Abstract
To analyze the assembly of herpes simplex virus type 1 (HSV1) by triple-label fluorescence microscopy, we generated a bacterial artificial chromosome (BAC) and inserted eukaryotic Cre recombinase, as well as β-galactosidase expression cassettes. When the BAC pHSV1(17+)blueLox was transfected back into eukaryotic cells, the Cre recombinase excised the BAC sequences, which had been flanked with loxP sites, from the viral genome, leading to HSV1(17+)blueLox. We then tagged the capsid protein VP26 and the envelope protein glycoprotein D (gD) with fluorescent protein domains to obtain HSV1(17+)blueLox-GFPVP26-gDRFP and -RFPVP26-gDGFP. All HSV1 BACs had variations in the a-sequences and lost the oriL but were fully infectious. The tagged proteins behaved as their corresponding wild type, and were incorporated into virions. Fluorescent gD first accumulated in cytoplasmic membranes but was later also detected in the endoplasmic reticulum and the plasma membrane. Initially, cytoplasmic capsids did not colocalize with viral glycoproteins, indicating that they were naked, cytosolic capsids. As the infection progressed, they were enveloped and colocalized with the viral membrane proteins. We then analyzed the subcellular distribution of capsids, envelope proteins, and nuclear pores during a synchronous infection. Although the nuclear pore network had changed in ca. 20% of the cells, an HSV1-induced reorganization of the nuclear pore architecture was not required for efficient nuclear egress of capsids. Our data are consistent with an HSV1 assembly model involving primary envelopment of nuclear capsids at the inner nuclear membrane and primary fusion to transfer capsids into the cytosol, followed by their secondary envelopment on cytoplasmic membranes.
Herpes simplex virus type 1 (HSV1) causes severe human diseases such as herpes encephalitis or herpes keratoconjunctivitis (18). Its double-stranded DNA genome of 152 kb that codes for more than 80 open reading frames is enclosed in an icosahedral capsid with a diameter of 125 nm. HSV1 is a spherical, enveloped virus with a diameter of about 250 nm. Between the capsid and the viral membrane, there is an amorphous, asymmetric layer, the tegument, which consists of about 20 different proteins (45, 74). HSV1 enters cells by fusion at the plasma membrane or after endocytosis by fusion with an endosomal membrane (19, 42, 66, 67, 82). After dynein-mediated transport along microtubules (32, 56, 59, 81), capsids reach the nuclear pore where the viral genome is released into the nucleoplasm (68) for viral transcription and DNA replication (74). Progeny viral genomes are packaged into preassembled nuclear capsids, which translocate to the inner nuclear membrane. The subsequent steps of herpesvirus morphogenesis are controversial (12, 13, 65).
HSV1 capsids can leave the nucleus by primary envelopment at the inner nuclear membrane (6, 64). According to the luminal or single-envelopment hypothesis, these enveloped virions present in the lumen of the nuclear envelope or the endoplasmic reticulum are further transported within the secretory pathway, and ultimately leave the infected cell upon fusion of a virion-containing vesicle or vacuole with the plasma membrane (12, 37, 55). Alternatively, according to the deenvelopment-reenvelopment hypothesis, the luminal virions shed their primary envelope by fusion with the outer nuclear membrane or membranes of the endoplasmic reticulum which is continuous with the nuclear envelope, resulting in cytosolic capsids (12, 37, 64). A third recent hypothesis suggested that nuclear capsids may also directly access the cytosol through dilated nuclear pores whose central channels with a regular width of about 50 nm may be widened sufficiently for direct egress of nuclear capsids (55, 94). Whatever their origin during egress, the cytosolic capsids can then be transported to the organelle of secondary envelopment, e.g., the trans-Golgi network (39, 89) or multivesicular bodies (11, 22). The resulting vesicle containing one or several virions then fuses with the plasma membrane, and releases virions into the extracellular medium (reviewed in references 12, 37, and 64).
To study the dynamics of entry, assembly or egress, viral particles can be labeled with fluorescent dyes or proteins to circumvent the limitations of immunolabeling. Since the direct attachment of fluorescence labels may alter the viral life cycle or the growth kinetics, electron as well as immunofluorescence microscopy are used to identify any potential differences between the wild type and the tagged strains (reviewed in references 31 and 78). There are several HSV1 strains in which fluorescent protein domains have been attached to structural proteins. In HSV1(KOS)-GFPVP26, the small capsid protein VP26 has been N-terminally labeled with green fluorescent protein (GFP) (29). The addition of GFP to VP26 interferes neither with cell entry and dynein-mediated microtubule transport to the nucleus (30) nor with dynein and dynactin binding to isolated capsids or capsid transport along microtubules in vitro (96). The addition of GFP to the envelope glycoprotein gB of HSV1 reduces plaque sizes by three- to fivefold and virus titers by 100-fold (70), whereas the addition of GFP or yellow fluorescent protein (YFP) to glycoprotein D (gD) seems to be better tolerated (66, 80).
One method to generate herpesvirus mutants relies on bacterial artificial chromosomes (BACs) as vectors to manipulate entire viral genomes in Escherichia coli (1). After bacterial mutagenesis in E. coli, such BAC-cloned viral genomes are transfected into eukaryotic cells, and the mutated viruses are recovered. Since the first cloning of a herpesvirus as a BAC (63), this technology has been used successfully to generate an ever growing number of viral mutants. The faithful construction and examination of viral mutants is facilitated, if the exact sequence of the targeted genome is known, as is the case for HSV1 strain 17+, which has been completely sequenced (60-62, 69) (GenBank accession no. NC_001806).
Here we describe a new BAC of HSV1(17+), in which the BAC sequences and a eukaryotic Cre recombinase expression cassette were flanked by loxP sites. We labeled the capsid protein VP26 and the envelope protein gD with fluorescent proteins to generate single-color and dual-color fluorescence viruses (cf. Table 1). Using these, we studied the subcellular itinerary of capsids and viral envelope proteins during the viral life cycle. Capsid egress from the nucleus commenced around 6 h postinfection (p.i.). The fluorescent viral capsids were followed through the cytosol to cytoplasmic membranes labeled by the fluorescent envelope protein, and dual-color fluorescence particles were secreted into the medium. Thus, the fluorescently labeled structural proteins faithfully reflected the intracellular trafficking of the wild-type HSV1 structural proteins during assembly and egress. Although some cells showed a dramatic reorganization of the nuclear pore network, changes in the nuclear pore architecture did not correlate with efficient nuclear capsid egress. Our data are most consistent with an HSV1 assembly model involving primary envelopment of nuclear capsids at the inner nuclear membrane and fusion at the outer nuclear membrane to transfer capsids into the cytosol, followed by secondary envelopment of cytosolic capsids on cytoplasmic membranes.
TABLE 1.
HSV1 strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| HSV1(F) | Wild type | ATCC VR-733 |
| HSV1(KOS) | Wild type | P. Spear, Northwestern University, Chicago, IL |
| HSV1(KOS)-tk12 | ΔUL23, lacZ | 92 |
| HSV1(KOS)-ΔVP26 | ΔVP26, lacZ | 28 |
| HSV1(KOS)-GFPVP26 | GFPVP26 | 29 |
| HSV1(17+) | Wild type | J. Subak-Sharpe, MRC, Glasgow, United Kingdom |
| HSV1(17+)blue | ΔUL23, lacZ oriL−, BAC genes | This study |
| HSV1(17+)blueLox | ΔUL23, lacZ oriL− | This study |
| HSV1(17+)blueLox-ΔVP26 | ΔUL23, lacZ oriL−, ΔVP26, rpsLneo | This study |
| HSV1(17+)blueLox-GFPVP26 | ΔUL23, lacZ oriL−, GFPVP26 | This study |
| HSV1(17+)blueLox-RFPVP26 | ΔUL23, lacZ oriL−, RFPVP26 | This study |
| HSV1(17+)blueLox-gDGFP | ΔUL23, lacZ oriL−, gDGFP | This study |
| HSV1(17+)blueLox-GFPVP26-gDRFP | ΔUL23, lacZ oriL−, GFPVP26, gDRFP | This study |
| HSV1(17+)blueLox-RFPVP26-gDGFP | ΔUL23, lacZ oriL−, RFPVP26, gDGFP | This study |
MATERIALS AND METHODS
Cells, viruses, and antibodies.
BHK-21 (ATCC CCL-10) and Vero (ATCC CCL-81) cells were grown in minimal essential medium with fetal calf serum, and HSV1 was propagated in BHK-21 cells and purified as described previously (30, 81). John Subak-Sharpe provided HSV1(17+), Pat Spear provided the HSV1(KOS) and HSV1(KOS)tk12 (92), Prashant Desai provided the HSV1(KOS)-ΔVP26 (HSV1-KΔ26Z [28]) and HSV1(KOS)-GFPVP26 (HSV1-K26GFP [29]), and the HSV1(F) was purchased from ATCC (VR-733; cf. Table 1). For single-step growth kinetics, BHK-21 cells were infected with a multiplicity of infection (MOI) of 5 PFU/cell for 48 h, and the resulting culture supernatants, as well as all virus preparations, were titrated on Vero cells (32). We used the rabbit polyclonal antibodies (PAb) α-NC1 directed against VP5 (21), α-VP26 against VP26 (28), R69 against gB (35), or R45 against gD (20), as well as the mouse monoclonal antibodies (MAb) 5C10 against VP5 (88), DL6 against gD (34), 7520 against gE (4), and LP11 against gH (9). Nuclear pore complexes were labeled with MAb 414 (Babco, Richmond, CA) (25).
Cloning of HSV1 as a BAC.
