Abstract
Hepatitis C virus (HCV) clearance has been associated with reduced viral evolution in targeted cytotoxic T-lymphocyte (CTL) epitopes, suggesting that HCV clearers may mount CTL responses with a superior ability to recognize epitope variants and prevent viral immune escape. Here, 40 HCV-infected subjects were tested with 406 10-mer peptides covering the vast majority of the sequence diversity spanning a 197-residue region of the NS3 protein. HCV clearers mounted significantly broader CTL responses of higher functional avidity and with wider variant cross-recognition capacity than nonclearers. These observations have important implications for vaccine approaches that may need to induce high-avidity responses in vivo.
Although broad and strong hepatitis C virus (HCV)-specific T-cell responses are considered a key factor in the clearance of HCV infection, the exact mechanisms mediating viral control remain unclear (7, 18). Work comparing host (HLA) genetics and viral sequence polymorphisms suggests that a substantial proportion of all observed sequence changes in the first 6 months after HCV infection are likely driven by CD8 T-cell-mediated immune pressure (4, 15). Furthermore, a number of reports have associated increased viral evolution with the establishment of chronic infection, whereas conservation of T-cell epitope sequences has been observed for subjects eventually clearing HCV (3). These findings are substantiated by studies using the chimpanzee model that have linked an improved disease outcome with the occurrence of fewer mutations in major histocompatibility complex class I-restricted T-cell epitopes (11, 28). Together, these studies suggest that the induction of an effective CTL response shortly after acute HCV infection may be able to control viral replication and prevent chronic infection, especially if potentially occurring T-cell epitope variants could be cross-recognized by the wild-type-specific CTL population.
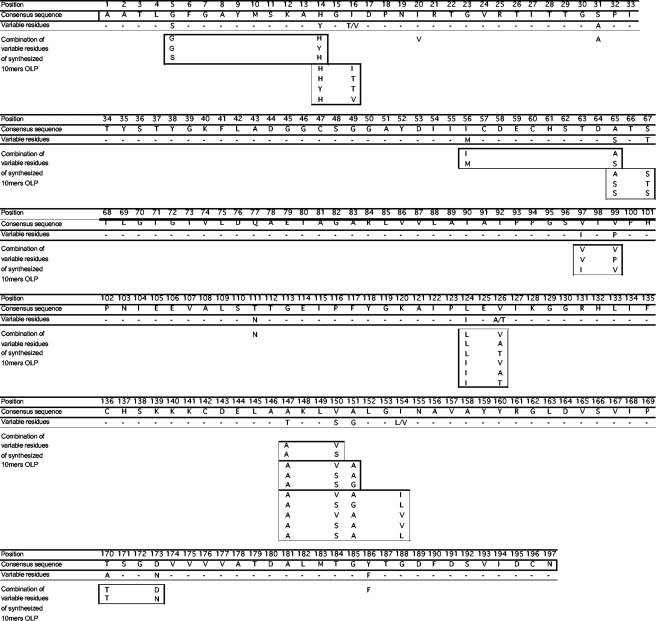
To directly test if the ability of epitope-variant cross-recognition is indeed associated with viral clearance, we tested HCV clearers and chronically infected subjects against an extensive set of variant 10-mer peptides covering a region of 197 amino acids in the moderately variable nonstructural protein 3 (NS3) of HCV. Overall, 92 NS3 sequences of genotypes 1a and 1b were obtained from the HCV database at Los Alamos National Laboratory (http://hcv.lanl.gov) and aligned with an additional set of 73 genotype 1a sequences generated in our laboratory (25). All unique 10-mer sequences that occurred in this alignment were determined (n = 2,050 individual 10-mers), and those 10-mers that were present in at least 5% of the aligned sequences were synthesized. This gave rise to a set of 406 10-mer peptides overlapping by 9 residues spanning the analyzed 197 amino acids of NS3 (Fig. 1). The 10-mer peptide length was chosen to reduce the chance that responses to multiple epitopes within the same test peptide would be detected and to increase the sensitivity for in vitro analyses over that with longer peptide sets (9, 10).
FIG. 1.
