Abstract
Small interfering RNAs (siRNAs) have been shown to effectively inhibit human immunodeficiency virus type 1 (HIV-1) replication in vitro. The mechanism(s) for this inhibition is poorly understood, as siRNAs may interact with multiple HIV-1 RNA species during different steps of the retroviral life cycle. To define susceptible HIV-1 RNA species, siRNAs were first designed to specifically inhibit two divergent primary HIV-1 isolates via env and gag gene targets. A self-inactivating lentiviral vector harboring these target sequences confirmed that siRNA cannot degrade incoming genomic RNA. Disruption of the incoming core structure by rhesus macaque TRIM5α did, however, provide siRNA-RNA-induced silencing complex access to HIV-1 genomic RNA and promoted degradation. In the absence of accelerated core disruption, only newly transcribed HIV-1 mRNA in the cytoplasm is sensitive to siRNA degradation. Inhibitors of HIV-1 mRNA nuclear export, such as leptomycin B and camptothecin, blocked siRNA restriction. All HIV-1 RNA regions and transcripts found 5′ of the target sequence, including multiply spliced HIV-1 RNA, were degraded by unidirectional 3′-to-5′ siRNA amplification and spreading. In contrast, HIV-1 RNA 3′ of the target sequence was not susceptible to siRNA. Even in the presence of siRNA, full-length HIV-1 RNA is still encapsidated into newly assembled viruses. These findings suggest that siRNA can target only a relatively “naked” cytoplasmic HIV-1 RNA despite the involvement of viral RNA at nearly every step in the retroviral life cycle. Protection of HIV-1 RNA within the core following virus entry, during encapsidation/virus assembly, or within the nucleus may reflect virus evolution in response to siRNA, TRIM5α, or other host restriction factors.
Even with formidable obstacles in delivery, there is high enthusiasm to develop RNA interference (RNAi)/small interfering RNA (siRNA) as an anti-human immunodeficiency virus (HIV) therapeutic strategy (11, 13, 27, 33, 41, 47). RNAi is a process of sequence-specific gene silencing initiated by the cleavage of double-stranded RNA (dsRNA) segments, common to many RNA viruses, into siRNAs by the cellular endonuclease Dicer. These siRNAs are incorporated into an RNase-containing RNA-induced silencing complex (RISC) of approximately 360 kDa and then target a complementary sequence in mRNA (10, 17, 18, 22, 25, 26, 29, 30, 45, 56). Synthetic siRNAs, introduced via transfection, can override the need for Dicer, target specific sequences, induce RNase degradation of mRNA, and in the case of HIV-specific siRNAs, inhibit HIV type 1 (HIV-1) replication (13, 20, 27, 41). Alternatively, constitutive expression of short 21- to 23-nucleotide (nt) transcripts or short hairpin RNAs from DNA plasmids can also invoke this process and inhibit a targeted gene or virus (27, 32). Once siRNA-RISC targets and degrades an RNA sequence, the siRNA can act as a primer for RNA-dependent RNA synthesis. This results in the generation of new dsRNA templates for Dicer and subsequent 3′-to-5′ multimerization of RNA degradation with new siRNA species (55).
Initial reports suggested that siRNAs can inhibit HIV-1 infection by specifically degrading transcribed HIV-1 mRNA as well as the incoming genomic RNA prior to integration (11, 27). However, these observations have been challenged by studies describing protection of genomic HIV-1 RNA from siRNA degradation (54). The exact or major siRNA target for HIV-1 inhibition has yet to be fully characterized, and the attempt to identify it poses a considerable challenge, considering the different HIV-1 mRNA and genomic RNA isoforms coexist during multiple phases of the HIV-1 life cycle. In addition to possibly degrading incoming genomic RNA, siRNA may impose a transcriptional block (9, 39) or degrade viral mRNA and/or nascent genomic RNA, thereby inhibiting HIV-1 gene expression and packaging into new virus particles (Fig. 1A). Aside from understanding the complex processing of dsRNA into siRNAs, understanding the mechanism(s) of siRNA inhibition of HIV-1 replication will be important for potential therapeutic use of siRNA.
FIG. 1.
Mapping the potentially siRNA degradation-susceptible HIV-1 RNA species during the retroviral life cycle. As illustrated in panel A, siRNA complexed to RISC may interfere with several stages of the HIV-1 life cycle to inhibit virus replication: siRNA may inhibit following virus entry by targeting HIV-1 genomic RNA (1); siRNA may inhibit transcription by directing chromatin modification (2); siRNA may degrade viral mRNA in the nucleus (3); siRNA may degrade viral mRNA following export from the nucleus to the cytoplasm (4); siRNA (through an miRNA form) may reduce translation of viral proteins (5); siRNA may interfere with the packaging of viral genomic RNA into a new virus particle or siRNA-RISC could be incorporated into the budding virus (6). PIC, pre-integration complex. Panel B delineates the sequence and target sites of the seven siRNAs designed to specifically inhibit HIV-1 v120-A (a subtype A virus) or v126-D (a subtype D virus). The locations and sequence complementarity of each siRNA in the HIV-1 env or gag genes are depicted in panel B. The starting nucleotide position (HXB2 numbering) is provided for each siRNA.
To address the mechanism(s) involved in siRNA inhibition of the HIV-1 replication, we compared the specific degradation of HIV-1 RNA by siRNA treatment during various RNA-related steps in the retroviral life cycle. These analyses involved the monitoring and modulation of HIV-1 genomic RNA prior to integration through the use of pseudotyped viruses containing a green fluorescent protein (GFP) reporter. Newly transcribed HIV-1 RNA and the GFP RNA were linked in the viral RNA genome but an independent transcriptional promoter drove expression of the GFP RNA and not the target HIV-1 RNA sequence in the infected cell. By use of this system, incoming genomic RNA appeared resistant to siRNA degradation unless the viral core was disrupted via a TRIM5α-mediated mechanism. Newly transcribed RNA from the HIV-1 long terminal repeat (LTR) was sensitive to siRNA degradation following release from the nucleus and prior to encapsidation on the inner plasma membrane. These findings suggest a very narrow window of siRNA-mediated degradation of HIV-1 mRNA following release from the nuclease and prior to encapsidation/virus assembly.
MATERIALS AND METHODS
Cell culture.
Peripheral blood mononuclear cells were isolated from blood samples from HIV-1-seronegative donors by Ficoll-Paque density centrifugation and cultured in a RPMI 1640 medium with 10% fetal bovine serum as described previously (19). The U87.CD4.CXCR4 cell line (obtained from the AIDS Research and Reference Reagent Program) and 293T cells (American Type Culture Collection) were grown in Dulbecco's modified Eagle's medium (DMEM; Cellgro) with 15% and 10% fetal bovine serum, respectively. All cells were grown at 37°C in 5% CO2.
Viruses.
The CXCR4-using primary HIV-1 isolates,v120-A (clade A) and v126-D (clade D) have been described previously (19). The gag, reverse transcriptase (RT) gene, and env sequences are available in GenBank. Tissue culture doses for 50% infectivity (TCID50) were determined using the Reed-Munch assay as previously described (2). Titers are expressed as infectious units per milliliter.
siRNA preparation.
Twenty-one-nucleotide dsRNAs were chemically synthesized as 2′ bis(acetoxyethoxy)-methyl ether-protected, desalted, and duplexed oligonucleotides by Dharmacon (Lafayette, CO). Seven siRNAs were designed according to the manufacturer's recommendations. siRNA120a, siRNA120b, siRNA120c, and siRNA120d specifically target upstream of env gene in v120-A; siRNA126a and siRNA126b specifically target downstream of env gene in v126-D; and siRNAgag1 specifically target gag gene in v120-A. See Fig. 1B for general locations.
Chemicals.
Both leptomycin B and camptothecin were purchased from Sigma, and lamivudine (3TC) was obtained from the NIH AIDS Reagent Repository. Camptothecin and 3TC were dissolved in dimethyl sulfoxide and prepared at appropriate concentration for use. The purchased leptomycin B was in a 7:3 mixture of methanol and water and ready for use.
Construction of self-inactivating lentiviral vectors.
