Abstract
Herpesvirus lytic DNA replication requires both the cis-acting element, the origin, and trans-acting factors, including virally encoded origin-binding protein, DNA replication enzymes, and auxiliary factors. Two lytic DNA replication origins (ori-Lyt) of Kaposi's sarcoma-associated herpesvirus (KSHV) have been identified, and two virally encoded proteins, namely, RTA and K8, have been shown to bind to the origins. In this study, we sought to identify cellular factors that associate with ori-Lyt by using DNA affinity purification and mass spectrometry. This approach led to identification of several cellular proteins that bind to KSHV ori-Lyt. They include topoisomerases (Topo) I and II, MSH2/6, RecQL, poly(ADP-ribose) polymerase I (PARP-1), DNA-PK, Ku86/70 autoantigens, and scaffold attachment factor A (SAF-A). RecQL appears to associate with prereplication complexes and be recruited to ori-Lyt through RTA and K8. Topoisomerases, MSH2, PARP-1, DNA-PK, and Ku86 were not detected in prereplication complexes but were present in replication initiation complexes on ori-Lyt. All these cellular proteins accumulate in viral replication compartments in the nucleus, indicating that these proteins may have a role in viral replication. Topo I and II appear to be essential for viral DNA replication as inhibition of their activities with specific inhibitors (camptothecin and ellipticine) blocked ori-Lyt-dependent DNA replication. Furthermore, inhibition of PARP-1 with chemical inhibitors (3-aminobenzamide and niacinamide) resulted in decreased ori-Lyt-dependent DNA replication, whereas hydroxyurea, which raises PARP-1 activity, caused an increase in the DNA replication, suggesting a positive role for PARP-1 in KSHV lytic DNA replication.
Kaposi's sarcoma-associated herpesvirus (KSHV), also referred to as human herpesvirus 8, is the etiologic agent of several AIDS-associated malignancies, including Kaposi's sarcoma (KS), primary effusion lymphoma, and multicentric Castleman's disease (1, 17, 33). In KS lesions, most spindle cells of endothelial origin are latently infected with KSHV, and a small percentage of these cells undergo spontaneous lytic replication (34, 35, 43). Increasing evidence suggests that the small percentage of cells experiencing viral lytic replication play important roles in viral pathogenicity. Treatment of KS patients or AIDS patients at risk for KS with antiherpesviral drugs, like foscarnet and ganciclovir, which are known to block lytic DNA replication, results in regression of KS lesions or a decease in the incidence of KS development. The lytic replication cycle directly contributes to viral tumorigenesis by spreading viruses to target cells and by providing paracrine regulation for KS development (11). A recent study by Grundhoff and Ganem (19) suggests a new role for lytic replication in sustaining the population of latently infected cells that otherwise would be quickly lost by segregation of latent viral episomes as spindle cells divide. Thus, unlike with other oncogenic viruses, where the latent life cycle is primarily responsible for the oncogenic potential of the virus, KSHV lytic replication and constant primary infection of fresh cells are crucial for viral tumorigenicity and pathogenesis.
Lytic DNA replication of a herpesvirus initiates at an origin (ori-Lyt) and requires trans-acting elements. Two duplicated copies of lytic DNA replication origin, referred to as ori-Lyt (L) and ori-Lyt (R), have been identified in the KSHV genome in our laboratory and others (3, 25). These ori-Lyts are located in the KSHV genome between K4.2 and K5 and between K12 and ORF71, respectively. These two ori-Lyts share almost identical 1.1-kb core component sequences and 600-bp GC-rich repeats, which are represented as 20-bp and 30-bp tandem arrays (25).
Like that of other herpesviruses, KSHV lytic DNA replication relies on virus-encoded proteins (2, 38, 41). Six core replication proteins, including a DNA polymerase (POL), a polymerase processivity factor (PPF), a single-stranded DNA binding protein (SSB), and a trivalent helicase-primase complex (HEL, PRI, and PAP) have been identified in the KSHV genome. These core replication proteins are conserved among all herpesviruses (40, 41). In addition, two virally encoded regulatory proteins, namely, K8 and RTA, were found to bind to ori-Lyt (37) and shown to be absolutely required for KSHV ori-Lyt-dependent DNA replication (2). RTA binds to a consensus RTA responsive element (RRE) in KSHV ori-Lyt, while K8 associates with the ori-Lyt DNA through interaction with C/EBP α molecules bound on a cluster of C/EBP binding motifs (37, 42). Our recent work showed that the six core machinery proteins plus K8 and RTA form a prereplication complex independent of the presence of ori-Lyt DNA. The complex is recruited to ori-Lyt DNA through K8 and RTA, which bind to their binding motifs (38).
In addition to virally encoded replication enzymes and factors, it is believed that herpesviruses also utilize cellular proteins in their DNA replication. To search for the cellular proteins that play roles in KSHV DNA replication, we designed a DNA affinity purification procedure to isolate proteins that bind to KSHV ori-Lyt DNA fragments. This study led to the identification of several cellular replication, repair, and recombination factors, such as topoisomerases (Topo) I and II, MSH2/6, RecQL, DNA-PK, poly(ADP-ribose) polymerase 1 (PARP-1), and Ku autoantigens. These cellular proteins accumulate in viral replication compartments (VRCs) during viral DNA replication, suggesting their possible roles in KSHV replication. Additionally, we found that a nuclear scaffold/matrix protein (scaffold attachment factor A, or SAF-A) bound to the viral ori-Lyt DNA, suggesting that attachment of ori-Lyt DNA to the nuclear scaffold/matrix structure may be necessary for efficient viral DNA replication.
MATERIALS AND METHODS
Cell culture.
The primary effusion lymphoma cell line BCBL-1, which carries latent KSHV and was established by Ganem and his colleagues (30), was obtained from the NIH AIDS Research and Reference Reagent Program. The cells were grown in RPMI 1640 medium (Gibco-BRL, Gaithersburg, MD) supplemented with 10% fetal bovine serum (Gibco-BRL), penicillin-streptomycin (50 units/ml), and fungizone (1.25 μg/ml amphotericin B and 1.25 μg/ml sodium deoxycholate).
Plasmids and constructs.
Plasmids pOri-A and its mutants (pOri-Δ15.7, pOri-M12, pOri-M1256, etc.) were described previously (37). pCR3.1-ORF50 was constructed by cloning the cDNA sequence of the ORF50 coding region into the pCR3.1 vector (Invitrogen). The construct was described in Lin et al. (25).
DNA affinity purification and assay.
Various biotinylated DNA fragments were synthesized using PCR with pOri-A DNA or its mutants as templates and two oligonucleotides as primers. The two oligonucleotides for 3F and its derivative DNA fragments were 3F (5′-CGGCAAAGCTAATTTGCATG-3′) and Biotin-7R (5′-biotin-ACTGGAATAGGGGCTGCGATGACTC-3′). The oligonucleotides for 9F and its derivative DNAs were 9F (5′-CAATTCTATAATTAAACAAGGTAGAA-3′) and Biotin-ID13R (5′-biotin-CGCCACCGAACAACCCCGTGGACAG-3′). The oligonucleotides for 11F and its derivative DNAs were 11F (5′-TAGGGCCCGATGAGTCATGGGGTT-3′) and Biotin24280R (5′-biotin-ACGGGTAAATCCAAGAGATCCGTCCC-3′). The resultant biotinylated PCR fragments were coupled to streptavidin-conjugated magnetic beads (Dynal, Oslo, Norway) and then mixed with nuclear extracts prepared from tetradecanoyl phorbol acetate (TPA)-induced (and uninduced) BCBL-1 cells. In each reaction mixture, 2/3 volume of DNA-coupled beads in a solution [20 mM HEPES, pH 7.9, 20% glycerol, 0.2 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.05% NP-40, 15 mM MgCl2, 75 μg/ml salmon sperm DNA, and 0.2 A260 unit/ml poly(dI-dC)] was mixed with 1/3 volume of the nuclear extract in buffer C (20 mM HEPES, pH 7.9, 25% glycerol, 0.2 mM EDTA, 0.42 M NaCl, 1 mM DTT, 0.05% NP-40, 1 mM PMSF, and a protease inhibitor tablet) and incubated at 25°C for 45 min. The bound material was washed four times in D150 buffer (20 mM HEPES, pH 7.9, 20% glycerol, 0.2 mM EDTA, 150 mM KCl, 1 mM DTT, 0.05% NP-40, 1 mM PMSF) and then progressively eluted with D300 (same as D150, except the KCl concentration was increased to 300 mM), D500 (500 mM KCl), and D1000 (1 M KCl). The affinity-purified materials were subjected to mass spectrometric or Western blot analyses.
Mass spectrometric analysis.
DNA affinity-purified proteins were resolved on 4 to 12% bis-Tris NuPAGE gels (Invitrogen) and stained with a colloidal Coomassie G-250 staining kit (Invitrogen). The protein bands were excised and subjected to trypsin digestion. A portion of the peptide digest was injected onto a nanocapillary reverse-phase high-performance liquid chromatograph coupled to a nanoelectrospray ionization source of an ion trap mass spectrometer (ThermoFinnigan LCQ). This mass spectrometer measures peptide masses and then fragments individual peptides to produce tandem mass spectrometry (MS/MS) spectra of fragments that reflect the peptide sequence. The MS/MS spectra were run against a sequence database by using the program SEQUEST. The mass spectrometry was carried out in the protein microchemistry/mass spectrometry facility at the Wistar Institute.
Coimmunoprecipitation assay.
