Abstract
The broadly neutralizing monoclonal antibody (MAb) 4E10 recognizes a linear epitope in the C terminus of the membrane-proximal external region (MPER) of gp41. This epitope is particularly attractive for vaccine design because it is highly conserved among human immunodeficiency virus type 1 (HIV-1) strains and neutralization escape in vivo has not been observed. Multiple env genes were cloned from an HIV-1 subtype C virus isolated from a 7-year-old perinatally infected child who had anti-MPER neutralizing antibodies. One clone (TM20.13) was resistant to 4E10 neutralization as a result of an F673L substitution in the MPER. Frequency analysis showed that F673L was present in 33% of the viral variants and in all cases was linked to the presence of an intact 2F5 epitope. Two other envelope clones were sensitive to 4E10 neutralization, but TM20.5 was 10-fold less sensitive than TM20.6. Substitutions at positions 674 and 677 within the MPER rendered TM20.5 more sensitive to 4E10 but had no effect on TM20.6. Using chimeric and mutant constructs of these two variants, we further demonstrated that the lentivirus lytic peptide-2 domain in the cytoplasmic tail affected the accessibility of the 4E10 epitope, as well as virus infectivity. Collectively, these genetic changes in the face of a neutralizing antibody response to the MPER strongly suggested immune escape from antibody responses targeting this region.
The membrane-proximal external region (MPER) of the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein comprises the last 23 amino acids, from residues 660 to 683, of the extracellular domain of gp41 just before the transmembrane domain. This region has attracted much attention in the field of HIV vaccinology due to some particular features: (i) it is the target of two of the few broadly neutralizing monoclonal antibodies (MAbs) against HIV-1, namely, 4E10 and 2F5; (ii) it has been shown to be important in the fusion process and therefore in viral entry (11, 28); and (iii) it is a highly conserved linear region among all HIV-1 subtypes.
MAb 4E10 recognizes an epitope containing the sequence NWF(D/N)IT (30, 38) in the tryptophan-rich region of gp41. Mutagenesis experiments have shown that residues W672, F673, and W680 are indispensable for 4E10 recognition (37). Crystal structures of the Fab 4E10 in complex with a peptide containing the epitope illustrate that residues W672, F673, I675, and T676 are the key residues in this interaction (7). A more recent study extended the 4E10 epitope to the motif WFx(I/L)(T/S)xx(L/I)W (residues 672 to 680), where the amino acids marked with an x do not play a major role in 4E10 binding (6). The sequence ELDKWA (residues 663 to 667) immediately N terminal to the 4E10 epitope is the target of the 2F5 MAb (21). Mutagenesis studies have revealed that the amino acid motif DKW is required for recognition by this MAb (37), and structural studies have demonstrated that these three residues are deeply buried in the interface with 2F5 (25). While 4E10 neutralizes viruses from all HIV-1 subtypes, 2F5 fails to neutralize subtype C and some subtype D viruses, and this can be directly correlated to changes in the antibody epitope (3, 14).
Despite the high level of conservation of the MPER and its importance in the fusion process, multiple studies have demonstrated that mutations in this region do not necessarily impair viral infectivity (5, 37). It has been proposed that this region is not targeted by the host immune response and therefore is not under diversifying selection pressure (36). Recent studies have addressed the question of whether HIV-1 infection induces the production of neutralizing antibodies that target the MPER. The presence of such antibodies was assessed using a novel strategy in which the HIV-1 MPER was engrafted onto a simian immunodeficiency virus (35) or HIV-2 envelope (F. Bibollet-Ruche et al., presented at the Keystone Symposium on HIV Vaccines, Keystone, CO, 2006). These studies indicated that antibodies with specificities such as those of 4E10 and 2F5 are rarely produced (35; J. M. Decker et al., presented at the Keystone Symposium on HIV Vaccines, 2006); however, other anti-MPER antibodies were detected in around one-third of HIV-1-infected patients (F. Bibollet-Ruche et al., presented at the Keystone Symposium on HIV Vaccines, 2006). The effect of such antibodies on the viral population remains unclear, as escape variants have not been described.
In this study we characterized HIV-1 subtype C viral quasispecies with different sensitivities to MAb 4E10. We explored the genetic determinants of these phenotypes as well as the anti-MPER antibody response that developed in the individual from whom this virus was isolated.
MATERIALS AND METHODS
Cloning of envelope genes and production of pseudovirions.
Proviral DNA extracted from in vitro-infected peripheral blood mononuclear cells was used to amplify full-length envelope genes using the primers envA and envM (13). The 3-kb PCR fragments were cloned into an expression vector and used to generate Env-pseudotyped viruses as previously described (14).
Envelope sequencing.
gp41 was amplified from viral RNA from culture supernatant by nested reverse transcription-PCR using published primers (9, 13) and cloned using the TOPO TA cloning kit (Invitrogen Corporation, Carlsbad, CA). The gp41 and gp160 clones were sequenced using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA) and resolved on an ABI 3100 automated genetic analyzer. The sequences were assembled and edited using Sequencher v.4.0 software (Genecodes, Ann Arbor, MI).
Single-cycle neutralization assay.
Neutralization was measured as a reduction in luciferase gene expression after a single round of infection of JC53bl-13 cells with Env-pseudotyped viruses (20). Briefly, 200 50% tissue culture infective doses of pseudoviruses in 50 μl were incubated with 100 μl of serially diluted MAbs, CD4-immunoglobulin G2 (CD4-IgG2), plasma, or T-20 (enfuvirtide) in Dulbecco modified Eagle medium with 10% fetal bovine serum in a 96-well plate in triplicate for 1 h at 37°C. A 100-μl quantity of JC53bl-13 cells (1 × 104 cells/well) containing 75 μg/ml DEAE-dextran (Sigma-Aldrich, St. Louis, MO) was added, and the cultures were incubated at 37°C in 5% CO2. Infection was determined 48 h later using the Bright Glo reagent (Promega, Madison, WI). Luminescence was measured in a Wallac 1420 Victor Multilabel counter (Perkin-Elmer, Norwalk, CT). Titers were calculated as the 50% inhibitory concentration (IC50), or reciprocal plasma dilution (ID50) causing 50% reduction of relative light units compared to the virus control (wells with no inhibitor) after subtracting the background (wells without virus infection).
