Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) spike glycoprotein (S) is a class I viral fusion protein that binds to its receptor glycoprotein, human angiotensin converting enzyme 2 (hACE2), and mediates virus entry and cell-cell fusion. The juxtamembrane domain (JMD) of S is an aromatic amino acid-rich region proximal to the transmembrane domain that is highly conserved in all coronaviruses. Alanine substitutions for one or two of the six aromatic residues in the JMD did not alter the surface expression of the SARS-CoV S proteins with a deletion of the C-terminal 19 amino acids (S Δ19) or reduce binding to soluble human ACE2 (hACE2). However, hACE2-dependent entry of trypsin-treated retrovirus pseudotyped viruses expressing JMD mutant S Δ19 proteins was greatly reduced. Single alanine substitutions for aromatic residues reduced entry to 10 to 60% of the wild-type level. The greatest reduction was caused by residues nearest the transmembrane domain. Four double alanine substitutions reduced entry to 5 to 10% of the wild-type level. Rapid hACE2-dependent S-mediated cell-cell fusion was reduced to 60 to 70% of the wild-type level for all single alanine substitutions and the Y1188A/Y1191A protein. S Δ19 proteins with other double alanine substitutions reduced cell-cell fusion further, from 40% to less than 20% of wild-type levels. The aromatic amino acids in the JMD of the SARS-CoV S glycoprotein play critical roles in receptor-dependent virus-cell and cell-cell fusion. Because the JMD is so highly conserved in all coronavirus S proteins, it is a potential target for development of drugs that may inhibit virus entry and/or cell-cell fusion mediated by S proteins of all coronaviruses.
Coronaviruses (CoVs) are large enveloped viruses with positive-sense RNA genomes that cause economically important respiratory or enteric diseases of many species, including humans. Entry of coronaviruses into host cells and virus-induced cell-cell fusion are mediated by interactions between the ∼200-kDa viral spike glycoprotein (S) and receptor glycoproteins. Trimers of S make up the characteristic spikes that form the “corona” seen in negatively stained CoVs. S is a class I viral membrane fusion glycoprotein (2, 11). Class I viral fusion proteins of diverse virus families, including Retroviridae, Filoviridae, Orthomyxoviridae, Paramyxoviridae, and Coronaviridae (also called type I viral fusion proteins), differ greatly in size and amino acid sequence, but their membrane-anchored domains share common structural features that are essential for membrane fusion, including two heptad repeats (called HR-1 and HR-2), preceded by a hydrophobic fusion peptide (11, 48). Upon receptor-binding and/or pH triggering, class I viral fusion proteins mediate both virus-cell and cell-cell fusion through conformational changes that trigger formation of intermediate conformations and finally the postfusion (six-helix bundle) conformation (7, 11). Peptides homologous to C-terminal HR-2 can inhibit coronavirus entry (1, 27, 44), similar to findings with other class I viral fusion proteins.
Class I fusion glycoproteins of coronaviruses and many other types of viruses also contain a short region in the ectodomain between the end of HR-2 and the beginning of the transmembrane domain (TM), called the juxtamembrane domain (JMD), which is rich in aromatic amino acids. Several possible roles in membrane fusion have been suggested for the JMDs of class I viral fusion proteins of human immunodeficiency virus (HIV) (33, 34, 42, 46, 47), feline immunodeficiency virus (FIV) (16), and Ebola virus (43) and for the class III fusion protein of vesicular stomatitis virus (VSV) (20, 21, 39). These JMDs may contribute to the conformational changes that occur during membrane fusion, interact with membranes, induce membrane destabilization, and/or facilitate membrane fusion. Therefore, the JMD of class I viral fusion proteins is a potential target for viral inhibitors. Entry inhibitors that target the JMD of FIV include a monoclonal antibody that binds to the FIV JMD and an octapeptide that mimics the FIV JMD (13, 15). In addition, two neutralizing monoclonal antibodies to the HIV gp41 JMD have been identified (29, 38).
Although the JMDs of fusion proteins of enveloped viruses are rich in aromatic amino acids, the number, spacing, and sequence of the aromatic amino acids are quite variable (Fig. 1A). To define the C-terminal boundaries of the JMDs of fusion proteins of viruses, it was necessary to identify the beginning of the TM. Because computer algorithms differ in their prediction of the beginning of the TM, we used TMPred to predict the beginning of the TMs of all fusion proteins but the coronavirus S proteins. We predicted the TM of coronaviruses using four algorithms (TMpred [http://www.ch.embnet.org/software/TMPRED_form.html], DAS, HMMTOP, and SOSUI [http://sosui.proteome.bio.tuat.ac.jp/about-sosui.html]). In contrast to the highly variable JMDs of other enveloped virus families, the JMDs of S glycoproteins of all three major phylogenetic groups of coronaviruses are highly conserved (Fig. 1). They contain 14 amino acids of which 6 are tyrosine and tryptophan residues (Fig. 1B). Several observations suggest that the JMD of the severe acute respiratory syndrome CoV (SARS-CoV) S protein may play a role in virus entry, membrane destabilization, and cell-cell fusion (3, 18). These processes are dependent upon binding of the spike to the receptor, human angiotensin-converting enzyme 2 (hACE2) (26). Retrovirus pseudotyped viruses containing chimeric spikes with the ectodomain of the SARS-CoV S glycoprotein and the JMD, TM, and endodomain of the VSV G glycoprotein form less-stable trimeric spikes and reduce entry and cell-cell fusion relative to results with the wild-type SARS-CoV S protein (3). Several synthesized peptides derived from the JMD of the SARS-CoV S protein destabilized liposomes, indicating that the peptides can directly interact with lipid bilayers (18, 45).
FIG. 1.
Aromatic amino acids in the JMDs of enveloped viruses. (A) Amino acid sequences of the JMDs were aligned according to the predicted TMs, predicted by several computer algorithms: TMpred (http://www.ch.embnet.org/software/TMPRED_form.html), DAS, HMMTOP, and SOSUI (http://sosui.proteome.bio.tuat.ac.jp/about-sosui.html). Aromatic amino acids are highlighted in bold. (B) Conserved amino acid sequences in the JMDs of diverse coronavirus spike glycoproteins were aligned based on the consensus KWPWYVWL sequence obtained from CLUSTALW. Identical amino acids are shaded, and aromatic amino acids are displayed in bold. GenBank accession numbers are as follows: SARS-CoV, P59594; murine hepatitis virus A59 (MHV-A59), AAR92026; infectious bronchitis virus (IBV), AAO34396; HIV type 1 (HIV-1), AAR05826; HIV type 2 (HIV-2), 2105299E; Moloney murine leukemia virus (MoMuLV), AAQ95285; Mason-Pfizer monkey virus (MPMV), NP_056894; Ebola virus, AAA96744; Marburg virus, ABE27015; VSV, NP_955548); influenza HA (Flu HA), ABP49228; Sendai virus, AAC82296; and Sindbis virus, CAA24684.
To examine the role of the six tyrosine and tryptophan residues in the JMD of SARS-CoV S protein in membrane fusion, we tested S proteins containing alanine substitutions for these residues for the ability to mediate receptor-dependent entry of pseudotyped murine leukemia virus (MLV) and cell-cell fusion. Each of the tryptophan residues within the SARS-CoV S JMD was required for efficient S-mediated entry and cell-cell fusion. Alanine substitution of multiple tryptophan residues further reduced both entry and cell-cell fusion. Each of the tyrosine residues within the JMD was necessary for efficient cell-cell fusion, although only one substitution, Y1197A, greatly reduced virus entry. Y1188A and Y1191A only slightly reduced cell-cell fusion alone or together. Thus, the six highly conserved aromatic amino acid residues in the JMD of coronavirus S proteins are critically important for the membrane fusion activity that leads to virus-cell and cell-cell fusion. In addition, several of the aromatic amino acids exhibit differences in their contribution to virus-cell fusion (virus entry) and cell-cell fusion (spread of infection), indicating subtle differences between the roles of the coronavirus JMD during entry and in cell-to-cell fusion.
MATERIALS AND METHODS
Cell culture and reagents.
HEK-293 (kindly provided by Jerry Schaak, University of Colorado Health Sciences Center), 293T, Vero E6, and BHK-21 cells (obtained from the ATCC) were grown in high-glucose Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and 2% penicillin-streptomycin-fungizone (Invitrogen) at 37°C with 5% CO2. HEK-293 cells stably expressing hACE2 were made by transfection of HEK-293 cells with pcDNA3.1 hACE2, selection with G418 (Invitrogen), and sorting for hACE2 expression with goat anti-human hACE2 AF933 (R&D Systems, Minneapolis, MN) and phycoerythrin (PE)-conjugated anti-goat immunoglobulin G (IgG) (Jackson Immunoresearch, West Grove, PA) antibodies. Flow cytometry was performed using a Beckman Coulter FC500 running CXP software in the University of Colorado Cancer Center Flow Cytometry Core.