To insert the BAC sequences together with a β-galactosidase expression cassette and a single loxP site into the UL23 locus of HSV1, the recombination plasmid pblueLox-HomTK was constructed. The kanamycin resistance gene from pEYFP-ER (Clontech, San Jose, CA) was amplified with primers Kan-fwd and Kan-rev (cf. Table 2; all primers were purchased from MWG Biotech, Ebersberg, Germany), treated with NotI and XbaI, and cloned into pUC18ΔNdeIlinker (84), resulting in pUC18ΔNdeIlinker-Kan. DNA sequences homologous to 2 kb of UL23 and its flanking regions were amplified from HSV1(17+) DNA isolated from viral capsids (57). The 3′ sequence (nucleotides [nt] 44591 to 46840) was amplified with 3′tk-fwd and 3′tk-rev, digested with XbaI and NheI, and ligated into pUC18ΔNdelinker-Kan cut with XbaI and NdeI. The 5′ sequence (nt 47561 to 49773) was amplified with 5′tk-fwd and 5′tk-rev, digested with Bsp120I and NdeI, and ligated into the NotI- and NdeI-cut product of the previous cloning step, resulting in pHomTK. The homology cassette was released from pHomTK with SalI and cloned into the SalI site of pblueLox (79), yielding the recombination plasmid pblueLox-HomTK.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| Kan-fwd | TAT AGC GGC CGC TAC AGG GCG CGT CAG GTG GC |
| Kan-rev | AGC ATC TAG ATT AAT TAA GTG ATG GCA GGT TGG GCG TCG CTT G |
| 3′tk-fwd | GGT GGC TTG AGC CAG CGC GTC CAG |
| 3′tk-rev | CTA GCT AGC GTC GAC ATG CAT GTC TTT ATC CTG GAT TAC GAC CAA TCG CC |
| 5′tk-fwd | AAA CAT ATG TCG ACG TAG ACG ATA TCG TCG CGC GAA CCC AGG |
| 5′tk-rev | AAG GGC CCT TAA TTA ACG TGG TGC ATC AGC GTG GCG ATC ACG ATG TGC |
| loxP-sense | AGC TGC GGC CGC ATA ACT TCG TAT AAT GTA TGC TAT ACG AAG TTA TTT AAT TAA TCT AGA GGG GCT AGC G |
| loxP-antisense | AAT TCG CTA GCC CCT CTA GAT TAA TTA AAT AAC TTC GTA TAG CAT ACA TTA TAC GAA GTT ATG CGG CCG C |
| Tet-fwd | NNN NGC TAG CTG ATG TGC TTA AAA ACT TAC TCA |
| Tet-rev | NNN NGC TAG CTG ATT CCC TTT GTC AAC AGC AAT |
| 5′BAC-sense | GGC CGC TCG ACA GCG ACA CAC TTG CAT CGG ATG CAG CCC GGT TAA CGT GCC GGC ACT A |
| 5′BAC-antisense | GGC CTA GTG CCG GCA CGT TAA CCG GGC TGC ATC CGA TGC AAG TGT GTC GCT GTC GAG C |
| 3′BAC-sense | CTA GAG GCC TGG GTA ACC AGG TAT TTT GTC CAC ATA ACC GTG CGC AAA ATG TTG TG |
| 3′BAC-antisense | CTA GCA CAA CAT TTT GCG CAC GGT TAT GTG GAC AAA ATA CCT GGT TAC CCA GGC CT |
| VP26rpsLneo-fwd | CGA CAC CCC CAT ATC GCT TCC CGA CCT CCG GTC CCG ATG GCC GTC CCG CAG GCC TGG TGA TGA TGG CGG GAT C |
| VP26rpsLneo-rev | CGC GCA TGC CAA GCG CCC GGA CGC TAT CGG TGG TAA CGG TGC TGG GGC GGT CAG AAG AAC TCG TCA AGA AGG |
| VP26Hom-fwd | ATG CCC GGC CGA TGA TGG |
| VP26Hom-rev | CGC CGT GCT GAC CAG CCT AC |
| gDGFP-fwd | CCC ACA TCC GGG AAG ACG ACC AGC CGT CCT CGC ACC AGC CCT TGT TTT ACG TGA GCA AGG GCG AGG AG |
| gDGFP-rev | CCC AAC CCC GCA GAC CTG ACC CCC CCG CAC CCA TTA AGG GGG GGT ATC TAC TTG TAC AGC TCG TCC ATG |
| gDRFP-fwd | CCC ACA TCC GGG AAG ACG ACC AGC CGT CCT CGC ACC AGC CCT TGT TTT ACG CCT CCT CCG AGG ACG TCA TC |
| gDRFP-rev | CCC AAC CCC GCA GAC CTG ACC CCC CCG CAC CCA TTA AGG GGG GGT ATC TAC AAG GCG CCG GTG GAG TG |
| BAC-fwd | CGT AGT CGA GCT AGT CGA TCG TAC GAT ACG TCA G |
| BAC-rev | TCG ATC GCT AGT CGA TCT ACG TCG TAG ATG CTA C |
| oriL-fwd | GTC TCC CGA GCG TCA AAA TC |
| oriL-rev | GCA AGG GCC TTG TTT GTC TG |
To insert BAC sequences into HSV1(17+), 0.3 μg of virus DNA and 5 μg of PacI-linearized pblueLox-HomTK were cotransfected into 106 Vero cells (MBS mammalian transfection kit; Stratagene, La Jolla, CA). After complete cytopathic effects had developed, cells and supernatant were harvested. Recombinant viruses were plaque purified three times. BHK-21 were inoculated with the recombinant virus HSV1(17+)blue at an MOI of 10 PFU/cell. Hirt extracts (48) were prepared at 2 h p.i. and used to transform E. coli DH10B (44) under chloramphenicol selection. Three clones of pHSV1(17+)blue were analyzed by restriction analysis. To introduce further mutations into the BACs, the Red recombination enzymes (98) were either induced from plasmid pKD46 (24) in E. coli DH10B harboring BACs or by heat shock in E. coli DY380, which has the red genes inserted into the bacterial genome (54).
Insertion of a Cre recombinase gene.
Two complementary oligonucleotides containing a loxP site flanked by NheI, PacI, XbaI, and NotI restriction sites (loxP-sense and loxP-antisense) were annealed and ligated into pUC18 digested with BamHI and HindIII, resulting in pUC18LoxP. A eukaryotic expression cassette for Cre recombinase with an intron was excised from pCreIn (79) with PacI and cloned into the PacI site of pUC18LoxP to yield pUC18LC. The tetracycline resistance cassette from pCP16 (16) was amplified with Tet-fwd and Tet-rev, cut with NheI, and cloned into the XbaI site of pUC18LoxP to construct pUC18LCT. 50-bp double-stranded DNA oligonucleotides homologous to sequences upstream of the BAC cassette were obtained by annealing complementary oligonucleotides (5′BAC-sense/5′BAC-antisense and 3′BAC-sense/3′BAC-antisense). These were ligated into the NheI and NotI sites of pUC18LCT, respectively, leading to pUC18LCTH. The complete 5.4-kb cassette was excised with NheI and NotI and inserted into the BAC pHSV1(17+)blue, resulting in pHSV1(17+)blueLox.
Reconstitution of HSV1(17+) strains from BACs.
For transfection, BAC-DNA was prepared from 500-ml overnight E. coli cultures using the NucleoBond BAC 100 kit (Macherey & Nagel, Düren, Germany). A total of 5 × 105 Vero cells were grown overnight, transfected with 2 μg of BAC-DNA (MBS mammalian transfection kit), and cultured for several days until cytopathic effects developed.
Fluorescence tagging of the HSV1 proteins VP26 and gD.
VP26 was N terminally tagged with GFP by using pK26GFP (29). Sequencing of pK26GFP and of the DNA of HSV1(KOS)-GFPVP26 (29) revealed that in addition to the first four original codons of VP26, 50 bases in the noncoding region upstream of the VP26 open reading frame (ORF) were missing. The deleted region began 55 bp after the stop codon of UL34 (data not shown; P. Desai, unpublished data). For pK26RFP, the GFP sequence in pK26GFP was removed and replaced with the sequence of mRFP1 from pRSETB-mRFP1 (14). For pHSV1(17+)-BAC mutagenesis, an rpsLneo cassette was amplified from pRpsLneo (Gene Bridges GmbH, Dresden, Germany) with VP26rpsLneo-fwd and VP26rpsLneo-rev. This cassette replaced codons 4 to 7 of VP26 and interrupted its reading frame, generating pHSV1(17+)blueLox-ΔVP26. The cassette was subsequently replaced with the PCR products obtained from plasmids pK26GFP or pK26RFP using the primers VP26Hom-fwd and VP26Hom-rev. The resulting BACs were named pHSV1(17+)blueLox-GFPVP26 or -RFPVP26.
gD was tagged with GFP or mRFP1 using an en-passant mutagenesis technique (87). The GFP insertion construct was amplified from pEP-EGFP-in (81) with the primers gDGFP-fwd and gDGFP-rev and introduced into pHSV1(17+)-BACs. The mRFP1 sequence was amplified from pEP-mRFP1-in (81) with gDRFP-fwd and gDRFP-rev. Positive clones were transformed with the plasmid pBAD-I-SceI (87). I-SceI expression was induced by l-arabinose, and Red enzyme expression was induced by heat shock. The bacteria were grown with chloramphenicol, ampicillin, and l-arabinose, and kanamycin-sensitive clones were screened by restriction analysis.
DNA electrophoresis and Southern blotting.
DNA fragments were separated in agarose gels, blotted onto nylon membranes, hybridized, and probed with digoxigenin (DIG)-labeled DNA probes (Roche, Mannheim, Germany). A probe against BAC sequences was generated by PCR with primers BAC-fwd and BAC-rev using the PCR DIG probe synthesis kit. A probe against the HSV1 replication origin oriL was produced with oriL-fwd and oriL-rev.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting.
Protein samples were separated on linear 5 to 15% polyacrylamide gradient gels (52). After transfer onto nitrocellulose membranes, the samples were incubated with primary antibodies, followed by secondary antibodies coupled to alkaline phosphatase (Dianova, Hamburg, Germany) and developed with BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium salt.
Light microscopy.
Vero cells grown on coverslips were infected with HSV1 at an MOI of 10 PFU/cell. In some experiments (Fig. 7 and 8), the cells were synchronously infected by inoculation with HSV1 for 2 h on ice in a small volume and then further incubated at 37°C (32, 81). After 1 h, the cells were washed with 40 mM citric acid-135 mM NaCl-10 mM KCl (pH 3) to inactivate extracellular virions (38, 95). Fresh medium was added, and at different time points the cells were fixed and immunofluorescence labeling was performed as described previously (81). The HSV1 Fc receptor (33) was blocked by using 10% (vol/vol) human serum of healthy, HSV1-seronegative volunteers (30). Secondary antibodies were coupled to lissamine-rhodamine sulfonyl chloride, fluorescein isothiocyanate (FITC), or Cy5 (Dianova). The specimens were analyzed on a Axiovert 200M microscope equipped with an LSM 510 META confocal laser scanning unit (Zeiss, Jena, Germany) using a Plan-Apochromatic ×63 oil immersion objective lens with a 1.4 numeric aperture. For a quantitative analysis of the HSV1 egress pathway, at least 85 randomly chosen cells were scored for several time points from 4 to 12 h p.i. Image acquisition and processing was performed by using the Zeiss LSM imaging software, ImageJ 1.35 (Wayne Rasband; National Institute of Health [http://rsb.info.nih.gov/ij/]), and Adobe Photoshop CS (Adobe Systems, San Jose, CA).
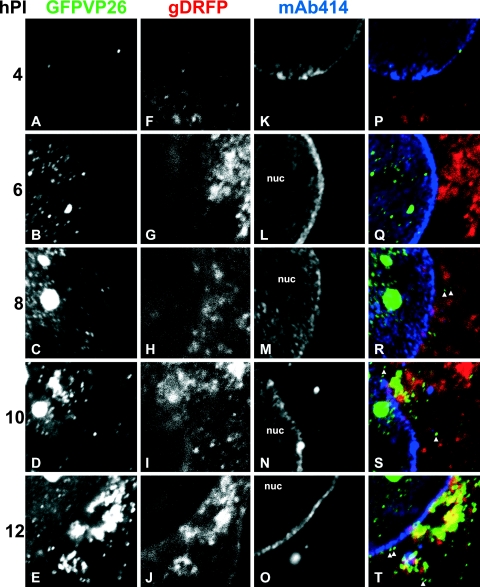
FIG. 7.
