Abstract
The molecular pattern of the human immunodeficiency virus (HIV) epidemic in Argentina provides an appropriate scenario to study cellular immune responses in patients with non-clade B infection. We aimed to map T-cell responses in patients infected with BF recombinant variants and compare them with those of clade B patients. Sixteen recently infected patients were enrolled and grouped by viral subtype. Nef-specific responses were evaluated with a peptide matrix-based gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assay using B and BF overlapping peptides. Cross-clade and clade-specific responses were found. A correlation between B versus BF Nef-specific responses was identified. Detailed analysis at the single-peptide level revealed that BF patients show a narrower response but greater magnitude. Nef immunodominant responses agreed with previous publications, although the B loop was targeted at an unexpectedly high frequency. The putative HLA allele(s) restricting each positive response was determined. Single-peptide level screening with two different peptide sets uncovered discordant responses (mostly caused by peptide offsetting) and allowed detection of increased breadth. Positive responses identified by ELISPOT assay were further studied by intracellular cytokine staining. These were almost exclusively mediated by CD8 T cells. Characterization of concordant responses revealed that cells show distinct functional profiles, depending on the peptide presented. Last, quality (in terms of polyfunctionality) of T cells was associated with better viral replication containment. Overall, interclade differences in the frequency of epitopes recognized, structural domains targeted, and magnitude of responses were identified. Screening T-cell responses with multiple sets increased sensitivity. Further support for the notion of polyfunctional CD8+ T-cell requirement to better control viral replication is also provided.
Human immunodeficiency virus type 1 (HIV-1)-specific cytotoxic T lymphocytes (CTLs) arising during the acute phase of infection are associated with containment of viral replication and resolution of acute-phase symptoms (5, 13, 14, 44). Detailed characterization of these cell populations and viral immunodominant epitopes will provide key data for developing and enhancing immunization strategies. In this sense, one major challenge is HIV variability, characterized by substantial inter- and intraclade sequence diversity. To date, most reports aimed at studying CTL responses are focused on HIV clade B- and C-infected patients. In Argentina, epidemiological studies revealed that early predominance of B subtype has been overshadowed by the emergence of BF recombinants (16, 23, 32, 33, 53). These reports indicate that near 50% of the HIV-infected people living in Argentina are infected either with the circulating recombinant form CRF12_BF or other BF recombinant forms related to CRF12_BF. Moreover, in other South American countries, such as Brazil and Uruguay, high prevalence of F subtype and BF recombinant variants are also found (16, 32, 34). These findings provide an adequate scenario to fill the gap concerning fine mapping of CTL responses in non-clade B- and C-infected patients.
Cross-clade CTL responses were studied by different groups using different approaches. The first publications used vaccinia virus-based constructs or peptide pools as stimulating factors (2, 19, 20, 27). Not until recently was fine mapping of these responses achieved by using individual peptides based on different subtypes (22, 28). The latter allows for a more detailed and comprehensive understanding of cross-clade reactive populations.
Here, cross-clade and clade-specific T-cell responses in B- and BF-infected patients from Argentina are reported. These assays were performed shortly after seroconversion in order to avoid the confounders associated with viral diversification, selection of escape mutants, and superinfection. Overlapping peptides spanning the Nef protein were chosen as antigens because of the following: (i) Nef is halfway along the spectrum of variability found in HIV proteins; (ii) during primary HIV infection, Nef-specific CTLs represent between 46% and 94% of the total magnitude of the HIV T-cell response (43); (iii) Nef has, together with Gag, the highest epitope density (1, 39). This study also contributes to minimizing the problem of breadth underestimation during CTL screening and provides insights into discrepancies of CD8 T-cell effector function.
(The research performed by Gabriela Turk was in partial fulfillment of her Ph.D. degree from the University of Buenos Aires, Buenos Aires, Argentina, 2008.)
MATERIALS AND METHODS
Samples.
Sixteen patients with acute/early primary HIV infection (within 6 months from seroconversion), two HIV-1 nonprogressors (HIV infection for at least 10 years, CD4 cell count of >500 cells/μl, asymptomatic, and low or undetectable viral load [VL] in the absence of any antiretroviral therapy [54]), and five seronegative donors were enrolled. The study was approved by the Ethics Committee of the School of Medicine, University of Buenos Aires, and all subjects provided informed consent before enrollment. All patients remained off therapy while included in this study.
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by Ficoll-Hypaque density gradient centrifugation (Amersham, Sweden). Plasma VL (Versant HIV-1 RNA 3.0 assay; Bayer) and CD4+ and CD8+ T-cell counts (flow cytometry double platform [Epics XL; Coulter]) were determined.
Viral subtyping.
Genomic DNA was used as a template to amplify the vpu and nef HIV-1 genes (nucleotides [nt] 5967 to 6570 and nt 8378 to 9414 of HXB2 clone, respectively) by nested PCR (23, 33). Cellular DNA was extracted from 2 × 106 PBMCs using QIAamp DNA kit (Qiagen GmbH, Germany). Amplicons were directly sequenced using the Big Dye Terminator sequencing kit (Amersham, Sweden) on an automatic sequencer (Applied Biosystems DNA sequencer 3100). Nucleotide sequences were analyzed and manually adjusted using Sequencher 4.0.5 software (Gene Codes Co.). Multiple alignments of the newly generated vpu and nef sequences together with selected reference sequences were performed using ClustalX and visually corrected with the BioEdit version 5.0.9 (http://www.mbio.ncsu.edu/bioedit/bioedit.html). Subtype ascribing of vpu and nef sequences was performed by constructing phylogenetic trees by neighbor joining using the Kimura two-parameter model with the MEGA v.3.0 program (http://megasoftware.net). Bootstrap analysis was done to assess the stability of the nodes. Recombination analysis was performed by bootscanning using the SimPlot v.2.5 program (http://www.welch.jhu.edu), and visual inspection of alignments was used in order to identify breakpoints in recombinant sequences. Reference sequences included in the analysis were those recommended by the Los Alamos HIV sequence database (http://www.hiv.lanl.gov/content/hiv-db).
HLA typing.
Genomic DNA was prepared as described above. HLA-A and HLA-B loci were typed, at the two-digit level, by using PCR-single-stranded oligonucleotide probes as previously described (17, 25).
Peptides and matrix design.
Overlapping synthetic peptides (13- to 15-mers, overlapping by 11 amino acids [aa]) were designed based on the Nef protein from CRF12_BF reference strain (GenBank accession number AF385936) using the PeptGen software (http://www.hiv.lanl.gov/content/hiv-db) and custom ordered from JPT Peptide Technologies (Germany). Overlapping synthetic peptides spanning the clade B Gag, Vif, Tat, and Nef proteins and the CEF (cytomegalovirus, Epstein-Barr virus, and influenza virus) peptide pool (21) were obtained from the NIH AIDS Reagent Program. Lyophilized peptides were dissolved in dimethyl sulfoxide (DMSO) at 20 μg/μl and stored at −20°C.
Nef B and BF peptides (referred to in the text as NefB39 to NefB87 and NefBF1 to NefBF36, respectively) were arrayed in two different peptide matrix systems as described elsewhere (1). Within a given protein matrix, each peptide was represented in two different peptide pools, allowing for the identification of the respective peptide by responses in the two corresponding pools. The final concentration of each peptide within a peptide pool was 1 μg/μl.
Also, pools of peptides spanning the whole NefB, Vif, p17, p24, p2p7p1p6, and Tat proteins were constructed. The final concentration of each peptide within each pool was 200 ng/μl.
ELISPOT assays.
Gamma interferon (IFN-γ)-secreting cells were detected using enzyme-linked immunospot (ELISPOT) assays conducted as described previously (22) with the following modifications: PBMCs were cryopreserved, and 1 day before the assay, they were thawed and rested overnight in RPMI medium supplemented with 10% fetal bovine serum (Gibco BRL), 2 mM l-glutamine (Gibco BRL), 100 U/ml penicillin (Gibco BRL), 100 mg/ml streptomycin (Gibco BRL), and 10 mM HEPES (Gibco BRL) (>95% viability checked by trypan blue exclusion after thawed and after overnight rest). Rested PBMCs were plated on sterile 96-well plates (MultiScreen IP plates; Millipore), previously coated with mouse anti-human IFN-γ monoclonal antibody (BD Biosciences) at 105 cells/well. Individual peptides or peptide pools were added (final concentration, 2 μg/ml of each peptide). Negative (peptide-free medium plus 0.05% DMSO) and positive (CEF; 2 μg/ml of each peptide) controls were included for each patient. Plates were developed using biotinylated anti-human IFN-γ monoclonal antibody, streptavidin-peroxidase complex, and AEC (3-amino-9-ethylcarbazole) substrate reagent set (BD Biosciences). Plates were scanned on an ImmunoSpot reader (Cellular Technology Ltd.). Specific spots were counted using the ImmunoSpot software. Results were expressed as spot-forming units (SFU)/106 PBMCs after subtraction of the negative-control values. Thresholds for positive responses for the test wells were defined as at least 50 SFU/106 PBMCs or as mean SFU greater than three times the mean SFU of the negative-control wells, whichever was higher (19, 36, 45, 46). Confirmation of individual peptide responses was performed in triplicate.
The breadth of the responses was assessed by considering every recognized peptide as a separate response. When two adjacent peptides were recognized, the response was counted as one. The total magnitude of responses was determined as the sum of the responses to all individual peptides.