Cloning.
The SARS S DNA sequence (amino acids 1 to 1255) was cloned from cDNA of the Urbani strain of SARS-CoV (kindly provided by W. Bellini, Centers for Disease Control and Prevention). The DNA encoding the S glycoprotein was subcloned into pcDNA3.1D TOPO (Invitrogen) using the manufacturer's protocol, generating pcDNA3.1 SARS-CoV S. A plasmid encoding the codon-optimized Urbani strain SARS-CoV S glycoprotein was obtained from D. Ambrosino (University of Massachusetts Medical School). Nucleotides encoding the C-terminal 19 amino acids of the codon-optimized SARS-CoV S glycoprotein were removed by PCR, generating pcDNA3.1 SARS-CoV S Δ19 by modification of a published protocol (17). Primers to the 5′ end (5′-CACCATGTTCATCTTCCTGC-3′) and the 3′ end (5′-CTAGCAGCAAGAGCCGC-3′) coding for a stop codon at codon position 1236 were used to amplify the region that codes for amino acids 1 to 1236. The PCR product was cloned into pcDNA3.1 TOPO using the manufacturer's protocol (Invitrogen). Site-directed mutagenesis (QuikChange kit; Stratagene, La Jolla, CA) was used to generate plasmids encoding mutant SARS-CoV S Δ19 glycoproteins. Plasmids encoding full-length hACE2 (pACE2-cq) and soluble hACE2 (shACE2) fused to the Fc domain of human IgG [shACE2-Fc protein; pACE2(H378N)-Fc)] were kindly provided by M. Farzan (Harvard University, Cambridge, MA). The DNA sequence encoding hACE2 was amplified by PCR from pACE2-cq using a forward primer that included a Kozak sequence (5′-CACCATGTCAAGCTCTTCCTGG-3′) and a reverse primer that eliminated the stop codon of hACE2 (5′-CTAAAAGGAGGTCTGAACATCATCAGT-3′). PCR fragments of hACE2 were cloned into the pcDNA3.1D TOPO V5/6His vector (Invitrogen) using the manufacturer's protocol to generate pcDNA3.1 ACE2. cDNA encoding the soluble SARS-CoV S Urbani strain (amino acids 1 to 1190) with a C-terminal six-histidine tag was subcloned into pcDNA3.1 from a plasmid encoding the full-length SARS-CoV Urbani S glycoprotein (pcDNA3.1 SARS-CoV S), which generated pcDNA3.1 S1190. The nucleotide sequences of all plasmids were confirmed by DNA sequencing at the CU Cancer Center DNA Sequencing and Analysis Core. Plasmids pcDNA3.1 MLV gagpol and pcDNA3.1 LTR-LacZ, used to generate murine leukemia virus (MLV) pseudotyped viruses, were kindly provided by W. Mothes (Yale University).
Purification of soluble SARS-CoV S1190.
S1190 RNA was expressed by RNA polymerase encoded by modified vaccinia virus Ankara strain T7 (kindly provided by B. Moss [4]). Subconfluent BHK-21 cells were inoculated with modified vaccinia virus Ankara strain T7 at a multiplicity of infection of 5. Three hours postinfection, each T150 flask of infected cells was transfected with 24 μg pcDNA3.1 S1190 using 48 μl Lipofectamine 2000 (Invitrogen). Supernatant medium was harvested at 72 h postinfection, clarified by centrifugation, and filtered (Millipore). The S1190 protein was purified by nickel affinity chromatography using HiTrap nickel affinity columns (Pharmacia, Piscataway, NJ). All fractions were analyzed by immunoblotting using rabbit anti-V5 tag IgG (Invitrogen). Goats were immunized with purified protein to generate goat anti-SARS-CoV S antibody no. 4473.
Expression of SARS-CoV S Δ19 glycoproteins.
Expression of wild-type and mutant SARS S Δ19 was detected by immunoblotting, flow cytometry, and immunofluorescence. BHK-21 cells in six-well plates were transiently transfected with 4 μg pcDNA3.1 SARS S Δ19, mutant SARS S Δ19 plasmids, or empty pcDNA3.1 vector using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). Cells detached with trypsin (2.5 mg/ml) containing 0.38 mg/ml EDTA (trypsin/EDTA) (Invitrogen) were resuspended in lysis buffer (50 mM Tris [pH 8.0], 62.5 mM EDTA, 0.4% deoxycholate, 1% NP-40). Lysates were spun at 10,000 × g for 10 min, and proteins in the supernatant were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene difluoride membranes, and visualized with polyclonal antipeptide rabbit anti-SARS S BL637 antibody raised to a peptide derived from SARS-CoV S Urbani amino acids 551 to 577 (Bethyl Laboratories, Montgomery, TX) or a polyclonal goat antibody raised to the purified soluble ectodomain of the SARS-CoV S protein (amino acids 1 to 1190), Gt 4473, followed by horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-goat IgG antibody (Jackson Immunoresearch). Proteins were visualized with Western Lighting Chemiluminescent Reagent Plus (Perkin-Elmer, Boston, MA).
Surface expression of wild-type or mutant SARS-CoV S Δ19 proteins was detected by flow cytometry. Cells were detached 24 h posttransfection with phosphate-buffered saline (PBS) containing 1 mM EDTA. S protein on the cell surface was labeled with monoclonal mouse anti-SARS S (Zymed, Invitrogen) and a PE-conjugated anti-mouse IgG (Jackson Immunoresearch). Cells were fixed with 4% paraformaldehyde in PBS and analyzed by flow cytometry. Graphs were prepared using the FlowJo software program (Ashland, OR), where the y axis sets the maximum fluorescence for each curve to 100%. Surface expression of wild-type and mutant SARS S Δ19 glycoproteins was also analyzed by immunofluorescence. Twenty-four hours posttransfection, BHK-21 cells were fixed with 4% paraformaldehyde (Ted Pella, Redding, CA) in PBS, labeled with monoclonal mouse anti-SARS S antibody and Alexafluor 488 conjugated anti-mouse IgG (Molecular Probes, Invitrogen), and visualized with a Zeiss Axiovert fluorescence microscope.
Soluble human ACE2-Fc binding assay.
293T cells in a T150 flask were transfected with 10 μg plasmid encoding soluble human ACE2-Fc [pACE2(H378N)-Fc)] using calcium phosphate. Seventy-two hours posttransfection, supernatant medium containing shACE2-Fc was harvested, filtered though a 0.22-μm filter (Fisher), and concentrated 10-fold through 30,000-molecular-weight-cutoff concentrators (Millipore, Billerica, MA). BHK-21 cells were transfected with plasmids encoding wild-type or mutant SARS S Δ19 or empty pcDNA3.1 vector using Lipofectamine 2000. The concentrated hACE2-Fc supernatants were incubated with cells expressing S proteins detached with trypsin-EDTA or PBS with 1 mM EDTA 24 h posttransfection. Bound shACE2-Fc was detected with PE-conjugated anti-human IgG antibody (Jackson Immunoresearch). Cells were fixed in 4% paraformaldehyde and analyzed by flow cytometry.
Pseudotype production, purification, and analysis.
MLV pseudotyped viruses were produced as previously reported (49). Briefly, BHK-21 cells in 10-cm2 dishes were transiently transfected with 4 μg pcDNA3.1 MLV gagpol, 4 μg pcDNA3.1 LTR-LacZ, and 4 μg of wild-type or mutant S Δ19 protein plasmid or empty pcDNA3.1 plasmid using 47 μl Fugene 6 (Roche, Indianapolis, IN). Twenty-four hours posttransfection, cells were detached with trypsin-EDTA (Gibco) or 1 mM EDTA (Sigma) and plated in T150 flasks in DMEM with 10% FBS. Forty-eight hours posttransfection, supernatant medium containing pseudotyped virus was harvested and filtered through 0.45-μm syringe filters (Millipore). Pseudotyped virus was concentrated by ultracentrifugation through 15% sucrose in PBS at 25,000 rpm at 4°C in an SW28 rotor for 1.5 h using a Beckman ultracentrifuge. Pseudotyped viruses were resuspended in DMEM and analyzed by electron microscopy. Alternatively, filtered medium containing pseudotyped viruses was spun at 20,800 × g (14,000 rpm in an Eppendorf centrifuge) for 30 min at 4°C (protocol adapted from reference 30). Pellets containing pseudotyped virus were resuspended in 4× Laemmli buffer (8% SDS, 40% glycerol, 0.008% bromophenol blue, 0.250 M Tris-HCl, 50 mM dithiothreitol), analyzed by SDS-PAGE, and transferred to polyvinylidene difluoride membranes. Immunoblots were performed with goat anti-MLV p30 (kindly provided by L. Albritton) or goat anti-SARS-CoV S no. 4473 and HRP-conjugated rabbit anti-goat IgG (Jackson Immunoresearch). Proteins were visualized with Western Lighting Chemiluminescent Reagent Plus (Perkin-Elmer).