Approximately 5 × 107 cells were collected by centrifugation and washed with cold phosphate-buffered saline (PBS). The cells were suspended in 0.5 ml of immunoprecipitation buffer (120 mM potassium acetate, 20 mM Tris-acetate [pH 7.9], 5 mM EDTA, 1 mM DTT, 10% glycerol, 0.1% Nonidet P-40, and one complete protease inhibitor cocktail tablet per 25 ml), placed on ice for 30 min, and sonicated for 5 seconds. The cell lysates were clarified by centrifugation at 4°C for 10 min. Immunoprecipitation was performed by addition of 2 μl of monoclonal anti-RTA (a gift from Keiji Ueda at Osaka University, Japan) or monoclonal anti-K8 antibodies to the cell lysates (0.5 ml) with gentle agitation at 4°C for 60 min. Then, 100 μl of protein G-coated paramagnetic beads (Dynal Biotech, Oslo, Norway) was added and the mixtures were incubated at 4°C overnight. A reaction mixture with 2 μl of mouse immunoglobulin G (IgG; Sigma) was included as a control. The beads were then washed five times with cold immunoprecipitation buffer. The precipitates were resuspended in 100 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, boiled for 10 min, and loaded onto SDS-PAGE gels. To eliminate DNA-mediated protein interactions, ethidium bromide (EtBr) was added to aliquots of cell lysates and maintained at a concentration of 50 or 100 μg/ml throughout the entire immunoprecipitation process, including wash steps.
Western blot analysis.
DNA affinity-purified or immunoprecipitated materials were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked in 5% dried milk in 1× PBS plus 0.2% Tween 20 and then incubated with diluted primary antibodies for 2 h at room temperature or overnight at 4°C. Anti-rabbit or anti-mouse IgG antibodies conjugated to horseradish peroxidase (Amersham) were used as the secondary antibodies. An enhanced chemiluminescence system (Amersham) was used for detection of antibody-antigen complexes.
Chromatin immunoprecipitation (ChIP) assay.
BCBL-1 cells were treated with 20 ng of TPA/ml for 48 h. The cells were fixed by addition of 1% formaldehyde to the medium for 10 min and collected by centrifugation. After being washed with cold PBS, the cells (107) were suspended in 1 ml of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.0]) supplemented with 0.5 mM PMSF, 1 μg of pepstatin A/ml, and 1 μg of leupeptin/ml and left to sit on ice for 10 min. Then, the cells were sonicated eight times for 10 s each, and the lysates were cleared by centrifugation for 10 min. The cell lysates were diluted 10-fold with dilution buffer (0.01% SDS, 1.1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 167 mM NaCl). Chromatin solution (1 ml for each reaction) was incubated with 2 μl of specific antibodies against different viral and cellular proteins overnight at 4°C. Immune complexes were collected on protein A beads preadsorbed with sonicated single-stranded DNA. Beads were washed sequentially twice each in low-salt-concentration wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 150 mM NaCl), high-salt-concentration wash buffer (500 mM NaCl), LiCl wash buffer (0.25 mM LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0]), and Tris-EDTA buffer (pH 8.0). Immune complexes were eluted by incubation in 150 μl of Tris-EDTA buffer-1% SDS. Cross-links were reversed by heating at 65°C for 7 h up to overnight, followed by digestion with proteinase K (25 μg/ml) for 2 h at 50°C. DNA was extracted with phenol-chloroform-isoamyl alcohol and ethanol precipitated. PCR analyses of immunoprecipitated DNAs were carried out using three pairs of primers. The oligonucleotides 3F (5′-CGGCAAAGCTAATTTGCATG-3′; nucleotides 23129 to 23148) and K4.2P5 (5′-GATGGGCCAATGGCGGCTCG-3′; nucleotides 23405 to 23386) were used to detect the region between nucleotides 23129 and 23405 and the oligonucleotides 12F (5′-ACGGGCCTGGAATCTCGCCTCTGG-3′; nucleotides 24020 to 24043) and 12R (5′-ATGGGCGTAACCGTAGGACAAGCTG-3′; nucleotides 24155 to 24131) to detect the region between nucleotides 24020 and 24155 in the ori-Lyt. A PCR with oligonucleotides ORF45-RACE1 (5′-GGCGTCCATGGGATGGGTTAGTCAGGAT-3′; nucleotides 68097 to 68070) and ORF45-RACE2 (5′-ACGTCCGGAGAGTTGGAACTGTCATCGC-3′; nucleotides 67813 to 67840) was performed as a control.
Antibodies and IFA.
The primary antibodies used in the immunofluorescence assay (IFA) included anti-K8 monoclonal IgG (raised using purified GST-K8 α fusion protein; 1:300 dilution), anti-SSB polyclonal antibody (provided by Gary Hayward at Johns Hopkins; 1:300 dilution), anti-RecQL polyclonal antibody (provided by Perry Blackshear at NIEHS; 1:300 dilution), anti-SAF-A monoclonal antibody 3G6 (provided by Gideon Dreyfuss at the University of Pennsylvania; 1:300 dilution), anti-Ku86 monoclonal antibody (provided by Harris Busch at Baylor College of Medicine; 1:100 dilution), anti-Topo I monoclonal antibody (provided by Daniel Simmons at the University of Delaware; 1:100 dilution), anti-Topo IIβ polyclonal antibody (provided by Gary Gorbsky at the Oklahoma Medical Research Foundation; 1:100 dilution), anti-MSH2 monoclonal antibody (purchased from BD Biosciences, San Diego, CA; 1:50 dilution), anti-PARP-1 monoclonal antibody (purchased from Trevigen, Inc., Gaithersburg, MD; 1:200 dilution), and goat anti-bromodeoxyuridine (anti-BrdU) antibody (purchased from Capralogics Inc., Hardwick, MA; 1:100 dilution).
BCBL-1 cells were induced with TPA (20 ng/ml) for 48 h. To visualize replication-active DNA in the nucleus, cells were incubated in the medium containing 10 μM BrdU (Sigma) for 60 min. The cells were washed with PBS and fixed with 2% paraformaldehyde in PBS for 20 min at room temperature and then incubated with 2 N HCl for 30 min at room temperature in order to expose incorporated BrdU residues. After four washes with PBS, the cells were incubated with primary antibodies specific to each cellular protein, together with rabbit polyclonal anti-SSB or mouse monoclonal anti-K8 as well as goat anti-BrdU antibodies, at room temperature for 50 min, followed by incubation with a combination of secondary antibodies (fluorescein isothiocyanate-conjugated anti-rabbit IgG, Cy5-conjugated anti-mouse IgG [Rockland Immunochemicals, Inc., Gilbertsville, PA], and Texas Red-conjugated anti-goat IgG [Vector Laboratories, Inc.]). All secondary antibodies were used at a dilution of 1:250.
The slides were examined with a Leica TCS SPII confocal laser scanning system. Three channels were recorded simultaneously or sequentially. Data acquisition was controlled for possible breakthrough between the green and red channels and between the blue and red channels.
Transient-transfection DNA replication assay.
To transfect BCBL-1 cells, 5 μg of pOri-A or its mutant plasmids and 5 μg of pCR3.1-ORF50 (or the pCR3.1 vector) were mixed with 107 cells in OPTI-MEM medium (Gibco-BRL) and electroporated (200 V, 960 μF) with a GenePulser II apparatus (Bio-Rad, Hercules, CA). Electroporated cells were then transferred to RPMI 1640 medium supplemented with 10% serum. In the experiments of chemical treatment, transfected cells were cultured in media containing various inhibitors of topoisomerases and PARP-1 at the indicated concentrations for 72 h.
Extrachromosomal DNAs were prepared from cells by using the Hirt DNA extraction method as follows. Cells were lysed in 700 μl lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM EDTA, and 0.6% SDS). Chromosomal DNA was precipitated at 4°C overnight by adding 5 M NaCl to give a final concentration of 0.85 M. Cell lysates were centrifuged at 4°C at 14,000 rpm for 30 min. The supernatant containing extrachromosomal DNA was subjected to phenol-chloroform extraction, followed by ethanol precipitation. The DNA was treated with RNase A at 25°C for 30 min and then with proteinase K at 50°C for 30 min. DNA (5 μg) was digested with KpnI/SacI or KpnI/SacI/DpnI (New England Biolabs). The DNAs were separated by electrophoresis on 1% agarose gels and transferred onto GeneScreen membranes (Perkin Elmer, Boston, MA). The Southern blots were hybridized with 32P-labeled pBluescript plasmid in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2× Denhardt's solution, 1% SDS, and 50 μg/ml denatured salmon sperm DNA at 68°C.
RESULTS
Identification of proteins that are associated with KSHV ori-Lyt DNA.
To find proteins that bind to KSHV ori-Lyt DNA, we designed a DNA affinity purification procedure. Three overlapping DNA fragments, representing the core domain of KSHV ori-Lyt, were synthesized and labeled with biotin at their 5′ ends (Fig. 1A). The DNAs were coupled to streptavidin-conjugated magnetic beads and then incubated with a nuclear extract derived from TPA-induced BCBL-1 cells. The bound material was washed with 150 mM KCl-containing buffer and sequentially eluted with 300 mM, 500 mM, and 1 M KCl. The affinity-purified materials were resolved on 4 to 12% SDS-PAGE gels and stained with colloidal Coomassie blue G-250. Figure 1B shows the patterns of proteins enriched by the DNA affinity procedure with the three DNA fragments and eluted with 500 mM KCl. The DNA affinity purification was repeated many times, and the results were reproducible.
FIG. 1.
Identification of cellular proteins that bind to KSHV ori-Lyt. (A) DNA affinity purification of BCBL-1 nuclear extract was performed with three overlapping DNA fragments that represent the core domain of KSHV ori-Lyt. (B) The eluates with 500 mM KCl were separated on a 4 to 12% bis-Tris NuPAGE gel and then stained with colloidal Coomassie G-250. The molecular masses are indicated at the left in kilodaltons. Prominent bands from each lane were excised, digested in gel with trypsin, and subjected to liquid chromatography-MS/MS analysis. The resultant MS/MS spectra were run against a sequence database with the SEQUEST program. Matched proteins are indicated at the right of each lane. (C) The binding of some of the mass spectrometry-identified proteins to ori-Lyt DNA were confirmed by Western bolts with specific antibodies.
