Abstract
The human cytomegalovirus (HCMV) UL37 exon 1 protein (pUL37x1), also known as vMIA, is the predominant UL37 isoform during permissive infection. pUL37x1 is a potent antiapoptotic protein, which prevents cytochrome c release from mitochondria. The UL37x1 NH2-terminal bipartite localization signal, which remains uncleaved, targets UL37 proteins to the endoplasmic reticulum (ER) and then to mitochondria. Based upon our findings, we hypothesized that pUL37x1 traffics from the ER to mitochondria through direct contacts between the two organelles, provided by mitochondrion-associated membranes (MAMs). To facilitate its identification, we cloned and tagged the human phosphatidylserine synthase 1 (huPSS-1) cDNA, whose mouse homologue localizes almost exclusively in the MAM. Using subcellular fractionation of stable HeLa cell transfectants expressing mEGFP-huPSS-1, we found that HCMV pUL37x1 is present in purified microsomes, mitochondria, and MAM fractions. We further examined the trafficking of the full-length UL37 glycoprotein cleavage products, which divergently traffic either through the secretory apparatus or into mitochondria. Surprisingly, pUL37NH2 and gpUL37COOH were both detected in the ER and MAM fraction, even though only pUL37NH2 is preferentially imported into mitochondria but gpUL37COOH is not. To determine the sequences required for MAM importation, we examined pUL37x1 mutants that were partially defective for mitochondrial importation. Deletion mutants of the NH2-terminal UL37x1 mitochondrial localization signal were reduced in trafficking into the MAM, indicating partial overlap of MAM and mitochondrial targeting signals. Taken together, these results suggest that HCMV UL37 proteins traffic from the ER into the MAM, where they are sorted into either the secretory pathway or to mitochondrial importation.
Human cytomegalovirus (HCMV) is the leading viral cause of congenital birth defects, including mental retardation and blindness, and the leading nongenetic cause of neurosensory hearing loss in developed countries (8, 29). In addition to its impact on neonatal health, HCMV is a significant pathogen in immunosuppressed adults, particularly in transplant recipients (7, 29).
Alternative processing of HCMV UL37 immediate-early (IE) pre-mRNA, one of the first viral products produced during infection, predominantly generates the unspliced UL37 exon 1 (UL37x1) RNA and 10 UL37 alternatively spliced RNAs (1, 18, 21, 47, 50). These transcripts are predicted to encode six UL37 protein isoforms (1, 18, 21). The UL37x1 protein (pUL37x1), also known as viral mitochondrion-localized inhibitor of apoptosis (vMIA) and a product of the UL37x1 unspliced RNA, is the predominant UL37 isoform produced during permissive HCMV infection (18, 25, 42). pUL37x1 induces Ca2+ release from endoplasmic reticulum (ER) stores, resulting in reorganization of the F-actin stress network and the characteristic cytopathology of permissive HCMV infection (35, 42).
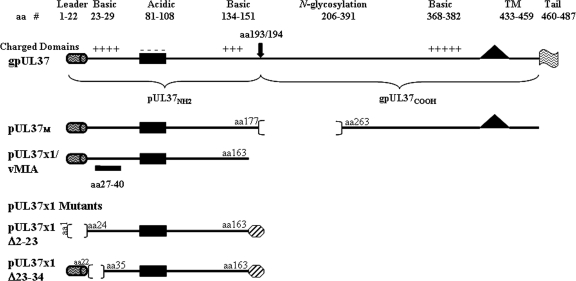
pUL37x1, the full-length UL37 glycoprotein (gpUL37), and the medium isoform (pUL37M) contain common NH2-terminal 163 amino acids (aa) encoded by UL37x1 (18, 21) (Fig. 1). These sequences include a strongly hydrophobic leader (aa 1 to 22) and a juxtaposed basic domain (aa 23 to 34), which jointly serve as a bipartite signal to target UL37 proteins to the ER and mitochondria (25, 27). This NH2-terminal targeting domain constitutes the first UL37x1 antiapoptotic domain, which when combined with a downstream domain (aa 118 to 147) are sufficient to confer its antiapoptotic activity (5, 18, 19, 28, 32, 34, 45). Between the UL37x1 antiapoptotic domains lies an acidic domain (aa 81 to 108), which plays a role in the transactivation of HCMV early gene promoters, whose products are needed for its oriLyt replication (13, 31, 52).
FIG. 1.
HCMV UL37 proteins and ORFs. The UL37x1 hydrophobic leader (aa 1 to 22) and juxtaposed basic (aa 23 to 29) and acidic (aa 81 to 108) domains are represented. The downstream UL37x2 and UL37x3 N-glycosylation domain, basic residues, transmembrane, and cytosolic tail are present in gpUL37 and pUL37M. The unique internal cleavage site of gpUL37 is indicated by the arrow at aa 193/194. The epitope for Ab1064 and DC35 (aa 27 to 40) and the C-terminal Flag tag (flag) are shown. For the pUL37x1 mutants, the sequences in pUL37x1 Δ2-23 and pUL37x1 Δ23-34 (19, 25), as well as the C-terminal Myc tag (hexagon), are shown.
pUL37x1, gpUL37, and pUL37M dually traffic to the ER and to the mitochondrial outer membrane in transfected and in HCMV-infected cells (2, 18, 19, 25-27, 42). Most mitochondrial proteins, including soluble and membrane-bound proteins, are synthesized in the cytosol, transported to mitochondria by heat shock proteins, and imported via translocation complexes in the outer and inner mitochondrial membranes (30, 36). Mitochondrial import requires that the cytosolic preprotein bind to cytosolic HSP70 and HSP90 and form a multichaperone complex, which then docks onto Tom70 (54). In stark contrast, we found that a gpUL37 cleavage site mutant was first translocated into the ER, where it was N glycosylated, and subsequently imported into mitochondria (27). Because this protein is anchored in the membrane by three transmembrane domains, we hypothesized its sequential trafficking from ER to mitochondria occurs through a continuous lipid bilayer connecting the two organelles. Although localization of proteins in the ER and in mitochondria is frequently observed (3, 9, 23), few cellular (12) and viral (27, 41) proteins are known to traffic sequentially from the ER to mitochondria. Consequently, this trafficking pathway connecting the ER to mitochondria is poorly understood.
High-resolution electron tomography has demonstrated that the ER and mitochondria are in close proximity and have numerous contacts (24). About 5 to 20% of the mitochondrial network surface within a cell is in close apposition to the ER (38). The mitochondrion-associated membrane (MAM), a subdomain of the ER, provides direct physical contact to mitochondria, as membrane bridges. Contacts between ER and mitochondria provide sites for transfer of lipids (46) and potentially of Ca2+ (17, 33, 49, 53). MAM has been fractionated to high purity from rat liver and is enriched in lipid synthetic enzymes (4, 6, 46, 51). MAM has also been less well characterized as a pre-Golgi compartment because of the presence of Golgi specific and endoglycosidase H (EndoH)-sensitive markers (40). Phosphatidylserine synthase types 1 and 2 (PSS-1 and PSS-2), fatty acid coenzyme A ligase 4 (FACL-4), ceramide synthase, and sphingolipid-specific glycotransferases are enriched in MAM, permitting direct delivery of their membrane-bound products to mitochondria (4, 6, 46, 51). Addition of an NH2-terminal Myc tag to murine PSS-1 did not alter traffic its trafficking pattern (46). Importation of phosphatidylserine into mitochondria occurs through contact points between the MAM and mitochondria and requires a mitochondrial membrane protein and energy (43, 46). The contacts between ER and mitochondria are partially stabilized by glucose regulated protein 75 (Grp75) (48) and by PACS-2 (44).
Because of the documented role of MAM in trafficking membrane-bound molecules from the ER to mitochondria, its potential role as a microdomain for Ca2+ transfer, and the sequential trafficking of membrane-anchored UL37 proteins from the ER to mitochondria, we tested whether HCMV pUL37x1 traffics into the MAM. To that end, we adapted a procedure to purify the MAM from the ER and mitochondria from transfected cells (10, 51). Using highly purified fractions, we found pUL37x1 trafficking into MAM in transfectants and identified the MAM targeting signals. To determine the MAM importation of other HCMV UL37 proteins, we also examined the trafficking of the cleavage products of gpUL37, only one of which traffics to the mitochondria. Surprisingly, both pUL37NH2 and gpUL37COOH trafficked into the MAM, suggesting that the MAM serves as a sorting compartment for HCMV UL37 proteins into the secretory apparatus or to mitochondria.
