Abstract
During the adenovirus infectious cycle, the early proteins E4orf6 and E1B55K are known to perform several functions. These include nuclear export of late viral mRNAs, a block of nuclear export of the bulk of cellular mRNAs, and the ubiquitin-mediated degradation of selected proteins, including p53 and Mre11. Degradation of these proteins occurs via a cellular E3 ubiquitin ligase complex that is assembled through interactions between elongins B and C and BC boxes present in E4orf6 to form a cullin 5-based ligase complex. E1B55K, which has been known for some time to associate with the E4orf6 protein, is thought to bind to specific substrate proteins to bring them to the complex for ubiquitination. Earlier studies with E4orf6 mutants indicated that the interaction between the E4orf6 and E1B55K proteins is optimal only when E4orf6 is able to form the ligase complex. These and other observations suggested that most if not all of the functions ascribed to E4orf6 and E1B55K during infection, including the control of mRNA export, are achieved through the degradation of specific substrates by the E4orf6 ubiquitin ligase activity. We have tested this hypothesis through the generation of a virus mutant in which the E4orf6 product is unable to form a ligase complex and indeed have found that this mutant behaves identically to an E4orf6− virus in production of late viral proteins, growth, and export of the late viral L5 mRNA.
The late phase of an adenoviral infection, typified by human adenovirus type 5 (Ad5), is characterized by a massive production of progeny virions. To support this production, the virus takes control of the cellular machinery to produce abundant amounts of its own late proteins to the detriment of the synthesis of cellular proteins. This host cell shutoff is achieved via several mechanisms. The translation of cellular mRNAs is blocked by the action of the L4-100K protein on the ribosomal machinery (15, 16) so that only the late viral mRNAs containing the tripartite leader sequence can be translated (26). At the same time, export of cellular mRNAs to the cytoplasm is blocked by the action of the early viral proteins E4orf6 and E1B55K (3, 6, 23, 42). These same proteins were also shown to be required for the export of the late viral mRNAs (4, 20, 23, 42, 53). The E4orf6 and E1B55K proteins were shown to interact during infection (48) and to function in the same pathway, as a viral mutant with defects in both of these products was seen to have essentially the same phenotype as mutants affecting only one of these species (3, 17, 23, 42). The role that E4orf6 and E1B55K proteins play in the control of mRNA export has yet to be defined.
It has been known for some time that the E4orf6 and E1B55K proteins play a role in the degradation of the p53 tumor suppressor, and recently the mechanism for such degradation has been elucidated by our group. Expression of the virus E1A protein results in increased levels of p53 (10, 34); however, during infection, in the presence of both E4orf6 and E1B55K products, p53 is degraded in a proteasome-dependent manner (12, 24, 43, 44, 46, 50). The E4orf6 polypeptide was shown to assemble with the cellular proteins cullin 5 (Cul5), Rbx1, and elongins B and C to form an E3 ubiquitin ligase that, in the presence of E1B55K, ubiquitinates p53 (7, 24). Although E4orf6 can bind p53 in the absence of E1B55K (18), the substrate specificity component of this complex appears principally to be E1B55K, which ensures efficient recruitment of p53 to the complex (7). The E4orf6/E1B55K complex also degrades the MRN double-stranded break repair complex, Mre11, and in this case E1B55K alone is responsible for recruitment of Mre11 (7, 11). Under normal conditions, 95% of E1B55K is associated with the ligase complex (24); however, little E1B55K was found to associate with E4orf6 mutant proteins that were unable to bind to elongins B and C (7), suggesting that the E4orf6/E1B55K interaction is probably not direct but rather occurs through the formation of the ligase complex. Thus, in infected cells, E4orf6 appears to highjack a cellular Cul5-based E3 ligase in order to ubiquitinate and degrade specific substrates brought to the complex by E1B55K.
As noted in our previous report (7), we have observed a perfect correlation between the ability of E4orf6 mutants to complement the growth of an E4− virus and their ability to form the E3 ligase complex. This observation suggested to us that the major functions ascribed to E4orf6 and E1B55K during infection, including the control of mRNA export, are achieved through ubiquitination by the E4orf6/E1B55K complex and subsequent degradation of specific protein substrates. This model is supported by the observation that active proteasomes are required in the early phase of infection for maximal virus growth (14). More recently, a study has been published in which similar conclusions were drawn when the ligase complex was blocked by the expression of a dominant-negative mutant of Cul5 (54).
In this report, we demonstrate that a virus mutant in which the BC boxes of the E4orf6 protein are nonfunctional exhibits the same defective phenotype as does a viral mutant completely defective for production of the E4orf6 product in terms of virus growth and nuclear export of viral mRNAs. These results strongly implicate the ligase activity of the E4orf6 complex in the majority of the E4orf6 and E1B55K functions during infection.
MATERIALS AND METHODS
Cell lines.
Human non-small-cell lung carcinoma H1299 cells (ATCC CRL-5803) carrying a homologous deletion of the p53 gene (37), human lung carcinoma A549 cells (ATCC CCL-185) containing wild-type (wt) p53, and W162 containing the whole Ad5 E4 region (52) were cultured in monolayers in Dulbecco's modified Eagle's medium (Gibco; catalog no. 11995) supplemented with 10% fetal calf serum (Gibco) and 100 U/ml of penicillin and streptomycin.
Antibodies.
Primary monoclonal and polyclonal antibodies used included E1B55K mouse monoclonal 2A6 (48), E4orf6 rabbit polyclonal 1807 (8), Mre11 rabbit polyclonal pNB 100-142 (Novus Biologicals, Inc.), elongin C mouse monoclonal SIII p15 (Transduction Laboratories), E4orf6 mouse monoclonal RSA3 (36), E1A mouse monoclonal M73 (25), E4orf3 rat monoclonal 6A11 (39), Ad5 capsid rabbit polyclonal L133 (30), and DBP (E2A) mouse monoclonal B6-8 (45). Secondary antibodies conjugated to horseradish peroxidase for detection in Western blotting were goat anti-mouse immunoglobulin G (IgG), goat anti-rabbit IgG, and goat anti-rat IgG (Jackson ImmunoResearch Laboratories).