Prominent bands were excised from each lane and subjected to in-gel trypsin digestion. A portion of each peptide digest was injected onto a nanocapillary reverse-phase high-performance liquid chromatograph coupled to a nanoelectrospray ionization source of an ion trap mass spectrometer (ThermoFinnigan LCQ). Mass spectrometry measures peptide masses and then fragments individual peptides to produce liquid chromatography-MS/MS spectra of fragments that reflect the peptide sequence. The MS/MS spectra were run against a nonredundant database with the program SEQUEST. The identities of the proteins that were identified by this proteomic approach are indicated in Fig. 1B. The mass spectrometry spectra and the sequences of the corresponding peptides of each of the proteins are listed in Table 1.
TABLE 1.
Tryptic peptides of KSHV ori-Lyt-associated cellular proteins identified by mass spectrometry
| Protein | Swiss-Prot no. | No. of amino acids | No. of peptides | Positions and sequences of peptidesa |
|---|---|---|---|---|
| Actin type 5 | gi/1703123 | 376 | 12 | 20-29, AGFAGDDAPR; 30-38, AVFPSIVGR; 52-62, DSYVGDEAQSK; 86-96, IWHHTFYNELR; 97-114, VAPEEHPVLLTEAPLNPK; 198-207, GYSFTTTAER; 217-239, LCYVALDFEQEMATAASSSSLEK; 240-255, SYELPDGQVITIGNER; 258-285, CPEALFQPSFLGMESCGIHETTFNSIMK; 293-313, DLYANTVLSGGTTMYPGIADR; 317-327, EITALAPSTMK; 361-373, QEYDESGPSIVHR |
| APOBEC-3C | gi/22907039 | 190 | 8 | 11-22, AMYPGTFYFQFK; 31-43, NETWLCFTVEGIK; 57-69, NQVDSETHCHAER; 70-85, CFLSWFCDDILSPNTK; 111-122, HSNVNLTIFTAR; 123-136, LYYFQYPCYQEGLR; 137-154, SLSQEGVAVEIMDYEDFK; 155-172, YCWENFVYNDNEPFKPWK |
| DNA-PK | gi/13654237 | 4,128 | 80 | 338-350, LQYFMEQFYGIIR; 380-391, DVDFMYVELIQR; 394-406, QMFLTQTDTGDDR; 477-489, NCISTVVHQGLIR; 811-820, NNWEVSALSR; 839-852, NLSSNEAISLEEIR; 855-868, VVQMLGSLGGQINK; 869-881, NLLTVTSSDEMMK; 914-924, VTELALTASDR; 945-963, ATQMPEGGQGAPPMYQLYK; 1076-1087, LGASLAFNNIYR; 1120-1136, SLGTIQQCCDAIDHLCR; 1203-1209, SPNLWLK; 1275-1292, TVGALQVLGTEAQSSLLK; 1293-1311, AVAFFLESIAMHDIIAAEK; 1341-1357, IMEFTTTLLNTSPEGWK; 1423-1445, ITAQSIEELCAVNLYGPDAQVDR; 1461-1489, AGLLHNILPSQSTDLHHSVGTELLSLVYK; 1574-1591, NLDLAVLELMQSSVDNTK; 1592-1606, MVSAVLNGMLDQSFR; 1618-1627, LATTILQHWK; 1713-1727, VLEQLIVAHFPMQSR; 1769-1783, EQQHVM*EELFQSSFR; 1789-1806, GSCVTQVGLLESVYEMFR; 1858-1869, LNESTFDTQITK; 1896-1913, INQVFHGSCITEGNELTK; 1918-1936, LCYDAFTENMAGENQLLER; 1961-1973, FYQGFLFSEKPEK; 1988, 2000, YNFPVEVEVPMER; 2073-2090, DPTVHDDVLELEMDELNR; 2091-2102, HECMAPLTALVK; 2107-2120, SLGPPQGEEDSVPR; 2149-2162, LVINTEEVFRPYAK; 2334-2347, KNILEESLCELVAK; 2378-2388, FMNAVFFLLPK; 2405-2416, VEGMTELYFQLK; 2453-2470, ELLNPVVEFVSHPSTTCR; 2486-2500, DPESETDNDSQEIFK; 2539-2549, LLALNSLYSPK; 2599-2619, STVLTPMFVETQASQGTLQTR; 2629-2636, WPVAGQIR; 2637-2653, ATQQQHDFTLTQTADGR; 2654-2683, SSFDWLTGSSTDPLVDHTSPSSDSLLFAHK; 2705-2715, LGLPGDEVDNK; 2763-2773, MKQDAQVVLYR; 2777-2786, HGDLPDIQIK; 2787-2800, HSSLITPLQAVAQR; 2900-2908, LLPAELPAK; 2941-2950, GIFTSEIGTK; 2951-2962, QITQSALLAEAR; 2979-2991, QDWVDGEPTEAEK; 3010-3029, SLEYCSTASIDSENPPDLNK; 3030-3046, IWSEPFYQETYLPYMIR; 3051-3067, LLLQGEADQSLLTFIDK; 3148-3158, QGNLSSQVPLK; 3173-3186, MDPMNIWDDIITNR; 3197-3217, LTPLPEDNSMNVDQDGDPSDR; 3218-3232, MEVQEQEEDISSLIR; 3290-3302, SQGCSEQVLTVLK; 3303-3318, TVSLLDENNVSSYLSK; 3325-3335, DQNILLGTTYR; 3336-3355, IIANALSSEPACLAEIEEDK; 3359-3372, ILELSGSSSEDSEK; 3426-3449, KEEENASVIDSAELQAYPALVVEK; 3475-3485, YPEETLSLMTK; 3656-3669, LSDFNDITNMLLLK; 3697-3708, NELEIPGQYDGR; 3764-3784, VEQLFQVMNGILAQDSACSQR; 3814-3825, DLLLNTMSQEEK; 3850-3860, HDVGAYMLMYK; 3962-3976, SDPGLLTNTMDVFVK; 3993-4007, SDPGLLTNTMDVFVK; 4023-4036, KGGSWIQEINVAEK; 4051-4070, LAGANPAVITCDELLLGHEK; 4060-4074, AQEPESGLSEETQVK; 4091-4105, AQEPESGLSEETQVK; 4106-4119, CLMDQATDPNILGR |
| DNA ligase III | gi/109113978 | 1,009 | 11 | 156-174, KIEDLTELEGWEELEDNEK; 175-187, EQITQHIADLSSK; 202-213, LTTTGQVTSPVK; 293-308, GSAGDGFHGDVYLTVK; 344-355, DLEQGDVSETIR; 390-403, EDEQQQALQDIASR; 428-442, HVLDALDPNAYEAFK; 454-468, VLHNAQEVEKEPGQR; 616-627, FLHDNMVEIPNR; 779-791, AAVWEITGAEFSK; 836-867, ADFTVVAGDEGSSTTGGSSEENKGPSGSAVSR |
| DNA polymerase β2 | gi/114619948 | 370 | 7 | 90-102, QDDTSSSINFLTR; 188-202, EEMLQMQDIVLNEVK; 203-217, KVDSEYIATVCGSFR; 219-241, GAESSGDMDVLLTHPSFTSESTK; 245-255, LLHQVVEQLQK; 256-265, VHFITDTLSK; 335-352, PLGVTGVAGEPLPVDSEK |
| Interferon-inducible protein 16 | gi/16877622 | 729 | 19 | 13-23, GLEVINDYHFR; 46-55, IQIADLMEEK; 68-83, IFEDIPTLEDLAETLK; 101-116, EVDATSPAPSTSSTVK; 144-165, VSEEQTQPPSPAGAGMSTAMGR; 171-186, TSLSAPPNTSSTENPK; 209-225, VLSTTKPFEYETPEMEK; 310-324, QASGNIVYGVFMLHK; 331-340, TTIYEIQDDR; 378-389, LISEMHSFIQIK; 404-427, LPQEQSQLPNPSEASTTFPESHLR; 428-443, TPQMPPTTPSSSFFTK; 504-522, LKTEPEEVSIEDSAQSDLK; 523-538, EVMVLNATESFVYEPK; 543-556, MFHATVATENEVFR; 628-639, GSFVNGVFEVHK; 644-657, GEFTYYEIQDNTGK; 666-678, LTTINCEEGDKLK; 712-729, KDILNPDSSMETSPDFFF |
| Kappa-B motif-binding protein | gi/1083569 | 464 | 13 | 22-34, RPAEDMEEEQAFK; 38-46, NTDEMVELR; 70-86, TDYNASVSVPDSSGPER; 140-148, GSDFDCELR; 149-163, LLIHQSLAGGIIGVK; 180-191, LFQECCPHSTDR; 192-201, VVLIGGKPDR; 208-219, IILDLISESPIK; 306-316, NLPLPPPPPPR; 378-396, GSYGDLGGPIITTQVTIPK; 397-405, DLAGSIIGK; 423-433, IDEPLEGSEDR; 434-456, IITITGTQDQIQNAQYLLQNSVK |
| Ku70 | gi4503841 | 609 | 21 | 10-31, TEGDEEAEEEQEENLEASGDYK; 36-46, DSLIFLVDASK; 81-92, DLLAVVFYGTEK; 95-100, NSVNFK; 101-114, NIYVLQELDNPGAK; 166-182, IMLFTNEDNPHGNDSAK; 195-206, DTGIFLDLMHLK; 207-218, KPGGFDISLFYR; 219-230, DIISIAEDEDLR, 231-244, VHFEESSKLEDLLR, 302-317, TFNTSTGGLLLPSDTK; 326-339, QIILEKEETEELKR, 340-351, FDDPGLMLMGFK, 405-424, NIPPYFVALVPQEEELDDQK; 425-443, IQVTPPGFQLVFLPFADDK; 475-488, SDSFENPVLQQHFR; 489-510, NLEALALDLMEPEQAVDLTLPK; 518-526, LGSLVDEFK, 527-539, ELVYPPDYNPEGK, 557-565, VEYSEEELK; 596-605, KQELLEALTK |