Sequence variability in NS3/1259-1455 covered by 406-10-mer peptide set. A consensus sequence was derived from the 165 aligned genotype 1a and 1b sequences spanning positions 1259 to 1455 of HCV NS3. Variable positions that were present in at least 5% of the aligned sequences are indicated and were included in the synthesis of a 406-peptide set of overlapping peptides (OLP) of 10-amino-acid length overlapping by 9 residues.
A cohort of 40 HCV-infected subjects, of which 30 were chronically infected and 10 were virus clearers, was tested. The most common infecting genotype was genotype 1, with 16 subjects infected by genotype 1a (of which 4 subjects had cleared the infection) and 11 individuals infected with genotype 1b (2 clearers). Additional subjects were infected with either HCV genotype 2b (n = 2; one clearer) or 3a (n = 4; no clearer) or by undetermined genotypes (n = 7; three clearers). Although 90% of subjects (36 out of the 40 tested individuals) were coinfected with HIV, none had a CD4 T-cell count of below 250 cells/μl, a threshold above which earlier studies have shown maintenance of HCV-specific CD8 T-cell activity (14). Peripheral blood mononuclear cells (PBMC) from all subjects were stimulated with 4 different peptide pools, each containing about 100 of the synthesized 406 peptides, and expanded for 2 weeks before being tested in gamma interferon enzyme-linked immunospot (ELISPOT) assays. ELISPOT assays were carried out using a matrix pool approach, as described in the past, with subsequent deconvolution to identify each individually targeted peptide (2). This allowed us to determine the breadth of each individual's response and the number of variants that were recognized among the 10-mers that spanned the same protein stretch. As a control, six healthy HIV- and HCV-negative donors were screened, following the same protocol, and showed only weak responses against two individual peptides (ALGINAVAYY and KLVALGINAV), which are different from the epitopes for which a frequent cross-reactivity between HCV and influenza or self proteins has been described (13, 27).
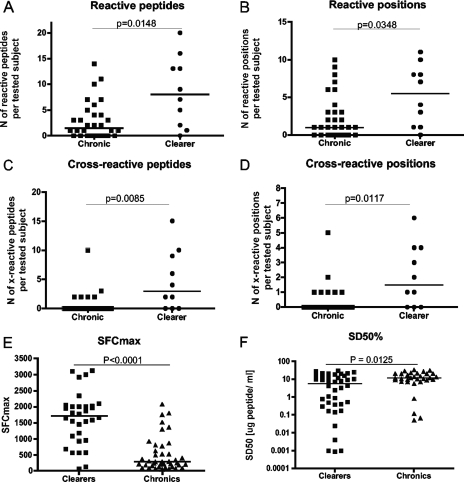
Altogether, 174 responses were identified for the 40 subjects tested and analyzed in two different ways, with or without considering cross-reactivity among 10-mer variants. In a first step, responses to the entire set of 406 individual peptides were compared between chronically infected subjects and HCV clearers, demonstrating that clearers mounted significantly broader responses (median of 8 responses; range, 0 to 20) than the chronically infected subjects (median, 1.5; range, 0 to 14; P = 0.0148) (Fig. 2A). These data are in line with those of earlier studies that have linked HCV clearance to the presence of broadly directed virus-specific T-cell activities (7, 16, 18). However, since responses to variants of individual 10-mer peptides were counted as independent events, the increased breadth of responses for clearers may partly be due to a greater rate of variant recognition for these subjects than for the chronically infected individuals. To correct for this, the analysis was repeated, this time counting responses to an individual 10-mer only once even if one or multiple variants of the specific 10-mer region were targeted. Still, the HCV clearers showed a significantly greater breadth of responses (median = 5.5; range, 0 to 11) than the chronics (median = 1.0; range, 0 to 10; P = 0.0349) (Fig. 2B). These data indicate that after comprehensively testing HCV-specific responses to even a relatively short stretch of the viral genome only, significantly broader responses can be detected for subjects who cleared the virus than for chronically infected individuals (8, 17, 20, 23, 24).
FIG. 2.
