopic cleavage sites were shuttled into the attTn7 site of pSM3-117K by Tn7-mediated site-specific transposition as described previously (11). Clones were screened for correct transposition using three PCRs. The reaction using primers TA1 and TA2 generates a 302-bp product from the parental pSM3-117K BAC that is lost upon insertion of the transposon, whereas reactions using primers TA1 and TA3 and primers TA2 and TA4, which span the left and right junctions between BAC and transposed sequences, generate 481- and 570-bp products, respectively, if transposition has occurred correctly. The sequences of these primers are given in Table 1.
TABLE 1.
Sequences of oligonucleotides
| Plasmid | Oligonucleotide | Sequence (5′ to 3′) |
|---|---|---|
| TA1 | GGTTTTCCCAGTCACGACGTTG | |
| TA2 | AGCTATACCATGATTACGCCAAGC | |
| TA3 | CCGCCACCTAACAATTCGTTC | |
| TA4 | CAGCCATACCACATTTGTAGAGG | |
| pMA30 | NOL 11 | CTAGAGGACAAAAATATAGCCCCCCCATCAAAATACCATGGTAGGGGGCC |
| NOL 12 | CCCTACCATGGTATTTTGATGGGGGGGCTATATTTTTGTCCT | |
| pMA51 | NOL 13 | CTAGAGGACAGTTAACTAGCCCCCC |
| NOL 14 | CATGGGGGGGCTAGTTAACTGTCCT | |
| pMA174 | MOL 160 | GGAATTCCAACTAGCAGCCCAACACCCA |
| MOL 162 | GGGGTACCCCTCAGCGCACTCTCAGGAGCA | |
| pMA220 | MOL 173 | CTAGAGGGCGCTGTTGCATCGCCTGGTAC |
| MOL 174 | CAGGCGATGCAACAGCGCCCT | |
| pMA215 | MOL 165 | AATTCGTCGACGGCCCAACACCCACTCCACGCCATTCA |
| MOL 166 | AATGGCGTGGAGTGGGTGTTGGGCTGCTAGTT | |
| pMA216 | MOL 167 | AATTCCAACTAGCATATACGTGCCCACTCCACGCCATTCA |
| MOL 168 | AATGGCGTGGAGTGGGCACGTATATGCTAGTTGG | |
| pMA217 | MOL 169 | AATTCCAACTAGCAGCCCAACATATGTCATACGCCATTCA |
| MOL 170 | AATGGCGTATGACATATGTTGGGCTGCTAGTTGG | |
| pMA218 | MOL 171 | AATTCCAACTAGCAGCCCAACACCCACTCCGATATCCGCA |
| MOL 172 | CGGATATCGGAGTGGGTGTTGGGCTGCTAGTTGG | |
| pMA229 | MOL 198 | TGCATGCGGGCGGGCTGCACTTTTTTGCCGCCGAGGGC |
| MOL 199 | GCCCTCGGCGGCAAAAAAGTGCAGCCCGCCCGCATGCATG | |
| pMA230 | MOL 200 | CTTGCATTTAATTAATGCACTTTTTTGCCGCCGAGGGC |
| MOL 201 | GCCCTCGGCGGCAAAAAAGTGCATTAATTAAATGCAAGTG | |
| pMA231 | MOL 202 | CTTGCATGGGCGGGCGTAGAGCGGTTGCCGCCGAGGGC |
| MOL 203 | GCCCTCGGCGGCAACCGCTCTACGCCCGCCCATGCAAGTG | |
| pMA232 | MOL 204 | CTTGCATGGGCGGGCTGCACTTTTCGTTTAAACAGGGC |
| MOL 205 | GCCCTGTTTAAACGAAAAGTGCAGCCCGCCCATGCAAGTG |
Plasmids.
The sequences of oligonucleotides used for plasmid construction are given in Table 1. Plasmid pMA30 was constructed by annealing oligonucleotides NOL 11 and NOL 12 and ligation into XbaI/ApaI-digested plasmid pON4074 (21). Plasmid pMA51 was constructed by annealing oligonucleotides NOL 13 and NOL 14 and ligation into XbaI/NcoI-digested pON4077 (21). For BAC-derived virus construction, ectopic cleavage sites were engineered in pFastbac1-based shuttle plasmids for subsequent transposition into pSM3-117K. MCMV cleavage/packaging sequences were initially derived from a 1.9-kb fusion of MCMV termini cloned as a HindIII/EcoRI fragment in pON4048 (21). Relevant restriction sites used for plasmid construction are shown below in Fig. 4. Plasmid pMA100 was constructed by excision of the 1.9-kb HindIII/EcoRI fragment from pON4048 and ligation into HindIII/EcoRI-restricted pFastbac1 (Invitrogen). Plasmid pMA101 was similarly constructed by excision of a 1.5-kb XhoI/HindIII fragment from pON4048 and ligation into XhoI/HindIII-restricted pFastbac1. Plasmid pMA111 was constructed by restricting pMA101 with NdeI/XhoI, blunt ending the resulting ends with T4 DNA polymerase with all four deoxynucleoside triphosphates, and blunt end ligation. Plasmids pMA170, pMA174, pMA175, pMA187, and pMA189 were similarly derived from pMA111 by double digestion with PpuMI/RsrII, BtrI/HindIII, XmaI/HindIII, PstI/NotI, and SpeI/BsgI, respectively; plasmid pMA177 was derived by HindIII/XbaI digestion of pMA170, plasmid pMA183 was derived by SpeI/BsgI digestion of pMA174, and plasmids pMA190 and pMA200 were derived by XbaI/HindIII digestion of pMA189 and pMA187, respectively. Plasmid pMA197 was constructed by EcoRI/KpnI digestion of a PCR product produced from amplification of pMA174 DNA using primers MOL160 and MOL162 and ligation into EcoRI/KpnI-restricted pFastbac1. Plasmid pMA220 was constructed by annealing oligonucleotides MOL173 and MOL174 and ligation into XbaI/KpnI-restricted pMA197. Plasmids pMA215, pMA216, pMA217, and pMA218 were constructed by annealing oligonucleotide pairs MOL165/MOL166, MOL167/MOL168, MOL169/MOL170, and MOL171/MOL172, respectively, and ligation into EcoRI/BsgI-restricted pMA197. Plasmids pMA229, pMA230, pMA231, and pMA232 were constructed by annealing oligonucleotide pairs MOL198/MOL199, MOL200/MOL201, MOL202/MOL203, and MOL204/MOL205, respectively, and ligation into SfoI/BsgI-restricted pMA215.
