Abstract
The transmembrane subunit (TM) of the trimeric retrovirus Env complex is thought to direct virus-cell membrane fusion by refolding into a cell membrane-interacting, extended form that subsequently folds back on itself into a very stable trimer of hairpin-like TM polypeptides. However, so far there is only limited evidence for the formation of a stable TM trimer during Env activation. Here we have studied the oligomer composition and stability of an intermediate and the fully activated form of Moloney murine leukemia virus (Mo-MLV) Env. Activation of Mo-MLV Env is controlled by isomerization of its intersubunit disulfide. This results in surface subunit (SU) dissociation and TM refolding. If activation is done in the presence of an alkylator, this will modify the isomerization-active thiol in the SU of Env and arrest Env at an intermediate stage, the isomerization-arrested state (IAS) of its activation pathway. We generated IAS and fully activated Envs in vitro and in vivo and studied their states of oligomerization by two-dimensional blue native polyacrylamide gel electrophoresis (PAGE) and nonreducing sodium dodecyl sulfate (SDS)-PAGE. The IAS Env was composed of trimers of SU-TM complexes, whereas the activated Env consisted of SU monomers and TM trimers. When the oligomers were subjected to mild SDS treatment the TM trimer was found to be 3.5 times more resistant than the IAS oligomer. Thus, this demonstrates that a structural conversion of TM takes place during activation, which results in the formation of a stable TM trimer.
Viral membrane fusion proteins such as the influenza virus hemagglutinin (HA) and retrovirus envelope protein (Env) are made as trimers of a type I transmembrane protein in the endoplasmic reticulum of the infected cell (13, 38, 39). They become potentiated for activation by limited proteolytic processing of their ectodomain by furin when they pass the Golgi complex on their way to the site of virus budding in the cell (5, 11, 17, 24, 28, 32, 37). The cleavage generates a surface subunit (SU) and a transmembrane subunit (TM) of each fusion protein precursor. The SUs carry the receptor binding activity and the TMs the fusion activity. Studies on HA have shown that the latter activity is expressed by refolding of the TMs (30). Analysis of the native HA crystal structure demonstrated that the TM (HA2) subunits had a long central α-helix with which they formed a triple-stranded coiled-coil core in the fusion protein (38). The N-terminal fusion peptide of HA2 was buried in a cavity on the surface of the HA stem and linked by a shorter α-helix and a loop to the N terminus of the long helix. Analysis of the crystal structure of a proteolytic product of activated HA showed that it consisted of a trimer of HA2 ectodomains devoid of the fusion peptide. The short helix and the loop, which connected the fusion peptide to the central helix of native HA2, extended the latter at its N-terminal end (1). Furthermore, in the middle of the central helix of native HA2 a loop had been formed that allowed chain reversal and packing of the C-terminal region of the HA2 polypeptide into the grooves of the coiled coil in an antiparallel orientation. From these data a model was created for how the HA2 subunits mediate membrane fusion. According to this, the HA2 subunits first extend the central helix. This will expel the fusion peptide on the top of the molecule, where it can interact with the cell membrane. At a second stage, the HA2 subunits make the back folding reaction. This will orient the C-terminal membrane anchors of the HA2 subunits into the same direction as their N-terminal fusion peptides and thereby facilitate approximation and fusion of the viral and the cell membranes.
The HA2 activation is suppressed in native HA by the associated SU HA1. Triggering follows when the HA1 subunit is displaced by the effects that the low pH has on the HA structure in endocytosed virus (10, 15). HA2 has been proposed to be arrested in its kinetic folding pathway in the native HA (2). Acid triggering overcomes the kinetic block and allows HA2 to fold into a stable conformation, e.g., the trimer of hairpin-like HA2 polypeptides. This has been supported by the increased thermostability of the activated TM trimer compared to native HA, the ability to trigger HA under neutral conditions by unspecific protein perturbation treatments such as heat and urea incubations, and the fact that recombinant HA2 ectodomain, produced in the absence of the HA1 subunit, folds into the low-pH form (2, 4, 26).
The Env of retrovirus is thought to control the membrane fusion by using its fusion protein SU and TM subunits in a similar way as influenza virus. This is corroborated by several findings. First, although there is no atomic structure of the native Env, analyses of crystals of ectodomain fragments of the retrovirus TM protein have demonstrated a similar thermostable complex of three hairpin-like polypeptide structures, as is found in the low-pH form of HA2 (3, 6, 16). Second, TM peptides corresponding to the interacting regions in the TM hairpin can effectively inhibit Env-mediated membrane fusion, suggesting a prehairpin-to-hairpin conversion of TM during Env activation (9, 14, 18, 22, 35). Third, Env, which normally is activated by receptor binding or by the receptor and subsequent acid treatment in the endosome, can also be triggered by nonspecific protein perturbation (31, 33). This suggests that TM is metastable in native Env but folds into a stable trimeric complex as a result of activation. However, so far there is only limited evidence for the actual formation of a stable TM trimer during the activation of the complete Env complex. Indeed, there is only one report showing that the TM of the avian leukosis virus (ALV) Env converts into a trimer that resists sodium dodecyl sulfate (SDS) at up to 60°C upon activation (21). Here we have studied the stability of the Env trimer of Moloney murine leukemia virus (Mo-MLV).
Mo-MLV Env activation is controlled by isomerization of the intersubunit disulfide (25, 34). This is linked to a C-X-X-C motif in SU, where the other Cys carries a free thiol. Upon receptor binding, the latter becomes activated to attack the intersubunit disulfide and rearrange it into a disulfide isomer within the motif. This results in dissociation of the SU, refolding of TM, and membrane fusion. However, activation in the presence of an alkylator will, after initial receptor-induced changes in Env, modify the thiol before it has time to attack the intersubunit disulfide. The Env will now be blocked in isomerization and arrested at an intermediate stage of its activation process (isomerization-arrested state [IAS]) (34, 36). However, the fusion activity of Env can be rescued by reducing the intersubunit disulfide with dithiothreitol (DTT). The native state of Env is stabilized by Ca2+ ions, and receptor-induced isomerization involves removal of the suppressing Ca2+ ions from Env (34). Consequently, isomerization, i.e., Env activation, can also be induced in vitro by Ca2+ depletion and unspecific protein destabilization such as incubation in Ca2+-free medium, lysis with a buffer containing a nonionic detergent and EDTA, SDS, or treatment with urea or heat.