HLA binding peptide predictions.
Web-based immunology tools were used to scan identified positive peptides for putative epitopes, according to each patient's HLA haplotype. BIMAS HLA Binding Prediction software (BioInformatics & Molecular Analysis Section [BIMAS], Center for Information Technology [CIT], National Institutes of Health [NIH] website http://www-bimas.cit.nih.gov/molbio/hla_bind/) ranks potential 8-mer, 9-mer, or 10-mer peptides based on a predicted half-time of dissociation to HLA class I molecules. The analysis is based on coefficient tables deduced from the published literature (42, 50). Also, data available at the Los Alamos Immunology HIV Database (http://www.hiv.lanl.gov/content/immunology/) were used as well as the MotifScan tool, available therein, which scans for HLA anchor residue motifs within protein sequences for the specified HLA haplotype.
Intracellular cytokine staining (ICS).
Thawed and overnight rested PBMCs were dispensed in 96-well U-bottom plates (5 × 105 cells/well). Cell viability was checked before and after overnight rest by trypan blue exclusion. Only samples with >95% were used for the assays. Costimulatory antibodies (anti-CD28 and anti-CD49d; 1 μg/ml; BD Biosciences), monensin (Golgistop, 0.7 μl/ml; BD Biosciences), brefeldin A (10 μg/ml; Sigma-Aldrich),, and the corresponding peptide (5 μg/ml) were added. An unstimulated (peptide-free medium plus 0.05% DMSO and costimulatory antibodies) and two positive controls (5 μg/ml CEF peptide pool and 10 ng/ml phorbol myristate acetate [PMA] plus 250 ng/ml ionomycin [Sigma-Aldrich]) were included in each assay. A mixture of anti-CD107a labeled with fluorescein isothiocyanate (anti-CD107a-FITC) and anti-CD107b-FITC antibodies (BD Biosciences) was added to one of the replicates. Cells were incubated for 6 hours at 37°C, washed, stained with surface antibodies (CD3-PECy7, CD4-PerCP, and CD8-APCCy7; BD Biosciences) for 30 min at 4°C, and then permeabilized and fixed using the Cytofix/Cytoperm kit (BD Biosciences). After the permeabilization/fixation step, one of the replicates was stained using anti-interleukin 2 antibody labeled with allophycocyanin (IL-2-APC), anti-tumor necrosis factor alpha (TNF-α) antibody labeled with FITC (TNF-α-FITC), and anti-IFN-γ labeled with phycoerythrin (IFN-γ-PE) (BD Biosciences) while the replicates already containing the anti-CD107 mix were stained with anti-IL-2-APC and anti-macrophage inflammatory protein 1β (Mip-1β) antibody labeled with PE (Mip-1β-PE). Cells were then washed and stored until acquired in a BD FACSCanto flow cytometer. Data acquisition and analysis was performed using the BD FACSDiva software. Instrument settings and fluorescence compensation were performed for each day of testing using unstained and single-stained samples. Stimulated cells in which surface molecules were stained and isotype controls corresponding to intracellular markers included were analyzed for each patient in order to accurately set negative populations.
Initial gating was performed on small lymphocytes in a forward scatter-versus-side scatter (SSC) plot. At least 75,000 events were acquired in the lymphocyte gate. CD3+ events were gated in an SSC versus CD3 plot prior to gating on CD3+ CD8+ and CD3+ CD4+ events. Following identification of these cells, a gate was made for each studied function and their combinations. For this purpose, “derived gate tools” available at the FACSDiva software was used. To study double- and triple-positive populations, intersections of two and three gates were created, respectively. Once the percentages of events were determined for each derived gate, the value of triple-positive events was subtracted from those of double-positive events and, in turn, double- and triple-positive events were subtracted from the total events positive for a given function. Samples with nonspecific backgrounds higher than 0.5% for any function (degranulation and cytokine/chemoquine production) were retested using a new vial of frozen cells. Data presented correspond to background-subtracted results using the CD28/49d stimulation. This was performed on a cytokine subset by cytokine subset basis, i.e., subtracting the result from the “CD28/49d-only” condition for a given cytokine subset to the same subset of a peptide-stimulated condition.
Two standard deviations (SDs) above background was set as the threshold for determining positive responses (as it was applied by other authors in equivalent determinations [12]). Values below this threshold were set at 0. Samples from five HIV seronegative donors were used to validate the assay as well as the criteria for positive responses.
Data analysis.
Statistical analyses were performed using GraphPad Prism 4 (GraphPad Software). All data except log10 VL and breadth of response were analyzed using nonparametric statistics. Wilcoxon and Mann-Whitney tests were used to compare intra- and interclade responses, respectively. Correlations were determined using Spearman's rank test. All tests were considered significant if the P value obtained was less than 0.05.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper were deposited in the GenBank database and are available through accession numbers EU312168 to EU312203.
RESULTS
Study subjects.
Sixteen recently infected HIV-1 subjects were enrolled within 6 months from seroconversion and grouped by viral subtype. Nine out of 16 patients were infected with clade B (56%; from now on referred as patients B1 to B9), while 7 were infected with BF forms (44%; from now on referred as patients BF1 to BF7) (Table 1). These frequencies agree with local epidemiological data (15, 23).
TABLE 1.
Characteristics of subjects studied
| Patient and/or group parametera | Gender | Viral genotype
|
VLb
|
No. of cells/μlc
|
HLA haplotype
|
Estimated time from infection (no. of days)d | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| vpu | nef | No. of RNA copies/ml | Log10 VL | CD4+ | CD8+ | HLA-A allele(s) | HLA-B allele(s) | |||
| B patients | ||||||||||
| B1 | Female | B | B | >500,000 | >5.7 | 416 | 1,900 | 24, 26 | 35, 3543 | 120 |
| B2 | Male | B | B | >500,000 | >5.7 | 981 | 5,886 | 2, 2 | 44, 35 | 156 |
| B3 | Male | B | B | 23,763 | 4.38 | 451 | 758 | 3, 26 | 13, 38 | 95 |
| B4 | Male | B | B | 9,701 | 3.99 | 373 | 713 | 2, 32 | 57, 44 | 45 |
| B5 | Male | B | B | 41,134 | 4.61 | 364 | 569 | 11, 24 | 44, 35 | 68 |
| B6 | Male | B | B | 151,662 | 5.18 | 439 | 1,602 | 2, 3 | 44, 45 | 145 |
| B7 | Male | B | B | 19,289 | 4.29 | 497 | 1,766 | 2, 31 | 40, 35 | 170 |
| B8 | Male | B | B | 46,762 | 4.67 | 465 | 630 | 2, 32 | 51, 35 | 32 |
| B9 | Male | B | B | 19,133 | 4.282 | 865 | 865 | 3, 29 | 7, 39 | 20 |
| B patient parameters | ||||||||||
| IQ25 | 19,211 | 4.28 | 393.8 | 735 | ||||||
| Median | 41,134 | 4.61 | 439 | 1,184 | ||||||
| IQ75 | 325,831 | 5.44 | 681 | 1,832 | ||||||
| BF patients | ||||||||||
| BF1 | Male | BF | F | 32,389 | 4.51 | 308 | 770 | 1, 2 | 35, 7 | 120 |
| BF2 | Male | BF | F | >500,000 | >5.7 | 306 | 1,152 | 3, 24 | 7, 50 | 20 |
| BF3 | Male | BF | F | >500,000 | >5.7 | 243 | 2,079 | 1, 68 | 49, 49 | 60 |
| BF4 | Female | BF | F | 36,668 | 4.56 | 612 | 774 | 2, 3 | 7, 44 | 135 |
| BF5 | Male | F | F | >500,000 | >5.7 | 209 | 1,229 | 2, 31 | 40, 35 | 152 |
| BF6 | Female | BF | F | 55,279 | 4.74 | 568 | 999 | 23, 66/26 | 58, 41 | 45 |
| BF7 | Male | BF | F | >500,000 | >5.7 | 227 | 385 | 2, 24 | 35, 35 | 35 |
| BF patient parameters | ||||||||||
| IQ25 | 36,668 | 4.56 | 227 | 770 | ||||||
| Median | >500,000 | 5.7 | 306 | 999 | ||||||
| IQ75 | >500,000 | 5.7 | 568 | 1,229 | ||||||
| Nonprogressors | ||||||||||
| NP1 | Female | B | B | <50 | <1.7 | 1,093 | 864 | 1, 31 | 35, 57 | >10 yr |
| NP2 | Male | B | B | 1,112 | 3.046 | 708 | 369 | 2, 3 | 15, 7 | >10 yr |
For the B and BF patient groups, the median and 25% and 75% interquartile values (IQ25 and IQ75, respectively) for VL, log10 VL, CD4 count, and CD8 count are shown. The differences observed between the values for both groups for VL (P = 0.142), log10 VL (P = 0.146), CD4 count (P = 0.071), and CD8 count (P = 0.6) were not statistically significant.
Viral load (VL) calculated by the Versant HIV-1 RNA 3.0 assay (Bayer AG). The detection threshold was 50 RNA copies/ml (1.7 log10).
Flow cytometry double platform (Epics XL; Coulter).
The estimated time from infection is shown in days unless specified otherwise.