MATERIALS AND METHODS
Cell culture, transfections, and infections.
Life-extended human foreskin fibroblasts (LE-HFFs) (11) and HeLa cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, glutamine (4 mM), nonessential amino acids (0.1 mM), and penicillin-streptomycin (100 U/ml/0.1 mg/ml). Cells were lipofected as previously described previously (27) with plasmid DNA expressing wild-type pUL37x1 (p327) (15), pUL37x1 Δ2-23-myc (19), pUL37x1 Δ23-34-myc (19), gpUL37-Flag (p816) (14), or empty vector DNA (p396, p796, or p790). Cells were harvested after transfection and fractionated as described below and as previously published (10).
Cloning and expression of human PSS-1 (huPSS-1) gene product.
Two gene specific primers (forward, 5′-CGGGATCCGGACGGGGAGGCGGGCCATG-3′; reverse, 5′-CGGGATCCTTTCTTTCCAACGCCATTGGT-3′) were designed by using the huPSS-1 gene sequence (GenBank accession number NM_014754) to include the full open reading frame (ORF). Both primers contained a BamHI restriction site used for subsequent cloning. poly(A)+ RNA was isolated (Ambion) from HeLa cells and used for the generation of a cDNA library by reverse transcription (47). A total of 100 ng of the HeLa cDNA library was used as a template for the PCR amplification of huPSS-1 cDNA. PCRs were performed for 35 cycles at 94°C for 1 min, 53°C for 30 s, and 72°C for 2 min and a final 10-min extension at 72°C. The PCR products were separated by electrophoresis in 1% agarose gels. The desired 1.4-kbp product was excised from the gel and isolated by using a gel extraction kit (Qiagen). The purified cDNA fragment was cloned into the pCR BLUNT 4 zero-TOPO plasmid vector (Invitrogen), and its sequence was verified by DNA sequencing as previously described (47). DNA sequence comparisons were carried out by using BLAST 2.0 at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST). The huPSS-1 cDNA was subcloned downstream of the monomeric enhanced green fluorescent protein (mEGFP) ORF in the pmEGFP-C1 plasmid vector, carrying the neomycin resistance gene, using the forward primer (5′-GGGACTCGAGCCATGGCGTCCT-3′), containing an XhoI restriction site, and the reverse primer (5′-CCCCCCGGGATCATTTCTTTCCAAC-3′), containing a SmaI restriction site, generating pmEGFP-huPSS-1 (p1420). HeLa cells were lipofected with 7 μg of pmEGFP-huPSS-1 DNA. Stable transfectants were selected by culturing cells in medium containing G418 (1,500 μg/ml). Individual colonies were isolated, and the clone expressing the highest levels of mEGFP-huPSS-1, designated PSS-120, was used for transfections and isolation of the MAM fraction. PSS-120 cells were cultured in a reduced concentration of G418 (500 μg/ml) throughout the experiments.
Fractionation of MAM, mitochondria, and microsomes from cultured cells.
MAM, mitochondria, and microsomes were isolated from transfected PSS-120 cells as previously described (51) with our modifications (10). Briefly, PSS-120 cells were washed twice in phosphate-buffered saline (PBS) and pelleted by centrifugation at 1,000 × g for 10 min. The pellet was resuspended in sucrose homogenization medium (0.25 M sucrose, 10 mM HEPES [pH 7.4]), and cells were lysed by using a motor-driven Potter-Elvehjem homogenizer. The homogenates were subjected at least twice to a centrifugation at 600 × g for 5 min at 4°C, and a sample of the supernatant was removed and used as the total in our analysis. The supernatant was then centrifuged at 10,300 × g for 10 min in order to separate the crude microsomal (microsomes and cytosol) from the crude mitochondrial (MAM and mitochondria) fraction. The crude microsomal fraction (supernatant) was subjected to an ultracentrifugation at 100,000 × g in a Beckman SW41 rotor for 60 min at 4°C to pellet the microsomes, while the supernatant was concentrated by using 4-ml centrifugal filter units (Amicon, PLBC Ultracel-3 Membrane, 3 kDa) by centrifugation at 3,000 × g for 1 h at 4°C and was used as cytosol in our analysis. The crude mitochondrial fraction (pellet) was resuspended in 300 μl of ice-cold mannitol buffer A (0.25 M mannitol, 5 mM HEPES, 0.5 mM EGTA) and layered on top of 10 ml of a 30% Percoll suspension in mannitol buffer B (0.225 M mannitol, 25 mM HEPES, 1 mM EGTA). Mitochondria and MAM fractions were separated during the formation of the self-generating Percoll gradient by ultracentrifugation at 95,000 × g in a Beckman SW41 rotor for 65 min at 4°C. Both isolated fractions were diluted five times in sucrose homogenization medium and subjected separately to a centrifugation at 6,300 × g for 10 min at 4°C. The pellet of the mitochondria centrifugation was used as the purified mitochondria in our analysis, while the supernatant of the MAM centrifugation was further separated by centrifugation at 100,000 × g in a Beckman SW41 rotor for 30 min at 4°C, and the pellet was used as the purified MAM fraction. All of the fractions were resuspended in sucrose homogenization medium and analyzed by Western blot analysis.
Protein concentration determination.
The protein concentration of each subcellular fraction was determined by using a BCA reagent kit (Pierce), as suggested by the manufacturer.
EndoH and PNGase treatment.
Proteins in purified MAM or microsomal fractions were treated with EndoH or peptide-N-glycosidase (PNGase) as previously described (26). After denaturation, 10 μg of MAM or 20 μg of microsomal proteins was treated with 500 U of the desired enzyme in the appropriate buffer for 30 min at 37°C. Deglycosylated proteins were precipitated using 80% cold acetone and resuspended in 1× sodium dodecyl sulfate (SDS) loading buffer prior to electrophoresis in 10% polyacrylamide gels.
Western blot analysis.
A total of 10 μg of total lysate and subcellular fractions was separated by SDS-polyacrylamide gel electrophoresis (PAGE) in 12% Bis-Tris NuPage gels (Invitrogen) and analyzed by Western analyses as previously described (10, 27). Blots were probed with the following primary antibodies: rabbit anti-UL37x1 (aa 27 to 40) antisera (DC35, 1:1,000), mouse anti-Myc (MAb9E10, Berkeley Antibody Company, 1:100), mouse anti-Flag (M2, 1:2,000; Covance), goat anti-dolichyl phosphate mannose synthase 1 (DPM1; I-20, 1:100; Santa Cruz Biotechnology), mouse anti-Grp75 (SPA-825, 1:1000; StressGen Biotechnologies), or mouse anti-GFP (sc-9996 at 1:100; Santa Cruz Biotechnology) and with corresponding horseradish peroxidase-conjugated secondary antibody (1:2,000 or 1:3,000). Protein bands were detected using an ECL detection kit (Pierce). Each blot was stripped as described before (27) and reprobed for the detection of the specific protein organelle markers. Digital images were generated by using Scan Wizard Pro version 1.21 and processed in Adobe Photoshop version 7.0.
Indirect immunofluorescence.
LE-HFFs, HeLa (1.6 × 105), or PSS-120 (105) cells were plated onto coverslips in six-well plates and transiently cotransfected with vectors expressing the mitochondrion resident form of Discosoma red fluorescent protein (dsRed-1-Mito, 0.5 μg; Invitrogen), mEGFP-huPSS-1 (p1420), mEGFP (pmEGFP-C1), and/or pUL37x1 (p327, 0.5 μg) and were harvested 24 h later by fixation in ice-cold methanol. LE-HFFs (105) cells were plated onto coverslips in six-well plates and transiently cotransfected with vectors expressing mEGFP-huPSS-1 and infected 24 h later with HCMV strain Ad169 at a multiplicity of 3 PFU/cell as indicated. Infected cells were harvested at 12 h postinfection in ice-cold methanol. Fixed cells were probed by staining with anti-UL37x1 (aa 27 to 40; Ab1064, 1:300), human autoimmune antimitochondrial antibody (1:50; ImmunoVision) (14), mouse anti-KDEL receptor (1:100; Stressgen), or anti-GFP (mouse anti-GFP; Santa Cruz) antibodies as previously described (27). The secondary antibodies were fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG; 1:250; Jackson Immunoresearch), Texas red (TR)-goat anti-rabbit IgG (1:250), TR-goat anti-mouse IgG (1:100), cyanine 5 (Cy5)-conjugated goat anti-rabbit IgG (1:50), Cy5-goat anti-mouse IgG (1:200), or Cy5-goat anti-human IgG (1:50).