Viruses.
The construction of plasmid pH5pg4100 and the transfer vector pE4-1155 has been described recently (22). The Ad5 genome in pH5pg4100 is inserted into the PacI site of the bacterial cloning vector pPG-S2 (22). It lacks nucleotides (nt) 28593 to 30471 (encompassing most of E3) and contains an additional unique endonuclease restriction site at nt 30955 (BstBI) (nucleotide numbering is according to the published Ad5 sequence from GenBank, accession no. AY339865). The transfer vector pE4-1155 contains Ad5 nt 32840 to 35934 in pPG-S2 (22). The entire sequences of pH5pg4100 (36,067 bp) and pE4-1155 (5,099 bp) are available upon request.
To generate the Ad5 E4orf6− virus and the mutant carrying amino acid changes in the BC1 and BC2 box motifs (see Fig. 1), point mutations were first introduced into the E4orf6 gene in pE4-1155 by site-directed mutagenesis with oligonucleotide primers 930 (5′-CGT GCG AGG TCT TCC TGC AGT GTG G-3′) and 931 (5′-CCA CAC TGC AGG AAG ACC TCG CAC G-3′), resulting in pE4-1298, and with primers 1076 (5′-GGA GGA TCA TCC GGG GCT GCC CGA ATG TAA CAC-3′), 1077 (5′-GTG TTA CAT TCG GGC AGC CCC GGA TGA TCC TCC-3′), 1078 (5′-GGT TAC GAG TCC TGG GCT TCC CAC TGT CAT TGT TCC-3′), and 1079 (5′-GGA ACA ATG ACA GTG GGA AGC CCA GGA CTC GTA ACC-3′), resulting in pE4-1435. Plasmid pE4-1298 contains a 1-bp deletion (nt 33870), and pE4-1435 carries two amino acid changes at positions 47 (L47G) and 122 (L122S). The mutations in both plasmid DNAs were verified by sequencing. Finally, the 5.1-kb BstBI fragment from pH5pg4100 was replaced with the corresponding fragments from plasmids pE4-1298 and pE4-1435 to generate adenoviral plasmids pH5pm4154 (orf6−) and pH5pm4139 (BC1/2), respectively. Recombinant plasmids were partially sequenced to confirm the mutations in the E4orf6 gene.
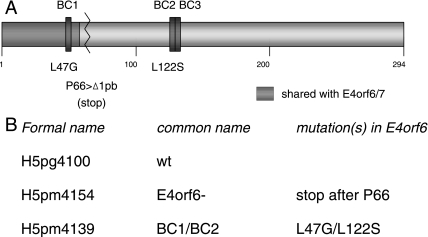
FIG. 1.
Virus mutants. (A) Schematic representation of the E4orf6 protein indicating the positions of the BC boxes and the mutations created, the region shared by E4orf6 with the E4orf6/7 protein, and the position of the deletion generated for the E4orf6− virus. (B) List of the viruses used with their respective mutations.
For the generation of mutant viruses H5pm4154 and H5pm4139 and their wt parent H5pg4100, the viral genomes were released from the recombinant plasmids by PacI digestion and transfected in the complementing W162 cells by the calcium phosphate procedure (21). After 5 days, cells were harvested and viruses were released by four freeze-thaw cycles. The viruses were propagated in W162 monolayer cells, and titers were determined by the fluorescence-forming unit (FFU) method (22). Briefly, serial dilutions of the viruses were used to infect A549 cells, which were fixed at 20 h postinfection (hpi). Infected cells were then identified by immunostaining with an antibody against the early E2A protein (B6-8). Viral DNA was isolated as described previously (49) from viral particles purified by cesium chloride equilibrium density centrifugation (22, 49) and analyzed by HindIII restriction endonuclease digestion. In addition, the viral DNA was partially sequenced to verify the presence of the E4orf6 mutations.
Infections, immunoprecipitations, time courses, and growth curves.
Cells were infected with viruses diluted in infection medium (0.2 mM CaCl2, 0.2 mM MgCl2, 2% serum in phosphate-buffered saline plus 100 U/ml of penicillin and streptomycin) for 90 min before its removal and addition of normal growth medium. For time course studies, cells were infected at a multiplicity of infection (MOI) of 5 or 50 FFU/cell depending on the experiment and harvested by scraping at different times postinfection. The cells were washed in phosphate-buffered saline and collected by centrifugation, and the pellets were frozen until completion of the experiment. Pellets were then lysed in RIPA buffer (10 mM sodium phosphate, pH 7.2, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM dithiothreitol) plus inhibitors (4 mM NaF, 2 mM NaPP, 500 μM sodium vanadate, 200 μg/ml phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, and 5 μg/ml leupeptin), freeze-thawed in liquid nitrogen, and sonicated three times for 20 s. Equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes. For immunoprecipitations, cells were infected at an MOI of 50 FFU/cell, and at 24 hpi, they were lysed in a less stringent buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 5% glycerol, 1 mM dithiothreitol) plus inhibitors (as above) on ice for 20 min. Equal protein amounts (1 mg) were immunoprecipitated with 1807 (E4orf6) antibodies, before the addition of protein G Sepharose (Upstate). Following extensive washing in lysis buffer plus phenylmethylsulfonyl fluoride, the beads were eluted in Laemmli buffer and run on SDS-polyacrylamide gels. For the viral growth curves, cells were infected at an MOI of 5 or 50 FFU/cell depending on the experiment and harvested at 24, 48, and 72 hpi. Viruses were released from the cells by three freeze-thaw cycles, and titers were determined by the fluorescent antibody assay. Total progeny virions in each sample were calculated, and values were plotted on a graph.
Rescue experiment.
Cells expressing E4orf6 were generated by cotransfecting H1299 cells with plasmid DNAs encoding either wt E4orf6 or the BC-box mutant L47G/L122S (BC1/BC2) (7) or with control pcDNA3 and a plasmid DNA including the puromycin resistance gene at a ratio of 10:1. These cells were selected with puromycin (2 μg/ml), and the resistant colonies were pooled and frozen. Cells were cultured as little as possible before use in experiments to maximize the retention of high levels of E4orf6 expression. These cells were then infected with either wt or mutant viruses and analyzed as described for growth curves.