No statistically significant differences were found in VL, CD4, and CD8 counts for both patient groups (Table 1). A wide range of HLA alleles whose frequencies agree with local population data were represented (www.allelefrequencies.net) (Table 1). Additionally, samples from two HIV-1-positive individuals defined as nonprogressors (NP1 and NP2) were obtained.
Total magnitude of the Nef-specific cellular immune response.
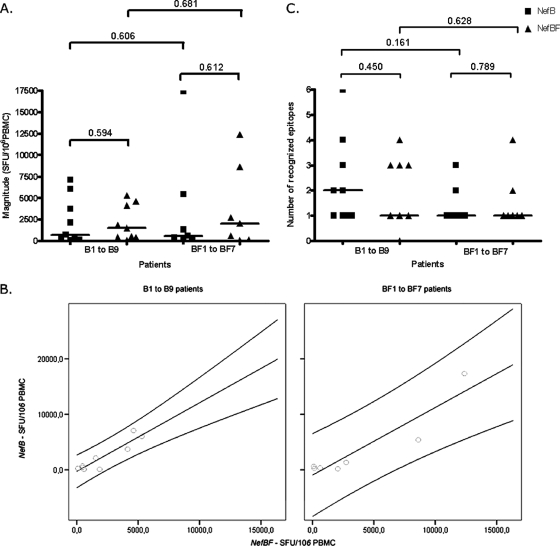
Previous studies have determined that during primary HIV infection Nef-specific CD8 T-cell responses represented a significant ratio (46% to 94%) of the total magnitude of the HIV-specific T-cell response (43). In our B/BF cohort, we characterized the Nef-specific T-cell response by employing a peptide matrix-based IFN-γ ELISPOT assay followed by confirmation of individual peptide response in a further ELISPOT assay. Peptide sets representing subtypes B and BF were used to accurately analyze the T-cell responses in this mixed subtype population. Homology between the peptide sequences and the patient autologous sequences ranged between 71 and 79% regardless of viral subtype. In Fig. 1A, the total magnitude of the responses detected against NefB and NefBF proteins, calculated as the sum of the responses found against each peptide, is detailed. In subtype B-infected subjects (n = 9), the magnitude of positive T-cell responses ranged from 80 to 7,150 SFU/106 PBMCs (median, 730) for NefB peptides, while it ranged from 110 to 5,295 SFU/106 PBMCs (median, 1,535) for NefBF peptides (P = 0.594). For subtype BF-infected patients (n = 7), the median values obtained were 2,060 (range, 90 to 12,385) and 600 (range, 220 to 17,375) SFU/106 PBMCs for NefBF and NefB peptides, respectively (P = 0.612 [Fig. 1A]). Although the responses of BF patients were of higher magnitude than those of B patients, the difference was not statistically significant. A positive correlation was observed between responses to NefB and NefBF peptides for patients infected with subtypes B (Spearman's r = 0.633) and BF (Spearman's r = 0.679) (Fig. 1B). Thus, the magnitudes of T-cell Nef-specific responses analyzed as a whole showed no significant differences between patients recently infected with subtypes B and BF.
FIG. 1.
T-cell responses directed at HIV-1 Nef protein from B and BF viral variants and detected using NefB- or NefBF-derived peptides in a matrix-based IFN-γ ELISPOT assay. (A) Total magnitude of anti-Nef T-cell responses, measured in SFU/106 PBMC, in B- and BF-infected patients directed at either NefB and NefBF peptides. Symbols show the values for the responses from individual patients. The horizontal black bars indicate the median values. Intra- and interclade differences were analyzed using Wilcoxon and Mann-Whitney tests, respectively. P values are shown above the brackets. (B) Positive correlations were observed in B-infected patients (r = 0.633) and BF-infected patients (r = 0.679) for antigens derived from clade B (NefB) and BF intersubtype variants (NefBF). Correlations are shown together with 95% confidence intervals. (C) Breadth of total anti-Nef responses detected in each patient and expressed as the number of epitopes recognized. Data were analyzed as described above for panel A. Symbols: ▪, NefB-infected patients; ▴, NefBF-infected patients.
Next, we analyzed the breadth of the anti-Nef response in subtype B- and BF-infected patients using both NefB and NefBF peptide sets. Analyzing the total number of epitopes recognized in both groups of patients, B-infected patients recognized a total of 34 different epitopes, while in BF-infected subjects 13 different Nef-specific responses were detected. All seven BF patients reacted to a minimum of one and a maximum of four or three NefBF and NefB peptides, respectively (means of 1.57 and 1.42, respectively; P = 0.789; Fig. 1C). In B-infected subjects, a broader response was detected. All individuals reacted to at least one NefB peptide (mean, 2.44; range, 1 to 6), while eight out of nine patients reacted to at least one NefBF peptide (mean, 1.89; range, 0 to 4) (P = 0.450). A trend toward a narrower response was observed in BF-infected patients, although the differences detected were not statistically significant (P = 0.161 and 0.628 for NefB and NefBF peptides, respectively [Fig. 1C]).
Clade-specific breadth and magnitude of responses targeted against different Nef domains.
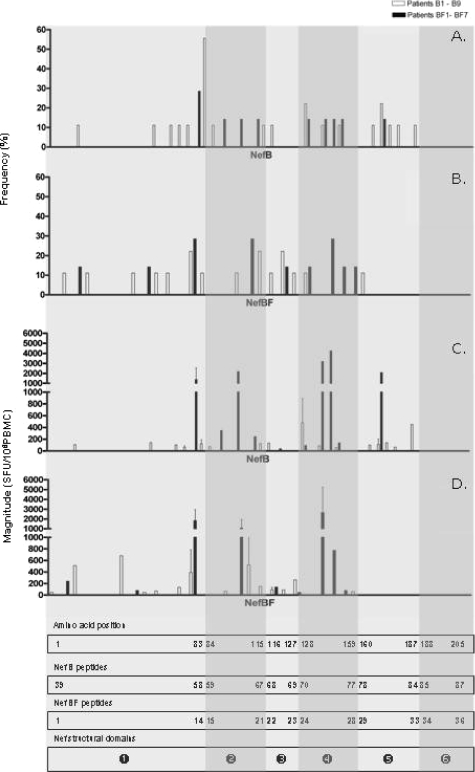
We then performed a detailed analysis of the peptides targeted and their location along the Nef protein (Fig. 2). The region between aa 66 and 148 contained immunodominant epitopes, with 63% and 82% of the responses found in subtype B- and BF-infected patients, respectively, being targeted toward this region (Fig. 2A and B). This observation has also been reported previously for subtype B and other subtypes (22, 26, 28, 35, 43, 45). However, in this study, clade-specific differences in targeted Nef domains were observed. The responses of B patients could be found within almost every domains, even when PBMCs were stimulated with peptides corresponding to the BF-derived protein. In contrast, BF patient responses were concentrated between the N-terminal core and central core domains, strengthening the idea that these patients elicit a narrower response than B patients, not only in terms of the number of epitopes recognized but also in the regions of Nef being recognized.
FIG. 2.
Histograms showing the frequency of respondents (A and B) and magnitude of responses (C and D) for all overlapping peptides. Histograms show all positive responses (by cutoff criteria described in Materials and Methods) obtained in patients infected with B (white bars) and BF (black bars) for peptides derived from both NefB (A and C) and NefBF (B and D). The median magnitude obtained for each peptide is shown together with SDs (error bars). The rectangles at the bottom of the figure show the amino acid numbering, peptide number within each peptide set (NefB and NefBF), and Nef structural domains. The HIV-1 Nef protein can be divided into six functional domains (depicted as white numbers of a solid black circle): the N-terminal domain (domain 1, aa 1 to 83), the N-terminal core (domain 2, aa 84 to 115), oligomerization domain (domain 3, aa 116 to 127), central core (domain 4, aa 128 to 159), loop (domain 5, aa 160 to 187), and C-terminal domain (domain 6, aa 188 to 205) (6). Regions encompassing each domain in each histogram are highlighted with different degrees of shading.
It is worth noting that the NefB protein flexible loop was targeted in an unexpected high frequency (21%), while only one positive response (from a B-infected patient) was found within the BF loop. Also, three out of four responses from B patients targeted toward the central core domain were missed when challenging PBMCs with the BF peptide set.
When the magnitude of the responses of subtype B- versus BF-infected subjects was split and analyzed into each Nef domain (Fig. 2C and D), we found that in the region encompassing the N-terminal core to the central core, the median response for BF subjects was 618 SFU/106 PBMCs (range, 50 to 4,530), while in B-infected patients, the median was 105 (range, 55 to 960; Mann-Whitney test P = 0.0071). Thus, analysis by domain revealed that there exist differences in the magnitude of the specific T-cell responses, between the two patient groups, that were previously hidden.
Fine mapping of Nef-specific response at the individual peptide level revealed concordant and discordant responses.