| Ku86 | gi/10863945 | 732 | 18 | 82-97, HLMLPDFDLLEDIESK; 131-141, HIEIFTDLSSR; 145-155, SQLDIIIHSLK; 185-195, LGGHGPSFPLK; 243-250, HSIHWPCR; 251-260, LTIGSNLSIR; 275-282, TWTVVDAK; 292-315, ETVYCLNDDDETEVLKEDIIQGFR; 401-413, ANPQVGVAFPHIK; 414-431, HNYECLVYVQLPFMEDLR; 444-465, YAPTEAQLNAVDALIDSMSLAK; 470-481, TDTLEDLFPTTK; 503-525, EPLPPIQQHIWNM*LNPPAEVTTK; 535-543, TLFPLIEAK; 545-565, KDQVTAQEIFQDNHEDGPTAK; 569-599, TEQGGAHFSVSSLAEGSVTSVGSVNPAENFR; 689-702, EEASGSSVTAEEAK; 709-732, DKPSGDTAAVFEEGGDVDDLLDMI |
| MSH2 | gi/4557761 | 934 | 26 | 7-21, ETLQLESAAEVGFVR; 40-55, GDFYTAHGEDALLAAR; 74-82, NLQSVVLSK; 114-122, ENDWYLAYK; 160-171, QVGVGYVDSIQR; 198-212, ECVLPGGETAGDMGK; 236-243, DIYQDLNR; 249-275, KGEQMNSAVLPEMENQVAVSSLSAVIK; 309-332, ALNLFQGSVEDTTGSQSLAALLNK; 360-373, LNLVEAFVEDAELR; 374-382, QTLQEDLLR; 407-423, LYQGINQLPNVIQALEK; 431-444, LLLAVFVTPLTDLR; 450-471, FQEMIETTLDMDQVENHEFLVK; 472-482, PSFDPNLSELR; 492-501, MQSTLISAAR; 502-509, DLGLDPGK; 513-524, LDSSAQFGYYFR; 538-546, NFSTVDIQK; 556-565, LTSLNEEYTK; 566-579, NKTEYEEAQDAIVK; 721-737, GVSTFMAEMLETASILR; 742-752, DSLIIIDELGR; 819-838, KGVCDQSFGIHVAELANFPK; 848-871, ALELEEFQYIGESQGYDIMEPAAK; 893-907, QMPFTEMSEENITIK |
| MSH6 | gi/1840467 | 1,360 | 32 | 34-58, AAAAPEASPSPGGDAAWSEAGPGPR; 79-99, SVAPAAPTSCDFSPGDLVWAK; 129-140, VHVQFFDDSPTR; 162-174, GGHFYSAKPEILR; 219-234, SEEDNEIESEEEVQPK; 250-270, VISDSESDIGGSDVEFKPDTK; 271-295, EEGSSDEISSGVGDSESEGLNSPVK; 339-361, AFSAPQNSESQAHVSGGGDDSSR; 385-411, RPDHPDFDASTLYVPEDFLNSCTPGMR; 418-428, SQNFDLVICYK; 454-468, GNWAHSGFPEIAFGR; 483-495, VEQTETPEMMEAR; 520-537, GTQTYSVLEGDPSENYSK; 584-598, TLVAHYPPVQVLFEK; 611-632, SSLSCSLQEGLIPGSQFWDASK; 636-644, TLLEEEYFR; 647-660, LSDGIGVMLPQVLK; 661-676, GMTSESDSIGLTPGEK; 792-804, LDAIEDLMVVPDK; 842-852, AIMYEETTYSK; 871-883, IIGIMEEVADGFK; 902-911, FPDLTVELNR; 931-945, AGFDSDYDQALADIR; 946-957, ENEQSLLEYLEK; 966-974, TIVYWGIGR; 977-988, YQLEIPENFTTR; 1015-1024, LANLINAEER; 1077-1092, PVILLPEDTPPFLELK; 1127-1140, AYCVLVTGPNMGGK; 1218-1233, GTATFDGTAIANAVVK; 1243-1263, TLFSTHYHSLVEDYSQNVAVR; 1305-1315, LSDGIGVMLPQVLK; 647-660, LANLPEEVIQK |
| NF-Yα | gi/109071077 | 244 | 2 | 149-167, IPLPGAEMLEEEPLYVNAK; 220-242, EKDSPHMQDPNQADEEAMTQIIR |
| Oct-1 | gi/21927972 | 755 | 7 | 13-42, MNNPSETSKPSMESGDGNTGTQTNGLDFQK; 267-284, TIAATPIQTLPQSQSTPK; 286-305, IDTPSLEEPSDLEELEQFAK; 314-327, LGFTQGDVGLAMGK; 328-340, LYGNDFSQTTISR; 361-390, WLNDAENLSSDSSLSSPSALNSPGIEGLSR; 409-432, SFLENQKPTSEEITMIADQLNMEK |
| PARP-1 | gi/130781 | 1,014 | 28 | 35-47, MAIMVQSPMFDGK; 66-78, HPDVEVDGFSELR; 109-119, TLGDFAAEYAK; 143-156, MVDPEKPQLGMIDR; 166-182, NREELGFRPEYSASQLK; 183-197, GFSLLATEDKEALKK; 208-221, RKGDEVDGVDEVAK; 240-253, AQNDLIWNIKDELK; 270-282, QQVPSGESAILDR; 283-305, VADGMVFGALLPCEECSGQLVFK; 306-320, SDAYYCTGDVTAWTK; 332-337, EWVTPK; 341-346, EISYLK; 453-467, VVSEDFLQDVSASTK; 487-496, AEPVEVVAPR; 529-548, GGAAVDPDSGLEHSAHVLEK; 552-564, VFSATLGLVDIVK; 572-582, LQLLEDDKENR; 601-616, LEQM*PSKEDAIEHFMK; 637-654, KFYPLEIDYGQDEEAVKK; 684-695, KAMVEYEIDLQK; 736-747, FYTLIPHDFGMK; 748-761, KPPLLNNADSVQAK; 780-796, GGSDDSSKDPIDVNYEK; 803-815, VVDRDSEEAEIIR; 820-838, NTHATTHNAYDLEVIDIFK; 866-878, TTNFAGILSQGLR; 894-903, GIYFADMVSK |
| RecQL | gi/14591902 | 649 | 3 | 153-167, QLGISATMLNASSSK; 292-306, QKPSNTEDFIEDIVK; 426-439, ISSMVVMENVGQQK |
| RNA binding protein KOC | gi/2105469 | 579 | 11 | 4-23, LYIGNLSENAAPSDLESIFK; 139-150, LNGFQLENFTLK; 151-169, VAYIPDEMAAQQNPLQQPR; 200-213, LLVPTQFVGAIIGK; 243-258, SITILSTPEGTSAACK; 310-325, ITISPLQELTLYNPER; 453-465, MVIITGPPEAQFK; 509-525, TVNELQNLSSAEVVVPR; 526-538, DQTPDENDQVVVK; 539-551, ITGHFYACQVAQR; 568-577, ALQSGPPQSR |
| hnRNP A1 | gi/114644273 | 412 | 8 | 55-70, LFIGGLSFETTDESLR; 71-86, SHFEQWGTLTDCVVMR; 95-114, GFGFVTYATVEEVDAAMNAR; 162-169, DYFEQYGK; 170-179, IEVIEIMTDR; 186-200, GFAFVTFDDHDSVDK; 272-304, GGGGYGGSGDGYNGFGNDGGYGGGGPGYSGGSR; 93-410, NQGGYGGSSSSSSYGSGR |
| hnRNP A2/B1 | gi/109134362 | 341 | 5 | 11-26, LFIGGLSFETTEESLR; 118-125, DYFEEYGK; 142-156, GFGFVTFDDHDPVDK; 227-254, GFGDGYNGYGGGPGGGNFGGSPGYGGGR; 314-338, NMGGPYGGGNYGPGGSGGSGGYGGR |
| hnRNP A3 isoform 3 | gi/109100194 | 332 | 8 | 36-52, KLFIGGLSFETTDDSLR; 58-68, WGTLTDCVVMR; 114-126, EDSVKPGAHLTVK; 128-143, IFVGGIKEDTEEYNLR; 144-151, DYFEKYGK; 152-161, IETIEVMEDR; 168-182, GFAFVTFDDHDTVDK; 309-330, SSGSPYGGGYGSGGGSGGYGSR |
| hnRNP C | gi/109082727 | 293 | 11 | 18-29, VFIGNLNTLVVK; 30-39, KSDVEAIFSK; 51-61, GFAFVQYVNER; 74-89, MIAGQVLDINLAAEPK; 100-117, SAAEMYGSSFDLDYDFQR; 175-185, LKGDDLQAIKK; 194-203, VDSLLENLEK; 217-230, NDKSEEEQSSSSVK; 238-262, MESEGGADDSAEEGDLLDDDDNEDR; 263-284, GDDQLELIKDDEKEAEEGEDDR; 276-293, EAEEGEDDRDSANGEDDS |
| hnRNP D0 | gi/109074343 | 289 | 2 | 184-197, IFVGGLSPDTPEEK; 200-218, EYFGGFGEVESIELPMDNK |
| hnRNP G | gi/3256007 | 391 | 6 | 10-22, LFIGGLNTETNEK; 23-30, ALEAVFGK; 50-63, GFAFVTFESPADAK; 81-93, VEQATKPSFESGR; 126-144, GGHMDDGGYSMNFNMSSSR; 204-210, DVYLSPR |
| hnRNP K | gi/109111925 | 438 | 11 | 22-34, RPAEDMEEEQAFK; 38-46, NTDEMVELR; 53-60, NAGAVIGK; 70-86, TDYNASVSVPDSSGPER; 104-115, IIPTLEEYQHYK; 125-139, LLIHQSLAGGIIGVK; 184-195, IILDLISESPIK; 353-371, |
| GSYGDLGGPIITTQVTIPK; 372-380, DLAGSIIGK; 398-408, IDEPLEGSEDR; 409-431, IITITGTQDQIQNAQYLLQNSVK | ||||
| SAF-A/hnRNP U | gi/14141161 | 806 | 4 | 237-246, GYFEYIEENK; 405-414, NGQDLGVAFK; 506-517, YNILGTNTIMDK; 557-571, NFILDQTNVSAAAQR |
| Topo I | gi/11225260 | 765 | 10 | 205-216, WWEEERYPEGIK; 224-239, GPVFAPPYEPLPENVK; 253-262, AEEVATFFAK; 292-299, NIITNLSK; 300-310, CDFTQMSQYFK; 489-508, AGNEKEEGETADTVGCCSLR; 550-558, NLQLFMENK; 576-587, HLQDLMEGLTAK; 591-603, TYNASITLQQQLK; 701-712, LEVQATDREENK |
| Topo IIβ | gi/19913408 | 1,621 | 18 | 100-112, IFDEILVNAADNK; 123-139, VSIDPESNIISIWNNGK; 247-257, LDKDIVALMTR; 293-303, DKLDETGVALK; 323-337, GFQQISFVNSIATTK; 341-352, HVDYVVDQVVGK; 467-482, HSLECTLILTEGDSAK; 516-528, QIMENAEINNIIK; 552-566, IMIMTDQDQDGSHIK; 583-595, HGFLEEFITPIVK; 678-688, KEWLTNFMEDR; 718-729, ELILFSNSDNER; 730-743, SIPSLVDGFKPGQR; 966-974, TPALISDYK; 1050-1063, KEWLVGMLGAESTK; 1182-1199, EDLAAFVEELDKVESQER; 1223-1235, LQLEETMPSPYGR; 1337-1351, SESDLEETEPVVIPR |
Amino acid positions of corresponding peptides in the matched protein and the sequences of these peptides.