The self-inactivating lentiviral vector pMND-GFPpre contains an internal MND promoter (a modified myeloproliferative sarcoma virus promoter) for GFP, the central polypurine tract/central termination sequence, and HIV-1 LTR with U3 deleted and a cytomegalovirus promoter added at the 5′ LTR (21). The env genes of v120-A and v126-D were PCR amplified and subcloned into the pMND.GFP vector to generate pGFPenv120-A and pGFPenv126-D (see below). Pseudotyped viruses were produced as described by Zielske et al. (57). Briefly, 293T cells were triple transfected with the packaging vector (pCMVΔR8.91), the vesicular stomatitis virus G protein (VSV-G) pseudotyping vector (pMD.G), and pMND.GFPpre, pGFPenv120-A, or pGFPenv126-D at a mass ratio of 3:1:3 by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). vGFP, vGFP-env120-A, or vGFP-env126-D virus was harvested after 24 to 48 h posttransfection and filtered (0.45-μm filter; Millipore) and stored at −80°C. Titers ranged from 6 × 106 to 3 × 107 expression units/ml.
To construct the full-length chimeric molecular clones pNL4-3/env120-A and pNL4-3/env126-D, the env fragments from v120-A and v126-D were PCR amplified and cloned into pRECenv/URA3 using homologous recombination of Saccharomyces cerevisiae (yeast) as previously described (35, 38). The env fragment (EcoRI-XhoI) from pRECenv120-A or pRECenv126-D was then subcloned into pNL4-3. The chimeric molecular clones were transfected into 293T cells to produce infectious viruses.
Primer sequences for semiquantitative RT-PCR and real-time PCR.
Primer sequences were as follows: Kpn1-env (sense), 5′-TGTGGGTCACAGTCTATTATGG (nt 6325 to 6346, HXB2 numbering); E80R1 (antisense), 5′-CACAATAATGTATGGGAATTGG (nt 6879 to 6858); E15, 5′-CTTGCTCTCCACCTTCTTCTTC (nt 8445 to 8424); E80 (sense), 5′-CCAATTCCCATACATTATTGTG (nt 6858 to 6879); E125 (antisense), 5′-CAATTTCTGGGTCCCCTCCTGAGG (nt 7338 to 7316); univ-GA4, 5′-TTGCCAAAGAGTGACCTGAGGGAA (nt 2273 to 2250); univ-GS2 (sense), 5′-GGGGGGACATCAAGCAGCCATGC (nt 1362 to 1384); GAD4 (antisense), 5′-CCACATTTCCAACAGCC (nt 2023 to 2039); LTR2, 5′-TCCCCCTGGCCTTAACCGAAT (nt 864 to 844); SUNS-8 (sense), 5′-TGACTCTGGTAACTAGAGATCCCTC (nt 576 to 600); LTR4 (antisense), 5′-GGGTAAGCTTTCCTTCTAGCCTCCGCTAGT (nt 794 to 765); SMS-7 (sense), 5′-CTTAGGCATCTCCTATGGCAGGAA (nt 5956 to 5979); LTR B CAT, 5′-GCACTCAAGGCAAGCTTTATTGAGGC (nt 547 to 522); LTR1-YG (sense), 5′-TGGAAGGGCTAATTTACTC (nt 1 to 19); β-actin-1 (sense), 5′-GCTCGTCGTCGACAACGGCTC; β-actin-2 (antisense), 5′-CAAACATGATCTGGGTCATCTTCTC; real-time LTR1 primer (sense), 5′-GCCTCAATAAAGCTTGCCTTGA (nt 522 to 543); real-time LTR2 primer (antisense), 5′-CTGAGGGATCTCTAGTTACCAGAGTCA (nt 602 to 576).
RPAs.
For probe preparation, the HIV-1 RNA template transcription plasmid pHIV-PBS was constructed by cloning a 947-bp BglII-PstI fragment containing both the primer binding sequence and repeat (R and U5) regions of the 5′ LTR (positions +473 to +1420 of pHXB2 cloned viral DNA) into the pSP72 transcription vector (Promega) (4). pHIV-PBS was linearized with BglII to yield a transcript of 250 nt (HIV primer binding sequence RNA template) under the control of an sp6 promoter which was used as a probe to detect the unspliced RNA. A human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) housekeeping template was purchased from BD, prepared according the provided protocol, and used as an internal control for RNase protection assay (RPA) detection for HIV-1 RNA.
Total RNAs from the pNL4-3/env120-A transfected cells with or without siRNA treatment were isolated using the RNeasy kit (Qiagen) and were used in the RPA. The RPA was performed with the BD RiboQuant RPA kit using the HIV RNA probe according to the manufacturer's instructions. Briefly, 20 μg of RNA was hybridized with a 32P-labeled RNA probe (3.6 × 105 cpm/sample) prepared using the RiboQuant in vitro transcription kit (BD Biosciences-Pharmingen). After hybridization, the samples were treated with RNases A and T1, and the RNases were inactivated by using a proteinase K mixture (390 μl of proteinase K buffer, 30 μl of proteinase K, and 15 μg of yeast tRNA/20 samples). Then, the samples were precipitated at −70°C for 30 min by 4 M ammonium acetate and ethanol and centrifuged at 14,000 × g at 4°C for 15 min. The pellets were suspended in 5 μl of sample buffer and subjected to 5% polyacrylamide gel electrophoresis as recommended by the manufacturer. Autoradiographs were obtained using high-performance autoradiographic film at −70°C for 4 days.
Production of stable TRIM5αrh-expressing cells and infections.
pLPCX-TRIM5αrh was obtained from the AIDS Research and Reference Reagent Program. This plasmid was transfected into a murine packaging cell line to produce the defective Moloney murine leukemia virus particles which would then deliver the rhesus macaque TRIM5α (TRIM5αrh) and puromycin resistance genes into 293T cells upon infection. The infected 293T cells were selected with puromycin-containing Dulbecco's modified Eagle's medium (see above) during seven days of culturing and then diluted to one cell per well of a 96-well plate for subsequent clonal selection. Each clonal cell line was then infected with vGFP-env120-A or vGFP-env126-D virus (see above) to measure the level of restriction. Two clones (clones 6 and 10) were selected based on a >90% reduction in vGFP-env120-A in 293TTRIM5α cells compared to that in 293T cells. In the experiment examining siRNA restriction of incoming HIV-1 RNA in 293TTRIM5α cells compared to that in 293T cells, cells were first treated with siRNAs (as described above) and then exposed to virus at a multiplicity of infection (MOI) of 2. Cells were washed at 2 h after virus exposure, and then GFP expression was detected by flow cytometry (BD LSR I instrument) at 48 h.
Detection of siRNA inhibition on primary and chimeric HIV-1 replication.
U87.CD4.CXCR4 cells were plated in 24-well plates (1.0 × 105/well) and, 24 h later, transfected with siRNA120, siRNA126, or siRNAgag1 by using Lipofectamine 2000 (Invitrogen) at various concentrations (see Fig. 2) or treated with 3TC at a concentration of 10 μM. The cells were then infected by v120-A or v126-D at an MOI of 0.001 after 16 h. Virus was removed 5 h postinfection, and cells were washed with phosphate-buffered saline three times; the wells were then refilled with fresh media. Supernatant and/or cells were collected at time points (see Fig. 2 and 3) for further detections. 293T cells (1 × 107) were plated in a 100-mm dish, cotransfected with 15 μg of pNL4-3/env120-A or pNL4-3/env126-D and 20 nM of siRNA120a, siRNA126a, or control siRNA (siRNAcon) by using Lipofectamine 2000 (Invitrogen), and then washed and resuspended in fresh media. Supernatant and cells were harvested at day 2 and day 5 posttransfection for further analyses. RT activity in the supernatant was monitored to determine relative virus production (36). For self-inactivating virus vector infection, 293T cells were plated in a 24-well plate (2 × 105/well) treated with siRNAs at 24 h (see above) and then transduced by pseudotyped viruses of vGFP-env120-A or vGFP-env126-D with an MOI of 2 with 6 μg/ml Polybrene. Cells were washed at 2 h after virus exposure, and then GFP expression was detected by flow cytometry (BD LSR I) at 48 h.
FIG. 2.