Confocal microscopy.
Imaging was performed by using a Bio-Rad MRC1024 confocal laser scanning microscope (Center for Microscopy and Image Analysis, GWU and Children's Mental Retardation and Developmental Disabilities Research Center) as previously described (14, 27). Confocal optical sections were obtained by using with a ×100 (NA = 1.35) lens and a 0.79-μm Z-dimension. Individual signals were captured sequentially to avoid spurious overlap of the emission signals. Images were generated by using Adobe Photoshop (version 7.0.1), Bio-Rad plug-ins, and Microsoft PowerPoint 2003.
RESULTS
HCMV pUL37x1 traffics into the MAM of transfected cells.
To determine whether HCMV UL37 proteins traffic into the MAM, PSS-120 cells stably expressing a tagged, known MAM marker (mEGFP-huPSS-1) were transiently transfected with pUL37x1 expression vector and fractionated (10). Purified microsomes, MAM, mitochondria, and cytosol were examined by Western analyses for the presence of pUL37x1 (Fig. 2). Consistent with our previous results (25-27), pUL37x1 was detected in microsomes and in mitochondria but not in the cytosol. Substantial amounts of pUL37x1 were also detected in the purified MAM fraction of transfected cells. The markers for mitochondria (Grp75), MAM (mEGFP-huPSS-1), and ER (DPM1) verified the identity and purity of the isolated subcellular fractions. Importantly, the markers for ER and mitochondria were faintly detectable in highly purified MAM, whereas pUL37x1 was readily detected in the MAM. These results demonstrate that the pUL37x1 localizes to the MAM, which is known to provide contact points between the ER and mitochondria, and its presence there does not result from mitochondrial or microsomal contamination of the MAM fraction.
FIG. 2.
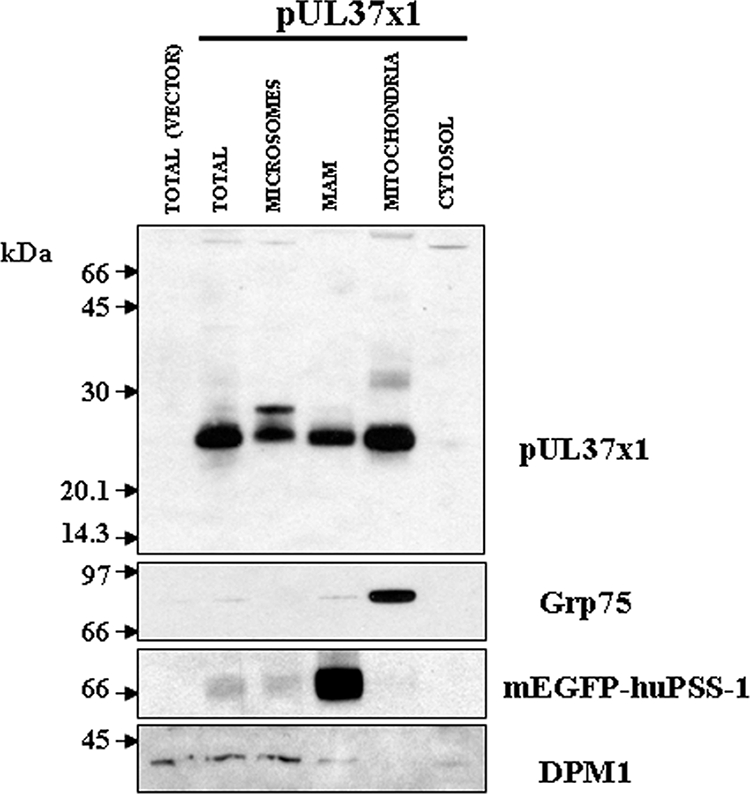
pUL37x1 traffics into microsomes, mitochondria, and MAM in transfected HeLa cells. PSS-120 cells that stably express the tagged MAM protein marker (mEGFP-huPSS-1) were transfected with either empty vector (p396) or vector expressing pUL37x1 (p327) and fractionated into microsomes, MAM, mitochondria, and cytosol by using Percoll gradient fractionation (10). A total of 10 μg of microsomes (0.7% of fraction), MAM (9.8%), mitochondria (9.7%), and cytosol (0.3%) was resolved by SDS-PAGE using 12% polyacrylamide, blotted, and reacted with a rabbit polyclonal antibody (DC35, 1:1,000) that recognizes UL37x1. The blots were stripped and reprobed with antibodies to specific protein markers for mitochondria (Grp75, mouse anti-Grp75 antibody, 1:1,000; Stressgen), MAM (mEGFP-huPSS-1, mouse anti-GFP antibody, 1:100; Santa Cruz), and ER (DPM1, goat anti-DPM1 antibody, 1:100; Santa Cruz) in order to verify the identity of purified fractions.
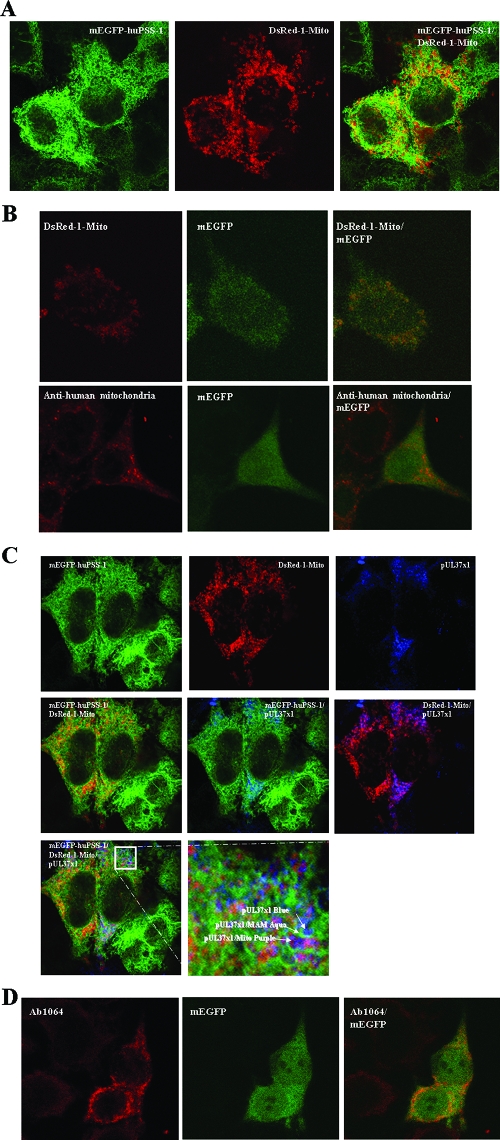
To verify the physical proximity of the MAM to mitochondria in the PSS-120 cells, we examined the colocalization of the MAM marker, mEGFP-huPSS-1 (green), with a second mitochondrial marker, dsRed-1-Mito (red) by confocal microscopy (Fig. 3A). Points of contact between MAM and mitochondria were observed by colocalization seen as yellow in the overlaid optical sections (right). In contrast, mEGFP was diffusely localized in control cells, transfected with pmEGFP expression vector and did not colocalize specifically with the mitochondrial marker, dsRed-1-Mito (Fig. 3B). Taken together, these results verify the close proximity of MAM and mitochondria in the PSS-120 cells, a finding consistent with the previously documented continuous membrane routes of transport between the ER through the MAM to mitochondria in purified subcellular fractions (46). Nonetheless, it is notable that the MAM domains distal to mitochondria were also observed as unaltered green in the overlaid image.
FIG. 3.