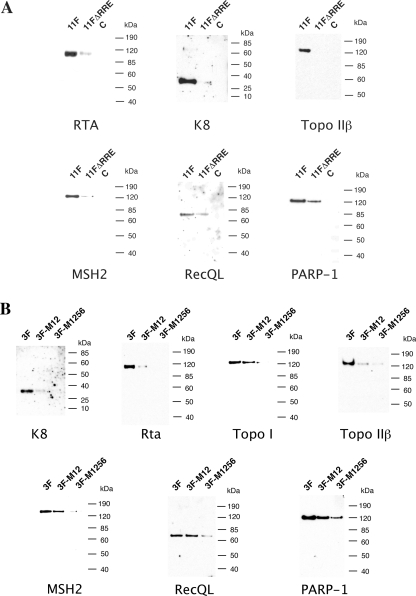
To confirm the identities of these ori-Lyt-bound proteins, Western analyses of the DNA affinity-purified materials were performed with antibodies specific to some of the identified proteins. As shown in Fig. 1C, the Western analysis proved that cellular Topo I, Topo II, MSH2, RecQL, PARP-1, DNA-PK, Ku86, and SAF-A were indeed enriched by DNA affinity purification. Furthermore, we observed an interesting phenomenon, that many cellular proteins, including the DNA repair proteins MSH2, RecQL, DNA-PK, and Ku86, bound both 3F and 11F DNAs. But Topo I and SAF-A can be detected only in 3F and 9F affinity-purified protein pools, respectively (Fig. 1C).
A formaldehyde cross-linking ChIP assay was performed to ensure that these identified cellular proteins are indeed associated with ori-Lyt DNA in the virus context in cells. In brief, protein-DNA cross-linking was induced by addition of formaldehyde to living TPA-induced BCBL-1 cells. Chromatin from these cells was fragmented by sonication, and the resulting material was immunoprecipitated with specific antibodies against each of the viral and cellular proteins to be tested. The protein-bound DNAs were quantified by PCR using two pairs of primers designed to amplify the KSHV sequences from nucleotides 23147 to 23405 and from nucleotides 24020 to 24155 within the ori-Lyt (L). The former region includes the K8 binding sites, and the latter region is near the RRE. Findings revealed that (i) both ori-Lyt sequences could be coprecipitated by antibodies against three viral proteins, namely, K8, RTA, and ORF57, which are known to be involved in lytic viral DNA replication, but not by preimmune sera (either mouse or rabbit) or antibodies against ORF45 and ORF64 (Fig. 2); (ii) all the cellular proteins tested were found to be associated with at least one of the regions in the ori-Lyt or both; and (iii) RecQL showed the strongest signals for binding to both sequences of the ori-Lyt among all the cellular proteins tested in the study, suggesting a proximate distance between the protein and ori-Lyt DNA in a replication complex (Fig. 2). Another pair of primers that amplify the viral DNA between nucleotides 67813 and 68097 (within the ORF45 coding region) was used as a control. The ORF45 sequence was not detected in the precipitate brought down by any antibodies used in this study (Fig. 2). Thus, the data of the ChIP assay provided additional evidence that the cellular proteins identified in our proteomics study indeed bind to the ori-Lyt DNA sequence in vivo.
FIG. 2.
Analysis of the association of the cellular proteins with the ori-Lyt region by use of a ChIP assay. The immunoprecipitates by specific mouse (m) or rabbit (r) antibodies against different viral and cellular proteins as indicated as well as mouse IgG and rabbit preimmune serum were analyzed by PCR designed to amplify KSHV DNA between nucleotides 23129 and 23405 (upper), between nucleotides 24020 and 24155 (middle), and between nucleotides 67813 and 68097 (lower).
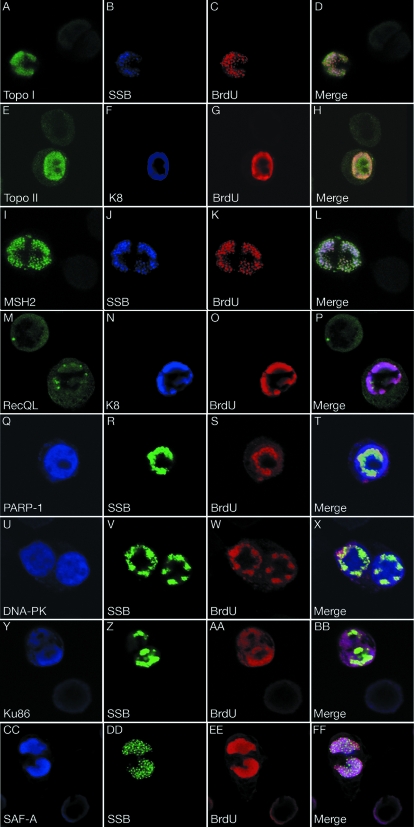
Colocalization of the cellular proteins with KSHV replication compartments.
Then, we asked whether these ori-Lyt-associated cellular proteins are recruited to VRCs during the DNA replication. VRCs are intranuclear domains which are formed when viral replication successively annexes a large portion of the nucleus and the location where viral DNA replication takes place in the host cell nucleus (29). In herpesviruses, VRCs are visible in IFAs as large, irregularly shaped nuclear domains. We used triple-label IFAs to examine the localization of several representative proteins during lytic DNA replication in BCBL-1 cells. BCBL-1 cells were treated with TPA for 48 h to induce lytic DNA replication. To visualize the sites of DNA replication, we pulse labeled the TPA-induced cells with BrdU for 60 min. The cells were fixed and stained with rabbit polyclonal antibody for SSB; mouse monoclonal antibodies against Topo I, MSH2, PARP-1, DNA-PK, Ku-86, or SAF-A; and goat polyclonal anti-BrdU antibody. For Topo II and RecQL, cells were stained with rabbit polyclonal anti-Topo IIβ or anti-RecQL, monoclonal anti-K8, and goat polyclonal anti-BrdU antibodies. Since TPA treatment of unsynchronized BCBL-1 cells is able to induce KSHV reactivation in only about 20% cells, this allowed us to compare the expression and distribution of these cellular proteins in the cells that were undergoing viral DNA replication and those that were not. Topo I, Topo II, MSH2, RecQL, DNA-PK, and PARP-1 were uniformly distributed in the nuclei of the cells where no viral DNA replication occurred, whereas Ku86 and SAF-A were localized mainly in the periphery area of the nucleus. In the cells in which viral lytic replication was successfully induced, replication compartment-like large nuclear domains were revealed by antibodies against viral DNA replication-associated proteins (SSB or K8) and BrdU. In these cells, the nuclear domains were colocalized with the sites of DNA replication as indicated by staining with anti-BrdU antibody, indicating that they are functional VRCs. In the cells with viral lytic DNA replication, the expression levels of Topo I, Topo II, MSH2, DNA-PK, Ku86, and SAF-A were significantly elevated in comparison to those in the cells without DNA replication. The newly synthesized cellular proteins were accumulated in the replication compartments. For example, the cells with viral DNA replication exhibited much higher MSH2 staining than those without viral DNA replication. In Fig. 3I to L, there are two cells in the field, one displaying a VRC (on the left) and the other showing no sign of viral DNA replication (on the right). The left cell shows a bright MSH2 staining in the replication compartment, but in the cell on the right, the dull green fluorescence can barely be seen in the nucleus. Similar phenomena were also seen in Topo I, Topo II, DNA-PK, Ku86, and SAF-A (Fig. 3). These data suggest that the expression of MSH2, as well as Topo I, Topo II, DNA-PK, Ku86, and SAF-A, was induced to higher levels during viral lytic replication. In contrast, no obvious increases of RecQL and PARP-1 expression were found in the cells where viral DNA replication was taking place. However, some of these proteins were enriched or relocated in the VRCs (Fig. 3M and Q). Together, the recruitment of these cellular proteins to VRCs argued that these cellular proteins may have roles to play in the viral DNA replication.
