Abstract
Coronavirus envelope (E) proteins play an important, not fully understood role(s) in the virus life cycle. All E proteins have conserved cysteine residues located on the carboxy side of the long hydrophobic domain, suggesting functional significance. In this study, we confirmed that mouse hepatitis coronavirus A59 E protein is palmitoylated. To understand the role of the conserved residues and the necessity of palmitoylation, three cysteines at positions 40, 44, and 47 were changed singly and in various combinations to alanine. Double- and triple-mutant E proteins resulted in decreased virus-like particle output when coexpressed with the membrane (M) protein. Mutant E proteins were also studied in the context of a full-length infectious clone. Single-substitution viruses exhibited growth characteristics virtually identical to those of the wild-type virus, while the double-substitution mutations gave rise to viruses with less robust growth phenotypes indicated by smaller plaques and decreased virus yields. In contrast, replacement of all three cysteines resulted in crippled virus with significantly reduced yields. Triple-mutant viruses did not exhibit impairment in entry. Mutant E proteins localized properly in infected cells. A comparison of intracellular and extracellular virus yields suggested that release is only slightly impaired. E protein lacking all three cysteines exhibited an increased rate of degradation compared to that of the wild-type protein, suggesting that palmitoylation is important for the stability of the protein. Altogether, the results indicate that the conserved cysteines and presumably palmitoylation are functionally important for virus production.
Coronaviruses are medically important viruses that cause primarily respiratory and enteric infections in humans and a broad range of animals. The emergence of severe acute respiratory syndrome coronavirus (SARS-CoV) and the recent identification of new human coronaviruses (HCoVs), HCoV-NL63 and HCoV-HKU1 (10, 21), have significantly increased the importance of understanding key interactions during virus assembly, since these interactions may provide insight into potential targets for antiviral and vaccine development.
Coronaviruses are enveloped, positive-stranded RNA viruses that belong to the Coronaviridae family in the Nidovirales order. The virion envelope contains at least three proteins: membrane (M), spike (S), and envelope (E). The genomic RNA is encapsidated by the phosphorylated nucleocapsid. Coronaviruses assemble at and bud into membranes of the endoplasmic reticulum Golgi intermediate compartment (ERGIC) (22, 46).
The focus of this paper is the E protein. Coronavirus E proteins are small (76 to 109 amino acids) integral membrane proteins bearing long hydrophobic domains. The protein plays an important, but not fully understood role(s) in virus production (7, 11, 24, 38). Virus-like particles (VLPs) are formed only when the E and M proteins are expressed (3, 5, 50). When E protein is expressed by itself, E protein-containing vesicles are released from cells (5, 30). Virus production is blocked when the E gene is removed from porcine transmissible gastroenteritis coronavirus (TGEV); however, deletion of the gene from mouse hepatitis coronavirus (MHV) does yield severely crippled virus (7, 24, 38). The E protein is not absolutely required for SARS-CoV production, but deletion of the gene results in virus yields that are 20- to 200-fold lower than those of the wild-type (WT) virus, depending on the cell type that is infected (8). Recently, it was demonstrated that E proteins of several coronaviruses, including MHV, are viroporins that exhibit ion channel activity (26, 29, 52, 53).
Coronavirus E proteins share little homology at the sequence level; however, two to three highly conserved cysteine residues are located adjacent to the carboxy side of the hydrophobic, membrane-associated domain of all E proteins (Fig. 1). The MHV A59 E protein specifically consists of 83 amino acids. The protein actually contains four cysteine residues. One residue is located in the hydrophobic domain at position 23, whereas three of these residues are present at positions 40, 44, and 47 adjacent to the hydrophobic domain. We investigated the functional significance of the conserved residues using reverse genetics. We hypothesized that conservation of the residues must be important for virus production either as targets for palmitoylation and/or to facilitate protein-protein interactions through disulfide bonding. Earlier studies that focused on MHV E palmitoylation produced conflicting results, with one suggesting from thioester cleavage experiments that the protein was modified, whereas another study suggested that the protein is not modified (40, 57). Studies with infectious bronchitis virus (IBV) and SARS-CoV show that the E proteins from virus-infected cells are palmitoylated (6, 27). Importantly, none of the earlier studies addressed the functional significance of the cysteine residues in the context of the virus. We have now analyzed the effect of the replacement of cysteines on VLP assembly and, more importantly, virus growth, release and entry, protein transport, and stability. The data strongly indicate that palmitoylation is important since mutant viruses lacking all three potential target sites are significantly crippled in virus production, but not in entry. E protein stability, but not transport, is affected by this lack, which likely contributes to diminished virus yields.
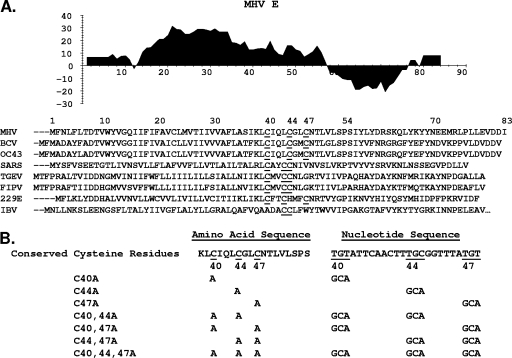
FIG. 1.
Coronavirus envelope protein alignments and alanine substitution mutants. (A) Kyte-Doolittle hydropathy plot of MHV A59 E protein. Hydrophobic and hydrophilic domains are plotted above and below the line, respectively. An alignment of representative E proteins from group II MHV, BCV, HCoV OC43, and SARS-CoV (SARS); group I TGEV, feline infectious peritonitis virus (FIPV), and HCoV 229E; and group III IBV is shown. Conserved cysteine residues are underlined. Amino acid numbers refer to those in MHV E. (B) Amino acid substitutions at positions 40, 44, and 47 were used to create seven mutant MHV E viruses. Nucleotide changes corresponding to respective codon changes are shown. Underlining indicates conserved cysteines and corresponding wild-type codons.
MATERIALS AND METHODS
Cells and viruses.
Mouse L2 cells and 17Cl1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 5% heat-inactivated fetal calf serum supplemented with glutamine and the antibiotics penicillin and streptomycin. Baby hamster kidney (BHK) cells expressing the MHV Bgp1a receptor (BHK-MHVR cells) were kindly provided by Ralph Baric, University of North Carolina at Chapel Hill (56). BHK-21 cells and BHK-MHVR cells were grown in Glasgow minimal essential medium containing 5% fetal calf serum supplemented with 10% tryptose phosphate broth, as described above. BHK-MHVR cells were maintained under selection with Geneticin (G418) for selection of cells expressing the receptor. Stocks of WT MHV A59 and infectious cloned viruses were grown in mouse 17Cl1 and L2 cells. Virus titers were determined for L2 cells.
Construction of alanine substitution mutants.
pScript-E, a pPCR-Script Amp SK(+) vector (Stratagene) containing the MHV A59 E gene, was used for mutagenesis. Site-directed alanine substitutions were made using primers containing the codon changes shown in Fig. 1B. Mutants were constructed by whole-plasmid PCR as described previously (55). The PCR products were incubated at 37°C for 2 h with DpnI to destroy methylated template DNA before transformation into Escherichia coli DH5α. Mutations were confirmed by sequencing the entire insert from pScript-E before subcloning into the MHV G clone at the SbfI and EcoRV restriction sites.
Generation of alanine substitution mutant viruses.