Concentrated pseudotyped viruses on carbon-coated formvar grids (Ted Pella) were fixed with 1% glutaraldehyde (Ted Pella), negatively stained with 2% sodium silicotungstate (Canemco, Canton de Gore, Quebec, Canada), and visualized using a FEI Technai G2 transmission electron microscope at 80 kV running Digital Micrograph software.
HEK-293/hACE2 and Vero E6 cells in 24-well plates were inoculated with 170 to 180 μl supernatant containing pseudotyped viruses or with concentrated pseudotyped viruses diluted in DMEM for 4 to 6 h at 37°C in 5% CO2. DMEM (0.5 ml) with 10% FBS and 2% penicillin-streptomycin-fungizone was added to each well, and cells were incubated for 48 h at 37°C in 5% CO2. Forty-eight hours postinoculation, cells were fixed in 0.5% glutaraldehyde (Ted Pella) in PBS and stained with 0.1 M K3Fe(CN)6-0.1 M K4Fe(CN)6 and 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (Invitrogen) in PBS containing 1 mM MgCl2. Titers of pseudotyped viruses were calculated by counting at least three 10× fields/well per dilution. Dilutions were plated in triplicate. The percentage of beta-galactosidase-positive cells per well was divided by the inoculum volume to obtain titers in transduction units per ml (TU/ml). Data were compared between replicate experiments as percentages of transduction by wild-type SARS-CoV S Δ19.
Cell fusion assays.
HEK-293 cells were transiently transfected with plasmids encoding wild-type or mutant SARS S Δ19 glycoproteins using Lipofectamine 2000. Twenty-four hours posttransfection, cells were labeled with 2.5 mM Celltracker Green (CMTMR) (Molecular Probes, Eugene, OR). HEK-293 cells stably expressing hACE2 were labeled with Hoechst 33342 (Sigma). S-expressing cells were detached with trypsin-EDTA and overlaid on a 50% confluent monolayer of HEK-293 cells stably expressing hACE2 at a ratio of one S-expressing cell to three hACE2-expressing cells and incubated in DMEM with 10% FBS for 1 to 6 h. Cells were fixed in PBS with 4% paraformaldehyde and visualized on a Nikon TE200 inverted epifluorescence microscope running MetaMorph 7.1 (Molecular Devices). The total number of nuclei in at least four ×20 fields for each SARS S Δ19 glycoprotein was determined, and the percentages of nuclei in syncytia of different sizes were scored. Data from replicate experiments are shown as percentages of wild-type SARS-CoV S Δ19 cell fusion.
RESULTS
Cloning and expression of wild-type and mutant SARS-CoV S proteins.
To explore the role of the aromatic amino acids in the JMD of SARS-CoV S in receptor-dependent membrane fusion, we introduced alanine substitutions for the tyrosine or tryptophan residues and studied the effects of these substitutions on virus-cell fusion (entry) and cell-cell fusion (syncytium formation). Coronaviruses bud from internal cell membranes, and S proteins are generally targeted to these internal membranes. The C-terminal tails of S proteins of SARS-CoV and several other coronaviruses contain endoplasmic reticulum retention and/or endosomal recycling motifs (28, 36, 57). C-terminal truncation of the S glycoprotein of murine hepatitis virus (MHV) increased the level of S expression on the cell surface (5). We retargeted the SARS-CoV S protein to the plasma membrane by deleting the C-terminal 19 amino acids (to generate SARS-CoV S Δ19) to enhance both incorporation of S into MLV pseudotyped viruses and S-mediated cell-cell fusion (Fig. 2A). Previously, deletion of 19 amino acids from the C terminus of the SARS-CoV S glycoprotein was shown to increase the yield of pseudotyped virus (14).
FIG. 2.
SARS-CoV S Δ19 protein and amino acid substitutions in the juxtamembrane domain. (A) Full-length SARS-CoV S protein and S Δ19 truncated S protein. The receptor binding domain (RBD), HR-1, HR-2, and JMD are labeled in gray. (B) Positions of alanine substitutions in SARS-CoV S Δ19. Dashes indicate amino acid residues identical to those of the wild type.
Flow cytometry showed that the level of expression of codon-optimized S Δ19 on the surface of transfected cells was threefold higher than that of the full-length codon-optimized S glycoprotein (Fig. 3). Although both codon-optimized full-length and S Δ19 glycoproteins of the Urbani strain of SARS-CoV were incorporated into pseudotyped viruses that infected hACE2-expressing HEK-293 cells (HEK-293/hACE2 cells), the yield of pseudotyped viruses containing codon-optimized S Δ19 was 5- to 10-fold greater than that of pseudotyped viruses containing the full-length non-codon-optimized S protein (data not shown). Therefore, pseudotyped viruses and cells expressing codon-optimized S Δ19 (hereafter called S Δ19) were used for all subsequent virus entry and cell fusion experiments.
FIG. 3.
Surface expression of codon-optimized SARS-CoV S glycoproteins. Cells detached with 1 mM EDTA were stained with mouse monoclonal antibody against the SARS-CoV S protein (Zymed; Invitrogen) and PE-conjugated donkey anti-mouse secondary IgG antibody. Cells expressing wild-type, codon-optimized, full-length SARS-CoV S (A) or cells expressing wild-type, codon-optimized SARS-CoV S Δ19 (B) are labeled with black lines. Gray peaks indicate mock-transfected cells similarly treated. Data are from two independent experiments. This figure was prepared using the FlowJo (Ashland, OR) software program.
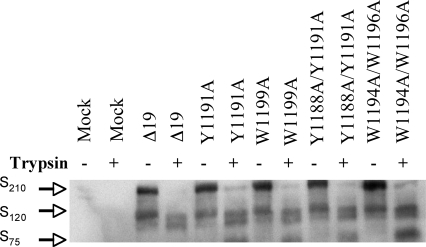
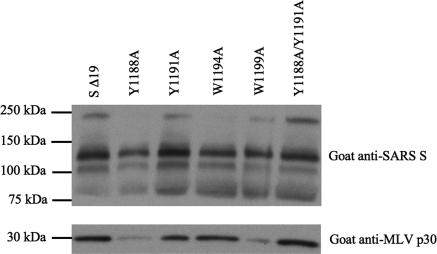
The boundaries of the TM and JMD in the SARS-CoV S protein were predicted using computer algorithms including TMPred (http://www.ch.embnet.org/software/TMPRED_form.html), DAS (41), HMMTOP (56), and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/). Based on these analyses, the TM is from amino acid 1201 to 1223 (GFIAGLIAIVMVTILLCCMTSCC) (Fig. 1A). All algorithms gave similar predictions, except TMPred, which predicted that W1199 lies within the TM. Single and double alanine substitutions for each conserved tyrosine (Y) or tryptophan (W) residue were introduced into the JMD of SARS-CoV S Δ19 to explore the contributions of these aromatic residues to hACE2-dependent membrane fusion and virus entry (Fig. 2B). To ensure that the mutant S Δ19 proteins were processed and transported in cells similarly to the wild-type S Δ19 protein, we transfected BHK-21 cells with plasmids encoding wild-type or mutant S Δ19 and compared the total amount of S Δ19 glycoprotein expression by immunoblotting. Wild-type and all mutant S Δ19 glycoproteins were expressed in cells at similar levels (Fig. 4). When cells expressing wild-type or mutant S Δ19 proteins were detached with EDTA alone, immunoblotting showed a strong band at ∼210 kDa, the SARS-CoV S monomer, and two weaker bands at ∼120 and ∼75 kDa (Fig. 4). In contrast, wild-type and mutant S Δ19 proteins in cells that were detached from the monolayer with trypsin-EDTA showed bands at ∼75 and ∼120 kDa (Fig. 4). The doublet at ∼120 kDa and the band at ∼75 kDa were likely due to cleavage of the S Δ19 glycoprotein on the cell surface by either exogenous trypsin or a cellular protease, such as furin.