Efficiency and specificity of siRNA-mediated inhibition of divergent HIV-1 isolates. Panel A provides the drug susceptibility curves after 5 days of v120-A (V120-UG, where UG is Uganda) replication in U87.CD4.CXCR4 cells in the presence of various concentrations (0.0002 to 20 nM) of siRNA120a, siRNA120b, siRNA120c, and siRNA120d (transfected by Lipofectamine 2000). (B) v126-D replication in U87.CD4.CXCR4 cells treated with siRNA126a and siRNA126b. (C) The specificity of siRNA120a was measured in U87.CD4.CXCR4 cells exposed to v120-A and v126-D (V126-UG). (D) Finally, the decay of antiviral activity of siRNA120a in U87.CD4.CXCR4 cells exposed to v120-A was determined over a 5- to 8-day period. Each infection and siRNA treatment were performed in triplicate; in order to improve the clarity of panels A and B, error bars of ∼10% variance are not shown.
FIG. 3.
Effect of specific siRNAs on viral RNA and cDNA synthesis. (A) v120-A RNA and proviral DNA were measured by env-specific RT-PCR and PCR (respectively) in siRNA-treated U87.CD4.CXCR4 cells at various time points after infection and treatment with siRNAcontrol (siRNAcon), 3TC, siRNA120a, and siRNAgag1. PCR amplifications of undiluted and diluted nucleic acid extracts from infected cells are shown on the left of panel A, and quantitation of these PCR products is shown on the right. The production of HIV-1 DNA following these infections was also measured by real-time PCR specific for the 5′ LTR region (panel B). Panel C provides the relative production of v120-A under these various conditions as measured by RT activity released in the supernatant at different time points postinfection. All experiments were performed in triplicate. Since variance was less than 10% for relative virus production and siRNA inhibition, RT-PCR amplifications were performed only on a set of serially diluted RNA samples.
Monitoring HIV-1 DNA reverse transcripts and mRNA transcripts in the presence of siRNA.
v120-A-infected U87.CD4.CXCR4 cells treated with siRNA120a, siRNAgag1, 3TC, or siRNAcon were harvested at various time points postinfection. HIV-1 proviral DNA was extracted by use of the QIAamp DNA blood mini kit (Qiagen), diluted, and subjected to semiquantitative PCR using primer pair Kpn1-env/E80R1 to amplify an env fragment harboring the siRNA120a target region. Total RNA was also extracted from lysates of transfected or infected cells at days 2 and 5 by using the RNeasy kit and Qiashredder spin columns (Qiagen). Viral RNA was converted into cDNA by using Moloney murine leukemia virus RT (Invitrogen) and PCR amplified. RT primer/PCR primer pairs (listed in parentheses) were employed to detect envelope C1-V2 RNA (E15 and Kpn1-env/E80R1), envelope C2-V3 RNA (E15 and E80-E125), gag (univGA4 and univ-GS2-GAD4), 5′ LTR HIV-1 RNA (LTR2 and SUNS8-LTR4), multiply spliced mRNA (E15 and SMS7-E15), and 3′ LTR HIV-1 RNA (LTRBCAT and LTR1YG-LTRBCAT). The cDNA of β-actin was synthesized with oligo(dT) (Invitrogen) and PCR amplified with β-actin-1/β-actin-2. For semiquantitative PCR, the cDNA templates were diluted 1:10, 1:100, 1:1,000, and 1:10,000. Real-time PCR detection of cDNA following reverse transcription of various HIV-1 RNA species was performed in triplicate with an ABI Prism 7000 sequence detection system using Power SYBR green reagents (ABI) according to the provided protocol. RT primer/PCR primer pairs (listed in parentheses) were employed to detect gag (univGA4 and Sk39-GAG6), 5′ LTR HIV-1 RNA (LTR2 and SUNS8-LTR4), and 3′ LTR HIV-1 RNA (LTRBCAT and LTR1YG-ENV N).
Real-time PCR was also performed to measure the level of HIV-1 DNA. These real-time PCR amplifications were performed in triplicate with an ABI Prism 7000 sequence detection system using a specific probe (nt 572 to 545; 5′-AACAGACGGGCACACACTACTTGAAGCA) containing a reporter dye at the 5′ end and a quencher dye at the 3′ end and using a primer pair (real-time LTR1/real-time LTR2) (52). The RPA was performed as described in the manufacturer's protocol.
Detection of HIV-1 protein by Western blotting.
siRNA knockdown of HIV-1 proteins was detected by Western blotting. Blotting analyses were performed on 293T cells treated with siRNA and transfected with pNL4-3/env120-A or pNL4-3/env126-D as well as on virus pelleted from supernatants. Virus-containing supernatants were clarified of cellular debris by centrifugation (2,500 rpm for 15 min), and virions were pelleted by centrifugation at 32,000 rpm for 1 h. Samples were resuspended in sodium dodecyl sulfate (SDS) lysis buffer (40 mM Tris-HCl [pH 6.8], 10% glycerol, 10% β-mercaptoethanol, 1% SDS) and heated to 95°C for 5 min, separated by SDS-10% polyacrylamide gel electrophoresis, and transferred to nitrocellulose membranes. Membranes were blocked with 5% milk and incubated with primary antibody. Envelope protein was detected using the B13 antibody (provided by George Lewis, Institute of Human Virology, Baltimore, MD, and Bruce Chesebro, NIAID, Hamilton, MT). Total viral proteins were detected using a 1:400 dilution of patient sera derived from individuals infected with HIV-1 from either subtype A (JLT02A) or subtype C (JCR12). Total protein loading was controlled by detection of actin (Santa Cruz Biotechnology, Santa Cruz, CA). Membranes were incubated in horseradish peroxidase-conjugated secondary antibodies, detected with the ECL Plus kit (Amersham Biosciences, Piscataway, NJ), and exposed to X-ray film.
Virus infectivity.
Progeny virus collected from v120-A-infected, siRNA120a-treated U87.CD4.CXCR4 cells was measured for TCID50, in comparison to the values for virus produced from untreated cells as previously described (2). Titer-equilibrated samples were used to infect untreated U87.CD4.CXCR4 cells in a second round. The supernatant was harvested at 2, 5, and 10 days postinfection, and output of siRNA-treated progeny virus was measured by RT activity.
RESULTS
Evaluation of HIV-1-specific siRNA.
To test the efficiency and specificity of siRNA inhibition on HIV-1 replication, we designed seven siRNAs targeting sequences in specific HIV-1 isolates. v120-A and v126-D virus strains were derived from treatment-naïve, HIV-1-infected pediatric patients in Kampala, Uganda, in 1996 (Fig. 1B) (19). Two siRNAs specific for the gp41 coding region targeted the v126-D virus (subtype D) (Fig. 1B), whereas five siRNAs were designed to target the C1 region of env or the MA region of gag (Fig. 1B). Inhibitory activity of all siRNAs was tested in U87.CD4.CXCR4 cells with replication competent, primary HIV-1 isolates (i.e., v120-A or v126-D) by measuring virus production in the supernatant using radioactive RT activity (2). siRNA120a, siRNA120d, siRNA126a, and siRNA126b (Fig. 1B) were specific for either v120-A or v126-D and could not cross-inhibit due to 4- to 6-nt differences in the target sequences (Fig. 1B). When siRNAs were transfected into U87.CD4.CXCR4 cells at 16 h prior to infection, we found that siRNA120a inhibited v120-A replication with the greatest efficiency, siRNA120b and siRNA120c were moderately efficient, and siRNA120d lacked significant inhibitory activity (Fig. 2A). Similarly, siRNA126a showed greater potency against v126-D than did siRNA126b (Fig. 2B). siRNAgag1 showed strong inhibition for v120-A (Fig. 3A) but not for v126-D (data not shown). Optimal siRNA inhibition was observed at five days postinfection and then decayed such that minimal siRNA inhibition was observed at day 8 postinfection (Fig. 2D).
siRNA inhibition has been demonstrated to be sequence specific and, as a result, virus specific. Our data are consistent with this previous observation: siRNA with a single-nucleotide mismatch from the target sequence reduced its potency, and four mismatched nucleotides result in a 100-fold reduction of inhibition efficiency. As shown in Fig. 2C, we observed that v120-A was susceptible to siRNA120a inhibition (50% inhibitory concentration, 0.16 ± 0.02 nM), but only a 50% reduction of v126-D production was evident with even 20 nM siRNA120a. Similar isolate-specific inhibition was observed, with siRNA126a and siRNA126b targeting a v126-D env sequence and with siRNA120gag1 targeting a v120-A gag sequence (data not shown).
siRNA influence on HIV-1 reverse transcription and general transcription.