Virus infectivity and envelope incorporation.
The amount of virus was normalized by the quantity of p24 pelleted through a 20% sucrose cushion for 2 h at 20,000 × g. JC53bl-13 cells were infected with 10 ng of p24 in the presence of 30 μg/ml DEAE-dextran, and cultures were incubated at 37°C in 5% CO2 for 48 h. Infection was monitored by evaluating the luciferase activity. Envelope incorporation was estimated by Western blotting. Viral proteins were resolved in a Criterion 4 to 15% Tris-HCl gradient gel (Bio-Rad Laboratories, Hercules, CA) and blotted onto a polyvinylidene difluoride nitrocellulose membrane (GE Healthcare Life Science, Piscataway, NJ). The membranes were blocked overnight with 5% milk in Tris-buffered saline-0.05% Tween 20; probed with the anti-gp120 (D7324), anti-gp41 (7B2), or anti-p24 (D7312) antibody; and developed with a horseradish peroxidase-labeled secondary antibody (Sigma) using the enhanced chemiluminescence detection system (GE Healthcare Life Science, Piscataway, NJ). To determine envelope expression levels, 293T cells were transfected with envelope constructs using the Fugene transfection reagent (Roche, Applied Science, Indianapolis, IN). The cells were harvested after 48 h and lysed with nondenaturing lysis buffer (1% Triton X-100, 300 mM NaCl, 5 mM EDTA, and 50 mM Tris-Cl [pH 7.4] with Complete protease inhibitor cocktail [Roche]). Western blotting was carried out as described above.
Site-directed mutagenesis.
Specific amino acid changes in the HIV-1 and HIV-2 envelope glycoproteins were introduced using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The presence of mutations was confirmed by sequence analysis.
Construction of chimeric envelope glycoproteins.
To construct the gp120-gp41 chimeras, the TM20.5 and TM20.6 env plasmids were digested with NdeI and the interchanged fragments ligated with T4 DNA ligase (Invitrogen Corporation). The correct orientation of the cloned fragments was determined by a colony PCR with primers T7 and envM. The ectodomain/cytoplasmic tail chimeras were constructing by overlapping PCR spanning the transmembrane domain using the primer pairs TM20CTfo (CAGATCCGTGAGATTAGTGAGCGGATTCTTAG)-envM and TM20CTre (CTAAGAATCCGCTCACTAATCTCACGGATCTG)-envA. The resultant amplicons was cloned into pcDNA3.1D (Invitrogen Corporation, Carlsbad, CA). Chimerism was confirmed by sequencing.
Anti-MPER neutralization assay.
The anti-MPER neutralization assay was adapted from that previously described by Decker et al. (presented at the Keystone Symposium on HIV Vaccines, 2006) using chimeric HIV-2/HIV-1 MPER constructs (15; F. Bibollet-Ruche et al., presented at the Keystone Symposium on HIV Vaccines, 2006). Briefly, 2,000 infectious units of virus preincubated with plasma/serum at a starting dilution of 1:20 and serially diluted 1:5 in the presence of normal human plasma/serum was added to 40% confluent JC53bl-13 cells that were seeded the day before. Infection was measured 48 h later by evaluating the luciferase activity as described above.
Nucleotide sequence accession numbers.
The GenBank database accession numbers for the three env clones described in this study are EU161643 to EU161645.
RESULTS
Discovery of a naturally occurring 4E10-resistant virus.
While characterizing functional envelope genes from HIV-1 subtype C viruses isolated from children (14), we identified an envelope clone, TM20.13, that was resistant to MAb 4E10. This was the result of an F-to-L mutation at position 673, which is known from mutagenesis studies to confer resistance to 4E10 (37). However, this phenotype has not been previously described in patient samples, and analysis of sequences in the Los Alamos HIV database suggested that this mutation is very rare in group M strains of HIV-1. This clone was derived from a virus isolated from a 7-year-old female who was infected perinatally and at the time of blood collection had a CD4 count of 334 cells/μl and a viral load of 34,310 copies/ml. Although this patient had lymphocytic interstitial pneumonitis, she was considered to be a slow progressor (8). The virus TM20 was isolated on donor peripheral blood mononuclear cells and used the CCR5 coreceptor with a V3 sequence typical of subtype C viruses (8). Only a single sample from this patient was available. Attempts to obtain additional samples either from this individual or from her mother were unsuccessful.
Frequency of the F673L mutation in the TM20 virus isolate.