FIG. 4.
Effects of trypsin on SARS-CoV S JMD mutant proteins. Immunoblot of SDS-PAGE gel of lysates from HEK-293 cells transiently expressing single or double alanine substitution mutant proteins lifted from the monolayer with either trypsin-EDTA or EDTA alone. Additional, smaller cleavage products of S were seen when cells were lifted with trypsin (data not shown).
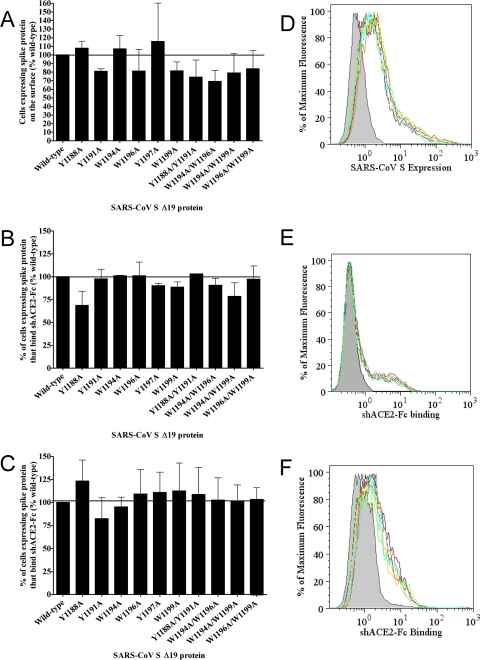
Interestingly, trypsin-EDTA cleavage of S Δ19 eliminated an unmapped epitope on the SARS S protein that is detected by a monoclonal mouse anti-SARS-CoV S antibody (Fig. 5); therefore, measurement of cell surface expression by flow cytometry was performed using cells detached with EDTA alone. The percentage of cells expressing each of the SARS-CoV S Δ19 proteins containing single alanine substitutions was comparable to that for wild-type S Δ19 (Fig. 6A). While the expression of the Y1188A/Y1191A double substitution mutant was comparable to that of wild-type S Δ19, a 20 to 40% reduction in the number of cells expressing the W1194A/W1196A, W1194A/W1199A, and W1196A/W1199A double mutants on the cell surface relative to that for the wild type was observed (Fig. 6A). In flow cytometry experiments, the levels of surface expression of wild-type and most mutant S Δ19 glycoproteins were similar (Fig. 6A).
FIG. 5.
Mouse monoclonal antibody only bound uncleaved SARS-CoV S protein. Cells expressing the wild-type SARS-CoV S Δ19 protein were detached with 1 mM EDTA (solid black line) or trypsin-EDTA (dashed line). Cells transiently transfected with empty vector were detached with trypsin-EDTA (gray). Cells were labeled with a mouse monoclonal antibody to SARS-CoV S (Zymed) and a secondary anti-mouse PE-conjugated antibody. The flow cytometry overlay shows detection of the SARS-CoV S glycoprotein on the surface of cells treated with EDTA alone (solid black line) but not on the surface of cells treated with trypsin-EDTA (dashed line).
FIG. 6.
Surface expression and soluble receptor binding of wild-type and mutant SARS-CoV S Δ19 proteins. (A) Cell surface expression of the wild-type or mutant SARS-CoV S Δ19 protein was measured by flow cytometry. (B and C) Binding of the soluble hACE2-Fc receptor protein by cells expressing wild-type or mutant SARS-CoV S Δ19 detached with trypsin-EDTA (B) or EDTA alone (C). (D to F) Representative flow cytometry overlays shown to the right of corresponding bar graphs. Mock-transfected cells are shown in dark gray, and SARS-CoV S Δ19 is shown by a black line; representative SARS-CoV S Δ19 mutant proteins are shown by color (SARS-CoV S Δ19 Y1188A, green; S Δ19 W1194A, red; S Δ19 Y1197A, orange; S Δ19 Y1188A/Y1191A, cyan; and S Δ19 W1194/W1199A, yellow, respectively). Error bars represent standard errors of percent wild-type levels between at least three independent experiments.
Binding of soluble hACE2-Fc fusion protein to wild-type and mutant SARS-CoV S Δ19 spike glycoproteins.
To determine whether alanine substitutions in the JMD of SARS-CoV S Δ19 alter binding of S to the SARS-CoV receptor, human ACE2, we used flow cytometry to measure binding of the shACE2-Fc fusion protein to cells expressing wild-type or mutant S Δ19 glycoproteins. Although trypsin cleavage of S decreased the amount of the shACE2-Fc protein bound to cells expressing wild-type or mutant S Δ19 (Fig. 6E versus 6F), the levels of shACE2 binding to wild-type or mutant S Δ19 proteins were comparable between cells detached with trypsin-EDTA (Fig. 6B) or EDTA alone (Fig. 6C). Thus, trypsin cleavage of spike proteins on the plasma membrane did not change the ability of S proteins to bind soluble receptor, and alanine substitutions within the JMD of the SARS-CoV S protein did not affect the receptor binding activity.
Characterization of MLV pseudotyped viruses expressing wild-type or JMD mutant SARS-CoV S Δ19 glycoproteins.
To study the effects of the JMD of SARS-CoV S proteins on virus entry, we produced recombinant MLVs pseudotyped with wild-type or mutant JMD S Δ19 glycoproteins (49). Pseudotyped viruses harvested from the supernatant of transfected cells were concentrated by ultracentrifugation and analyzed by immunoblotting (Fig. 7) and transmission electron microscopy (data not shown). In general, immunoblotting of concentrated pseudotyped viruses with an anti-MLV capsid antibody showed that similar amounts of the MLV p30 capsid protein were present in wild-type and mutant S Δ19 pseudotyped viruses (Fig. 7). Immunoblotting with an anti-SARS-CoV S antibody showed that mutant S Δ19 proteins were incorporated into pseudovirions as efficiently as wild-type S Δ19 proteins. S proteins in concentrated pseudotyped viruses produced in cells treated with trypsin-EDTA migrated at ∼210, 120, and 75 kDa (Fig. 7), similar to the S protein expressed in cells treated with trypsin-EDTA (Fig. 4). Cleavage of S did not affect the infectious pseudovirus yield (average yields from three experiments [standard error], trypsin-EDTA-detached cells, 2.3 × 103 [± 2.4 × 102] TU/ml versus EDTA detached cells, 4.6 × 103 [±8.6 × 102] TU/ml). Electron microscopy of negatively stained, concentrated pseudotyped virus generated in cells detached with either trypsin or EDTA showed the characteristic 200-Å-long coronavirus S proteins on the viral envelope (data not shown). No spikes were seen on control viruses generated in cells that did not express the SARS-CoV S protein (data not shown). As seen with the spike protein of murine coronavirus MHV, trypsin cleavage did not change the morphological appearance of the SARS-CoV S protein present on the pseudotyped virus (25). Thus, immunoblotting and electron microscopy demonstrated that wild-type and mutant SARS-CoV S Δ19 glycoproteins were incorporated into pseudotyped virus similarly.
FIG. 7.
Characterization of wild-type or mutant pseudotyped viruses. SDS-PAGE gel of representative concentrated MLV pseudotyped viruses containing wild-type or mutant SARS-CoV S Δ19 proteins that were produced in cells detached with trypsin-EDTA 24 h posttransfection were concentrated using a protocol adapted from reference 30. Immunoblots of concentrated pseudotyped viruses with goat anti-SARS-CoV S no. 4473 (top) or goat anti-MLV p340 capsid protein (bottom) followed by an HRP-conjugated secondary antibody are shown.
Effects on virus entry of alanine substitutions in the JMD of the SARS-CoV S Δ19 protein.