Viruses containing alanine substitutions in the MHV E protein were generated by using an MHV A59 full-length infectious clone (56). Full-length cDNA clones were assembled, transcribed, and electroporated into BHK-MHVR cells as described previously (51). At 40 to 72 h after electroporation, the medium was harvested and an aliquot was used to infect 17Cl1 cells. Total RNA was extracted from cells remaining on the flasks by using an RNAqueous-4PCR extraction kit (Ambion). The extracted RNA was treated with DNase prior to being used as the template for reverse transcription (RT) with an oligo(dT) primer. The RT product was subjected to 30 cycles of PCR amplification using Ambion's SuperTaq Plus with forward primer (5′-CAGAACTGTCCAACAGGCCGTTAGCAAG-3′) and reverse primer (5′-GCAACCCAGAAGACACCTTCAATGC-3′) to obtain E and M cDNA gene products. PCR products were cleaned up using Qiagen MiniElute columns and sequenced directly for E and M genes using the reverse primers (5′-CGGTACCTTTCATATCTATAC-3′ and 5′-AGTCTGCTTTGGCTGATTCCTTC-3′), respectively. Viruses were plaque purified from the electroporated medium on L2 cells and passaged five times in 17Cl1 cells. RNA was extracted from infected cells at passages 1 and 5 for RT-PCR. The E and M gene cDNA products were sequenced each time to determine the stability of the mutations and to identify any potential compensating changes.
Growth analysis of viruses.
Growth kinetic experiments were carried out with 17Cl1 cells infected with passage 5 virus stocks at a multiplicity of infection (MOI) of 0.1 or 0.01 PFU/cell. Cell culture supernatants were collected at various times after infection. Titers were determined by a plaque assay on mouse L2 cells. For all plaque assay experiments, agarose/medium overlays were removed at 72 to 96 h postinfection (p.i.) and the cells were fixed and stained with crystal violet in ethanol.
Determination of MHV E disulfide linkage.
Mouse 17Cl1 cells were either mock treated or infected with WT MHV-A59 at an MOI of 0.001 PFU/cell. Cells were harvested at 24 h p.i. in lysis buffer containing 100 mM Tris, 100 mM NaCl, 0.5% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride. A fraction of the lysate was incubated in Laemmli sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer either with or without 2-mercaptoethanol to generate reducing and nonreducing conditions, respectively. Samples were run on a 15% SDS-PAGE gel, and Western blot analysis was performed probing for MHV E using the rabbit anti-E antibody 9410 (L. A. Lopez and B. G. Hogue, submitted for publication). After incubation with appropriate secondary antibodies, the blot was visualized by chemiluminescence (Pierce).
Metabolic labeling for determining palmitoylation of MHV E.
Mouse 17Cl1 cells in 60-mm-diameter plates were either mock treated or infected with WT MHV-A59 virus at an MOI of 2 PFU/cell. At 6 h p.i., a set of mock and infected cells was labeled with 150 μCi/ml [9,10(n)-3H]palmitic acid (Perkin-Elmer) in DMEM supplemented with 5 mM sodium pyruvate for 6 h. Another set of mock and infected cells was labeled with 100 μCi/ml of EXPRE35S35S protein labeling mix (Perkin-Elmer) for 6 h in DMEM. At 12 h p.i., cytoplasmic lysates were harvested using radioimmunoprecipitation assay lysis buffer (1% TritonX-100, 1% deoxycholate, 0.3% SDS, 150 mM NaCl, 50 mM Tris-HCl, pH 7.6, 20 mM EDTA) containing 1 mM phenylmethylsulfonyl fluoride. Lysates were precleared by incubation with protein A-Sepharose at 4°C for 1 h with rocking. MHV E and S proteins from both 3H- and 35S-labeled lysates were immunoprecipitated by incubation with antibodies against either MHV E 9410 or MHV S J7.5 (14) overnight at 4°C. Immunoprecipitated protein complexes were isolated by incubation with protein A-Sepharose for 2 h at 4°C with constant rocking. Immunoprecipitates were washed six times with radioimmunoprecipitation assay buffer prior to elution in SDS-PAGE sample buffer by heating them at 95°C for 5 min, and we analyzed them by SDS-PAGE on a 5 to 20% gradient gel. Prior to being dried, the gels were incubated for 30 min at room temperature with Amplify fluorographic reagent (GE Healthcare Life Sciences). Proteins were detected by fluorography.
Subcloning of mutant genes into pCAGGS for VLP analysis.
Mutant E genes were subcloned from the MHV G fragment used to make the infectious clone and shuttled into the expression vector pCAGGS under the control of the chicken β-actin promoter (33). PCR was used with the forward primer (5′-GGGCCACGGAATTCGAAAGAAATG-3′) and the reverse primer (5′-CGATCGATTTAGGTTCTCAACAATGCGGTGTCCG-3′) to introduce an EcoRI and ClaI restriction site on either side of the E gene to allow cloning into pCAGGS. All genes were cloned into pCAGGS in the correct orientation and sequenced to confirm that gene sequence integrity was maintained following subcloning.
BHK-21 cells were transfected with pCAGGS DNA by using the TransIT-LT1 reagent (Mirus) as recommended by the manufacturer. Plasmids containing either WT or mutated E genes singly and in combination with the WT M gene were used at 5 μg of total DNA per transfection. At 24 h posttransfection, cells were lysed on ice in a buffer containing 100 mM Tris, 100 mM NaCl, 0.5% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride. The medium was clarified at 14,000 × g for 10 min at 4°C. VLPs were collected by pelleting the clarified medium through a 30% sucrose cushion by ultracentrifugation for 3 h at 4°C in a Beckman SW55Ti rotor at 30,000 rpm. Pellets were resuspended directly in Laemmli SDS-PAGE sample loading buffer. Intracellular and extracellular samples were analyzed by SDS-PAGE. Proteins were transferred to polyvinylidene difluoride membranes, analyzed with mouse anti-MHV M A03 (kindly provided by Kathryn Holmes, University of Colorado Health Sciences) and the rabbit anti-MHV E antibody 9410, and visualized by chemiluminescence, as described above.
Indirect immunofluorescence analysis.
Mouse 17Cl1 cells were infected with mutant viruses and analyzed in parallel with WT MHV to determine the localization of the E and M proteins. Cells were seeded on Lab-Tek chamber slides (Nunc) 1 day prior to infection at an MOI of 0.1 PFU/ml. At 6 h p.i., cells were washed with phosphate-buffered saline (PBS) and fixed with 100% methanol for 15 min at 20°C. Fixed cells were washed with PBS and then blocked with 0.2% gelatin in PBS overnight. Slides were incubated with a mixture of anti-MHV E 9410 polyclonal and anti-MHV M J1.3/2.7 monoclonal (12) primary antibodies for 2 h at room temperature. Cells were washed multiple times with 0.2% gelatin in PBS before incubation with donkey Alexa Fluor 488- and 594-labeled secondary antibodies. Cells were washed extensively with PBS containing 0.2% gelatin and then once with PBS alone. Slides were mounted in ProLong Gold antifade reagent (Invitrogen). Laser scanning confocal microscopy was performed using the Zeiss LSM 510 META microscope (magnification, ×63) and software (Carl Zeiss, Inc., Thornwood, NY).
Analysis of virus release.
Mouse 17Cl1 cells were infected with WT and mutant viruses at an MOI of 0.01 PFU/cell. At 10, 20, or 30 h p.i., the supernatant and cells were harvested separately. The supernatant containing extracellular virus was clarified by centrifugation to remove cell debris. Medium was added to the cell monolayer followed by three freeze-thaw cycles. Virus titers for both fractions were determined by a plaque assay with mouse L2 cells. The calculation of percentage of virus release was determined by dividing the amount of extracellular virus by the total (intracellular plus extracellular) amount of virus.
Evaluation of virus entry.
Mouse 17Cl1 cells were seeded on slides 1 day prior to infection. Cells were infected with WT and triple-mutant viruses at an MOI of 0.001 PFU/cell. At either 6 or 12 h p.i., cells were fixed in 100% methanol for 15 min at −20°C. Fixed cells were then blocked overnight in PBS containing 0.2% gelatin, and indirect immunofluorescence was carried out as described above. The primary antibody used was a rabbit polyclonal anti-MHV N (4). Donkey anti-rabbit Alexa Fluor 488 was used as the secondary antibody. Fluorescing cells were viewed with laser scanning confocal microscopy, as described above, using either a 20× or 63× objective. At each time point, the total number of individual fluorescent cells or fusion foci was counted for the entire field (4 cm2).