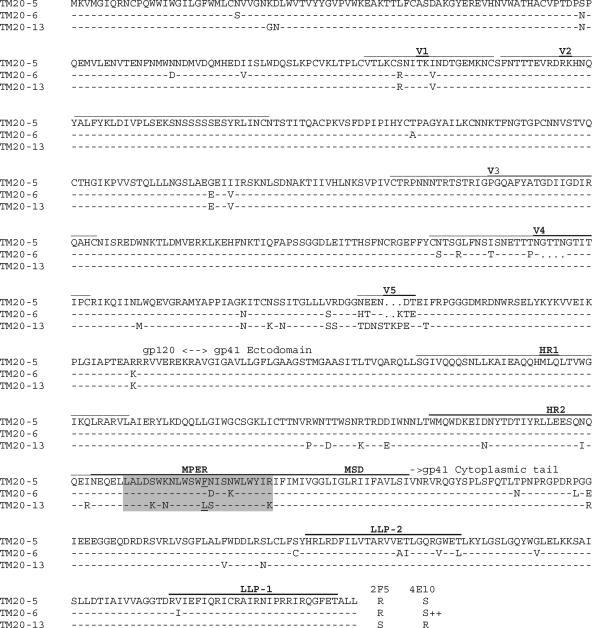
To determine the frequency of the F673L mutation in this cultured virus isolate, we analyzed the gp41 sequences of 43 molecular clones. The molecular clones were generated from three independent reverse transcription-PCRs from the viral RNA to ensure that the mutation was not introduced by PCR error. The mutation F673L was observed in 14 of the 43 (33%) molecular clones sequenced (Fig. 1). In total, six different quasispecies were identified. Interestingly, the F673L mutation was always present in conjunction with the mutation S665K in the 2F5 epitope, as well as with a group of associated mutations within the heptad repeat 2 (HR2) domain and the cytoplasmic tail of gp41 (Fig. 1 and data not shown).
FIG. 1.
Frequency analysis of substitutions in the MPER regions of 43 gp41 molecular clones obtained from the TM20 isolate. The substitutions K665S and F673L, associated with 2F5 and 4E10 resistance, respectively, are underlined and in bold. The functional envelope clones corresponding to some of these genotypes are indicated to the left of the sequences.
Generation and characterization of functional TM20 envelope clones.
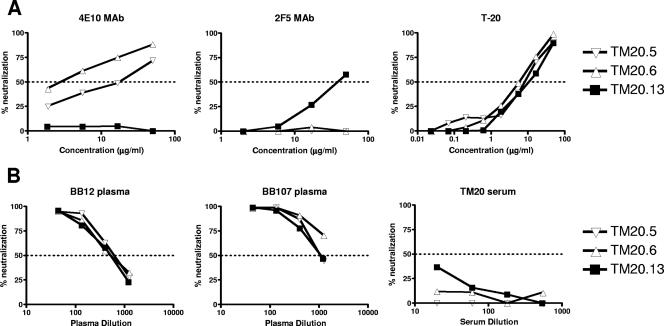
Full-length envelope genes were amplified from the TM20 isolate and cloned into an expression vector to generate functional clones. Three envelope clones were pseudotyped and characterized for their sensitivity to the broadly neutralizing MAbs 4E10 and 2F5 and the entry inhibitor T-20 (enfuvirtide, Fuzeon, DP178) (Fig. 2A). As predicted from the sequence analysis, clone TM20.13 was not neutralized by 4E10, but it was sensitive to 2F5. The other two clones were sensitive to 4E10 and resistant to 2F5, but TM20.6 (IC50 = 1.5 μg/ml) showed at least 10-fold-higher sensitivity to 4E10 than clone TM20.5 (IC50 = 17.5 μg/ml). There were no differences in the neutralization sensitivities of the three clones to T-20, which binds to the HR1 region of gp41. In addition, all clones showed similar sensitivity to the HIV-1-positive plasma samples BB12 and BB107 (Fig. 2B), suggesting that the increased sensitivity of clone TM20.6 to 4E10 was not due to a generally neutralization-sensitive phenotype. All three clones were resistant to the contemporaneous autologous serum (below 50% neutralization), which is typical in HIV-1 infection.
FIG. 2.
Neutralization of TM20 envelope clones. The three functional clones were tested for neutralization by MAbs 4E10 and 2F5 and the entry inhibitor T-20 (enfuvirtide) (A) and by polyclonal antibodies from two broadly cross-reactive HIV-1-positive plasma specimens and autologous contemporaneous sera (B). The dotted lines indicate 50% neutralization, with only those values above the line considered positive.
Sequence analysis of functional clones from TM20.
The full-length envelope genes of the three functional clones were sequenced (Fig. 3). The genetic variation observed between the clones was consistent with the 7-year duration of infection; however, the extent of variability from the C1 to C3 region of gp120 was surprisingly low. Since these clones were amplified from in vitro-grown virus, we cannot exclude the possibility that quasispecies bearing different sequences in this region of the envelope existed in vivo. Clone TM20.13 had an F673L mutation in the 4E10 epitope as well as the DKW motif intrinsic to the 2F5 epitope, which correlated with the observed phenotypes. TM20.5 and TM20.6 were identical in the gp41 ectodomain, except for two amino acids in the MPER at positions 674 and 677. Multiple other differences between clones TM20.5 and TM20.6 were observed along gp120 and the cytoplasmic tail of gp41. All three clones had conserved HR1 and V3 regions, which are involved in T-20 sensitivity (10, 27), consistent with the observed equivalent sensitivity to this compound.
FIG. 3.
Full-length amino acid sequences of the functional envelope clones TM20.5, TM20.6, and TM20.13. Variable regions, HR domains (HR1 and HR2), the MPER, the membrane-spanning domain (MSD), and LLPs (LLP-1 and LLP-2) are indicated. The mutation F673L in the MPER is underlined. The sensitivity of each of the clones to 2F5 and 4E10 neutralization is indicated, with R denoting resistance and S denoting sensitivity. TM20.6 was extremely sensitive to 4E10 neutralization and is indicated with S++.
Changes in the MPER are partially responsible for the differential sensitivity of TM20.5 and TM20.6 to 4E10.