The use of siRNA to inhibit HIV-1 has been considered a therapeutic option because of many factors, including the high dependence of HIV-1 on RNA throughout the replication cycle. siRNAs have the potential to degrade incoming genomic RNA and newly transcribed mRNA, to inhibit translation, and to possibly disrupt HIV-1 genome encapsidation. It has been reported that HIV-1 RNA released into the cell following virus-cell membrane fusion is sensitive to siRNA-mediated degradation (11, 27, 41), but others suggest that incoming genomic RNA is not accessible to siRNA as part of the RISC (24, 53, 54). Using semiquantitative RT-PCR for the HIV-1 env C1-C2, we detected similar amounts of viral RNA in the presence or absence of siRNA120a or siRNAgag1 at 2 h after virus exposure of U87.CD4.CXCR4 cells (MOI, 0.1) (Fig. 3A). RNA of v120-A was not modulated by the control siRNA, siRNA126a. However, production of viral RNA was substantially reduced with siRNA treatment at 36 h postinfection (up to 91% by siRNA120a and 93% by siRNAgag1) (Fig. 3A), providing an unclear picture of the susceptibility of incoming viral genomic RNA to siRNA-mediated cleavage. However, treatment of the cells with siRNA120a or siRNAgag1 prior to infection had no effect on proviral DNA synthesis (detected by real-time PCR) until 60 h postinfection, whereas 3TC, which blocks reverse transcription on the incoming RNA genome, had an immediate inhibitory effect (Fig. 3A and B). From this experiment, it appears that the ability of siRNA to inhibit proviral DNA levels at 60 h postinfection is likely related to the block in HIV-1 mRNA synthesis (as observed at 36 h) (Fig. 3A) from the integrated genome and a subsequent prevention of virus spread in the culture (Fig. 3C).
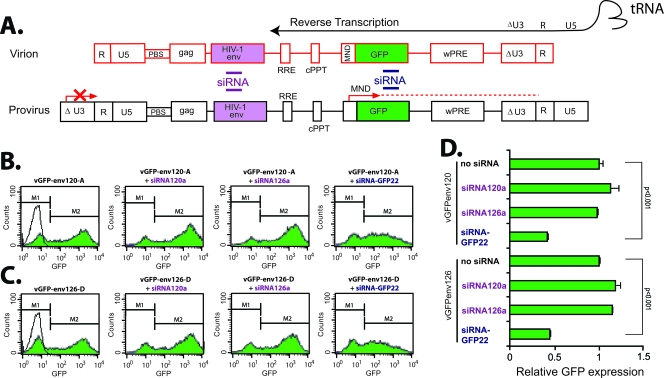
To exclusively investigate whether siRNA can degrade HIV-1 genomic RNA, we developed a system that can specifically map siRNA inhibition to degradation of the incoming HIV-1 RNA genome as opposed to de novo HIV-1 mRNA synthesis. An env fragment from HIV-1 v120-A or v126-D (containing the target region of siRNA120a or siRNA126a) was cloned into the self-inactivating lentiviral vector pMND-GFPpre, which also contains an internal MND promoter driving GFP expression (Fig. 4A) (57). Virus produced from the 293T packaging cells (vGFP-env120-A or vGFP-env126-D) can integrate into the host chromosome in newly infected cells but lacks a functional U3 promoter/enhancer and thus cannot drive lentiviral mRNA transcription or express the siRNA-targeting RNA. However, the GFP mRNA is successfully transcribed through the independent and internal MND promoter. Thus, GFP expression, as measured by flow cytometry, can be inhibited only if the siRNA120a or siRNA126a degrades the incoming genomic RNA and, as a result, blocks reverse transcription and subsequent proviral DNA integration (Fig. 4A). Here, we showed that 20 nM of siRNA120a or siRNA126a did not decrease GFP expression in 293T cells infected with VSV envelope-pseudotyped virus (containing env siRNA target sequence). However, GFP expression in these cells was significantly inhibited by siRNA-GFP22 targeting GFP mRNA (Fig. 4B, C, and D). It is important to note that 20 nM of siRNA120a or siRNA126a resulted in >90% inhibition of the respective v120-A and v126-D HIV-1 isolates (Fig. 2). These data suggest that incoming genomic HIV-1 RNA cannot be degraded by siRNA, and it suggests that the viral core and possible stearic hindrance is responsible for this resistance.
FIG. 4.
Determining the sensitivity of incoming genomic HIV-1 RNA to siRNA degradation. (A) The env gene of v120-A or v126-D was inserted into the self-inactivating lentiviral vector pMND-GFPpre, which contains an internal MND promoter driving GFP, the central polypurine tract (cPPT)/central termination sequence, and the HIV-1 LTR with U3 deleted and cytomegalovirus promoter added at the 5′ LTR. PBS, primer binding sequence; wPRE, Woodchuck hepatitis virus posttranscriptional regulatory element. The virus particles derived from triple transfections of 293T cells with the packaging vector (pCMVΔR8.91), the VSV-G-pseudotyping vector (pMD.G), and pMND.GFPpre, pGFPenv120-A (containing the env target sequence of v120-D), or pGFPenv126-D (containing the env target sequence of v126-D). The viral RNA in these virus particles can be reverse transcribed, and the proviral DNA can be integrated into the target 293T cells. However, the LTR in the integrated virus genome cannot drive transcription and thus does not express viral RNA with the env target sequence. An internal promoter in the integrated virus genome can transcribe an mRNA encoding enhanced GFP. Eight graphs describe the flow cytometry results gated for 293T cells expressing GFP (M2) from the vGFP-env120-A (B) and vGFP-env126-D (C) infections in the presence or absence of siRNA120a or siRNA126a. Quantitative results from this experiment are shown in panel D. All experiments were performed in triplicate.
It is important to note that the VSV-G-pseudotyped viruses enter the cell through an endocytic pathway followed by a low-pH-triggered virus-endosome membrane fusion to release the HIV-1 core. However, there are no significant delays in reverse transcription and the other steps in the retroviral life cycle by this pathway of virus entry as opposed to virus-cell membrane fusion mediated by HIV-1 env/host cell receptors. In addition, subsequent experiments confirm that the virus core is impervious to siRNA-RISC regardless of the method of virus entry and core expulsion into the cytoplasm.
Accessing siRNA degradation of genomic HIV-1 RNA following disruption of the virus core.
In order to investigate the possible protection of the incoming HIV-1 RNA from siRNA degradation by the virus core, we attempted to disrupt the HIV-1 core structure following virus entry into the host cell. TRIM5αrh has been shown to inhibit HIV-1 replication, to target the viral capsid (46, 49), and to possibly disrupt the incoming virus cores (50). We transfected 293T cells with pLPCX-TRIM5αrh and then used 1 μg/ml of puromycin and selected for a clonal cell line stably expressing high levels of TRIM5αrh (293TTRIM5αrh) and efficiently inhibited >90% infection by HIV-1 pseudotyped with VSV G protein (data not shown).
siRNA120a treatment of 293TTRIM5αrh cells exposed to the VSV-pseudotyped virus of vGFP-env120-A did not result in an additive or synergistic effect on virus inhibition mediated by TRIM5αrh (>90% inhibition in the absence of siRNA) (Fig. 5A). To investigate the fate of the HIV-1 RNA following virus entry and possible core disruption by TRIM5α, we reverse transcribed and PCR amplified an env fragment of the viral RNA in vGFP-env120-A that was targeted by siRNA120a. Significant reductions in the amount of viral RNA in the 293TTRIM5α cells at 2 and 8 h postinfection compared to the amount of viral RNA in 293T cells treated with siRNA120a (P < 0.01) (Fig. 5C) were observed. Furthermore, we observed a significant enhancement of siRNA120a-mediated degradation of HIV-1 RNA in the cells with TRIM5αrh compared with the degradation in the nonspecific siRNA126a- or mock-treated cells (Fig. 5C). Again, in the absence of TRIM5α, the incoming, encapsidated viral RNA was not degraded by siRNA (Fig. 5B). Thus, viral genomic RNA can be degraded by siRNAs when the core is disrupted by the TRIM5α process. Typically, the viral RNA is inaccessible to siRNA-RISC during the normal process of core expulsion into the cytoplasm and subsequent reverse transcription following host cell entry.