RNA analysis.
Nuclear and cytoplasmic mRNA was extracted from virus-infected cells using an RNA purification kit (Qiagen) according to the manufacturer's instructions. RNA samples (0.5 μg) were separated by denaturing electrophoresis in formaldehyde-agarose gels and transferred to positively charged nylon membranes (Roche Molecular Biochemicals) by capillary transfer with 20× SSC (0.3 M sodium citrate, 3 M sodium chloride, pH 7.2) for 6 h. RNAs were cross-linked to membranes by exposure to 120,000 mJ UV light using a UV Stratalinker (Stratagene). Analysis of the methylene blue-stained 18S and 28S rRNA bands was used to verify that equal amounts of RNA were loaded onto each lane. The blot was prehybridized at 68°C for 30 min in prewarmed hybridization buffer (DIG-Easy Hyb; Roche Molecular Biochemicals). The prehybridization solution was then replaced with prewarmed hybridization buffer containing 50 ng/ml digoxigenin (DIG)-labeled L5 fiber probe and hybridized at 68°C overnight. The membranes were washed two times for 10 min at room temperature with 2× SSC-0.1% (wt/vol) SDS and three times for 15 min at 68°C with 0.1× SSC-0.1% (wt/vol) SDS. Probe detection was performed using the DIG Luminescent Detection Kit (Roche Molecular Biochemicals) according to the manufacturer's protocol. In brief, blots were incubated in blocking solution for 30 min and then in antibody solution containing anti-DIG, alkaline phosphatase-conjugated antibody for 30 min at room temperature, followed by two washes in washing buffer. After equilibration in detection buffer, blots were incubated with chemiluminescent substrate CSPD and exposed to CL-XPosure film (Pierce). Signals corresponding to L5 fiber transcripts were quantitated using ImageJ software (1).
To generate L5 fiber-specific probes, cytoplasmic RNA from late H5pg4100-infected A549 cells was isolated as described above and single-stranded complementary cDNA was prepared using 12- to 18-mer oligo(dT) primers and Superscript III reverse transcriptase (Invitrogen). A ∼600-bp L5 fiber cDNA fragment was then PCR amplified with primers 996-forward (5′-CTCTCGAGAAAGGCGTCTAACCAG-3′) and 1218-reverse (5′-CCATTTTGAGCGCAAGCATGC-3′), cloned into the pCRII-TOPO vector (Invitrogen), and sequenced. Primers 996-forward and 1218-reverse correspond to Ad5 wt nt 9697 to 9720 and 31215 to 31235, respectively. For the synthesis of the DIG-labeled L5 fiber-specific RNA probe the plasmid was linearized with XbaI and antisense labeled RNA was generated in the presence of DIG-labeled UTP (DIG-UTP) as runoff transcripts with SP6 RNA polymerase. Labeling efficiency was checked by detection of oligonucleotides spotted on a positively charged nylon membrane (Roche Molecular Biochemicals) with a DIG detection system (Roche Molecular Biochemicals).
RESULTS
Generation of mutant viruses.
To evaluate further the functional role of the ligase activity associated with E4orf6 during infection, a viral mutant was generated that contained specific point mutations in the E4orf6 sequence encoding BC boxes 1 and 2 that had been shown previously in DNA transfection experiments to almost completely abolish the association of E4orf6 with the Cul5-containing ligase complex (7). Using the wt H5pg4100 sequence as template (30), point mutations in each of the first two BC boxes (L47G/L122S) were introduced, creating virus mutant H5pm4139, also referred to as mutant BC1/BC2. The altered E4orf6 product encoded by this mutant thus corresponds to that of the previously published “mutant 5” from our plasmid-based BC-box mutant series (7). A virus mutant in the same genetic background was also generated, which produced a severely truncated E4orf6 product (E4orf6−) through the deletion of 1 base pair following the codon for proline 66, resulting in the creation of an opal stop codon shortly after the coding sequence shared with the alternatively spliced product E4orf6/7. This virus, named H5pm4154 (also referred to as E4orf6−), should express normal E4orf6/7 protein but an unstable or nonfunctional product of E4orf6 containing only the first 66 residues. These mutant viruses are depicted in Fig. 1.
Characterization of the E4orf6 virus mutants.
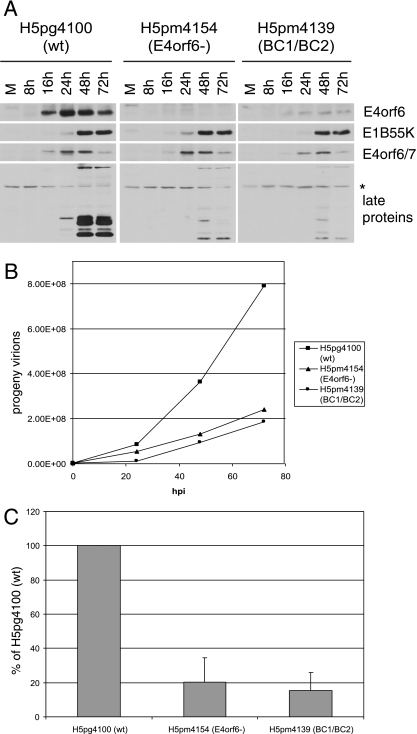
The E4orf6 virus mutants were first tested in a time course assay to compare the expression of both early and late adenovirus gene products with that of wt virus (Fig. 2A). H1299 cells were infected at the low MOI of 5 FFU/cell, and cells were harvested at different time points, as indicated. Equal amounts of protein were then analyzed by SDS-PAGE followed by Western blotting using appropriate antibodies. Figure 2A shows that wt virus (H5pg4100, left panel, as indicated) produced high levels of E4orf6 protein whereas none was seen with the null mutant E4orf6− (H5pm4154, middle panel). Interestingly, reduced levels of E4orf6 protein relative to wt were observed with the BC1/BC2 mutant (H5pm4139, right panel). This was surprising, as reduced stability of the E4orf6 mutant protein was not seen previously in experiments in which the equivalent E4orf6 mutant (“mutant 5”) and wt E4orf6 were expressed following transfection with equal amounts of appropriate plasmid cDNAs (7). Figure 2A shows that expression of other early viral proteins in cells infected with either the E4orf6− or BC1/BC2 mutant was comparable with wt virus infection. While the expression of E1B55K and E4orf6/7 proteins was similar to that of the wt with both viral mutants, such was not the case for the late proteins. E4orf6 (along with E1B55K) is required for the export of the late viral mRNAs (4, 20, 23, 42, 53) and consequently the expression of the late viral proteins. Figure 2A shows that infection by E4orf6− virus indeed resulted in a decrease in production of the late proteins. Interestingly a similar decrease in late protein levels was also observed with the BC1/BC2 (H5pm4139) virus mutant.