FIG. 4.
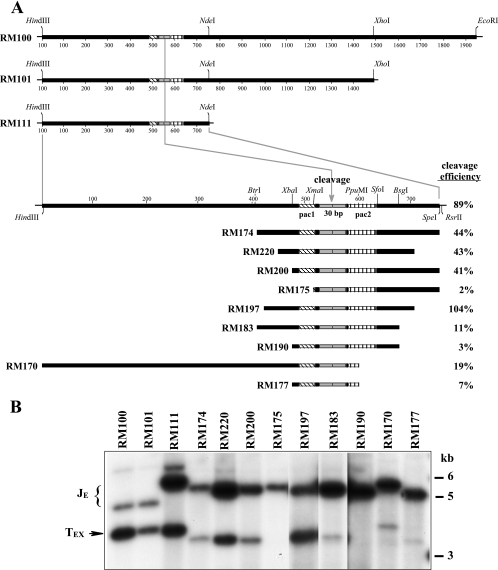
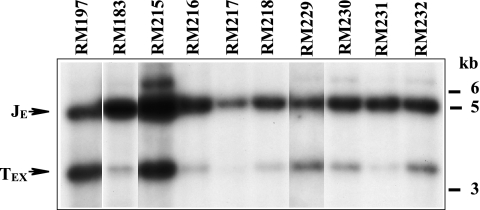
Deletional analysis of ectopic cleavage sites in BAC-derived viruses. (A) Sequences comprising the ectopic cleavage sites in each of the viruses analyzed are illustrated along with their corresponding cleavage efficiencies. Diagonally hatched boxes represent pac1, gray boxes represent the 30-bp terminal repeats, and vertically hatched boxes represent sequences from pac2 to the CGCGGCG motif. The restriction sites shown were used in construction of certain ectopic site mutations/deletions. Note that although two 30-bp repeats are shown, MCMV cleavage sites can contain either one or two copies of the 30-bp repeat (16); the proportions of single and double 30-bp repeats at the various ectopic cleavage sites described here have not been determined. (B) DNAs extracted from extracellular virions produced by the indicated viruses were digested with XhoI, separated by agarose electrophoresis, transferred to a nylon membrane, and hybridized with sequences cloned in plasmid pMA34. The positions of molecular mass markers are shown on the right. Ectopic junction (JE) and ectopic terminal (TEX) fragments are indicated. Densitometric measurements of the signal intensities for JE and TEX were used to calculate ectopic cleavage efficiencies for each virus, as indicated in panel A (see Materials and Methods for details).
Construction of recombinant viruses by homologous recombination.
Viruses RMA30 and RMA51 were constructed by homologous recombination using plasmids pMA30 and pMA51, respectively, gpt selection, and limiting dilution purification as described previously (21).
Reconstitution of BAC-derived viruses.
Viruses were reconstituted by transfection of 2 to 4 μg of BAC DNA into murine NIH 3T3 cells (ATCC CRL 1658) in six-well plates as described previously (11). The correct sequence for each ectopic site was confirmed by Sanger dideoxy sequencing of DNA isolated from virions as previously described (21). Viruses were named using the prefix RM followed by the plasmid number corresponding to the ectopic cleavage site that the virus contains (e.g., RM174 contains the same ectopic cleavage site sequences as plasmid pMA174).
Analysis of ectopic cleavage site function.
Cells were infected at a multiplicity of infection (MOI) of ∼0.1. Ten days after infection, virion DNAs were purified from extracellular viral particles, restricted with XhoI, separated by agarose electrophoresis, transferred to nylon membranes, and hybridized using a 32P-labeled probe consisting of gel-purified viral sequences derived from plasmids pMA34 or pON4048 as described previously (21). Autoradiographs of hybridized membranes were scanned as transparencies using an Epson Expressions 1680 scanner equipped with a transparency adapter. Fragment intensities were quantitated as follows using NIH Image software. For each lane containing data for a given virus, a box was drawn to include the JE fragment. The total image density within the box was then recorded. The box was then shifted to the blank region between JE and TEX, and the density in this region was recorded as the background for data in this lane. Finally, the box was shifted to contain TEX, and a third measurement was taken. Percent cleavage efficiencies were calculated using the following formula: [(TEX − background)/(JE − background)] − 100. For the experiment in Fig. 3, below, TE signals were first corrected by subtraction of the JE signals that remained after double digestion.
FIG. 3.
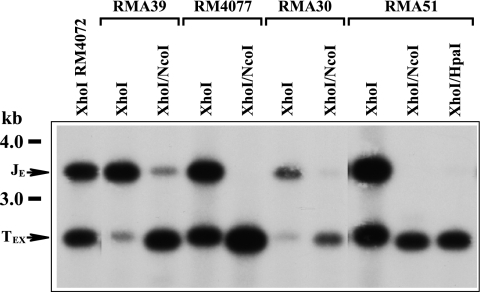
Cleavage at ectopic sites containing substitutions in pac1. DNAs extracted from extracellular virions produced by the indicated viruses were digested with XhoI alone or double digested with XhoI/NcoI or XhoI/HpaI, separated by agarose electrophoresis, transferred to a nylon membrane, and hybridized with sequences cloned in plasmid pMA34 (Fig. 1). The positions of molecular mass markers are indicated to the left. Arrows identify ectopic junction (JE) and ectopic terminal (TEX) fragments.
FIGE.