Time course of HSV1 nuclear egress and assembly. Vero cells were inoculated with 10 PFU/cell of HSV1(17+)blueLox-GFPVP26-gDRFP for 2 h at 4°C to allow efficient binding and synchronous infection. Unbound virus was removed by extensive washing, the cells were transferred to 37°C, and fixed at 4 h p.i (A, F, K, and P), 6 h p.i (B, G, L, and Q), 8 h p.i (C, H, M, and R), 10 h p.i (D, I, N, and S), or 12 h p.i (E, J, O, and T). GFPVP26 (A to E; green areas in panels P to T) and gDRFP (F to J; red areas in panels P to T) were detected by their intrinsic fluorescence. After permeabilization, the cells were labeled with MAb 414 against nucleoporins (K to O; blue areas in panels P to T) and analyzed by confocal laser scanning microscopy. To evaluate the relative subcellular localization of capsids (GFPVP26), viral envelope proteins (gDRFP), and nuclear pores (MAb 414), the signals were overlaid and displayed in color (P, Q, R, S, and T). The nuclei (nuc) and some cytoplasmic capsids, which do not colocalize with gD and thus appear green in the overlays (arrowheads in panels Q and R), are indicated. The specimens were analyzed by confocal laser scanning microscopy.
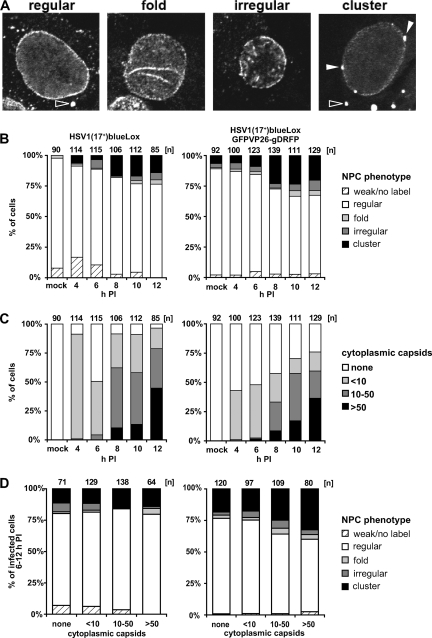
FIG. 8.
Changes in the nuclear pore network during HSV1 nuclear capsid egress and assembly. Vero cells were inoculated with 10 PFU/cell of HSV1(17+)blueLox or of HSV1(17+)blueLox-GFPVP26-gDRFP for 2 h at 4°C to allow efficient binding and a rather synchronous infection. Unbound virus was removed by extensive washing, and the cells were transferred to 37°C and fixed at 4, 6, 8, 10, or 12 h p.i. After permeabilization, they were labeled with the antibody directed against nuclear pore proteins and analyzed by confocal laser scanning microscopy. (A) Representative confocal sections through the nucleus. Different NPC labeling phenotypes were classified as “regular,” “folded” (fold), “irregular,” or “cluster.” Open or filled arrowheads indicate cytoplasmic or nuclear NPC aggregates, respectively. (B) Quantification of NPC phenotypes in infected cells at different times postinfection. Note that between 6 h and 8 h p.i., the number of cells characterized by altered nuclear pore architecture increased for both viruses. (C) Quantification of capsid egress from the nucleus. The cells with were classified according to their concentration of cytoplasmic capsids into “none,” “less than 10” (<10), “10 to 50” (10-50), and more than 50 (>50). Note that between 6 h and 8 h p.i. the amount of cytoplasmic capsids increased for both viruses. (D) The extent of nuclear capsid release into the cytoplasm did not correlate to the different nuclear pore architectures. For all time points between 6 and 12 h p.i., the cells were pooled and classified according to the extent of capsid egress and assigned to different NPC phenotypes.
RESULTS
Cloning of HSV1 strain 17+ as a BAC.
The genes needed for replication in E. coli (cat, oriS, repE, parB, and parA) were inserted together with a single loxP site and a β-galactosidase expression cassette driven by the simian virus 40 promoter (79) into the thymidine kinase locus (UL23) of HSV(17+) by homologous recombination in Vero cells (Fig. 1A). Vero cells were infected with the resulting plaque-purified, recombinant HSV1(17+)blue (Fig. 1B and Table 1), and viral genome replication intermediates were isolated for the transformation of E. coli.
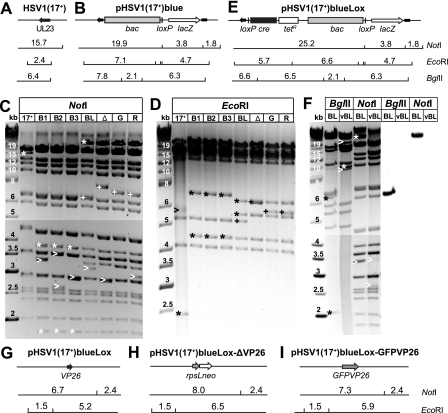
FIG. 1.
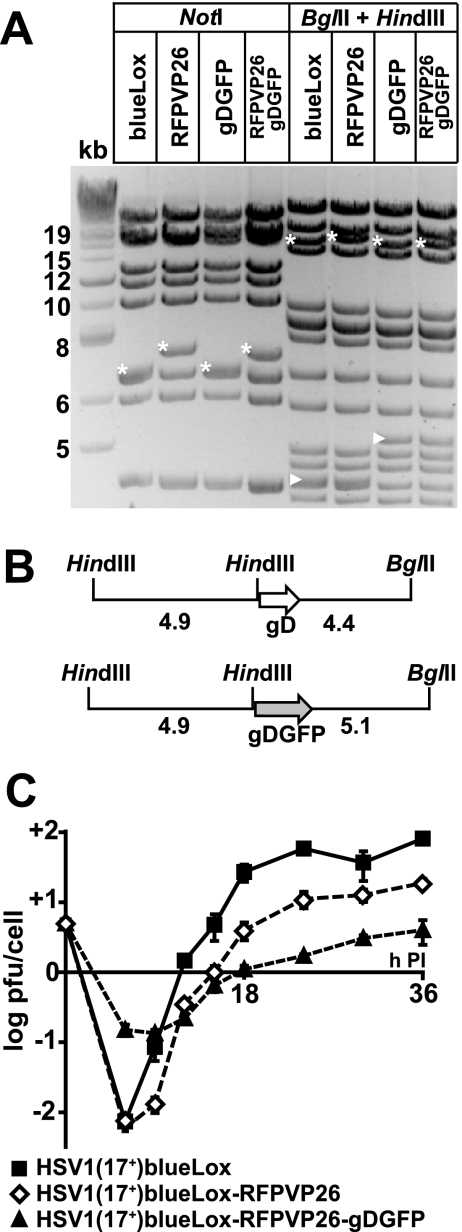
Generation of pHSV1(17+)-BACs and tagging of VP26. (A and B) BAC sequences, a loxP site, and a β-galactosidase expression cassette were inserted into the UL23 locus of HSV1(17+) (A) to clone pHSV1(17+)blue (B). (C and D) Restriction endonuclease digestions of HSV1(17+) and three different clones of pHSV1(17+)blue BAC (B1/B2/B3) with NotI or EcoRI. Lanes: BL, pHSV1(17+)blueLox; Δ, pHSV1(17+)blueLox-ΔVP26; G, pHSV1(17+)blueLox-GFPVP26; R, pHSV1(17+)blueLox-RFPVP26. Changes resulting from the BAC and Cre-insertion (*), different terminal and internal joint restriction fragments (>), and band shifts caused by modifying VP26 (+) are indicated. (E) A Cre recombinase expression cassette and a second loxP site were inserted under tetracycline selection to construct pHSV1(17+)blueLox. (F) Efficient excision of the BAC sequences by Cre recombinase. BglII or NotI digestions of pHSV1(17+)blueLox (BL) and HSV1(17+)blueLox (vBL). Differences in restriction fragments resulting from Cre-mediated excision (*) and the terminal as well as joint fragments (>) are indicated. The DNA fragments were transferred to a nylon membrane and hybridized with a DIG-labeled DNA probe against the BAC-encoded chloramphenicol resistance. (G to I) Fluorescence tagging of the capsid. The UL35 ORF encoding for the capsid protein VP26 was disrupted by inserting an rpsLneo selection/counterselection cassette into pHSV1(17+)blueLox (G), resulting in pHSV1(17+)blueLox-ΔVP26 (H). In pHSV1(17+)blueLox-GFPVP26 (I) and -RFPVP26 (not shown), this cassette was replaced by sequences for GFP or mRFP1 at the 5′ end of the VP26 ORF. The sizes of the restriction fragments and molecular weight markers are given in kilobases.
The HSV1 genome consists of a unique long (UL) and unique short (US) region which are flanked by inverted repeats (reviewed in references 6 and 74). At the genome termini and the junction between the long and short regions of the genome, the a-sequences are located, which contain DNA packaging signals. During replication, four isomers of viral DNA with regard to the relative orientation of the UL and US region are recovered in equimolar amounts. The restriction fragments spanning the connection between the long and short region were termed “joint fragments.” Please note that a circular BAC contains two, but linear viral DNA contains only one joint fragment. HindIII treatment revealed that the genome configuration of UL and US was antiparallel in clones 1 and 3 and parallel in clone 2 (data not shown).
The pHSV1(17+)blue clones were analyzed by restriction enzyme digestion. Expected fragment sizes were calculated according to the published HSV1(17+) sequence (GenBank accession no. NC_001806). Novel fragments resulting from the BAC insertion were detected after NotI and EcoRI digestion (Fig. 1B and asterisks in Fig. 1C and D, lanes B1, B2, and B3). For the viral DNA (Fig. 1C, lane 17+), the genome termini and the NotI joint fragment were not recovered as distinct bands. Instead, there was a very weak, diffuse signal at around 3 kb (not shown) due to the varying numbers of the repetitive 400-bp a-sequence in HSV1 (27, 74). Furthermore, the a-sequence itself can contain various numbers of direct repeats (27). In pHSV1(17+)blue, the two joint fragments had definite sizes that varied between clones (Fig. 1C, arrowheads in lanes B1, B2, and B3). According to the published sequence, both NotI joint fragments have a size of 2,918 bp, as in one clone (Fig. 1C, lane B3), whereas other clones (Fig. 1C, lanes B1 and B2) showed permutations of the joint fragment. Thus, the joint fragments were fixed as definite structures in the BACs recovered from E. coli but varied in length in the HSV1 genomes isolated from eukaryotic cells. Due to the circular organization, the 3′-terminal EcoRI fragment of linear HSV1-DNA forming a diffuse band around 5.6 kb (Fig. 1D, arrowhead in lane 17+) was absent in the BAC-DNAs. In all clones, the 2.0-kb BamHI fragment spanning the transition between the US region and the adjoining inverted repeat was reduced by about 50 bp (not shown). Variations in a highly repetitive sequence in this noncoding region have been observed before for individual HSV1 isolates (90).