FIG. 2.
Characterization of virus mutants—low MOI. (A) H1299 cells were infected with H5pg4100 (wt, left panels), H5pm4154 (E4orf6−, middle panels), or H5pm4139 (BC1/BC2, right panels) virus at the low MOI of 5 FFU/cell, and cells were harvested at several time points, as indicated. Cell extracts were analyzed by SDS-PAGE followed by Western blotting using appropriate antibodies as indicated to the right. (B) H1299 cells were infected with indicated viruses at an MOI of 5 FFU/cell and were collected at indicated times. The collected cells were broken by freeze-thaw cycles, and the progeny virions were titrated by immunofluorescence, as described in Materials and Methods to generate growth curves. A representative experiment is presented. (C) The 48-h time points of six experiments were converted to percentage of yield of wt virus (H5pg4100) and charted with error bars (standard errors of the means).
The effect of the BC1/BC2 mutation on the growth of the virus was verified by growth curve experiments. H1299 cells were infected at an MOI of 5 FFU/cell, and at different time points thereafter the cells were harvested and lysed by successive freeze-thaw cycles and the amount of virus produced was quantified, as described in Materials and Methods. The results of a representative experiment are shown in Fig. 2B. In some experiments the growth of the viruses (in particular the wt virus) decreased at 72 h. For this reason, the 48-h time point was chosen to perform standard deviation analysis, and these analyses are shown in Fig. 2C (n = 6). In all experiments, the growth of E4orf6− (H5pm4154) virus was found to be significantly reduced compared to that of wt virus. Importantly, the growth of the BC1/BC2 virus (H5pm4139) was equally defective, reflecting the results obtained for the expression of late viral proteins (Fig. 2A).
The defect of the BC1/BC2 virus (H5pm4139) is a consequence of the E4orf6 mutations.
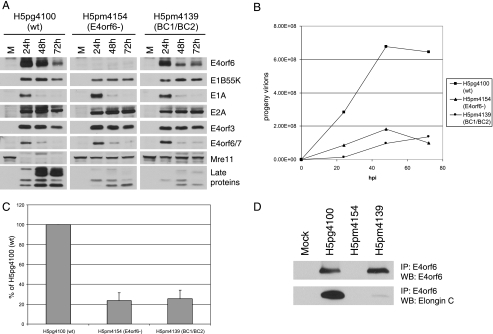
Due to the lower level of expression of E4orf6 protein with the BC1/BC2 mutant virus (H5pm4139), it was not possible to determine if the effect obtained with this mutant was the result of the disruption of the ligase function of E4orf6 or of the low level of mutant E4orf6 protein expression. To evaluate the role of the ligase function during infection, two complementary approaches were taken. The first approach was to infect cells at higher MOIs to increase expression of mutant E4orf6 protein while saturating the expression of wt protein, thus reducing the difference of expression. Experiments similar to those in Fig. 2 were done at an MOI of 50 (Fig. 3A, B, and C). Although the expression of mutant E4orf6 protein in the BC1/BC2 virus did not quite reach the levels of wt E4orf6, especially at 48 h (Fig. 3A), the levels were much more similar than at MOI 5 (Fig. 2A). Figure 3A also shows that the expression levels of the early proteins E1B55K, E1A, E2A, E4orf3, and E4orf6/7 were very similar in all three infections. As expected, the cellular substrate of the E4orf6/E1B55K ligase complex, Mre11, was degraded in the wt-infected cells but not in E4orf6− or in BC1/BC2-infected cells. As in the case of the low-MOI experiments, the expression of the late viral proteins was much reduced in the infections with the mutant viruses at high MOIs. This reduction of late proteins was consistent with the effect of the mutations on the growth of their viruses as shown in Fig. 3B. As was done in Fig. 2C, standard deviation analyses were performed on the 48-h time point, and the results are shown in Fig. 4C (n = 4).
FIG. 3.
Characterization of virus mutants—high MOI. (A) H1299 cells were infected as for Fig. 2A but with a high MOI of 50 FFU/cell. Cell extracts were analyzed by SDS-PAGE followed by Western blotting using appropriate antibodies as indicated to the right. (B) H1299 cells were infected with indicated viruses at an MOI of 50 FFU/cell and were collected at indicated times. The collected cells were broken by freeze-thaw cycles, and the progeny virions were titrated by immunofluorescence, as described in Materials and Methods to generate growth curves. A representative experiment is presented. (C) The 48-h time points of four experiments were converted to percentage of yield of wt virus (H5pg4100) and charted with error bars (standard errors of the means). (D) H1299 cells were infected at an MOI of 50 FFU/cell, and at 24 hpi equal amounts of proteins were immunoprecipitated with antibodies to E4orf6 (1807), separated on SDS gels, and immunoblotted with antibodies against either E4orf6 or elongin C.