Stability of E proteins.
Mouse 17Cl1 cells were infected with WT and triple-mutant stock viruses at an MOI of 0.01. At 23 h p.i., a set of dishes for each virus was lysed, representing time zero prior to drug treatment. Separate infected dishes were treated with 50 μg/ml of cycloheximide (Sigma) to stop translation. Treatments of infected cells proceeded for 1, 3, and 5 h prior to harvesting. Cell lysates were analyzed by SDS-PAGE and Western blotting. Rabbit anti-MHV E (9410) was used to detect E proteins. Mouse anti-β-actin (Abcam) was used to detect actin as a loading control. Bands on X-ray film were quantified using Alpha Innotech software (San Leandro, CA).
Alignment of coronavirus E genes.
The E genes from different coronaviruses were used in an alignment to compare conservation of cysteine residues. MHV (AAF69347), bovine coronavirus (BCV) (AAA42914), OC43-CoV (AAT84364), SARS-CoV (AAP13443), TGEV (NP058426), feline infectious peritonitis virus (AAY32597), 229E-CoV (AAG48595), and IBV (AAY24436) amino acid sequences were used in the alignment.
RESULTS
MHV E is palmitoylated and does not form disulfide-linked oligomers.
When we initiated this study, we considered that the conserved cysteine residues in coronavirus E proteins might function as targets for palmitoylation and/or possibly participate in the formation of disulfide linkages to form protein oligomers. To gain insight into this possibility, we used MHV A59 E protein as a model to study the significance of the conserved residues. Initially, infected cell lysates were analyzed by SDS-PAGE under reducing and nonreducing conditions. Under these conditions, no disulfide-linked oligomers were observed (Fig. 2A). It should be noted that the E protein migrates slower than expected for its predicted molecular mass of approximately 9.7 kDa, likely due to the highly hydrophobic nature of the protein.
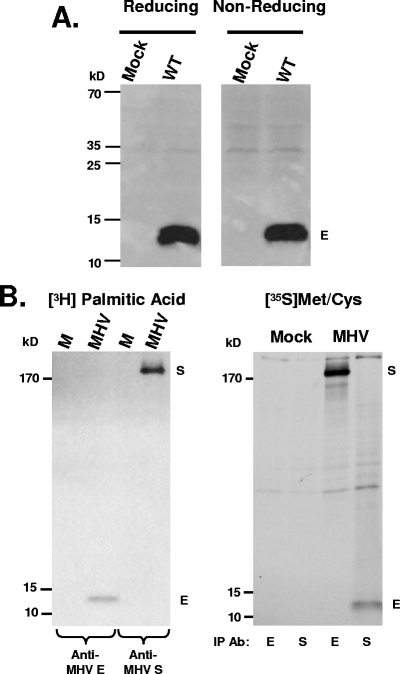
FIG. 2.
MHV E is not disulfide bond linked, but is palmitoylated. (A) Mock and MHV-A59-infected cell lysates were analyzed by SDS-PAGE under either reducing or nonreducing conditions. The position of the E protein between the 10- and 15-kDa markers is indicated. (B) Radiolabeling of MHV-A59-infected cells with [3H]palmitic acid and [35S]methionine-cysteine. Lysates were immunoprecipitated (IP) with antibodies (Ab) against MHV E or S and analyzed on gradient gels. Proteins were detected by fluorography. Positions of the E and S proteins are indicated.
To determine whether the MHV E protein is palmitoylated, infected cells were labeled with [3H]palmitic acid in parallel with [35S]methionine-cysteine. Lysates from mock-treated and infected cells were immunoprecipitated for either the E or S proteins. The S protein served as a labeling control for palmitic acid, since it was recently shown to be palmitoylated (45). Both proteins were labeled with palmitic acid (Fig. 2B). This result confirms that MHV A59 E protein is palmitoylated, thus resolving previously conflicting reports on the protein (40, 57).
Conserved cysteines are important for virus production.
To study the significance and role of the conserved cysteines, site-directed mutagenesis was performed to systematically remove these residues from the protein. Alanine substitution was used to replace cysteines at positions 40, 44, and 47 singly and in various combinations (Fig. 1B). A total of seven mutant E genes with single, double or triple substitutions were shuttled into a full-length MHV A59 infectious clone (56). Full-length RNAs were transcribed for each mutant or WT virus. Following electroporation into BHK-MHVR cells, centers of fusion were observed for all viruses. It was noted that the triple-cysteine mutant virus exhibited significantly fewer centers of fusion compared with those of the other viruses. Medium from electroporated cells was passaged in 17Cl1 mouse cells. RT-PCR and sequence analysis confirmed the integrity of the introduced mutations and that no additional changes were present in either the E or M gene.
All mutant viruses were plaque purified on mouse L2 cells, and multiple isolated plaques were passaged five times in 17Cl1 cells. At passage 5, RT-PCR and sequence analysis of each mutant virus confirmed the genetic stability of the introduced mutations and showed that no additional changes were present in the remainder of the E gene or in the M gene, with one exception. One of the plaque purified viruses with the triple-cysteine substitution (triple virus number 5) had a codon change within E, where serine 55 was changed to phenylalanine. Initial analysis of the virus revealed no growth advantage over the viruses that did not contain the additional change; thus, this virus was not studied further.
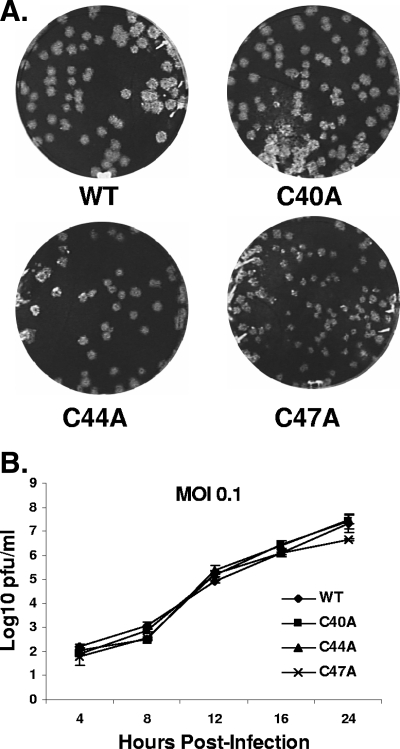
After the fifth passage, viruses were analyzed for their plaque phenotype and growth characteristics. The single-mutant viruses C40A and C44A produced plaques very similar in size and shape to the WT virus, whereas the C47A virus yielded smaller plaques (Fig. 3A). Growth kinetic analysis was performed on mouse 17Cl1 cells for the single mutants at a MOI of 0.1 PFU/cell. All of the single-substitution mutants exhibited growth patterns very similar to that of the WT virus (Fig. 3B). The data indicate that the loss of single cysteines from the MHV A59 E protein does not have a significant effect on virus growth.
FIG. 3.
Plaque morphologies and growth kinetics of single-substitution mutants. (A) Plaque sizes and morphologies of WT, C40A, C44A, and C47A substitution viruses were analyzed by a plaque assay of mouse L2 cells. (B) Mouse 17Cl1 cells were infected with WT and mutant viruses at an MOI of 0.1 PFU/cell. Plaque titrations from the indicated time points were carried out with mouse L2 cells. Error bars indicate deviations from the average results of two independent experiments.
