Abstract
We have previously reported that immortalized human hepatocytes (IHH) support the generation of infectious hepatitis C virus (HCV) genotype 1a (clone H77). In the present study, we have investigated the growth of HCV genotype 1a (clone H77) through serial passages and accompanying changes in IHH in response to infection. Eleven serial passages of HCV genotype 1a (clone H77) in IHH were completed. Virus replication was ascertained from the presence of HCV-specific sequences, the detection of core antigen, the virus genome copy number, and the virus titer in IHH culture fluid. Electron microscopy suggested that HCV infection induces autophagic vacuole formation in IHH. Fluorescence microscopy displayed localization of autophagic markers, microtubule-associated protein-1 light chain-3 and Apg5, on the vacuoles of HCV-infected hepatocytes. Taken together, our results suggested that HCV genotype 1a (clone H77) can be serially passaged in IHH and that HCV infection induces an autophagic response in hepatocytes.
Hepatitis C virus (HCV) is primarily hepatotropic and is classified within the Flaviviridae family. HCV infection causes a spectrum of diseases ranging from an asymptomatic carrier state to end-stage liver disease, which includes cirrhosis and hepatocellular carcinoma (11, 14, 19). The current therapies for chronic HCV infection do not often result in viral clearance. The chimpanzee is the only animal model which has been used extensively to study HCV infection (46); however, spontaneous viral clearance occurs in some chimpanzees following inoculation of HCV (2). A small chimeric animal model using HCV-infected human liver transplanted into immune-deficient mice has been developed (17, 32). However, this experimental animal model has limitations due to a short life span. Different groups of investigators, including our group, have reported the generation of infectious HCV (genotypes 1a and 2a) in cell lines of human hepatocyte origin (6, 15, 22, 30, 57, 59, 61). Recently, lipid droplets formed in hepatocytes have been shown to play an important function in the assembly of HCV genotype 2a (34).
Autophagy is often triggered during infection, and several bacteria and viruses have evolved strategies to use components of the autophagic pathway to facilitate their own replication (50). Induction of autophagy has been observed in response to infection by herpes simplex virus (HSV), coronavirus, and poliovirus (18, 25, 44, 49, 52, 54). Infection of human cells with poliovirus induces the proliferation of double-membrane cytoplasmic vesicles whose surfaces are used as sites for viral RNA replication and whose origin is unknown (18, 58). The double-membrane structures provide membranous supports for viral RNA replication complexes, possibly enabling the nonlytic release of cytoplasmic contents (including progeny virions) from infected cells. Components of the cellular machinery of autophagosome formation are proposed to be subverted to promote poliovirus and rhinovirus replication. Severe acute respiratory syndrome-associated coronavirus (SARS-CoV) replicase proteins have been shown to localize to cytoplasmic complexes containing markers for autophagosome membranes (44). Similar rearrangements are seen in cells in response to protein aggregation, where aggresomes and autophagosomes are produced to facilitate protein degradation. A connection between pathways that regulate autophagy and apoptosis exists (8). Several molecules, such as the Bcl-2 family proteins, the Bcl-2-interacting protein Beclin 1, caspase 8, caspase 9, PI3K class I, and the sphingolipid ceramide are involved in the control of both apoptosis and autophagy. Autophagy has also been described as a component of the innate immune response that degrades intracellular pathogens and for endogenous major histocompatibility complex class II processing (10, 20, 28, 40). In the present study, we have shown that HCV genotype 1a (clone H77) generated in immortalized human hepatocytes (IHH) can be serially passaged. Hepatocytes expressing HCV proteins displayed autophagic vacuole formation and the induction of autophagic markers.
(This work was presented in part at the 14th International Symposium on Hepatitis C Virus and Related Viruses, Glasgow, Scotland, United Kingdom, 9 to 13 September 2007.)
MATERIALS AND METHODS
Cells and virus.
Human hepatoma cells (Huh-7.5) were maintained in Dulbecco's modified Eagle's medium (Cambrex, Walkersville, MD) containing 10% fetal bovine serum, 200 U/ml penicillin G, and 200 μg/ml streptomycin at 37°C in an atmosphere of 5% CO2. IHH were generated and maintained in SABM (Cambrex) supplemented with 5% chemically denatured serum, as described previously (47). HCV genotypes 1a and 2a (clones H77 and JFH1, respectively) were grown in cell culture as described previously (22).
Antibodies and reagents.
Rabbit anti-Apg5 and Beclin 1 (BECN 1) antibodies were purchased (Santa Cruz Biotechnology, Santa Cruz, CA). Anticore (C750) mouse monoclonal antibody was kindly provided by Darius Moradpour and Jack R. Wands (Harvard University, MA). Anti-NS4-fluorescein isothiocyanate-conjugated antibody was purchased (Biodesign International, ME). Mouse anti-NS3 monoclonal antibody to HCV genotype 2a was kindly provided by George Luo (University of Kentucky, KY). All secondary antibodies used for immunofluorescence were procured as Alex Fluor conjugates from Molecular Probes (Eugene, OR).
Isolation of RNA, reverse transcription (RT)-PCR, and real-time PCR.