FIG. 5.
siRNA degradation of genomic HIV-1 RNA following disruption of the virus cores by TRIM5αrh. lipo, Lipofectamine 2000. (A) VSV envelope-pseudotyped virus vGFP-env120-A was used to infect 293T or 293TTRIM5αrh cells in the presence or absence of 20 nM siRNA120a or siRNA126a. GFP-expressing cells were measured by flow cytometry. Semiquantitative RT-PCR was performed on extracted RNA from the cells exposed to vGFP-env120-A with or without siRNAs. Panels B and C show the quantified results from the experiments using the 293T or 293TTRIM5αrh cells, performed in triplicate. Panel D displays the relative amounts of HIV env mRNA that were RT-PCR amplified at 8 h and 24 h or of HIV-1 env DNA that were PCR amplified at 36 h and 48 h after virus addition.
It is important to note that the relative amount of incoming HIV-1 RNA in 293T cells was nearly equivalent at 2 and 8 h after virus exposure (Fig. 5B), but in 293TTRIM5α cells (Fig. 5C), TRIM5αrh-mediated degradation was ∼90% at 2 h but only 40% at 8 h. Nonetheless, the remaining HIV-1 RNA in the cores exposed to TRIM5αrh was nearly completely degraded by the specific siRNA120a but not by siRNA126a (Fig. 5C). Although TRIM5αrh was not directly mediating RNA degradation, TRIM5αrh must still bind to the virus core and disrupt core structure, since the siRNA-RISC could mediate >95% degradation of HIV-1 RNA, compared to <10% degradation in the absence of TRIM5αrh (Fig. 5, compare panels B and C). HIV-1 RNA was degraded within 24 h in the absence of TRIM5αrh and siRNA, suggesting that HIV-1 RNA template for reverse transcription is likely undergoing an RNase H-mediated degradation via RT (as well as RNA degradation-mediating cellular nucleases) (Fig. 5C).
Finally, we examined whether a disruption of reverse transcription by TRIM5αrh, rather than accelerated core dissolution, was the primary mechanism responsible for siRNA-mediated degradation of incoming HIV-1 RNA. For this experiment, 293T cells were treated with siRNA120a or siRNA126a (as a control) with or without 3TC prior to infection with virus vGFP-env120-A. At 8 h, we observed similar levels of HIV-1 RNA (by RT-PCR) in the presence or absence of 3TC and specific siRNA120a or nonspecific siRNA126a (Fig. 5D). HIV-1 DNA that was PCR amplified at 36 and 48 h postinfection clearly indicated that 3TC was inhibiting proviral DNA synthesis and that siRNA120a was not having an additive effect (Fig. 5D). We were unable to detect GFP expression or viral RNA in the presence of 3TC beyond 24 h postinfection (data not shown).
Degradation of newly transcribed HIV-1 mRNA degradation by siRNA.
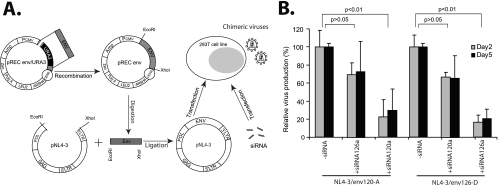
Once the HIV-1 genomic cDNA has been integrated into the host chromosome, the newly transcribed viral mRNA species, as well as the unspliced genomic RNA, can be potential targets for RNAi. To determine the effects of siRNA on provirally derived HIV-1 RNA transcripts, the full-length env genes of v120-A and v126-D (i.e., containing the siRNA target sequences) were cloned into the NL4-3 backbone through a yeast-based recombination/HIV-1 cloning system (Fig. 6A) (35, 38). The chimeric infectious molecular clones (e.g., pNL4-3/env120-A or pNL4-3/env126-D) were then transfected into 293T cells treated with siRNA120a or siRNA126a. On day 2 posttransfection, NL4-3/env120-A and NL4-3/env126-D virus production (derived from transfected DNA) by siRNA120a and by siRNA126a reached 77.7% and 83.7%, respectively. In contrast, virus production was not significantly inhibited by the heterologous siRNA (e.g., 30% inhibition of NL4-3/env126-D by siRNA120a) (Fig. 6B).
FIG. 6.
siRNA inhibition of HIV-1 transcription for cells transfected with proviral DNA vectors. Panel A provides a schematic representation of the general cloning strategy to replace the env gene in pNL4-3 with the env genes of v120-A and v126-D. The resulting plasmids were then transfected in 293T cells in the presence or absence of siRNAs. Amp, ampicillin resistance cassette; PCMV, cytomegalovirus promoter; Zeo, Zeocin resistance cassette. (B) The relative virus production as measured by RT activity was monitored in supernatants at days 2 and 5 after transfection and siRNA treatment.
Six sets of HIV-1-specific primers were then used to reverse transcribe and PCR amplify specific HIV-1 RNA regions representing different HIV-1 mRNA species: the RU5 region or CA-NC gag genes for genomic RNA, the env C1-C2 or env C2-V3 region for singly spliced as well as unspliced genomic RNA, the first Tat exon through the second Tat exon for multiply spliced RNA, and the U3R region for all the HIV-1 mRNA species (Fig. 7A). These sets of primers also provide the relative degradation of HIV-1 genomic regions at various distances upstream and downstream of the initial target sequence. With RT-PCR specific for the target region (C1-C2), a >90% loss in signal suggests that the siRNA120a can mediate degradation of both singly spliced and full-length HIV-1 mRNA.
FIG. 7.
siRNA degradation on different HIV-1 mRNA species in the presence or absence of inhibitors of nuclear mRNA export. Panel A provides a schematic illustration of the location of the RT-PCR primers in the HIV-1 genome and in specific HIV-1 unspliced and multiply spliced mRNA species. Images of the agarose gels containing the various amplified HIV-1 RNA products are shown in panel B. RNA for these RT-PCRs was extracted from pNL4-3/env120-A-transfected 293T cells exposed to siRNA120a in the presence or absence of inhibitors of the CRM-1 nuclear export pathway (leptomycin B [LepB] and camptothecin [Camp]). The relative levels of these various HIV-1 RNA messages (measured by RT-PCR as described for panels A and B) in the absence of siRNAs (set to 100%) were compared to levels in the presence of siRNA and with no inhibitor of nuclear export (C), leptomycin B (D), or camptothecin (E). All experiments whose results are shown in panels A through E were performed in triplicate. Since variance was less than 10% for relative virus production and siRNA inhibition, RT-PCR amplifications were performed only on a set of serially diluted RNA samples. Panel F provides a comparison of HIV-1 RNA levels determined by using semiquantitative PCR and real-time PCR methods (SYBR green). (G) The levels of HIV-1 RNA were determined by real-time PCR for each replicate in this triplicate experiment. The levels of HIV-1 RNA were measured in the absence of nuclear export inhibitor, with leptomycin B, or with camptothecin by using the RNA protection method described in Materials and Methods. Two probes specific for HIV-1 RU5 (unspliced RNA) or U3R products were used in this RPA. semi-quant, semiquantitative.
RT-PCR amplification of the MA gag region (∼4,300 nt upstream of the target sequence or 5′ of the target in the HIV-1 RNA) and the RU5 LTR (∼5,600 nt) indicated a 85% and 87% reduction (respectively) by siRNA120a (Fig. 7B and C). Multimerization of siRNA from the target sequence in the 3′-to-5′ direction (in HIV-1 genomic RNA) also efficiently degrades 97% of the multiply spliced RNA (∼370 nt upstream of target). In contrast, we observed less degradation (71%) of the HIV-1 C2-V3 env region located just ∼420 nt downstream of the target sequence (Fig. 7B and C). The U3R region at the extreme 3′ end of the HIV-1 genome was resistant to siRNA120a-mediated cleavage and was located ∼2,600 nt downstream of the C1 env target sequence. To confirm observations derived from semiquantitative RT-PCR, HIV-1 RU5 and U3R RNAs (extreme ends of the RNA genome) levels were measured using both quantitative real-time RT-PCR and RPAs. Both methods confirmed limited RNA degradation ∼2,600 nt downstream (U3R) compared to ∼5,600 nt upstream (RU5) of the siRNA120a target sequence (Fig. 7C).
We have previously observed that the “semiquantitative” PCR method is in fact quite accurate and sensitive (5, 6) as well as being more flexible in terms of primer/probe design than the real-time PCR technique. Due to the diversity among the subtype A and D env genes and our standard real-time PCR primers based on subtype B, we first utilized the semiquantitative PCR. However, the levels of unspliced (5′ LTR), gag, and 3′ LTR mRNA was confirmed by utilizing the same cDNA for both semiquantitative and real-time PCRs (Fig. 7F). Using both PCR methods, we observed greater than 80% reduction of unspliced and gag HIV-1 mRNA but no significant reduction of 3′ LTR HIV-1 RNA. This again confirmed the multimerization of siRNA from the target sequence in the 3′-to-5′ direction (in HIV-1 genomic RNA).
siRNA degradation occurs in cytoplasm.