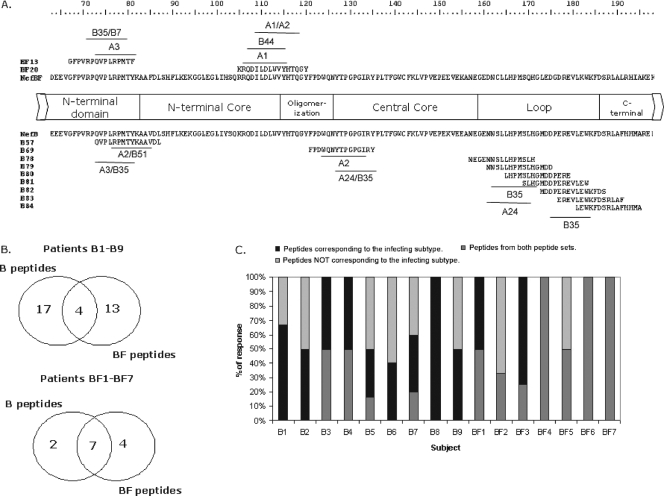
The individual peptides most frequently recognized were NefB57 (71-QVPLRPMTYKAAVDL) and NefBF13 (67-GFPVRPQVPLRPMTF). The NefB57 and NefBF13 peptides were recognized by 56% and 51% of patients, respectively, and both peptides share an 8-aa portion of their primary sequence (QVPLRPMT [Fig. 3A]). Other frequently recognized peptides, although to a lesser extent, were NefB69 (121-FPDWQNYTPGPGIRY), NefB78 (157-NEGENNSLLHPMSLH), and NefBF20 (105-KRQDILDLWVYHTQGY [Fig. 3A]), all recognized in 37% of cases. HLA restriction for NefB57 and NefB69 is widely described in the literature (26, 39), but not for NefB78. As described above, positive responses were found toward epitopes located inside the NefB flexible loop. The primary structures of these B peptides are also shown in Fig. 3A.
FIG. 3.
Fine mapping of responses, evaluated at the single-peptide level. (A) Nef domains encompassing aa 163 to 198 are shown together with NefB and NefBF primary structures. The most frequently recognized peptides are indicated. Also, the primary structures of subtype B peptides spanning the NefB loop are shown. This domain was targeted in an unexpected high frequency of patients (21%). Close to each peptide, putative restricting HLA alleles are shown as predicted using Web-based immunological tools. (B) Venn diagrams showing the absolute number of responses obtained using each peptide set for each patient group. Overlapping regions denote the number of responses detected with both peptide sets (i.e., concordant responses). (C) Bar graph showing the relative contribution of concordant and discordant responses to the total breadth in all patients enrolled. Dark gray portions stand for recognition of peptides corresponding to the peptide set derived from the same subtype than the infecting virus; light gray portions stand for recognition of peptides derived from the peptide set not corresponding to the infecting subtype, and black portions represent concordant responses.
Using the HLA haplotype of each patient, we attempted to determine the putative HLA allele(s) restricting each positive response in every patient. We used Web-based immunological tools to bring together both pieces of data. First, the Los Alamos HIV Immunology Database was screened for previous reported restriction for those subtype B peptides toward which positive responses were detected in our cohort. For those peptides for which no previous data were available, as well as for NefBF peptides, we used the BIMAS epitope prediction software to identify 8- to 10-mer epitopes within each peptide capable of binding HLA alleles expressed in each patient. This analysis revealed that putative restricting alleles for those peptides more frequently recognized in our cohort were as follows: HLA-A2, -A3, -B35, and -B51 for NefB57; HLA-A2, -A24, and -B35 for NefB69; HLA-A24 for NefB78; HLA-A3, -B35, and -B7 for NefBF13; and HLA-A1, -A2, and -B44 for NefBF20 (Fig. 3A).
Then, the cohort was examined for the number of responses detected using one set of peptides, the other set of peptides, or both sets. The three possible situations were found. Given a B patient, for instance, responses could be found toward (i) a B peptide but not to its BF counterpart, (ii) a BF peptide but not to its B counterpart, and (iii) a B peptide and its BF counterpart. The first two situations account for discrepant responses, while the third accounts for concordant responses. Overlapping Venn diagrams (Fig. 3B) were used to display the number of the responses accounting for discrepant responses (i.e., detected by each individual peptide set) and for concordant responses (central overlap of Venn diagrams). In B patients, 17/34 (50%) and 13/34 (38%) of the responses were detected with one peptide set or the other peptide set (discrepant responses), while the minority of the responses (4/34 [12%]) were concordant. The opposite situation was found for BF patients, as nearly 54% (7/13) of the responses were concordant, while 2/13 (15%) and 4/13 (31%) were detected either with the NefB and NefBF peptide set, respectively. Peptides responsible for concordant responses were mainly contained within epitope-rich zones. The latter analyses led to the conclusion that if only the peptide pool corresponding to each individual infecting subtype had been used, an underestimation of up to 66% of the breadth of response would have been obtained. In other words, the use of two different peptide sets enhanced the sensitivity of T-cell response detection between 33% and 66%. This can be observed for each patient in Fig. 3C.
Last, we focused our analysis on discordant responses. We aimed at determining the putative reason(s) for non recognition of one of the peptides within a discordant pair. In order to perform this analysis, we first attempted to determine the HLA allele restricting each positive response in every patient using Web-based immunological tools as described above. Once the putative restricting allele and minimal epitope within each peptide were identified, we aligned each positive peptide with their nonrecognized counterparts from the other peptide set. Figure 4 depicts three representative examples of such alignments. For instance, patient BF2 has HLA alleles A3, A24, B7, and B50. A positive response to peptide NefB77 was found in the ELISPOT assay. Putative restricting HLA alleles for this peptide are A24 and B7. Then, this peptide was aligned with the autologous viral sequence and the overlapping peptides of the NefBF peptide set (peptides NefBF28 and NefBF29). It can be observed that the sequences of these peptides are identical to that of the autologous viral sequence (except for 1 aa, which is not predicted to affect binding affinity to HLA-A24 and -B7 molecules). However, the predicted minimal epitope is in a different position in each peptide (i.e., it is offset). Therefore, we ascribe the lack of recognition of peptides NefBF28 and NefBF29 to the offsetting of the minimal epitope within those peptides. Similarly, we found that other reasons for nonrecognition were polymorphism within the optimal epitope (which may affect interaction to the T-cell receptor and thus effector response) and on epitope anchor positions. Examples of each situation are shown in Fig. 4. Analysis of all discordant responses revealed that offsetting of peptides is the major reason accounting for lack of recognition within a discordant pair of peptides, as it was found as a putative cause of nonrecognition in 67% (24/36) of the discordant responses identified in our cohort. On the other hand, polymorphisms within the optimal epitope or at HLA anchor positions accounted for 19% (7/36) or 14% (5/36) of the discordant responses found in our cohort, respectively.
FIG. 4.
Reasons driving discordant responses. Three reasons (a, b, and c) were identified to explain discordant responses. Here, examples for each of the reasons are shown. Patient identification, HLA haplotype, and autologous viral sequence (numbers at the left indicate the amino acid positions) are shown together with the primary sequences of reactive and nonreactive peptides. Putative minimal epitopes are shown boxed, and the putative restricting HLA alleles are also indicated. In panel c, the anchor position is denoted with an asterisk.
Cellular responses in NP subjects.
A weak Nef-specific response was detected (165 SFU/106 PBMCs) as a response in the nonprogressor NP1 subject. Using the matrix approach, it was possible to show that this response was directed toward three different peptides located within the N-terminal domain, the N-terminal core, and the oligomerization domains. No specific anti-Nef response was detected in subject NP2.
Peptide pools were used to identify responses directed toward other viral proteins in these patients. Vif- and p24-specific responses were also observed in NP1 (100 and 265 SFU/106 PBMCs, respectively) while in patient NP2, anti-Gag reactive populations were exclusively detected (p17, 120 SFU/106 PBMCs; p24, 690 SFU/106 PBMCs; p2p7p1p6, 195 SFU/106 PBMCs).
Phenotypic and functional profiles of positive T-cell responses.
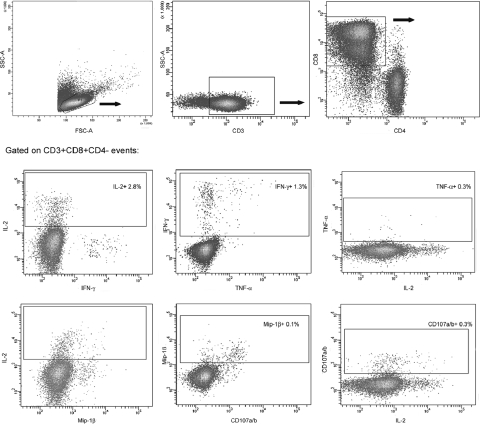
In order to further characterize cross-reactive T-cell populations identified by ELISPOT assay, the phenotype and functionality of these cells were studied in a subset of seven patients. Recently, a flow cytometry-based assay was developed to measure the vast complexity of the HIV-specific T-cell responses (12, 41, 51). We used a modification of this assay which was adapted to our resources. Cells were stimulated with the corresponding peptide, as described in Materials and Methods, then surface molecules (CD3, CD4, and CD8) were stained, and five different functions were studied in the CD3+ CD8+ population (degranulation [10, 11) and IFN-γ, Mip-1β, IL-2, and TNF-α secretion) (12). The gating strategy, illustrated using data from one representative patient, is shown in Fig. 5. Two different positive controls were included: polyclonal stimulation with a mix of PMA and ionomycin and the control peptide pool CEF. Mean ± SD percentages of positive CD3+ CD8+ cells for each function were 22.52 ± 6.02 (IL-2), 44.6 ± 16.28 (Mip-1β), 38.6 ± 5.37 (TNF-α), 35.1 ± 4.24 (IFN-γ), and 42.65 ± 3.32 (CD107a/b) for PMA-ionomycin and 0.97 ± 0.35 (IL-2), 2.26 ± 1.62 (Mip-1β), 1.67 ± 1.17 (TNF-α), 1.1 ± 0.81 (IFN-γ), and 1.3 ± 0.43 (CD107a/b) for the CEF pool.