All three pHSV1(17+)blue clones were infectious after transfection into Vero cells, and clone B1 was chosen for further analysis and mutagenesis.
Stability of pHSV(17+)-BACs.
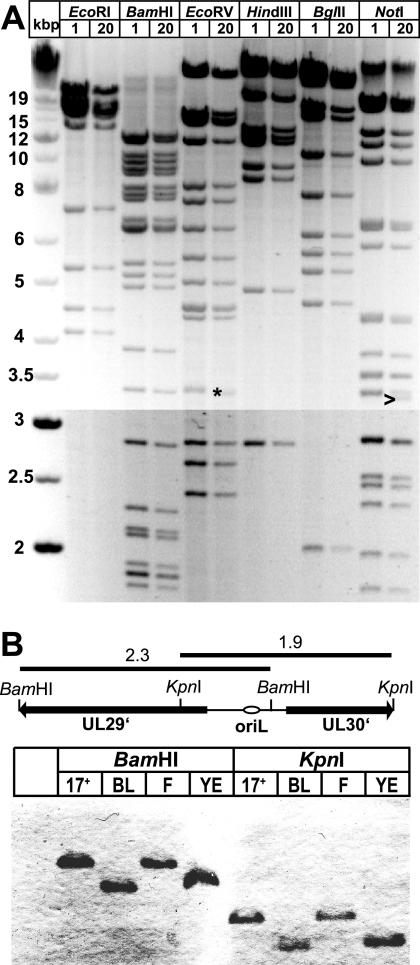
Since repetitive regions could provide targets for unwanted recombination in bacteria, the stability of the HSV1-BACs was tested. After 20 passages of pHSV1(17+)blue in E. coli, no major changes were observed in the restriction fragment patterns using six different enzymes (Fig. 2A). However, one of the NotI joint fragments had decreased in size (Fig. 2A, arrowhead), suggesting that a deletion had occurred in the highly repetitive a-sequences. Moreover, an EcoRV fragment also slightly shifted to a lower size (Fig. 2A, asterisk), probably by shortening of a repetitive sequence in a noncoding region upstream of UL1 (see also reference 90). However, BAC DNA prepared from both passages 1 and 20 induced cytopathic effects with comparable kinetics after transfection into Vero cells (data not shown). The HSV1 replication origin oriL consists of a 144-bp palindrome which, in addition, contains several direct repeats (47). All of the HSV1-BACs we tested, including a BAC of HSV1(F) (86), had lost their oriL (Fig. 2B). In contrast, a PCR revealed no changes in oriS between the HSV1(17+) wild type and the corresponding BACs (data not shown). Thus, with the exception of the indicated changes, the BAC pHSV1(17+)blue and its derivatives were stably maintained in E. coli and retained their infectivity upon transfection into eukaryotic cells.
FIG. 2.
The pHSV1(17+)-BACs were stable in E. coli but had lost the oriL. (A) HSV1-BAC pHSV1(17+)blue clone B1 was passaged for 20 generations in E. coli, and DNA of the first (lanes 1) and 20th (lanes 20) generation was digested with EcoRI, BamHI, EcoRV, HindIII, BglII or NotI. The changes in a EcoRV (asterisk) and NotI (arrowhead) fragment are indicated. (B) The palindromic oriL was lost after BAC cloning. DNA from HSV1(17+), pHSV1(17+)blueLox (BL), HSV1(F), or the HSV1(F)-BAC pYEbac102 (YE) was digested with BamHI or KpnI, transferred to a nylon membrane, and probed with a probe specific for oriL and the neighboring sequences. Restriction fragment sizes are indicated in kilobases.
Growth kinetics of HSV1-BAC derived viruses.
The insertion of foreign genes can attenuate the virus. Therefore, the BAC sequences are often removed after transfection into eukaryotic cells (79, 97). This can be done by flanking the BAC sequences with loxP sites and providing, with the BAC itself, a eukaryotic Cre expression cassette containing an intron, which prevents expression of functional Cre in bacteria but results in self-excision in eukaryotic cells (79). We therefore generated pHSV1(17+)blueLox (Fig. 1E and cf. Table 1) by inserting a second loxP site and a Cre expression cassette into pHSV1(17+)blue. Restriction analysis revealed no genomic changes, apart from the intended insertion, and a shift in the NotI joint fragment (Fig. 1E and asterisks in Fig. 1C and D, lanes BL). As for all other BAC modifications, we also performed restriction digestions with the enzyme BamHI, EcoRV, HindIII, or BglII, which generated the predicted fragment sizes (data not shown). After transfection of pHSV1(17+)blueLox, plaques and cytopathic effects developed. The viral DNA of HSV1(17+)blueLox (Table 1) no longer contained a discrete NotI joint fragment but diffuse bands at 2.6 and 3.5 kb (Fig. 1F). Upon replication in eukaryotic cells, the region containing the repetitive a-sequences had regained its size variability, and the Cre recombinase had efficiently removed the BAC sequences (Fig. 1F).
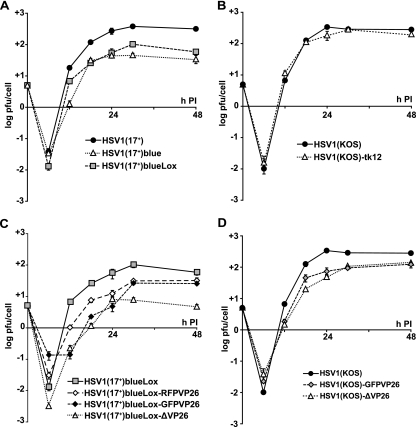
Compared to HSV1(17+), the BAC-derived virus HSV1(17+)blue exhibited a moderate growth defect (Fig. 3A). HSV1(17+)blueLox was slightly less attenuated than HSV1(17+)blue, with its titers being reduced ∼5-fold compared to HSV1(17+) (Fig. 3A). Thus, HSV1(17+)blueLox was attenuated compared to the wild type, but the excision of the BAC sequences was beneficial. Similar to HSV1(17+)blueLox, the mutant HSV1(KOS)-tk12 (92) also contains a β-galactosidase expression cassette instead of UL23, but HSV1(KOS)-tk12 showed no growth defect in BHK cells compared to HSV1(KOS) (Fig. 3B).
FIG. 3.
Growth kinetics of pHSV1(17+)-BAC derived viruses. For single-step growth kinetics, BHK-21 cells were inoculated with 5 PFU/cell. At the indicated time points, the supernatant was harvested and titrated on Vero cells in triplicates. (A) A comparison of HSV1(17+), HSV1(17+)blue, and HSV1(17+)blueLox showed that the BAC-derived HSV1(17+) strains were attenuated compared to the wild type and that deleting the BAC sequences partially reversed this attenuation. (B) There was no difference in replication between HSV1(KOS)tk12 (tk− lacZ+) and its parental strain HSV1(KOS). (C) The titers of HSV1(17+)blueLox-RFPVP26 and HSV1(17+)blueLox-GFPVP26 were reduced about fivefold and the titer of HSV1(17+)blueLox-ΔVP26 was reduced tenfold compared to their parental strain HSV1(17+)blueLox. (D) The titers of HSV1(KOS)GFPVP26 and HSV1(KOS)ΔVP26 were reduced by 50% compared to their parental strain HSV1(KOS).
Fluorescence tagging of HSV1-VP26.
To analyze the subcellular distribution of HSV1 capsids, we replaced amino acids 1 to 7 of the capsid protein VP26 with GFP or red fluorescence protein (RFP). First, the VP26 ORF was disrupted by inserting a rpsLneo cassette (Fig. 1H), which was subsequently replaced by GFPVP26 or RFPVP26 (Fig. 1J). Restriction analysis of the resulting BACs pHSV1(17+)blueLox-ΔVP26, -GFPVP26, and -RFPVP26 showed no alterations except the desired modifications and, again, permutations in the NotI joint fragments (Fig. 1C and D, lanes Δ, G, and R). HSV1(17+)blueLox-GFPVP26 and -RFPVP26 secreted less infectious virus compared to HSV1(17+)blueLox but produced higher titers than HSV1(17+)blueLox-ΔVP26 (Fig. 3C). Similarly, HSV1(KOS)-GFPVP26 produced lower titers than HSV1(KOS), but there was no growth difference between HSV1(KOS)-ΔVP26 and HSV1(KOS)-GFPVP26 (Fig. 3D).
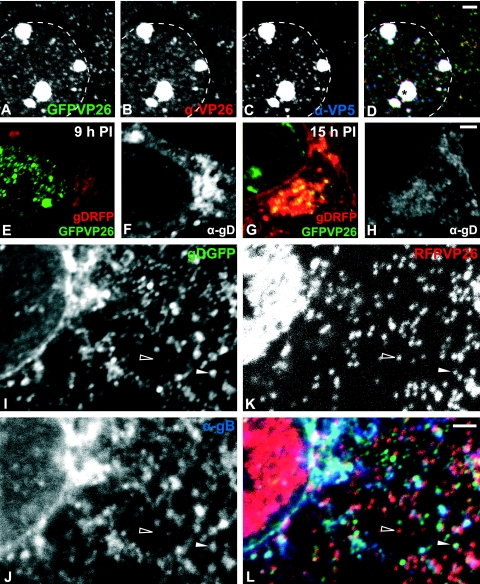
Vero cells were then infected with HSV1(17+)blueLox-GFPVP26 (Fig. 4A to D) or -RFPVP26 (data not shown), fixed, and labeled with anti-VP26 and with anti-VP5, directed against the major capsid protein. At 9 h p.i., GFPVP26 (Fig. 4A) was mostly localized in the nucleus, either on single capsids or on capsid aggregates, but there were also many capsids in the cytoplasm. The GFPVP26 signal (Fig. 4A and green area in Fig. 4D) colocalized with the anti-VP26 labeling (Fig. 4B and red area in Fig. 4D) on single capsids and nuclear capsid aggregates (yellow area in Fig. 4D). VP5 was mostly detected on nuclear capsids and aggregates (Fig. 4C and blue area in Fig. 4D), where it colocalized with GFPVP26 and anti-VP26 (white area in Fig. 4D). Some GFPVP26-containing nuclear capsids were stronger labeled by anti-VP5 (blue area in Fig. 4D) than other nuclear and cytoplasmic capsids (green and yellow areas in Fig. 4D). Similar results were obtained with HSV1(17+)blueLox-RFPVP26 (data not shown). Thus, GFPVP26 and RFPVP26 were recruited onto newly synthesized nuclear capsids. Cytoplasmic and nuclear capsids contained comparable amounts of GFPVP26 or RFPVP26, since the intensity of their fluorescence was rather homogeneous. In contrast, the antibodies to VP26 and VP5 had less access to the surface of cytoplasmic capsids, probably due to tegumentation and envelopment.
FIG. 4.