(A) Partial colocalization of mEGFP-huPSS-1 and dsRed-1-Mito markers. PSS-120 cells expressing mEGFP-huPSS-1 were transiently transfected with a plasmid expression vector for DsRed-1-Mito. After 24 h they were fixed and probed with mouse anti-GFP (1:200). The secondary antibody was FITC-goat anti-mouse (1:250, green). The cells were imaged for emissions at both 520 nm (FITC, left) and 615 nm (TR, middle) by using confocal microscopy. The overlaid image is on the right. (B) Diffuse mEGFP localization in transfected HeLa cells. HeLa cells were transfected with plasmids expressing mEGFP (pmEGFP-C1) and DsRed-1-Mito. Cells were fixed 24 h later and examined as in panel A for the presence of mEGFP and DsRed-1- mito. Fixed cells were examined for DsRed (top) or probed with human autoimmune anti-mitochondria antibodies (1:50) (bottom). Secondary antibody was TR-goat anti-human IgG (1:50). Confocal images were taken sequentially through a ×100 objective lens. Zeiss Lasersharp 2000 Acquisition software was used for additional magnifications. (C) Colocalization of pUL37x1 with mEGFP-huPSS-1 and with dsRed-1-Mito. PSS-120 cells expressing mEGFP-huPSS-1 were transiently transfected with DsRed-1-Mito expression vector and a pUL37x1 expression plasmid (p327). After 24 h they were fixed and probed with mouse anti-GFP (1:200) and rabbit anti-UL37x1 (Ab1064, 1:250). The secondary antibodies were FITC-goat anti-mouse (1:250, green) and Cy5-Goat anti-rabbit (1:50, blue). Cells were imaged by using confocal microscopy at 520 nm (FITC, top row, left panel), 615 nm (TR, top row, middle panel), and 670 nm (Cy5, top row, right panel). Panels also show the overlaid images of green-red (center row, left), green-cyan (center row, middle), red-cyan (center row, right) and green-red-cyan (bottom row, left). The bottom right panel shows detail of the boxed area from the triple overlay. (D) Lack of colocalization of mEGFP and pUL37x1 in transfected HeLa cells. HeLa cells were transfected with expression plasmid for mEGFP (pmEGFP-C1) and for pUL37x1 (p327). Cells were fixed 24 h later and examined as panel C for the presence of EGFP and for pUL37x1 by using Ab1064 (1:300) and TR-goat anti-rabbit IgG (1:300). Cells were imaged by using confocal microscopy at 520 nm (FITC, left panel) and 615 nm (TR, middle panel). The overlaid image is on the right.
To independently verify the presence of pUL37x1 in the MAM detected by subcellular fractionation, we examined the localization of pUL37x1 with the MAM marker and mitochondrial markers (Fig. 3C). Consistent with the findings above (Fig. 3A), physical proximity of the MAM and mitochondria was detected by the yellow signal in the overlay between mEGFP-huPSS1 (green) and dsRed-1-Mito (red) (Fig. 3C, center row, left panel). Moreover, pUL37x1 (blue) colocalized with the MAM marker (mEGFP-huPSS1) as shown by the aquamarine signal (center row, middle panel). Colocalization of pUL37x1 with the MAM (aquamarine) and of pUL37x1 with mitochondria (purple) was detected in proximal sites (bottom row), suggesting potential sites of exchange. To verify the specificity of the colocalization, control cells expressing mEGFP and pUL37x1 were examined by confocal microscopy (Fig. 3D). As observed above, mEGFP distribution in the transfected cells was broad and did not colocalize specifically with pUL37x1. These results independently establish the presence of pUL37x1 in the MAM in regions of close proximity with mitochondria. Because colocalizations of pUL37x1 with the MAM marker and with the mitochondrial marker are separable, they verify that the MAM-localized pUL37x1 species observed previously in subcellular fractionation does not arise from mitochondrial contamination of the MAM fraction.
Partial colocalization of pUL37x1 and the MAM marker in HCMV-infected cells.
To determine whether pUL37x1 could be visually detected in the MAM of permissively infected cells, we examined HCMV-infected LE-HFFs transiently expressing mEGFP-huPSS-1 (Fig. 4A). mEGFP-huPSS-1 in infected LE-HFFs was broadly distributed, suggestive of a reticular pattern of MAM, similar to the MAM morphology described in rat and mouse cells using anti-PSS-1 antibodies probes (46) and similar to that seen herein with PSS120 cells (Fig. 3A and B). As we previously found (42), pUL37x1 was detected in a reticular pattern in the cytosol of infected cells also expressing IE1/2 at 12 hpi. Partial colocalization of pUL37x1 with the MAM marker was detected in small, circumscribed regions of the infected cell, suggesting its transient presence in the MAM.
FIG. 4.
pUL37x1 partially colocalizes with mEGFP-huPSS-1 in permissive cells. LE-HFFs were transiently transfected with vectors expressing mEGFP-huPSS-1. (A) Cells were HCMV infected (3 PFU/cell) 24 h later, harvested at 12 h postinfection, and stained with Ab1064 (anti-pUL37x1) and MAb810 (anti-IE1/IE2). Cy5-Goat anti-mouse IgG (1:100) or TR-goat anti-rabbit IgG (1:100) secondary antibodies were used for visualization. Green (upper left), red (upper right), cyan (lower left), and overlaid channels (lower right) are shown. (B) Uninfected LE-HFFs, transiently expressing mEGFP-huPSS-1, were stained with human autoimmune anti-mitochondria (1:50) or (C) with mouse anti-KDELR (1:100). Cy5-goat anti-human IgG (1:50) or TR-goat anti-mouse IgG (1:100) secondary antibodies were used for visualization. In panel B, the green channel is at the top, red channel is in the middle, and the overlay is at the bottom. In panel C, the green channel is on the left, the red channel is in the middle, and the overlay is on right.
To determine whether MAM are in proximity of mitochondria in LE-HFFs, we performed colocalization studies with markers for each compartment. LE-HFFs, transiently expressing mEGFP-huPSS-1, were stained with anti-human mitochondrial antibody (Fig. 4B). The broad distribution of MAM, observed in infected cells at 12 h postinfection, was also observed in these uninfected cells. Partial colocalization of the MAM and mitochondrial markers was observed, suggesting their proximity in permissive cells.
Since the MAM have also been reported to be pre-Golgi compartments (40), we examined the colocalization of MAM compartment and a cis-Golgi marker, KDEL receptor (Fig. 4C). As above, the MAM was broadly distributed. Moreover, it was distinguishable from the cis-Golgi although some colocalization was observed between the two compartments.
Both gpUL37 cleavage products, pUL37NH2 and gpUL37COOH, traffic into MAM of transfected cells.
The HCMV full-length UL37 glycoprotein, gpUL37, is internally cleaved at its unique ER signal peptidase I cleavage site between aa 193 and 194 (Fig. 1) (26, 27). Its stable NH2-terminal fragment traffics into mitochondria, whereas the C-terminal fragment, gpUL37COOH, is preferentially retained in the secretory apparatus (26, 27). To determine whether these UL37 proteins also traffic into the MAM, we examined microsomes, MAM, mitochondria, and cytosol of cells transfected with a vector expressing gpUL37-Flag (Fig. 5). Similarly to pUL37x1, pUL37NH2 was detected in microsomes, MAM, and mitochondria but not in the cytosol (Fig. 5A, left top panel). After a stripping step, the blot was reacted with antibody against the C-terminal fragment. Surprisingly, gpUL37COOH, which preferentially traffics through the secretory apparatus, was enriched in the MAM fraction, as detected by its Flag tag of the microsomes, MAM and mitochondria (right panel). These results indicate that multiple HCMV UL37 proteins traffic into the MAM, but only a subset (pUL37x1 and pUL37NH2) of these traffics into mitochondria. These results further suggest that the MAM serves as a sorting compartment for HCMV UL37 proteins, which traffic divergently upon exiting the MAM subcompartment.
FIG. 5.