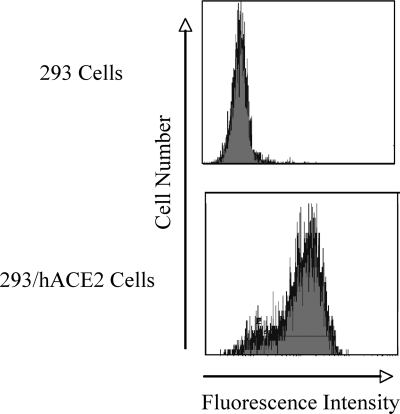
To determine the functional significance of the aromatic amino acids in the JMD of the SARS-CoV S protein, we assessed the ability of recombinant viruses pseudotyped with wild-type or mutant SARS-CoV S Δ19 to transduce HEK-293 cells overexpressing hACE2. The HEK-293/hACE2 stable cell line was generated to express very high levels of hACE2 in order to maximize pseudotyped-virus infection (Fig. 8). Pseudotyped viruses containing wild-type or mutant SARS-CoV S Δ19 glycoproteins and carrying the beta-galactosidase reporter were used to transduce HEK-293/hACE2 cells. Transduction, which corresponds to entry, was quantitated by counting cells that expressed the beta-galactosidase reporter gene. Wild-type S Δ19 efficiently transduced HEK-293 cells overexpressing hACE2 but not HEK-293 cells alone. Interestingly, alanine substitution of tyrosine or tryptophan residues closer to the TM had a greater effect on reduction of entry, indicating an effect of amino acid position on virus entry. The W1194A substitution reduced transduction by 50%, while single W1196A, Y1197A, and W1199A substitutions reduced transduction by 70 to 85% (Fig. 9). All three double tryptophan-to-alanine substitutions reduced transduction by 90 to 95% (Fig. 9). Our data also suggest that tyrosine- or tryptophan-to-alanine substitutions have synergistic effects on reduction of virus entry. For example, although each of the single tyrosine-to-alanine substitutions, Y1188A or Y1191A, partially reduced pseudotyped virus entry, the double substitution Y1188A/Y1191A nearly eliminated viral entry and transduction (Fig. 9). Data from pseudotyped virus entry experiments of filtered supernatant medium (Fig. 9) were run in parallel with pseudotyped viruses concentrated by ultracentrifugation, which gave similar results with somewhat higher variability (data not shown). Alanine substitution for the aromatic residues W1194, W1196, Y1197, or W1199 or both Y1188 and Y1191 profoundly reduced the ability of S to mediate virus entry by 10 to 60%.
FIG. 8.
Surface expression of hACE2. HEK-293 cells stably expressing high levels of hACE2 were selected by flow cytometry.
FIG. 9.
Alanine substitutions in the JMD of the SARS-CoV S Δ19 protein reduce receptor-dependent entry of pseudotyped viruses. Relative transduction activity of pseudotyped viruses with wild-type or mutant SARS-CoV S Δ19 proteins is shown. Pseudotypes in filtered medium were used to transduce HEK-293/hACE2 cells. Beta-galactosidase expression was analyzed 2 days posttransduction. Transduction units per ml were calculated {TU/ml = [(% cells expressing reporter/100) × total number of cells per well]/ml inoculum}. Data are represented as percent transduction activity of wild-type SARS-CoV S Δ19 proteins and are representative of three to six experiments. Error bars represent standard errors. Student's t test was used to evaluate the significance of differences between the transducing activities of pseudotyped viruses with mutant or wild-type proteins. (*, P = 0.01 to 0.05; **, P = 0.01 to 0.001; ***, P ≤ 0.0001).
Fusion of cells expressing wild-type or mutant SARS-CoV S Δ19 glycoproteins with cells expressing hACE2.
To investigate the role of the JMD in receptor-mediated cell-cell fusion, we tested the membrane fusion activities of the wild-type or mutant S Δ19 glycoproteins with HEK-293 cells stably expressing hACE2. S-expressing cells were detached from the monolayer with trypsin-EDTA and added to a 50% confluent monolayer of receptor-expressing cells. Cell-cell fusion was quantitated by the percentage of total nuclei present in syncytia and by the number of nuclei in each syncytium after 30 min, 1 h, 2 h, and 3 h. Cell-cell fusion was detected after 1 h and increased markedly by 2 h (Fig. 10). By 3 h, many large syncytia had detached from the monolayer. Although low background levels of spontaneous cell-cell fusion (up to 15%) were observed in control HEK-293 cells, wild-type S Δ19 glycoproteins induced much higher levels of cell-cell fusion (55 to 60%).
FIG. 10.
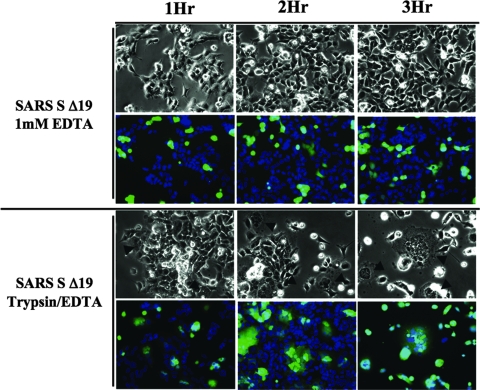
Trypsin enhances receptor-dependent cell-cell fusion induced by wild-type SARS-CoV S Δ19 protein. The time course of syncytium formation was determined after 30 min, 1 h, 2 h, or 3 h. HEK-293 cells transfected with cDNA encoding the wild-type SARS-CoV S Δ19 glycoprotein (green) were stained with Celltracker Green (Molecular Probes) and were detached from the monolayer with EDTA alone or trypsin-EDTA and added to a 50% confluent monolayer of HEK-293/hACE2 cells (blue) stained with Hoechst 33342 (Sigma) at a ratio of one S-expressing cell to three hACE2-expressing cells. Cells were fixed with paraformaldehyde 2 h postmixing. Pictures were taken with a ×20 objective. Pictures are representative of two independent experiments..
Previously, receptor-dependent cell-cell fusion induced by the S protein of SARS-CoV was reported to proceed slowly, but furin or trypsin cleavage of SARS-CoV S accelerated the rate of cell-cell fusion (12, 32, 51). To determine the effect of trypsin treatment on the cell fusion activity of the SARS-CoV S Δ19 glycoprotein, we compared membrane fusion mediated by S Δ19-expressing cells treated with trypsin-EDTA versus treatment with EDTA alone. By 2 h after treatment with trypsin-EDTA, extensive receptor- and spike-dependent cell-cell fusion was observed, but treatment with EDTA alone resulted in very little fusion up to 3 h postmixing (Fig. 10). Uncleaved S Δ19 caused little cell-cell fusion even after 3 h, while cleaved S Δ19 mediated such extensive fusion that many syncytia spontaneously detached after three or more hours postmixing (Fig. 10).
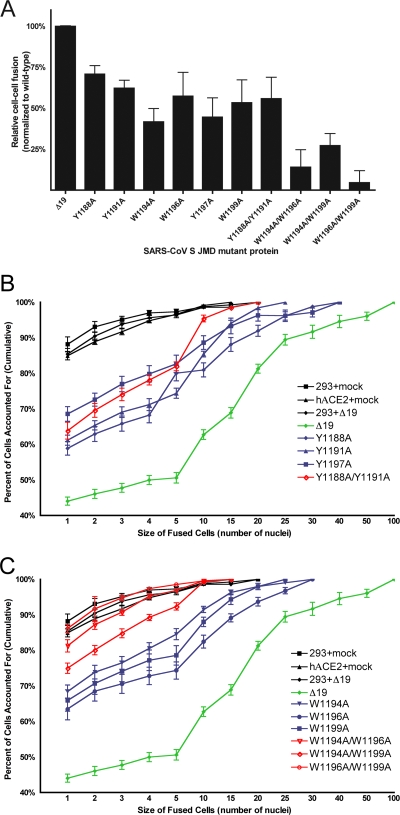
Each of the JMD mutant S Δ19 glycoproteins had less receptor-dependent cell-cell fusion activity than the wild-type S Δ19 glycoprotein (Fig. 11A). All of the single tyrosine- or tryptophan-to-alanine mutants and the Y1188A/Y1191A double mutant reduced fusion to 40 to 60% of the wild-type level, while each of the other double alanine substitutions in the JMD reduced cell-cell fusion to nearly background levels. To compare the cell-cell fusion activities of the mutant S Δ19 proteins, 2 h after mixing cells expressing spike with cells expressing receptor, we counted the nuclei in each syncytium and plotted the cumulative extent of fusion (percentage of nuclei in syncytia of any size) versus the size of syncytia (the number of nuclei in each syncytium) (Fig. 11B and C). This sensitive way of displaying the cell-cell fusion data (40) shows that spontaneous fusion of the HEK-293 cells affected less than 15% of cells and primarily yielded syncytia with two to four nuclei. In contrast, the wild-type S Δ19 protein induced fusion of 55% of cells and generated much larger syncytia that contained up to 100 nuclei (Fig. 11B and C). All single tryptophan-to-alanine substitutions (W1194A, W1196A, and W1199A) reduced cell-cell fusion levels (Fig. 11A). All of the single and double tyrosine-to-alanine substitutions (Y1188A, Y1191A, Y1197A, and Y1188A/Y1191A) also caused reduced levels of cell fusion (Fig. 11A). Plotting the sizes of syncytia induced by single or double alanine mutant proteins against the cumulative percentage of nuclei in syncytia revealed that all single alanine substitution proteins and the double substitution Y1188A/Y1191A formed syncytia of intermediate size compared to results with the wild type (Fig. 11B and C). This, and the intermediate levels of fusion mediated by the Y1188A/Y1191A double-substitution protein, indicated that there was no synergistic effect of the N-terminal tyrosines Y1188 and Y1191 during cell-cell fusion, unlike the synergistic effects observed on virus entry (Fig. 11A and B). Interestingly, the Y1188A/Y1191A double-substitution protein appeared to mediate slightly smaller-sized syncytia than any single tyrosine or tryptophan substitution (Fig. 11B). In contrast to results with the Y1188A/Y1191A substitutions, the three double tryptophan-to-alanine substitutions reduced cell-cell fusion to background levels (Fig. 11A and C), indicating a synergistic effect of tryptophan-to-alanine substitutions.