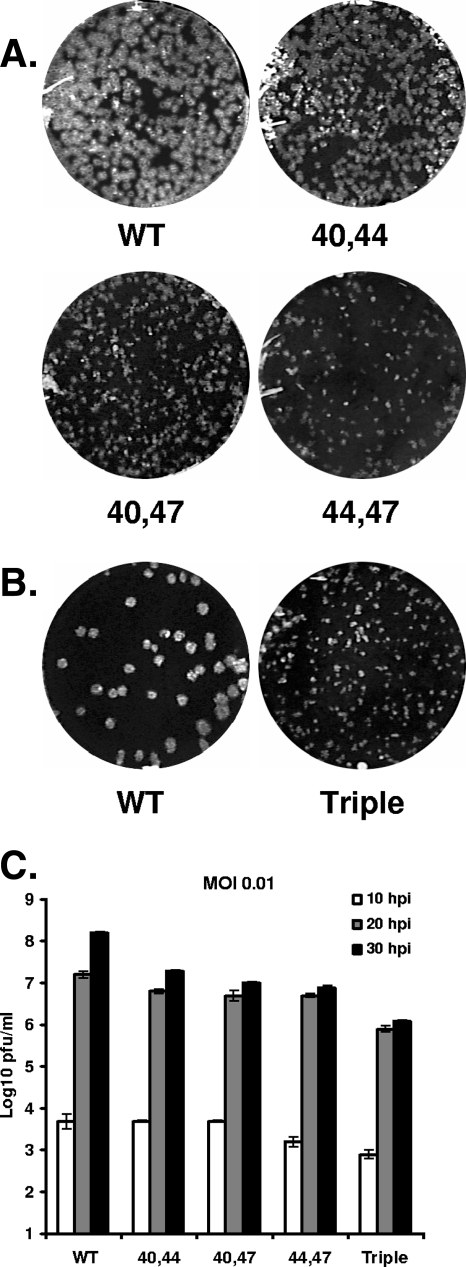
The viruses with replacement of cysteine pairs were also analyzed for their plaque sizes and growth characteristics (Fig. 4). The C40,44A, C40,47A, and C44,47A viruses yielded titers about a log lower than that of the WT virus at 30 h p.i. (Fig. 4C). The viruses also produced distinctly different plaque sizes, with the C40,44A mutant having a slightly smaller plaque phenotype (Fig. 4A). The C40,47A and C44,47A viruses gave rise to even smaller plaques (Fig. 4A). The triple-mutant virus exhibited the most crippled phenotype of the series. The virus produced small plaques even at 4 days p.i. (Fig. 4B), with a titer that was approximately 2.0 logs lower than the WT virus at 30 h p.i. (Fig. 4C).
FIG. 4.
Plaque morphologies and growth characteristics of double- and triple-substitution mutants. (A) Plaque sizes and morphologies of WT, C40,44A, C40,47A, and C44,47A double-substitution viruses were analyzed by a plaque assay on mouse L2 cells. (B) Separately, the plaque sizes and morphologies of WT and the triple-substitution mutant virus were analyzed by plaque assay. (C) Mouse 17Cl1 cells were infected with WT and mutant viruses at an MOI of 0.01 PFU/cell. At 10, 20, and 30 h p.i., media were removed from cells and used to infect mouse L2 cells for plaque assay. Error bars represent the deviations from the average of two separate experiments.
Altogether, the initial characterization of the cysteine mutant viruses pointed to the importance of having at least one cysteine residue present on the carboxy side of the hydrophobic domain of the E protein. The removal of all three conserved cysteines significantly cripples the virus, suggesting that at least one cysteine is necessary for the E protein to efficiently carry out its functional role(s).
Envelope formation is impaired in mutant E proteins.
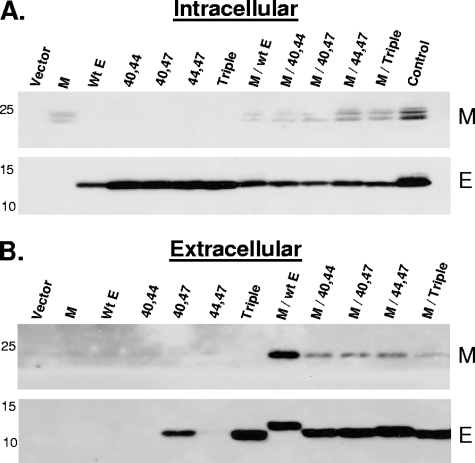
To begin gaining insight into the functional role of the conserved cysteine residues, several parameters were analyzed. First, the ability of mutant E proteins to function in the formation of the viral envelope was assessed. A number of previous studies have clearly demonstrated that coronavirus VLPs form when the E and M proteins are coexpressed, but not when M is expressed alone (3, 5, 50). The double- or triple-mutant E genes and WT M genes were expressed together under the control of the chicken beta actin promoter in the pCAGGS vector (33) in BHK-21 cells. Both intracellular and extracellular fractions were assayed for the release of VLPs. All mutant E proteins promoted release of VLPs into the medium as indicated by the presence of the M protein in the extracellular medium (Fig. 5B). However, the yield of extracellular VLPs was reduced with the mutant E proteins, even though the amounts of intracellular E protein were comparable. The amount of VLPs released into the medium was less with expression of the triple mutant than with expression of the double mutants. The triple mutant appeared to be the least efficient in promoting the formation of VLPs since the level of M in the medium was only slightly above the background level (Fig. 4B, top panel). Extracellular E was also detected, as previously reported (5, 30), even though this result was not observed in all cases when the E proteins were expressed alone (Fig. 4B, lower panel). What accounts for this variability is not understood. Overall, the results clearly demonstrate that all of the mutants can support VLP assembly. However, results with the triple mutant suggest that efficiency of envelope formation and/or release is dependent, at least in part, on the presence of at least one cysteine residue in the E protein.
FIG. 5.
Effect of cysteine substitutions on VLP production. BHK-21 cells were transfected with pCAGGS plasmids containing the WT, double-substitution mutants, and triple-mutant E genes singly and in combination with the WT M gene. Proteins from control cells transfected with empty vector were analyzed in lane 1 of each panel. A lysate from infected mouse 17Cl1 cells was used as a blotting control. Intracellular lysates (A) and pelleted extracellular VLPs (B) were run on a 5 to 20% gradient SDS-PAGE gel, where half of the pelleted VLP fractions and 8% of the total intracellular fractions were analyzed. Western blotting was performed by probing for M and E using antibodies specific for each protein.
Mutant E proteins correctly colocalize with M during infection.
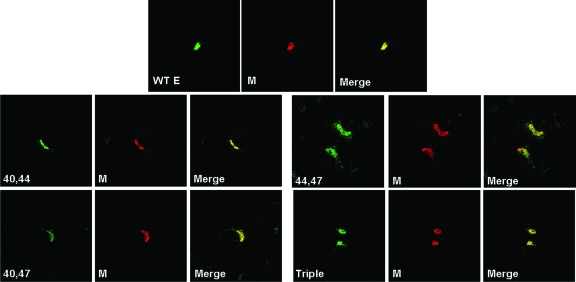
Palmitoylation has been shown to play a role in protein sorting and proper trafficking of viral proteins to either the plasma membrane or the Golgi (25, 34, 41, 59). We therefore tested the ability of mutant E proteins to properly traffic within virus-infected cells. We previously determined that during infection, the E protein colocalizes with M in the ERGIC/Golgi region (Lopez and Hogue, unpublished data), the site of coronavirus assembly (22, 46). To determine whether the mutant E proteins localize correctly, virus-infected cells were analyzed by confocal microscopy. All of the mutant E proteins colocalized with the M protein in the ERGIC/Golgi in virus-infected cells similar to the WT protein (Fig. 6). These results indicate that replacement of the cysteine residues and, more specifically, a total lack of cysteines and palmitoylation, in the case of the triple-mutant virus, do not affect the ability of the E protein to traffic to the site of assembly with the M protein.
FIG. 6.
Colocalization of E and M proteins in infected cells. Mouse 17Cl1 cells were infected with WT, double-, and triple-substitution mutant viruses at an MOI of 0.1 PFU/cell. Cells were fixed in 100% methanol at 6 h p.i. and analyzed by immunofluorescence using mouse and rabbit antibodies against the M and E proteins, respectively. Alexa Fluor 488- and Alexa Fluor 594-conjugated donkey secondary antibodies were used to visualize the localized proteins by confocal microscopy. The colocalization of M and E proteins is shown as yellow in the merged images.