Both gpUL37 cleavage products traffic into microsomes, mitochondria, and MAM in transfected HeLa cells. PSS-120 cells that stably express the tagged MAM protein marker (mEGFP-huPSS-1) were transfected with either empty vector (p790) or vector expressing gpUL37-Flag (p816) and fractionated into microsomes, MAM, mitochondria, and cytosol by using Percoll gradient fractionation (10). A total of 10 μg of microsomes (0.6% of fraction), MAM (2.1%), mitochondria (1.6%), and cytosol (0.3%) were resolved by SDS-PAGE using 12% polyacrylamide and blotted. The blot was reacted with DC35 (anti-UL37x1, aa 27 to 40, 1:1,000) (left panel), stripped, and then reacted with anti-Flag antibody (M2, 1:2,000; Covance) (right panel) that recognizes the C-terminal tag of gpUL37. Blots were stripped and reprobed with antibodies against specific protein markers for mitochondria (Grp75, mouse anti-Grp75 antibody, 1:1,000; StressGen), MAM (mEGFP-huPSS-1, mouse anti-GFP antibody, sc-9996, 1:100; Santa Cruz), and ER (DPM1, goat anti-DPM1 antibody 1:100; Santa Cruz) to verify the identity of the purified fractions. In the left panel, the pUL37NH2 monomeric (arrowhead) and multimeric (filled circle) species are marked. The asterisk marks a background band. On the right, the multiple N-glycosylated gpUL37COOH species are indicated. (B) Endoglycosidase sensitivity of gpUL37COOH in MAM fraction. The microsomal (10 μg) and MAM (20 μg) proteins from panel A were treated with either PNGase (+) or EndoH (+). Control reactions were treated with buffer but lacked the added enzyme. Samples were then resolved by SDS-PAGE, blotted, and reacted with anti-Flag antibody as described above.
Because of this unanticipated finding that the gpUL37COOH fragment also trafficked into MAM, we examined whether the MAM was targeted after ER translocation but before Golgi trafficking. To that end, we examined the N glycosylation of the gpUL37COOH using endoglycosidases (Fig. 5B). In addition to its role as a direct contact with mitochondria, MAM has been characterized as a pre-Golgi compartment in that EndoHs proteins traffic through it in their trafficking to the plasma membrane (40). Treatment of banded MAM fractions from cells expressing gpUL37 with EndoH or peptide-N-glycosidase resulted in the reduction in the mass of gpUL37COOH detected in the MAM fraction, similar to the species present in the microsomal fraction. These results indicate that gpUL37COOH in the MAM is N glycosylated and EndoHs, a finding consistent with its previously characterized role of the MAM as a pre-Golgi compartment.
The MAM importation signals span UL37x1 aa 2 to 34.
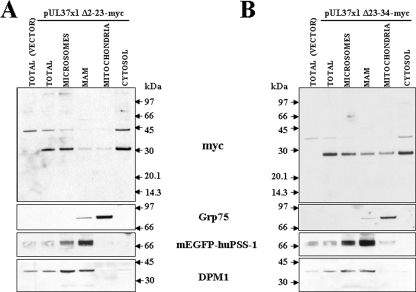
To determine the UL37x1 targeting signal for the MAM, we examined the trafficking of two C-terminally tagged pUL37x1 deletion mutants, known to be partially defective in mitochondrial importation (19). PSS-120 cells were transfected with a vector expressing pUL37x1 Δ2-23-myc and fractionated. Microsomes, MAM, and mitochondria were examined for the presence of pUL37x1 Δ2-23-myc (Fig. 6A). The importation of pUL37x1 Δ2-23-myc into both MAM and mitochondria was severely reduced compared to other fractions. As we previously found (25), the Myc-tagged UL37x1 mutant was aberrantly detected in the cytosol. However, in contrast to our previous finding of its absence from the ER fraction using a sonication protocol and banded ER (25), pUL37x1 Δ2-23-myc was detected here in the microsomal fraction. This detection likely results from use of this gentler homogenization lysis and suggests peripheral (but not integral) association of the pUL37x1 Δ2-23-myc mutant with the ER membrane, because of the lack of the strong hydrophobic leader sequence. The markers used verified the identity and purity of the mitochondria (Grp75), MAM (mEGFP-huPSS-1), and microsomes (DPM1).
FIG. 6.
Inefficient trafficking of pUL37x1 NH2-terminal deletion mutants to both MAM and mitochondria subcellular fractions. PSS-120 cells were lipofected with empty vector (p796) or vector expressing (A) pUL37x1 Δ2-23-myc (p856) or (B) pUL37x1 Δ23-34-myc (p857) and fractionated into microsomes, MAM, mitochondria, and cytosol. A total of 10 μg of p856 transfected microsomes (2.1% of fraction), MAM (3.3%), mitochondria (8.7%), and cytosol (1.5%) and 10 μg of p857 transfected microsomes (2% of fraction), MAM (4%), mitochondria (7.9%), and cytosol (1.2%) were analyzed by Western blot analysis by using a mouse antibody to the C-terminal Myc tag (1:100, MAb9E10; BabCo). Membranes were stripped and reprobed with antibodies to organelle-specific markers as described in Fig. 2.
We then examined a second pUL37x1 deletion mutant, which lacks the juxtaposed basic domain (aa 23 to 34) is selectively defective for mitochondrial importation (Fig. 6B). Transfected HeLa cells expressing pUL37x1 Δ23-34-myc were fractionated and examined for the presence of tagged mutant protein. As previously found (25), mitochondrial importation of pUL37x1 Δ23-34-myc was reduced compared to the wild-type pUL37x1. Although trafficking of pUL37x1 Δ23-34-myc into MAM was reduced, this reduction was comparable to that observed in trafficking of the wild-type pUL37x1 into the MAM. Again, the tagged mutant was observed in the cytosol as previously found (25). The markers for mitochondria (Grp75), MAM (mEGFP-huPSS-1), and microsomes (DPM1) verified the identity and purity of the fractionated organelles. Taken together, these results suggest that the pUL37x1 hydrophobic leader and, to a lesser degree, its juxtaposed basic domain play a role in the importation of pUL37x1 protein into the MAM.
DISCUSSION
The findings that some HCMV UL37 isoforms sequentially traffic from the ER to mitochondria and remain anchored to membranes (27) led us to examine whether the MAM might serve as a connection for their trafficking to mitochondria. To that end, we modified a Percoll gradient fractionation protocol to carefully separate MAM from mitochondria. We report here that UL37 proteins traffic into the MAM fraction of transfected cells. To verify the identity of the MAM fraction, we cloned, NH2-terminally tagged, and characterized the human homologue of the PSS-1 gene. The mouse PSS-1 gene product is known to localize almost exclusively to the MAM of rodent cells (46). mEGFP-huPSS-1 and other known cellular markers substantiated the efficient separation of the MAM fraction from mitochondria and from ER during fractionation. Fluorescent colocalization of pUL37x1 with mEGFP-huPSS-1 independently verified the physical presence of pUL37x1 in the MAM. Finally, by the subcellular fractionation, we found that the NH2-terminal UL37x1 targeting signals required for its mitochondrial importation also target importation of pUL37x1 into the MAM.
Since the hydrophobic leader is not cleaved, pUL37x1 and pUL37NH2 are anchored by their NH2 termini to membranes during trafficking and localization in ER and mitochondria. Consistent with this membrane anchoring is the finding that wild-type, untagged pUL37x1 is not detectable in the cytosol fraction of either transfected or HCMV-infected cells. However, two Myc-tagged pUL37x1 deletion mutants, defective in their trafficking to MAM and mitochondria, were aberrantly detected in the cytosol (19, 25).
Because of their sequential trafficking from the ER to mitochondria and their integral membrane association, we examined the trafficking of UL37 proteins into the MAM, which is known to serve as a venue for the translocation of membrane-bound phospholipids and glycosphingolipids into mitochondria. We report that HCMV pUL37x1 and the cleavage products of gpUL37, pUL37NH2, and gpUL37COOH target the MAM, presumably as a trafficking pathway from the ER into either the secretory pathway (gpUL37COOH) or the mitochondria (pUL37x1 or pUL37NH2). All of the HCMV UL37 proteins examined here (pUL37x1, pUL37NH2, and gpUL37COOH) were detected in the MAM fractions, suggesting that the MAM subcompartment may represent a key juncture for sorting of UL37 proteins into mitochondria or the secretory apparatus. This suggestion is supported by the finding that the gpUL37 cleavage fragments were both readily detected in the microsomes and in the MAM, whereas only pUL37NH2 trafficked efficiently into mitochondria. This possibility is consistent with the previous characterization of the MAM both as a pre-Golgi compartment of the secretory route through which albumin and apoproteins B traffic (40) and as a membrane bridge between the ER and mitochondria, which transfers lipids (46, 51) and HCV core protein (41) into mitochondria. Although pUL37x1 trafficking into mitochondria occurs efficiently, it, too, can traffic through the secretory apparatus, and we have previously detected pUL37x1 in the Golgi apparatus (14).