Cells were infected with recombinant viruses at an MOI of 0.1 and incubated for 4 days. The culture medium was then removed and clarified of cell debris by two rounds of low-speed centrifugation (800 × g for 5 min). Virion particles were pelleted from the supernatants by ultracentrifugation at 20,000 × g for 30 min. Infected cells were washed with phosphate-buffered saline-EDTA, trypsinized, resuspended in 10 ml phosphate-buffered saline-EDTA, and pelleted by low-speed centrifugation (800 × g for 5 min). Virion or cell pellets were resuspended in 50 μl molten (52°C) 1% SeaPlaque agarose (FMC) in Tris-EDTA, cast into molds, and cooled on ice. The solidified agarose plugs were suspended in SE (0.5 M EDTA, 1% Sarkosyl) with 1 mg/ml proteinase K, incubated at 52°C for 48 h, dialyzed three times for 2 h with Tris-EDTA, and stored at 4°C. Agarose plugs were inserted into the loading wells of 11- by 14-cm 1% Seakem agarose (FMC) gels and separated by field inversion gel electrophoresis (FIGE) in 0.5× Tris-borate electrophoresis buffer for 36 h at 120 V and 14°C. The pulse times began at 5 s and increased to 60 s with a 3:1 forward-to-backward ratio. DNA species were visualized by ethidium bromide staining and UV light and then transferred to Nytran nylon membranes (Schleicher & Schuell) and hybridized to specifically detect MCMV DNA using 32P-labeled DNA from plasmid pON4048 as previously described (19).
RESULTS
Site-directed mutations in pac1.
Herpesvirus pac1 elements are comprised of short A- or T-rich regions flanked on the left and right by poly(C) tracts (21). We previously found that in MCMV the left poly(C) tract was critically important for cleavage site function while, curiously, the sequence of the central A-rich region was not (21). We were concerned, however, that a second “distal A-rich” tract just to the left of pac1 (Fig. 2A) could be functionally redundant with the central A-rich region such that cleavage might only be affected when both A-rich regions were simultaneously mutated. We also wished to extend the mutagenesis of pac1 by evaluating a substitution mutant in the right poly(C) tract. To address these questions, two additional viruses were constructed as described previously (21). In virus RMA30, the right poly(C) tract within pac1 was mutated to introduce an NcoI site, whereas in virus RMA51 two mutations were introduced: one identical to the mutation previously evaluated in virus RM4077 (21) targeting the central A-rich region and creating an NcoI site, and the second targeting the distal A-rich region and creating a HpaI site (Fig. 2A). Both mutations were engineered within the context of the 1.9-kb ectopic site and inserted in direct orientation relative to the native terminal sequences, as illustrated in Fig. 1. Ectopic cleavage efficiencies for RMA30 and RMA51 were measured as described before (21), and previously analyzed viruses with a wild-type ectopic cleavage site (RM4072), a pac1 left poly(C) mutation (RMA39), and a central A-rich mutation (RM4077) (21) were included for comparison (the sequences of these mutations are included in Fig. 2A). Briefly, extracellular virion DNAs from each virus were digested with XhoI or double digested with XhoI/NcoI or XhoI/HpaI, separated by agarose gel electrophoresis, and hybridized with probe pMA34, which hybridizes with equal efficiency to JE or TEX fragments (Fig. 1).
FIG. 2.
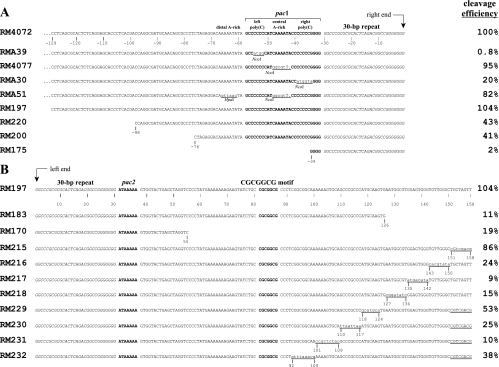
Sequences of ectopic cleavage sites. Sequences close to the point of cleavage are shown to illustrate relevant nucleotide differences between the ectopic cleavage sites of different viruses. Corresponding cleavage site efficiencies are given to the right of each sequence. (A) Substitutions and nested deletions on the pac1 side of the cleavage site with predicted right ends aligned to the right. (B) Nested deletions and scanning substitutions on the pac2 side of the cleavage site with predicted left ends aligned to the left. Known functional cis elements (pac1, pac2, and the CGCGGCG motif) are in bold and set off by spaces; 30-bp terminal repeats are in italics. Nucleotide positions are numbered negatively to the left of right-end termini and positively to the right of left-end termini. Substitution mutations are underlined, nucleotide changes are in lower case, and a dot indicates a missing base. Restriction sites created by mutations are indicated. Note that although a single 30-bp terminal repeat is shown at each terminus, pac2-containing termini sometimes lack a 30-bp repeat (16); the disposition of 30-bp repeats at ectopic termini described here has not been determined.
To quantitate ectopic cleavage site efficiencies, the signal intensities of JE and TE fragments were measured densitometrically. However, as previously noted (21), RMA39 exhibited a small amount of recombinational repair, as evidenced by residual JE signal in the XhoI/NcoI double digestion (Fig. 3). This reflects JE fragments that have lost the NcoI site introduced by the mutation. A similar amount of repair was also detected in RMA30 but not in RM4077 or RMA51 (Fig. 3). As a portion of TE fragments presumably arise from cleavage of repaired JE junctions, the TE signals for RMA39 and RMA30 were corrected by subtraction of the JE signals measured in the double digestions. The cleavage efficiencies of ectopic sites were then calculated as the TE/JE ratios for each virus. They are listed in Fig. 2A.
The results for RM4072, RMA39, and RM4077 were essentially as reported before (21). The wild-type ectopic site in RM4072 was cleaved efficiently, giving rise to equimolar amounts of JE and TEX (Fig. 3). The 0.8% cleavage efficiency measured for the left poly(C) mutation in RMA39 again confirmed the critical importance of this cis element. The 20% efficiency of the right poly(C) mutation in RMA30 was not so profound but still indicated an important role for this region. Interestingly, the efficiency of the double A-rich mutation in RMA51 (83%) was not significantly different from that of the matching single mutation in RM4077 (95%), clearly demonstrating that neither the central nor the distal A-rich regions are important for cleavage site function.
Ectopic cleavage in BAC-derived viruses.