FIG. 11.
JMD mutations reduce receptor-dependent cell-cell fusion induced by SARS S Δ19 proteins. (A) Fusion of HEK-293 cells expressing wild-type or mutant SARS-CoV S Δ19 proteins or empty vector with HEK-293/hACE2 cells. Fusion was scored as the percentage of nuclei present in syncytia of any size at 2 h after mixing cells expressing spike and receptor. Background levels of fusion up to 15% were seen in HEK-293 cells. Data are expressed as a percentage of wild-type SARS-CoV S Δ19 fusion with background levels of fusion subtracted. Data are representative of three independent experiments. P values are ≤0.02 for all mutant proteins relative to results for wild-type SARS-CoV S Δ19. (B and C) Analysis of syncytium size induced by tyrosine-to-alanine (B) or tryptophan-to-alanine (C) mutants of the SARS-CoV S Δ19 protein. Each individual syncytium was scored for size (number of nuclei). Results are plotted as the cumulative percentage of cells containing a given number of nuclei or less.
DISCUSSION
The class I viral fusion proteins of many enveloped viruses mediate fusion of the viral envelope with host cell membranes, and some of these glycoproteins also induce cell-cell fusion. Binding of the murine coronavirus spike glycoprotein to its cellular receptor, soluble murine carcinoembryonic antigen-related cellular adhesion molecule 1a, at 37°C initiates conformational changes in the MHV spike protein that likely lead to membrane fusion (24, 31, 58). Similarly, conformational changes in SARS-CoV S are induced by binding of hACE2 and possibly protease cleavage (25). The importance in membrane fusion of the heptad repeats (HR-1 and HR-2) and the fusion peptide of class I viral fusion proteins is well known (11). In this study, we explored the role in virus-cell and cell-cell fusion of the aromatic amino acids tyrosine and tryptophan in the SARS-CoV S JMD, which is highly conserved in all coronavirus S proteins. First, we defined the boundaries of the TM and JMD in coronavirus S proteins using several different computer algorithms. All algorithms gave nearly identical predictions, except TMPred, which predicted that W1199 lies within the TM. In previous studies of SARS-CoV S, some predictions of the TM included a portion of the JMD (3, 18, 36).
To compare the receptor-dependent cell-cell fusion activities of the JMD mutant spike proteins, we optimized several factors: surface expression of the receptor, hACE2, expression of the SARS-CoV S protein, and the trypsin cleavage state of the spike. We selected a stably transfected HEK-293 cell line that expressed a high level of hACE2 on the cell surface (Fig. 8). C-terminal truncations of the cytoplasmic tail of the lentivirus Env and paramyxovirus F glycoproteins markedly increase virus-induced cell-cell fusion (22, 35, 53, 55). In order to optimize cell-cell fusion assays and transport the SARS-CoV S protein to the surface, an S protein with a 19-amino-acid deletion was engineered that lacks both a potential endoplasmic reticulum retention signal and an endosomal recycling motif (36, 57). Trypsin activates cell-cell fusion mediated by SARS-CoV S or S Δ19 (Fig. 10) (51); however, we found that trypsin did not affect entry of virus pseudotyped with the SARS-CoV S Δ19 protein. This rapid fusion may be useful for studies on inhibition of cell-cell fusion induced by SARS-CoV S. In contrast, with the full-length SARS S protein, trypsin treatment of virions prior to adsorption reduces virus entry (32, 51). It is generally believed that the S glycoprotein of SARS-CoV is cleaved by cathepsin L after internalization of virions in acidified endosomes; however, the cleavage site remains unmapped (19, 50). In addition, serine proteases have been shown to activate cell-cell fusion induced by many class I viral fusion glycoproteins (8, 10, 52, 59), including several coronaviruses, i.e., MHV A59 (58) and SARS-CoV (12, 32, 51). Our observations that trypsin cleavage of the SARS-CoV S protein enhanced receptor-dependent cell-cell fusion but not virus-cell fusion are in agreement with those of Follis et al., which showed that insertion of a prototypic furin recognition site in the SARS-CoV S Δ19 protein increased cell-cell fusion but not virus-cell fusion (12).
Studies of the JMDs of several unrelated fusion glycoproteins, VSV G, HIV type 1 env, and FIV env, indicate that aromatic amino acids in the JMD play a role in membrane fusion (16, 20, 46, 54). Aromatic amino acids, such as tyrosine and tryptophan, are likely to partition into the lipid-water interface of lipid bilayers (23). Aromatic amino acids in the JMD of coronaviruses may destabilize membranes during virus-cell and cell-cell fusion (18, 45). Studies using synthetic peptides have shown that the coronavirus S JMD can partition into and destabilize liposomes (18, 45), indicating that the JMD may function by destabilizing the virus and/or the cell membrane during membrane fusion. In addition, Broer et al. investigated the role(s) of the coronavirus JMD and TM in virus-cell and cell-cell fusion using chimeric S proteins containing the ectodomain of SARS-CoV S and the JMD, TM, and C-terminal endodomain of either the S protein of the coronavirus MHV or the G protein of the rhabdovirus VSV (3). Replacement of a portion of the JMD (SARS-CoV amino acids 1193 to 1199), TM, and endodomain of the SARS-CoV S protein with the orthologous region of MHV reduced virus-cell and cell-cell fusion by 40 to 50%. This suggested that some sequence-dependent element(s) required for fusion lie within the JMD, TM, and endodomain of coronavirus S proteins (3). Broer et al. also showed that replacement of the C-terminal endodomain of the SARS-CoV S protein with that of VSV G reduced virus-cell and cell-cell fusion mediated by the SARS-CoV S protein by 40 to 50% and that replacement of the JMD and TM of SARS-CoV S in addition to the endodomain with that of the VSV G protein reduced virus-cell fusion by over 90% and reduced cell-cell fusion by 70% (3). Broer et al. showed that the JMD plus the TM of SARS-CoV are necessary for efficient virus-cell fusion and for cell-cell fusion. Our study further identified specific aromatic amino acids within the JMD of SARS-CoV S that were important for virus-cell (Fig. 9) and cell-cell (Fig. 11) fusion: Y1188A, Y1191A, and W1194A.
In contrast, other single-amino-acid mutations within the S JMD reduced virus-cell fusion more than cell-cell fusion. The single alanine substitutions W1196A, Y1197A, and W1199A reduced virus-cell fusion dramatically (Fig. 9). In contrast to the dramatic reduction of virus-cell fusion, these single alanine substitutions reduced cell-cell fusion to intermediate levels (55 to 75% of the wild-type level [Fig. 11A; summarized in Fig. 12]). Interestingly, alanine substitutions for aromatic amino acids closer to the TM domain inhibited virus-cell fusion more than substitutions in the N-terminal region of the JMD (Fig. 12). This position effect is seen with the tyrosine substitutions (Y1188A and Y1191A versus Y1197A) and with the tryptophan substitutions (W1194A versus W1199A) during virus-cell fusion (Fig. 9) but not cell-cell fusion (Fig. 11A; summarized in Fig. 12). During cell-cell fusion, this position effect may not occur because the large contact area between cells increases the number of spikes interacting with receptors and may overcome the reduced fusion activity of alanine mutant S Δ19 proteins.
FIG. 12.