Total cellular RNA or RNA from cell culture medium was extracted after HCV infection, using a specific isolation kit (Qiagen, CA). To detect HCV genome sequences, RNA and random hexamers were used for cDNA synthesis using a SuperScriptIII first-strand synthesis system (Invitrogen, CA), following the supplier's protocol. PCR amplification was performed with cDNA as the template, using the sense (5′-TCTGCGGAACCGGTGAGTA-3′) and antisense (5′-TCAGGCAGTACCACAAGGC-3′) primers from the 5′ untranslated region (UTR) or the sense (5′-TGTGGAGCTGAGATCACTGG-3′) and antisense (5′-CCGCCTTATCTCCACGTATT-3′) primers for NS5A, denaturing at 94°C for 30 s, annealing at 55°C for 60 s, and extension at 72°C for 90 s. PCR products were subjected to electrophoresis on a 1.8% agarose gel. HCV-specific RNA was quantitated by real-time PCR as the increase in fluorescence of SYBR Green I on an ABI PRISM 7700 (Applied Biosystems, CA), as described previously (23). The housekeeping gene product GAPDH was used as a control for the normalization of intracellular RNA. Each real-time PCR assay was performed in triplicate. Virus genome copies/ml were calculated as described previously (22). The lowest limit for the detection of HCV genome was 103 to 104 copies/ml. The relative HCV RNA level was calculated from the HCV RNA copy number in the cell culture fluid at the first passage, which was arbitrarily taken as 1 for the comparison of changes in subsequent virus passage. GAPDH was used for the normalization of intracellular HCV RNA. Similarly, the relative intracellular HCV RNA level with respect to GAPDH RNA in the first passage was arbitrarily taken as 1 for the comparison of changes in subsequent virus passages.
Focus formation assay.
Hepatocytes were fixed for 3 days after infection with 3.7% formaldehyde and incubated at room temperature for 1 h with a NS4-fluorescein isothiocyanate-conjugated mouse monoclonal antibody. Nuclear staining was performed with TO-PRO3-iodide (Molecular Probes). Cells were mounted for fluorescence microscopy (Bio-Rad 1024; Bio-Rad, CA), and focus-forming units per milliliter (ffu/ml) were counted as described previously (22). The lowest limit of detection of infectious titer was approximately 100 ffu/ml.
ELISA.
The HCV-infected IHH culture supernatant was examined for core protein detection by enzyme-linked immunosorbent assay (ELISA) using a commercially available kit (Ortho Diagnostics, Japan), following the supplier's protocol. The limit of detection for the HCV core protein was 44 to 3,600 fmol/liter.
Electron microscopy.
Immunogold labeling was performed for the localization of HCV-like particles in IHH. IHH harboring HCV for 4 days in culture were detached from collagen-coated petri dishes by a brief trypsin treatment, pelleted in a microcentrifuge, and fixed in 4% paraformaldehyde and 1% glutaraldehyde in phosphate-buffered saline (PBS) for 16 h at 4°C. After cell pellets were washed with PBS, they were washed in distilled water, dehydrated in ethanol, and infiltrated with London Resin White resin (London Resin Co., United Kingdom). Cell pellets were transferred to BEEM capsules (Ted Pella, Inc., CA) containing fresh London Resin White resin and polymerized at −20°C under UV light. Thin sections were cut from blocks, collected on Formvar-coated nickel grids, and soaked section side down on drops of PBS for 5 min. The grids were next floated on drops of 0.02 M glycine in distilled water to block unreacted aldehyde groups, washed in PBS, and blocked with 1% fish gelatin and 1% bovine serum albumin (BSA) in PBS for 10 min. Sections were incubated for 2 h in a 1:100 dilution (titrated beforehand for the best results) of monoclonal antibody to E1 glycoprotein (305/C3), kindly provided by Michael Houghton (Chiron Corporation, CA), or in normal mouse immunoglobulin G in PBS containing 0.1% BSA, washed in PBS containing 0.1% BSA, and incubated for 1 h in protein A-10-nm colloidal gold diluted at a ratio of 1:200 in PBS containing 0.1% BSA. After grids were washed with PBS, they were fixed for 3 min in glutaraldehyde, washed in distilled water, stained with uranyl acetate and lead citrate, and examined with a JEOL 100 CX electron microscope.
Confocal microscopy.
Formaldehyde (3.7%)-fixed cells were incubated with anti-HCV (core, NS3, or NS4 protein) antibody or anti-Apg5 serum and stained with a fluorochrome-conjugated secondary antibody (Molecular Probes). After cells were stained, they were mounted for confocal microscopy (Bio-Rad MRC 1024). Whenever necessary, the images were merged digitally to monitor colocalization, in which two different colors produce a distinct color, whereas physically separate signals retain their individual colors.
Transfection of GFP-LC3.
The green fluorescent protein-light chain 3 (GFP-LC3) construct was generated from MIGhCD8t-GFP-LC3 (kindly provided by Craig Thompson, University of Pennsylvania, PA). The MIGhCD8t-GFP-LC3 construct was digested by EcoRI and XhoI, and the GFP-LC3 cDNA was cloned into the eukaryotic expression vector pCI-neo (Promega, WI). Mock-infected or HCV-infected cells were transfected after 3 days with 2 or 4 μg of pCI-GFP-LC3, using Lipofectamine (Invitrogen). Cells were examined after they were incubated for 24 to 36 h at 37°C. Mock-infected or HCV-infected cells following transfection with GFP-LC3 were visualized by confocal microscopy for intracellular localization of LC3.
Western blotting analysis.
Cells were harvested using sodium dodecyl sulfate sample buffer. Proteins were subjected to electrophoresis on a polyacrylamide gel and transferred onto a nitrocellulose membrane. The membrane was probed with an antibody to the autophagic markers (Apg5 or Beclin 1) and for actin (Santa Cruz Biotechnology, Santa Cruz, CA) to estimate the relative protein load in each lane. Proteins were identified by using a secondary antibody conjugated to horseradish peroxidase (Promega), and the bands were visualized by using an enhanced chemiluminescence detection kit (Amersham Pharmacia, NJ). X-ray film was scanned by an image analyzer for densitometry of the protein bands using ImageQuant software (Amersham Molecular Dynamics, CA).
RESULTS AND DISCUSSION
Replication of HCV genotype 1a after serial passage in IHH.