HCV clearers mount broader CTL responses with greater cross-reactivity potential and higher magnitude and functional avidity than chronically infected individuals. CTL immunes responses to the 406 OLP were assessed by gamma interferon ELISPOT assay using in vitro-expanded PBMC (2). (A) Reacting peptides among the 406 OLP were counted individually, showing that cells from HCV clearers recognized significantly more peptides than those from chronic subjects. (B) Given the variable number of peptide variants in a 10-mer stretch, only reactive positions were taken into account, i.e., responses to a specific 10-mer window were scored only once without considering if or how many different variant 10-mers were reactive, still showing statistically significantly broader responses in clearers. (C) Responses to each 10-mer region were also analyzed individually and the level of variant cross-recognition determined. Even though cells from HCV clearers did not target regions of higher variability than those from chronics (i.e., comparable numbers of variants per 10-mer region targeted were tested in either group), they recognized more individual variant peptides than cells from chronically infected subjects. (D) Since the inclusion of different numbers of variants for a specific 10-mer region (i.e., entropy [29]) could have biased the analysis in favor of the HCV clearers, the number of 10-mer regions for which some level of cross-reactive responses were seen was also compared between clearers and chronics, again showing a broader variant recognition by the HCV clearer group. (E) The maximal magnitude of the responses, i.e., frequency of the maximal SFC per million input cells, was determined for 40 and 41 responses, respectively, and compared between HCV clearers and chronically infected subjects. (F) The peptide concentrations needed to elicit half-maximal SFC (SD50) were compared between chronically infected subjects and HCV clearers, revealing responses of significantly higher functional avidity for the latter (all Mann-Whitney test).
To more directly assess whether or not the response seen in clearers targeted a greater proportion of variants than that of nonclearers, the detected responses were further studied on an individual 10-mer basis and the number of simultaneously recognized 10-mer variants was determined for each response. This analysis is different from that described above, where responses to one or multiple variant of the same 10-mer were counted only once, and it was performed to answer the question of whether cells from HCV clearers had a superior ability to recognize different variants of a targeted 10-mer compared to those from chronically infected subjects. The data indeed show that cells from clearers were able to recognize a significantly larger number of 10-mer variants (median = 3.0; range, 0 to 15) than those from the chronically infected individuals (median = 0; range, 0 to 5; P = 0.0085) (Fig. 2C). However, these results could theoretically be biased if cells of HCV clearers preferentially targeted regions of higher variability than did those of the chronically infected individuals. This would have led us to test more variant peptides in the clearer group, giving them an unfair advantage to react with more variants. However, subsequent analyses showed that the 10-mer regions targeted by cells from clearers overall did not contain more variant sequences than the 10-mers targeted by those from the chronically infected subjects (data not shown) (P = 0.6875). In line with this, responses detected for HCV clearers were generally more likely to react with at least one other 10-mer variant (median, 1.5 cross-reactive responses per subject; range, 0 to 6) than responses detected for chronically infected subjects (median = 0; range 0 to 5; P = 0.0117) (Fig. 2D).
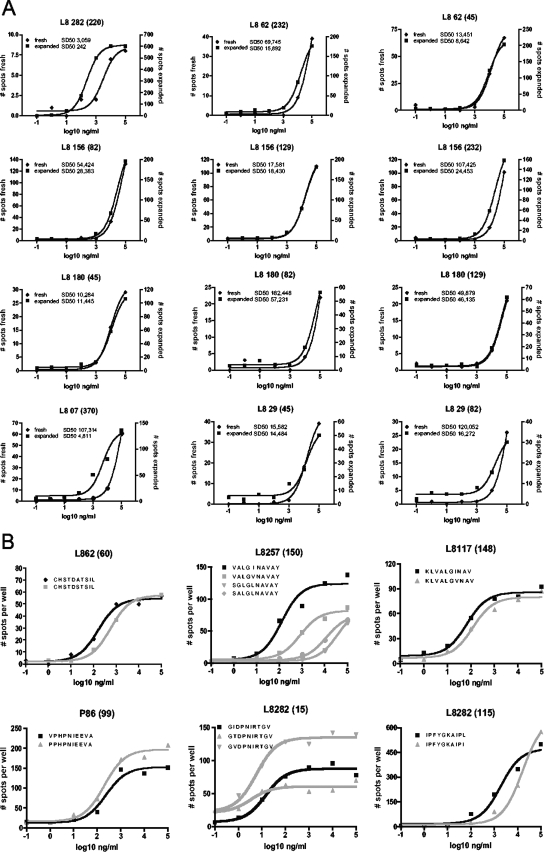