In the present work we activated Mo-MLV Env using solubilization of virus particles with mild detergent and EDTA in the presence or absence of alkylator to generate Env in the IAS or fully activated Env and determined their oligomeric state and stability toward dissociation by a low concentration of SDS. The native Env trimer could not be analyzed in this assay because SDS is a potent inducer of the disulfide isomerization reaction. We found that Env in the IAS was composed of SU-TM trimers and the fully activated form of TM trimers. Significantly, the TM trimers were 3.5 times more resistant toward dissociation than the SU-TM trimers of the IAS form of Env. This suggests that the TM of Mo-MLV refolds during Env activation into a form that supports stable trimerization.
MATERIALS AND METHODS
Cells and virus.
XC (ATCC CCL-165, LGC Promochem, Borås, Sweden) and MOV-3 (G. Schmidt, GSF-National Research Center for Environment and Health, Neuherberg, Germany) cells were maintained in Dulbecco's modified Eagle's medium (GIBCO BRL) (1 g/liter glucose for XC cells and 4.5 g/liter glucose for MOV-3 cells) supplemented with 10% fetal calf serum, 20 mM HEPES, and l-glutamine (growth medium). [35S]Cys-labeled wild-type Mo-MLV was prepared in MOV-3 cells by overnight labeling with 90 μCi/ml [35S]Cys (GE Healthcare Biosciences, Buckinghamshire, United Kingdom) in the presence of 25 μM unlabeled cysteine as described previously (23). Virus in culture supernatant was purified by sedimentation in a step gradient of 1 ml 50% (wt/wt) and 4.5 ml 20% (wt/wt) sucrose in 50 mM HEPES, 100 mM NaCl (pH 7.4) (HN buffer) containing 1.8 mM CaCl2 for 1 h at a relative centrifugal force at maximum radius of 2.2 × 105 × g (26 krpm) and 4°C in a Beckman SW 41 rotor.
In vitro activation of MLV-Env, generation of IAS, DTT treatment, and cross-linking.
Gradient-purified [35S]Cys-labeled Mo-MLV was activated in HN buffer containing one of the following combinations: 0.15% Triton X-100 and 9 mM EDTA, 0.15% Triton X-100 and 1.8 mM CaCl2, 9 mM EDTA only, 1.8 mM CaCl2 only, 36 mM octyl-β-d-glucopyranoside (octylglucoside [OG]) and 9 mM EDTA, or 36 mM OG only. Incubation was for 0, 15, 30, 60, 90, or 180 min at 37°C. At the indicated time 20 mM N-ethylmaleimide (NEM) was added and the incubation continued up to 180 min at 37°C. Env blocked at the IAS was generated by including 20 mM NEM prior to activation incubation at 37°C. In order to reduce the intrasubunit disulfide bond of the TM subunit, TM trimers were generated from [35S]Cys-labeled Mo-MLV by incubation in HN buffer containing 0.15% Triton X-100 and 9 mM EDTA for 40 min at 37°C, and then 33 mM DTT was added, the incubation was continued for 20 min at 37°C, and finally 80 mM NEM was added. Cross-linking of activated Env and Env in the IAS was performed on [35S]Cys-labeled Mo-MLV that had been incubated for 20 min at 37°C in HN containing 36 mM OG and 9 mM EDTA without and with 20 mM NEM, respectively. Dithiobis(succinimidyl)propionate (DSP) (Pierce Co., Rockford, IL) in the range from 0 to 800 μM was added and the samples incubated for 30 min at room temperature. Excessive reagent was blocked by addition of 45 mM glycine and 5 min at room temperature. The oligomeric state of the proteins was analyzed, without further purification, by blue native polyacrylamide gel electrophoresis (BN-PAGE) in the absence or presence of an excess of SDS and by nonreducing SDS-PAGE.
Receptor-induced isomerization.
Virus was bound to receptor-carrying XC cells grown on 24-well dishes (BD Falcon, Franklin Lakes, NJ) by spin inoculation. Briefly, the cells were washed once in phosphate-buffered saline (PBS), gradient-purified [35S]Cys-labeled Mo-MLV diluted in growth medium supplemented with 8 μg/ml Polybrene was added (250 μl/well), and the plates were centrifuged at 4°C for 1 h at a relative centrifugal force at maximum radius of 1.3 × 103 × g (2,900 rpm) in a Beckman JS 5.9 rotor. The cells were washed once in PBS, 1 ml growth medium was added, and the cells were incubated for 30 min at 37°C to allow receptor-induced activation of the Env complex. The cells were washed twice in ice-cold PBS and extracted with a buffer containing 0.15% Triton X-100, 0.75 M 6-amino-hexanoic acid, 9 mM EDTA, and 10 mM NEM for 10 min at room temperature (100 μl/well). The cells were then transferred to an Eppendorf tube, insoluble matter was removed by low-speed centrifugation, and the supernatant was incubated for 10 min at 37°C.
Limited SDS treatment of Env protein complexes.
Aliquots of virus-XC cell extracts, in vitro-activated Env (with or without DTT treatment), or Env in the IAS were incubated in the presence or absence of SDS (1.5 mol SDS/mol Triton X-100) for 10 min at room temperature. The samples were then split in two and prepared directly for BN-PAGE and nonreducing SDS-PAGE.
Electrophoresis and quantitations.