We investigated the generation of infectious HCV particles during serial passage in IHH, a hepatocyte cell line immortalized by HCV core protein (47). For this, virus grown in IHH by transfection of full-length RNAs from HCV genotype 1a (clone H77) was used as the initial passage for subsequent infection, with following passages done utilizing culture medium as the infection source. The presence of HCV in IHH culture medium during passage was examined. After 5 days of incubation for each passage, culture medium from virus-infected cells was filtered through a 0.45-μm cellulose acetate membrane (Nalgene, NY). Filtered culture medium was used for the infection of naïve IHH. The HCV 5′ UTR and the NS5A genomic sequences were detected by RT-PCR from each passage to confirm the presence of viral RNA. Intracellular RNA extracted from virus-infected cells was also examined for HCV-specific sequences. HCV 5′ UTR and NS5A sequences were detected in cell culture medium after virus infection but not from uninfected IHH used as a negative control. No significant variations were observed, using limited sequence analysis of the 5′ UTR and the NS5A region, between the RNA extracted from HCV-infected cell culture fluid and that of the infected cells (data not shown). Further analysis of nucleotide changes of the entire HCV genome for a limited passage number with a significant change in virus titer will be performed to determine adaptive mutation hot spots, if any are generated during passage in IHH.
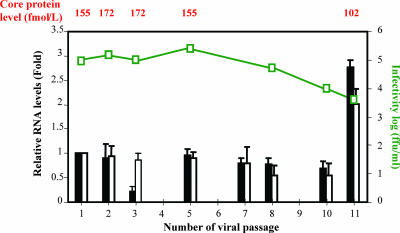
Next, we determined whether infectious virus particles could be recovered from the culture medium of HCV-infected IHH during the serial passage. For this, virus culture medium was serially 10-fold diluted and inoculated into naïve IHH. Cells were incubated for 6 h, washed three times, and incubated with fresh medium for 5 days before being tested by real-time PCR, by core ELISA, and for ffu/ml, as described previously (22). The results provide evidence for HCV replication and infectious virus particle secretion in culture medium. Representative results following the serial passage of HCV into IHH are shown (Fig. 1). The virus was passaged up to 11 times, and limited HCV genomic sequences were detected up to the last passage, suggesting HCV growth during each passage. We obtained approximately 1 × 108 to 4 × 108 HCV genome copies/ml of culture medium using real-time RT-PCR. However, the presence of HCV RNA may not always reflect infectious virus production, and virus titer varied between 5.0 × 103 and 3.2 × 105 ffu/ml. The serial passage of cell culture-grown HCV, the presence of nucleotide sequences, and the core protein in the culture medium clearly indicated genome replication and assembly into infectious virus particles. Although IHH was generated by the introduction of the HCV core gene, viral protein expression in this cell line was extremely weak (3). We could not detect core antigen by ELISA in the culture medium of uninfected IHH. At present, we cannot clarify the discrepancy between the drop in extracellular RNA at passage 3 and the maintenance of the other parameters, as observed in relation to those of previous and subsequent passages. However, the extracellular RNA level was recovered in the subsequent passage. We cannot rule out several possibilities, including the restriction of virus growth from autophagy or the generation of defective interfering particles or antiviral cytokines. As in the classic von Magnus experiments, these inhibitory factors can vary (or cycle) from one passage to the next. On the other hand, neither the HCV RNA level nor the core protein level correlated well with the infectious HCV titer, especially with passage 11. This could be due to the detection of higher genome copy numbers in culture medium from noninfectious or degraded virus particles and the release of HCV core protein from IHH harboring HCV as well as virus-associated core protein. A lack of correlation among these parameters has also been observed with HCV genotype 2a or chimeric virus (6, 24, 26, 43). A decrease in virus replication or infectious titer could be due to selective pressure on HCV growth by autophagy. Further work is in progress to determine if additional passages of the virus increase the infectious HCV titer.
FIG. 1.
Replication of HCV genotype 1a (clone H77) after serial passage in IHH. The HCV replication and secretion of virus in culture medium were examined using different parameters. The relative copy number of HCV RNA at the intracellular level and in culture supernatant were measured by real-time PCR and calculated as described in Materials and Methods. The results are presented with standard errors from three independent experiments. Mock-infected or HCV-infected IHH culture supernatant was tested for core protein detection by ELISA. HCV titer (ffu/ml) was determined by the immunofluorescence of IHH 3 days after infection with filtered culture medium for the detection of NS4 protein expression. Relative HCV RNA in cell culture fluid (black bars) and HCV GAPDH ratio in cellular RNA (white bars) are shown. Infectivity (ffu/ml) is shown as a green line, and core protein level is shown on top.
HCV infection induces autophagic vacuole formation.
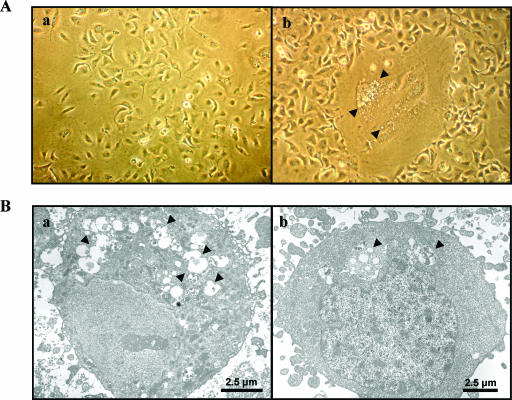
We initially examined cellular changes in virus-infected hepatocytes by light microscopy. HCV-infected cells were larger and contained extensive vacuole formation than mock-infected control IHH (Fig. 2A). These larger cells developed over 3 to 5 days of incubation following HCV infection and lost anchorage from the collagen-coated plate. These characteristics were increasingly prevalent in HCV-infected IHH as the duration of infection increased. Approximately 20% of infected cells had the enlarged phenotype, although the majority of cells were infected with HCV, as detected by fluorescence microscopy (data not shown).