The viruses described above were constructed by homologous recombination between plasmid and viral sequences followed by gpt selection, limiting dilution cloning, and screening for loss of lacZ (21). With the advent of infectious BAC clones of herpesvirus genomes (23), it became feasible to engineer viral genomes in Escherichia coli and reconstitute recombinant viruses by transfection of BAC DNA into appropriate mammalian host cells. We therefore sought to adapt a rapid method for introducing foreign sequences into BACs via site-specific Tn7-mediated transposition. Allelic exchange was used to modify the pSM3 infectious BAC clone of the MCMV genome (23) to contain a lacZα-mini-attTn7 site at the junction between HindIII L and J fragments, giving rise to BAC pSM3-117K (11). Because transposition occurs in a fixed orientation, care was taken to ensure that all ectopic sites were cloned into shuttle plasmids in the orientation that would, upon transposition, give rise to ectopic sites that are in direct orientation relative to sequences at the genomic termini, as shown in Fig. 1. Ectopic sites transposed into pSM3-117K would thus be in the same orientation and location in the genome as viruses that were previously constructed by homologous recombination.
However, by necessity the sequences that flank transposed ectopic sites differ from those that flank sites constructed by homologous recombination, raising the concern that the altered context might impact the functionality of ectopic cleavage sites. To address this concern, we cloned the same 1.9-kb ectopic site found in RM4072 into a transposition shuttle to generate plasmid pMA100 and then transposed this cleavage site into the attTn7 site of pSM3-117K and transfected the resulting BAC DNA into mouse NIH 3T3 cells to reconstitute recombinant virus RM100. Extracellular virion DNA from RM100 was analyzed for ectopic cleavage using the same assay as before. Surprisingly, the TEX signal intensity for RM100 was nearly threefold higher than that of JE (Fig. 4B), suggesting that the ectopic cleavage site in RM100 is about three times more efficient than the natural site. Curiously, this enhanced efficiency was subsequently lost as flanking sequences were progressively deleted (see below). These results suggested that the natural cleavage site of MCMV is not fully optimized and that flanking context can moderately influence cleavage site efficiency. Nevertheless, efficient cleavage of the ectopic site demonstrated the feasibility of the Tn7 transposition system as a platform with which to perform further mutagenic analyses.
Deletional mutagenesis of the ectopic cleavage site.
Nested deletions of the 1.9-kb ectopic site were next constructed and evaluated as described above. Figure 4A shows the relevant restriction sites that were used for constructing deletions and illustrates the ectopic cleavage sites in each virus and the positions of pac1, pac2, and the 30-bp terminal repeats. The precise sequences of selected cleavage sites are shown in Fig. 2. The autoradiographs of XhoI-digested virion DNAs from this series of deletion mutants are shown in Fig. 4B, and the cleavage efficiencies calculated from these results are listed in Fig. 4A and also in Fig. 2.
Removal of sequences to the right of the XhoI site to create the 1.5-kb cleavage site in virus RM101 resulted in a slight decrease in efficiency, but ectopic cleavage was still nearly twice that of natural sites. Deletion of sequences to the right of the NdeI site in virus RM111 decreased efficiency to 89%, and further deletions of sequences to the left of pac1 in viruses RM174, RM220, and RM200 exhibited only modest additional decreases in efficiency (44%, 43%, and 41%, respectively). However, ectopic cleavage was essentially eliminated by deletion to the XmaI site in RM175, which removed virtually all of pac1 (Fig. 2A). Taken together, these results indicate that the 76 bp on the pac1 side of the cleavage site in virus RM200 (Fig. 2A) are sufficient for efficient cleavage.
Additional nested deletions removing sequences from the pac2 side of the cleavage site were also constructed and evaluated for cleavage efficiency. Effects of flanking context were again noted, as removal of flanking sequences caused an increase in efficiency from 44% for RM174 to 104% for RM197 (Fig. 4). However, ectopic cleavage efficiency dropped precipitously as additional sequences were deleted from the pac2 side. In virus RM183, efficiency dropped ninefold to 11%, and further deletions in viruses RM170 and RM177 produced similar efficiencies of 19% and 7%, respectively (Fig. 4). These results indicated that on the pac2 side of the cleavage site a surprisingly large region of sequence extending to 158 bp from the cleavage point is needed for efficient cleavage. Moreover, the profound drop in efficiency between RM197 and RM183 strongly suggested the existence of important cis sequences between nucleotides 126 and 158 (Fig. 2B).
Scanning mutagenesis of nucleotides 92 to 158 distal to pac2.
Using the RM197 ectopic site as a common background, scanning site-directed mutations were constructed spanning the region from nucleotides 92 (just distal to the CGCGGCG motif) to 158 (Fig. 2B) and analyzed by Southern hybridization for cleavage efficiency (Fig. 5). Cleavage efficiency of virus RM215 was slightly reduced relative to RM197 (86% versus 104%), while efficiencies of viruses RM216, RM217, and RM218 were profoundly inefficient, to a level similar to that of RM183. For reasons of technical convenience, viruses RM229 to -232 were constructed in the RM215 background; therefore, their cleavage efficiencies should be evaluated relative to RM215. The additional mutation in RM229 caused only a slight decrease in efficiency relative to RM215 (53% versus 86%). In contrast, RM231 exhibited a substantial drop in efficiency (10%), while the two flanking mutations in RM230 and RM232 exhibited intermediate efficiencies of 25% and 38%, respectively (Fig. 5).
FIG. 5.
Ectopic cleavage site efficiencies of scanning substitution mutants. The indicated viruses were analyzed as described in the legend for Fig. 4B. Note that in addition to scanning mutations, viruses RM229 to RM232 also contain a common substitution of nucleotides 151 to 158 (Fig. 2B) that in virus RM215 has only a modest impact on efficiency.
The poor cleavage efficiencies of viruses with mutations spanning positions 127 to 150 (RM216, RM217, and RM218; 24%, 9%, and 15%, respectively) contrasted with the relatively unimpaired cleavage efficiencies of viruses with mutations that flank this region (RM215 and RM229; 86% and 53%, respectively) (Fig. 2B), suggesting the existence of a novel cis element between nucleotides 127 and 150. Similarly, the low efficiencies of viruses with mutations spanning positions 101 to 117 (RM230 and RM231; 10% and 25%, respectively) contrasted with the somewhat more efficient cleavage of flanking mutations in viruses RM229 and RM232 (53% and 38%, respectively) (Fig. 2B), further suggesting the possibility of another cis element in the bp 101 to 117 region. However, the current results do not clearly differentiate these regions from each other or from the CGCGGCG motif. Thus, it remains uncertain whether the region from nucleotides 81 to 150 represents a single “pac2-distal” cis element or contains multiple cis elements that are spatially and perhaps functionally distinct.