Summary of wild-type and JMD mutant SARS-CoV S Δ19-induced virus-cell and cell-cell fusion. Summary of virus-cell (virus entry) and cell-cell fusion induced by wild-type or mutant S Δ19 proteins (expressed as a percentage of wild-type levels.) The standard error for at least three independent experiments is shown. Bold indicates virus-cell fusion that is at least 25% less than corresponding cell-cell fusion.
While double tryptophan-to-alanine substitutions severely reduced both virus entry (Fig. 9) and cell-cell fusion (Fig. 11), the double tyrosine substitution Y1188A/Y1191A reduced virus entry by 90% (Fig. 9) but mediated intermediate levels of cell-cell fusion (Fig. 11A and B). Although the Y1188A/Y1191A substitution reduced virus entry by 90%, individually the Y1188A or Y1191A substitution only partially reduced entry (Fig. 9). This synergistic reduction of virus entry and cell-cell fusion by double substitutions was also seen for all double tryptophan-to-alanine substitutions (W1194A/W1196A, W1194A/W1199A, and W1196A/W1199A [Fig. 9 and 11A and C]).
Tryptophan-to-alanine substitutions in the JMD also had a synergistic effect on syncytia size (Fig. 11C) and percentage of cells fused (Fig. 11A). The three single tryptophan-to-alanine substitutions induced syncytia of intermediate size, while all three double tryptophan-to-alanine substitutions failed to increase cell-cell fusion over background levels (Fig. 11C). Interestingly, no synergistic reduction of cell-cell fusion was mediated by the Y1188A/Y1191A protein (Fig. 11A and B), although this mutant protein did induce slightly smaller syncytia during cell-cell fusion than either the Y1188A or Y1191A mutant (Fig. 11B). The lack of a synergistic effect during cell-cell fusion mediated by tyrosine-to-alanine substitutions may indicate a difference in the contribution of the tyrosine side chain to virus-cell versus cell-cell fusion (Fig. 12). These data suggest that the contribution of tryptophan to cell-cell fusion may be greater than that of tyrosine residues in the N-terminal region of the JMD. We postulate that substitution of phenylalanine, rather than alanine, might have a lesser effect on S Δ19-induced membrane fusion.
Our data show that the highly tryptophan- and tyrosine-rich JMD of SARS-CoV S Δ19 is required for virus-cell and cell-cell fusion. Because the JMD sequences of S proteins of all coronaviruses are so highly conserved (Fig. 1B), the aromatic residues in all coronavirus JMDs are likely to be important in membrane fusion. Since all of the JMD mutants bound soluble hACE2-Fc, the effects of the JMD mutations on membrane fusion must occur after S1 binds to the receptor.
The coronavirus JMD is much more conserved than the JMDs of other class I viral membrane fusion glycoproteins (Fig. 1). Because coronaviruses bud at the endoplasmic reticulum-Golgi intermediate compartment (6), coronavirus S proteins exist in radically different lipid environments in the viral envelope versus the plasma membrane. Coronavirus envelopes include ∼8% semilysobisphosphatidic acid (SLBPA) and ∼40% phosphatidylcholine, ∼10% sphingomyelin, and ∼12% phosphatidylinositol (6). SLBPA is not found in the plasma membrane but is enriched in the Golgi network (6). SLBPA has an unusual structure, consisting of a small, negatively charged head group and three acyl chains (6). Related lipids can induce high levels of membrane curvature, and SLBPA may also do this (37). These different lipid environments during virus-cell and cell-cell fusion may affect the pre- or postfusion activities of the JMD. Perhaps the unique lipid composition of the coronavirus envelope is responsible, in part, for the extremely high conservation of the JMD among coronaviruses.
Class I viral fusion proteins mediate membrane fusion by transitioning from the trimeric prefusion conformation to a postfusion six-helix bundle, bringing the lipid bilayer of the virus envelope into close apposition with the cell membrane (11). The SARS-CoV JMD apparently contributes to virus-cell and cell-cell fusion after receptor binding, perhaps by affecting the receptor-induced conformational changes that lead to membrane fusion. The JMD may function as a flexible hinge that facilitates the large conformational rearrangements of HR-2. Because peptides derived from the SARS-CoV S JMD directly interact with lipid bilayers, the JMD may destabilize membranes to facilitate hemifusion and fusion pore formation during S-mediated membrane fusion (18, 45). In the prefusion conformation of S, the JMD may be held away from the virus membrane. Perhaps the aromatic residues in the JMD can interact with the lipid bilayer only following receptor-induced conformational changes.
The JMD of class I viral fusion proteins is a potential target for the development of drugs that prevent virus entry and/or cell-cell fusion. Peptides of the FIV JMD or a monoclonal antibody that targets the HIV JMD reduces virus infection in vitro and in vivo by an unknown mechanism (9, 15, 38). For the SARS-CoV S protein, two short peptides derived from the JMD induce membrane destabilization in liposome leakage assays (18), suggesting that the SARS-CoV JMD may play a role in membrane destabilization during membrane fusion. Peptides or antibodies that target the conserved coronavirus JMD may reduce virus entry and cell-cell fusion induced by S proteins of all coronaviruses.
Acknowledgments
We thank Robert Hodges, Tanya Miura, Laura Gillim-Ross, Zhaohui Qian, and Susanna McReynolds for many helpful discussions and critical reviews of the manuscript.
This research was supported by NIH grant PO1-AI 59576. Megan W. Howard was supported in part by NIH training grant T32 AI 52066. Flow cytometry and DNA sequencing cores are supported by NIH/NCI Cancer Center core support grant P30 CA046934.
Footnotes
Published ahead of print on 16 January 2008.
REFERENCES
- 1.Bosch, B. J., B. E. Martina, R. Van Der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. Heck, R. De Groot, A. D. Osterhaus, and P. J. Rottier. 2004. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA 1018455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 778801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broer, R., B. Boson, W. Spaan, F. L. Cosset, and J. Corver. 2006. Important role for the transmembrane domain of severe acute respiratory syndrome coronavirus spike protein during entry. J. Virol. 801302-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll, M. W., and B. Moss. 1997. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology 238198-211. [DOI] [PubMed] [Google Scholar]
- 5.Chang, K. W., Y. Sheng, and J. L. Gombold. 2000. Coronavirus-induced membrane fusion requires the cysteine-rich domain in the spike protein. Virology 269212-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cluett, E. B., E. Kuismanen, and C. E. Machamer. 1997. Heterogeneous distribution of the unusual phospholipid semilysobisphosphatidic acid through the Golgi complex. Mol. Biol. Cell 82233-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4309-319. [DOI] [PubMed] [Google Scholar]
- 8.Diederich, S., M. Moll, H. D. Klenk, and A. Maisner. 2005. The nipah virus fusion protein is cleaved within the endosomal compartment. J. Biol. Chem. 28029899-29903. [DOI] [PubMed] [Google Scholar]
- 9.D'Ursi, A. M., S. Giannecchini, C. Esposito, M. C. Alcaro, O. Sichi, M. R. Armenante, A. Carotenuto, A. M. Papini, M. Bendinelli, and P. Rovero. 2006. Development of antiviral fusion inhibitors: short modified peptides derived from the transmembrane glycoprotein of feline immunodeficiency virus. Chembiochem 7774-779. [DOI] [PubMed] [Google Scholar]
- 10.Dutch, R. E., T. S. Jardetzky, and R. A. Lamb. 2000. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Biosci. Rep. 20597-612. [DOI] [PubMed] [Google Scholar]
- 11.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 28525-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Follis, K. E., J. York, and J. H. Nunberg. 2006. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology 350358-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freer, G., S. Giannecchini, A. Tissot, M. F. Bachmann, P. Rovero, P. F. Serres, and M. Bendinelli. 2004. Dissection of seroreactivity against the tryptophan-rich motif of the feline immunodeficiency virus transmembrane glycoprotein. Virology 322360-369. [DOI] [PubMed] [Google Scholar]
- 14.Fukushi, S., T. Mizutani, M. Saijo, S. Matsuyama, N. Miyajima, F. Taguchi, S. Itamura, I. Kurane, and S. Morikawa. 2005. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 862269-2274. [DOI] [PubMed] [Google Scholar]
- 15.Giannecchini, S., M. C. Alcaro, P. Isola, O. Sichi, M. Pistello, A. M. Papini, P. Rovero, and M. Bendinelli. 2005. Feline immunodeficiency virus plasma load reduction by a retroinverso octapeptide reproducing the Trp-rich motif of the transmembrane glycoprotein. Antivir. Ther. 10671-680. [PubMed] [Google Scholar]
- 16.Giannecchini, S., F. Bonci, M. Pistello, D. Matteucci, O. Sichi, P. Rovero, and M. Bendinelli. 2004. The membrane-proximal tryptophan-rich region in the transmembrane glycoprotein ectodomain of feline immunodeficiency virus is important for cell entry. Virology 320156-166. [DOI] [PubMed] [Google Scholar]