A number of past studies with humans and animal models of viral infections have suggested that control of viral replication may be associated with the magnitude and functional avidity of the epitope-specific T-cell population (5, 6, 21, 22). We thus assessed whether the rate of variant cross-reactivity correlated with the functional avidity of the detected responses and whether responses in clearers were overall of higher magnitude and avidity than those in chronics. Indeed, and although results were potentially biased by the proliferative capacity of these cells during the in vitro expansion, significantly stronger responses were seen for clearers (median, 1,715 spot-forming cells [SFC] per million input cells; range, 70 to 3,125) than for chronics (median, 290 SFC/106 PBMC; range, 50 to 2075; P < 0.0001) (Fig. 2E). Similarly, when the functional avidity (referring to the peptide concentration [sensitizing dose] required to elicit half-maximal response rates [SD50]) (1) for 81 of the 174 positive responses was determined and compared between the two groups, cells from clearers elicited responses with overall significantly higher functional avidity than those from chronically infected individuals (P = 0.0125) (Fig. 2F). As these data were obtained with T-cell lines after antigen-specific in vitro stimulation and expansion, the measured functional avidity may have been biased toward the selective expansion of high-avidity responses. Although such a bias would likely have affected SD50 determinations for both groups (chronics and clearers), titration curves for 20 responses were repeated using unexpanded PBMC samples. Indeed, the SD50 of the 12 responses that were detectable ex vivo was significantly increased (i.e., had lower functional avidity) over that of the same responses assessed after in vitro expansion of cells (P = 0.0034) (Fig. 3A). However, in 9 of the 12 cases, the difference in SD50 was less than fivefold, and no statistically significant difference was noted when comparing the “fold” increases in SD50 for responses tested in PBMC from either clearers or chronically infected individuals, suggesting that antigen-specific in vitro culture generally drives expansion of high(er)-avidity cells, regardless of whether or not the cells were obtained from individuals with or without persisting viremia. Together, the present data thus associate the increased breadth, magnitude, and functional avidity of HCV-specific T-cell responses of HCV clearers with a superior ability to cross-recognize epitope variants compared to responses of chronically infected subjects. Of note, peptide titration analyses on six peptides and their variants also showed that variant recognition was not simply due to spurious variant reactivity at the highest peptide concentration but indeed was mediated by high-avidity recognition of the peptide variants (Fig. 3B), further confirming that functional avidity and variant epitope recognition may be crucial factors in the outcome of HCV infection.
FIG. 3.
Higher functional avidity after in vitro expansion and responses of high functional avidity to peptide variants. (A) The functional avidity of responses elicited by cells tested directly ex vivo (gray lines) or after in vitro expansion (black lines) was assessed by peptide titration analyses. Graph titles indicate subject identifier and, in parentheses, the peptide(s) tested. Subjects L807 and L829 are chronically infected individuals; the remainder are HCV clearers. (B) Titration curves against peptides and their variants are shown for four individuals who showed responses to consensus and variant peptides in the initial screening using highest peptide concentrations and from whom sufficient cells were available to test for the full range of decreasing variant peptide concentrations. Numbers of spots per well represent averages for duplicate wells; black lines and symbols show the 10-mer variant that was most common in the aligned sequences (consensus), and symbols and lines in gray show responses to variant peptides.