Detection of termini and internal junctions in BAC-derived viruses.
An underlying premise of the ectopic cleavage site system is that viruses with ectopic cleavage sites will form two genome types. Those cleaved at natural sites will have natural termini and will contain uncleaved ectopic junctions, whereas those cleaved at ectopic sites will have ectopic termini and will contain uncleaved natural junctions. In support of this we previously showed that RM4072 virion DNA contains a mixture of ectopic and natural termini as well as uncleaved ectopic and natural junctions (21). However, given that the context of the ectopic site is somewhat different in BAC-derived viruses and that the mode of virus construction is very different, it was important to confirm that a BAC-derived virus with a functional ectopic site also forms the two predicted genome types.
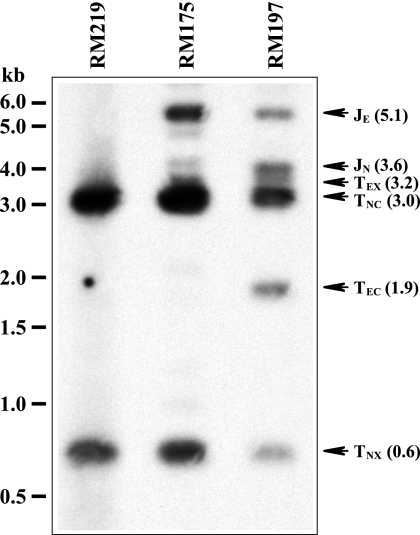
Virus RM197 was selected as a representative virus containing an efficiently cleaved ectopic site, and virus RM175 was selected as one in which the ectopic site is cleaved with extremely low efficiency. A third virus, RM219, contains irrelevant sequences transposed into the attTn7 site and hence serves as a negative control that lacks an ectopic site (J. B. Wang and M. McVoy, unpublished data). Extracellular virion DNAs were digested with XhoI and hybridized with a probe consisting of the 1.9-kb fusion of genomic termini cloned in pON4048, which contains sequences from both ends of the viral genome and hence was expected to hybridize to all six potential fragments (Fig. 1). The assembled sequence of RM197 was used to calculate the sizes predicted for XhoI fragments derived from the natural termini TNX (0.6 kb) and TNC (3.0 kb), the ectopic termini TEX (3.2 kb) and TEC (1.9 kb), and the ectopic and natural junctions JE (5.1 kb) and JN (3.6 kb). In the RM219 control virus, the probe detected the expected natural terminal fragments TNX and TNC, and their sizes exactly matched the sequence-predicted sizes (Fig. 6). JN was neither detected nor expected in RM219, because this virus lacks an ectopic cleavage site and hence all natural junctions should be cleaved into termini. JE was also neither detected nor expected, as the transposed sequences in RM219 lack homology to the probe. RM197 contained the same natural terminal fragments TNX and TNC but also contained the ectopic terminal fragments TEX and TEC. Their sizes again exactly matched the sequence-based predictions (Fig. 6). Moreover, both junction fragments, JN and JE, were clearly present and, again, matched the sequence-predicted sizes (Fig. 6).
FIG. 6.
Detection of natural termini and junctions. Virion DNAs from the indicated viruses were digested with XhoI, separated by agarose electrophoresis, transferred to a nylon membrane, and hybridized with sequences from the left and right ends of the MCMV genome cloned in plasmid pON4048 (Fig. 1). Arrows indicate the positions of relevant fragments. Their sequence-predicted sizes (in kb) are shown in parentheses.
RM175 DNA contained the natural terminal fragments and an ectopic junction (JE) but clearly lacked the 1.9-kb TEC ectopic terminal fragment, consistent with little or no cleavage at its ectopic site (Fig. 6). Curiously, several additional unexplained faint fragments were detected. One of these migrated with a molecular weight very similar to the 3.2-kb ectopic terminal fragment TEX (Fig. 6), but given that not even a trace amount of TEX was detected in RM175 DNA using the pMA34 probe (Fig. 4B), this fragment is unlikely to be TEX. A second faint fragment was similar in size to the natural junction JN but on close inspection migrated slightly slower (Fig. 6), again suggesting that it is not the genuine natural junction. At least four other faint fragments of ∼1.0, 1.2, 2.1, and 4.9 kb were also unexplained but unique to RM175 (Fig. 6). Thus, the ectopic site of RM175 may undergo aberrant cleavages or rearrangements at very low levels.
The results demonstrate that all four terminal fragments and both junctions are present in RM197 DNA. This finding supports the premise that when a functional ectopic cleavage site is present, cleavage occurs at both natural and ectopic cleavage sites and that cleavage at ectopic sites generates genomes in which natural junctions remain uncleaved.
Accuracy and stability of ectopic site-containing viruses.
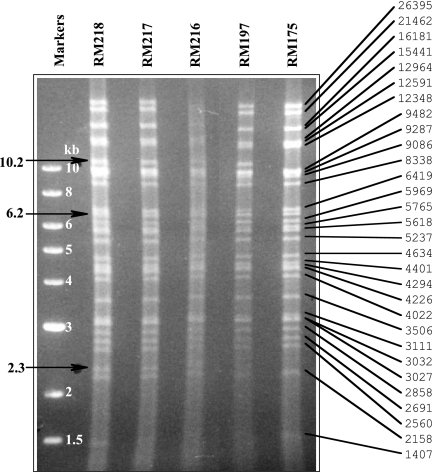
The faint unexplained fragments detected in RM175 raised concerns that the presence of duplicated sequences at the ectopic site might promote unexpected changes in genome structure that could potentially impact the ectopic cleavage assay. We therefore examined the restriction patterns of virion DNAs from a panel of viruses that exhibited ectopic cleavage that was inefficient (RM175, RM217, and RM218), moderately efficient (RM216), or highly efficient (RM197). Virion DNAs were double digested with HindIII and NheI, and the resulting fragments were separated electrophoretically and visualized with UV light and ethidium bromide staining. With the exception of two fragments of 2.3 and 10.2 kb that were not present in RM175 but were present in the other four viruses, the restriction patterns for all five viruses appear to be identical (Fig. 7). These two fragments result from inclusion of a HindIII site within the ectopic sites of viruses RM197 and RM216 to -218 that is not present in the ectopic site of RM175 (Fig. 4A). Hence, in RM175 a 12.5-kb fragment results that cannot be easily distinguished from 12.3- and 12.9-kb fragments that derive from elsewhere in the genome (Fig. 7). The observed pattern differences are therefore entirely consistent with expected differences in the viral genome sequences. Moreover, with the exception of a 6.2-kb fragment, the observed restriction fragments correlated exactly with the fragment sizes predicted from sequence files (Fig. 7). That this fragment was common to all five viruses suggests that the discrepancy likely arises from inaccuracies in the sequence file, which indeed lacks small regions that have not been sequenced at both ends of the BAC origin. The results indicate that genomes of viruses containing ectopic cleavage sites are essentially as predicted by the assembled sequence data and are not unstable or prone to rearrangements.