BN-PAGE was performed essentially as described previously (20, 27). Solubilized samples were mixed with an equal volume of 2× BN sample buffer containing 100 mM morpholinepropanesulfonic acid (MOPS), 100 mM Tris-HCl, 40% glycerol, and 0.1% Serva blue G (pH 7.7) and incubated at room temperature for 10 min. The indicated samples from the cross-linking experiments were supplemented with 12 g/liter SDS and incubated for 3 min at 70°C. All samples were separated on 4.5 to 16% acrylamide (of which 2.6% comprised bisacrylamide) gradient gels containing 50 mM bis-Tris (pH 7.0) and 0.5 M 6-amino-hexanoic acid at 4°C min at 200 V for 80 with 50 mM MOPS-50 mM Tris-HCl (pH 7.7) containing 0.002% Serva blue G as the cathode buffer and the same buffer without Serva blue G as the anode buffer. When cell extracts or SDS-containing samples were analyzed, 0.01% Serva blue G was used in the cathode buffer. 14C-methylated molecular mass marker proteins (CFA 626) and unlabeled standard proteins (HMW-native) from GE Healthcare Biosciences (Buckinghamshire, United Kingdom) were used. For two-dimensional (2D) BN-PAGE/nonreducing SDS-PAGE a sample was separated on BN-PAGE (0.75-mm by 10-cm by 8-cm gel), and the middle part of the lane was cut out and incubated first in 1 ml HN buffer containing 0.1 M NEM for 30 min on ice and then in 1 ml sample buffer containing 120 g/liter SDS, 0.19 M Tris-HCl (pH 8.0), 93 g/liter sucrose, 14 mM EDTA, 0.6 g/liter bromophenol blue, 0.4 g/liter methionine, and 20 mM NEM for 30 min at room temperature. The gel strip was then fitted on an SDS-polyacrylamide gel (1 mm by 10 cm by 10.5 cm) (13% acrylamide, of which 2.6% comprised bisacrylamide) and electrophoresed under nonreducing conditions. All gels were dried and exposed to phosphorimager screens (BAS-MS2025; Fujifilm, Science Imaging Scandinavia, Nacka, Sweden) and the labeled proteins visualized (and quantitated) using a Molecular Imager FX and the QuantityOne program from Bio-Rad Laboratories (Hercules, CA). The extent of Env isomerization was estimated as described previously (29). The percentage of oligomeric TM was calculated as TMoligo = (TMtri + TM2xtri)/(TMmono + TMtri + TM2xtri) × 100 in the presence (TMoligo+SDS) or absence (TMoligo-SDS) of SDS (see Fig. 4 for identities of spots). The percentage of oligomeric SU-TM was calculated in the corresponding way. The difference in SDS sensitivity between the two complexes (S) was estimated as S = (SU-TMoligo−SDS − SU-TMoligo+SDS)/(TMoligo−SDS − TMoligo+SDS).
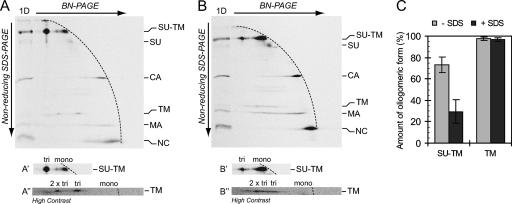
FIG. 4.
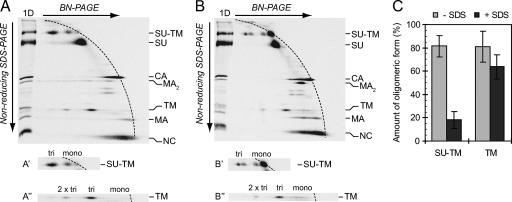
The TM trimer of activated Env is more stable than the trimer of IAS Env. [35S]Cys-labeled Mo-MLV was lysed in Triton X-100-EDTA HN buffer for 20 min at 37°C to partially activate Env. NEM was added, and the remaining native Env was chased into the IAS form. One half of the sample was subjected to mild treatment with SDS and the other half left untreated. The samples were analyzed by 2D BN-PAGE/nonreducing SDS-PAGE. Shown are phosphorimages of the gel analyses of untreated (A) and SDS-treated (B) samples. Viral proteins and their oligomeric state in the first dimension (BN-PAGE) are indicated to the right. (A′, A″, B′, and B″) Cutouts with the oligomeric state of the proteins in the first dimension indicated. (C) Relative amounts of the trimeric forms of the SU-TM complexes and the TM subunits in the two samples as percentages of the total SU-TM and TM, respectively (mean ± standard deviation; n = 6).
RESULTS
Lysis-induced SU-TM disulfide isomerization.