FIG. 3.
Distribution of cellular proteins in TPA-induced BCBL-1 cells. BCBL-1 cells were treated with TPA for 48 h and pulse labeled with BrdU for 60 min. The cells were subjected to triple-label IFA using mouse monoclonal antibodies against cellular proteins as indicated (except RecQL and Topo IIβ), rabbit polyclonal anti-SSB antibody, and goat anti-BrdU antibody. For RecQL and Topo IIβ, cells were labeled with rabbit polyclonal anti-RecQL and Topo IIβ antibodies, a mouse monoclonal anti-K8 antibody, and goat anti-BrdU antibody. The slides were examined with a confocal laser scanning system, and three channels were recorded simultaneously or sequentially. The triple-label IFA shows that these cellular proteins are colocalized with core replication machinery proteins (SSB or K8) as well as newly synthesized DNA (BrdU incorporated) in VRC in TPA-induced BCBL-1 cells.
Association of RecQL with prereplication complexes.
Topo II, MSH2/6, RecQL, PARP-1, DNA-PK, and Ku-86/70 bound to both 3F and 11F DNA (Fig. 1C), similar to the pattern in which the proteins of viral core replication machinery complexes bind to ori-Lyt DNA (38). Viral core replication machinery complexes composed of at least six virally encoded DNA replication enzymes (SSB, PRI, HEL, PAF, POL, and PPF), K8, and RTA were found to be assembled as a prereplication complex and loaded on ori-Lyt DNA with two contact points, K8 binding motifs and RRE. Thus, these viral core replication machinery proteins can be brought down by either 3F or 11F DNA in DNA affinity assays (38). The binding of even the cellular DNA replication and repair proteins to 3F and 11F raised a possibility that these cellular proteins also could be associated with the viral core replication machinery complexes and hence be recruited to ori-Lyt DNA together through K8 and RTA. To test this hypothesis, we first examined whether these cellular proteins associate with viral core replication complexes by coimmunoprecipitation assays. BCBL-1 cells, induced with TPA for 48 h, were lysed in immunoprecipitation buffer and subjected to immunoprecipitation with mouse monoclonal anti-RTA and anti-K8 antibodies in the absence and presence of EtBr. Immunoprecipitated material was resolved by SDS-PAGE, followed by Western blot analysis with antibodies specific to some of these cellular proteins (Table 2). The results showed that only RecQL was able to be coimmunoprecipitated by both K8 and RTA in the presence and absence of EtBr, suggesting that this protein is an integral component of the prereplication complex (Fig. 4A and B). To further confirm the presence of RecQL in the complexes, we performed an additional coimmunoprecipitation with an antibody against PPF, a viral core replication protein. The result shows that RecQL can be coprecipitated with PPF, suggesting that RecQL is in the same complex with K8, RTA, and PPF (Fig. 4C). The prereplication complexes appear to form in solution rather than on ori-Lyt DNA because the complexes were found to be tighter in the presence of EtBr in coimmunoprecipitation assays (Fig. 4). To rule out the possibility that RecQL can be independently associated with PTA, K8, and PPF, rather than being in the same complex with these three viral proteins, we tested the binary interactions of RecQL with each of the viral proteins. 293T cells were transfected with the expression vector for each of the three viral proteins (pCR3.1-K8α, pCR3.1-ORF50, and pCR3.1-ORF59). At 48 hours posttransfection, the cells were collected and subjected to a coimmunoprecipitation assay with anti-K8, anti-RTA, and anti-PPF antibodies. The results show that RecQL is able to interact with RTA but not with K8 and PPF in the absence of a prereplication complex (Fig. 5). PARP-1 can be precipitated with anti-RTA antibody but not with anti-K8 antibody (Table 2). We believe that PARP-1 is complexed with RTA as previously reported (20) but is not in a prereplication complex. SAF-A can be coprecipitated with anti-K8 antibody. MSH2 and other cellular proteins were not coprecipitated with either K8 or RTA (Table 2).
TABLE 2.
Summary of properties of viral and cellular proteins in binding to ori-Lyt and interactions with prereplication complex proteins
| Protein | DNA affinity
|
Coimmunoprecipitation with indicated antibody and EtBr concn (μg/ml)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Anti-K8
|
Anti-RTA
|
Anti-PPFa
|
||||||||||
| 3F | 9F | 11F | 0 | 50 | 100 | 0 | 50 | 100 | 0 | 50 | 100 | |
| Viral | ||||||||||||
| RTA | + | − | + | + | + | + | + | + | + | + | + | + |
| K8 | + | − | + | + | + | + | + | + | + | + | + | + |
| SSB | + | − | + | + | + | + | + | + | + | + | + | + |
| PPF | + | − | + | + | + | + | + | + | + | + | + | + |
| ORF57 | + | − | + | + | + | + | + | + | + | + | + | + |
| Cellular | ||||||||||||
| Topo I | + | − | − | − | − | − | − | − | − | NA | NA | NA |
| Topo IIβ | + | − | + | − | − | − | − | − | − | NA | NA | NA |
| RecQL | + | − | + | + | + | + | + | + | + | + | + | + |
| MSH2 | + | − | + | − | − | − | − | − | − | NA | NA | NA |
| PARP-1 | + | − | + | − | − | − | + | + | + | NA | NA | NA |
| DNA-PK | + | − | + | − | − | − | − | − | − | NA | NA | NA |
| Ku86 | + | − | + | − | − | − | − | − | − | NA | NA | NA |
| SAF-A | − | + | − | + | + | + | − | − | − | NA | NA | NA |
NA, not applicable.
FIG. 4.

Coimmunoprecipitation of RecQL with viral DNA replication complexes. Immunoprecipitations were performed with TPA-induced BCBL-1 lysates by using specific antibodies against K8 (α-K8), RTA, and PPF in the presence and absence of EtBr. Immunoprecipitation with mouse IgG was included as negative controls. Precipitation samples were separated on SDS-PAGE gel, followed by Western analyses using antibodies as indicated.
FIG. 5.

Coimmunoprecipitation of RecQL with RTA. 293T cells were transfected with pCR3.1-K8α, pCR3.1-ORF50, pCR3.1-ORF59, and empty pCR3.1 plasmids. At 48 hours posttransfection, whole-cell extracts (WCE) were prepared and subjected to immunoprecipitation (IP) with corresponding antibodies against K8, RTA, and PPF. Immunoprecipitation with rabbit normal serum was included as a negative control. Precipitation samples were separated on SDS-PAGE gel, followed by Western analyses using anti-RecQL antibody (upper panel) and a combination of antibodies against RTA, K8, and PPF (lower panel).
Since RecQL is recruited to ori-Lyt as a part of a viral prereplication complex, we further speculated that binding of RecQL to ori-Lyt relies on K8 and RTA binding motifs, like that of other core replication proteins in the complex. To test this hypothesis, a mutagenesis experiment was performed. The result showed that mutations of K8 binding motifs or deletion of RRE from the ori-Lyt DNA greatly reduced the binding of RecQL to the ori-Lyt DNA (Fig. 6). Taken together, our data strongly suggest that at least one cellular protein, RecQL, associates with prereplication complexes and is recruited to ori-Lyt DNA, together with the core viral replication proteins, through K8 and RTA.
FIG. 6.
Effect of mutations of the C/EBP motifs in the ori-Lyt fragment (3F) and deletion of RRE from the ori-Lyt fragment (11F) on the recruiting of cellular proteins to ori-Lyt DNA. (A) Biotinylated DNA fragments were prepared by PCR, with pOri-A (wild type) DNA and an RRE deletion derivative, which was described in Wang et al. (37), as templates. (B) Similarly, biotinylated DNA fragments were prepared by PCR with the pOri-A (wild type) DNA template and pOri-M12 and pOri-M1256, mutations on certain C/EBP motifs, which was described in Wang et al. (37). TPA-induced BCBL-1 nuclear extract was incubated with the DNA fragments conjugated on magnetic beads, washed, and eluted with D500 elution buffer. Samples were assayed by Western blotting with antibodies as indicated. C, an irrelevant DNA was included as a control.
Recruitment of replication-associated cellular proteins to ori-Lyt DNA by K8 and RTA proteins.
MSH2, Topo II, PARP-1, DNA-PK, and Ku antigens were not coimmunoprecipitated with prereplication complexes but bind to both 3F and 11F DNA, similar to the behavior of K8, RTA, and other core replication machinery proteins (38). We were curious as to whether recruitment of these cellular proteins to ori-Lyt is also dependent on K8 and RTA binding motifs. Therefore, we investigated the effects of deletion of the RRE motif from 11F DNA and mutations of the K8 binding motifs in 3F DNA on the association of these cellular proteins with DNA. Wild-type (11F) and RRE deletion mutant (11FΔRRE) DNAs were used for DNA affinity purification with nuclear extract from TPA-induced BCBL-1 cells. As shown in Fig. 6A, RTA, K8, MSH2, Topo II, and PARP-1 were able to bind to wild-type 11F but not to RRE-deleted DNA or bound the mutant DNA with reduced affinities. Similarly, wild-type 3F and its mutants, in which the C/EBP cluster was mutated, were used in the DNA affinity assay. The results showed that not only K8, but also RTA, MSH2, Topo II, and PARP-1, failed to bind to the M1256 mutant (Fig. 6B). These observations can be explained as follows. The cellular proteins, such as MSH2/6 heterodimer and DNA-PK/Ku86/70 heterotrimer, are not integral components of the prereplication complex, but recruitment of these proteins to ori-Lyt DNA is dependent on the presence of prereplication complexes on ori-Lyt. The binding of the core replication protein complex to ori-Lyt may cause structural changes in ori-Lyt DNA that facilitate the recruitment of MSH2/6 and DNA-PK/Ku86/70 to ori-Lyt. Together, these data suggest that binding of MSH2/6, DNA-PK/Ku86/70, and other cellular proteins to ori-Lyt DNA occurs after a prereplication complex lands on the ori-Lyt DNA, and recruitment of prereplication complexes as well as MSH2/6 and DNA-PK/Ku86/70 complexes to ori-Lyt leads to the formation of a replication initiation complex.