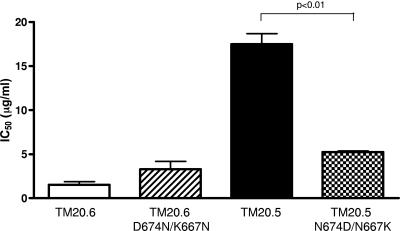
As noted above, TM20.5 was 10-fold less sensitive to 4E10 than TM20.6. These two clones differed in two amino acids in the 4E10 epitope (Fig. 3). In order to determine if these residues were associated with 4E10 sensitivity, we introduced the mutations D674N and K677N into TM20.6 by site-directed mutagenesis. These changes did not render the TM20.6 mutant more resistant to 4E10 neutralization (Fig. 4). Conversely, when the mutations N674D and N677K were introduced in TM20.5, the 4E10 IC50 dropped threefold, suggesting that in some contexts these positions may have an effect on antibody binding affinity.
FIG. 4.
Changes in positions 674 and 677 in the MPER affect 4E10 neutralization. TM20.6, TM20.5, and mutants TM20.6 D674N/K677N and TM20.5 N674D/N677K were tested for their sensitivity to neutralization by 4E10. Neutralization was scored as the antibody concentration required to reduce infectivity by 50% (IC50). The graph shows the means and standard deviations from three independent experiments. P values are indicated when statistically significant differences between the means were observed in a Mann-Whitney nonparametric t test analysis.
4E10 neutralization of envelope chimeras.
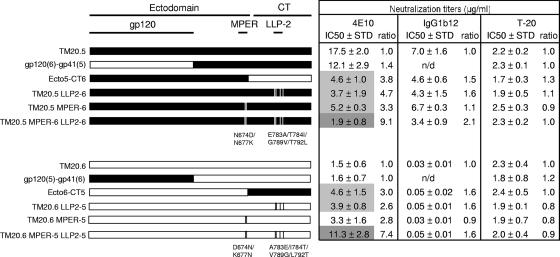
These apparently discordant results suggested that regions outside the MPER might influence the sensitivity to 4E10. We therefore constructed chimeras in which large regions of the envelope gene were interchanged between the TM20.5 and TM20.6 clones (Fig. 5). For both clones, the gp120 subunit had very little to no effect on the neutralization sensitivity, indicating that the major determinants of 4E10 sensitivity were in the gp41 region. When the cytoplasmic tail from TM20.6 was combined with the ectodomain of TM20.5, which included gp120 and the external region of gp41, the resulting chimera, Ecto5-CT6, was around fourfold more sensitive than TM20.5. Conversely, Ecto6-CT5 was threefold less sensitive than TM20.6. Altogether these results suggest that while the major determinants of 4E10 sensitivity are in the MPER, the cytoplasmic tail can have a tangible impact on the exposure of this epitope.
FIG. 5.
Changes in the cytoplasmic tail affect neutralization sensitivity. Schematic representations of the chimeras, constructed by exchanging gp120 or cytoplasmic tail segments of TM20.5 and TM20.6, and LLP-2 and MPER mutants are shown. All the constructs were tested for 4E10, IgG1b12, and T-20 neutralization. The mean IC50s from three independent experiments are indicated on the right. The IC50s of the chimeras and mutants were compared to those of the parental clone TM20.5 or TM20.6, and the IC50 ratio is shown in each case. Statistically significant differences between means of the parental and chimeric/mutant IC50s with P values of <0.05 and <0.01 in a Mann-Whitney t test are highlighted in light and dark gray, respectively.
Mutations in the LLP-2 domain affect sensitivity to neutralization.
The cytoplasmic tail sequences of clones TM20.5 and TM20.6 differed by 10 amino acid residues (Fig. 3), 4 of which were in the lentivirus lytic peptide-2 (LLP-2) domain. Frequency analysis showed that the LLP-2 sequence of TM20.6 was present in 17% of the quasispecies, while 36% had the TM20.5 LLP-2 sequence. To determine if variations in LLP-2 were responsible for the changes in 4E10 sensitivity in the cytoplasmic tail chimeras, we exchanged the LLP-2 regions of TM20.5 and TM20.6 by introducing the mutations E783A, T784I, G789V, and T792L in TM20.5 (TM20.5 LLP2-6) and the converse changes in TM20.6 (TM20.6 LLP2-5). We tested all constructs for their sensitivity to neutralization by 4E10 and T-20 as well as IgG1b12 and CD4-IgG2, which bind to gp120 and block interactions with CD4.
As observed for the chimera Ecto5-CT6, the mutant TM20.5 LLP2-6 became more sensitive to neutralization by 4E10 than the parental TM20.5 virus (Fig. 5). Similarly, these changes in the LLP-2 explained the decrease in sensitivity to 4E10 in the chimera Ecto6-CT5 compared to the parental TM20.6. Thus, in both cases, the four amino acids in the LLP-2 domain accounted for the altered changes in sensitivity to 4E10 neutralization. These changes also had a minor effect on the neutralization sensitivity for IgG1b12 (Fig. 5) and CD4-IgG2 (data not shown) but had no effect on T-20 neutralization.
In order to look at the combined effects of changes in the MPER and LLP-2, we introduced these four mutations into the two MPER mutants to form TM20.5 MPER-6 LLP2-6 and TM20.6 MPER-5 LLP2-5. For TM20.5 the resultant clone showed a ninefold increase in neutralization sensitivity to 4E10, and the IC50 titer decreased to a level similar to that obtained for the TM20.6 clone (Fig. 5). The opposite mutant, TM20.6 containing the MPER and LLP-2 of TM20.5, became sevenfold more resistant to neutralization, but the IC50 titer remained slightly lower than that for TM20.5 (11.3 versus 17.5 μg/ml), suggesting that perhaps other areas of the envelope may influence this phenotype. These double mutants showed similar sensitivities to IgG1b12 and CD4-IgG2 as the corresponding LLP-2-only mutants, corroborating that the MPER plays no role in determining sensitivity to these reagents.
Effect of gp120 and cytoplasmic tail switching on envelope incorporation and viral infectivity.