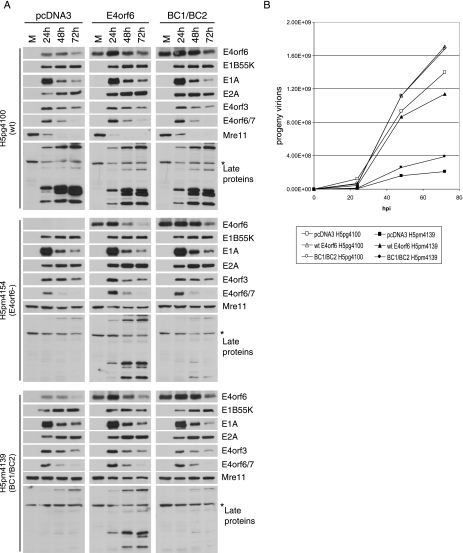
FIG. 4.
Growth of H5pm4139 cannot be rescued by overexpression of the BC-box mutant E4orf6 protein. (A) Time course. Semistable H1299 cell lines expressing either wt E4orf6 or the BC1/BC2 mutant E4orf6 (BC1/BC2) or containing a control vector only were infected with either wt, E4orf6−, or mutant BC1/BC2 viruses at an MOI of 5 FFU/cell. Cells were harvested at several time points, as indicated, and cell extracts were analyzed by SDS-PAGE followed by Western blotting using appropriate antibodies as indicated to the right. (B) Growth curves. Semistable H1299 cell lines expressing either wt E4orf6 or the BC1/BC2 mutant E4orf6 (BC1/BC2) or containing a control vector only were infected with either wt or mutant BC1/BC2 virus at an MOI of 5 FFU/cell. Growth curves were generated as described for Fig. 2B.
In previous studies in which the BC1/BC2 mutant E4orf6 protein was expressed in cells from a cDNA following transfection, it was found to be incapable of forming the E3 ligase as well as binding E1B55K (7). To confirm that this phenotype was also exhibited in the context of virus infections, H1299 cells were infected at an MOI of 50 FFU/cell and at 24 hpi cell lysates were immunoprecipitated with antibodies against E4orf6 (1807) under conditions where the antibody was limiting. As seen in Fig. 3D (top panel), equal amounts of E4orf6 proteins were immunoprecipitated from lysates of cells infected with wt virus and from lysates of cells infected with the BC1/BC2 mutant virus. The ability of wt or mutant E4orf6 to assemble the ligase complex was determined by looking at its ability to coimmunoprecipitate elongin C (7). As seen in Fig. 3D (bottom), the mutant E4orf6 protein produced by the BC1/BC2 virus had a drastically reduced ability to associate with the complex compared to the E4orf6 protein from the wt virus. Thus, the mutant E4orf6 protein produced by the BC1/BC2 virus behaved just like the identical protein generated during transfection experiments (7).
The second approach used to evaluate the role of the ligase activity of E4orf6 during infection was to perform a rescue experiment. Semistable H1299 cell lines were created that express either wt or BC1/BC2 mutant E4orf6 protein from plasmid cDNAs, and a control line that contained the pcDNA3 vector alone was also created. These three cell lines were infected with the wt (H5pg4100, top set in Fig. 4A), E4orf6− (H5pm4154, middle set), or BC1/BC2 (H5pm4139, bottom set) viruses, and time course analyses were performed as described for Fig. 2A and 3A. Figure 4A shows the expression levels of both early and late viral proteins as well as the degradation state of the cellular substrate Mre11.
E4orf6 expression levels of the cell lines can be seen in the mock lanes of E4orf6 and BC1/BC2 cell lines. The mutant E4orf6 protein was expressed at slightly higher levels than was wt E4orf6 protein while no exogenous E4orf6 protein was produced. In all cases the expression patterns of the early genes E1B55K, E1A, E2A, E4orf3, and E4orf6/7 were similar, indicating that the lack of E4orf6 expression or expression of the E4orf6 mutant does not affect the early phase of the virus infection. Unlike the expression pattern of the early products, expression of the late viral proteins was highly dependent on the cell lines infected. While high levels of late proteins were detected in all cell lines infected with wt (H5pm4100) virus, for both mutant viruses, significant amounts of late proteins were detected only following infection in the cell line expressing wt E4orf6 protein (middle column). Infection of cells expressing mutant E4orf6 protein (right column) resulted only in a slight increase of expression of the late proteins. These results indicated the existence of a strong defect in the late phase of the virus infection of both mutant viruses. Results obtained on the degradation of Mre11 were also of interest. The expression of exogenous E4orf6 (wt or mutant) did not affect the ability of the wt virus to cause the degradation of Mre11 (top set); however, considering the degree of rescue of the late protein expression in the wt E4orf6 cell line, it was surprising to observe only a slight increase in the degradation of Mre11 following infection by both mutant viruses (middle column, middle and bottom sets). This slight increase of degradation was specific for wt E4orf6, as it was not observed in the cell line expressing mutant E4orf6 protein (right column, middle and bottom sets).
These three cell lines were also used to study the growth of wt (H5pg4100) and BC1/BC2 (H5pm4139) viruses in a fashion similar to that of Fig. 2B and 3B. Figure 4B shows that while wt virus grew well in all cell lines, such was not the case for the BC1/BC2 mutant virus (H5pm4139). Expression of wt E4orf6 during infection in the semistable E4orf6 cell line greatly rescued growth of the mutant virus (wt E4orf6 H5pm4139); however, expression of similar levels of mutant BC1/BC2 protein (BC1/BC2 H5pm4139) was unable to rescue the growth of this virus. Together these results strongly suggest that the defect observed with the BC1/BC2 mutant virus (H5pm4139) is indeed caused by the mutations in the BC boxes of E4orf6, which prevent the assembly of the ligase complex.
Control of mRNA export is defective in the BC1/BC2 (H5pm4139) virus mutant.