Similar to the case of functional avidity, the observed magnitudes of responses may not directly reflect the in vivo frequency of these cells either, since the assays were performed using in vitro-expanded PBMC. Thus, the observed differences in the magnitudes may be a consequence of either a truly higher frequency of the specific cells in vivo, an enhanced ability of epitope-specific cells to proliferate upon antigen stimulation, or, most probably, a combination of both. Although the ex vivo titration data suggest that chronically infected individuals can expand their most avid epitope-specific cell populations, their reduced breadth of responses after in vitro expansion may also reflect some missed responses that, even after expansion, fell under the detection limit of the ELISPOT assay. Testing this with directly ex vivo-isolated cells could potentially help to discriminate between these different mechanisms; however, with current techniques, such analyses will likely still be biased toward the detection of strongest responses, which in turn may be characterized by high avidity. Regardless of this potential limitation and although recent data showing that functional impairment of HCV-specific cells in chronically infected subjects could impact the analysis of in vitro-expanded cells, our data confirm earlier findings that have linked higher-magnitude responses with cleared infection (16). How these factors translate, for instance, into T-cell receptor breadth will require detailed T-cell receptor analyses of the same responses in chronically infected subjects and in HCV clearers. While this will be highly informative for vaccine design and for the understanding of in vivo control of infections other than HCV, it will likely be complicated by the low magnitude of ex vivo responses in the chronic group. Finally, although the relatively high peptide concentrations needed to elicit some of these responses may call into question their physiological relevance, the data are in line with those of earlier reports showing an overall low-avidity T-cell activity for HCV-derived HLA class I-restricted CTL epitopes when compared, for instance, to HIV- or Epstein-Barr virus-specific CTL responses (1, 26). Whether these low-avidity responses are adequately effective in recognizing virally infected cells in the solid tissue of an infected liver, and thus not necessarily directly comparable to responses to other viruses, remains to be clarified in future studies.
Further complicating the analyses of protective immunity in HCV infection is the uncertainty of whether or not the CTL responses detected for clearers and chronically infected subjects corresponded to the first line of responses after acute infection. In particular, the lower avidity of responses for chronically infected subjects could reflect a gradual decrease in affinity over time due to elimination of high-avidity T-cell populations in the presence of continuous antigenic stimulation. While some evidence for such a scenario exists for HIV and recent data in chronic HCV infection are in support of such an interpretation as well, few data are available on direct ex vivo analyses in acute HCV infection and on longitudinal samples for chronics and HCV clearers (12, 19). The design of such studies, with approaches sufficiently sensitive to allow for direct ex vivo detection of even low-magnitude T-cell responses, are thus urgently needed to further confirm our present findings and to guide future HCV vaccine design. Aside from informing vaccine design, however, the present data may also help in interpreting earlier data that have associated sequence conservation within HCV-encoded CTL epitopes with viral clearance (4). The present data suggest that epitope variant recognition may be crucial early in infection to prevent the virus from testing different CTL epitope variants that could mediate effective escape. Ultimately, detailed analyses of the sequence and quasispecies distribution of the infecting viral population will be required to determine whether the induction of high-avidity responses with broad variant recognition capacity is the cause of sequence conservation or whether the introduction of particularly homogeneous or heterogeneous viral populations could influence the variant recognition patterns observed. For vaccine design, however, it appears critical to aim for the induction of strong, highly avid responses that could possibly protect from a challenge with heterologous virus showing sequence polymorphisms in the targeted epitope.
Footnotes
Published ahead of print on 9 January 2008.