Fluorescence tagging of HSV1-VP26 and HSV1-gD. (A to D) Vero cells were infected with 10 PFU/cell of HSV1(17+)blueLox-GFPVP26 for 9 h and labeled using antibodies directed against VP26 (B) or VP5 (MAb 5C10; C). GFPVP26 (A; green) colocalized with the anti-VP26 (B, red) and anti-VP5 (C; blue) in the nucleus and the cytoplasm (white area in panel D). Nuclear capsids were stronger labeled with anti-VP5 than cytoplasmic capsids (blue in panel D). Note the intranuclear capsid aggregates (asterisk in panel D). The nuclear rim is indicated by a dashed line. (E to H) Vero cells were infected with 10 PFU/cell of HSV1(17+)blueLox-GFPVP26-gDRFP for 9 h (E and F) or 15 h (G and H). The images were scaled identical for 9 h and 15 h p.i. to reflect the increase in viral proteins. After fixation and permeabilization, the cells were labeled with anti-gD (MAb DL6; F and H) which colocalized with gDRFP (red areas in panels E and G). Note the higher concentration of cytoplasmic capsids (G) and the weaker anti-gD labeling (compare panels F and H) at 15 h p.i. and the colocalization of gDRFP with the anti-gD labeling. At 15 h, many capsids colocalized with gDRFP (yellow area in panel G). (I to L) Vero cells were infected with 10 PFU/cell of HSV1(17+)blueLox-RFPVP26-gDGFP for 9 h. After fixation and permeabilization, the cells were labeled with anti-gB (PAb R69; J) which colocalized with gDGFP (I). Note the cytosolic capsids (red areas and open arrowhead in panel L), which do neither colocalize with gDGFP (green and truquoise areas in panel L) nor with gB (blue and turquoise areas in panel L). In contrast, there was a large degree of colocalization between gDGFP and gB (bright blue/turquoise areas in panel L). There were also several cytoplasmic capsids that colocalized with both gDGFP and gB or either glycoprotein (filled arrowhead in panel L). All specimens were analyzed by laser scanning confocal fluorescence microscopy. Scale bar, 2 μm.
Tagging of HSV1-gD.
To determine the subcellular localization of envelope proteins, we added GFP or RFP to the C terminus of gD as described previously (66, 80; Charles Whitbeck, unpublished data) and introduced these tags into pHSV1(17+)blueLox-RFPVP26 (Fig. 5A and B) or pHSV1(17+)blueLox-GFPVP26. The dual-color strains reached lower titers and released infectious particles more slowly (Fig. 5C). Cells were infected with HSV1(17+)blueLox-GFPVP26-gDRFP and labeled with the MAb DL6 (Fig. 4F and H) directed against luminal amino acids 272 to 279 of gD located close to the membrane (50). At 9 h p.i., gDRFP was mainly localized in several cytoplasmic clusters (red area in Fig. 4E), which most likely corresponded to the host membranes implicated in secondary envelopment. The gDRFP pattern (red area in Fig. 4E) colocalized with the anti-gD labeling (Fig. 4F). At 15 h p.i., gDRFP was distributed almost in the entire cytoplasm, had accumulated in juxtanuclear regions and the nuclear envelope, and was also detected in plasma membrane protrusions (red area in Fig. 4G). Again, gDRFP colocalized very well with the anti-gD signal (Fig. 4H); however, the relative intensity of gDRFP increased from 9 to 15 h p.i. (compare the red area in Fig. 4E to the red area in Fig. 4G), whereas the labeling efficiency of the antibody decreased (compare Fig. 4F to Fig. 4H).
FIG. 5.
Characterization of HSV1(17+)blueLox-RFPVP26-gDGFP. The sequence for GFP was added to the 3′ end of the US6 ORF to tag the C terminus of gD. (A) Restriction analysis of pHSV1(17+)blueLox (blueLox), -RFPVP26, -gDGFP, and -RFPVP26-gDGFP with NotI or BglII/HindIII. The addition of GFP to VP26 was indicated by a NotI bandshift from 6.7 to 7.3 kb (cf. Fig. 1C and J) and a BglII/HindIII bandshift from 17.2 to 17.9 kb (*). (B) A BglII/HindIII bandshift from 4.4 to 5.1 kb (arrowheads) revealed the addition of GFP to gD. Restriction fragment sizes are indicated in kilobases. (C) For single-step growth kinetics, BHK-21 cells were inoculated at an MOI of 5 PFU/cell. Samples were taken from the supernatant and titrated on Vero cells in triplicates.
The subcellular localization of GFPVP26 after infection with the dual-color fluorescence strains was the same as for the single-color virus. At earlier time points, the nuclei contained single capsids and capsid aggregates (green area in Fig. 4E), whereas later many capsids had left the nucleus and accumulated in a juxtanuclear region that coincided with a high concentration of gDRFP (yellow area in Fig. 4G). Thus, efficient capsid export from the nucleus had occurred between 9 and 15 h p.i., and the majority of cytoplasmic capsids colocalized with that region of the cytoplasm which had accumulated the highest concentration of gD.
Since the detection of GFP is more sensitive than that of RFP, we also analyzed HSV1-(17+)-RFPVP26-gDGFP (Fig. 4I to L). At 9 h p.i., there were numerous cytoplasmic capsids of which a large portion did not colocalize with either glycoprotein and thus most likely represented unenveloped, naked cytosolic capsids (open arrowhead and red area in Fig. 4L). There were also cytoplasmic membranes which contained gDGFP (green area in Fig. 4L) and/or gB (blue area in Fig. 4L; colocalization turquoise in Fig. 4L) but no capsids. Other capsids colocalized with gDGFP and gB (filled arrowhead and white area in Fig. 4L), suggesting that RFPVP26, gDGFP and gB (as well as GFPVP26 and gDRFP) had been correctly targeted to the cytoplasmic site of HSV1 assembly.
Dual-color fluorescence HSV1 virions.
We next analyzed the protein composition of viral particles secreted from infected cells (Fig. 6A). An anti-VP26 antibody detected a single band at the expected molecular weight in wild-type and HSV1(17+)blueLox but no signal in HSV1(17+)blueLox-ΔVP26. The bands at around 40 kDa represented the fluorescent VP26 fusion proteins, which were incorporated in full length into HSV1 particles. Probing with an anti-gD antibody showed an increase in the molecular weight of gD, indicating the incorporation of the full-length fusion protein into virions. A fraction of gDRFP was most likely cleaved at the gD-RFP-junction, since we still observed a minor signal at the original gD position (Fig. 6A, lane GR). Similar results were obtained for HSV1(17+)blueLox-RFPVP26-gDGFP (data not shown).
FIG. 6.
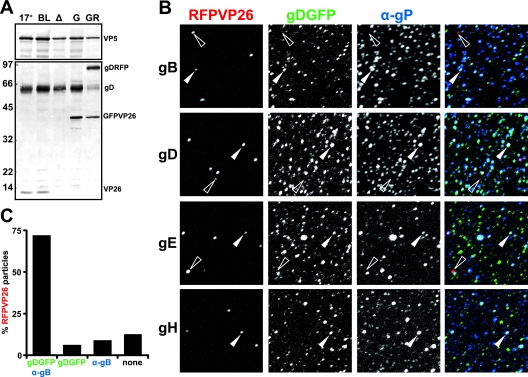
Secretion of dual-color fluorescence HSV1 virions. (A) Virions were sedimented from the medium of cells infected with HSV1(17+), HSV1(17+)blueLox (BL), -ΔVP26 (Δ), -GFPVP26 (G), or -GFPVP26-gDRFP (GR), and analyzed by immunoblotting. An antibody directed against VP26 detected no protein in HSV1(17+)blueLox-ΔVP26 but fusion proteins of the expected molecular weight in HSV1(17+)blueLox-GFPVP26 and -GFPVP26-gDRFP. An antibody directed against gD (PAb R45) detected gD and gDRFP at the expected molecular weights. Possibly, the fusion protein was cleaved to some extent, leading to an additional gD signal in HSV1(17+)blueLox-GFPVP26-gDRFP. The capsid protein VP5 (PAb α-NC1) served as a loading control. (B) BHK-21 cells were infected with 0.01 PFU/cell of HSV1(17+)blueLox-RFPVP26-gDGFP. Particles secreted from the infected cell were harvested at 71 h p.i.; adsorbed to glass coverslips; labeled with antibodies against the viral membrane glycoproteins gB, gD, gE, or gH (α-gP); and analyzed by fluorescence microscopy. Most RFPVP26-positive particles colocalized with gDGFP and the respective glycoprotein (filled arrowhead), but some lacked detectable gDGFP or glycoprotein labeling (red areas and open arrowhead). The gDGFP particles, which did not colocalize with RFPVP26, but with the respective other glycoprotein may represent L-particles (blue/turquoise areas in the overlay). (C) Quantification of the colocalization of RFPVP26 particles with gDGFP and gB.
Immunolabelings demonstrated that most, if not all RFPVP26 particles with gDGFP also contained other HSV1 envelope proteins such as gB, gE, and gH (Fig. 6B). More than 80% of RFPVP26 particles colocalized with gDGFP (Fig. 6C and filled arrowhead in Fig. 6B), and more than 70% colocalized with both gDGFP and gB. There was a small fraction of RFPVP26 particles that lacked detectable amounts of any glycoprotein (open arrowheads in Fig. 6B) and thus were most likely nonenveloped capsids, which are also released after infection with HSV1 wild type (30). There were also numerous particles with fluorescent envelope but no fluorescent capsid protein (turquoise areas in Fig. 6B). These were most likely L-particles, which are secreted in considerable amounts from infected BHK cells. L-particles contain HSV1 envelope and tegument proteins but no capsids and thus on linear gradients band at a lighter position than virions (73, 85). In summary, we generated dual-color fluorescence HSV1(17+) strains, which produced virions incorporating both tagged structural proteins.
Time course of HSV1 nuclear egress and assembly.
The nuclear pores are the gateways of the nuclear envelope that control macromolecular transport between the cytosol and the nucleoplasm. Recent studies suggested that nuclear pore complexes can become dilated during a herpesvirus infection and that HSV1 capsids with their diameter of 125 nm might also directly traverse through such pores to the cytosol (55, 65, 94). To obtain an overview of the HSV1 induced changes in the nuclear pore network, we infected Vero cells with 10 PFU/cell of either HSV1(17+)blueLox or of HSV1(17+)blueLox-GFPVP26-gDRFP and analyzed them from 2.5 to 12 h p.i. For the first, the small capsid protein VP26 was labeled with specific antibodies (data not shown [but cf. Fig. 4 and 8]), whereas for the latter VP26 (Fig. 7A to E) and gD (Fig. 7F to J) were detected by their intrinsic fluorescence. The nuclear pores were labeled with MAb 414 (25), which recognizes phenylalanine-glycine (FG) repeats present in several proteins of the nuclear pore complexes (26) (Fig. 7K to O).