Preliminary data suggest that the incoming encapsidated HIV-1 RNA is resistant to siRNA degradation, whereas newly transcribed HIV-1 RNA appears sensitive and mediates virus inhibition. Slow spliceosome formation on HIV-1 hnRNA will eventually process mRNA into multiply spliced messages and transport it out of the nucleus for translation of various accessory proteins (e.g., Tat and Rev proteins) (Fig. 1A) (40). The Rev proteins are transported back to the nucleus, bind to the Rev-responsive element (RRE), and then rescue unspliced and singly spliced mRNA by interfering with spliceosome formation and mediating rapid nuclear export through the CRM-1 pathway. Leptomycin B (10 ng/ml) and camptothecin (100 mM) were used to block the nuclear export of HIV-1 mRNA and to determine the relative siRNA sensitivity of nuclear and cytoplasmic HIV-1 mRNAs. The various HIV-1 RNA transcripts (described above) were detected by RT-PCR in the leptomycin B- or camptothecin-treated 293T cells cotransfected with pNL4-3/env120-A and siRNA120a. Both camptothecin and leptomycin B counteracted most of the siRNA120a-mediated degradation of all the HIV-1 RNA messages (Fig. 7D, E, and F). Again, this observation was confirmed by using RPAs specific for RU5 and U3R regions, i.e., there was a lack of siRNA-mediated degradation when the HIV-1 mRNA was trapped in the nucleus. These studies also provide some support to the hypothesis that the majority of HIV-1 RNA must be rapidly exported to the cytoplasm due to the sensitivity of HIV-1 RNA to siRNA degradation in the absence of nuclear export inhibitors and its relative protection from siRNA when trapped in the nucleus.
Inhibition of HIV-1 protein production by siRNA.
Inhibition of subsequent virus protein production was assessed by Western blot detection of total viral proteins. Expression of p24, p41, and p51 of NL4-3/env120-A in 293T cells were inhibited by 72%, 87%, and 64.3%, respectively, by siRNA120a on day 5 posttransfection with pNL4-3/env120-A. Similar results were obtained for pNL4-3/env126-D transfections treated with siRNA126a (p24, p41, and p51 were inhibited by 69%, 92.5%, and 80.6%, respectively) (Fig. 8C). Production of virion-associated gp120 in NL4-3/env120-A was reduced by siRNA120a by 82% compared with the control (in the absence of siRNA), while virion-associated p24 was inhibited by 72% (Fig. 8A and B). These results are consistent with the reduction in virus production by siRNAs as measured by RT activity in the cell-free supernatant (Fig. 6B).
FIG. 8.
Measuring HIV-1 protein production in cells and supernatant in the presence of siRNAs. CA p24 or unprocessed p55gag was monitored by Western blotting for cells (C) and supernatants (A and B), at days 2 and 5, of 293T cells transfected with pNL4-3/env120-A or pNL4-3/env126-D and treated with siRNA120a or siRNA126a or mock treated. Both panels A and C employed a 1:400 dilution of patient sera derived from individuals infected with HIV-1 of either subtype A (JLT02A) or subtype C (JCR12) as a primary anti-p24 antibody, followed by peroxidase-conjugated secondary antibodies. (C) The anti-gp120 B13 antibody was employed in Western blot analyses with virus lysates obtained from day 5 supernatants of 293T cells transfected with pNL4-3/env and mock treated or treated with siRNA120a or siRNA126a. The B13 anti-gp120 antibody binds to a highly conserved linear epitope found in most HIV-1 gp120 sequences of most subtypes (panel B) (1)
Interestingly, the expressions of p24, p41, p51, and gp120 of NL4-3/env120-A were also inhibited by siRNA126a up to 45%, 46%, 42%, and 45%, respectively. It is important to note that 20 nM siRNA120a did not appreciably affect v126-D HIV-1 mRNA levels as measured by RT-PCR but did inhibit v126-D replication by 50% (Fig. 2C). These findings suggest that mismatched siRNA could still function as microRNA (miRNA) and mediate inhibition of HIV-1 replication as opposed to cleavage of the mRNA.
Influence of siRNA on progeny HIV virions.
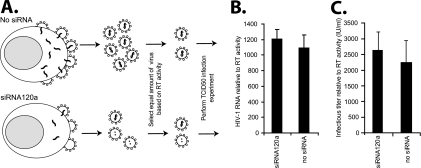
Finally, we determined whether siRNA activity could affect HIV-1 RNA packaging and decrease infectivity of virus progeny. Virus v120-A was collected from infected U87.CD4.CXCR4 cells treated with or without siRNA120a (20 nM). Virus titers were determined by measuring RT activity in the supernatant and using a TCID50 assay (2, 35). We suspect that siRNA did not alter the virus structure but could have partially/fully degraded the HIV-1 genomic RNA in the budding virus (Fig. 9A). In addition, there is a possibility that the siRNA in association with RISC could be packaged in the virion. Using real-time RT-PCR, we estimated the amount of HIV-1 RNA (RU5) in the virus particles relative to RT activity. siRNA treatment did not significantly affect the relative encapsidation of HIV-1 RNA. Virus produced in the presence or absence of siRNA treatment (equalized for RT activity, i.e., “virtual” TCID50 values) was then added to U87.CD4.CXCR4 cells in a limiting dilution for infection assay (or actual TCID50 assay) (Fig. 9A). Again, the ratios of virus particles (as measured by RT activity) to infectious virions (as measured by TCID50) for virus produced in the presence of siRNA and in the absence of siRNA were identical (Fig. 9B and C). These findings suggest that HIV-1 RNA is protected from siRNA-mediated degradation during the process of virus assembly.
FIG. 9.
siRNA effects on packaging of genomic HIV-1 RNA and on the infectivity of newly produced virus. Panel A provides a schematic of virus production in U87.CD4.CXCR4 cells in the presence and absence of siRNAs. Although siRNAs inhibit 80 to 90% of virus production, the v120-A released in the supernatant in the presence or absence of siRNA120a was serially diluted and quantified by RT activity to determine a “virtual” TCID50 (36). (B) Equalized infectious titers (based on RT activity) of virus (with or without siRNA) were again serially diluted and used to infect U87.CD4.CXCR4 cells in the absence of siRNA to determine if siRNA have any inhibitory effects on infectivity of newly produced virus. The HIV-1 RNA levels in virus particles equalized for RT activity following production from cells infected in the presence or absence of siRNA. HIV-1 RNA levels were measured with real-time PCR using an ABI Prism 7000 sequence detection system. All conditions were performed in triplicate.
DISCUSSION
In this study, we explored the mechanism(s) by which siRNA inhibits HIV-1 replication. The sensitivity of each HIV-1 RNA species to siRNA degradation was measured during its timed appearance in the retroviral life cycle. In order to replicate, HIV-1 requires unspliced or genomic RNA as well as singly spliced and multiply spliced mRNAs amounting to over 40 different viral mRNA species derived from HIV-1 hnRNA transcript. Approximately half of the HIV-1 RNA transcripts are not spliced and serve as mRNA and as genomic RNA in the cytoplasm (48). In our study, encapsidated HIV-1 genomic RNA, expelled after virus-cell membrane fusion, was completely resistant to siRNA activity unless the viral core was disrupted by an extrinsic mechanism, e.g., binding of core proteins by TRIM5αrh. In absence of this artificial disruption, only the newly transcribed HIV-1 RNA (except 3′ LTR) from the integrated proviral DNA was sensitive to siRNA degradation but only after the mRNA was exported out of the nucleus. Furthermore, this cytoplasmic HIV-1 RNA was labile only to siRNA degradation prior to encapsidation. Preliminary data in our laboratory suggest that disruption of the interaction between Gag proteins and HIV-1 RNA through mutations in nucleocapsid (NC) (16, 43) will increase sensitivity of the mRNA to siRNA-mediated digestion (data not shown). As previously described, siRNA inhibition was highly sequence specific and even a 4-nt mismatch between siRNA and a primary HIV-1 isolate can abolish inhibition. However, in some cases, we found that even with this mismatch there is still limited inhibition, which may suggest a miRNA process. miRNAs are incorporated into a RISC-like complex and, depending on their degree of complementarity to the target mRNA, elicit either translational repression or mRNA cleavage (8, 23).