Lack of E protein palmitoylation does not significantly affect virus release.
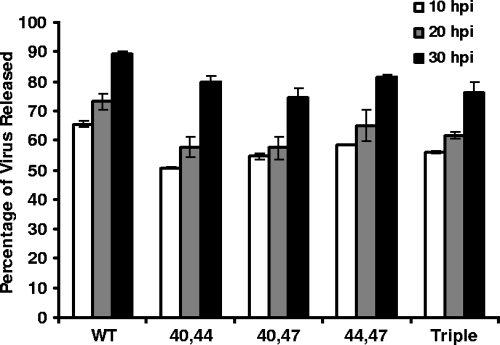
To test whether the lack of palmitoylation on the E protein affects virus release, the amounts of intracellular and extracellular virus were measured for each mutant virus and compared with the WT virus output. Mouse 17Cl1 cells were infected at an MOI of 0.01 PFU/cell, and at 10, 20, and 30 h p.i., media were removed and both the intracellular and extracellular fractions were analyzed by a plaque assay with mouse L2 cells as described previously (55). The amount of virus released into the media ranged from approximately 75% for the C40,47A and triple-mutant viruses to about 90% for the WT virus (Fig. 7). Across the time course, 10 to 20% less virus was released from cells infected with the double- or triple-mutant viruses compared with the WT virus. The data suggest that palmitoylation of the E protein does not dramatically affect release of the virus from cells, even though it appears to contribute to efficient release.
FIG. 7.
Virus release from WT and substitution mutant viruses. Mouse 17Cl1 cells were infected with WT or mutant viruses at an MOI 0.01 PFU/cell. At 10, 20, and 30 h p.i., titers of intracellular and extracellular virus for all mutants were determined by the plaque assay. The percentage of virus released was calculated by dividing the extracellular virus by the total (intracellular plus extracellular) virus. Extracellular titers are from the experiment described in Fig. 4C. Error bars represent the deviations from the averages of two independent measurements.
Palmitoylation is not responsible for virus entry.
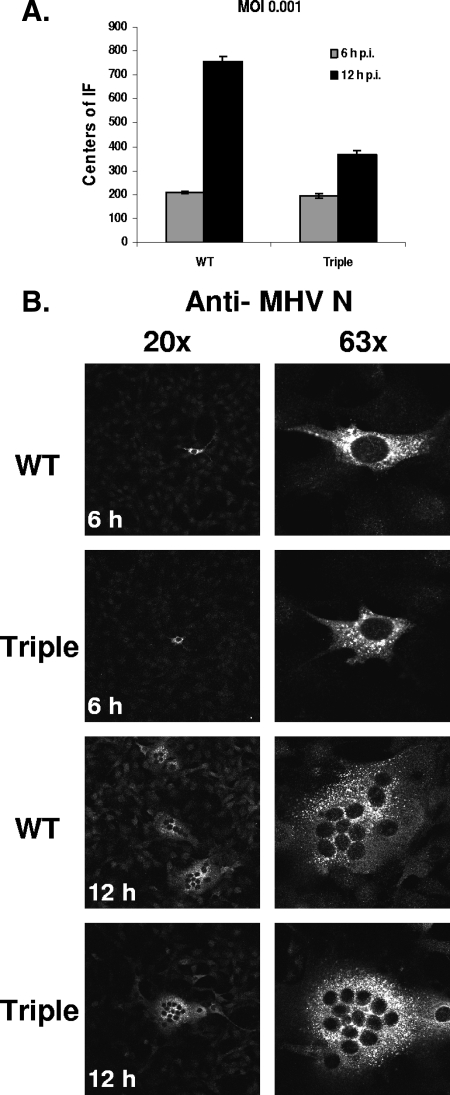
While our data supported the involvement of palmitoylation in the production of coronaviruses, we could not rule out the possibility that entry might be affected by the lack of modification of the E protein. To test the ability of the triple-mutant virus to efficiently enter cells, an immunofluorescence approach was used. Mouse 17Cl1 cells were infected with WT or the triple-mutant virus at an MOI of 0.001 PFU/cell. At 6 and 12 h p.i., cells were fixed and indirect immunofluorescence was used to probe for the nucleocapsid protein (Fig. 8). At 6 h p.i., essentially equal numbers of singly infected cells were observed for both the WT and triple-mutant viruses, suggesting that infectivity and entry of the mutant virus are as efficient as those of the WT virus (Fig. 8A). At 12 h p.i., the number of centers of fusion in the WT virus-infected cells had increased significantly compared to that in the triple-mutant cells (Fig. 8A). There were at least 50% more WT than mutant virus centers of fusion. While this result does not provide insight regarding entry because of virus spread by this later time point, the data strongly supports our other results, demonstrating that the triple-mutant virus is crippled in virus production.
FIG. 8.
Analysis of virus entry. Mouse 17Cl1 cells were infected with WT and the triple-mutant virus at an MOI of 0.001 PFU/cell. At either 6 or 12 h p.i., cells were fixed in 100% methanol and immunofluorescence was carried out using a rabbit polyclonal antibody against the MHV N protein. An Alexa Fluor 488-conjugated donkey secondary antibody was used to visualize the cells by confocal microscopy. (A) For cells fixed at 6 h p.i., the number of individual fluorescing cells (IF) was counted per square (4 cm2 area). For cells fixed at 12 h p.i., the individual fusion foci were counted per square. Each separate multinucleated foci was counted as one. Two squares were counted for each virus, and the average is shown. Error bars represent the deviation of the two measurements. (B) Magnifications of ×20 and ×63 of infected cells are shown on the left and right, respectively. At 6 h p.i., only single cells were infected, but by 12 h, infection had spread to form large fusion foci.
Palmitoylation provides stability for E proteins.
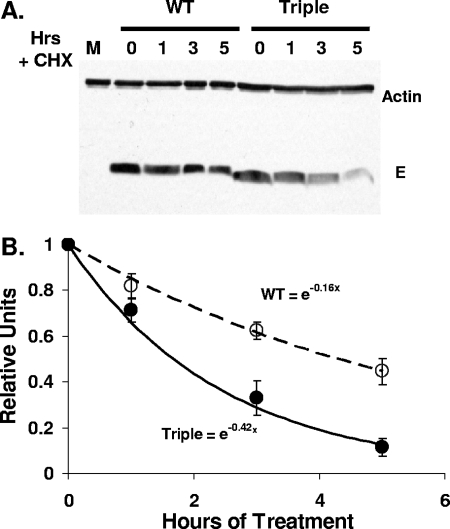
Palmitoylation has been shown to increase stability of proteins (36, 47). To determine whether this is the case for the E protein, stabilities of both the WT and the triple mutant were determined by treating infected cells with the translation inhibitor cycloheximide at 23 h p.i. The amount of E protein in infected cells after 1, 3, and 5 h of treatment with cycloheximide was analyzed by SDS-PAGE and Western blotting and quantification of the protein over the time course (Fig. 9). Interestingly, a continual decrease in the amount of protein was observed for both WT and the triple mutant, as indicated by the rates of degradation (Fig. 9). However, without palmitoylation, the triple-mutant protein was degraded 2.5 times faster than the WT E protein. The results indicate that the addition of palmitic acid is important to prevent more rapid turnover of the protein.
FIG. 9.
Stability of nonpalmitoylated E protein. (A) 17Cl1 mouse cells were infected with WT and triple-mutant viruses at an MOI of 0.01. At 23 h p.i., cells were treated with 50 μg/ml of the translation inhibitor cycloheximide (CHX). At 1, 3, and 5 h of treatment, cells were harvested. Lysates were run on SDS-PAGE gels and probed for both MHV E and actin. Due to differences in virus production between these two viruses, only 5% of total lysate was run for WT, while 16% of lysate was run for the triple mutant. Mock (M)-infected cells were not treated with drug. (B) E protein band intensities were quantified. The means and standard deviations (error bars) of four independent measurements for the WT (open circles) and triple-mutant (filled circles) viruses are shown. The data were fit with exponentials constrained to 1 relative unit at time zero when cycloheximide was added.