FIG. 5.
Gating scheme used for the identification of specific T cells. Data shown are from cells derived from one representative patient, stimulated with an HIV peptide. Initial gating was performed on small lymphocytes in a forward scatter (FSC)-versus-SSC plot. CD3+ events were gated in an SSC-versus-CD3 plot prior to gating on CD3+ CD8+ and CD3+ CD4+ events. Then a gate was made for each studied function and their combinations. Only the functional profile of cells gated on CD3+ CD8+ events is shown, but a similar analysis was also performed on CD3+ CD4+ cells. For each patient, two different positive controls were included: polyclonal stimulation with a mix of PMA-ionomycin and the control peptide pool CEF. Mean ± SD percentages of positive CD3+ CD8+ cells for each function were 22.52 ± 6.02 (IL-2), 44.6 ± 16.28 (Mip-1β), 38.6 ± 5.37 (TNF-α), 35.1 ± 4.24 (IFN-γ), and 42.65 ± 3.32 (CD107a/b) for the PMA-ionomycin mixture and 0.97 ± 0.35 (IL-2), 2.26 ± 1.62 (Mip-1β), 1.67 ± 1.17 (TNF-α), 1.1 ± 0.81 (IFN-γ) and 1.3 ± 0.43 (CD107a/b) for the CEF pool.
(i) IFN-γ responses detected by the ELISPOT assay were predominately due to CD8 T cells.
First, we aimed at correlating the data obtained by the ELISPOT assay and the results of the ICS assay. We observed that the hierarchy of SFU/106 PBMCs correlated with the sum of CD8+ CD3+ and CD4+ CD3+ IFN-γ-secreting cells (data not shown). As expected based on a previous report (21), no CD4+ CD3+ CEF-specific IFN-γ-secreting cells were detected in the ICS assay. Out of the 18 HIV-specific IFN-γ-secreting responses analyzed by the ICS assay, only 3 responses were mediated mainly by CD3+ CD4+ cells (between 64% and 100% of the total response), while in the remaining 15 responses, CD3+ CD8+ IFN-γ-producing cells were almost exclusively responsible (between 62% and 100% of the total response) for the results observed by ELISPOT assay. These data are consistent with previous studies indicating that the vast majority of these types of responses are due to CD8+ T cells (7, 8, 24).
(ii) Correlation of polyfunctional CD8+ T-cell responses with better clinical status of the patients.
Out of the seven patients studied by ICS, clinical follow-up data suggested that three patients (patients B6, B8, and BF6) had improved control of primary infection (lower viral set point, CD4+ cell count recovery, containment of symptoms [data not shown]), while the other three patients (B5, BF3, and BF4) presented a regular resolution of acute phase. The seventh patient evaluated was NP2. As reported in the literature, longitudinal evaluation of Nef-specific T-cell response (evaluated by ELISPOT assay) in these patients revealed no association between the breadth or magnitude of the Nef-specific response and CD4 count, VL, or clinical status (primary infection versus nonprogressing status [data not shown]).
Recent observations suggest that the role of polyfunctional CD8+ T-cell responses may play a role in controlling virus replication (3, 12, 30, 56, 60). Given these reports, we then focused on studying the polyfunctional profile of the responses detected in this subset of seven patients and aimed to correlate it to the capacity to contain viral replication. We individually evaluated degranulation (CD107a and CD107b mobilization) and IFN-γ, Mip-1β, IL-2, and TNF-α secretion, as well as the following combinations of effector functions: IL-2 plus IFN-γ, IL-2 plus TNF-α, IFN-γ plus TNF-α, IL-2 plus Mip-1β, IL-2 plus degranulation, Mip-1β plus degranulation, IL-2 plus IFN-γ plus TNF-α, and IL-2 plus Mip-1β plus degranulation.
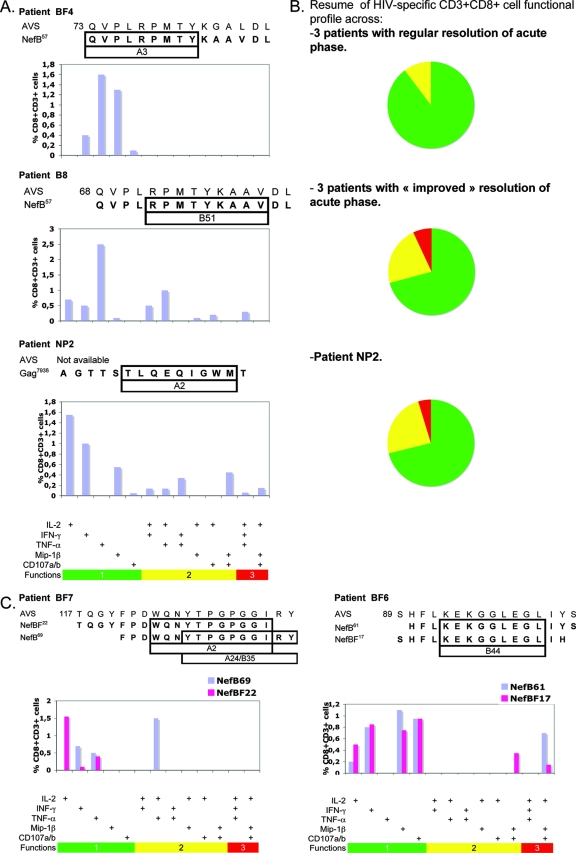
Figure 6A shows representative data for one patient from each group, while Fig. 6B summarizes the data for each patient group and for patient NP2. It was found that bi- and trifunctional CD8+ cells together (yellow and red sections in Fig. 6B) account for almost 30% of the total specific response in patients with improved control of primary viremia. This functionality pattern was strikingly similar to what was observed for NP2. On the other hand, patients with regular resolution of acute phase showed limited responses with respect to the functions detected. In these patients, most of the specific cells detected were monofunctional, only 10% (on average) were bifunctional, and no specific CD3+ CD8+ cells with three simultaneous functions were detected in any of these patients.
FIG. 6.
Functional profiles of specific CD8+ T cells. (A) Specific CD8 T-cell responses of a representative patient with regular resolution of acute phase (patient BF4), of a patient with “improved” control of acute phase (patient B8), and of patient NP2. For each patient, the peptide used as stimulus is shown aligned to the autologous viral sequence (AVS) (numbers to the left of the sequence indicate the amino acid positions within the Nef protein) and putative restricting HLA allele. Vertical bars represent the frequency of CD8+ T cells expressing the particular combination of functions indicated on the x axis. Mono-, bi-, and trifunctional combinations are further denoted by green, yellow, and red horizontal bars, respectively. Responses shown correspond to background subtracted results using the CD28/49d control. (B) Summary of functional profile in each group of patients. Each pie chart represents the mean (of all patients within a group) contribution of each effector function to the total response. Green sectors stand for CD8+ cells positive for only one function, yellow sectors stand for bifunctional CD8+ cells, and red sectors stand for trifunctional cells, matching the color code used in panel A. (C) ICS evaluation of ELISPOT assay concordant responses. Here, both peptides used in turn as stimulus are shown aligned to the AVS. The frequency of CD8+ T cells detected using both peptides is shown in each plot.
These results suggested a correlation between a better clinical status of the patients and the polyfunctionality of the CD8 T-cell response. The situation observed for subject B8 is particularly interesting. The first sample obtained from this subject was 15 days after the beginning of acute symptoms. The VL was 4.67 log10 units and had 465 CD4+ cells/μl. Fifteen days later, the subject fully seroconverted, and the VL dropped 2 log10 units. The patient currently remains asymptomatic and off highly active antiretroviral therapy. This patient shows a narrow Nef-specific response, as only cells reactive against only one peptide (NefB57) were found. Evaluation of specific immune responses against Gag was also performed, although no specific cellular immune responses were found. In this patient, the high quality of the CD8 response (in terms of functionality) may be one of the factors contributing to the excellent control of viral replication. Longitudinal studies of subjects B6, B8, BF6, and others undergoing a similar situation will allow determination of whether polyfunctional cell populations present at the acute phase of infection are predictors of nonprogression. Also of interest, patient B6 shows a strikingly similar profile of functionality (even with similar magnitudes) in different CD8 populations specific for two different peptides (one from the NefB peptide set and one from the NefBF peptide set) corresponding to different Nef domains.
(iii) Discrepant functionality patterns observed in ELISPOT assay concordant responses.
Last, we examined the polyfunctional profile obtained for both peptides within the concordant cross-clade responses identified by ELISPOT assay. It was observed that when peptides within a given concordant pair are offset, different functional profiles are observed. Results obtained for patient BF7 are shown in Fig. 6C to illustrate this fact. On the other hand, this contrasts with what was observed when peptides are fully overlapping (Fig. 6C, patient BF6), where an agreement in functional profile is observed. These phenomena were systematically observed for every concordant response analyzed.