Topo I is also not found in the prereplication complex and cannot be coprecipitated by K8 and RTA. However, Topo I is present in replication initiation complexes on ori-Lyt and binds to 3F DNA. The binding of Topo I to ori-Lyt is also dependent on the presence of core replication proteins and K8/RTA on the ori-Lyt DNA (Fig. 6B).
Roles of topoisomerases I and II in KSHV lytic DNA replication.
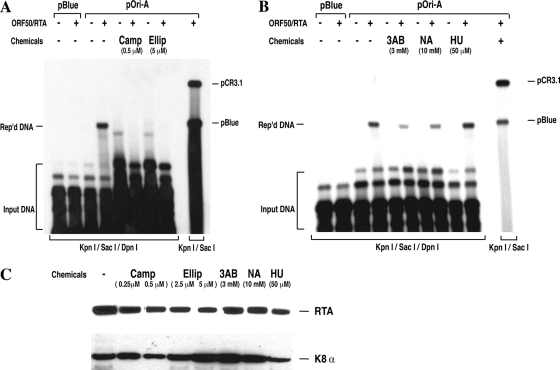
The next question is whether the ori-Lyt-associated cellular proteins play any role in ori-Lyt-dependent DNA replication. Cellular Topo I and II bind to KSHV ori-Lyt DNA and accumulate in VRCs, suggesting their involvement in viral DNA replication. Topo I is an enzyme needed to release the topological stress created by DNA unwinding during DNA replication by nicking and religating DNA ahead of the replication fork (4). Topo II also modulates the topological state of DNA by making transient double-stranded breaks in DNA, which is believed to be needed for converting replicating intermediates into a mature replication product (31). To assess the role of these two topoisomerases in KSHV DNA replication, we studied the effects of specific topoisomerase inhibitors on viral DNA replication in cells. BCBL-1 cells were transfected with an ori-Lyt-containing plasmid (pOri-A). Lytic DNA replication was induced by cotransfection with an RTA expression vector. The ori-Lyt-dependent DNA replication was measured by a DpnI assay (25, 37). Replicated DNA was detected in the cells that were cotransfected with pOri-A and RTA expression vector (Fig. 7). Addition of 0.5 μM camptothecin, a specific inhibitor of mammalian Topo I, completely abolished ori-Lyt-dependent DNA replication (Fig. 7A). Similarly, when 5 μM of ellipticin, a specific inhibitor of mammalian topoisomerase II, was used, the ori-Lyt-dependent DNA replication became undetectable (Fig. 7A). Camptothecin at 0.5 μM and ellipticin at 5 μM did not significantly inhibit gene expression in cells, as we monitored the expression of RTA and K8 genes in the presence of these drugs at these concentrations (Fig. 7C). These data demonstrate that both topoisomerase I and II activities are essential for KSHV lytic DNA replication.
FIG. 7.
(A) Inhibition of Topo I and Topo II activities with chemical inhibitors abolishes KSHV ori-Lyt-associated DNA replication. BCBL-1 cells were transfected with ori-Lyt-containing plasmid (pOri-A) and RTA expression vector (pCR3.1-ORF50). Transfected cells were cultured in the absence and presence of 0.5 μM of camptothecin (Camp) or 5 μM ellipticine (Ellip). After 48 h of incubation, Hirt DNAs were extracted from the cells and subjected to a viral DNA replication assay as described in Materials and Methods. DpnI-resistant products of DNA replication (Rep'd DNA) were detected by Southern blotting with 32P-labeled pBluescript plasmid. (B) Effects of chemicals that affect PARP-1 activity on KSHV ori-Lyt-associated DNA replication. Transfected cells were cultured in the absence and presence of the PARP-1 inhibitors 3-AB and NA as well as HU, which raises PARP-1 activity. The DNA replication assay was performed as described above. (C) Effects of the chemicals used in the replication assays on RTA and K8 gene expression. BCBL-1 cells were induced with TPA and incubated in the presence of the chemicals indicated for 48 h. Cells were subjected to Western analyses with monoclonal anti-anti-RTA and K8 antibodies.
Roles of PARP-1 in KSHV lytic DNA replication.
To examine the effect of PARP-1 in KSHV lytic DNA replication, we investigated DNA replication in the presence of the PARP-1 inhibitors 3-aminobenzamide (3-AB) and niacinamide (NA) as well as hydroxyurea (HU), which raises PARP-1 activity. The results are shown in Fig. 7B. When the cells cotransfected with pOri-A and RTA expression vector were cultured in the presence of 3-AB and NA, DNA replication efficiency decreased by 70% and 50%, respectively. While in the presence of HU, DNA replication appeared to be enhanced by 40%. All three drugs did not induce DNA replication in the absence of RTA expression. The effects of these drugs on gene expression (RTA and K8) were also monitored (Fig. 7C). The results suggest a positive role for PARP-1 in KSHV ori-Lyt-dependent DNA replication.
DISCUSSION
Because of the limited sizes of viral genomes, viruses cannot possibly encode all the functions needed for their life cycle. Thus, viruses have to use host cellular factors in many biological processes in their life cycle. To address the questions as to whether KSHV recruits any cellular proteins for viral lytic replication and, if so, what roles these cellular proteins may play in viral DNA replication, we designed an affinity purification procedure combined with proteomic analyses to identify proteins that bind to KSHV ori-Lyt. The study led to isolation of a number of cellular proteins that bind viral ori-Lyt DNA at different regions. The identities of these proteins were determined by mass spectrometric analyses and further confirmed by Western analyses using specific antibodies. A ChIP assay confirmed the association of these cellular proteins with ori-Lyt DNA in the viral context in cells. Furthermore, we demonstrated that all the cellular proteins tested accumulate in VRCs and the expression levels of some of the proteins were found to be dramatically elevated when viral lytic DNA replication occurred. We also initiated studies to analyze the functional roles of these cellular proteins in KSHV lytic DNA replication by inhibiting the enzymatic activities of some of the cellular proteins with specific inhibitors, and for the future, we plan to extend the studies by depleting specific host factors, using RNA interference techniques.
Cellular proteins in viral prereplication complexes and replication initiation complexes.
Our previous studies suggested that viral core replication machinery proteins, together with K8 and RTA, form a prereplication complex regardless of the presence of ori-Lyt DNA; the prereplication complex loads on ori-Lyt DNA through K8 and RTA and becomes a replication initiation complex (38). In this study, DNA affinity and coimmunoprecipitation assays were used to investigate whether these ori-Lyt-associated cellular proteins are present in any viral DNA replication complexes. The results of the studies divided these cellular proteins in three categories. The first category is the proteins that are integrated into the prereplication complex. RecQL is the only cellular protein found in this category. RecQL could be coimmunoprecipitated with K8, RTA, and PPF, and recruitment of this protein to ori-Lyt was dependent on the presence of K8 and RTA. The proteins in the second category contain MSH2/6 and DNA-PK/Ku86/70 complexes. Herein, these complexes are not associated with a prereplication complex in solution but recruitment of these complexes to ori-Lyt relies on the presence of a core replication protein complex on ori-Lyt. It is possible that binding of the core replication protein complex to ori-Lyt results in structural changes in ori-Lyt DNA, such as looping, distortion, and unwinding of the ori-Lyt DNA, that facilitate the recruitment of MSH2/6 and DNA-PK/Ku86/70 to ori-Lyt. SAF-A represents the proteins in the third category, which directly interact with ori-Lyt DNA. Figure 8 presents a model for incorporating the cellular proteins of different categories into a viral DNA replication complex.
FIG. 8.
Model for recruitment of cellular proteins to KSHV DNA replication complexes and ori-Lyt. KSHV ori-Lyt-associated cellular proteins can be divided into three categories. RecQL, as a protein of the first category, is likely to be a component of the prereplication complex and recruited to ori-Lyt, together with viral core replication proteins in the complex, through K8 and RTA. The loading of the prereplication complex on ori-Lyt may cause structural changes of ori-Lyt DNA that facilitate the recruiting of the proteins of the second category, including MSH2/6 and DNA-PK/Ku86/70 complexes, to the ori-Lyt. SAF-A, a protein of the third category, binds directly to ori-Lyt DNA. It is proposed that SAF-A may tether the ori-Lyt DNA to the nuclear scaffold or matrix and the attachment of ori-Lyt to the nuclear matrix may be an important event for efficient DNA replication.
Does RecQL unwind viral origin DNA?