DISCUSSION
The role of coronavirus E proteins in virus biogenesis is of significant interest. The protein is a minor component of virions, but it is very clear that the small protein is important for efficient virus production. Deletion of the E gene from the genome of several coronaviruses is either lethal or results in severely crippled virus (7, 8, 24, 38). A recent study with TGEV showed that virus release is reduced when the E gene is not expressed (37). Alteration of charged residues in the carboxy end of MHV E protein results in thermolabile viruses with distorted morphologies (11). A recent report from our lab showed that the MHV E hydrophobic domain and, more specifically, positioning of residues within the predicted transmembrane domain are important for virus production (55). The transmembrane domain of IBV has also been shown to be important for efficient virus release (28). Thus, at least two regions of the protein, the hydrophobic and charged carboxy domains, are important for virus assembly and release. In the study reported here, we have extended insight into the significance of the E protein through our analysis of highly conserved cysteine residues that are located on the carboxy side of the long hydrophobic domain in all coronavirus E proteins.
One important result from our study is the direct demonstration that the MHV E protein is palmitoylated. This result is significant since an earlier study showed that MHV E exhibits faster mobility when analyzed by SDS-PAGE following hydroxylamine treatment which cleaves cysteine-palmitoyl thioester linkages on proteins (57). This finding strongly suggested that the protein is modified by the addition of palmitic acid on cysteine residues. However, another study was unable to confirm the results through either thioester bond cleavage or palmitic acid labeling (40). Our direct confirmation that indeed the protein is palmitoylated is consistent with a recent demonstration that both IBV and SARS-CoV E proteins are modified by palmitic acid addition (6, 27). Thus, palmitoylation is likely common to all coronavirus E proteins. Our results appear to also exclude the possibility that the protein forms oligomers through disulfide linkage, since we were unable to detect any slower-migrating species when electrophoresis was performed under nonreducing conditions.
Our results clearly demonstrate that the cysteine residues on the carboxy side of the MHV hydrophobic domain and, presumably, their palmitoylation are important for virus production. Replacement of each residue individually gave rise to viruses with phenotypes essentially identical to the WT parental virus. The mutant viruses with double substitutions exhibited intermediate phenotypes with slightly smaller plaques and virus titer yields that were at least a log lower than those of the wild-type. The most dramatic effect on the virus was seen when all three cysteine residues were replaced. The triple-substitution virus exhibited a significantly crippled phenotype with decreased growth properties based on smaller plaques and maximum virus yields that were at least 2 logs lower than those of the WT virus. Our results suggest that the loss of more than one cysteine affects the formation of the viral envelope, since VLP production was reduced when all three cysteines were removed from the E protein. The presence of at least one cysteine appeared to increase the amount of extracellular VLPs in the case of the mutants lacking two of the three cysteines. The impairment of envelope formation could account for the decreased virus yields that we observed with both the double- and triple-substitution viruses. Palmitoylation of other viral proteins has been shown to affect virus assembly. The removal of cysteines from alphavirus 6K protein resulted in slower virus release from cells and viruses with aberrant morphology, suggesting that palmitoylation is important for virus assembly and budding (13). Palmitoylation of HIV-1 gp41 is required to target the protein to lipid rafts to allow the assembly and budding of virions (1).
How might palmitoylation of the MHV E protein affect virus production? Palmitoylation can occur at different organelles along the secretory pathway (32, 48), and the modification is known to affect protein trafficking in some cases (16, 41, 42). The addition of palmitic acid is important for correct localization of some viral proteins (1, 34, 36). Our results indicate that this is not the case for MHV E since the triple-cysteine mutant protein localizes correctly with the M protein in the ERGIC/Golgi region in infected cells. Further studies are needed to show whether E is palmitoylated when expressed alone, outside the context of infected cells, and to determine whether trafficking is linked with the modification of the protein.
Our results indicate that MHV entry is not affected when the E protein is not palmitoylated. An equivalent number of infected cells were detected when the triple-mutant and WT viruses were compared at an early time following infection. Thus, decreased infectivity does not account for the observed decreased virus production.
Palmitoylation has been shown to increase protein stability. For Rous sarcoma virus, the removal of palmitoylated cysteines resulted in increased degradation of the envelope protein (36). The yeast protein Tlg1 is protected from ubiquitination by palmitoylation (47). We observed that MHV E protein stability is decreased in the triple-mutant virus-infected cells. The unpalmitoylated E protein decayed with a half-life of ∼1.7 h, whereas the WT protein's half-life was ∼4.3 h in infected 17Cl1 cells. While these results support the conclusion that the addition of palmitic acid to MHV E contributes to its stability, we can only speculate at this time on the significance of this during virus assembly. It is possible that decreased virus yield from the triple-mutant virus is a secondary effect of the increased turnover of the E protein.
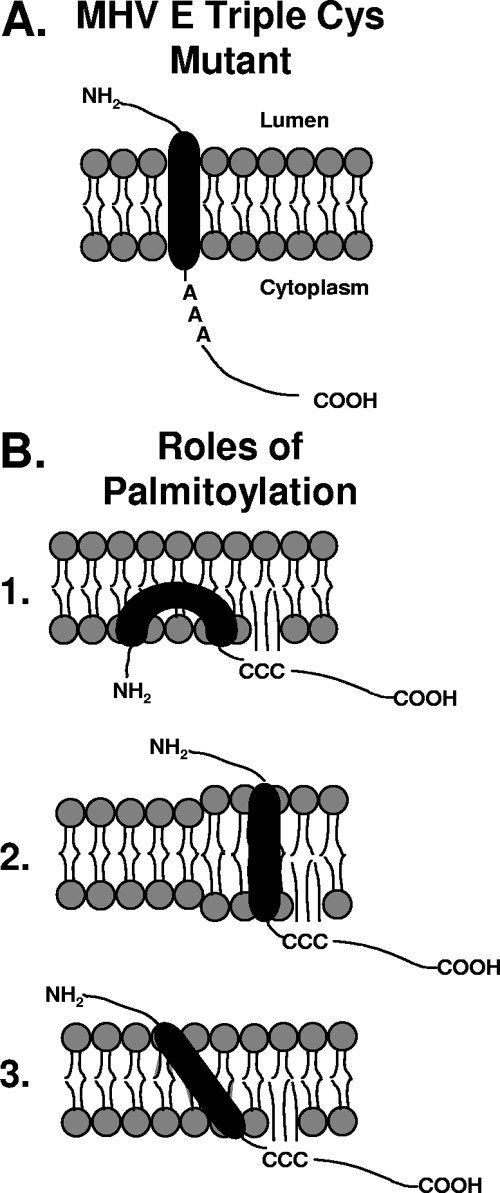
It is possible that palmitoylation impacts how the E protein associates with the membrane. The addition of palmitic acid adjacent to the hydrophobic domain should increase the affinity of the region for the membrane (Fig. 10). This interaction may stabilize or alter association of the protein with the membrane. While such alterations might contribute to routing of the protein to a degradation pathway, it is equally possible that protein-protein interactions, either between E molecules or between E and another protein (viral or cellular) during assembly and/or possibly oligomerization, are disrupted. Such interactions might not be possible without palmitoylation (Fig. 10A).
FIG. 10.