At 2.5 h (results not shown) and at 4 h p.i., the cells contained almost no nuclear capsids, but some incoming capsids derived from the inoculum were still located on the nuclear rim (Fig. 7A and green area in Fig. 7P) (cf. references 31, 32, and 81). As the infection progressed, many cells contained increasing numbers of nuclear capsids, and capsid aggregates developed in more and more nuclei (cf. Fig. 7C). At 8 h p.i., and more prominently at 10 h p.i., newly synthesized capsids had reached the cytoplasm (arrowheads in Fig. 7R and S) and later often accumulated there in clusters (Fig. 7E and T). Newly synthesized gDRFP was detected as early as 4 h p.i. (Fig. 7F). From 6 to 12 h p.i., more and more gD was concentrated in cytoplasmic clusters (Fig. 7G and J). At 10 h p.i. and more so at 12 h p.i., the cytoplasmic capsids colocalized with gD in these clusters (yellow areas in Fig. 7S and T).
Labeling of the DNA (data not shown) or the nuclear pores enabled a distinction of nuclear from cytoplasmic capsids by confocal fluorescence microscopy. Nuclear capsids rarely colocalized with gD and, if so, only did so in the nuclear periphery (results not shown). Several cytoplasmic capsids did not colocalize with gD (arrowheads in Fig. 7R, S, and T), but as the infection progressed many had translocated to those regions of the cytoplasm that also contained a high concentration of gD (Fig. 7T and cf. Fig. 4L). According to the luminal pathway hypothesis (12, 37, 55), HSV1 capsids acquire their final envelope at the inner nuclear membrane, and one would expect a stronger colocalization of GFPVP26 with gDRFP at the nuclear envelope and throughout the entire cytoplasm. However, our data were most consistent with the deenvelopment-reenvelopment hypothesis of HSV1 assembly (12, 37, 64), which predicts that initially cytosolic capsids do not colocalize with gDRFP, but upon secondary envelopment in the cytoplasm acquire their secondary, final envelope containing gDRFP.
Changes in the nuclear pore network during HSV1 infection.
The nuclear pores in mock-infected cells were mostly randomly distributed over the entire nuclear surface, yielding a ring-like, rim appearance of the nuclear pore network around the nucleus in confocal images (Fig. 8A). Moreover, several cytoplasmic spots contained a high concentration of nuclear pore antigens (open arrowhead in Fig. 8A). Some cells showed a particularly weak labeling, and others showed a particularly strong labeling, which most likely reflected cell-cycle-dependent changes in the concentration of nuclear pores and in the organization of the nuclear pore had (10, 58). At 8 h p.i., the morphology of the nuclear pore network had changed in some cells. Occasionally, the nuclear envelope appeared to be extended or invaginated into the nuclear interior, as suggested by a dotted line appearance of nuclear pore complexes within the boundaries of the ring-like labeling of the outer nuclear diameter (Fig. 8A, “fold”). In some cells, the labeling of the ring-like and the apparently invaginated nuclear envelopes was interrupted by large gaps (Fig. 8A, “irregular”). The most striking phenotype displayed nuclei with a ring-like labeling of rather low intensity, and instead the nuclear pores had been clustered on the nuclear envelope (Fig. 8A, “cluster,” filled arrowhead).
Since the morphology of the nuclear pore network was heterogeneous after HSV1 infection, we randomly sampled cells for several time points p.i. and sorted them, depending on their nuclear pore labeling, into five different classes: weak or no label, regular, fold, irregular, and cluster (Fig. 8A). For the mock-infected cells, almost 90% of the nuclei were characterized by a “regular” morphology (Fig. 8B). At 8 h p.i., ca. 20 to 25% of the cells had acquired the described changes in the nuclear pore network (fold, irregular, and cluster). The fraction of cells characterized by changes in the nuclear pore architecture then did not change further up to 12 h p.i. The different distribution of the nuclear pore phenotypes in the uninfected, mock-treated cells reflected the heterogeneity in the nuclear pore architecture among different cell populations (compare HSV1(17+)blueLox to HSV1(17+)blueLox-GFPVP26-gDRFP in Fig. 8B).
For all time points, the infected cells were also categorized into four classes according to the stage of the HSV1 infection as judged by the amount of cytoplasmic capsids (Fig. 8C). At 4 h p.i., almost 90% of the cells infected with HSV1(17+)blueLox and ca. 40% of those infected with HSV1(17+)blueLox-GFPVP26-gDRFP contained fewer than 10 cytoplasmic capsids but no nuclear capsids (cf. Fig. 7A and P), suggesting that most if not all of these cytoplasmic capsids were derived from the inoculum (30, 81). At 8 h p.i., ca. 25 to 50% of the cells contained more than 10 cytoplasmic capsids. This proportion increased at later time points. However, particularly at 12 h p.i., many cells had been lost due to cytopathic effects and detachment. Overall, more cytosolic capsids of HSV1(17+)blueLox than of HSV1(17+)blueLox-GFPVP26-gDRFP were detected, which most likely reflected that the infection with the dual-color fluorescence HSV1 strain progressed slower (cf. Fig. 5C).
Intriguingly, the number of cells characterized by changes in the nuclear pore network increased (Fig. 8B) with kinetics similar to those of the accumulation of cytoplasmic capsids (Fig. 8C). We therefore sought to determine whether, independent of the time point, reorganization of the nuclear pore network correlated with the amount of capsids that had egressed from the nucleus to the cytoplasm. All cells recorded from 6 to 12 h p.i. were therefore sorted into the four classes depending on the concentration of cytoplasmic capsids (none, fewer than 10, 10 to 50, and more than 50), and further into the five classes depending on the morphology of the nuclear pore network but irrespective of the time point postinfection analyzed (Fig. 8D). Of the cells containing a high concentration of cytoplasmic capsids (i.e., more than 10), 80 or 60% had an apparently regular nuclear pore network after infection with HSV1(17+)blueLox or HSV1(17+)blueLox-GFPVP26-gDRFP, respectively. However, ca. 20 to 25% of all cells showing no cytoplasmic capsids were characterized by HSV1 induced changes in the architecture of the nuclear pore network (fold, irregular, and cluster). This proportion remained rather constant for HSV1(17+)blueLox and increased up to ca. 40% for HSV1(17+)blueLox-GFPVP26-gDRFP as the concentration of cytoplasmic capsids increased to more than 50 per cell. The amount of cytoplasmic capsids was not higher in cells in which the nuclear pore architecture was not regular. In another experiment with a slower infection at a lower MOI, 50 to 75% of the cells with a high amount of cytoplasmic capsids showed major changes in the nuclear pore architecture (data not shown). Thus, surprisingly the organization of the nuclear pore network was less heterogeneous among cells infected with a higher MOI.
In summary, the percentage of cells characterized by HSV1 induced alterations in the nuclear pore architecture increased around 8 h p.i. irrespective of whether VP26 and gD were tagged or not. Nevertheless, irrespective of the virus dose or strain, the HSV1-induced changes in the nuclear pore network were not a prerequisite for HSV1 nuclear capsid egress, since many cytoplasmic capsids were present in cells with an apparently regular nuclear pore distribution.
DISCUSSION
Characterization of HSV1(17+) BACs.
We constructed dual-color fluorescence HSV1(17+) strains to analyze the subcellular localization of capsid, envelope, and host structures during assembly and egress. Although HSV1 has been cloned as a BAC before, these were either based on strain F (49, 86), whose sequence has not been published, or had a packaging signal at an ectopic position (76, 83). We decided, like Gierasch et al. (43), to clone the fully sequenced genome of HSV1(17+). We obtained infectious BAC clones containing internal and terminal joint fragments of different length. Whereas in eukaryotic cells the terminal and internal fragments of the HSV1 genome display heterogeneity with respect to the number of a-sequences (27, 74), the joint fragments in the BACs were frozen in a definite arrangement. Recombination between direct repeats has also been reported for another BAC-cloned herpesvirus (3). Such undesired mutations may not only occur during BAC mutagenesis but also after constructing HSV1 mutants by homologous recombination in cell culture and sometimes remained undetected for decades, even with recombinant strains used in clinical trials (23). Another important question is whether these alterations are of functional relevance. The copy number and composition of the a-sequence varies between different HSV1 strains (6, 27, 74), isolates from the same individual during different episodes of recurrence (90), and within the population of a specific HSV1 strain (27, 74, 91). HSV1 BAC genomes with different a-sequences from E. coli can be used to study the biological fitness of such clones. Notably, all HSV1 BACs analyzed in the present study remained infectious, and the DNA fragments harboring the a-sequences immediately regained heterogeneity after transfecting the genomes back into eukaryotic cells.
All HSV1-BAC clones we analyzed lacked the 144-bp oriL, which displays a notorious instability upon cloning (93). This palindromic sequence has only been maintained in an E. coli strain that lacks several DNA repair and recombination genes and thus increases the stability of inverted repeats and palindromes (47). However, such a strain is not suitable for BAC mutagenesis, since the oriL most likely would become unstable as soon as recombination enzymes required for mutagenesis would be expressed. Since oriL is not required for replication in cell culture, its absence will not hamper most experiments. However, deleting the oriL results in reduced mortality and pathogenicity in a corneal mouse in vivo infection model (7). For such studies, it may become necessary to construct HSV1 mutants by combining BAC mutagenesis with recombination in eukaryotic cells. Mutagenesis may be performed in an HSV1 BAC lacking oriL and one of its neighboring essential genes. The respective DNA fragment harboring the lacking elements may be maintained in the special E.coli strain and, after cotransfection with the mutated BAC into eukaryotic cells, replication-competent HSV1 mutants with intact oriL may be generated.
The expression of Cre recombinase led to the deletion of the BAC sequences after transfection into eukaryotic cells and faster replication, as reported for pseudorabies virus, human cytomegalovirus, and murine gammaherpesvirus 68 (2, 79, 97). The lower infectivity of HSV1(17+)blueLox compared to HSV1(17+) was probably neither due to the insertion of a β-galactosidase expression cassette nor to the deletion of UL23 but might have been caused by the deletion of oriL. We cannot exclude that there were additional differences between the HSV1(17+)-BACs and HSV1(17+). HSV1(strain F)-BAC viruses are not attenuated compared to the wild type (86). Interestingly, already the mere addition of a single loxP site into HSV1(17+) can result in a threefold titer reduction (43). Despite its slight attenuation, we considered the BAC pHSV1(17+)blueLox a useful base for the construction of dual-fluorescence strains.
Dual-color fluorescence HSV1(17+).