- 17.Giroglou, T., J. Cinatl, Jr., H. Rabenau, C. Drosten, H. Schwalbe, H. W. Doerr, and D. von Laer. 2004. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J. Virol. 789007-9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillen, J., A. J. Perez-Berna, M. R. Moreno, and J. Villalain. 2005. Identification of the membrane-active regions of the severe acute respiratory syndrome coronavirus spike membrane glycoprotein using a 16/18-mer peptide scan: implications for the viral fusion mechanism. J. Virol. 791743-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, I. C., B. J. Bosch, F. Li, W. Li, K. H. Lee, S. Ghiran, N. Vasilieva, T. S. Dermody, S. C. Harrison, P. R. Dormitzer, M. Farzan, P. J. Rottier, and H. Choe. 2006. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2813198-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeetendra, E., K. Ghosh, D. Odell, J. Li, H. P. Ghosh, and M. A. Whitt. 2003. The membrane-proximal region of vesicular stomatitis virus glycoprotein G ectodomain is critical for fusion and virus infectivity. J. Virol. 7712807-12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeetendra, E., C. S. Robison, L. M. Albritton, and M. A. Whitt. 2002. The membrane-proximal domain of vesicular stomatitis virus G protein functions as a membrane fusion potentiator and can induce hemifusion. J. Virol. 7612300-12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston, P. B., J. W. Dubay, and E. Hunter. 1993. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J. Virol. 673077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Killian, J. A., and G. von Heijne. 2000. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25429-434. [DOI] [PubMed] [Google Scholar]
- 24.Lewicki, D. N., and T. M. Gallagher. 2002. Quaternary structure of coronavirus spikes in complex with carcinoembryonic antigen-related cell adhesion molecule cellular receptors. J. Biol. Chem. 27719727-19734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, F., M. Berardi, W. Li, M. Farzan, P. R. Dormitzer, and S. C. Harrison. 2006. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 806794-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, S., G. Xiao, Y. Chen, Y. He, J. Niu, C. R. Escalante, H. Xiong, J. Farmar, A. K. Debnath, P. Tien, and S. Jiang. 2004. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 363938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lontok, E., E. Corse, and C. E. Machamer. 2004. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 785913-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorizate, M., A. Cruz, N. Huarte, R. Kunert, J. Perez-Gil, and J. L. Nieva. 2006. Recognition and blocking of HIV-1 gp41 pre-transmembrane sequence by monoclonal 4E10 antibody in a Raft-like membrane environment. J. Biol. Chem. 28139598-39606. [DOI] [PubMed] [Google Scholar]
- 30.Lu, C. W., and M. J. Roth. 2003. Role of the mutation Q252R in activating membrane fusion in the murine leukemia virus surface envelope protein. J. Virol. 7710841-10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuyama, S., and F. Taguchi. 2002. Receptor-induced conformational changes of murine coronavirus spike protein. J. Virol. 7611819-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuyama, S., M. Ujike, S. Morikawa, M. Tashiro, and F. Taguchi. 2005. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. USA 10212543-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreno, M. R., M. Giudici, and J. Villalain. 2006. The membranotropic regions of the endo and ecto domains of HIV gp41 envelope glycoprotein. Biochim. Biophys. Acta 1758111-123. [DOI] [PubMed] [Google Scholar]
- 34.Moreno, M. R., R. Pascual, and J. Villalain. 2004. Identification of membrane-active regions of the HIV-1 envelope glycoprotein gp41 using a 15-mer gp41-peptide scan. Biochim. Biophys. Acta 166197-105. [DOI] [PubMed] [Google Scholar]
- 35.Mulligan, M. J., G. V. Yamshchikov, G. D. Ritter, Jr., F. Gao, M. J. Jin, C. D. Nail, C. P. Spies, B. H. Hahn, and R. W. Compans. 1992. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J. Virol. 663971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petit, C. M., J. M. Melancon, V. N. Chouljenko, R. Colgrove, M. Farzan, D. M. Knipe, and K. G. Kousoulas. 2005. Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion. Virology 341215-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell, G. L., and S. W. Hui. 1996. Tetraoleoylpyrophosphatidic acid: a four acyl-chain lipid which forms a hexagonal II phase with high curvature. Biophys. J. 701402-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, A. Jungbauer, et al. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 101651-1658. [DOI] [PubMed] [Google Scholar]
- 39.Roche, S., S. Bressanelli, F. A. Rey, and Y. Gaudin. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313187-191. [DOI] [PubMed] [Google Scholar]
- 40.Roos, D. S., C. S. Duchala, C. B. Stephensen, K. V. Holmes, and P. W. Choppin. 1990. Control of virus-induced cell fusion by host cell lipid composition. Virology 175345-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rost, B., P. Fariselli, and R. Casadio. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 51704-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saez-Cirion, A., J. L. Arrondo, M. J. Gomara, M. Lorizate, I. Iloro, G. Melikyan, and J. L. Nieva. 2003. Structural and functional roles of HIV-1 gp41 pretransmembrane sequence segmentation. Biophys. J. 853769-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saez-Cirion, A., M. J. Gomara, A. Agirre, and J. L. Nieva. 2003. Pre-transmembrane sequence of Ebola glycoprotein. Interfacial hydrophobicity distribution and interaction with membranes. FEBS Lett. 53347-53. [DOI] [PubMed] [Google Scholar]
- 44.Sainz, B., Jr., E. C. Mossel, W. R. Gallaher, W. C. Wimley, C. J. Peters, R. B. Wilson, and R. F. Garry. 2006. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infectivity by peptides analogous to the viral spike protein. Virus Res. 120146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sainz, B., Jr., J. M. Rausch, W. R. Gallaher, R. F. Garry, and W. C. Wimley. 2005. The aromatic domain of the coronavirus class I viral fusion protein induces membrane permeabilization: putative role during viral entry. Biochemistry 44947-958. [DOI] [PubMed] [Google Scholar]
- 46.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 732469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schibli, D. J., R. C. Montelaro, and H. J. Vogel. 2001. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry 409570-9578. [DOI] [PubMed] [Google Scholar]
- 48.Schibli, D. J., and W. Weissenhorn. 2004. Class I and class II viral fusion protein structures reveal similar principles in membrane fusion. Mol. Membr. Biol. 21361-371. [DOI] [PubMed] [Google Scholar]
- 49.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4785-801. [DOI] [PubMed] [Google Scholar]
- 50.Simmons, G., D. N. Gosalia, A. J. Rennekamp, J. D. Reeves, S. L. Diamond, and P. Bates. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 10211876-11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simmons, G., J. D. Reeves, A. J. Rennekamp, S. M. Amberg, A. J. Piefer, and P. Bates. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 1014240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sjoberg, M., M. Wallin, B. Lindqvist, and H. Garoff. 2006. Furin cleavage potentiates the membrane fusion-controlling intersubunit disulfide bond isomerization activity of leukemia virus Env. J. Virol. 805540-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spies, C. P., G. D. Ritter, Jr., M. J. Mulligan, and R. W. Compans. 1994. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J. Virol. 68585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suarez, T., W. R. Gallaher, A. Agirre, F. M. Goni, and J. L. Nieva. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 748038-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tong, S., M. Li, A. Vincent, R. W. Compans, E. Fritsch, R. Beier, C. Klenk, M. Ohuchi, and H. D. Klenk. 2002. Regulation of fusion activity by the cytoplasmic domain of a paramyxovirus F protein. Virology 301322-333. [DOI] [PubMed] [Google Scholar]
- 56.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283489-506. [DOI] [PubMed] [Google Scholar]
- 57.Youn, S., E. W. Collisson, and C. E. Machamer. 2005. Contribution of trafficking signals in the cytoplasmic tail of the infectious bronchitis virus spike protein to virus infection. J. Virol. 7913209-13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zelus, B. D., J. H. Schickli, D. M. Blau, S. R. Weiss, and K. V. Holmes. 2003. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37 degrees C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 77830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhirnov, O. P., I. V. Vorobjeva, A. V. Ovcharenko, and H. D. Klenk. 2003. Intracellular cleavage of human influenza A virus hemagglutinin and its inhibition. Biochemistry (Moscow) 681020-1026. [DOI] [PubMed] [Google Scholar]