Effect of palmitoylation on MHV E. (A) Model of how the triple-substitution mutant E protein might be orientating within the membrane. Removing conserved cysteine residues near the transmembrane domain may alter the positioning of the E carboxy tail relative to the membrane which may not allow the protein to interact with itself, with another viral protein such as M, or with cellular factors that are required for the function of the protein. (B) Possible ways palmitoylation might be assisting in the positioning of E to allow it to function properly (1). If the E protein forms a hairpin conformation within the membrane, palmitoylation may help anchor the protein in this position (2). The hydrophobic domain of MHV E is slightly larger than what is needed to form a single-span alpha helix. The mismatch in length between the E hydrophobic domain and the lipid bilayer may be relieved by positioning palmitoylated cysteine residues adjacent to the transmembrane domain. Palmitoylation may help influence the protein to move laterally within the membrane to regions where the lipid bilayer is thicker (3). Palmitoylation may act to anchor the E protein firmly in the membrane and to generate a tilt in the α-helix that may be optimal for proper protein function. In each of these examples, palmitoylation would aid in bringing the cytoplasmic tail closer to the membrane, making the tail more readily accessible for additional interactions.
Two membrane topologies have been predicted for coronavirus E protein (for a review, see reference 18). IBV E protein appears to have a single transmembrane domain with a cytoplasmic (inside the virion) tail (5). A “hairpin” conformation has also been predicted for MHV and SARS E proteins (31, 58). Palmitoylation is a dynamic posttranslational modification (2). Thus, the extent to which the E protein is palmitoylated may determine how the protein is positioned in the membrane, allowing both orientations to exist (Fig. 10B, panels 1 and 2). The long hydrophobic domain of the protein makes the “hairpin” orientation a feasible possibility. Lateral movement of the E protein to thicker membrane regions able to support the long domain is another possibility (Fig. 10B, panel 2).
Our lab previously demonstrated the importance of the MHV E transmembrane α-helix. Maintenance of the correct pitch of the helix is important for virus production (55). Palmitoylation could help tilt the helix within the membrane to relieve possible hydrophobic mismatch between E and the lipid bilayer within the ERGIC/Golgi (Fig. 10B3) (20). Tilting of the α-helix is one way a protein can adjust the length of its transmembrane domain to match the hydrophobic thickness of the lipid bilayer (9, 35). Palmitoylation of a cysteine residue(s) on E might also alter the positioning of the α-helix, thus allowing the protein to achieve a conformation required for further interaction either with itself or another protein.
It is possible that palmitoylation helps segregate or cluster E into particular lipid microdomains that are important for virus assembly or vesicle formation for transport of assembled virions out of the cell from the point of assembly. Palmitoylated proteins have an affinity for cholesterol-rich domains (41). Several viruses use palmitoylation to specifically target their envelope proteins to lipid rafts, where they can drive the assembly and budding of virions (1, 25, 59). The role of E in release of virions from cells is not understood. The functional impact of removal of the cysteine residues on the modest decrease (approximately 10 to 20%) that we observed in virus release for our double- and triple-mutant viruses remains to be determined since our results suggest that there is some contribution to efficient release.
Palmitoylation is one of the most common posttranslation modifications that often plays roles in membrane-protein interactions and protein sorting (16, 41, 42). There is no clear consensus sequence for this modification; however, palmitoylated cysteines are often found with or in close proximity to transmembrane domains (17, 39, 44, 49, 54). Placement of the conserved cysteines in coronavirus E proteins is consistent with this finding (Fig. 1). It should be noted that while positioning of cysteines relative to the hydrophobic domain is conserved among all coronavirus E proteins the actual number of residues is not absolutely fixed (Fig. 1A). IBV E has only two cysteines, whereas the other sequences examined bear three residues. A recent study by Kuo et al. reported successful replacement of MHV A59 E with the counterparts from coronavirus groups 2 and 3 (23). This result indicates that a specific number of cysteines are not required. Our data further support the contention that the actual number of cysteine residues may not be critical as long as at least one palmitoylated residue is present. Exactly which residues are palmitoylated at any given time remains to be determined. It is possible that multiple residues represent evolutionary redundancy to assure that at least one is modified. It should also be noted that group 1 HCoV 229E and group 2 MHV, BCV, and HCoV OC43 each have an additional cysteine located within the hydrophobic domain. Our preliminary analysis of a mutant virus suggests that the hydrophobic domain cysteine in MHV E can be replaced with alanine without significant effect on the virus (L. A. Lopez and B. G. Hogue, unpublished data).
Coronavirus E proteins have recently been added to a growing list of viral proteins called viroporins (26, 29, 52, 53). Viroporins are small highly hydrophobic proteins that assemble into oligomers which form hydrophilic pores or ion channels. The proteins play roles in promoting the release of virus particles, but they also exhibit effects on the cellular vesicular system, glycoprotein transport, and membrane permeability (15). The role(s) of viroporin activity in coronavirus-infected cells is not known. We recently showed that disruption of the putative transmembrane domain of MHV E is important for ion channel activity (55). A very well-characterized viroporin, the influenza virus A M2 protein, is also posttranslationally modified by palmitoylation. Mutagenesis of the palmitoylated cysteine on the M2 protein appears to have no significant impact on the ion channel activity of the protein (19). Another recognized viroporin, the nonstructural protein from reovirus called p10, is also palmitoylated on two cysteines flanking the transmembrane domain of the protein (43). These cysteines were shown to be essential for p10-induced membrane fusion. While the mutant proteins were not evaluated for their ion channel function, the authors suggested that palmitoylated cysteines in p10 may influence the proposed viroporin activity of the protein via interactions with membranes. Palmitoylation of the alphavirus 6K viroporin protein is important for virus assembly and release, as summarized above (13). Preliminary analysis of our triple-cysteine mutant virus in the presence of the sodium ion channel inhibitor hexamethylene amiloride suggests that the channel activity is decreased. Since hexamethylene amiloride inhibition is an indirect measure of ion channel activity, it will be important to analyze our mutant E proteins, especially the triple mutant, for ion channel activity in future studies.
The data presented here reveal that the conserved cysteines adjacent to the hydrophobic domain of the MHV E protein play an important role in virus production. Palmitoylation of the protein clearly enhances its stability. Future studies will focus on the impact of decreased protein stability on virus production and on understanding whether palmitoylation plays a role in helping to determine the orientation and positioning of the E protein in membranes, since this is likely a key component in helping to drive the assembly and release of virions.
Acknowledgments
The work was supported by Public Health Service AI53704 from the National Institute of Allergy and Infectious Diseases to B.G.H. L.A.L. was supported in part by the American Society for Microbiology Robert D. Watkins Graduate Research Fellowship. A.J.R. was supported by the National Institute of Diabetes and Digestive and Kidney Diseases STEP-UP Program.
We thank all members of the Hogue lab for helpful discussions throughout the study and especially Sandhya Verma and Valerie Bednar for help with the construction, isolation, and preliminary analysis of the single substitution mutant viruses.
Footnotes
Published ahead of print on 9 January 2008.