The LLP-2 domain is a highly conserved cationic, amphipathic α-helix with calmodulin-binding properties (29) that is involved in cell-to-cell fusion (17, 33), neutralization sensitivity (16), envelope cell surface expression (4), and envelope incorporation into virions and virus infectivity (24). To explore the mechanism behind the effect of the cytoplasmic tail on 4E10 neutralization sensitivity, we compared the viral infectivities and envelope incorporations of the two TM20 clones and their chimeras.
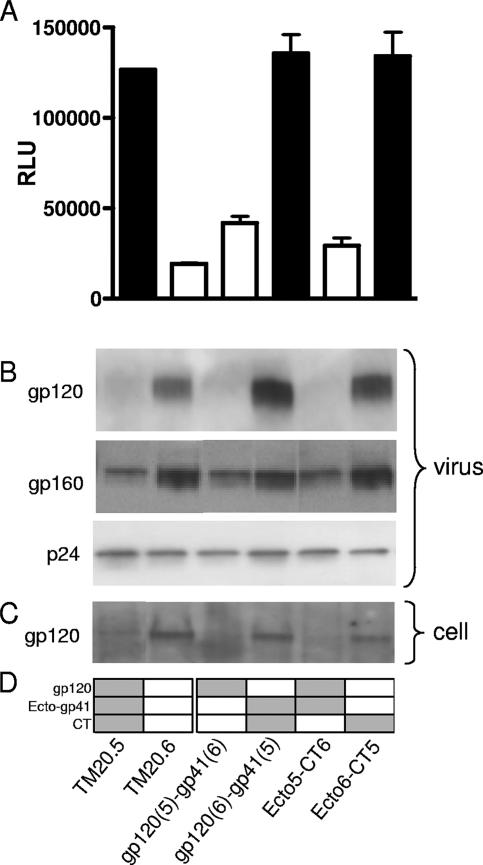
Chimeras bearing the cytoplasmic tail from TM20.6 [i.e., gp120(5)-gp41(6) and Ecto5-CT6] displayed low infectivity similar to that of the parental virus (Fig. 6A). The TM20.5 cytoplasmic tail conferred an infectivity advantage to the variants carrying it [i.e., gp120(6)-gp41(5) and Ecto6-CT5]. Furthermore, the LLP-2 mutants of TM20.5 displayed the same reduced infectivity as Ecto5-CT6, while TM20.6 LLP-2 mutants, like Ecto6-CT5, were more infectious (data not shown). This suggested that the increased infectivity conferred by the TM20.5 cytoplasmic tail was largely determined by four amino acids in LLP-2.
FIG. 6.
Roles of gp120 and the cytoplasmic tail in infectivity and envelope incorporation. (A) JC53bl-13 cells were infected with equal amounts of p24 (10 ng) of each parental and chimeric Env-pseudotyped virus. Infectivity was determined by luciferase expression measured as relative light units (RLU). The bars are color coded according to the cytoplasmic tail carried by the construct, black for TM20.5 and white for TM20.6. Error bars indicate standard deviations. (B and C) Pelleted virions (B) and env-transfected cells (C) were lysed, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and visualized by Western blotting with anti-gp120 (D7312), anti-gp41 (7B2), or anti-p24 (D7312) antibodies. (D) Schematic representation of the gp120, gp41 ectodomain, and cytoplasmic tail encoded by the chimeric constructs. Regions derived from clone TM20.5 are shaded gray, and regions derived from TM20.6 are in white.
The levels of envelope incorporation into virus particles and envelope expression were determined by Western blotting. The virus input was standardized by using equal quantities of p24 antigen, which was confirmed by p24 detection on the blots. The amount of envelope incorporated into virions was assessed by staining with the anti-gp120 antibody D7324 and the anti-gp41 MAb 7B2 to detect gp160 (Fig. 6B). In addition, envelope-transfected cells were lysed, and the levels of envelope expression were determined. Surprisingly, the less infectious TM20.6 clone showed greater envelope expression and incorporation into virions than clone TM20.5. This appeared to be determined by gp120, as all chimeras containing TM20.6 gp120 showed higher levels of envelope irrespective of the gp41 sequence. Thus, the differential neutralization sensitivity of the clones could not be explained by the levels of envelope on virus particles. These results agree with other studies showing that the LLP-2 region can affect viral infectivity without changes in envelope incorporation (17, 24).
Evaluation of anti-MPER neutralizing antibodies in TM20 serum.
The variability within the MPER among the quasispecies and the concomitant variations in 4E10 sensitivity suggested the presence of immunological pressure targeting this region. To determine if this was the case, we tested the serum of this patient for antibodies to the MPER. We measured the neutralization activity of this serum against chimeric HIV-2 viruses engrafted with complete or partial HIV-1 MPER sequences (F. Bibollet-Ruche et al., presented at the Keystone Symposium on HIV Vaccines, 2006). Using this method, we were able to detect anti-MPER antibodies in TM20 serum (Fig. 7). This serum was able to neutralize viruses that carried the full MPER sequence of either subtype B (7312A-C1) or C (7312A-C1C) or a more similar match to the autologous MPER sequence where positions 671 and 676 were mutated to serine (7312A-C1Cm). This neutralization response was mapped to the second half of the MPER as evidenced by the neutralization of C4, C4GW, and C8, but not C3 or C7. It has previously been shown that the 4E10 MAb neutralizes the chimeras C1, C4, and C6 at similar antibody concentrations (J. M. Decker et al., presented at the Keystone Symposium on HIV Vaccines, 2006). Since TM20 serum failed to neutralize C6, this suggested the absence of 4E10-like antibodies. However, it partially neutralized the C4 chimera, which, in contrast to C6, includes the W680 residue, which has been described to be important for 4E10 recognition (37). Taken together, our results suggest that the serum from this individual contained anti-MPER antibodies that recognize an epitope closely related but not identical to the 4E10 epitope.