REFERENCES
- 1.Bihl, F., N. Frahm, L. Di Giammarino, J. Sidney, M. John, K. Yusim, T. Woodberry, K. Sango, H. S. Hewitt, L. Henry, C. H. Linde, J. V. Chisholm III, T. M. Zaman, E. Pae, S. Mallal, B. D. Walker, A. Sette, B. T. Korber, D. Heckerman, and C. Brander. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 1764094-4101. [DOI] [PubMed] [Google Scholar]
- 2.Bihl, F. K., E. Loggi, J. V. Chisholm III, H. S. Hewitt, L. M. Henry, C. Linde, T. J. Suscovich, J. T. Wong, N. Frahm, P. Andreone, and C. Brander. 2005. Simultaneous assessment of cytotoxic T lymphocyte responses against multiple viral infections by combined usage of optimal epitope matrices, anti-CD3 mAb T-cell expansion and “RecycleSpot.” J. Transl. Med. 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox, A. L., T. Mosbruger, G. M. Lauer, D. Pardoll, D. L. Thomas, and S. C. Ray. 2005. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology 42104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox, A. L., T. Mosbruger, Q. Mao, Z. Liu, X. H. Wang, H. C. Yang, J. Sidney, A. Sette, D. Pardoll, D. L. Thomas, and S. C. Ray. 2005. Cellular immune selection with hepatitis C virus persistence in humans. J. Exp. Med. 2011741-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davenport, M. P., C. Fazou, A. J. McMichael, and M. F. Callan. 2002. Clonal selection, clonal senescence, and clonal succession: the evolution of the T cell response to infection with a persistent virus. J. Immunol. 1683309-3317. [DOI] [PubMed] [Google Scholar]
- 6.Derby, M., M. Alexander-Miller, R. Tse, and J. Berzofsky. 2001. High-avidity CTL exploit two complementary mechanisms to provide better protection against viral infection than low-avidity CTL. J. Immunol. 1661690-1697. [DOI] [PubMed] [Google Scholar]
- 7.Diepolder, H. M., R. Zachoval, R. M. Hoffmann, M. C. Jung, T. Gerlach, and G. R. Pape. 1996. The role of hepatitis C virus specific CD4+ T lymphocytes in acute and chronic hepatitis C. J. Mol. Med. 74583-588. [DOI] [PubMed] [Google Scholar]
- 8.Diepolder, H. M., R. Zachoval, R. M. Hoffmann, E. A. Wierenga, T. Santantonio, M. C. Jung, D. Eichenlaub, and G. R. Pape. 1995. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet 3461006-1007. [DOI] [PubMed] [Google Scholar]
- 9.Draenert, R., M. Altfeld, C. Brander, N. Basgoz, C. Corcoran, A. G. Wurcel, D. R. Stone, S. A. Kalams, A. Trocha, M. M. Addo, P. J. Goulder, and B. D. Walker. 2003. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J. Immunol. Methods 27519-29. [DOI] [PubMed] [Google Scholar]
- 10.Draenert, R., C. Brander, X. G. Yu, M. Altfeld, C. L. Verrill, M. E. Feeney, B. D. Walker, and P. J. Goulder. 2004. Impact of intrapeptide epitope location on CD8 T cell recognition: implications for design of overlapping peptide panels. AIDS 18871-876. [DOI] [PubMed] [Google Scholar]
- 11.Erickson, A. L., Y. Kimura, S. Igarashi, J. Eichelberger, M. Houghton, J. Sidney, D. McKinney, A. Sette, A. L. Hughes, and C. M. Walker. 2001. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity 15883-895. [DOI] [PubMed] [Google Scholar]
- 12.Golden-Mason, L., B. Palmer, J. Klarquist, J. A. Mengshol, N. Castelblanco, and H. R. Rosen. 2007. Upregulation of Pd-1 expression on circulating and intrahepatic hepatitis C virus-specific Cd8+ T cells associated with reversible immune dysfunction. J. Virol. 819249-9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kammer, A. R., S. H. van der Burg, B. Grabscheid, I. P. Hunziker, K. M. Kwappenberg, J. Reichen, C. J. Melief, and A. Cerny. 1999. Molecular mimicry of human cytochrome P450 by hepatitis C virus at the level of cytotoxic T cell recognition. J. Exp. Med. 190169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, A. Y., G. M. Lauer, K. Ouchi, M. M. Addo, M. Lucas, J. Schulze Zur Wiesch, J. Timm, M. Boczanowski, J. E. Duncan, A. G. Wurcel, D. Casson, R. T. Chung, R. Draenert, P. Klenerman, and B. D. Walker. 2005. The magnitude and breadth of hepatitis C virus-specific CD8+ T cells depend on absolute CD4+ T-cell count in individuals coinfected with HIV-1. Blood 1051170-1178. [DOI] [PubMed] [Google Scholar]