FIG. 2.
Cellular changes following HCV genotype 1a (clone H77) infection of IHH. (A) Comparison of the cellular changes, by using light microscopy, between the uninfected (panel a) and the HCV-infected (panel b) IHH. Arrowheads in the photomicrograph indicate vacuole-like inclusions in larger cells. (B) Cellular changes were also compared by electron microscopy of uninfected (panel a) and HCV-infected (panel b) IHH. Numerous autophagic vacuoles were observed in the virus-infected cells compared to the uninfected cells and are indicated by arrows.
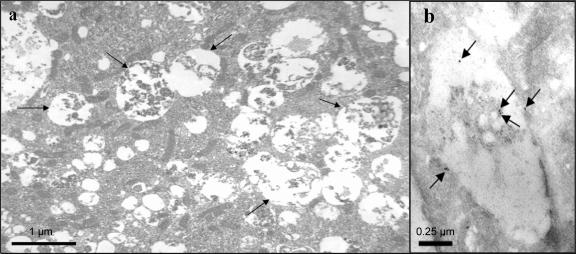
We next examined cellular changes at the ultrastructural level, using electron microscopy. The infection of IHH with HCV induced the formation of abundant autophagic vacuoles with cytoplasmic contents (Fig. 2B). In contrast, we observed a significantly lower number of autophagic vacuoles with mock-infected cells. The cytoplasmic content of the autophagosomes associated with HCV-infected IHH could be observed from electron photomicrographs (Fig. 3a). Anti-E1 HCV protein immunogold labeling revealed the presence of gold particles within and associated with the limiting membrane of the autophagic vacuoles, probably representing degraded HCV coat proteins (Fig. 3b). On the other hand, uninfected control IHH incubated with monoclonal antibody to E1 glycoprotein did not exhibit immunogold labeling. Other negative controls were labeled with normal mouse immunoglobulin G and were not incubated with the primary antibody (not shown in figure). We have used HCV RNA-transfected and cell culture-grown HCV-infected IHH from different passage levels (between passages 2 to 6) and obtained similar levels of autophagic vacuole formation. These observations indicated that autophagic vacuole formation was induced upon HCV infection.
FIG. 3.
Electron micrographs showing cytoplasmic contents in autophagic vacuoles. IHH were infected with HCV genotype 1a (clone H77) and incubated for 5 days. Infected cells were subjected to electron microscopy. IHH showing cytoplasmic contents in autophagosome are indicated by arrows (panel a). Uninfected cells were treated similarly to the negative control cells and did not exhibit any major changes (not shown). Immunogold label showing localization for E1 protein inside and on the vacuolar membrane is indicated by arrows (panel b).
HCV-infected hepatocytes display accumulation of autophagic marker proteins on vacuole membranes.
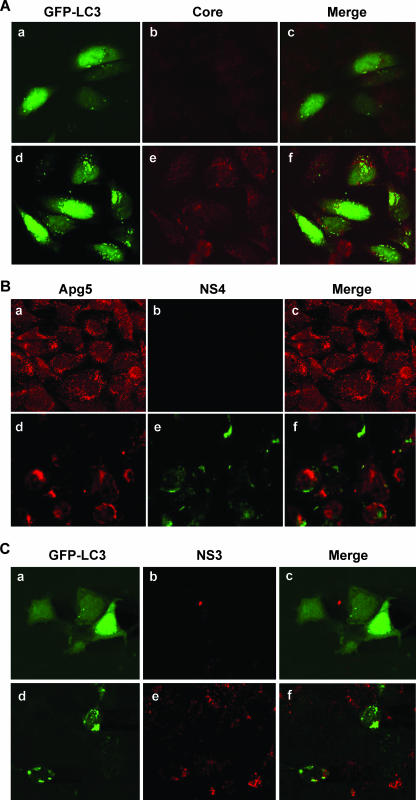
Endogenous microtubule-associated LC3 exists in the form LC3I, which shows a diffuse distribution within the cytoplasm. When autophagy is induced, LC3I is lipidated to form LC3II, which integrates into the autophagic vacuole membrane (21). The LC3 modification is one of the early steps in cellular autophagy (56). The exogenous marker GFP-LC3 behaves similarly and can be easily observed as distinct punctate dots on an autophagic vacuole by fluorescence microscopy (1). To examine autophagy, mock-infected or HCV-infected IHH were transfected with GFP-LC3. Infection of these hepatocytes with HCV genotype 1a (clone H77) displayed a large number of GFP-LC3 punctate dots on the autophagic vacuole (Fig. 4A, panel d), while uninfected cells expressing GFP-LC3 exhibited a clear difference in localization of GFP signals characterized by a diffuse pattern of immunofluorescence (Fig. 4A, panel a). Cells were stained with anticore antibody, and HCV core protein did not colocalize with autophagy marker proteins.
FIG. 4.
HCV-infected hepatocytes display characteristic localization of autophagic markers. (A) Confocal microscopy examination for subcellular localization of GFP-LC3 visualized by intrinsic fluorescence (panel a), HCV core protein (panel b), and merged image (panel c) of uninfected IHH control cells. Fluorescence from HCV genotype 1a (clone H77)-infected IHH are shown (panels d, e, and f). Punctate localization of LC3 on autophagic vacuoles in HCV-infected cells was clearly visible. Confocal microscopy did not suggest colocalization of GFP-LC3 and HCV core protein. (B) Similar experiments with endogenous Apg5 also suggested that the autophagy marker did not colocalize with NS4 protein in HCV genotype 1a (clone H77)-infected IHH. Results from uninfected control cells (panels a, b, and c) and HCV-infected cells (panels d, e, and f) are shown. (C) Huh-7.5 cells infected with HCV genotype 2a (clone JFH1) also displayed autophagic vacuoles with localization of GFP-LC3 marker protein. Results from uninfected Huh-7.5 cells (panels a, b, and c) and HCV-infected cells (panels d, e, and f) are shown. Colocalization of GFP-LC3 and HCV NS3 protein was not observed upon merging these two images.