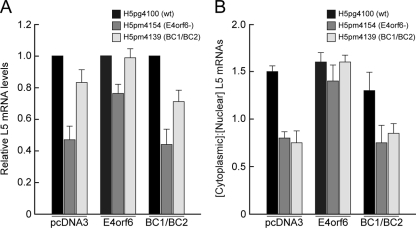
One well-known function of the E4orf6/E1B55K complex is to promote the export of late viral mRNAs. If, as we believe, most functions of E4orf6 during infection are achieved by degradation of specific substrates, then the BC-box virus mutant BC1/BC2 (H5pm4139) should behave identically to E4orf6− (H5pm4154) and exhibit reduced ability to export late viral mRNAs. To address this possibility, we used the stable cell line system to analyze the subcellular localization of the late L5 mRNAs. The L5 fiber mRNA was chosen for this analysis since among all viral late mRNA families cytoplasmic accumulation of this transcript is the most dependent on E1B55K functions (19, 31, 42) and, more importantly, on E1B55K/E4orf6 ubiquitin-protein ligase activity (54). The three cell lines were infected with the three viruses for 48 h, the cells were then fractionated into nuclear and cytoplasmic extracts, and the RNA was extracted. Figure 5A shows the relative amount of the late mRNA L5 as detected and quantified by Northern analysis. As was seen in previously published results (23, 47), infection with the E4orf6− virus in the pcDNA3 cell line resulted in a reduced level of late mRNAs (pcDNA3 line, H5pm4154); however, infection with the BC1/BC2 virus (H5pm4139) exhibited a much lower reduction of late mRNAs even though, with this virus, there was a clear reduction in the level of expression of the mutant E4orf6 protein (pcDNA3 control cell line [Fig. 4A]). This reduction of late mRNA expression was also observed in cells expressing mutant E4orf6, but it was not apparent following infection of cells expressing wt E4orf6, indicating that the wt E4orf6 protein rescued this phenotype.
FIG. 5.
Effect of the BC1/BC2 mutation in E4orf6 on nuclear export efficiency of viral late L5 fiber mRNA. (A) L5 fiber mRNA levels. Semistable H1299 cell lines expressing either wt E4orf6 or the mutant E4orf6 (BC1/BC2) or containing a control vector only were infected at an MOI of 5 FFU/cell with wt and E4orf6 mutant viruses. At 48 h after infection, total RNA was isolated from both the cytoplasmic and nuclear fractions, and L5 fiber transcript levels were determined by Northern blotting and quantified as described in Materials and Methods. L5 fiber signals were corrected using the 18S rRNA internal control (41) and are expressed relative to the wt value, which was set at 1.0. The results shown are averages obtained from three independent experiments. Error bars indicate the standard errors of the means. (B) Cytoplasmic-to-nuclear ratio of L5 fiber transcripts. Steady-state concentrations of RNAs in both the cytoplasm and the nucleus were determined by Northern blotting at 48 h after infection of the cell lines with wt or the E4orf6 mutant viruses. Signals were corrected using the 18S rRNA as described and used to calculate the ratios of cytoplasmic to nuclear L5 fiber mRNAs. The results shown are averages obtained from three independent experiments. Error bars indicate the standard errors of the means.
To determine the ability of the BC1/BC2 mutant E4orf6 protein to facilitate nuclear export of late viral mRNAs, the cytoplasmic-to-nuclear ratio of late mRNAs was measured. As seen in Fig. 5B, in the control pcDNA3 cell line, infection with both mutant viruses resulted in an approximately twofold reduction in the cytoplasmic-to-nuclear ratio for the late viral mRNA L5 compared with wt virus. This defect of mRNA export with both mutant viruses was rescued by the expression of wt E4orf6 in the cell line but not by the expression of the BC1/BC2 E4orf6 mutant. These results clearly suggest that the ligase function of E4orf6 is required for the ability of E4orf6 to function in concert with E1B55K to control the export of late viral mRNAs.
Together the results presented here support the conclusion that most functions attributed to E4orf6 during infection, at least those required for efficient virus growth, including late viral mRNA export, are mediated through the formation of the ubiquitin E3 ligase.
DISCUSSION
In this report we have taken a genetic approach to study the importance of the ubiquitin ligase activity of E4orf6 during productive viral infection. It was already clear that this activity is crucial for E4orf6, with E1B55K, to destabilize p53 and Mre11; however, some of our earlier work had suggested that other events requiring E4orf6 may also be dependent on formation of the ligase complex (7), and indeed recent studies that were published while our work was being completed have now suggested that such may be the case concerning control of mRNA export (54). We decided to address this question directly in the context of the virus infection by generating a virus mutant in which the BC boxes of the E4orf6 protein that interact with elongins B and C had been altered to render the protein incapable of forming the E3 ligase complex. This point mutant approach has the advantage over a deletion approach in that the E4 region remains largely intact, including all the splice donor and acceptor sites. In addition, the mutant protein is expressed from the endogenous viral E4 promoter, which is subjected to normal virus-mediated regulation. BC boxes in cellular proteins contain two highly conserved residues, a leucine and a cysteine (27, 28). We have shown previously that E4orf6 contains two such BC boxes (7), but more recently we have also shown the existence of a third modified BC box (13), similar to the one found in the human immunodeficiency virus type 1 Vif protein (56). Our previous work involving DNA transfection experiments demonstrated that alteration of the first two BC boxes of E4orf6 was sufficient for the near-complete abolition of E3 ligase complex formation (7). In making mutant viruses, we chose to generate a double leucine mutant (equivalent to “mutant 5”) (7) without changing the conserved cysteine residue because this mutant also exhibited near-complete abolition of E3 ligase activity in DNA transfection experiments (7). We also wanted to avoid altering cysteine residues which could be part of a functionally important zinc finger motif (9). In all but one of our experiments and in all of the cell lines and primary cells tested (data not shown), our BC-box mutant virus BC1/BC2 (H5pm4139) behaved exactly like mutant E4orf6− (H5pm4154), which produced only a very short nonfunctional amino-terminal E4orf6 peptide. Thus, these results strongly support our hypothesis that the ligase activity assembled by the E4orf6 protein is required for most of the functions ascribed to the E4orf6/E1B55K complex, including the control of mRNA export.