FIG. 7.
Restriction pattern analysis of selected viruses. Virion DNAs from the indicated viruses were double digested with HindIII/NheI, separated by agarose electrophoresis, and stained with ethidium bromide. On the right, the observed restriction fragments from RM175 are correlated with fragment sizes predicted from an assembled sequence file. Arrows on the left indicate fragments of interest that are discussed in the text.
FIGE analysis of viral DNA species.
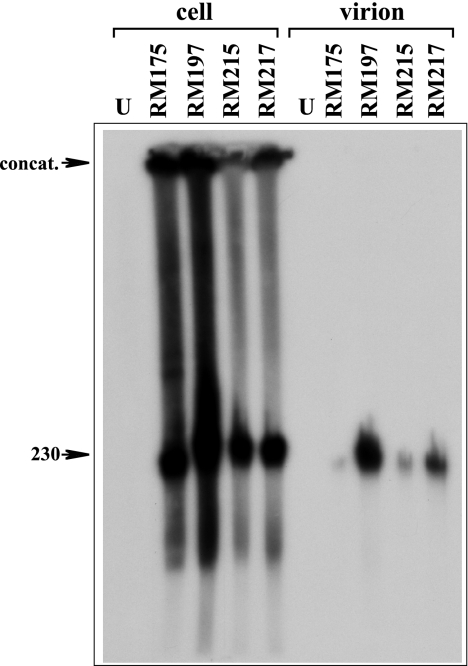
While results described above confirmed that not all natural or ectopic cleavage sites are cleaved, the possibility remained that some proportion of cleavage events might occur at adjacent sites within concatemers, giving rise to subgenomic DNA species of 191 and 45 kb. Packaging of such subgenomic DNAs within extracellular virions could potentially compromise the validity of our cleavage site assay, as terminal fragments on subgenomic DNAs would contribute to the observed signals but would not represent authentic genome cleavage/packaging events. To address this concern, the viral DNA species found in infected cells and packaged in extracellular virions were evaluated by FIGE. Four representative viruses were selected: two containing ectopic sites that are efficiently cleaved (RM197 and RM215) and two containing ectopic sites that are nonfunctional due to mutations on either side of the point of cleavage (RM175 and RM217).
Cells were infected with the four viruses and incubated for 4 days. DNA was prepared for FIGE from both the infected cells and from virions pelleted from the culture supernatants. The DNA species were separated by FIGE, transferred to a nylon membrane, and hybridized with probe pON4048, which contains both X and C sequences and hence should detect both of the postulated subgenomic fragments. The infected cell samples contained similar amounts of concatemeric DNA and genome-length 230-kb DNA (Fig. 8). Lower-molecular-weight smears of viral DNA were detected in samples from all four viruses and appeared similar to those observed previously for human cytomegalovirus (HCMV) (18) and guinea pig cytomegalovirus (24). Within these smears, no distinct 191- or 45-kb species could be distinguished, nor were differences evident between viruses containing functional or nonfunctional cleavage sites. Thus, if subgenomic species are selectively formed within cells infected with viruses containing functional ectopic sites, they do not accumulate to discernible levels.
FIG. 8.
FIGE analysis of intra- and extracellular viral DNA species. Replicate cell cultures were uninfected (U) or infected with the indicated viruses at an MOI of 0.1 and incubated for 4 days. DNAs prepared from infected cells (left) or from extracellular virions (right) were separated by FIGE, transferred to a nylon membrane, and hybridized with pON4048 to detect only viral DNA species. Arrows indicate the positions of unit-length genomes (230) and concatemeric viral DNA (concat.).
Virion samples from all four viruses contained only genome-length 230-kb viral DNA. The amounts varied significantly, but we believe this is likely due to sample loss during virion preparation (we have not observed any differences in replication for any of the viruses used in this study). No subgenomic DNA species were detected, even in the viruses that contain functional cleavage sites, RM197 and RM215 (Fig. 8). We therefore conclude that intracellular subgenomic DNA species, if they are formed at all, are not packaged into extracellular virions.
DISCUSSION
In our previous study we demonstrated that a 1.9-kb ectopic cleavage site functions with an efficiency approximately equal to that of the natural cleavage site. Substantial declines in cleavage site efficiencies caused by site-directed mutations introduced within the 1.9-kb sequence confirmed the importance of pac1, pac2, and the CGCGGCG motif for genome cleavage and packaging (21). In the present study two additional mutations within pac1 were evaluated, and the importance of sequences distal to pac1 and pac2 were assessed by deletional and substitution mutagenesis. The results revealed that sequences distal to pac1 are dispensable. Within pac1, both poly(C) tracts are important for cleavage site function, yet the intervening poly(A) tract and a similar poly(A) tract to the left of pac1 are unimportant (reference 21 and this study). While the sequences between pac1 and the point of cleavage have not as yet been mutagenized in the MCMV system, studies using herpes simplex virus type 1 (HSV-1) suggest that such proximal sequences are largely dispensable (13, 28). Thus, pac1 may be the sole cis-acting cleavage/packaging signal on the pac1 side of the cleavage site.