FIG. 7.
Anti-MPER neutralization activity present in TM20 serum. The HIV-1 MPER sequences introduced into the 7321A HIV-2 chimeric or mutant viruses used in the neutralization assay are highlighted in gray. The bold letters represent the sequence of the intact 2F5 and 4E10 epitope. The mutations N671S and T676S in C1Cm and F673L in C1CF/L are underlined. The ID50 titers are indicated on the right, with those showing activity highlighted in gray.
Neutralization of C1C F673L by TM20 serum.
The mutation F673L observed in clone TM20.13 was introduced in the HIV-2/HIV-1 chimeric virus C1C by site-directed mutagenesis (C1C F/L). This single mutation conferred resistance to the 4E10 MAb relative to the sensitive C1C parental virus (data not shown). The TM20 serum was unable to neutralize the C1C F/L mutant virus (Fig. 7), suggesting that the F673L mutation observed in clone TM20.13 could indeed be an escape mutation from the anti-MPER response developed by this patient.
DISCUSSION
In the present study we describe three viral quasispecies from an HIV-1 subtype C-infected child with different sensitivities to neutralization by the broadly cross-reactive MAb 4E10. Neutralization resistance was conferred by a rare mutation, F673L, in the 4E10 epitope. In addition, moderate changes in sensitivity between clones were modulated by secondary positions in this epitope and motifs in the cytoplasmic tail. The presence of anti-MPER neutralizing antibodies in this individual supports the hypothesis that escape from antibody-mediated immune pressure was driving these changes.
MAb 4E10 has been shown to be the most broadly cross-reactive antibody against HIV-1 (3); however, it is not a particularly potent antibody, with >1 μg/ml needed to neutralize most viruses at 50%. In this study, clone TM20.13 was resistant to neutralization at up to 100 μg/ml of 4E10 due to an F673L mutation in the MPER. This is consistent with a report by Zwick and coworkers, who showed that the introduction of the F673A mutation conferred resistance to 4E10 neutralization (37). The mutation F673L was present in one-third of the quasispecies in this virus isolate. However, given that they were derived from an in vitro-cultured virus, it is not clear whether this mutation was present at a higher or lower frequency in vivo, where neutralizing antibodies were present. While L673 is common among HIV-1 group O envelope sequences, this substitution is very rare among HIV-1 group M viruses in the Los Alamos HIV sequence database. The highly conserved nature of F673 suggests that this site either performs an important function or is not accessible to immune attack, as has been proposed previously (36). However, we found that virus infectivity was not obviously compromised by F673L (data not shown), similar to what was observed for the F673A mutant (37). This raises the question as to why this position is so highly conserved and under such strong negative selection pressure. Interestingly, in our study the F673L mutation was linked with the mutation S665K, which confers 2F5 sensitivity. All other clones were resistant to 2F5, due to the absence of a K at position 665, which is typical of subtype C viruses (3, 14). Furthermore, these mutations were accompanied in all cases by other mutations in gp41. HIV-1 is known to undergo considerable recombination during in vivo and in vitro proliferation (18), so the link between these changes may simply be the result of a recombination event between two divergent viral quasispecies. However, they could also represent compensatory changes associated with the mutation F673L that are required by the virus for survival in vivo. A recent study on in vitro escape from 4E10 neutralization showed the appearance of F673L/V mutations in the resistant virus, but these viruses had impaired infectivity (19), which may explain why 4E10 escape variants were not observed in passive immunization studies with this antibody (22, 31). However, these viruses had no other genetic changes in this region, such as those shown here, that might have compensated for this loss of function (19).
TM20.5 and TM20.6 showed a 10-fold difference in sensitivity to 4E10. These clones were identical in the ectodomain of gp41 except for two positions (674 and 677) within the 4E10-binding domain. Previous studies have shown that these two residues are dispensable for 4E10 recognition (7, 37). However, we observed that the introduction of mutations N674D/N677K into TM20.5 increased 4E10 sensitivity by threefold, suggesting that these positions can influence the presentation of the 4E10 epitope. On the other hand, the TM20.6 clone did not become more resistant to 4E10 when these two positions were changed. Therefore, TM20.6 D674N/K677N, which had an MPER identical to that of TM20.5, showed a fivefold difference in neutralization sensitivity to 4E10. This was also noted in another study where viruses with identical 4E10 epitopes had different IC50s (3), suggesting that factors outside the MPER affect accessibility of this epitope. Indeed, substitutions in the HR1 region of gp41 as well as other factors that affect fusion kinetics have been shown to influence sensitivity to this MAb (26, 37). Thus, the observation that TM20.5 showed enhanced infectivity compared to TM20.6 may explain why only TM20.5 was sensitive to changes at positions 674 and 677 in the MPER, possibly as a result of a limited window of opportunity for neutralization.
We demonstrated that the cytoplasmic tail can modulate sensitivity to neutralization, in agreement with other studies (12, 16, 32, 34). Furthermore, we were able to identify four amino acids in the LLP-2 domain that affect both neutralization and infectivity. Such changes could have an impact on the amphipathicity of the LLP-2 α-helix and therefore its membrane association, resulting in phenotypic differences, as observed here and by others (16, 24). It has been shown that deletion of the cytoplasmic tail or changes in this region, in particular the LLP-2, determine the fusogenicity of the envelope glycoprotein (1, 17, 24, 33), which could result in changes in sensitivity to some entry inhibitors (1) and MAbs (16). Abrahamyan and coworkers suggested that the cytoplasmic tail hinders the folding of the trimeric coiled coil into the six-helix bundle, resulting in greater inhibition by peptides that target the fusion intermediates (1). The fact that we did not observe changes in sensitivity to T-20, which also targets this conformation, suggests that the mutations in the LLP-2 did not influence this stage of the fusion process. On the other hand, early events may have been affected, such as the differential exposure of neutralizing epitopes in the native structure or, more likely, a higher kinetic rate from the CD4-bound conformation to the trimeric coiled coil. This may preferentially affect 4E10 neutralization, as this antibody is capable of binding in the postattachment stage (2). In agreement with this, we found that 4E10 neutralization was more affected by changes in the LLP-2 than neutralization by IgG1b12 and CD4-IgG2.