The inability of HIV-specific siRNA to inhibit incoming HIV-1 RNA may be related to three mechanisms: (i) stearic hindrance based on the occlusion of RISC by the core structure, (ii) resistance of HIV-1 genomic RNA to siRNA degradation, or (iii) the possibility that the HIV-1 core never encounters the cellular compartment harboring the active RNAi machinery (3). Potential resistance of genomic RNA is excluded based on the degradation of HIV-1 mRNA by siRNA following de novo transcription and release from the nucleus. Stearic hindrance preventing penetration by RISC is favored, considering the fact that a disruption of the HIV-1 core by TRIM5αrh did augment siRNA degradation of HIV-1 RNA. This finding also suggests that the virus genome is not protected beyond the core and that the dissolved core is located within the appropriate cellular compartment or at least in an area accessible to siRNA-RISC.
Our findings also point to a direct HIV-1 RNA target rather than a modification to proviral DNA during translocation of the preintegration complex to the nucleus. dsRNA and proteins of the RNAi machinery can direct modification to homologous DNA sequences to induce transcriptional gene silencing or, in extreme cases, DNA elimination (9, 37). However, these studies have been performed mostly with plants and fungi, and in research with mammals, studies have reached conflicting conclusions. Two studies indicated that siRNAs that target the promoters of endogenous genes can induce transcriptional silencing in human cells (28, 39), while others have shown that long dsRNA that knocked down expression of a target gene by RNAi in mouse oocytes did not induce de novo methylation of the corresponding DNA region. Our data here support the conclusion that siRNA cannot interfere with proviral DNA synthesis and block transcription. Nevertheless, further work is required to determine the generality of these results. Even if dsRNA or synthetic siRNAs do not normally trigger RNA-dependent DNA methylation in mammals, other noncoding and antisense RNAs have been implicated in chromatin-based regulation (37).
Previous work with Caenorhabditis elegans and plants suggested that RNAi can be amplified and the targeted region spread to neighboring sequences. This phenomenon was termed “transitive” RNAi (12) or siRNA amplification, in which an RNA-dependent RNA polymerase uses the guide strand of siRNA as a primer on the target mRNA to initiate an RNA polymerization reaction, generating further distinct dsRNA substrates for Dicer and thus more siRNAs to degrade in the 3′-to-5′ direction on the RNA (26, 34). In our study, even though multiply spliced RNA does not contain the target region of siRNA120a, it is the most susceptible HIV-1 message to siRNA120a degradation, suggesting that new siRNAs are generated in the 3′-to-5′ direction. Our study also showed that siRNA-directed RNA degradation is toward the regions 5′ or upstream but not 3′ or downstream of the target sequence.
Nuclear export of HIV-1 RNA requires binding by the viral regulatory protein Rev to the structured RRE within the HIV-1 env gene. Rev multimerized on the RRE also engages the CRM-1 pathway for nuclear export (48). Leptomycin B, an unsaturated, branched-chain fatty acid, is a specific inhibitor of the CRM-1 pathway for nuclear export. In addition to inhibiting DNA topoisomerase I, camptothecin blocks the CRM-1/exportin I pathway that results in a retention of Rev in the nucleus and a block in the export of unspliced HIV-1 mRNA out of the nucleus. In this study, we found that both leptomycin B and camptothecin can effectively suppress siRNA degradation of all HIV-1 mRNA, suggesting that active RNAi machinery exists in the cytoplasm but does not access the nucleus or is nonfunctional in that compartment. Multiply spliced HIV-1 mRNA, as well as all HIV-1 RNA species (except 3′ LTR RNA), is efficiently degraded by siRNA120a treatment in the absence of CRM-1 inhibitors. However, these CRM-1 inhibitors do not block the nuclear export of multiply spliced HIV-1 mRNA. In this case, the target in HIV-1 RNA is trapped in the nucleus such that siRNA amplification cannot be initiated and, as a result, the multiply spliced HIV-1 mRNA cannot be degraded by siRNA120a.
Taken together, our results show that siRNAs can target the newly transcribed HIV-1 genomic RNA but not the incoming HIV-1 genomic RNA, suggesting that this protection from siRNA degradation is related to viral capsid. During coevolution of pathogen and host, various RNA viruses, including retroviruses, have developed a tightly regulated process for genome replication in the face of endogenous nucleases, such as those directed to RNA cleavage by siRNA. In response to this pressure, RNA viruses have encapsidated or protected their genomes with least one protein layer. In HIV-1, these protein layers comprising the core are restructured into complexes necessary for reverse transcription and proviral DNA transport but remain impervious to siRNA or RNase cleavage. Similar to this slow “unwrapping” process following virus entry, the newly transcribed HIV-1 genomic RNA is thought to be wrapped up or encapsidated by the Gag precursor proteins being translated in cis and, as result, protected from siRNA activity. siRNA-RISC might have access to HIV-1 mRNA only after release from the nucleus and prior to ribosome association/translation. With other RNA viruses, such as alphavirus and picornovirus families, protection during virus assembly may be mediated by the formation of intracellular “factories” for virus assembly and the exclusion of nonessential host proteins (31, 42). Finally, reoviruses may represent the extreme for protection against siRNA, since only single-stranded RNA messages are transcribed out of the core, while the dsRNA genomes are always maintained within a well-organized and protective core (7, 14). Again, the host may have evolved to counter this protective capsid by localizing the siRNA machinery to the nuclear pore and polyribosome sites and to gain access to the most labile form of viral RNA. The RNAi machinery is reported to be associated with the translational factor eukaryotic initiation factor 2c, suggesting that ribosomal association might play a role in RNAi (15, 44, 51). We are now exploring the question of whether uncapped HIV-1 RNA (lacking a 5′ methyl cap) may be highly sensitive to siRNA degradation (not shown) due to a reduced ability to associate with the ribosomes. We are also examining whether specific mutations in the Gag precursor protein can disrupt binding to genomic HIV-1 RNA and as result, enhance sensitivity to siRNA degradation during virus assembly. Preliminary results suggest only a minimal increase in siRNA degradation of HIV-1 RNA with viruses harboring nucleocapsid mutations that reduce viral genomic RNA packaging (43). As observed with our experiments with core dissolution following virus entry, slight perturbations of virus core structure may still be insufficient for siRNA-RISC to gain access to the viral RNA genome.
Acknowledgments
This work was supported by NIH grants AI436402, AI49170, and AI46283.
Clinical samples were processed by the Ugandan Laboratory Core, whereas all virus work was performed in the biosafety level 2 and 3 facilities of the Case Western Reserve/University Hospitals Center for AIDS Research (AI36219).
Footnotes
Published ahead of print on 16 January 2008.