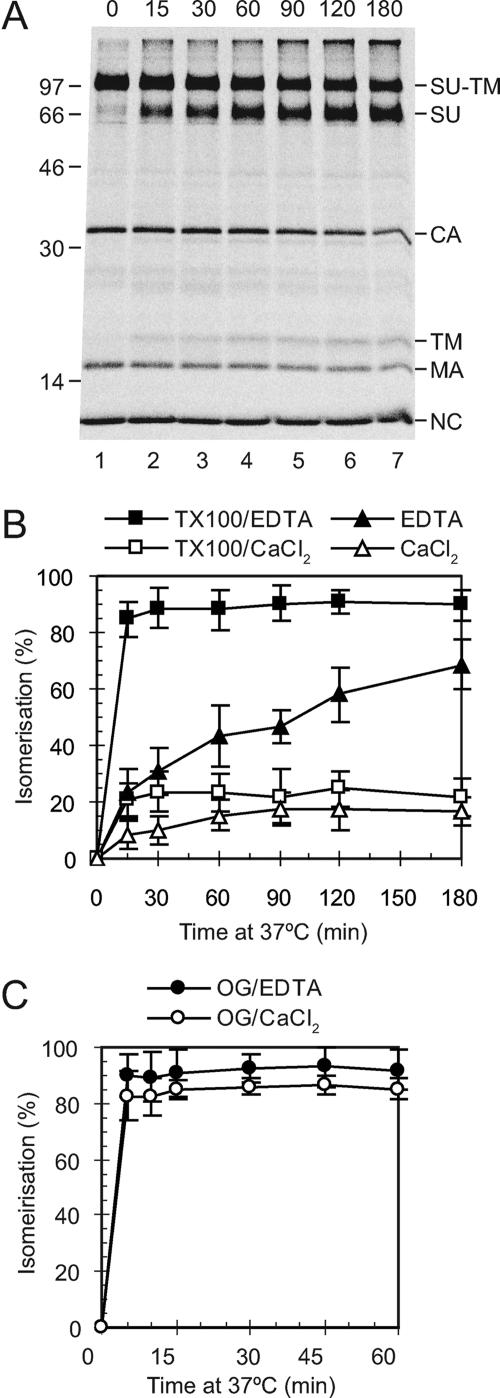
We chose to activate viral Env in our in vitro experiments by membrane lysis. We first studied the separate effects of Triton X-100 and EDTA in HN buffer on the intersubunit disulfide isomerization reaction. For this purpose, [35S]Cys-labeled Mo-MLV was prepared in MOV-3 cells and purified by ultracentrifugation in a sucrose step gradient. The virus was then incubated in HN buffer with either Triton X-100 and EDTA, Triton X-100 and Ca2+, or EDTA only for 0 to 180 min at 37°C. As a control we used incubation in HN buffer containing Ca2+ (Fig. 1). At the indicated times, further isomerization was blocked by adding NEM and all samples incubated for a total of 180 min. The viral proteins were analyzed directly, i.e., without immunoisolation, by nonreducing SDS-PAGE. This revealed the disulfide isomerization as SU and TM subunits released from the covalently linked SU-TM complexes (Fig. 1A). The analysis showed that the EDTA treatment was much more effective at causing isomerization than Triton X-100 (Fig. 1B). Indeed, incubation with the nonionic detergent induced only slightly more isomerization than incubation in the control buffer. However, the greatest effect was obtained by combining Triton X-100 and EDTA in the HN buffer. A similar pattern was found for the Triton X-100-related detergent Nonidet P-40 (data not shown). We also tested OG, an unrelated mild detergent, in the HN buffer. This was shown to be much more efficient than Triton X-100 and caused efficient isomerization even in the absence of EDTA (Fig. 1C). We concluded that mild detergents differed significantly in their effects on Env activation. In subsequent in vitro experiments we activated Env using HN buffer containing EDTA and either Triton X-100 or OG.
FIG. 1.
In vitro activation of SU-TM disulfide isomerization. [35S]Cys-labeled Mo-MLV was incubated in HN buffer containing either Triton X-100 and EDTA, Triton X-100 and Ca2+, EDTA, OG and EDTA, or OG and Ca2+ for 0 to 180 min at 37°C. A control incubation was done in HN buffer containing Ca2+. NEM was added, all samples were incubated for a total of 180 min, and viral proteins were analyzed by nonreducing SDS-PAGE. Shown are a phosphorimage of a gel with samples incubated in HN buffer with Triton X-100 and EDTA (A) and quantifications of the isomerization efficiencies under all conditions tested (B and C). In the gel analyses SU-TM complexes and free SU and TM subunits are indicated together with other viral proteins to the right and molecular weight standards to the left. The isomerization efficiencies (±standard deviations; n = 6) are given as percentages of complete isomerization.
Subunit structures of IAS and activated Envs.
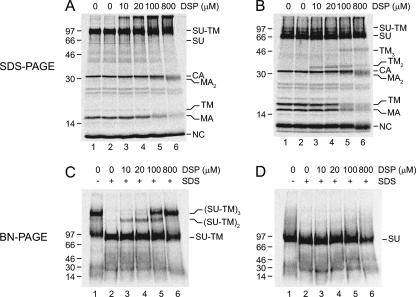
The oligomeric structures of IAS and activated Envs were compared by BN-PAGE. In contrast to SDS-PAGE, BN-PAGE preserves many noncovalent interactions between subunits in oligomeric complexes (27). Thus, [35S]Cys-labeled Mo-MLV was lysed in OG-EDTA HN buffer in the presence or absence of NEM for 20 min at 37°C to generate IAS and activated Envs. The samples were then analyzed by BN-PAGE and nonreducing SDS-PAGE. The latter analyses showed the isomerization-arrested SU-TM complexes of IAS Env (Fig. 2A, lanes 1 and 2) and the isomerization-released SU and TM subunits of the activated Env (Fig. 2B, lanes 1 and 2). In the BN-PAGE analysis, most of the IAS Env migrated as a large complex and the rest as apparent SU-TM complex monomers (Fig. 2C, lane 1). The corresponding analysis of activated Env revealed the SU as monomers, whereas the TM could not be identified (Fig. 2D, lane 1). To further characterize the oligomeric complexes, we cross-linked the lysed samples with increasing concentrations of DSP and subsequently subjected them to SDS treatment. SDS treatment of un-cross-linked IAS Env lysate dissociated all large SU-TM oligomers seen in the BN-PAGE into apparent SU-TM complex monomers (Fig. 2C, lane 2). These migrated slightly faster than the monomers of the corresponding sample that had not been treated with SDS (Fig. 2C, lane 1). Upon cross-linking with increasing DSP concentrations, the SU-TM complex was found as increasing amounts of apparent dimers and trimers (Fig. 2C, lanes 3 to 6). This was reflected by the formation of very slowly migrating material in the nonreducing SDS-PAGE (Fig. 2A, lanes 3 to 6). The BN-PAGE analysis of the cross-linked and SDS-treated activated Env lysate showed only SU monomers with slightly increased mobility (Fig. 2D, lanes 3 to 6). However, the nonreducing SDS-PAGE analysis resolved the TM subunit as dimers and trimers with increasing cross-linking (Fig. 2B, lanes 3 to 6). We concluded that IAS Env was a trimer of the SU-TM complex and that the activated Env dissociated into SU monomers and a trimer of TM. The nonreducing SDS-PAGE analyses of the cross-linked samples also showed that the viral matrix (MA) protein was present as apparent dimers in virus lysed with or without NEM (Fig. 2A and B, lanes 5 and 6). This was confirmed by 2D nonreducing SDS-PAGE/reducing SDS-PAGE (data not shown).