REFERENCES
- 1.Bhattacharya, J., P. J. Peters, and P. R. Clapham. 2004. Human immunodeficiency virus type 1 envelope glycoproteins that lack cytoplasmic domain cysteines: impact on association with membrane lipid rafts and incorporation onto budding virus particles. J. Virol. 785500-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bijlmakers, M. J., and M. Marsh. 2003. The on-off story of protein palmitoylation. Trends Cell Biol. 1332-42. [DOI] [PubMed] [Google Scholar]
- 3.Bos, E. C., W. Luytjes, H. V. van der Meulen, H. K. Koerten, and W. J. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 21852-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cologna, R., J. F. Spagnolo, and B. G. Hogue. 2000. Identification of nucleocapsid binding sites within coronavirus-defective genomes. Virology 277235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 744319-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corse, E., and C. E. Machamer. 2002. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 761273-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 761422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeDiego, M. L., E. Alvarez, F. Almazan, M. T. Rejas, E. Lamirande, A. Roberts, W. J. Shieh, S. R. Zaki, K. Subbarao, and L. Enjuanes. 2007. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 811701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Planque, M. R., and J. A. Killian. 2003. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol. Membr. Biol. 20271-284. [DOI] [PubMed] [Google Scholar]
- 10.Donnelly, C. A., A. C. Ghani, G. M. Leung, A. J. Hedley, C. Fraser, S. Riley, L. J. Abu-Raddad, L. M. Ho, T. Q. Thach, P. Chau, K. P. Chan, T. H. Lam, L. Y. Tse, T. Tsang, S. H. Liu, J. H. Kong, E. M. Lau, N. M. Ferguson, and R. M. Anderson. 2003. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet 3611761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 727885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming, J. O., S. A. Stohlman, R. C. Harmon, M. M. Lai, J. A. Frelinger, and L. P. Weiner. 1983. Antigenic relationships of murine coronaviruses: analysis using monoclonal antibodies to JHM (MHV-4) virus. Virology 131296-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaedigk-Nitschko, K., M. X. Ding, M. A. Levy, and M. J. Schlesinger. 1990. Site-directed mutations in the Sindbis virus 6K protein reveal sites for fatty acylation and the underacylated protein affects virus release and virion structure. Virology 175282-291. [DOI] [PubMed] [Google Scholar]
- 14.Gilmore, W., J. O. Fleming, S. A. Stohlman, and L. P. Weiner. 1987. Characterization of the structural proteins of the murine coronavirus strain A59 using monoclonal antibodies. Proc. Soc. Exp. Biol. Med. 185177-186. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, M. E., and L. Carrasco. 2003. Viroporins. FEBS Lett. 55228-34. [DOI] [PubMed] [Google Scholar]
- 16.Greaves, J., and L. H. Chamberlain. 2007. Palmitoylation-dependent protein sorting. J. Cell Biol. 176249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hausmann, J., D. Ortmann, E. Witt, M. Veit, and W. Seidel. 1998. Adenovirus death protein, a transmembrane protein encoded in the E3 region, is palmitoylated at the cytoplasmic tail. Virology 244343-351. [DOI] [PubMed] [Google Scholar]
- 18.Hogue, B. G., and C. E. Machamer. 2007. Coronavirus structural proteins and virus assembly, p. 179-200. In S. Perlman, T. Gallagher, and E. J. Snijder (ed.), Nidoviruses. ASM Press, Washington, DC.
- 19.Holsinger, L. J., M. A. Shaughnessy, A. Micko, L. H. Pinto, and R. A. Lamb. 1995. Analysis of the posttranslational modifications of the influenza virus M2 protein. J. Virol. 691219-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joseph, M., and R. Nagaraj. 1995. Interaction of peptides corresponding to fatty acylation sites in proteins with model membranes. J. Biol. Chem. 27016749-16755. [DOI] [PubMed] [Google Scholar]
- 21.Kahn, J. S. 2006. The widening scope of coronaviruses. Curr. Opin. Pediatr. 1842-47. [DOI] [PubMed] [Google Scholar]
- 22.Krijnse-Locker, J., M. Ericsson, P. J. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 12455-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo, L., K. R. Hurst, and P. S. Masters. 2007. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J. Virol. 812249-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 774597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, M., C. Yang, S. Tong, A. Weidmann, and R. W. Compans. 2002. Palmitoylation of the murine leukemia virus envelope protein is critical for lipid raft association and surface expression. J. Virol. 7611845-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao, Y., J. Lescar, J. P. Tam, and D. X. Liu. 2004. Expression of SARS-coronavirus envelope protein in Escherichia coli cells alters membrane permeability. Biochem. Biophys. Res. Commun. 325374-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao, Y., Q. Yuan, J. Torres, J. P. Tam, and D. X. Liu. 2006. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavrius envelope protein. Virology 349264-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machamer, C. E., and S. Youn. 2006. The transmembrane domain of the infectious bronchitis virus E protein is required for efficient virus release. Adv. Exp. Med. Biol. 581193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madan, V., M. J. Garcia, M. A. Sanz, and L. Carrasco. 2005. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 5793607-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda, J., A. Maeda, and S. Makino. 1999. Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology 263265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda, J., J. F. Repass, A. Maeda, and S. Makino. 2001. Membrane topology of coronavirus E protein. Virology 281163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLaughlin, R. E., and J. B. Denny. 1999. Palmitoylation of GAP-43 by the ER-Golgi intermediate compartment and Golgi apparatus. Biochim. Biophys. Acta 145182-92. [DOI] [PubMed] [Google Scholar]
- 33.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108193-199. [DOI] [PubMed] [Google Scholar]
- 34.Nozawa, N., T. Daikoku, T. Koshizuka, Y. Yamauchi, T. Yoshikawa, and Y. Nishiyama. 2003. Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J. Virol. 773204-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyholm, T. K., S. Ozdirekcan, and J. A. Killian. 2007. How protein transmembrane segments sense the lipid environment. Biochemistry 461457-1465. [DOI] [PubMed] [Google Scholar]
- 36.Ochsenbauer-Jambor, C., D. C. Miller, C. R. Roberts, S. S. Rhee, and E. Hunter. 2001. Palmitoylation of the Rous sarcoma virus transmembrane glycoprotein is required for protein stability and virus infectivity. J. Virol. 7511544-11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortego, J., J. E. Ceriani, C. Patino, J. Plana, and L. Enjuanes. 2007. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology 368296-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 7611518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ponimaskin, E., and M. F. Schmidt. 1998. Domain-structure of cytoplasmic border region is main determinant for palmitoylation of influenza virus hemagglutinin (H7). Virology 249325-335. [DOI] [PubMed] [Google Scholar]
- 40.Raamsman, M. J., J. K. Locker, A. de Hooge, A. A. De Vries, G. Griffiths, H. Vennema, and P. J. Rottier. 2000. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 742333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Resh, M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 14511-16. [DOI] [PubMed] [Google Scholar]
- 42.Resh, M. D. 2006. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2584-590. [DOI] [PubMed] [Google Scholar]
- 43.Shmulevitz, M., J. Salsman, and R. Duncan. 2003. Palmitoylation, membrane-proximal basic residues, and transmembrane glycine residues in the reovirus p10 protein are essential for syncytium formation. J. Virol. 779769-9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smotrys, J. E., and M. E. Linder. 2004. Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73559-587. [DOI] [PubMed] [Google Scholar]
- 45.Thorp, E. B., J. A. Boscarino, H. L. Logan, J. T. Goletz, and T. M. Gallagher. 2006. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J. Virol. 801280-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tooze, J., S. Tooze, and G. Warren. 1984. Replication of coronavirus MHV-A59 in sac− cells: determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 33281-293. [PubMed] [Google Scholar]
- 47.Valdez-Taubas, J., and H. Pelham. 2005. Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J. 242524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veit, M., and M. F. Schmidt. 1993. Timing of palmitoylation of influenza virus hemagglutinin. FEBS Lett. 336243-247. [DOI] [PubMed] [Google Scholar]
- 49.Veit, M., M. F. Schmidt, and R. Rott. 1989. Different palmitoylation of paramyxovirus glycoproteins. Virology 168173-176. [DOI] [PubMed] [Google Scholar]
- 50.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 152020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verma, S., V. Bednar, A. Blount, and B. G. Hogue. 2006. Identification of functionally important negatively charged residues in the carboxy end of mouse hepatitis coronavirus A59 nucleocapsid protein. J. Virol. 804344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, L., P. Gage, and G. Ewart. 2006. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 353294-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson, L., C. McKinlay, P. Gage, and G. Ewart. 2004. SARS coronavirus E protein forms cation-selective ion channels. Virology 330322-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, C., and R. W. Compans. 1996. Palmitoylation of the murine leukemia virus envelope glycoprotein transmembrane subunits. Virology 22187-97. [DOI] [PubMed] [Google Scholar]
- 55.Ye, Y., and B. G. Hogue. 2007. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J. Virol. 813597-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 7611065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, X., W. Bi, S. R. Weiss, and J. L. Leibowitz. 1994. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology 2021018-1023. [DOI] [PubMed] [Google Scholar]
- 58.Yuan, Q., Y. Liao, J. Torres, J. P. Tam, and D. X. Liu. 2006. Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells. FEBS Lett. 5803192-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 744634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]