DISCUSSION
Screening of the T-cell responses in patients infected with different subtypes and intersubtype recombinant forms of HIV has become a major goal to help inform vaccine design. Initial reports included only subtype B- or C-infected patients (1, 47-49), but more recently other subtypes and CRF recombinant forms are being studied (18, 22, 27, 28, 35). On the other hand, the immunoprotective role of HIV-specific vaccine-induced T cells may be limited by the large genetic distance between viral strains circulating in different locations and the vaccine strain. Thus, it is important to determine whether it would be advantageous to include peptides based on multiple clades, including those based on variants circulating in a specific geographic area.
Here, we report results from T-cell screening on a cohort of subjects with primary HIV infection. We chose this cohort on the basis that immune responses arising during this period are more likely to be relevant for immune control than those found during chronic infection. Recently, it was shown that subsequent broadening of the specific T-cell response is not associated with effective control of viral replication but with viral escape from temporarily effective immune responses (37).
To accurately evaluate Nef-specific CD8+ T-cell responses in this mixed-subtype cohort, two panels of overlapping peptides were used: a clade B-derived peptide set and a BF-derived set. Cross-clade responses were identified on the basis of IFN-γ production. Single-peptide-based screening of T-cell responses allowed fine mapping, which revealed broad cross-clade T-cell populations, although clade-specific magnitude, breadth, and domain targeting were observed. Immunodominance of the Nef central region was demonstrated in our cohort, in accordance with other studies (22, 26, 28, 35, 43, 45). Most of the cross-reactive peptides clustered in this conserved region of the protein. Peptides more frequently recognized contained epitopes restricted by HLA alleles highly prevalent in our cohort. Surprisingly, the NefB protein flexible loop was targeted in high frequency (three B patients and one BF patient). The fact that our population shows different HLA allele frequencies compared to other Caucasian populations (www.allelefrequencies.net), added to amino acid polymorphisms found in Argentinean B HIV isolates compared to B isolates from other countries, may explain this observation. Studies demonstrating that the distribution and nature of CTL responses depend on the predominant circulating virus and genetic background of the population (58, 59) support this idea. Also, the high frequency of specific responses to the B loop might be influenced by the short time since infection. It has been shown in longitudinal studies that responses to certain epitopes in Nef dominated early but were later replaced by responses to other Nef epitopes (7).
It is widely known that the use of peptides based on HIV-1 reference strains in order to screen CTL responses fails to detect the true breadth and magnitude of the response (4). The data presented here indicate that the use of two peptide sets based on the more prevalent subtypes circulating in our region increased the rate of detected responses. Considering these results together with recent publications (22, 28), it can be stated with certainty that the use of more than one peptide set profoundly impacts the detection rate of HIV-specific CTL responses for a wide range of subtypes (A, B, BF, C, D, and F), and presumably, these results could be extrapolated to all viral subtypes. Currier et al. (22) suggested that, in the particular case of Nef, the use of the homologous subtype-derived peptide set would be sufficient for screening specific responses. Our results do not agree with this statement, and more than one peptide set should be used to maximize breadth detection, irrespective of the viral protein being targeted by those responses.
Additionally, in these studies (18, 22, 28) and our study, concordant and discordant responses are found. This phenomenon can be explained by (i) amino acid substitutions within the optimal epitope, (ii) substitutions in close proximity to the optimal epitope, and (iii) the relative position of the optimal epitope within a given peptide (peptide offsetting). The third explanation was found as the main putative cause accounting for the lack of recognition of one of the peptides within a discordant pair. On the other hand, recent studies have shown that while different variants of a given epitope may be recognized by T cells, variation in the epitope sequence has a profound impact on the ability of CTLs to recognize and clear infected cells (9, 45). Here, we demonstrated that there exist significant differences in the quality (cytokine secretion profile and cytotoxic activity) and magnitude of the CD8 response to different epitope variants. Significant differences in the avidity of the T-cell responses (46), which can influence the cytokine response profiles (40, 52) and cytotoxic activity of CD8+ T cells (31), were demonstrated to have an important impact on virus control. Taken together, these data will help discern the best antigens to be included in future HIV vaccines that induce T-cell populations with optimal effector functions. This also has important implications in the selection of reagents for screening T-cell responses.
It was not until very recently that the correlates of HIV protection (or more accurately, the correlates of nonprogression) have started to be elucidated. Although the importance of CD8 T cells in controlling HIV replication is widely accepted, earlier reports have failed to show either a positive or negative correlation between viral load and the breadth or magnitude of CTL responses. Now it is known that Gag-specific responses are associated with low viremia (29, 38, 61), which can be explained by the capacity of Gag-specific CTLs to kill very recently infected cells, even before the viral genome integrates into the host genome (55). Also, it was shown that the functional profile of HIV-specific CD8 T cells in progressors is limited compared to that of nonprogressors, who consistently maintain highly functional CD8 T cells (12). In line with these studies, no correlation between the magnitude or breadth of the Nef-specific T-cell responses and the patient's viral or immune parameters was found. However, patients from our cohort showing an enhanced control of initial viremia also showed a high proportion of bi- and trifunctional specific CD8+ cells. Of these patients, only BF6 had an HLA allele (B58) associated with delayed progression to AIDS (57). Our results add further support to the notion that multiple functions are required in the specific T-cell repertoire to control viral replication and suggest that the proportion of multifunctional CD8+ cells may predict clinical outcome and that a multifunctional vaccine-induced T-cell response should be induced to provide protection.
To summarize, we determined the frequency and magnitude of recognition of epitopes within the Nef protein derived from BF viral variants and compared the pattern of immunodominance in Argentinean patients (whether infected with B or BF forms) to those described previously. Cross-clade and clade-specific responses between B subtype and BF variants were found. Detailed analysis at the single-peptide level revealed that BF patients show a narrower response (both in terms of the number of epitopes recognized and the Nef domains targeted) but greater in magnitude. Underestimation of the response was reduced to a minimum by challenging cells from both patient groups with both B and BF peptide sets. Functional differences in CD8+ T-cell population were observed depending on variations within the peptide presented (primary structure and offsetting). Also, different qualities of effector function were observed with different clinical resolutions of the acute phase. As a whole, our data suggest that the obstacle of genetic diversity in screening T-cell responses can be overcome by using multiple sets of peptides corresponding to regionally circulating HIV variants. Functional profiling of T-cell responses contributes significantly to dissection of the immune response.
Acknowledgments
This research has been funded by the Argentinean Agency for the Promotion of Science and Technology (ANPCYT) (grant 14104). G. Turk is supported by the Argentinean National Research Council (CONICET). J. H. Cox is supported by the U.S. Army Medical Research and Materiel Command and its Cooperative Agreement (DAMD17-98-2-7007) with the Henry M. Jackson Foundation for the Advancement of Military Medicine.
The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting true views of the Department of the Army or the Department of Defense of the United States.
We are very grateful to Bonnie Mathieson for encouraging the performance of this kind of study in Argentina and for all her support. We also thank Jeffrey Currier for helpful comments on the manuscript and Sergio Mazzini for assistance during its preparation.