HSV1(KOS)-GFPVP26, in which GFP has been added to VP26 (29), has the same cell entry characteristics as HSV1(KOS) (30). Sufficient amounts of GFP-labeled VP26 remained bound to capsids, so that their subcellular localization and motility could be analyzed in living cells (J. Janus, K. Döhner, A. Wolfstein, F. Büttner, S. Schmidt, and B. Sodeik, unpublished data). Moreover, GFPVP26-labeled capsids were used to study microtubule mediated transport of HSV1 capsids in vitro (96). Using antibodies specific to VP26 and VP5, we confirmed that the GFPVP26 and RFPVP26 signals represented HSV1 capsids (30; the present study) and that the fusion proteins gDRFP and gDGFP colocalized with a gD labeling, as recently also described for gDYFP (80). Adding GFP to gD reduced the amount of infectious virus secreted from BHK-21 cells. The addition of YFP to the cytosolic tail of gD was less detrimental to virus growth in Vero cells, whereas, after amplification in a neuroblastoma cell line, the titers are decreased (80). Thus, manipulating gD has different effects in different cell lines. The cytoplasmic tail of gD is dispensable for growth in cell culture (41), but adding GFP or RFP to it may reduce its affinity to tegument or envelope proteins (17, 40). However, ultimately HSV1 particles that incorporated both fluorescent fusion proteins and contained several other envelope proteins were secreted. Thus, the trafficking of the fluorescence-labeled gD was similar to untagged HSV1 glycoproteins. These dual-color fluorescence HSV1 strains provide promising tools for the analysis of assembly and egress (the present study), as well as for the study of cell entry (K. Döhner, C.-H. Nagel, and B. Sodeik, unpublished data). Moreover, these strains allow the detection of all subviral particles containing tagged VP26 and gD, irrespective of their interaction and possible masking by host factors or other HSV1 structural proteins.
Nuclear pore network during HSV1 nuclear capsid egress and assembly.
During HSV1 infection, many host organelles and also the cytoskeleton change their morphology and subcellular organization. These HSV1-induced changes are collectively summarized under the term cytopathic effects. For example, the membranes containing antigens of the Golgi apparatus are scattered over the entire cytoplasm rather than being localized in a tight juxtanuclear cluster, and the microtubules are marginalized to the cell periphery (5, 36, 51). Moreover, the nuclear envelope is severely modified during HSV1 infection (8, 46, 53, 77), resulting also in a reorganization of the nuclear pore network (55, 71). Together with the recent electron microscopy observations that some nuclear pores appear to become dilated during infection with alphaherpesviruses, this has led to the hypothesis that HSV1 induces changes in the nuclear pore network, and the widening of the lumen of individual nuclear pores might generate a channel large enough to allow a passage of nuclear capsids directly into the cytosol (55, 94). In this scenario, the capsids would step aside and bypass the barriers posed by the nuclear lamina and the inner and outer nuclear membranes.
Simultaneous recording of the subcellular localization of nuclear pore complexes, capsids, and envelope proteins allowed us to distinguish both nuclear from cytoplasmic capsids and virions in vesicles from cytosolic capsids. In accordance with Leuzinger et al. (55), the nuclear pore network was modified as the HSV1 infection progressed. However, in the majority of cells containing many cytoplasmic capsids, the nuclear pore architecture was not changed, suggesting that changes in the nuclear pore architecture were not required for efficient nuclear egress. Our data are consistent with studies using HSV1 deletion mutants. Without US3 (72) or the glycoproteins gB and gH (40) capsids can acquire a primary envelope at the inner nuclear membrane but are stuck in the perinuclear space between inner and outer nuclear membrane, presumably due to an inability to catalyze primary fusion. In the absence of UL31 (15) or UL34 (75), capsids are defective in primary budding, and nuclear capsids accumulate but are also unable to use potentially dilated nuclear pores as gateways to the cytosol. Leuzinger et al. (55) proposed that the changes in the nuclear pore labeling observed by fluorescence microscopy may reflect an extension of the luminal channel of individual nuclear pores as detected by electron microscopy. However, the apparent holes in the nuclear envelope may not contain bona fide nuclear pore proteins at their rims. If they would indeed lack nuclear pore proteins, they may not have arisen from dilation of preexisting pore complexes but might have been generated by HSV1-catalyzed fusion of the inner with the outer nuclear membrane. Moreover, the nuclei with irregular or clustered nuclear pore architectures might have clustered several nuclear pore complexes within a small area of the nuclear envelope rather than indeed enlarged the nuclear pore channel. The latter scenario agrees well with many electron microscopy studies that reported that the integrity of individual nuclear pores appeared intact until late in herpesvirus infection (40, 46, 64, 65, 72).
Our data are consistent with nuclear capsids leaving the nucleus via primary budding and primary fusion to release capsids into the cytosol rather than with a direct passage through a dilated nuclear pore. In addition, only later in infection gD had accumulated in the membranes surrounding the nucleus, whereas gD was rather prominent on cytoplasmic membranes, where it often colocalized with capsids. The cytoplasmic capsids that did not colocalize with gD and gB most likely represented cytosolic capsids rather than capsids inside virions. The resulting cytosolic capsids would then be transported to cytoplasmic membranes to acquire a final envelope by secondary envelopment. Our results are expected for capsids that follow the deenvelopment-reenvelopment pathway, whereas the luminal pathway for HSV1 egress predicts that capsids should already acquire gD and gB at the inner nuclear membrane and that all cytoplasmic capsids colocalize with all viral membrane proteins.
These dual-color fluorescence strains will be valuable tools for evaluating the fate of other host organelles and the cytoskeleton during the viral life cycle, for studying the dynamics and sequence of events in HSV1 assembly and entry in living cells, and for further characterizing the phenotypes of mutations in HSV1 structural proteins. Such studies will result in further insights into the HSV1-induced cytopathic effects and whether these are kinetically linked to HSV1 nuclear capsid egress, assembly, and secretion of infectious particles or whether they constitute bona fide bystander effects of viral infection.
Acknowledgments
We thank Rudi Bauerfeind (Department of Cell Biology and Confocal Laser Scanning Microscopy Facility, Hannover Medical School) for his continuous support, as well as Julia Schipke, Kerstin Radtke, Nadine Müther, Kathrin Rode, Kristina Theusner, and Christof Bohnen (Institute of Virology, Hannover Medical School) and Joel Baines (Cornell University), Prashant Desai (Johns Hopkins University), and Charles Whitbeck (University of Pennsylvania) for helpful discussions. We are grateful to Lynn Enquist (Princeton University), Nikolaus Osterrieder (Free University, Berlin), Greg Smith (Northwestern University), and Karsten Tischer (University of Kiel, Kiel, Germany) for assistance with BAC mutagenesis; Prashant Desai (Johns Hopkins University), Pat Spear (Northwestern University), John Subak-Sharpe (MRC Virology Unit, Glasgow, United Kingdom), and Roger Tsien (University of California, San Diego) for providing plasmids and viruses; and Jay Brown (University of Virginia), Roselyn Eisenberg and Gary Cohen (University of Pennsylvania), and Anthony Minson (University of Cambridge, Cambridge, United Kingdom) for donating antibodies.
This study was supported by the German Research Council (DFG; So403/2, So403/3 to B.S.), by the EU 6th Framework Program (EU-NEST AXON SUPPORT, contract number 12702 to B.S.), and by the Wilhelm-Sander-Foundation (grant 2004.075.1 to M.M.). C.-H.N. received a fellowship from the Hannover Biomedical Research School (DFG-GK745 on Mucosal Host-Pathogen Interaction).
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Adler, H., M. Messerle, and U. H. Koszinowski. 2003. Cloning of herpesviral genomes as bacterial artificial chromosomes. Rev. Med. Virol. 13111-121. [DOI] [PubMed] [Google Scholar]
- 2.Adler, H., M. Messerle, and U. H. Koszinowski. 2001. Virus reconstituted from infectious bacterial artificial chromosome (BAC)-cloned murine gammaherpesvirus 68 acquires wild-type properties in vivo only after excision of BAC vector sequences. J. Virol. 755692-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 746964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alconada, A., U. Bauer, B. Sodeik, and B. Hoflack. 1999. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J. Virol. 73377-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avitabile, E., S. Di Gaeta, M. R. Torrisi, P. L. Ward, B. Roizman, and G. Campadelli-Fiume. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 697472-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines, J. D., and S. K. Weller. 2005. Cleavage and packaging of herpes simplex virus 1 DNA, p. 135-150. In C. E. Catalano (ed.), Viral genome packaging machines: genetics, structure, and mechanism. Kluwer Academic/Plenum Publishers, New York, NY.
- 7.Balliet, J. W., and P. A. Schaffer. 2006. Point mutations in herpes simplex virus type 1 oriL, but not in oriS, reduce pathogenesis during acute infection of mice and impair reactivation from latency. J. Virol. 80440-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibor-Hardy, V., M. Suh, M. Pouchelet, and R. Simard. 1982. Modifications of the nuclear envelope of BHK cells after infection with herpes simplex virus type 1. J. Gen. Virol. 6381-94. [DOI] [PubMed] [Google Scholar]
- 9.Buckmaster, E. A., U. Gompels, and A. Minson. 1984. Characterisation and physical mapping of an HSV-1 glycoprotein of approximately 115 × 103 molecular weight. Virology 139408-413. [DOI] [PubMed] [Google Scholar]
- 10.Burke, B., and J. Ellenberg. 2002. Remodeling the walls of the nucleus. Nat. Rev. Mol. Cell. Biol. 3487-497. [DOI] [PubMed] [Google Scholar]
- 11.Calistri, A., P. Sette, C. Salata, E. Cancellotti, C. Forghieri, A. Comin, H. Gottlinger, G. Campadelli-Fiume, G. Palu, and C. Parolin. 2007. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 8111468-11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campadelli-Fiume, G. 2007. The egress of alphaherpesviruses from the cell, p. 151-163. In A. Arvin, G. Campadelli-Fiume, E. Mocarski, P. S. Moore, B. Roizman, R. Whitley, and K. Yamanishi (ed.), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 13.Campadelli-Fiume, G., B. Roizman, P. Wild, T. C. Mettenleiter, and A. Minson. 2006. The egress of herpesviruses from cells: the unanswered questions. (Letter and Author Reply.) J. Virol. 806716-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 997877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the U(L)31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 718307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1589-14. [DOI] [PubMed] [Google Scholar]
- 17.Chi, J. H., C. A. Harley, A. Mukhopadhyay, and D. W. Wilson. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86253-261. [DOI] [PubMed] [Google Scholar]
- 18.Cleator, M. C., and P. E. Klapper. 2004. Herpes simplex, p. 27-51. In A. J. Zuckerman, J. E. Banatvala, J. R. Pattison, P. D. Griffiths, and B. D. Schoub (ed.), Principles and practice of clinical virology, 5th ed. John Wiley & Sons, Inc., New York, NY.