In contrast, deletional mutagenesis on the pac2 side of the cleavage site demonstrated an important role for sequences distal of pac2 up to 150 bp from the point of cleavage. Substitution mutations had previously confirmed the critical importance of the pac2 A-rich region (nucleotides 31 to 37) and the CGCGGCG motif (nucleotides 81 to 87), whereas a second A-rich region (nucleotides 60 to 69) was fully dispensable (21). In this study, scanning substitution mutagenesis revealed clear evidence for additional cis-acting sequences between nucleotides 127 and 150 and also in the 92 to 117 region. Thus, the pac2 side of the MCMV cleavage site appears to be complex and may be comprised of several important sequence elements spanning 150 nucleotides. As these sequences presumably interact with multiple proteins or protein complexes, our results should facilitate efforts to further define the protein components involved genome maturation.
Evidence that the cleavage sites of other herpesviruses contain analogous distal packaging sequences is mostly inferential. Zimmermann et al. found that in Epstein-Barr virus (EBV) sequences just distal of the pac2 A-rich motif (nucleotides 36 to 106) (Fig. 9A) are essential for packaging plasmid amplicon DNA (33). In varicella-zoster virus (VZV), an 88-bp duplication of pac2-containing L-terminal sequences renders internal L/S junctions identical to natural cleavage sites up to 88 bp from the point of cleavage (Fig. 9B). Thus, the fact that L/S junctions are inefficiently utilized for genome packaging suggests that sequences of >88 bp from the VZV L terminus are important for genome maturation (5). In HSV-1, substitution mutagenesis showed that the nucleotide sequence of the bp 40 to 77 region (as numbered in Fig. 9C) is not important for packaging plasmid-amplicon DNA but that a deletion removing nucleotides 40 to 70 caused a substantial decrease in packaging efficiency (13). This suggests that nucleotides 40 to 70 may serve to provide proper spacing between the pac2 A-rich motif and a putative distal cis element. By exclusion, this putative element must lie within nucleotides 78 to 101, because the sequence of nucleotides 40 to 77 is not important and sequences beyond position 101 are dispensable (13).
FIG. 9.
pac2-containing terminal sequences from other herpesviruses. (A) EBV (33). (B) The L-terminal sequences of VZV are aligned with sequences from the VZV L/S junction to illustrate the 88-bp region of identity (5). (C) ClustalW was used to align the pac2-containing terminal sequences from alphaherpesviruses, including HSV-1 (6), HSV-2 (6), bovine herpesvirus 1 (BoHV-1) (12), equine herpesvirus 1 (EHV-1) (4), VZV (5), and pseudorabies virus (PRV) (9). (D) ClustalW alignment of pac2-containing terminal sequences from cytomegaloviruses, including HCMV (strain Merlin) (10), rat cytomegalovirus (RCMV) (31), guinea pig CMV (GPCMV) (20), and MCMV (16). In all panels, genome ends are to the left and proposed pac2 A-rich and CGCGGCG motifs (7, 16, 20-22, 26, 31, 33) are shown in bold.
In silico analyses failed to identify sequence homologies to MCMV nucleotides 92 to 150 near the pac2 termini of other herpesviruses (not shown); however, alignment of pac2 terminal sequences from several alphaherpesviruses revealed a conserved G/C-rich block spanning nucleotides 75 to 93 (Fig. 9C). This block coincides with the nucleotide 78 to 101 region of HSV-1 discussed above, and at VZV L/S junctions it is bisected by the breakpoint of the 88-bp duplication (Fig. 9B). However, the distal cis elements of MCMV, located either 92 to 150 bp from the termini of genomes that have one copy of the 30-bp repeat or 97 to 120 bp from the termini of those that lack the 30-bp repeat (16), are more distal than the G/C-rich block, or indeed, than the distal limits of sequences that are necessary for amplicon packaging by either EBV (106 bp) (33) or HSV-1 (101 bp) (13). Thus, although cumulatively the above findings make a strong case for the existence of distal cleavage/packaging sequences in other herpesviruses, it remains uncertain whether such sequences are functionally analogous to those that we have identified in MCMV.
Surprisingly, alignments of pac2 terminal sequences from different cytomegaloviruses did not reveal any clustering of G/C richness or sequence conservation in regions distal to pac2 (Fig. 9D). Moreover, HCMV lacks the typical pac2 A-rich motif (Fig. 9D) and thus the sequences comprising pac2 in HCMV remain an enigma (22). These observations suggest that divergence of cis sequences involved in DNA packaging on the pac2 side of the cleavage site has been more pronounced among the cytomegaloviruses than the alphaherpesviruses. Additional mutagenic studies using other viral systems will be required to determine the importance of the alphaherpesvirus-conserved G/C-rich regions as well as the existence of distal cis elements in cytomegaloviruses other than MCMV.
The functional roles of individual cis elements comprising herpesvirus cleavage sites are poorly understood. As our studies examined the final product—extracellular virions—they reveal only that both pac1 and pac2 as well as distal sequences associated with pac2 are essential for the complete process. However, other studies suggest that pac2 or adjacent sequences may direct the initiation of DNA packaging, perhaps by binding proteins that mediate docking to procapsid portals, and that pac1 and pac2 independently govern formation of their respective termini (13, 22, 26, 30). In addition, terminal sequences have been implicated as having a role in the duplication of terminal repeats (21) and in postpackaging capsid maturation events, such as the sealing or stabilization of DNA-containing capsids or their targeting for nuclear egress (13, 24). Indeed, using an HSV-1 amplicon system, Hodge et al. found that mutation of pac1 sequences did not prevent packaging of DNA into intracellular capsids but rather gave rise to packaged amplicon genomes that appeared normal at their Uc (pac2) ends but lacked Ub (pac1) ends. That these genomes were inefficiently propagated upon serial passage suggested that pac1 sequences may play a role in a post-DNA-packaging step of virion maturation (13). In our analysis of extracellular genomes formed by virus RM175, which lacks pac1 at its ectopic site, we failed to detect either TEX (pac1) or TEC (pac2) termini (Fig. 4 and 6). Thus, if such termini are formed on intracellular DNA species, they are tightly restricted from maturing into the extracellular virion population. Future studies examining packaged and unpackaged intracellular viral DNA species formed by our mutants should help to resolve this question and may yield additional clues to the individual functions of specific cis-acting sequences.