Footnotes
Published ahead of print on 9 January 2008.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 772081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addo, M. M., X. G. Yu, E. S. Rosenberg, B. D. Walker, and M. Altfeld. 2002. Cytotoxic T-lymphocyte (CTL) responses directed against regulatory and accessory proteins in HIV-1 infection. DNA Cell Biol. 21671-678. [DOI] [PubMed] [Google Scholar]
- 3.Almeida, J. R., D. A. Price, L. Papagno, Z. A. Arkoub, D. Sauce, E. Bornstein, T. E. Asher, A. Samri, A. Schnuriger, I. Theodorou, D. Costagliola, C. Rouzioux, H. Agut, A. G. Marcelin, D. Douek, B. Autran, and V. Appay. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 2042473-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altfeld, M., M. M. Addo, R. Shankarappa, P. K. Lee, T. M. Allen, X. G. Yu, A. Rathod, J. Harlow, K. O'Sullivan, M. N. Johnston, P. J. Goulder, J. I. Mullins, E. S. Rosenberg, C. Brander, B. Korber, and B. D. Walker. 2003. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J. Virol. 777330-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altfeld, M., E. S. Rosenberg, R. Shankarappa, J. S. Mukherjee, F. M. Hecht, R. L. Eldridge, M. M. Addo, S. H. Poon, M. N. Phillips, G. K. Robbins, P. E. Sax, S. Boswell, J. O. Kahn, C. Brander, P. J. Goulder, J. A. Levy, J. I. Mullins, and B. D. Walker. 2001. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J. Exp. Med. 193169-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arold, S. T., and A. S. Baur. 2001. Dynamic Nef and Nef dynamics: how structure could explain the complex activities of this small HIV protein. Trends Biochem. Sci. 26356-363. [DOI] [PubMed] [Google Scholar]
- 7.Bansal, A., E. Gough, S. Sabbaj, D. Ritter, K. Yusim, G. Sfakianos, G. Aldrovandi, R. A. Kaslow, C. M. Wilson, M. J. Mulligan, J. M. Kilby, and P. A. Goepfert. 2005. CD8 T-cell responses in early HIV-1 infection are skewed towards high entropy peptides. AIDS 19241-250. [PubMed] [Google Scholar]
- 8.Bansal, A., S. Sabbaj, B. H. Edwards, D. Ritter, C. Perkins, J. Tang, J. J. Szinger, H. Weiss, P. A. Goepfert, B. Korber, C. M. Wilson, R. A. Kaslow, and M. J. Mulligan. 2003. T cell responses in HIV type 1-infected adolescent minorities share similar epitope specificities with whites despite significant differences in HLA class I alleles. AIDS Res. Hum Retrovir. 191017-1026. [DOI] [PubMed] [Google Scholar]
- 9.Bennett, M. S., H. L. Ng, M. Dagarag, A. Ali, and O. O. Yang. 2007. Epitope-dependent avidity thresholds for cytotoxic T-lymphocyte clearance of virus-infected cells. J. Virol. 814973-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betts, M. R., J. M. Brenchley, D. A. Price, S. C. De Rosa, D. C. Douek, M. Roederer, and R. A. Koup. 2003. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods 28165-78. [DOI] [PubMed] [Google Scholar]
- 11.Betts, M. R., and R. A. Koup. 2004. Detection of T-cell degranulation: CD107a and b. Methods Cell Biol. 75497-512. [DOI] [PubMed] [Google Scholar]
- 12.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 1074781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 686103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3205-211. [DOI] [PubMed] [Google Scholar]
- 15.Carobene, M. G., A. E. Rubio, M. G. Carrillo, G. E. Maligne, G. H. Kijak, J. F. Quarleri, and H. Salomon. 2004. Differences in frequencies of drug resistance-associated mutations in the HIV-1 pol gene of B subtype and BF intersubtype recombinant samples. J. Acquir. Immune Defic. Syndr. 35207-209. [DOI] [PubMed] [Google Scholar]
- 16.Carr, J. K., M. Avila, M. Gomez Carrillo, H. Salomon, J. Hierholzer, V. Watanaveeradej, M. A. Pando, M. Negrete, K. L. Russell, J. Sanchez, D. L. Birx, R. Andrade, J. Vinoles, and F. E. McCutchan. 2001. Diverse BF recombinants have spread widely since the introduction of HIV-1 into South America. AIDS 15F41-F47. [DOI] [PubMed] [Google Scholar]
- 17.Cereb, N., P. Maye, S. Lee, Y. Kong, and S. Y. Yang. 1995. Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, -B, and -C alleles. Tissue Antigens 451-11. [DOI] [PubMed] [Google Scholar]
- 18.Chen, J., K. Hong, M. Jia, H. Liu, Y. Zhang, S. Liu, X. Zhang, H. Zhao, H. Peng, P. Ma, H. Xing, Y. Ruan, K. L. Williams, X. G. Yu, M. Altfeld, B. D. Walker, and Y. Shao. 2007. Human immunodeficiency virus type 1 specific cytotoxic T lymphocyte responses in Chinese infected with HIV-1 B'/C recombinant (CRF07_BC). Retrovirology 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Currier, J. R., M. deSouza, P. Chanbancherd, W. Bernstein, D. L. Birx, and J. H. Cox. 2002. Comprehensive screening for human immunodeficiency virus type 1 subtype-specific CD8 cytotoxic T lymphocytes and definition of degenerate epitopes restricted by HLA-A0207 and -CW0304 alleles. J. Virol. 764971-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Currier, J. R., W. E. Dowling, K. M. Wasunna, U. Alam, C. J. Mason, M. L. Robb, J. K. Carr, F. E. McCutchan, D. L. Birx, and J. H. Cox. 2003. Detection of high frequencies of HIV-1 cross-subtype reactive CD8 T lymphocytes in the peripheral blood of HIV-1-infected Kenyans. AIDS 172149-2157. [DOI] [PubMed] [Google Scholar]
- 21.Currier, J. R., E. G. Kuta, E. Turk, L. B. Earhart, L. Loomis-Price, S. Janetzki, G. Ferrari, D. L. Birx, and J. H. Cox. 2002. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J. Immunol. Methods 260157-172. [DOI] [PubMed] [Google Scholar]
- 22.Currier, J. R., U. Visawapoka, S. Tovanabutra, C. J. Mason, D. L. Birx, F. E. McCutchan, and J. H. Cox. 2006. CTL epitope distribution patterns in the Gag and Nef proteins of HIV-1 from subtype A infected subjects in Kenya: use of multiple peptide sets increases the detectable breadth of the CTL response. BMC Immunol. 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dilernia, D. A., A. M. Gomez, L. Lourtau, R. Marone, M. H. Losso, H. Salomón, and M. Gómez-Carrillo. 2007. HIV type 1 genetic diversity surveillance among newly diagnosed individuals from 2003 to 2005 in Buenos Aires, Argentina. AIDS Res. Hum. Retrovir. 231201-1207. [DOI] [PubMed] [Google Scholar]
- 24.Edwards, B. H., A. Bansal, S. Sabbaj, J. Bakari, M. J. Mulligan, and P. A. Goepfert. 2002. Magnitude of functional CD8+ T-cell responses to the Gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J. Virol. 762298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Vina, M. A., A. M. Lazaro, C. Y. Marcos, C. Nulf, E. Raimondi, E. J. Haas, and P. Stastny. 1997. Dissimilar evolution of B-locus versus A-locus and class II loci of the HLA region in South American Indian tribes. Tissue Antigens 50233-250. [DOI] [PubMed] [Google Scholar]
- 26.Frahm, N., B. T. Korber, C. M. Adams, J. J. Szinger, R. Draenert, M. M. Addo, M. E. Feeney, K. Yusim, K. Sango, N. V. Brown, D. SenGupta, A. Piechocka-Trocha, T. Simonis, F. M. Marincola, A. G. Wurcel, D. R. Stone, C. J. Russell, P. Adolf, D. Cohen, T. Roach, A. St. John, A. Khatri, K. Davis, J. Mullins, P. J. Goulder, B. D. Walker, and C. Brander. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 782187-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geels, M. J., S. A. Dubey, K. Anderson, E. Baan, M. Bakker, G. Pollakis, W. A. Paxton, J. W. Shiver, and J. Goudsmit. 2005. Broad cross-clade T-cell responses to Gag in individuals infected with human immunodeficiency virus type 1 non-B clades (A to G): importance of HLA anchor residue conservation. J. Virol. 7911247-11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geldmacher, C., J. R. Currier, M. Gerhardt, A. Haule, L. Maboko, D. Birx, C. Gray, A. Meyerhans, J. Cox, and M. Hoelscher. 2007. In a mixed subtype epidemic, the HIV-1 Gag-specific T-cell response is biased towards the infecting subtype. AIDS 21135-143. [DOI] [PubMed] [Google Scholar]
- 29.Geldmacher, C., J. R. Currier, E. Herrmann, A. Haule, E. Kuta, F. McCutchan, L. Njovu, S. Geis, O. Hoffmann, L. Maboko, C. Williamson, D. Birx, A. Meyerhans, J. Cox, and M. Hoelscher. 2007. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J. Virol. 812440-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genesca, M., T. Rourke, J. Li, K. Bost, B. Chohan, M. B. McChesney, and C. J. Miller. 2007. Live attenuated lentivirus infection elicits polyfunctional simian immunodeficiency virus Gag-specific CD8+ T cells with reduced apoptotic susceptibility in rhesus macaques that control virus replication after challenge with pathogenic SIVmac239. J. Immunol. 1794732-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillespie, G. M., R. Kaul, T. Dong, H. B. Yang, T. Rostron, J. J. Bwayo, P. Kiama, T. Peto, F. A. Plummer, A. J. McMichael, and S. L. Rowland-Jones. 2002. Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. AIDS 16961-972. [DOI] [PubMed] [Google Scholar]
- 32.Gomez-Carrillo, M., S. Pampuro, A. Duran, M. Losso, D. R. Harris, J. S. Read, G. Duarte, R. De Souza, L. Soto-Ramirez, and H. Salomon. 2006. Analysis of HIV type 1 diversity in pregnant women from four Latin American and Caribbean countries. AIDS Res. Hum. Retrovir. 221186-1191. [DOI] [PubMed] [Google Scholar]
- 33.Gomez Carrillo, M., M. Avila, J. Hierholzer, M. Pando, P. L. Martinez, F. E. McCutchan, and J. K. Carr. 2002. Mother-to-child HIV type 1 transmission in Argentina: BF recombinants have predominated in infected children since the mid-1980s. AIDS Res. Hum. Retrovir. 18477-483. [DOI] [PubMed] [Google Scholar]
- 34.Hierholzer, J., S. Montano, M. Hoelscher, M. Negrete, M. Hierholzer, M. M. Avila, M. G. Carrillo, J. C. Russi, J. Vinoles, A. Alava, M. E. Acosta, A. Gianella, R. Andrade, J. L. Sanchez, G. Carrion, J. L. Sanchez, K. Russell, M. Robb, D. Birx, F. McCutchan, and J. K. Carr. 2002. Molecular epidemiology of HIV type 1 in Ecuador, Peru, Bolivia, Uruguay, and Argentina. AIDS Res. Hum. Retrovir. 181339-1350. [DOI] [PubMed] [Google Scholar]
- 35.Inwoley, A., P. Recordon-Pinson, M. Dupuis, J. Gaston, M. Genete, A. Minga, F. Letourneur, F. Rouet, J. Choppin, H. Fleury, J. G. Guillet, and M. Andrieu. 2005. Cross-clade conservation of HIV type 1 Nef immunodominant regions recognized by CD8+ T cells of HIV type 1 CRF02_AG-infected Ivorian (West Africa). AIDS Res. Hum. Retrovir. 21620-628. [DOI] [PubMed] [Google Scholar]
- 36.Janetzki, S., J. H. Cox, N. Oden, and G. Ferrari. 2005. Standardization and validation issues of the ELISPOT assay. Methods Mol. Biol. 30251-86. [DOI] [PubMed] [Google Scholar]
- 37.Karlsson, A. C., A. K. Iversen, J. M. Chapman, T. de Oliviera, G. Spotts, A. J. McMichael, M. P. Davenport, F. M. Hecht, and D. F. Nixon. 2007. Sequential broadening of CTL responses in early HIV-1 infection is associated with viral escape. PLoS ONE. 2e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 1346-53. [DOI] [PubMed] [Google Scholar]
- 39.Korber, B. T. M., C. Brander, B. F. Haynes, R. Koup, J. P. Moore, B. D. Walker, and D. I. Watkins (ed.). 2005. HIV molecular immunology 2005. Los Alamos National Laboratory publication no. LA-UR 06-0036. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM.