The intracellular localization of Apg proteins appears close to the autophagic vacuoles (38, 53). To determine if HCV replicase proteins (for example, NS4) colocalized with the autophagic marker protein Apg5, specific antibody was used with dual-labeling experiments (Fig. 4B). HCV genotype 1a (clone H77)-infected IHH were labeled for the NS4 and Apg5 proteins. In uninfected IHH, we did not observe major ultrastructural changes. In HCV-infected cells, dual labeling with anti-NS4 and anti-Apg5 displayed two clear staining patterns which did not indicate colocalization (Fig. 4B). We also used antibody to the NS3 protein of HCV and obtained similar results.
HCV genotype 2a (clone JFH1) was first shown to grow in Huh-7 cells (57). Confocal microscopy was also performed with Huh-7.5 cells transfected with GFP-LC3, followed by HCV genotype 2a (clone JFH1) infection. Infected cells displayed localization of the LC3 autophagy marker as punctate dots (Fig. 4C), similar to HCV genotype 1a (clone H77)-infected IHH. The GFP-LC3 marker protein did not colocalize with HCV NS3 protein. Therefore, our results suggested that HCV replication may not be associated with autophagic vacuole formation, as suggested with other RNA viruses (5, 60), and additional studies are in progress to verify our observation. Taken together, these data suggest that autophagy is induced in two different hepatocyte cell lines infected by both of the available cell culture-grown HCV genotypes.
HCV-infected hepatocytes exhibit enhanced Apg5 complex proteins and Beclin expression.
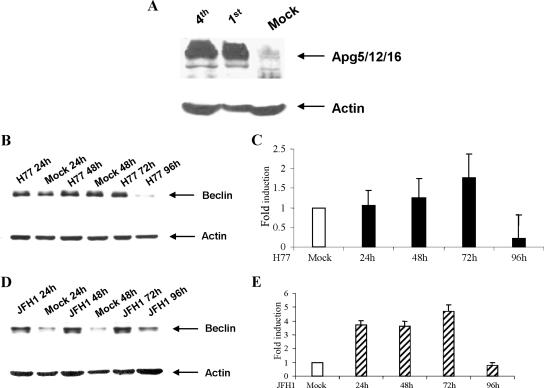
A novel preautosomal structure has been described in which Apg1p, Apg2p, Apg5p, aut7p, and Apg16p assemble in Saccharomyces cerevisiae (53). Hammond et al. (13) cloned hApg5 as an apoptosis-specific protein and suggested that the 45-kDa band of Apg5 appeared at a late stage during or as a result of apoptosis, possibly due to posttranslational regulation. A different protein, Apg12, was also discovered and shown to conjugate to a human Apg5 homologue (35). In mammalian cells, an Apg12p-Apg5p conjugate transiently associates with the membranes of precursor autophagosomes (36). We examined the autophagy conjugation system by detecting Apg5 using a specific rabbit polyclonal antibody directed to amino acids 1 to 275, representing full-length Apg5 of human origin (Santa Cruz). Western blotting analysis was performed to compare the expression of the autophagic marker (the Apg5 complex) in uninfected control and the HCV-infected IHH at two different passage levels. Cell lysates in sample-reducing buffer were subjected to Western blotting analysis, and autophagy marker protein Apg5 complex was detected. IHH that supported HCV genotype 1a replication displayed the expression of Apg5 associated with other Apg proteins as a high-molecular-mass (>200-kDa) complex migrating closer to the application well of the polyacrylamide gel (Fig. 5A). On the other hand, mock-infected IHH showed a much lower or undetectable level of Apg5 complex formation. The blots were stripped and reprobed using a mouse monoclonal antibody to actin. A mammalian system-based study has indicated that the Apg12-Apg5 conjugate forms a high-molecular-mass (∼800-kDa) complex with the novel WD-repeat protein Apg16 (37). A different study using the yeast system has shown that the Apg12-Apg5 conjugate and Apg16 form a multimeric complex mediated by the Apg16 homo-oligomer, and the formation of the ∼350-kDa complex is required for autophagy in yeast (27). Our results indicated that infection with HCV genotype 1a (clone H77) induces a high-molecular-mass Apg5 complex formation in IHH.
FIG. 5.
Expression of autophagy marker proteins in HCV-infected hepatocytes. (A) Western blotting analysis for autophagic marker Apg5, using a specific antiserum from mock-infected and HCV genotype 1a-infected IHH from passages 1 and 4. Apg appeared as a high-molecular-mass protein complex (>200 kDa), shown by an arrow on the right. (B and D) Western blotting analysis was performed for Beclin 1, using mock-infected or HCV genotype 1a (clone H77)-infected IHH or HCV genotype 2a (clone JFH1)-infected Huh-7.5 cells at the indicated time. Beclin 1 appeared as ∼50 kDa and is shown by arrows on the right. The blots were reprobed with an antibody to actin to ascertain the level of protein load in each lane. The molecular masses of the protein bands were verified from the migration of protein molecular mass markers (Cambrex, Rockland, ME). (C and E) The relative levels of Beclin 1 were estimated by densitometric scanning after normalization against actin and are shown as bar diagrams with H77-infected IHH and JFH1-infected Huh-7.5 cells. Error bars represent standard errors from three independent experiments.