Sensitivity to 4E10 neutralization was due to the combined effects of motifs in the MPER and LLP-2. The two amino acid changes in the MPER and the four amino acid changes in LLP-2 were sufficient to confer the 4E10-sensitive phenotype to TM20.5. However, engineering the more resistant phenotype appeared to require other sites in addition to these six amino acids. The fact that the TM20.6 MPER-5 LLP2-5 mutant showed the same sensitivity to 4E10 as the gp120(6)-gp41(5) construct, neither of which reached IC50 levels shown by TM20.5, suggested that perhaps sites in gp120 were also involved (Fig. 5). Interestingly, the two mutations in the TM20.6 MPER, which failed to show any effect on their own, appeared to contribute to 4E10 resistance when coexpressed with the four LLP-2 mutations that increased infectivity, again highlighting the role that infectivity plays in determining overall neutralization sensitivity.
The presence of an anti-MPER neutralization response in this patient's serum supports the hypothesis that the observed 4E10-resistant envelope quasispecies constituted escape variants. Though the antibodies elicited against the MPER in this patient were not clearly 4E10-like as evidenced by the inability to neutralize C6, their epitopes overlapped, and as for 4E10, residues F673 and W680 were important for recognition. A recent study with the antibody Z13e1 showed that the mutation D674N, as observed in TM20.5, eliminates antibody binding (23). Therefore, it is also possible that antibodies similar to Z13, where the amino acid at position 674 constitutes a critical residue, might be involved in this anti-MPER neutralization response.
This study strongly suggests that the MPER is indeed immunogenic and accessible to antibodies that can drive the evolution of the virus toward escape variants. However, such an argument is demonstrable only in the context of a longitudinal study where the evolution of changes in the envelope glycoprotein can be tracked over time and in parallel to the development of the neutralization response. Considering the interest in the MPER as a vaccine target, it is important that the potential of in vivo escape from anti-MPER antibodies be clarified in such longitudinal studies.
Acknowledgments
We thank Maria Salazar from the University of Alabama at Birmingham for her help with the sequencing of the HIV-2 mutants and Isaac Choge for his technical support. We also thank James Robinson for providing MAb 7B2 and Dennis Burton and Ralph Pantophlet from the Neutralizing Antibody Consortium (NAC) of the International AIDS Vaccine Initiative (IAVI) for providing MAbs 2F5 and 4E10 and IgG1b12. Progenics Pharmaceuticals, Inc., and Roche Palo Alto kindly provided the CD4-IgG2 and enfuvirtide, respectively.
This work was funded by the South African AIDS Vaccine Initiative (SAAVI), the Center for HIV-AIDS Vaccine Immunology (CHAVI), and the Southern African Fogarty AIDS Training Program (TWO-02).
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Abrahamyan, L. G., S. R. Mkrtchyan, J. Binley, M. Lu, G. B. Melikyan, and F. S. Cohen. 2005. The cytoplasmic tail slows the folding of human immunodeficiency virus type 1 Env from a late prebundle configuration into the six-helix bundle. J. Virol. 79106-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binley, J. M., C. S. Cayanan, C. Wiley, N. Schulke, W. C. Olson, and D. R. Burton. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 775678-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 7813232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bultmann, A., W. Muranyi, B. Seed, and J. Haas. 2001. Identification of two sequences in the cytoplasmic tail of the human immunodeficiency virus type 1 envelope glycoprotein that inhibit cell surface expression. J. Virol. 755263-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, J., L. Bergeron, E. Helseth, M. Thali, H. Repke, and J. Sodroski. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J. Virol. 672747-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardoso, R. M., F. M. Brunel, S. Ferguson, M. Zwick, D. R. Burton, P. E. Dawson, and I. A. Wilson. 2007. Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10. J. Mol. Biol. 3651533-1544. [DOI] [PubMed] [Google Scholar]
- 7.Cardoso, R. M., M. B. Zwick, R. L. Stanfield, R. Kunert, J. M. Binley, H. Katinger, D. R. Burton, and I. A. Wilson. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22163-173. [DOI] [PubMed] [Google Scholar]
- 8.Choge, I., T. Cilliers, P. Walker, N. Taylor, M. Phoswa, T. Meyers, J. Viljoen, A. Violari, G. Gray, P. L. Moore, M. Papathanosopoulos, and L. Morris. 2006. Genotypic and phenotypic characterization of viral isolates from HIV-1 subtype C-infected children with slow and rapid disease progression. AIDS Res. Hum. Retroviruses 22458-465. [DOI] [PubMed] [Google Scholar]
- 9.Cilliers, T., P. Moore, M. Coetzer, and L. Morris. 2005. In vitro generation of HIV type 1 subtype C isolates resistant to enfuvirtide. AIDS Res. Hum. Retroviruses 21776-783. [DOI] [PubMed] [Google Scholar]