- 40.La Gruta, N. L., S. J. Turner, and P. C. Doherty. 2004. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: correlation of cytokine profile and TCR avidity. J. Immunol. 1725553-5560. [DOI] [PubMed] [Google Scholar]
- 41.Lamoreaux, L., M. Roederer, and R. Koup. 2006. Intracellular cytokine optimization and standard operating procedure. Nat. Protoc. 11507-1516. [DOI] [PubMed] [Google Scholar]
- 42.Leslie, A., D. Kavanagh, I. Honeyborne, K. Pfafferott, C. Edwards, T. Pillay, L. Hilton, C. Thobakgale, D. Ramduth, R. Draenert, S. Le Gall, G. Luzzi, A. Edwards, C. Brander, A. K. Sewell, S. Moore, J. Mullins, C. Moore, S. Mallal, N. Bhardwaj, K. Yusim, R. Phillips, P. Klenerman, B. Korber, P. Kiepiela, B. Walker, and P. Goulder. 2005. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J. Exp. Med. 201891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lichterfeld, M., X. G. Yu, D. Cohen, M. M. Addo, J. Malenfant, B. Perkins, E. Pae, M. N. Johnston, D. Strick, T. M. Allen, E. S. Rosenberg, B. Korber, B. D. Walker, and M. Altfeld. 2004. HIV-1 Nef is preferentially recognized by CD8 T cells in primary HIV-1 infection despite a relatively high degree of genetic diversity. AIDS 181383-1392. [DOI] [PubMed] [Google Scholar]
- 44.Lichterfeld, M., X. G. Yu, S. Le Gall, and M. Altfeld. 2005. Immunodominance of HIV-1-specific CD8+ T-cell responses in acute HIV-1 infection: at the crossroads of viral and host genetics. Trends Immunol. 26166-171. [DOI] [PubMed] [Google Scholar]
- 45.Malhotra, U., F. Li, J. Nolin, M. Allison, H. Zhao, J. I. Mullins, S. Self, and M. J. McElrath. 2007. Enhanced detection of human immunodeficiency virus type 1 (HIV-1) Nef-specific T cells recognizing multiple variants in early HIV-1 infection. J. Virol. 815225-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malhotra, U., J. Nolin, J. I. Mullins, and M. J. McElrath. 2007. Comprehensive epitope analysis of cross-clade Gag-specific T-cell responses in individuals with early HIV-1 infection in the US epidemic. Vaccine 25381-390. [DOI] [PubMed] [Google Scholar]
- 47.Masemola, A., T. Mashishi, G. Khoury, P. Mohube, P. Mokgotho, E. Vardas, M. Colvin, L. Zijenah, D. Katzenstein, R. Musonda, S. Allen, N. Kumwenda, T. Taha, G. Gray, J. McIntyre, S. A. Karim, H. W. Sheppard, and C. M. Gray. 2004. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J. Virol. 783233-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masemola, A. M., T. N. Mashishi, G. Khoury, H. Bredell, M. Paximadis, T. Mathebula, D. Barkhan, A. Puren, E. Vardas, M. Colvin, L. Zijenah, D. Katzenstein, R. Musonda, S. Allen, N. Kumwenda, T. Taha, G. Gray, J. McIntyre, S. A. Karim, H. W. Sheppard, and C. M. Gray. 2004. Novel and promiscuous CTL epitopes in conserved regions of Gag targeted by individuals with early subtype C HIV type 1 infection from southern Africa. J. Immunol. 1734607-4617. [DOI] [PubMed] [Google Scholar]
- 49.Novitsky, V., H. Cao, N. Rybak, P. Gilbert, M. F. McLane, S. Gaolekwe, T. Peter, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2002. Magnitude and frequency of cytotoxic T-lymphocyte responses: identification of immunodominant regions of human immunodeficiency virus type 1 subtype C. J. Virol. 7610155-10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker, K. C., M. A. Bednarek, and J. E. Coligan. 1994. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 152163-175. [PubMed] [Google Scholar]
- 51.Precopio, M. L., M. R. Betts, J. Parrino, D. A. Price, E. Gostick, D. R. Ambrozak, T. E. Asher, D. C. Douek, A. Harari, G. Pantaleo, R. Bailer, B. S. Graham, M. Roederer, and R. A. Koup. 2007. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8+ T cell responses. J. Exp. Med. 2041405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price, D. A., J. M. Brenchley, L. E. Ruff, M. R. Betts, B. J. Hill, M. Roederer, R. A. Koup, S. A. Migueles, E. Gostick, L. Wooldridge, A. K. Sewell, M. Connors, and D. C. Douek. 2005. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 2021349-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quarleri, J. F., A. Rubio, M. Carobene, G. Turk, M. Vignoles, R. P. Harrigan, J. S. Montaner, H. Salomon, and M. Gomez-Carrillo. 2004. HIV type 1 BF recombinant strains exhibit different pol gene mosaic patterns: descriptive analysis from 284 patients under treatment failure. AIDS Res. Hum. Retrovir. 201100-1107. [DOI] [PubMed] [Google Scholar]
- 54.Rodes, B., C. Toro, E. Paxinos, E. Poveda, M. Martinez-Padial, J. M. Benito, V. Jimenez, T. Wrin, S. Bassani, and V. Soriano. 2004. Differences in disease progression in a cohort of long-term non-progressors after more than 16 years of HIV-1 infection. AIDS 181109-1116. [DOI] [PubMed] [Google Scholar]
- 55.Sacha, J. B., C. Chung, E. G. Rakasz, S. P. Spencer, A. K. Jonas, A. T. Bean, W. Lee, B. J. Burwitz, J. J. Stephany, J. T. Loffredo, D. B. Allison, S. Adnan, A. Hoji, N. A. Wilson, T. C. Friedrich, J. D. Lifson, O. O. Yang, and D. I. Watkins. 2007. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J. Immunol. 1782746-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sadagopal, S., R. R. Amara, D. C. Montefiori, L. S. Wyatt, S. I. Staprans, N. L. Kozyr, H. M. McClure, B. Moss, and H. L. Robinson. 2005. Signature for long-term vaccine-mediated control of a simian and human immunodeficiency virus 89.6P challenge: stable low-breadth and low-frequency T-cell response capable of coproducing gamma interferon and interleukin-2. J. Virol. 793243-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stephens, H. A. 2005. HIV-1 diversity versus HLA class I polymorphism. Trends Immunol. 2641-47. [DOI] [PubMed] [Google Scholar]
- 58.Yu, X. G., M. M. Addo, B. A. Perkins, F. Wej, A. Rathod, S. C. Geer, M. Parta, D. Cohen, D. R. Stone, C. J. Russell, G. Tanzi, S. Mei, A. G. Wurcel, N. Frahm, M. Lichterfeld, L. Heath, J. I. Mullins, F. Marincola, P. J. Goulder, C. Brander, T. Allen, Y. Cao, B. D. Walker, and M. Altfeld. 2004. Differences in the expressed HLA class I alleles effect the differential clustering of HIV type 1-specific T cell responses in infected Chinese and Caucasians. AIDS Res. Hum. Retrovir. 20557-564. [DOI] [PubMed] [Google Scholar]
- 59.Zhao, S., S. Zhai, Y. Zhuang, S. Wang, D. Huang, W. Kang, X. Li, X. G. Yu, B. D. Walker, M. A. Altfeld, and Y. Sun. 2007. Inter-clade cross-reactivity of HIV-1-specific T cell responses in human immunodeficiency virus type 1 infection in China. Curr. HIV Res. 5251-259. [DOI] [PubMed] [Google Scholar]
- 60.Zimmerli, S. C., A. Harari, C. Cellerai, F. Vallelian, P. A. Bart, and G. Pantaleo. 2005. HIV-1-specific IFN-gamma/IL-2-secreting CD8 T cells support CD4-independent proliferation of HIV-1-specific CD8 T cells. Proc. Natl. Acad. Sci. USA 1027239-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuñiga, R., A. Lucchetti, P. Galvan, S. Sanchez, C. Sanchez, A. Hernandez, H. Sanchez, N. Frahm, C. H. Linde, H. S. Hewitt, W. Hildebrand, M. Altfeld, T. M. Allen, B. D. Walker, B. T. Korber, T. Leitner, J. Sanchez, and C. Brander. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 803122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]