- 15.Kuntzen, T., J. Timm, A. Berical, L. L. Lewis-Ximenez, A. Jones, B. Nolan, J. Schulze Zur Wiesch, B. Li, A. Schneidewind, A. Kim, R. T. Chung, B. D. Walker, G. M. Lauer, and T. M. Allen. 2007. Viral sequence evolution 1 in acute HCV infection. J. Virol. 8111658-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauer, G. M., E. Barnes, M. Lucas, J. Timm, K. Ouchi, A. Y. Kim, C. L. Day, G. K. Robbins, D. R. Casson, M. Reiser, G. Dusheiko, T. M. Allen, R. T. Chung, B. D. Walker, and P. Klenerman. 2004. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology 127924-936. [DOI] [PubMed] [Google Scholar]
- 17.Lechner, F., N. H. Gruener, S. Urbani, J. Uggeri, T. Santantonio, A. R. Kammer, A. Cerny, R. Phillips, C. Ferrari, G. R. Pape, and P. Klenerman. 2000. CD8+ T lymphocyte responses are induced during acute hepatitis C virus infection but are not sustained. Eur. J. Immunol. 302479-2487. [DOI] [PubMed] [Google Scholar]
- 18.Lechner, F., D. K. Wong, P. R. Dunbar, R. Chapman, R. T. Chung, P. Dohrenwend, G. Robbins, R. Phillips, P. Klenerman, and B. D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp. Med. 1911499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lichterfeld, M., X. G. Yu, S. K. Mui, K. L. Williams, A. Trocha, M. A. Brockman, R. L. Allgaier, M. T. Waring, T. Koibuchi, M. N. Johnston, D. Cohen, T. M. Allen, E. S. Rosenberg, B. D. Walker, and M. Altfeld. 2007. Selective depletion of high-avidity human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T cells after early HIV-1 infection. J. Virol. 814199-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Missale, G., R. Bertoni, V. Lamonaca, A. Valli, M. Massari, C. Mori, M. G. Rumi, M. Houghton, F. Fiaccadori, and C. Ferrari. 1996. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J. Clin. Investig. 98706-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price, D. A., J. M. Brenchley, L. E. Ruff, M. R. Betts, B. J. Hill, M. Roederer, R. A. Koup, S. A. Migueles, E. Gostick, L. Wooldridge, A. K. Sewell, M. Connors, and D. C. Douek. 2005. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 2021349-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sedlik, C., G. Dadaglio, M. F. Saron, E. Deriaud, M. Rojas, S. I. Casal, and C. Leclerc. 2000. In vivo induction of a high-avidity, high-frequency cytotoxic T-lymphocyte response is associated with antiviral protective immunity. J. Virol. 745769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thimme, R., J. Bukh, H. C. Spangenberg, S. Wieland, J. Pemberton, C. Steiger, S. Govindarajan, R. H. Purcell, and F. V. Chisari. 2002. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc. Natl. Acad. Sci. USA 9915661-15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 1941395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Timm, J., B. Li, M. G. Daniels, T. Bhattacharya, L. L. Reyor, R. Allgaier, T. Kuntzen, W. Fischer, B. E. Nolan, J. Duncan, J. Schulze zur Wiesch, A. Y. Kim, N. Frahm, C. Brander, R. T. Chung, G. M. Lauer, B. T. Korber, and T. M. Allen. 2007. Human leukocyte antigen-associated sequence polymorphisms in hepatitis C virus reveal reproducible immune responses and constraints on viral evolution. Hepatology 46339-349. [DOI] [PubMed] [Google Scholar]
- 26.Urbani, S., B. Amadei, E. Cariani, P. Fisicaro, A. Orlandini, G. Missale, and C. Ferrari. 2005. The impairment of CD8 responses limits the selection of escape mutations in acute hepatitis C virus infection. J. Immunol. 1757519-7529. [DOI] [PubMed] [Google Scholar]
- 27.Wedemeyer, H., E. Mizukoshi, A. R. Davis, J. R. Bennink, and B. Rehermann. 2001. Cross-reactivity between hepatitis C virus and influenza A virus determinant-specific cytotoxic T cells. J. Virol. 7511392-11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiner, A., A. L. Erickson, J. Kansopon, K. Crawford, E. Muchmore, A. L. Hughes, M. Houghton, and C. M. Walker. 1995. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc. Natl. Acad. Sci. USA 922755-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yusim, K., C. Kesmir, B. Gaschen, M. M. Addo, M. Altfeld, S. Brunak, A. Chigaev, V. Detours, and B. T. Korber. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 768757-8768. [DOI] [PMC free article] [PubMed] [Google Scholar]