FIG. 2.
Subunit structures of IAS and activated Envs. Env from several [35S]Cys-labeled Mo-MLV samples was triggered but arrested at the IAS by lysis in OG-EDTA HN buffer in the presence of NEM for 20 min at 37°C or allowed to isomerize by lysis in the absence of NEM (activated Env). The samples were then cross-linked with 10 to 800 μM DSP or left without cross-linking, treated with an excess of SDS or left untreated, and analyzed by nonreducing SDS-PAGE and by BN-PAGE. Shown are the phosphorimages of the gels with IAS Env (A and C) and activated Env (B and D), respectively. The oligomeric states of viral proteins and SU-TM complexes are indicated.
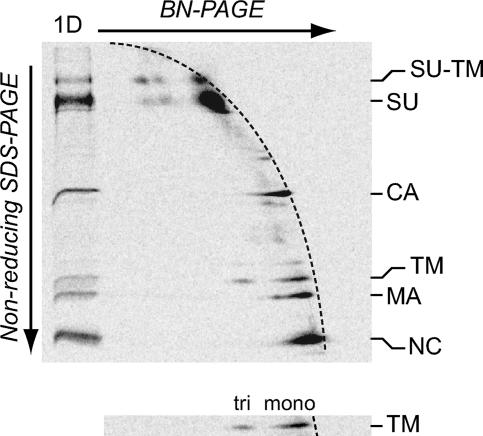
Nonreducing SDS-PAGE analysis in the second dimension reveals the TM trimer in BN-PAGE of activated Env.
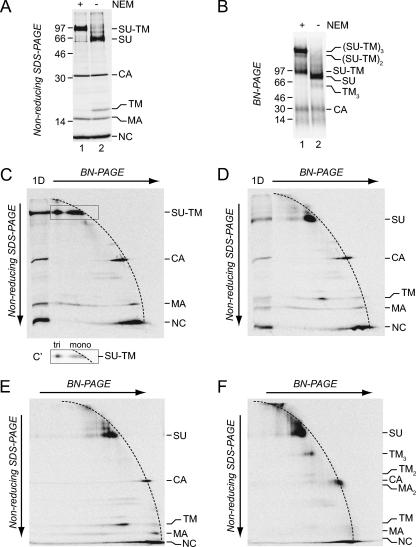
Our aim was to compare the stabilities of the trimeric interaction in SU-TM trimers of IAS Env and in TM trimers of activated Env by a limited SDS treatment. An obvious approach would be to treat the respective trimers with a small amount of SDS, cross-link, and analyze the pattern of cross-linked oligomers compared to that in untreated controls by nonreducing SDS-PAGE. Unfortunately, mild SDS treatments proved to affect cross-linking per se more than the oligomeric status of the proteins. Therefore, we investigated whether BN-PAGE could be used for the separation and detection of not only the IAS Env trimer but also the TM trimer in its native state if the analysis was complemented with nonreducing SDS-PAGE in the second dimension. In this gel system proteins that keep the same oligomeric state through both dimensions will, in the nonreducing SDS-PAGE, form a diagonal front of faster- or slower-migrating spots depending on their size. Because BN-PAGE, in contrast to nonreducing SDS-PAGE, uses a gradient gel, the front will have a curved appearance. In contrast, proteins that are oligomeric in BN-PAGE and dissociate into subunits in nonreducing SDS-PAGE will migrate faster and form spots below the curved front. Thus, samples of IAS and activated Envs, prepared as described above, were subjected to separate BN-PAGE and nonreducing SDS-PAGE and to 2D BN-PAGE/nonreducing SDS-PAGE. The nonreducing SDS-PAGE showed the complete formation of isomerization-arrested SU-TM complexes in IAS Env and free SU and TM subunits in activated Env (Fig. 3A, lanes 1 and 2). The BN-PAGE demonstrated that most of the SU-TM complexes were associated into trimers, whereas only free SU and apparent capsid (CA) were clearly detectable in activated Env (Fig. 3B, lanes 1 and 2). However, occasionally, as in this experiment, we were able to discriminate a diffuse band in front of SU which was not present in the IAS Env sample and thus fit TM trimers. Indeed, we found that almost all apparent TM in the second-dimension gel of the 2D BN-PAGE/nonreducing SDS-PAGE migrated as being derived from such an oligomer (Fig. 3D). Monomeric TM subunits were expected to migrate in the lower part of the curved front formed by the spots of the apparently monomeric SU, CA, MA, and nucleocapsid proteins of the virus. The identities of the apparent TM and SU spots were confirmed by analyses of the IAS Env, which contained only covalently linked SU-TM complexes (Fig. 3C). These were mostly migrating below the 2D gel front, as expected for oligomers in the BN-PAGE dimension (Fig. 3C′ [lower contrast]). The composition of the TM oligomer was determined by cross-linking activated Env and subjecting it together with an un-cross-linked control sample to 2D BN-PAGE/nonreducing SDS-PAGE (Fig. 3E and F). We found that the predominant TM spot in the second-dimension gel was virtually absent and instead a strong spot appeared above, which migrated slightly slower than the 46-kDa marker protein (not shown), between SU and CA. This fit a trimer of the 15-kDa TM. Below the apparent TM trimer spot there was a weak spot, migrating slightly slower than CA, which fit TM dimers. The cross-linked TM trimer migrated somewhat slower than would be expected in the first dimension. This could be due either to the shape of the complex or to lower-than-normal Coomassie blue stain binding. The 2D gel analysis of the cross-linked sample also revealed the apparent MA dimers described above, now comigrating with CA monomers. We concluded that virtually all TM of the activated Env formed TM trimers and that the 2D gel analysis could be used as an assay to reliably detect trimeric TM in its non-cross-linked form.