The accumulation of autophagy gene products during cell death, and their relationship with members of the Bcl-2 family, is a promising area for better understanding how autophagy is controlled during cell death. The interaction between Beclin 1 and Bcl-2/Bcl-xL is probably a key event in turning the accumulation of autophagosomes on or off (8, 9, 48, 51). Beclin 1 is involved in the early steps of the signaling cascade preceding autophagic vacuolization, although the precise mechanism of Beclin 1 accumulation remains to be understood. Beclin 1-mediated autophagy has also been proposed to protect against viral infection. Overexpression of Beclin 1 protects mice against Sindbis virus infection-induced encephalitis (29). Autophagy is also involved in the clearance of HSV type 1 (HSV-1) from infected host cells, and the HSV-1 neurovirulence protein ICP34.5 inhibits autophagy through its binding to Beclin 1 (39). This observation suggests that endogenous amounts of Beclin 1 are important in protecting against viral invasion. We investigated the accumulation of Beclin 1 after HCV infection by Western blotting analysis (Fig. 5B and D). Beclin 1 expression with IHH increased (∼twofold) between 24 and 72 h of HCV genotype 1a (clone H77) infection compared to mock-infected control IHH, and decreased at 96 h (Fig. 5B). On the other hand, Beclin 1 expression with Huh-7.5 cells significantly increased and remained unregulated (∼4.5-fold) up to 72 h following HCV genotype 2a (clone JFH1) infection and decreased at 96 h (Fig. 5D). Beclin 1 physically interacts with Bcl-2 or Bcl-XL, two antiapoptotic proteins predominantly localized to the mitochondrial membrane that prevent the loss of mitochondrial membrane potential and cytochrome C release. The proautophagic consequences of Beclin 1 upregulation may be associated with Bcl-2 and the related gene family in these two different cell types. However, Beclin 1 depletion may activate the mitochondrial apoptosis pathway, and the interaction of Beclin 1 with either Bcl-2 or Bcl-XL is essential for the antiapoptotic effect of Beclin 1 (7). We have previously observed modulation of Bcl-2 family of proteins (33, 51). Therefore, the differences in Beclin 1 expression between IHH and that of Huh-7.5 following infection with HCV probably reflect the status of Bcl-2-related gene expression.
We showed that HCV genotype 1a (clone H77) replicates during serial passage in naïve IHH, and we noted an induction of autophagy in infected hepatocytes. Autophagy is an essential process for physiological homeostasis, but its role in viral infection is only beginning to emerge. To our knowledge, the induction of autophagy has not previously been demonstrated in association with HCV infection of hepatocytes. Eleven serial HCV passages in IHH have been completed, and virus replication was ascertained from the presence of HCV-specific nucleotide sequences, the detection of core protein, the virus genome copy number, and the virus titer from cell culture supernatants. We did not observe any significant increase in virus replication parameters during the passage in IHH. The functional role of the cell culture-grown HCV genotype 1a (clone H77) was supported from neutralization by specific antibodies (4), interferon induction in naïve IHH (23), and interleukin-10 production in monocyte-derived dendritic cells by HCV-infected hepatocyte culture supernatant (K. Saito and R. Ray, unpublished results). We have observed that HCV infection induces autophagy in hepatocytes, although the colocalization of HCV core or nonstructural proteins with the autophagic markers LC3 or Apg5 was not observed. The use of the drug 3-methyladenine as an inhibitor of autophagy also did not suggest a significant inhibition of HCV replication by real-time PCR (data not shown). Thus, our results suggest that the HCV replication complex may not be associated with autophagic vacuoles in hepatocytes, and we do not know whether HCV growth is suppressed by the induction of an autophagic response.
A recent review (50) has eloquently discussed three main outcomes of interactions between viruses and the autophagic machinery: (i) autophagy successfully limits viral replication, for example, tobacco mosaic virus and Sindbis virus (29, 31); (ii) viruses, such as some members of the herpes virus family, inhibit autophagy to avoid the restriction of their replication (39, 41, 54); and (iii) viruses use accumulated autophagosomes for their replication, for example, poliovirus, rhinoviruses, mouse hepatitis virus, SARS-CoV, and equine arteritis virus (18, 42, 44, 45). Our results suggest that autophagy develops in HCV-infected hepatocytes, but the biological relevance of this outcome remains to be understood. Cell cycle arrest is often accompanied by an induction of autophagy, a key survival mechanism during stress conditions (16). The tremendous degradative capacity of autophagy allows this process to function in both cell survival and cell death.
Studies of the accumulation of autophagy gene products during cell death and their relationship with members of the Bcl-2 family may provide information for a better understanding of how autophagy is controlled during cell death (51). The interaction between Beclin 1 and Bcl-2/Bcl-xL88 is probably a key event in the accumulation of autophagosomes. Autophagy can be activated during endoplasmic reticulum (ER) stress. HCV replication in the ER generates ER stress (55), which may also be a mechanism for the induction of autophagy. It is important to mention that the IHH cell line was generated by transfection with an HCV core genomic sequence (47). These hepatocytes also displayed vacuole formation but to a much lesser extent than those infected with HCV. However, we did not observe a similar characteristic vacuole formation with Huh-7 cells. IHH display an extremely weak level of HCV core protein expression (3). However, we cannot rule out the possibility of HCV core protein in the induction of autophagic vacuole formation in IHH, and an accumulation of Beclin 1 protein in these cells may indicate a role of this protein. Unraveling the molecular basis of autophagy would be a crucial step in further understanding the role of autophagy in controlling cellular homeostasis. Autophagy delivers viral protein to late endosomal compartments where autophagy substrates are degraded by lysosomal hydrolases with the molecular machinery to load antigenic fragments onto major histocompatibility complex class II molecules for presentation to CD4+ T cells. (8, 50). Here, we noted the presence of HCV E1 in the autophagic vacuole, supporting this process.