- 19.Clement, C., V. Tiwari, P. M. Scanlan, T. Valyi-Nagy, B. Y. Yue, and D. Shukla. 2006. A novel role for phagocytosis-like uptake in herpes simplex virus entry. J. Cell Biol. 1741009-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen, G., M. Katze, C. Hydrean-Stern, and R. Eisenberg. 1978. Type-Common CP-1 antigen of herpes-simplex-virus is associated with a 59,000-molecular-weight envelope glycoprotein. J. Virol. 27172-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. J. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crump, C. M., C. Yates, and T. Minson. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 817380-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dambach, M. J., J. Trecki, N. Martin, and N. S. Markovitz. 2006. Oncolytic viruses derived from the gamma34.5-deleted herpes simplex virus recombinant R3616 encode a truncated UL3 protein. Mol. Ther. 13891-898. [DOI] [PubMed] [Google Scholar]
- 24.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis, L. I., and G. Blobel. 1986. Identification and characterization of a nuclear pore complex protein. Cell 45699-709. [DOI] [PubMed] [Google Scholar]
- 26.Davis, L. I., and G. Blobel. 1987. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. USA 847552-7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davison, A. J., and N. M. Wilkie. 1981. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J. Gen. Virol. 55315-331. [DOI] [PubMed] [Google Scholar]
- 28.Desai, P., N. A. DeLuca, and S. Person. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247115-124. [DOI] [PubMed] [Google Scholar]
- 29.Desai, P., and S. Person. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 727563-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Döhner, K., K. Radtke, S. Schmidt, and B. Sodeik. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 808211-8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Döhner, K., and B. Sodeik. 2004. The role of the cytoskeleton during viral infection. Curr. Top. Microbiol. Immunol. 28567-108. [DOI] [PubMed] [Google Scholar]
- 32.Döhner, K., A. Wolfstein, U. Prank, C. Echeverri, D. Dujardin, R. Vallee, and B. Sodeik. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 132795-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubin, G., I. Frank, and H. M. Friedman. 1990. Herpes simplex virus type 1 encodes two Fc receptors which have different binding characteristics for monomeric immunoglobulin G (IgG) and IgG complexes. J. Virol. 642725-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisenberg, R. J., D. Long, M. Ponce de Leon, J. T. Matthews, P. G. Spear, M. G. Gibson, L. A. Lasky, P. Berman, E. Golub, and G. H. Cohen. 1985. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J. Virol. 53634-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenberg, R. J., M. Ponce de Leon, H. M. Friedman, L. F. Fries, M. M. Frank, J. C. Hastings, and G. H. Cohen. 1987. Complement component C3b binds directly to purified glycoprotein C of herpes simplex virus types 1 and 2. Microb. Pathog. 3423-435. [DOI] [PubMed] [Google Scholar]
- 36.Elliott, G., and P. O'Hare. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 726448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1998. Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51237-347. [DOI] [PubMed] [Google Scholar]
- 38.Farnham, A. E., and A. A. Newton. 1959. The effect of some environmental factors on herpesvirus grown in HeLa cells. Virology 7449-461. [DOI] [PubMed] [Google Scholar]
- 39.Farnsworth, A., and D. C. Johnson. 2006. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread. J. Virol. 803167-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farnsworth, A., T. W. Wisner, and D. C. Johnson. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81319-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feenstra, V., M. Hodaie, and D. C. Johnson. 1990. Deletions in herpes simplex virus glycoprotein D define nonessential and essential domains. J. Virol. 642096-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gianni, T., G. Campadelli-Fiume, and L. Menotti. 2004. Entry of herpes simplex virus mediated by chimeric forms of nectin1 retargeted to endosomes or to lipid rafts occurs through acidic endosomes. J. Virol. 7812268-12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gierasch, W. W., D. L. Zimmerman, S. L. Ward, T. K. Vanheyningen, J. D. Romine, and D. A. Leib. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135197-206. [DOI] [PubMed] [Google Scholar]
- 44.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 874645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grünewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. M. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 3021396-1398. [DOI] [PubMed] [Google Scholar]
- 46.Haines, H., and R. J. Baerwald. 1976. Nuclear membrane changes in herpes simplex virus-infected BHK-21 cells as seen by freeze-fracture. J. Virol. 171038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hardwicke, M. A., and P. A. Schaffer. 1995. Cloning and characterization of herpes simplex virus type 1 oriL: comparison of replication and protein-DNA complex formation by oriL and oriS. J. Virol. 691377-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26365-369. [DOI] [PubMed] [Google Scholar]
- 49.Horsburgh, B. C., M. M. Hubinette, D. Qiang, M. L. MacDonald, and F. Tufaro. 1999. Allele replacement: an application that permits rapid manipulation of herpes simplex virus type 1 genomes. Gene Ther. 6922-930. [DOI] [PubMed] [Google Scholar]
- 50.Isola, V. J., R. J. Eisenberg, G. R. Siebert, C. J. Heilman, W. C. Wilcox, and G. H. Cohen. 1989. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J. Virol. 632325-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kotsakis, A., L. E. Pomeranz, A. Blouin, and J. A. Blaho. 2001. Microtubule reorganization during herpes simplex virus type 1 infection facilitates the nuclear localization of VP22, a major virion tegument protein. J. Virol. 758697-8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 53.Leach, N., S. L. Bjerke, D. K. Christensen, J. M. Bouchard, F. Mou, R. Park, J. Baines, T. Haraguchi, and R. J. Roller. 2007. Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J. Virol. 8110792-10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 7356-65. [DOI] [PubMed] [Google Scholar]
- 55.Leuzinger, H., U. Ziegler, E. M. Schraner, C. Fraefel, D. L. Glauser, I. Heid, M. Ackermann, M. Mueller, and P. Wild. 2005. Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 7913047-13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mabit, H., M. Y. Nakano, U. Prank, B. Saam, K. Döhner, B. Sodeik, and U. F. Greber. 2002. Intact microtubules support adenovirus and herpes simplex virus infections. J. Virol. 769962-9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacLean, A. R. 1998. Preparation of HSV-DNA and production of infectious virus, p. 19-25. In S. M. Brown and A. R. MacLean (ed.), Herpes simplex virus protocols. Humana Press, Inc., Totowa, NJ. [DOI] [PubMed]
- 58.Maeshima, K., K. Yahata, Y. Sasaki, R. Nakatomi, T. Tachibana, T. Hashikawa, F. Imamoto, and N. Imamoto. 2006. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J. Cell Sci. 1194442-4451. [DOI] [PubMed] [Google Scholar]
- 59.Marozin, S., U. Prank, and B. Sodeik. 2004. Herpes simplex virus type 1 infection of polarized epithelial cells requires microtubules and access to receptors present at cell-cell contact sites. J. Gen. Virol. 85775-786. [DOI] [PubMed] [Google Scholar]
- 60.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 691531-1574. [DOI] [PubMed] [Google Scholar]
- 61.McGeoch, D. J., A. Dolan, S. Donald, and D. H. Brauer. 1986. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 141727-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGeoch, D. J., A. Dolan, S. Donald, and F. J. Rixon. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 1811-13. [DOI] [PubMed] [Google Scholar]
- 63.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 9414759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9423-429. [DOI] [PubMed] [Google Scholar]
- 65.Mettenleiter, T. C., T. Minson, and P. Wild. 2006. Egress of alphaherpesviruses. J. Virol. 801610-1612. (Letter and Author Reply.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Milne, R. S., A. V. Nicola, J. C. Whitbeck, R. J. Eisenberg, and G. H. Cohen. 2005. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J. Virol. 796655-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nicola, A. V., J. Hou, E. O. Major, and S. E. Straus. 2005. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J. Virol. 797609-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ojala, P. M., B. Sodeik, M. W. Ebersold, U. Kutay, and A. Helenius. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 204922-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perry, L. J., and D. J. McGeoch. 1988. The DNA sequences of the long repeat region and adjoining parts of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 692831-2846. [DOI] [PubMed] [Google Scholar]
- 70.Potel, C., K. Kaelin, I. Gautier, P. Lebon, J. Coppey, and F. Rozenberg. 2002. Incorporation of green fluorescent protein into the essential envelope glycoprotein B of herpes simplex virus type 1. J. Virol. Methods 10513-23. [DOI] [PubMed] [Google Scholar]
- 71.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 758803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 768939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rixon, F. J., C. Addison, and J. McLauchlan. 1992. Assembly of enveloped tegument structures (L particles) can occur independently of virion maturation in herpes simplex virus type 1-infected cells. J. Gen. Virol. 73277-284. [DOI] [PubMed] [Google Scholar]
- 74.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fundamental virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 75.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J. Virol. 74117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saeki, Y., T. Ichikawa, A. Saeki, E. A. Chiocca, K. Tobler, M. Ackermann, X. O. Breakefield, and C. Fraefel. 1998. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum. Gene Ther. 92787-2794. [DOI] [PubMed] [Google Scholar]
- 77.Simpson-Holley, M., R. C. Colgrove, G. Nalepa, J. W. Harper, and D. M. Knipe. 2005. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 7912840-12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith, G. A., and L. W. Enquist. 2002. Break ins and break outs: viral interactions with the cytoskeleton of mammalian cells. Annu. Rev. Cell Dev. Biol. 18135-161. [DOI] [PubMed] [Google Scholar]
- 79.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 974873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Snyder, A., B. Bruun, H. M. Browne, and D. C. Johnson. 2007. A herpes simplex virus gD-YFP fusion glycoprotein is transported separately from viral capsids in neuronal axons. J. Virol. 818337-8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1361007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spear, P. G. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol. 6401-410. [DOI] [PubMed] [Google Scholar]
- 83.Stavropoulos, T. A., and C. A. Strathdee. 1998. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J. Virol. 727137-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strive, T., E. Borst, M. Messerle, and K. Radsak. 2002. Proteolytic processing of human cytomegalovirus glycoprotein B is dispensable for viral growth in culture. J. Virol. 761252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Szilagyi, J. F., and C. Cunningham. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J. Gen. Virol. 72661-668. [DOI] [PubMed] [Google Scholar]
- 86.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 771382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40191-197. [DOI] [PubMed] [Google Scholar]
- 88.Trus, B. L., W. W. Newcomb, F. P. Booy, J. C. Brown, and A. C. Steven. 1992. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc. Natl. Acad. Sci. USA 8911508-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Turcotte, S., J. Letellier, and R. Lippe. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 798847-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Umene, K., and T. Kawana. 2003. Divergence of reiterated sequences in a series of genital isolates of herpes simplex virus type 1 from individual patients. J. Gen. Virol. 84917-923. [DOI] [PubMed] [Google Scholar]
- 91.Umene, K., and H. Sakaoka. 1991. Homogeneity and diversity of genome polymorphism in a set of herpes simplex virus type 1 strains classified as the same genotypic group. Arch. Virol. 11953-65. [DOI] [PubMed] [Google Scholar]
- 92.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246179-189. [DOI] [PubMed] [Google Scholar]
- 93.Weller, S. K., A. Spadaro, J. E. Schaffer, A. W. Murray, A. M. Maxam, and P. A. Schaffer. 1985. Cloning, sequencing, and functional analysis of oriL, a herpes simplex virus type 1 origin of DNA synthesis. Mol. Cell. Biol. 5930-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wild, P., M. Engels, C. Senn, K. Tobler, U. Ziegler, E. M. Schraner, E. Loepfe, M. Ackermann, M. Mueller, and P. Walther. 2005. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 791071-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilson, D. W., N. Davis-Poynter, and A. C. Minson. 1994. Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J. Virol. 686985-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wolfstein, A., C. H. Nagel, K. Radtke, K. Döhner, V. Allan, and B. Sodeik. 2006. The inner tegument promotes herpes simplex virus transport along microtubules in vitro. Traffic 71-11. [DOI] [PubMed] [Google Scholar]
- 97.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 762316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20123-128. [DOI] [PubMed] [Google Scholar]