FIG. 3.
Detection of TM trimers of activated Env by 2D BN-PAGE/nonreducing SDS-PAGE. Samples of [35S]Cys-labeled Mo-MLV were lysed in OG-EDTA HN buffer in the presence or absence of NEM for 20 min at 37°C to generate IAS Env and activated Env. A portion of the latter sample was subsequently cross-linked with DSP. Samples were subjected to 1D and 2D PAGE. (A and B) Phosphorimages of nonreducing SDS-PAGE and BN-PAGE of IAS Env (lanes 1) and activated Env (lanes 2). (C and D) 2D BN-PAGE/nonreducing SDS-PAGE of IAS Env (C) and activated Env (D). (E and F) 2D BN-PAGE/nonreducing SDS-PAGE of un-cross-linked (E) and cross-linked (F) activated Env. (C′) Cutout of the marked region of the second-dimension gel with SU-TM complexes at lower contrast. The oligomeric states of the complexes in the first dimension (BN-PAGE) are indicated. The marker lane (1D) in the 2D BN-PAGE/nonreducing SDS-PAGE contains the respective sample separated in nonreducing SDS-PAGE only. Viral proteins and their oligomeric state (C) in the first dimension (BN-PAGE) are indicated.
The TM trimer of activated Env is more stable than the SU-TM trimer of IAS Env.
To compare the stabilities of the SU-TM trimer of IAS and the TM trimer of activated Env, we prepared a virus lysate with Env partially in the IAS and partially in the activated form and subjected them to a limited SDS treatment. However, this was not possible with samples lysed with OG because of the high critical micellar concentration of this detergent. Therefore we had to switch to HN buffer with Triton X-100, which has a much lower critical micellar concentration. Thus, a labeled virus sample was partially activated in Triton X-100-EDTA HN buffer before NEM was added, and the remaining unactivated Env was chased into the IAS by further incubation. One half of the sample was then subjected to limited SDS treatment and the other half left untreated as a control. The two samples were analyzed by 2D BN-PAGE/nonreducing SDS-PAGE to reveal the SDS stability of the SU-TM trimer of IAS Env and the TM trimer of activated Env. The TM in the control sample without SDS migrated in a position indicating that it was derived from trimers in the BN-polyacrylamide gel, as described above (Fig. 4A and A″). A minor amount was also seen at a position indicating higher oligomers, possibly a dimer of the trimers. Most of the SU-TM complexes of the IAS Env were also migrating faster than the front of monomeric proteins, indicating their presence as oligomers, (SU-TM)2-3, in the BN-polyacrylamide gel (Fig. 4A and A′). A minor amount was seen as monomeric SU-TM complexes. Analysis of the SDS-treated sample in the second dimension showed that the vast majority of the SU-TM complexes now migrated with the front as monomeric SU-TM complexes (Fig. 4B and B′). These SDS-treated monomers moved slightly faster than the nontreated ones, hence giving the double spot appearance for the monomers. Most significantly, the TMs of the activated Env mostly remained in their original position, indicating that they, in contrast to the SU-TM complex oligomer of IAS Env, were resistant to dissociation by the limited SDS treatment of the lysate (Fig. 4B and B″). Quantifications showed that the TM trimer was about 3.5 times more resistant than the SU-TM complex oligomer under these detergent conditions (Fig. 4C).
The stability of the TM trimer of activated Env is dependent on its intrasubunit disulfide.
The TM subunit of MLV contains a single disulfide between C86 and C93 (7). This covalent linkage is necessary for the high thermostability of the trimer of hairpins found in the crystallized TM ectodomain fragment, which is presumed to reflect the structure of TM in fully activated Env (6, 7). Therefore we tested whether DTT treatment of the TM trimers of in vitro-activated Env would destabilize the oligomer. We prepared a virus lysate that was fully activated in vitro and then incubated with DTT before it was subjected to mild SDS treatment. The sample was analyzed by 2D BN-PAGE/nonreducing SDS-PAGE, which showed that more than half of the TM had shifted from the oligomer location into monomers (Fig. 5). This indicated that the stability of the in vitro-activated TM trimer was dependent on the intrasubunit disulfide and suggested structural similarities with the crystallized form.
FIG. 5.
Reduction of an intrasubunit disulfide bond makes the TM trimer unstable. [35S]Cys-labeled Mo-MLV was lysed in Triton X-100-EDTA HN buffer for 40 min at 37°C, DTT was added, and the incubation was continued for 20 min at 37°C. The sample was then subjected to mild treatment with SDS and analyzed by 2D BN-PAGE/nonreducing SDS-PAGE. Shown is a phosphorimage of the dried gel with a cutout of the TM region below. The marker lane (1D) and labeling are as in Fig. 4.
Receptor activation of Env results in the formation of a stable TM trimer.
Incubation of Mo-MLV with receptor-positive cells, such as rat XC cells, results in activation of the fraction of viral Envs that interact with the cell receptor and a concomitant isomerization of their intersubunit disulfide. To find out whether this natural triggering of Env also results in the formation of stable TM trimers, we used our assay with SDS treatment and 2D BN-PAGE/nonreducing SDS-PAGE. Thus, labeled Mo-MLV was bound by centrifugation to a culture of XC cells at 4°C and then incubated for 30 min at 37°C. NEM was added and the virus-cell samples lysed with Triton X-100-EDTA in HN buffer. Lysis in the presence of alkylator converted all viral Envs that had not became activated by the receptor into the IAS. One half of the sample was then subjected to mild SDS treatment, whereas the other one was left untreated, and both were analyzed by 2D BN-PAGE/nonreducing SDS-PAGE. The analysis of the sample that was not SDS treated showed that most Env was present as IAS SU-TM complexes. These were mostly in a region of the gel that indicated that they migrated as oligomers of SU-TM in the BN-polyacrylamide gel (Fig. 6A and A′). However, there was also some free TM present, suggesting that a fraction of the Envs had been activated by the receptors. The corresponding free SU was mostly dissociated from the receptors into the cell medium during the incubation and was therefore not present in the virus-cell sample analyzed (M. Sjöberg, unpublished observation). The majority of the free TM was migrating in a position suggesting that it was derived from TM trimers in the BN-PAGE, but some was also migrating as being derived from a higher oligomer (Fig. 6A and A″). The analysis of the SDS-treated sample showed that most of the SU-TM complexes now migrated as monomers in the BN-polyacrylamide gel (Fig. 6B and B′). However, the TM did not change its oligomer-indicating position in the second dimension, although the two-spot appearance of the non-SDS-treated sample was more spread out in the SDS-treated one (Fig. 6B and B″). We conclude that receptor triggering of Env also resulted in formation of stable TM trimers.