Deletion of E4orf6 resulted in a twofold decrease in the ratio of cytoplasmic to nuclear concentrations of the late mRNA L5, indicating a block in its export. This effect was clearly as great with the BC1/BC2 mutant, which is defective only in the formation of the E3 ligase. In addition normal L5 mRNA transport was restored following infection of cells expressing wt E4orf6 protein but not with those expressing mutant E4orf6 protein. These results formally confirmed in a virus context similar conclusions presented in recent studies that addressed the question from a very different approach in which the ligase activity of E4orf6 was inhibited by coinfecting cells with a viral vector expressing a dominant-negative mutant of Cul5 (54). Thus, these results also strongly imply that the degradation of a specific substrate(s) by the E4orf6/E1B55K ligase complex is responsible for the control of mRNA export.
The only experiment for which the BC1/BC2 virus behaved differently from the E4orf6− virus was with the relative accumulation of the late viral mRNA L5 (Fig. 5A). Unlike the E4orf6− virus the BC1/BC2 virus did not exhibit a reduced accumulation of the late mRNAs. These results imply that, unlike mRNA transport, the accumulation of late mRNA dependent on E4orf6 is not achieved through the ligase function. They also suggest that the abundant accumulation of late mRNA is not sufficient for efficient virus growth (at least in tissue culture), as the BC1/BC2 mutant virus did not grow better than the E4orf6− virus.
Three substrates for the E4orf6 ligase complex have thus far been identified: p53, Mre11, and DNA ligase IV (5, 7, 11, 12, 24, 43, 44, 46, 50). Although inactivation of these proteins is probably important for the virus, as p53 and Mre11 have been found to be inactivated by more than one pathway (2, 12, 18, 29, 33, 35, 38, 55), they are probably not the only substrates of the complex. In the context of the present studies, it is difficult to imagine how degradation of p53, Mre11, or DNA ligase IV would contribute to the block of cellular mRNA export and enhancement of late viral mRNA export. The effects of E4orf6 and E1B55K are also seen in p53− cells (H1299), and E4orf3 is also present to redistribute the MRN complex, which prevents formation of viral genome concatemers (51). Obviously it will be of great interest to identify the precise substrates involved.
Expression of the mutant E4orf6 protein by the BC1/BC2 virus was reduced relative to that of wt at low MOIs. This effect was partially diminished by infection at higher MOIs. This observation was somewhat surprising, as such lower levels were not evident with the equivalent mutation (“mutant 5”) in DNA transfection experiments in the absence of the other viral products (7). The reasons for this observation are still unclear. It could be that in the presence of other viral proteins, the mutant E4orf6 protein somehow becomes less stable. Perhaps E4orf6 can regulate the cytoplasmic export of its own mRNA. Although intriguing, no observations so far support this possibility, but the subnuclear localization of the E4orf6 mRNA has not been determined in the BC1/BC2 mutant. Another possibility could be that in the context of expression from the E4 promoter, wt E4orf6 is capable, dependent on its ligase activity, of affecting the alternative splicing of the E4 RNA, thus increasing expression of its own mRNA. It is of interest that in experiments done in another cell line (A549 [data not shown]), we observed a decrease of expression of the other E4 gene products in addition to E4orf6 itself with both virus mutants. E4orf6 (as well as E4orf3) is thought to have an effect on the splicing of the major late promoter (40) as well as on some alternatively spliced chimeric β-globin RNA (40). Thus, perhaps it also plays some role in the splicing of other RNAs.
It also was surprising to observe the effect of the E4orf6-expressing cell lines on the degradation of Mre11. Although Mre11 was clearly degraded in all cell lines when they were infected with the wt viruses, infection by the mutant viruses of cells expressing wt E4orf6 protein, which rescued production of late protein as well as growth of the viruses, resulted only in partial degradation of Mre11. The reasons for this effect are also not clear. It could be possible that E4orf6 expressed outside the context of the virus infection (in the absence of other viral proteins) is not properly localized. Overexpressed E4orf6 is also known to change the subnuclear localization of E1B55K, resulting in a reduced association of E1B55K with the nuclear matrix (32). Perhaps under these conditions, E1B55K is less efficient at recruiting Mre11 to the E3 ligase formed by E4orf6.
In summary, the present studies have indicated that in addition to the degradation of the known substrates, functions of E4orf6 critical for efficient virus growth, including the regulation of viral mRNA export from the nucleus, and possibly additional functions attributed to E4orf6 and E1B55K, are dependent on the E4orf6-associated E3 ligase. It will be of great interest to identify the substrates involved in these activities to understand how these functions are elicited.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research and the Canadian Cancer Society through the National Cancer Institute of Canada (P.E.B.) and from the Deutsche Forschungsgemeinschaft (T.D.). Paola Blanchette had a Fonds de la recherche en santé du Québec (FRSQ) fellowship.