Acknowledgments
We are grateful to Martin Messerle for the kind gift of bacmid pSM3.
This work was supported by Public Health Service grants R01AI46668 and R21AI43527 (to M.A.M.) and KO8AI01435 (to D.E.N.).
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Bataille, D., and A. Epstein. 1994. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology 203384-388. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Porat, T. 1983. Replication of herpesvirus DNA, p. 81-106. In B. Roizman (ed.), The herpesviruses. Plenum Press, New York, NY.
- 3.Brown, J. C., M. A. McVoy, and F. L. Homa. 2002. Packaging DNA into herpesvirus capsids, p. 111-153. In E. Bogner and A. Holzenburg (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum Publishers, London, England.
- 4.Chowdhury, S. I., H. J. Buhk, H. Ludwig, and W. Hammerschmidt. 1990. Genomic termini of equine herpesvirus 1. J. Virol. 64873-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davison, A. J. 1984. Structure of the genome termini of varicella-zoster virus. J. Gen. Virol. 651969-1977. [DOI] [PubMed] [Google Scholar]
- 6.Davison, A. J., and N. M. Wilkie. 1981. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J. Gen. Virol. 55315-331. [DOI] [PubMed] [Google Scholar]
- 7.Deiss, L. P., J. Chou, and N. Frenkel. 1986. Functional domains within the a sequence involved in the cleavage packaging of herpes simplex virus DNA. J. Virol. 59605-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deiss, L. P., and N. Frenkel. 1986. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J. Virol. 57933-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeMarchi, J. M., Z. Q. Lu, G. Rall, S. Kupershmidt, and T. Ben-Porat. 1990. Structural organization of the termini of the L and S components of the genome of pseudorabies virus. J. Virol. 644968-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolan, A., C. Cunningham, R. D. Hector, A. F. Hassan-Walker, L. Lee, C. Addison, D. J. Dargan, D. J. McGeoch, D. Gatherer, V. C. Emery, P. D. Griffiths, C. Sinzger, B. P. McSharry, G. W. Wilkinson, and A. J. Davison. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 851301-1312. [DOI] [PubMed] [Google Scholar]
- 11.Hahn, G., M. Jarosch, J. B. Wang, C. Berbes, and M. A. McVoy. 2003. Tn7-mediated introduction of DNA sequences into bacmid-cloned cytomegalovirus genomes for rapid recombinant virus construction. J. Virol. Methods 107185-194. [DOI] [PubMed] [Google Scholar]
- 12.Hammerschmidt, W., H. Ludwig, and H. J. Buhk. 1986. Short repeats cause heterogeneity at genomic terminus of bovine herpesvirus 1. J. Virol. 5843-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodge, P. D., and N. D. Stow. 2001. Effects of mutations within the herpes simplex virus type 1 DNA encapsidation signal on packaging efficiency. J. Virol. 758977-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob, R. J., L. S. Morse, and B. Roizman. 1979. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J. Virol. 29448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marks, J. R., and D. H. Spector. 1984. Fusion of the termini of the murine cytomegalovirus genome after infection. J. Virol. 5224-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marks, J. R., and D. H. Spector. 1988. Replication of the murine cytomegalovirus genome: structure and role of the termini in the generation and cleavage of concatenates. Virology 16298-107. [DOI] [PubMed] [Google Scholar]
- 17.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 702075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McVoy, M. A., and S. P. Adler. 1994. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J. Virol. 681040-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McVoy, M. A., and D. E. Nixon. 2005. Impact of 2-bromo-5,6-dichloro-1-beta-D-ribofuranosyl benzimidazole riboside and inhibitors of DNA, RNA, and protein synthesis on human cytomegalovirus genome maturation. J. Virol. 7911115-11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McVoy, M. A., D. E. Nixon, and S. P. Adler. 1997. Circularization and cleavage of guinea pig cytomegalovirus genomes. J. Virol. 714209-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McVoy, M. A., D. E. Nixon, S. P. Adler, and E. S. Mocarski. 1998. Sequences within the herpesvirus-conserved pac1 and pac2 motifs are required for cleavage and packaging of the murine cytomegalovirus genome. J. Virol. 7248-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McVoy, M. A., D. E. Nixon, J. K. Hur, and S. P. Adler. 2000. The ends on herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences. J. Virol. 741587-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 9414759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nixon, D. E., and M. A. McVoy. 2004. Dramatic effects of 2-bromo-5,6-dichloro-1-beta-d-ribofuranosyl benzimidazole riboside on the genome structure, packaging, and egress of guinea pig cytomegalovirus. J. Virol. 781623-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 1048-1066. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, and B. Roizman (ed.), Fundamental virology. Raven Press, New York, NY.
- 26.Schynts, F., M. A. McVoy, F. Meurens, B. Detry, A. L. Epstein, and E. Thiry. 2003. The structures of bovine herpesvirus 1 virion and concatemeric DNA: implications for cleavage and packaging of herpesvirus genomes. Virology 314326-335. [DOI] [PubMed] [Google Scholar]
- 27.Severini, A., A. R. Morgan, D. R. Tovell, and D. L. Tyrrell. 1994. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology 200428-435. [DOI] [PubMed] [Google Scholar]
- 28.Smiley, J. R., J. Duncan, and M. Howes. 1990. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J. Virol. 645036-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamashiro, J. C., D. Filpula, T. Friedmann, and D. H. Spector. 1984. Structure of the heterogeneous L-S junction region of human cytomegalovirus strain AD169 DNA. J. Virol. 52541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varmuza, S. L., and J. R. Smiley. 1985. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell 41793-802. [DOI] [PubMed] [Google Scholar]
- 31.Vink, C., E. Beuken, and C. A. Bruggeman. 1996. Structure of the rat cytomegalovirus genome termini. J. Virol. 705221-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, X., S. Efstathiou, and A. Simmons. 1994. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology 202530-539. [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann, J., and W. Hammerschmidt. 1995. Structure and role of the terminal repeats of Epstein-Barr virus in processing and packaging of virion DNA. J. Virol. 693147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]