REFERENCES
- 1.Abacioglu, Y. H., T. R. Fouts, J. D. Laman, E. Claassen, S. H. Pincus, J. P. Moore, C. A. Roby, R. Kamin-Lewis, and G. K. Lewis. 1994. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retrovir. 10371-381. [DOI] [PubMed] [Google Scholar]
- 2.Abraha, A., R. M. Troyer, M. E. Quinones-Mateu, and E. J. Arts. 2005. Methods to determine HIV-1 ex vivo fitness. Methods Mol. Biol. 304355-368. [DOI] [PubMed] [Google Scholar]
- 3.Arhel, N. J., S. Souquere-Besse, S. Munier, P. Souque, S. Guadagnini, S. Rutherford, M. C. Prevost, T. D. Allen, and P. Charneau. 2007. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 263025-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arts, E. J., X. Li, Z. Gu, L. Kleiman, M. A. Parniak, and M. A. Wainberg. 1994. Comparison of deoxyoligonucleotide and tRNA(Lys-3) as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction. J. Biol. Chem. 26914672-14680. [PubMed] [Google Scholar]
- 5.Arts, E. J., J. Mak, L. Kleiman, and M. A. Wainberg. 1994. DNA found in human immunodeficiency virus type 1 particles may not be required for infectivity. J. Gen. Virol. 751605-1613. [DOI] [PubMed] [Google Scholar]
- 6.Arts, E. J., J. P. Marois, Z. Gu, S. F. Le Grice, and M. A. Wainberg. 1996. Effects of 3′-deoxynucleoside 5′-triphosphate concentrations on chain termination by nucleoside analogs during human immunodeficiency virus type 1 reverse transcription of minus-strand strong-stop DNA. J. Virol. 70712-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bamford, D. H. 2002. Those magnificent molecular machines: logistics in dsRNA virus transcription. EMBO Rep. 3317-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. [DOI] [PubMed] [Google Scholar]
- 9.Bayne, E. H., and R. C. Allshire. 2005. RNA-directed transcriptional gene silencing in mammals. Trends Genet. 21370-373. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409363-366. [DOI] [PubMed] [Google Scholar]
- 11.Capodici, J., K. Kariko, and D. Weissman. 2002. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 1695196-5201. [DOI] [PubMed] [Google Scholar]
- 12.Cerutti, H. 2003. RNA interference: traveling in the cell and gaining functions? Trends Genet. 1939-46. [DOI] [PubMed] [Google Scholar]
- 13.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 769225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diprose, J. M., J. N. Burroughs, G. C. Sutton, A. Goldsmith, P. Gouet, R. Malby, I. Overton, S. Zientara, P. P. Mertens, D. I. Stuart, and J. M. Grimes. 2001. Translocation portals for the substrates and products of a viral transcription complex: the bluetongue virus core. EMBO J. 207229-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doi, N., S. Zenno, R. Ueda, H. Ohki-Hamazaki, K. Ui-Tei, and K. Saigo. 2003. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 1341-46. [DOI] [PubMed] [Google Scholar]
- 16.Dorfman, T., J. Luban, S. P. Goff, W. A. Haseltine, and H. G. Gottlinger. 1993. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 676159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411494-498. [DOI] [PubMed] [Google Scholar]
- 18.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391806-811. [DOI] [PubMed] [Google Scholar]
- 19.Gao, Y., E. Paxinos, J. Galovich, R. Troyer, H. Baird, M. Abreha, C. Kityo, P. Mugyenyi, C. Petropoulos, and E. J. Arts. 2004. Characterization of a subtype D human immunodeficiency virus type 1 isolate that was obtained from an untreated individual and that is highly resistant to nonnucleoside reverse transcriptase inhibitors. J. Virol. 785390-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gitlin, L., and R. Andino. 2003. Nucleic acid-based immune system: the antiviral potential of mammalian RNA silencing. J. Virol. 777159-7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halene, S., L. Wang, R. M. Cooper, D. C. Bockstoce, P. B. Robbins, and D. B. Kohn. 1999. Improved expression in hematopoietic and lymphoid cells in mice after transplantation of bone marrow transduced with a modified retroviral vector. Blood 943349-3357. [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond, S. M., A. A. Caudy, and G. J. Hannon. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2110-119. [DOI] [PubMed] [Google Scholar]
- 23.Hannon, G. J., and J. J. Rossi. 2004. Unlocking the potential of the human genome with RNA interference. Nature 431371-378. [DOI] [PubMed] [Google Scholar]
- 24.Hu, W. Y., C. P. Myers, J. M. Kilzer, S. L. Pfaff, and F. D. Bushman. 2002. Inhibition of retroviral pathogenesis by RNA interference. Curr. Biol. 121301-1311. [DOI] [PubMed] [Google Scholar]
- 25.Hutvágner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293834-838. [DOI] [PubMed] [Google Scholar]
- 26.Hutvágner, G., and P. D. Zamore. 2002. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 12225-232. [DOI] [PubMed] [Google Scholar]
- 27.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawasaki, H., and K. Taira. 2004. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431211-217. [DOI] [PubMed] [Google Scholar]
- 29.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 152654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knight, S. W., and B. L. Bass. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 2932269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20500-505. [DOI] [PubMed] [Google Scholar]
- 33.Lee, N. S., and J. J. Rossi. 2004. Control of HIV-1 replication by RNA interference. Virus Res. 10253-58. [DOI] [PubMed] [Google Scholar]
- 34.Lipardi, C., Q. Wei, and B. M. Paterson. 2001. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107297-307. [DOI] [PubMed] [Google Scholar]
- 35.Marozsan, A. J., and E. J. Arts. 2003. Development of a yeast-based recombination cloning/system for the analysis of gene products from diverse human immunodeficiency virus type 1 isolates. J. Virol. Methods 111111-120. [DOI] [PubMed] [Google Scholar]
- 36.Marozsan, A. J., E. Fraundorf, A. Abraha, H. Baird, D. Moore, R. Troyer, I. Nankja, and E. J. Arts. 2004. Relationships between infectious titer, capsid protein levels, and reverse transcriptase activities of diverse human immunodeficiency virus type 1 isolates. J. Virol. 7811130-11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matzke, M. A., and J. A. Birchler. 2005. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 624-35. [DOI] [PubMed] [Google Scholar]
- 38.Moore, D. M., E. J. Arts, Y. Gao, and A. J. Marozsan. 2005. A yeast recombination-based cloning system to produce chimeric HIV-1 viruses and express HIV-1 genes. Methods Mol. Biol. 304369-385. [DOI] [PubMed] [Google Scholar]
- 39.Morris, K. V., S. W. Chan, S. E. Jacobsen, and D. J. Looney. 2004. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 3051289-1292. [DOI] [PubMed] [Google Scholar]
- 40.Nekhai, S., and K. T. Jeang. 2006. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: role of cellular factors for Tat and Rev. Future Microbiol. 1417-426. [DOI] [PubMed] [Google Scholar]
- 41.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8681-686. [DOI] [PubMed] [Google Scholar]
- 42.Novoa, R. R., G. Calderita, R. Arranz, J. Fontana, H. Granzow, and C. Risco. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97147-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poon, D. T., J. Wu, and A. Aldovini. 1996. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 706607-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasaki, T., A. Shiohama, S. Minoshima, and N. Shimizu. 2003. Identification of eight members of the Argonaute family in the human genome small star, filled. Genomics 82323-330. [DOI] [PubMed] [Google Scholar]
- 45.Sharp, P. A. 2001. RNA interference—2001. Genes Dev. 15485-490. [DOI] [PubMed] [Google Scholar]
- 46.Shi, J., and C. Aiken. 2006. Saturation of TRIM5 alpha-mediated restriction of HIV-1 infection depends on the stability of the incoming viral capsid. Virology 350493-500. [DOI] [PubMed] [Google Scholar]
- 47.Song, E., P. Zhu, S. K. Lee, D. Chowdhury, S. Kussman, D. M. Dykxhoorn, Y. Feng, D. Palliser, D. B. Weiner, P. Shankar, W. A. Marasco, and J. Lieberman. 2005. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 23709-717. [DOI] [PubMed] [Google Scholar]
- 48.Stoltzfus, C. M., and J. M. Madsen. 2006. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr. HIV Res. 443-55. [DOI] [PubMed] [Google Scholar]
- 49.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427848-853. [DOI] [PubMed] [Google Scholar]
- 50.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. USA 1035514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99123-132. [DOI] [PubMed] [Google Scholar]
- 52.Toossi, Z., H. Mayanja-Kizza, J. Baseke, P. Peters, M. Wu, A. Abraha, H. Aung, A. Okwera, C. Hirsch, and E. Arts. 2005. Inhibition of human immunodeficiency virus-1 (HIV-1) by beta-chemokine analogues in mononuclear cells from HIV-1-infected patients with active tuberculosis. Clin. Exp. Immunol. 142327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterhouse, P. M., M. B. Wang, and T. Lough. 2001. Gene silencing as an adaptive defence against viruses. Nature 411834-842. [DOI] [PubMed] [Google Scholar]
- 54.Westerhout, E. M., O. ter Brake, and B. Berkhout. 2006. The virion-associated incoming HIV-1 RNA genome is not targeted by RNA interference. Retrovirology 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zamore, P. D. 2002. Ancient pathways programmed by small RNAs. Science 2961265-1269. [DOI] [PubMed] [Google Scholar]
- 56.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 10125-33. [DOI] [PubMed] [Google Scholar]
- 57.Zielske, S. P., and S. L. Gerson. 2002. Lentiviral transduction of P140K MGMT into human CD34(+) hematopoietic progenitors at low multiplicity of infection confers significant resistance to BG/BCNU and allows selection in vitro. Mol. Ther. 5381-387. [DOI] [PubMed] [Google Scholar]