Footnotes
Published ahead of print on 9 January 2008.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 1136-42. [Google Scholar]
- 2.Araujo, F. D., T. H. Stracker, C. T. Carson, D. V. Lee, and M. D. Weitzman. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 7911382-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babiss, L. E., and H. S. Ginsberg. 1984. Adenovirus type 5 early region 1b gene product is required for efficient shutoff of host protein synthesis. J. Virol. 50202-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babiss, L. E., H. S. Ginsberg, and J. E. Darnell, Jr. 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 52552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 817034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltz, G. A., and S. J. Flint. 1979. Inhibition of HeLa cell protein synthesis during adenovirus infection. Restriction of cellular messenger RNA sequences to the nucleus. J. Mol. Biol. 131353-373. [DOI] [PubMed] [Google Scholar]
- 7.Blanchette, P., C. Y. Cheng, Q. Yan, G. Ketner, D. A. Ornelles, T. Dobner, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 249619-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boivin, D., M. R. Morrison, R. C. Marcellus, E. Querido, and P. E. Branton. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 731245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer, J. L., and G. Ketner. 2000. Genetic analysis of a potential zinc-binding domain of the adenovirus E4 34k protein. J. Biol. Chem. 27514969-14978. [DOI] [PubMed] [Google Scholar]
- 10.Braithwaite, A., C. Nelson, A. Skulimowski, J. McGovern, D. Pigott, and J. Jenkins. 1990. Transactivation of the p53 oncogene by E1a gene products. Virology 177595-605. [DOI] [PubMed] [Google Scholar]
- 11.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 226610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cathomen, T., and M. D. Weitzman. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 7411407-11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng, C. Y., P. Blanchette, and P. E. Branton. 2007. The adenovirus E4orf6 E3 ubiquitin ligase complex assembles in a novel fashion. Virology 36436-44. [DOI] [PubMed] [Google Scholar]
- 14.Corbin-Lickfett, K. A., and E. Bridge. 2003. Adenovirus E4-34kDa requires active proteasomes to promote late gene expression. Virology 315234-244. [DOI] [PubMed] [Google Scholar]
- 15.Cuesta, R., Q. Xi, and R. J. Schneider. 2000. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 193465-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuesta, R., Q. Xi, and R. J. Schneider. 2004. Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kilodalton protein. J. Virol. 787707-7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cutt, J. R., T. Shenk, and P. Hearing. 1987. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J. Virol. 61543-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 2721470-1473. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez, R., W. Huang, R. Finnen, C. Bragg, and S. J. Flint. 2006. Adenovirus E1B 55-kilodalton protein is required for both regulation of mRNA export and efficient entry into the late phase of infection in normal human fibroblasts. J. Virol. 80964-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez, R. A., and S. J. Flint. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 764507-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52456-467. [DOI] [PubMed] [Google Scholar]
- 22.Groitl, P., and T. Dobner. 2007. Construction of adenovirus type 5 early region 1 and 4 virus mutants, p. 29-39. In W. S. Wold and A. E. Tollefson (ed.), Adenovirus methods and protocols, 2nd ed., vol. 1. Humana Press Inc., Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 23.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harada, J. N., A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 769194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harlow, E., B. R. Franza, Jr., and C. Schley. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 55533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes, B. W., G. C. Telling, M. M. Myat, J. F. Williams, and S. J. Flint. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 642732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamura, T., D. Burian, Q. Yan, S. L. Schmidt, W. S. Lane, E. Querido, P. E. Branton, A. Shilatifard, R. C. Conaway, and J. W. Conaway. 2001. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 27629748-29753. [DOI] [PubMed] [Google Scholar]
- 28.Kamura, T., S. Sato, D. Haque, L. Liu, W. G. Kaelin, Jr., R. C. Conaway, and J. W. Conaway. 1998. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 123872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao, C. C., P. R. Yew, and A. J. Berk. 1990. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology 179806-814. [DOI] [PubMed] [Google Scholar]
- 30.Kindsmüller, K., P. Groitl, B. Härtl, P. Blanchette, J. Hauber, and T. Dobner. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein is regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. USA 1046684-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leppard, K. N., and T. Shenk. 1989. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 82329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lethbridge, K. J., G. E. Scott, and K. N. Leppard. 2003. Nuclear matrix localization and SUMO-1 modification of adenovirus type 5 E1b 55K protein are controlled by E4 Orf6 protein. J. Gen. Virol. 84259-268. [DOI] [PubMed] [Google Scholar]
- 33.Liu, Y., A. Shevchenko, and A. J. Berk. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 7914004-14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowe, S. W., and H. E. Ruley. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7535-545. [DOI] [PubMed] [Google Scholar]
- 35.Martin, M. E., and A. J. Berk. 1998. Adenovirus E1B 55K represses p53 activation in vitro. J. Virol. 723146-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marton, M. J., S. B. Baim, D. A. Ornelles, and T. Shenk. 1990. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J. Virol. 642345-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitsudomi, T., S. M. Steinberg, M. M. Nau, D. Carbone, D. D'Amico, S. Bodner, H. K. Oie, R. I. Linnoila, J. L. Mulshine, J. D. Minna, et al. 1992. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7171-180. [PubMed] [Google Scholar]
- 38.Nevels, M., S. Rubenwolf, T. Spruss, H. Wolf, and T. Dobner. 1997. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc. Natl. Acad. Sci. USA 941206-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nevels, M., B. Tauber, E. Kremmer, T. Spruss, H. Wolf, and T. Dobner. 1999. Transforming potential of the adenovirus type 5 E4orf3 protein. J. Virol. 731591-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nordqvist, K., K. Ohman, and G. Akusjarvi. 1994. Human adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol. Cell. Biol. 14437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Shea, C. C., L. Johnson, B. Bagus, S. Choi, C. Nicholas, A. Shen, L. Boyle, K. Pandey, C. Soria, J. Kunich, Y. Shen, G. Habets, D. Ginzinger, and F. McCormick. 2004. Late viral RNA export, rather than p53 inactivation, determines ONYX-015 tumor selectivity. Cancer Cell 6611-623. [DOI] [PubMed] [Google Scholar]
- 42.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 153104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 713788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128480-484. [DOI] [PubMed] [Google Scholar]
- 46.Roth, J., C. Konig, S. Wienzek, S. Weigel, S. Ristea, and M. Dobbelstein. 1998. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J. Virol. 728510-8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandler, A. B., and G. Ketner. 1989. Adenovirus early region 4 is essential for normal stability of late nuclear RNAs. J. Virol. 63624-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120510-517. [DOI] [PubMed] [Google Scholar]
- 49.Schmid, S. I., and P. Hearing. 1999. Adenovirus DNA packaging. Construction and analysis of viral mutants, p. 47-59. In W. S. Wold (ed.), Adenovirus methods and protocols, vol. 21. Humana Press Inc., Totowa, NJ. [Google Scholar]
- 50.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16349-357. [DOI] [PubMed] [Google Scholar]
- 51.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418348-352. [DOI] [PubMed] [Google Scholar]
- 52.Weinberg, D. H., and G. Ketner. 1983. A cell line that supports the growth of a defective early region 4 deletion mutant of human adenovirus type 2. Proc. Natl. Acad. Sci. USA 805383-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinberg, D. H., and G. Ketner. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J. Virol. 57833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yew, P. R., X. Liu, and A. J. Berk. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8190-202. [DOI] [PubMed] [Google Scholar]
- 56.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 182867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]