HCV infection is often associated with a compromised innate immune response. Several proteins involved in the interferon signaling pathways are linked to the regulation of autophagy (12). A component of the autophagic machinery was suggested to block innate antiviral immune responses, thereby contributing to RNA virus replication in host cells (20). We do not know whether the induction of autophagy by HCV infection modulates immune responses at this time. Exploration of the novel signaling pathways relevant to autophagy could lead to the development of new therapeutic strategies for persistent HCV infection.
Acknowledgments
We thank Craig Thompson for the GFP-LC3 clone, Charles Rice for the HCV H77 clone, Takaji Wakita for the HCV JFH1 clone, Michael Houghton for the monoclonal antibody to E1, George Luo for the NS3 specific monoclonal antibody to JFH1 clone, and Darius Moradpour and Jack Wands for anticore monoclonal antibody. We thank Lin Cowick for preparation of the manuscript.
This work was supported by research grants from the Foundation for Advancement of International Science, Japan (T.K.), and by grants AI45144 (R.B.R.) and AI068769 (R.R.) from the National Institutes of Health.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Bampton, E. T., C. G. Goemans, D. Niranjan, N. Mizushima, and A. M. Tolkovsky. 2005. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy 123-36. [DOI] [PubMed] [Google Scholar]
- 2.Bassett, S. E., D. L. Thomas, K. M. Brasky, and R. E. Lanford. 1999. Viral persistence, antibody to E1 and E2, and hypervariable region 1 sequence stability in hepatitis C virus-inoculated chimpanzees. J. Virol. 731118-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basu, A., K. Meyer, R. B. Ray, and R. Ray. 2002. Hepatitis C virus core protein is necessary for the maintenance of immortalized human hepatocytes. Virology 29853-62. [DOI] [PubMed] [Google Scholar]
- 4.Basu, A., T. Kanda, A. Beyene, K. Saito, K. Meyer, and R. Ray. 2007. Sulfated homologues of heparin inhibit hepatitis C virus entry into mammalian cells. J. Virol. 813933-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brabec-Zaruba, M., U. Berka, D. Blass, and R. Fuchs. 2007. Induction of autophagy does not affect human rhinovirus 2 production. J. Virol. 8110815-10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, Z., C. Zhang, K. S. Chang, J. Jiang, B. C. Ahn, T. Wakita, T. J. Liang, and G. Luo. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 7913963-13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, Y., and D. J. Klionsky. 2007. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 17839-849. [DOI] [PubMed] [Google Scholar]
- 8.Codogno, P., and A. J. Meijer. 2005. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2(Suppl 12)1509-1518. [DOI] [PubMed] [Google Scholar]
- 9.Daniel, F., A. Legrand, D. Pessayre, N. Vadrot, V. Descatoire, and D. Bernuau. 2006. Partial Beclin 1 silencing aggravates doxorubicin- and Fas-induced apoptosis in HepG2 cells. World J. Gastroenterol. 122895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deretic, V. 2006. Autophagy as an immune defense mechanism. Curr. Opin. Immunol. 18375-382. [DOI] [PubMed] [Google Scholar]
- 11.Di Bisceglie, A. M., R. L. Carithers, and G. J. Gorse. 1998. Hepatocellular carcinoma. Hepatology 281161-1165. [DOI] [PubMed] [Google Scholar]
- 12.Espert, L., P. Codogno, and M. Biard-Piechaczyk. 2007. Involvement of autophagy in viral infections: antiviral function and subversion by viruses. J. Mol. Med. 85811-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond, E. M., C. L. Brunet, G. D. Johnson, J. Parkhill, A. E. Milner, G. Brady, C. D. Gregory, and R. J. Grand. 1998. Homology between a human apoptosis specific protein and the product of APG5, a gene involved in autophagy in yeast. FEBS Lett. 425391-395. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi, J., H. Aoki, Y. Arakawa, and O. Hino. 1999. Hepatitis C virus and hepatocarcinogenesis. Intervirology 42205-210. [DOI] [PubMed] [Google Scholar]
- 15.Heller, T., S. Saito, J. Auerbach, T. Williams, T. R. Moreen, A. Jazwinski, B. Cruz, N. Jeurkar, R. Sapp, G. Luo, and T. J. Liang. 2005. An in vitro model of hepatitis C virion production. Proc. Natl. Acad. Sci. USA 1022579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, J., and D. J. Klionsky. 2007. Autophagy and human disease. Cell Cycle 61837-1849. [DOI] [PubMed] [Google Scholar]
- 17.Ilan, E., J. Arazi, O. Nussbaum, A. Zauberman, R. Eren, I. Lubin, O. Neville Ben-Moshe, A. Kischitzky, A. Litchi, I. Margalit, J. Gopher, S. Mounir, W. Cal, N. Daudi, A. Eid, O. Jurim, A. Czerniak, E. Galun, and S. Dagan. 2002. The hepatitis C virus (HCV)-trimera mouse: a model for evaluation of agents against HCV. J. Infect. Dis. 185153-161. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, W. T., T. H. Giddings, Jr., M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS. Biol. 3e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeffers, L. 2000. Hepatocellular carcinoma: an emerging problem with hepatitis C. J. Natl. Med. Assoc. 92369-371. [PMC free article] [PubMed] [Google Scholar]
- 20.Jounai, N., F. Takeshita, K. Kobiyama, A. Sawano, A. Miyawaki, K. Q. Xin, K. J. Ishii, T. Kawai, S. Akira, K. Suzuki, and K. Okuda. 2007. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. USA 10414050-14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 195720-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanda, T., A. Basu, R. Steele, T. Wakita, J. S. Ryerse, R. Ray, and R. B. Ray. 2006. Generation of infectious hepatitis C virus in immortalized human hepatocytes. J. Virol. 804633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanda, T., R. Steele, R. Ray, and R. B. Ray. 2007. Hepatitis C virus infection induces interferon-β signaling pathway in immortalized human hepatocytes. J. Virol. 8112375-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato, T., T. Matsumura, T. Heller, S. Saito, R. K. Sapp, K. Murthy, T. Wakita, and T. J. Liang. 2007. Production of infectious hepatitis C virus of various genotypes in cell cultures. J. Virol. 814405-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkegaard, K., M. P. Taylor, and W. T. Jackson. 2004. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2301-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 805308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuma, A., N. Mizushima, N. Ishihara, and Y. Ohsumi. 2002. Formation of the approximately 350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 27718619-21865. [DOI] [PubMed] [Google Scholar]