FIG. 6.
Receptor triggering of Env results in the formation of stable TM trimers. [35S]Cys-labeled Mo-MLV was bound by centrifugation to cultures of XC cells at 4°C and then incubated for 30 min at 37°C. NEM was added and the virus-cell samples lysed with Triton X-100-EDTA HN buffer. One sample was then mildly treated with SDS and the other one left untreated. Both samples were analyzed by 2D BN-PAGE/nonreducing SDS-PAGE. Shown are phosphorimages of the gel analyses of the untreated sample (A) and the SDS-treated sample (B). Marker lanes (1D), cutouts, and labeling as in Fig. 4. Note that A″ and B″ are shown in higher contrast to better visualize the TM subunit. (C) Relative amounts of the oligomeric forms of the SU-TM complexes and the TM subunits in the two samples as percentages of the SU-TM and TM, respectively (mean ± standard deviation; n = 4).
DISCUSSION
Our present analyses show that activation of the Mo-MLV Env results in the formation of a TM trimer, which was much more resistant toward SDS dissociation than the trimer of the IAS activation-intermediate form of Env. This provides direct biochemical evidence that a structural conversion of TM takes place during activation, which results in the formation of a stable TM trimer. Corresponding evidence has earlier been obtained only for the TM of the ALV Env (21). Together with previous structural analyses of TM ectodomain fragments and of TM peptide dependent inhibition studies, our analyses of oligomer stability provide strong support for an HA-like activation model for retrovirus Env also.
Earlier we showed that Mo-MLV Env in the IAS, in contrast to native Env and Env arrested at a late stage by lipid membrane modification, was able to complex with externally added TM peptides and thus inhibit reactivation of the IAS upon reduction of the intersubunit disulfide with DTT (35). Additionally, the native Env, in contrast to the IAS Env, has been found to hide the isomerization-active CXXC-thiol and intersubunit disulfide for external modification (34). These data together with the present study showing that the oligomeric interactions of IAS Env are significantly weaker than those of the activated TM trimer suggest that IAS Env is an open structure with its TM subunits possibly in an extended prehairpin conformation. This might correspond to the postulated HA activation intermediate where the fusion peptide connecting the α-helix and loop have extended the central helix of HA2 and expelled the fusion peptide, but the jackknife-like turn of HA2 has not yet taken place (30). Although we have not yet localized the fusion peptide in the Mo-MLV IAS Env, such an intermediate might represent a crucial step, especially in Mo-MLV Env activation. The intermediate would allow the TM to establish an interaction with the target membrane before the contact via SU is lost by isomerization of the intersubunit disulfide and before the TM undergoes its backfolding reaction into a stable trimer. In the case of ALV, which is activated by a combination of receptor binding at the cell surface and subsequent acid treatment in the endosomes, an Env activation intermediate has been found after activation with soluble receptors under neutral conditions (12, 21, 22). This intermediate bound TM peptides and exposed the fusion peptide for interactions with added liposomes. Notably, the TM subunits of this ALV Env intermediate had already obtained a conformation that supported SDS-resistant trimer interactions. However, an additional SDS-resistant TM oligomer has been characterized in receptor- and acid-activated ALV that did not bind TM peptides, suggesting incomplete TM hairpin formation in the receptor-only-activated virus (19).
Recently the native form of the Mo-MLV Env was studied by cryo-electron tomography (8). The Env reconstruction revealed a head domain that was connected to the membrane by three legs. These splayed outward toward the membrane, enclosing a central cavity below the head. The legs appeared to be composed of an inner helix that was continuous with the membrane anchor and an outer helix that was proposed to represent the fusion peptide-proximal part of the TM. Thus, according to this model, the TM subunits of native Env would form “reverse” hairpins that might contact only at the region of polypeptide turn and intersubunit disulfide with the SU. This organization would be quite different from the central coiled-coil structure of HA2 in native HA. Activation of the Mo-MLV Env might then initially involve SU rearrangements that allow the external, fusion peptide-proximal TM parts to relocate and form a central coiled coil according to the model proposed for IAS Env and the first step of HA2 activation. It is evident that the natures of the TM interactions in the native and the IAS Envs of Mo-MLV remain challenging questions for future research. Most likely they can be understood only by structural studies that reveal the polypeptide fold in the two forms of the Env trimer.
Acknowledgments
We thank Michael Wallin for critically reading the manuscript.