Our purification of low-density MAM relied on the previous fractionation methods initially developed for rat liver MAM (51). We increased the resolution between densities spanning the MAM and mitochondria in the self-generating Percoll gradients (10) and obtained highly purified MAM. Critical to the verification of the MAM fraction was the use of suitable known markers. As predicted from its identity as an ER subcompartment, MAM, banded in Percoll gradients, from human cells contained calreticulin, DPM1, and huPSS-1 (10). huPSS-1 tagged at its NH2 terminus with monomeric EGFP provided an invaluable tool for monitoring the MAM fractionation because it is highly enriched in the MAM. Murine PSS-1 protein is found almost exclusively in the MAM and is not detectable in the ER, and its low abundance in the microsomal fraction is attributable to residual MAM in the microsomal fraction (46). Moreover, unlike HCMV UL37 proteins, the tagged human homologue, mEGFP-huPSS-1, is selectively retained in the MAM and does not traffic into mitochondria and therefore served to distinguish these two closely apposed subcellular fractions.
Several lines of evidence suggest that the presence of pUL37x1 in MAM does not result from mitochondrial contamination. First, the abundance of mitochondrial marker (Grp75) is exceedingly low in banded MAM and does not correlate directly with the observed abundance of pUL37x1 in the MAM fraction. Because they do not correlate, the detected presence of pUL37x1 in the MAM is unlikely to result from the very low levels of mitochondrial contamination in transfected cells. In addition to fractionation studies, direct fluorescent colocalization of pUL37x1 with the MAM marker, mEGFP-huPSS-1, independently verified the presence of pUL37x1 in the MAM. As the Grp75 is concerned, it was recently reported (48) that an extramitochondrial Grp75 pool resides in the low-density MAM fraction and participates in high-molecular-weight complexes with ER and outer mitochondrial membrane proteins. Using confocal microscopy with the corresponding markers, the overlapping signals of the MAM (EGFP-huPSS1) and pUL37x1 (Ab1064) were distinguishable from those of mitochondria (dsRed-1-Mito) and pUL37x1 (Ab1064). Our findings are consistent with the identification of MAMs as direct contacts between the ER and mitochondria. Thus, the imaging studies show that some MAM domains are adjacent to mitochondria, likely sites of contact and transfer of lipids and UL37 proteins.
pUL37x1 traffics into the MAM of HCMV-infected HFFs, devoid of mitochondrial markers, as determined by subcellular fractionation (our unpublished results). In contrast, the colocalization of pUL37x1 in HCMV-infected cells was scantly detectable by confocal microscopy throughout infection (Fig. 3A) (C. D. Williamson and A. M. Colberg-Poley, unpublished results), suggesting that trafficking of this low-abundance HCMV protein through the MAM is rapid and transient in HCMV-infected cells and difficult to visualize with the highly expressed, transfected MAM marker mEGFP-huPSS-1.
To identify the sequences needed for targeting the MAM subcompartment, we made use of pUL37x1 deletion mutants, known to be defective in mitochondrial trafficking (19, 25). By the subcellular fractionation, we found that one of these mutants was also defective in MAM importation, indicating that the NH2-terminal UL37x1 mitochondrial localization signal also dictates importation of pUL37x1 into MAM. The lower level of pUL37x1Δ23-34 in the MAM was comparable to the levels observed with wild-type pUL37x1. These results suggest that the hydrophobic leader plays a more critical role than the basic domain for entry into the MAM. The fact that gpUL37COOH that lacks this signal also traffics into MAM implies that the two cleavage products (pUL37NH2 and gpUL37COOH) could remain associated, possibly through intermolecular interactions or by joint chaperone escort, after cleavage in the rough ER. Thus, the NH2-terminal importation signals could dictate the simultaneous importation of both products into the MAM subcompartment, from which they eventually traffic into different cellular locations. Alternatively, the UL37 C-terminal sequences unique to gpUL37COOH can independently mediate MAM trafficking.
Although numerous proteins localize to both the ER and the mitochondria (bcl-2, cytochrome), sequential trafficking of proteins from the ER to mitochondria has been rarely found (12, 27, 41). More often, the case is that separate but competitive trafficking of proteins occurs into either secretory apparatus or mitochondria (16, 22, 39). Cytochrome b5, NADH-cytochrome b5 reductase, and cytochrome P450 2E1 have all been reported to traffic alternatively into either the secretory pathway or the mitochondria (16, 22, 39). In these competitive pathways, modification of the targeting signals oftentimes dictates selective importation into either organelle. Sequential ER to mitochondria trafficking has been reported in very few cases. Spiro and coworkers (12) reported the translocation in rat liver cells of an unidentified glycoprotein from the ER into mitochondria. A portion of the hepatitis C virus core protein has also been shown to associate with the ER, mitochondria, and stable membrane bridges connecting the two, suggesting sequential trafficking from one organelle to the other (41).
Because correct protein targeting is necessary for proper function, these studies examining pUL37x1 trafficking provide insight into the requirements for pUL37x1 localization and resulting function in cells. pUL37x1 traffics unconventionally through the MAM, which serves as the ER-mitochondrion interface, en route from the ER to mitochondria, the site of its antiapoptotic activity. Importantly, the potent antiapoptotic effects of pUL37x1 are a downstream consequence of ER translocation and trafficking through the MAM. This particular routing suggests a required function. It has been suggested that the MAM serves as a microdomain for Ca2+ transfer from ER to mitochondria (17, 33, 48, 49, 53). HCMV pUL37x1 colocalizes with SERCA (a sarcoplasmic reticulum Ca2+ ATPase), induces Ca2+ release from the ER, and causes cytoskeletal changes during infection (35, 42). Trafficking of pUL37x1 from the ER through the MAM compartment will predictably affect several of its key known functions, including calcium release and antiapoptosis. Consistent with that suggestion is the finding that pUL37x1 mutants (Δ2-23 and Δ23-34) that are defective in trafficking through the MAM (the present study) and to mitochondria are defective in antiapoptosis (19, 25). HCMV UL37x1 viral mutants, defective in antiapoptotic activities, are also defective in their growth (37, 42). Taken together, these studies are important because they provide insight into how UL37 proteins traffic from the ER to mitochondria, where they play important roles for the survival of the infected cell.
In a broader sense, these studies with viral proteins substantiated a more global role of the MAM as a sorting compartment to both mitochondria and the Golgi apparatus. HCMV UL37 proteins represent useful tools for the characterization of the poorly understood MAM. Furthermore, the UL37 bipartite leader sequences could potentially allow scientists investigating mitochondrial dynamics or apoptosis a novel way to dynamically visualize mitochondria that have interacted with ER subdomains.
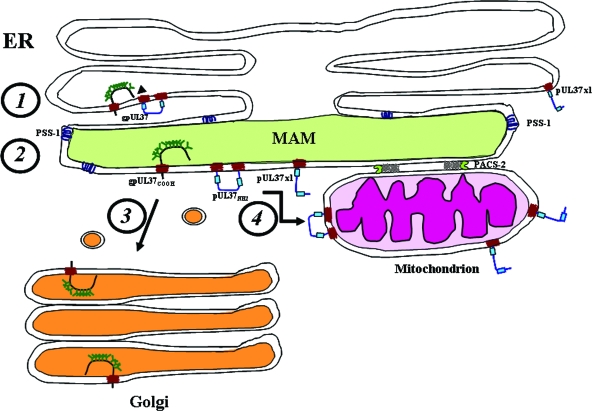
Based upon our results and those of others, we propose a model (Fig. 7) in which UL37 proteins (represented by gpUL37 and pUL37x1) are synthesized in the rough ER, processed, and remain anchored to the ER membrane by transmembrane domains (Fig. 7, step 1). Our previous findings of ER processing of UL37 proteins, including signal peptidase I cleavage, prior to mitochondrial importation (27) are consistent with step 1. We further propose that UL37 proteins traffic to the MAM compartment of the ER, where commitment to one of two pathways is chosen (Fig. 7, step 2). Detection of both gpUL37 proteolytic fragments and of pUL37x1 in the MAM is consistent with this trafficking in step 2. We then suggest that gpUL37COOH, which is preferentially retained in the secretory pathway (26, 27), and low levels of other UL37 proteins commit to the secretory pathway using the MAM as a pre-Golgi compartment (Fig. 7, step 3). The finding that gpUL37COOH is EndoHs is consistent with the previous description of MAM as a pre-Golgi compartment (40). This suggestion is further supported by the finding that gpUL37 produced during HCMV infection becomes EndoH resistant, supporting its subsequent processing in the Golgi (2), and by its detection at the plasma membrane of transfected cells (14). pUL37x1, which preferentially traffics to mitochondria, does to a lesser degree traffic through the secretory pathway and has been detected in the Golgi apparatus (14). PACS-2, which is known to affect the trafficking of polycystin-2 through the secretory apparatus (20), may likewise affect gpUL37COOH continued trafficking through the secretory apparatus.