- 28.Levine, B. 2005. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120159-162. [DOI] [PubMed] [Google Scholar]
- 29.Liang, X. H., L. K. Kleeman, H. H. Jiang, G. Gordon, J. E. Goldman, G. Berry, B. Herman, and B. Levine. 1998. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 728586-8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-636. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Y., M. Schiff, K. Czymmek, Z. Talloczy, B. Levine, and S. P. Dinesh-Kumar. 2005. Autophagy regulates programmed cell death during the plant innate immune response. Cell 121567-577. [DOI] [PubMed] [Google Scholar]
- 32.Mercer, D. F., D. E. Schiller, J. F. Elliott, D. N. Douglas, C. Hao, A. Rinfret, W. R. Addison, K. P. Fischer, T. A. Churchill, J. R. Lakey, D. L. Tyrrell, and N. M. Kneteman. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7927-933. [DOI] [PubMed] [Google Scholar]
- 33.Meyer, K., A. Basu, K. Saito, R. B. Ray, and R. Ray. 2005. Inhibition of hepatitis C virus core protein expression in immortalized human hepatocytes induces cytochrome c independent increase in Apaf-1 and caspase-9 activation for cell death. Virology 336198-207. [DOI] [PubMed] [Google Scholar]
- 34.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 91089-1097. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima, N., H. Sugita, T. Yoshimori, and Y. Ohsumi. 1998. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 27333889-33892. [DOI] [PubMed] [Google Scholar]
- 36.Mizushima, N., A. Yamamoto, M. Hatano, Y. Kobayashi, Y. Kabeya, K. Suzuki, T. Tokuhisa, Y. Ohsumi, and T. Yoshimori, T. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizushima, N., A. Kuma, Y. Kobayashi, A. Yamamoto, M. Matsubae, T. Takao, T. Natsume, Y. Ohsumi, and T. Yoshimori. 2003. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 1161679-1688. [DOI] [PubMed] [Google Scholar]
- 38.Mizushima, N. T., Yoshimori, and Y. Ohsumi. 2003. Role of the Apg12 conjugation system in mammalian autophagy. Int. J. Biochem. Cell. Biol. 35553-561. [DOI] [PubMed] [Google Scholar]
- 39.Orvedahl, A., D. Alexander, Z. Talloczy, Q. Sun, Y. Wei, W. Zhang, D. Burns, D. Leib, and B. Levine. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 123-25. [DOI] [PubMed] [Google Scholar]
- 40.Paludan, C., D. Schmid, M. Landthaler, M. Vockerodt, D. Kube, T. Tuschl, and C. Munz. 2005. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307593-596. [DOI] [PubMed] [Google Scholar]
- 41.Pattingre, S., A. Tassa, X. Qu, R. Garuti, X. H. Liang, N., Mizushima, M. Packer, M. D. Schneider, and B. Levine, B. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122927-939. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 732016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 1037408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 27910136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 789977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purcell, R. H. 1994. Hepatitis viruses: changing patterns of human disease. Proc. Natl. Acad. Sci. USA 912401-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ray, R. B., K. Meyer, and R. Ray. 2000. Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology 271193-204. [DOI] [PubMed] [Google Scholar]
- 48.Saeki, K., A. You, E. Okuma, Y. Yazaki, S. A. Susin, G. Kroemer, and F. Takaku. 2000. Bcl-2 down-regulation causes autophagy in a caspase-independent manner in human leukemic HL60 cells. Cell Death Differ. 71263-1269. [DOI] [PubMed] [Google Scholar]
- 49.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 706576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmid, D., and C. Munz. 2007. Innate and adaptive immunity through autophagy. Immunity 2711-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizu, S., T. Kanaseki, N. Mizushima, T. Mizuta, S. Arakawa-Kobayashi, C. B. Thompson, and Y. Tsujimoto. 2004. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 61221-1228. [DOI] [PubMed] [Google Scholar]
- 52.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 748953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki, K., T. Kirisako, Y. Kamada, N. Mizushima, T. Noda, and Y. Ohsumi. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 205971-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talloczy, Z., W. Jiang, H. W. Virgin, D. A. Leib, D. Scheuner, R. J. Kaufman, F. L. Eskelinen, and B. Levine. 2002. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. USA 99190-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tardif, K. D., K. Mori, and A. Siddiqui. 2002. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 767453-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor, M. P., and K. Kirkegaard. 2007. Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 8112543-12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wileman, T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312875-878. [DOI] [PubMed] [Google Scholar]
- 59.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 1032310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, H., C. E. Monken, Y. Zhang, J. Lenard, N. Mizushima, E. C. Lattime, and S. Jin. 2006. Cellular autophagy machinery is not required for vaccinia virus replication and maturation. Autophagy 291-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 1029294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]