- 10.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 748358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimitrov, A. S., S. S. Rawat, S. Jiang, and R. Blumenthal. 2003. Role of the fusion peptide and membrane-proximal domain in HIV-1 envelope glycoprotein-mediated membrane fusion. Biochemistry 4214150-14158. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hoxie, R. W. Doms, and F. Baribaud. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 762683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, F., S. G. Morrison, D. L. Robertson, C. L. Thornton, S. Craig, G. Karlsson, J. Sodroski, M. Morgado, B. Galvao-Castro, H. von Briesen, S. Beddows, J. Weber, P. M. Sharp, G. M. Shaw, B. H. Hahn, et al. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J. Virol. 701651-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray, E. S., T. Meyers, G. Gray, D. C. Montefiori, and L. Morris. 2006. Insensitivity of paediatric HIV-1 subtype C viruses to broadly neutralising monoclonal antibodies raised against subtype B. PLoS Med. 3e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray, E. S., P. L. Moore, I. A. Choge, J. M. Decker, F. Bibollet-Ruche, H. Li, N. Leseka, F. Treurnicht, K. Mlisana, G. M. Shaw, S. S. Abdool Karim, C. Williamson, and L. Morris. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 816187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalia, V., S. Sarkar, P. Gupta, and R. C. Montelaro. 2005. Antibody neutralization escape mediated by point mutations in the intracytoplasmic tail of human immunodeficiency virus type 1 gp41. J. Virol. 792097-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalia, V., S. Sarkar, P. Gupta, and R. C. Montelaro. 2003. Rational site-directed mutations of the LLP-1 and LLP-2 lentivirus lytic peptide domains in the intracytoplasmic tail of human immunodeficiency virus type 1 gp41 indicate common functions in cell-cell fusion but distinct roles in virion envelope incorporation. J. Virol. 773634-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy, D. N., G. M. Aldrovandi, O. Kutsch, and G. M. Shaw. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. USA 1014204-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manrique, A., P. Rusert, B. Joos, M. Fischer, H. Kuster, C. Leemann, B. Niederost, R. Weber, G. Stiegler, H. Katinger, H. F. Gunthard, and A. Trkola. 2007. In vivo and in vitro escape from neutralizing antibodies 2G12, 2F5, and 4E10. J. Virol. 818793-8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montefiori, D. C. 2004. Evaluating neutralizing antibodies againts HIV, SIV and SHIV in luciferase reporter gene assays, p. p12.11.1-p12.11.15. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, W. Strober, and R. Coico (ed.), Current protocols in immunology. John Wiley & Sons, New York, NY. [DOI] [PubMed]
- 21.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 676642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakowitsch, S., H. Quendler, H. Fekete, R. Kunert, H. Katinger, and G. Stiegler. 2005. HIV-1 mutants escaping neutralization by the human antibodies 2F5, 2G12, and 4E10: in vitro experiments versus clinical studies. AIDS 191957-1966. [DOI] [PubMed] [Google Scholar]
- 23.Nelson, J. D., F. M. Brunel, R. Jensen, E. T. Crooks, R. M. Cardoso, M. Wang, A. Hessell, I. A. Wilson, J. M. Binley, P. E. Dawson, D. R. Burton, and M. B. Zwick. 2007. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J. Virol. 814033-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman, J. T., T. J. Sturgeon, P. Gupta, and R. C. Montelaro. 2007. Differential functional phenotypes of two primary HIV-1 strains resulting from homologous point mutations in the LLP domains of the envelope gp41 intracytoplasmic domain. Virology 367102-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ofek, G., M. Tang, A. Sambor, H. Katinger, J. R. Mascola, R. Wyatt, and P. D. Kwong. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 7810724-10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reeves, J. D., F. H. Lee, J. L. Miamidian, C. B. Jabara, M. M. Juntilla, and R. W. Doms. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 794991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 732469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srinivas, S. K., R. V. Srinivas, G. M. Anantharamaiah, R. W. Compans, and J. P. Segrest. 1993. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J. Biol. Chem. 26822895-22899. [PubMed] [Google Scholar]
- 30.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 171757-1765. [DOI] [PubMed] [Google Scholar]
- 31.Trkola, A., H. Kuster, P. Rusert, B. Joos, M. Fischer, C. Leemann, A. Manrique, M. Huber, M. Rehr, A. Oxenius, R. Weber, G. Stiegler, B. Vcelar, H. Katinger, L. Aceto, and H. F. Gunthard. 2005. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat. Med. 11615-622. [DOI] [PubMed] [Google Scholar]
- 32.Vzorov, A. N., K. M. Gernert, and R. W. Compans. 2005. Multiple domains of the SIV Env protein determine virus replication efficiency and neutralization sensitivity. Virology 33289-101. [DOI] [PubMed] [Google Scholar]
- 33.Wyss, S., A. S. Dimitrov, F. Baribaud, T. G. Edwards, R. Blumenthal, and J. A. Hoxie. 2005. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J. Virol. 7912231-12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuste, E., W. Johnson, G. N. Pavlakis, and R. C. Desrosiers. 2005. Virion envelope content, infectivity, and neutralization sensitivity of simian immunodeficiency virus. J. Virol. 7912455-12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuste, E., H. B. Sanford, J. Carmody, J. Bixby, S. Little, M. B. Zwick, T. Greenough, D. R. Burton, D. D. Richman, R. C. Desrosiers, and W. E. Johnson. 2006. Simian immunodeficiency virus engrafted with human immunodeficiency virus type 1 (HIV-1)-specific epitopes: replication, neutralization, and survey of HIV-1-positive plasma. J. Virol. 803030-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zwick, M. B. 2005. The membrane-proximal external region of HIV-1 gp41: a vaccine target worth exploring. AIDS 191725-1737. [DOI] [PubMed] [Google Scholar]
- 37.Zwick, M. B., R. Jensen, S. Church, M. Wang, G. Stiegler, R. Kunert, H. Katinger, and D. R. Burton. 2005. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol. 791252-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 7510892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]