FIG. 7.
Model of UL37 protein sorting through the MAM to the Golgi or mitochondria. (Step 1) UL37 proteins are synthesized anchored to the ER membrane. The UL37x3 sequences are translocated into the ER lumen, where they are cleaved by signal peptidase I and N glycosylated. (Step 2) pUL37x1 and both cleavage products of gpUL37 traffic into the MAM. The presence of PSS-1 in the MAM is shown. Sorting of UL37 proteins from the MAM to the Golgi apparatus (step 3) or into mitochondria (step 4) is represented. Mitochondrial importation likely occurs through direct membrane contacts between the MAM and mitochondria, which are stabilized by PACS-2.
We further propose in the model that pUL37x1 and pUL37NH2 commit to a second trafficking pathway to mitochondria (Fig. 7, step 4). The presence of pUL37x1 and pUL37NH2 that traffic efficiently into mitochondria in highly purified MAM is consistent with step 4 in Fig. 7. When the bridges transiently form between the MAM and mitochondria, UL37 proteins could traffic into the mitochondrial outer membrane, where they have antiapoptotic activity. Its topology and anchoring to membranes enables pUL37x1 to preferentially interact with the membrane-bound proapoptotic Bax protein, inducing its oligomerization and leading to its functional neutralization (5, 32). Nonetheless, the finding that multiple HCMV UL37 proteins traffic into the MAM, whereas only a subset of these is specifically committed into mitochondrial importation, shows that MAM localization is insufficient to commit UL37 proteins into mitochondrial importation. Rather, our results suggest that multiple trafficking fates can be achieved from the MAM subcompartment. Thus, there appears to be a bifurcation in the trafficking pattern of HCMV UL37 proteins to either continue in secretory apparatus or transfer into mitochondria from the secretory apparatus at a decisive step within the MAM.
Acknowledgments
We thank Jennifer Lippincott-Schwartz for the gift of pmEGFP expression vector.
These studies were funded, in part, by NIH R01 AI057906 and by funds from Children's Research Institute, the CNMC Board of Visitors, and the Kiwanis Clubs of Virginia. The confocal microscopy imaging was supported by a core grant (1P30HD40677) to the Children's Mental Retardation and Developmental Disabilities Research Center.
Footnotes
Published ahead of print on 16 January 2008.
REFERENCES
- 1.Adair, R., G. W. Liebisch, and A. M. Colberg-Poley. 2003. Complex alternative processing of human cytomegalovirus UL37 pre-mRNA. J. Gen. Virol. 843353-3358. [DOI] [PubMed] [Google Scholar]
- 2.Al-Barazi, H. O., and A. M. Colberg-Poley. 1996. The human cytomegalovirus UL37 immediate-early regulatory protein is an integral membrane N-glycoprotein which traffics through the endoplasmic reticulum and Golgi apparatus. J. Virol. 707198-7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anandatheerthavarada, H. K., G. Biswas, J. Mullick, N. B. Sepuri, L. Otvos, D. Pain, and N. G. Avadhani. 1999. Dual targeting of cytochrome P4502B1 to endoplasmic reticulum and mitochondria involves a novel signal activation by cyclic AMP-dependent phosphorylation at ser128. EMBO J. 185494-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardail, D., I. Popa, J. Bodennec, P. Louisot, D. Schmitt, and J. Portoukalian. 2003. The mitochondria-associated endoplasmic-reticulum subcompartment (MAM fraction) of rat liver contains highly active sphingolipid-specific glycosyltransferases. Biochem. J. 3711013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnoult, D., L. M. Bartle, A. Skaletskaya, D. Poncet, N. Zamzami, P. U. Park, J. Sharpe, R. J. Youle, and V. S. Goldmacher. 2004. Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. USA 1017988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bionda, C., J. Portoukalian, D. Schmitt, C. Rodriguez-Lafrasse, and D. Ardail. 2004. Subcellular compartmentalization of ceramide metabolism: MAM (mitochondria-associated membrane) and/or mitochondria? Biochem. J. 382527-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeckh, M., and W. G. Nichols. 2004. The impact of cytomegalovirus serostatus of donor and recipient before hematopoietic stem cell transplantation in the era of antiviral prophylaxis and preemptive therapy. Blood 1032003-2008. [DOI] [PubMed] [Google Scholar]
- 8.Boppana, S. B., K. B. Fowler, R. F. Pass, L. B. Rivera, R. D. Bradford, F. D. Lakeman, and W. J. Britt. 2005. Congenital cytomegalovirus infection: association between virus burden in infancy and hearing loss. J. Pediatr. 146817-823. [DOI] [PubMed] [Google Scholar]
- 9.Borgese, N., I. Gazzoni, M. Barberi, S. Colombo, and E. Pedrazzini. 2001. Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol. Biol. Cell 122482-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bozidis, P., C. D. Williamson, and A. M. Colberg-Poley. 2007. Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts. Curr. Protocols Cell Biol. 3.271-23. [DOI] [PubMed] [Google Scholar]
- 11.Bresnahan, W. A., G. E. Hultman, and T. Shenk. 2000. Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J. Virol. 7410816-10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra, N. C., M. J. Spiro, and R. G. Spiro. 1998. Identification of a glycoprotein from rat liver mitochondrial inner membrane and demonstration of its origin in the endoplasmic reticulum. J. Biol. Chem. 27319715-19721. [DOI] [PubMed] [Google Scholar]
- 13.Colberg-Poley, A. M., L. Huang, V. E. Soltero, A. C. Iskenderian, R. F. Schumacher, and D. G. Anders. 1998. The acidic domain of pUL37x1 and gpUL37 plays a key role in transactivation of HCMV DNA replication gene promoter constructions. Virology 246400-408. [DOI] [PubMed] [Google Scholar]
- 14.Colberg-Poley, A. M., M. B. Patel, D. P. Erezo, and J. E. Slater. 2000. Human cytomegalovirus UL37 immediate-early regulatory proteins traffic through the secretory apparatus and to mitochondria. J. Gen. Virol. 811779-1789. [DOI] [PubMed] [Google Scholar]
- 15.Colberg-Poley, A. M., L. D. Santomenna, P. P. Harlow, P. A. Benfield, and D. J. Tenney. 1992. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J. Virol. 6695-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colombo, S., R. Longhi, S. Alcaro, F. Ortuso, T. Sprocati, A. Flora, and N. Borgese. 2005. N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. J. Cell Biol. 168735-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filippin, L., P. J. Magalhaes, G. Di Benedetto, M. Colella, and T. Pozzan. 2003. Stable interactions between mitochondria and endoplasmic reticulum allow rapid accumulation of calcium in a subpopulation of mitochondria. J. Biol. Chem. 27839224-39234. [DOI] [PubMed] [Google Scholar]
- 18.Goldmacher, V. S., L. M. Bartle, A. Skaletskaya, C. A. Dionne, N. L. Kedersha, C. A. Vater, J. W. Han, R. J. Lutz, S. Watanabe, E. D. Cahir McFarland, E. D. Kieff, E. S. Mocarski, and T. Chittenden. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. USA 9612536-12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayajneh, W. A., A. M. Colberg-Poley, A. Skaletskaya, L. M. Bartle, M. M. Lesperance, D. G. Contopoulos-Ioannidis, N. L. Kedersha, and V. S. Goldmacher. 2001. The sequence and antiapoptotic functional domains of the human cytomegalovirus UL37 exon 1 immediate-early protein are conserved in multiple primary strains. Virology 279233-240. [DOI] [PubMed] [Google Scholar]
- 20.Kottgen, M., T. Benzing, T. Simmen, R. Tauber, B. Buchholz, S. Feliciangeli, T. B. Huber, B. Schermer, A. Kramer-Zucker, K. Hopker, K. C. Simmen, C. C. Tschucke, R. Sandford, E. Kim, G. Thomas, and G. Walz. 2005. Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. EMBO J. 24705-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouzarides, T., A. T. Bankier, S. C. Satchwell, E. Preddy, and B. G. Barrell. 1988. An immediate-early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology 165151-164. [DOI] [PubMed] [Google Scholar]