Swedish Science Foundation grant 2778 and Swedish Cancer Foundation grant 0525 to H.G. supported this work.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 37137-43. [DOI] [PubMed] [Google Scholar]
- 2.Carr, C. M., C. Chaudhry, and P. S. Kim. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. USA 9414306-14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89263-273. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J., S. A. Wharton, W. Weissenhorn, L. J. Calder, F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1995. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc. Natl. Acad. Sci. USA 9212205-12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, J. Y., J. W. Dubay, L. G. Perez, and E. Hunter. 1992. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein define a requirement for dibasic residues for intracellular cleavage. J. Virol. 66865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 3465-469. [DOI] [PubMed] [Google Scholar]
- 7.Fass, D., and P. S. Kim. 1995. Dissection of a retrovirus envelope protein reveals structural similarity to influenza hemagglutinin. Curr. Biol. 51377-1383. [DOI] [PubMed] [Google Scholar]
- 8.Forster, F., O. Medalia, N. Zauberman, W. Baumeister, and D. Fass. 2005. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. USA 1024729-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5276-279. [DOI] [PubMed] [Google Scholar]
- 10.Godley, L., J. Pfeifer, D. Steinhauer, B. Ely, G. Shaw, R. Kaufmann, E. Suchanek, C. Pabo, J. J. Skehel, D. C. Wiley, et al. 1992. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell 68635-645. [DOI] [PubMed] [Google Scholar]
- 11.Hallenberger, S., V. Bosch, H. Angliker, E. Shaw, H. D. Klenk, and W. Garten. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360358-361. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez, L. D., R. J. Peters, S. E. Delos, J. A. Young, D. A. Agard, and J. M. White. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 1391455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter, E. 1997. Viral entry and receptors, p. 71-119. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 14.Jinno, A., Y. Haraguchi, H. Shiraki, and H. Hoshino. 1999. Inhibition of cell-free human T-cell leukemia virus type 1 infection at a postbinding step by the synthetic peptide derived from an ectodomain of the gp21 transmembrane glycoprotein. J. Virol. 739683-9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemble, G. W., D. L. Bodian, J. Rose, I. A. Wilson, and J. M. White. 1992. Intermonomer disulfide bonds impair the fusion activity of influenza virus hemagglutinin. J. Virol. 664940-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. USA 964319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobigs, M., and H. Garoff. 1990. Fusion function of the Semliki Forest virus spike is activated by proteolytic cleavage of the envelope glycoprotein precursor p62. J. Virol. 641233-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lombardi, S., C. Massi, E. Indino, C. La Rosa, P. Mazzetti, M. L. Falcone, P. Rovero, A. Fissi, O. Pieroni, P. Bandecchi, F. Esposito, F. Tozzini, M. Bendinelli, and C. Garzelli. 1996. Inhibition of feline immunodeficiency virus infection in vitro by envelope glycoprotein synthetic peptides. Virology 220274-284. [DOI] [PubMed] [Google Scholar]
- 19.Matsuyama, S., S. E. Delos, and J. M. White. 2004. Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein. J. Virol. 788201-8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore, P. L., E. T. Crooks, L. Porter, P. Zhu, C. S. Cayanan, H. Grise, P. Corcoran, M. B. Zwick, M. Franti, L. Morris, K. H. Roux, D. R. Burton, and J. M. Binley. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 802515-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103679-689. [DOI] [PubMed] [Google Scholar]
- 22.Netter, R. C., S. M. Amberg, J. W. Balliet, M. J. Biscone, A. Vermeulen, L. J. Earp, J. M. White, and P. Bates. 2004. Heptad repeat 2-based peptides inhibit avian sarcoma and leukosis virus subgroup a infection and identify a fusion intermediate. J. Virol. 7813430-13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opstelten, D. J., M. Wallin, and H. Garoff. 1998. Moloney murine leukemia virus envelope protein subunits, gp70 and Pr15E, form a stable disulfide-linked complex. J. Virol. 726537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortmann, D., M. Ohuchi, H. Angliker, E. Shaw, W. Garten, and H. D. Klenk. 1994. Proteolytic cleavage of wild type and mutants of the F protein of human parainfluenza virus type 3 by two subtilisin-like endoproteases, furin and Kex2. J. Virol. 682772-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinter, A., R. Kopelman, Z. Li, S. C. Kayman, and D. A. Sanders. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J. Virol. 718073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruigrok, R. W., A. Aitken, L. J. Calder, S. R. Martin, J. J. Skehel, S. A. Wharton, W. Weis, and D. C. Wiley. 1988. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J. Gen. Virol. 692785-2795. [DOI] [PubMed] [Google Scholar]
- 27.Schagger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199223-231. [DOI] [PubMed] [Google Scholar]
- 28.Scheid, A., and P. W. Choppin. 1974. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 57475-490. [DOI] [PubMed] [Google Scholar]
- 29.Sjoberg, M., M. Wallin, B. Lindqvist, and H. Garoff. 2006. Furin cleavage potentiates the membrane fusion-controlling intersubunit disulfide bond isomerization activity of leukemia virus Env. J. Virol. 805540-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69531-569. [DOI] [PubMed] [Google Scholar]
- 31.Smith, J. G., W. Mothes, S. C. Blacklow, and J. M. Cunningham. 2004. The mature avian leukosis virus subgroup A envelope glycoprotein is metastable, and refolding induced by the synergistic effects of receptor binding and low pH is coupled to infection. J. Virol. 781403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stieneke-Grober, A., M. Vey, H. Angliker, E. Shaw, G. Thomas, C. Roberts, H. D. Klenk, and W. Garten. 1992. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 112407-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallin, M., M. Ekstrom, and H. Garoff. 2005. The fusion-controlling disulfide bond isomerase in retrovirus Env is triggered by protein destabilization. J. Virol. 791678-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallin, M., M. Ekstrom, and H. Garoff. 2004. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 2354-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallin, M., Ekström, M., and H. Garoff. 2006. Receptor triggered but alkylation-arrested Env of murine leukaemia virus reveals the transmembrane subunit in a prehairpin conformation. J. Virol. 809921-9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallin, M., R. Loving, M. Ekstrom, K. Li, and H. Garoff. 2005. Kinetic analyses of the surface-transmembrane disulfide bond isomerization-controlled fusion activation pathway in Moloney murine leukemia virus. J. Virol. 7913856-13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward, C. D., R. G. Paterson, and R. A. Lamb. 1995. Mutants of the paramyxovirus SV5 fusion protein: regulated and extensive syncytium formation. Virology 209242-249. [DOI] [PubMed] [Google Scholar]
- 38.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289366-373. [DOI] [PubMed] [Google Scholar]
- 39.Yin, H. S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 43938-44. [DOI] [PMC free article] [PubMed] [Google Scholar]