- 22.Kuroda, R., T. Ikenoue, M. Honsho, S. Tsujimoto, J. Y. Mitoma, and A. Ito. 1998. Charged amino acids at the carboxyl-terminal portions determine the intracellular locations of two isoforms of cytochrome b5. J. Biol. Chem. 27331097-31102. [DOI] [PubMed] [Google Scholar]
- 23.Lithgow, T., R. van Driel, J. F. Bertram, and A. Strasser. 1994. The protein product of the oncogene bcl-2 is a component of the nuclear envelope, the endoplasmic reticulum and the outer mitochondrial membrane. Cell Growth Differ. 5411-417. [PubMed] [Google Scholar]
- 24.Marsh, B. J., D. N. Mastronarde, K. F. Buttle, K. E. Howell, and J. R. McIntosh. 2001. Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc. Natl. Acad. Sci. USA 982399-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mavinakere, M. S., and A. M. Colberg-Poley. 2004. Dual targeting of the human cytomegalovirus UL37 exon 1 protein during permissive infection. J. Gen. Virol. 85323-329. [DOI] [PubMed] [Google Scholar]
- 26.Mavinakere, M. S., and A. M. Colberg-Poley. 2004. Internal cleavage of the human cytomegalovirus UL37 immediate-early glycoprotein and divergent trafficking of its proteolytic fragments. J. Gen. Virol. 851989-1994. [DOI] [PubMed] [Google Scholar]
- 27.Mavinakere, M. S., C. D. Williamson, V. S. Goldmacher, and A. M. Colberg-Poley. 2006. Processing of human cytomegalovirus UL37 mutant glycoproteins in the endoplasmic reticulum lumen prior to mitochondrial importation. J. Virol. 806771-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCormick, A. L., V. L. Smith, D. Chow, and E. S. Mocarski. 2003. Disruption of mitochondrial networks by the human cytomegalovirus UL37 gene product viral mitochondrion-localized inhibitor of apoptosis. J. Virol. 77631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. II. Wolters Kluwer Health, Lippincott/The Williams & Wilkins Co., Philadelphia, PA. [Google Scholar]
- 30.Neupert, W. 1997. Protein import into mitochondria. Annu. Rev. Biochem. 66863-917. [DOI] [PubMed] [Google Scholar]
- 31.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 672575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauleau, A.-L., N. Larochette, F. Giordanetto, S. R. Scholz, D. Poncet, N. Zamzami, V. S. Golmacher, and G. Koemer. 2007. Structure-function analysis of the interaction between Bax and the cytomegalovirus-encoded protein vMIA. Oncogene 267067-7080. [DOI] [PubMed] [Google Scholar]
- 33.Pinton, P., D. Ferrari, E. Rapizzi, F. Di Virgilio, T. Pozzan, and R. Rizzuto. 2001. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. EMBO J. 202690-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poncet, D., N. Larochette, A. L. Pauleau, P. Boya, A. A. Jalil, P. F. Cartron, F. Vallette, C. Schnebelen, L. M. Bartle, A. Skaletskaya, D. Boutolleau, J. C. Martinou, V. S. Goldmacher, G. Kroemer, and N. Zamzami. 2004. An antiapoptotic viral protein that recruits Bax to mitochondria. J. Biol. Chem. 27922605-22614. [DOI] [PubMed] [Google Scholar]
- 35.Poncet, D., A. L. Pauleau, G. Szabadkai, A. Vozza, S. R. Scholz, M. Le Bras, J. J. Briere, A. Jalil, R. Le Moigne, C. Brenner, G. Hahn, I. Wittig, H. Schagger, C. Lemaire, K. Bianchi, S. Souquere, G. Pierron, P. Rustin, V. S. Goldmacher, R. Rizzuto, F. Palmieri, and G. Kroemer. 2006. Cytopathic effects of the cytomegalovirus-encoded apoptosis inhibitory protein vMIA. J. Cell Biol. 174985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rapaport, D. 2003. Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep. 4948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reboredo, M., R. F. Greaves, and G. Hahn. 2004. Human cytomegalovirus proteins encoded by UL37 exon 1 protect infected fibroblasts against virus-induced apoptosis and are required for efficient virus replication. J. Gen. Virol. 853555-3567. [DOI] [PubMed] [Google Scholar]
- 38.Rizzuto, R., P. Pinton, W. Carrington, F. S. Fay, K. E. Fogarty, L. M. Lifshitz, R. A. Tuft, and T. Pozzan. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 2801763-1766. [DOI] [PubMed] [Google Scholar]
- 39.Robin, M. A., H. K. Anandatheerthavarada, G. Biswas, N. B. Sepuri, D. M. Gordon, D. Pain, and N. G. Avadhani. 2002. Bimodal targeting of microsomal CYP2E1 to mitochondria through activation of an N-terminal chimeric signal by cAMP-mediated phosphorylation. J. Biol. Chem. 27740583-40593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rusiñol, A. E., Z. Cui, M. H. Chen, and J. E. Vance. 1994. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 26927494-27502. [PubMed] [Google Scholar]
- 41.Schwer, B., S. Ren, T. Pietschmann, J. Kartenbeck, K. Kaehlcke, R. Bartenschlager, T. S. Yen, and M. Ott. 2004. Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif. J. Virol. 787958-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharon-Friling, R., J. Goodhouse, A. M. Colberg-Poley, and T. Shenk. 2006. Human cytomegalovirus pUL37x1 induces the release of endoplasmic reticulum calcium stores. Proc. Natl. Acad. Sci. USA 10319117-19122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiao, Y. J., B. Balcerzak, and J. E. Vance. 1998. A mitochondrial membrane protein is required for translocation of phosphatidylserine from mitochondria-associated membranes to mitochondria. Biochem. J. 331(Pt. 1)217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmen, T., J. E. Aslan, A. D. Blagoveshchenskaya, L. Thomas, L. Wan, Y. Xiang, S. F. Feliciangeli, C. H. Hung, C. M. Crump, and G. Thomas. 2005. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 24717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, G., and E. S. Mocarski. 2005. Contribution of GADD45 family members to cell death suppression by cellular bcl-xL and cytomegalovirus vMIA. J. Virol. 7914923-14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stone, S. J., and J. E. Vance. 2000. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J. Biol. Chem. 27534534-34540. [DOI] [PubMed] [Google Scholar]
- 47.Su, Y., R. Adair, C. N. Davis, N. L. DiFronzo, and A. M. Colberg-Poley. 2003. Convergence of RNA cis elements and cellular polyadenylation factors in the regulation of human cytomegalovirus UL37 exon 1 unspliced RNA production. J. Virol. 7712729-12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szabadkai, G., K. Bianchi, P. Varnai, D. De Stefani, M. R. Wieckowski, D. Cavagna, A. I. Nagy, T. Balla, and R. Rizzuto. 2006. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 175901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szabadkai, G., and R. Rizzuto. 2004. Participation of endoplasmic reticulum and mitochondrial calcium handling in apoptosis: more than just neighborhood? FEBS Lett. 567111-115. [DOI] [PubMed] [Google Scholar]
- 50.Tenney, D. J., and A. M. Colberg-Poley. 1991. Human cytomegalovirus UL36-38 and US3 immediate-early genes: temporally regulated expression of nuclear, cytoplasmic, and polysome-associated transcripts during infection. J. Virol. 656724-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vance, J. E. 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 2657248-7256. [PubMed] [Google Scholar]
- 52.Yee, L. F., P. L. Lin, and M. F. Stinski. 2007. Ectopic expression of HCMV IE72 and IE86 proteins is sufficient to induce early gene expression but not production of infectious virus in undifferentiated promonocytic THP-1 cells. Virology 363174-188. [DOI] [PubMed] [Google Scholar]
- 53.Yi, M., D. Weaver, and G. Hajnoczky. 2004. Control of mitochondrial motility and distribution by the calcium signal: a homeostatic circuit. J. Cell Biol. 167661-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young, J. C., N. J. Hoogenraad, and F. U. Hartl. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 11241-50. [DOI] [PubMed] [Google Scholar]