RecQL was not only found to bind to KSHV ori-Lyt but also detected in prereplication complexes that are believed to form in solution prior to being recruited to ori-Lyt DNA (38). RecQL (also referred to as RecQ1 or RecQL1) is a member of the RecQ DNA helicase family, which is highly conserved in evolution. Five members of the family have been identified in human cells, and three are associated with human genetic disorders, namely, Bloom's syndrome, Werner's syndrome, and Rothmund-Thomson syndrome (5). RecQL is a major DNA helicase in human cells, and the function of this helicase is not known. No genetic disorder has been linked to this helicase. This protein has received our special attention for two reasons. First, RecQL is the only cellular protein that was detected in the prereplication complex, suggesting a unique function of the protein in initiation of viral DNA replication. Second, we are particularly interested in whether this cellular helicase is involved in initiation of KSHV lytic DNA replication. By analogy with other replication systems, a helicase activity is expected to unwind DNA at the origin during initiation of DNA replication. In herpes simplex virus type 1 (HSV-1), the origin binding protein UL9 is proposed to carry out this activity. UL9 is known to possess ATPase and helicase activities in addition to its origin binding property (8, 26). Genetic analysis showed that the conserved helicase motifs in UL9 are essential both in vivo and in vitro for viral DNA replication (27, 28). However, the functional analogues of UL9 are not found in KSHV or in other gammaherpesviruses, including Epstein-Barr virus (EBV) (15, 16). Although KSHV encodes a heterotrimeric helicase-primase complex, the complex is conserved among all herpesviruses and believed to unwind duplex viral DNA at replication forks and to prime lagging strand synthesis (12). Recently, cellular RecQL4, another member of the RecQ helicase family, was shown to promote loading of replication factors at origins after prereplication complexes were formed, suggesting that RecQL4 plays a role in unwinding DNA at the replication origin (32). Taken together, the identification of RecQL in the KSHV replication complex raises an exciting hypothesis, that the cellular RecQ helicase may be the long-sought helicase that unwinds origin DNA in the initiation of KSHV and EBV lytic DNA replication. The model regarding the function of RecQL in viral DNA replication is currently being tested in our laboratory.
Cellular DNA repair and recombination proteins in KSHV replication.
Many ori-Lyt-associated cellular proteins are DNA repair proteins, including proteins of the mismatch repair (MMR) and the nonhomologous end-joining (NHEJ) pathways. The MSH2/MSH6 heterodimer is important for the MMR DNA repair mechanism (9), and DNA-PK, Ku70/Ku86, and PARP-1 are major components of the NHEJ pathway (23, 39). These DNA repair mechanisms are also integrated with other cellular processes, including DNA replication. The MMR and NHEJ proteins that were detected in the KSHV DNA replication complex were also found in HSV-1 DNA replication complexes (36). It is conceivable that during coevolution with their host, viruses have evolved to interact with cellular repair/recombination machinery and developed fascinating ways to either subvert or benefit from these cellular responses and pathways. For example, human cytomegalovirus DNA replication activates the checkpoint response to DNA double-stranded breaks, but the virus responds by redirecting checkpoint proteins to the cytoplasm and thereby inhibiting the signaling pathway (18). On the other hand, in HSV-1 infection, several members of the cellular DNA damage-sensing machinery are activated and accumulate at sites of viral DNA replication. When this cellular response is abrogated, formation of HSV-1 replication centers is retarded, and viral production is compromised, suggesting that the virus exploits the cellular DNA damage response that aids viral replication (24). Identification of DNA repair/recombination proteins in the KSHV replication process provides an avenue for pursuing the questions as to whether the DNA repair pathways are involved in KSHV DNA replication and what their contributions to viral DNA replication are.
Roles of topoisomerases in KSHV lytic DNA replication.
All herpesviruses encode a DNA helicase-primase holoenzyme that can unwind double-stranded DNA as a DNA replication fork moves. But none of the herpesviruses encodes a DNA-relaxing enzyme to release the topological stress created by DNA unwinding. Therefore, cellular DNA topoisomerases I and II are reportedly required for HSV-1, cytomegalovirus, and EBV lytic DNA replication (6, 13, 22). In this report, Topo I and II were found to bind to KSHV ori-Lyt DNA, suggesting their involvement in viral DNA replication. The result, that inhibition of cellular Topo I and II by specific inhibitors abolished KSHV DNA replication in cells, demonstrates that both topoisomerase activities are essentially required for KSHV lytic DNA replication. Since Topo I and Topo II inhibitors are new classes of anticancer agents (14), our results suggest possibilities for treatment of KSHV-associated malignancies with topoisomerase inhibitors.
Roles of PARP-1 in KSHV lytic DNA replication.
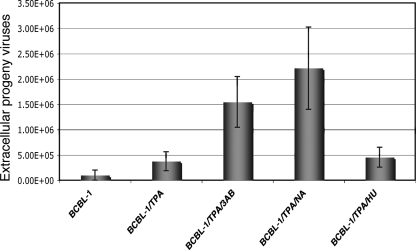
PARP-1 is a nuclear enzyme involved in the DNA damage surveillance network. This protein responds to DNA damage by transferring 50 to 200 molecules of ADP-ribose to various nuclear proteins, including transcription factors, DNA replication/repair factors, and histones (10). PARP-1 has been reported to be associated with LANA and RTA of KSHV and murine gammaherpesvirus 68. The association with RTA leads to poly(ADP-ribosyl)ation of the protein and represses RTA-mediated transcriptional activity. It was also shown that murine gammaherpesvirus 68 lytic replication was enhanced in PARP-1-null cells (20). Conversely, our results suggest a positive role for PARP-1 in KSHV DNA replication. To clarify the inconsistency, we analyzed effects of PARP-1 inhibitors and enhancer on KSHV virion production in BCBL-1 cells. The result showed that treatment with PARP-1 inhibitors enhanced virion production (Fig. 9), which is consistent with the data reported by Gwack et al. (20). Thus, these data suggest that (i) PARP-1 positively regulates KSHV lytic DNA replication, and (ii) PARP-1 may also have a negative regulatory role in capsid assembly or virion maturation, but (iii) viral DNA replication is not the rate-limiting step in virion production, and therefore, (iv) PARP-1 does not enhance but reduces KSHV virion production.
FIG. 9.
Effects of drugs that affect PARP-1 activity on production of extracellular progeny KSHV. BCBL-1 cells were induced with TPA in the presence of 3-AB (3 mM), NA (10 mM), and HU (50 μM) for 4 days. The media were replaced once, after 48 h of treatment, with fresh media containing the drugs. Viruses in the media were collected and concentrated 100-fold. Virus stocks (200 μl) were treated with Turbo DNase I for 1 h at 37°C, and viral DNAs were extracted. Viral DNAs were analyzed by a real-time PCR assay using primers to LANA. A serial dilution of a known amount of BAC36 DNA was used to construct a standard curve. Copy numbers were normalized and expressed per milliliter of supernatant.
Binding of SAF-A to ori-Lyt DNA and possible role of the nuclear matrix attachment in KSHV DNA replication.
The finding that scaffold attachment factor A (SAF-A) binds to an ori-Lyt DNA fragment spanning nucleotides 23705 to 24001 suggests the existence of a scaffold/matrix attachment region (S/MAR) in the ori-Lyt. The matrix protein SAF-A may tether the ori-Lyt DNA to the nuclear scaffold or matrix through the S/MAR. This hypothesis is very intriguing because increasing evidence suggests that the nuclear skeleton may act as a dynamic support for many specialized reactions, including DNA replication and transcription. For instance, S/MARs are frequently found near DNA replication origins and transcription enhancers or promoters and these attachment regions have clear effects on gene expression, chromatin accessibility, and DNA replication (7). Recently, it was demonstrated that a 155-bp minimal S/MAR DNA module linked to an upstream transcription unit is sufficient for the replication and mitotic stability of an episomal vector. This vector associates with the nuclear matrix by means of specific interaction of the S/MAR with SAF-A (21). This result suggests that the activation of a DNA replication origin depends on epigenetic events, such as binding of transcription factors, chromatin structure, and nuclear localization. However, the epigenetic events required for eukaryotic DNA replication and the molecular mechanism behind the effects are far from being understood.
Acknowledgments
We thank all members of the Yuan laboratory for suggestions and critical reading of the manuscript. We thank Keiji Ueda at Osaka University for anti-RTA monoclonal antibody, Gary Hayward at Johns Hopkins for anti-SSB polyclonal antibody, Perry Blackshear at NIEHS for anti-RecQL polyclonal antibody, Gideon Dreyfuss at the University of Pennsylvania for anti-SAF-A monoclonal antibody, Benigno Valdez at Baylor College of Medicine for anti-Ku86 monoclonal antibody, Daniel Simmons at the University of Delaware for anti-Topo-I monoclonal antibody, and Gary Gorbsky at the Oklahoma Medical Research Foundation for polyclonal anti-Topo-IIβ antibody. We thank Kaye Speicher and Thomas Beer of the Wistar Institute proteomics facility for superb mass spectrometric analyses.
This work was supported by funding from the National Institutes of Health (R01AI52789 and R01CA86839).
Footnotes
Published ahead of print on 16 January 2008.
REFERENCES
- 1.Antman, K., and Y. Chang. 2000. Kaposi's sarcoma. N. Engl. J. Med. 3421027-1038. [DOI] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318542-555. [DOI] [PubMed] [Google Scholar]
- 3.AuCoin, D. P., K. S. Colletti, Y. Xu, S. A. Cei, and G. S. Pari. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 767890-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avemann, K., R. Knippers, T. Koller, and J. M. Sogo. 1988. Camptothecin, a specific inhibitor of type I DNA topoisomerase, induces DNA breakage at replication forks. Mol. Cell. Biol. 83026-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachrati, C. Z., and I. D. Hickson. 2003. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 374577-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, J. D., and E. S. Huang. 1988. Two specific topoisomerase II inhibitors prevent replication of human cytomegalovirus DNA: an implied role in replication of the viral genome. J. Virol. 624797-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bode, J., S. Goetze, H. Heng, S. A. Krawetz, and C. Benham. 2003. From DNA structure to gene expression: mediators of nuclear compartmentalization and dynamics. Chromosome Res. 11435-445. [DOI] [PubMed] [Google Scholar]
- 8.Boehmer, P. E., M. S. Dodson, and I. R. Lehman. 1993. The herpes simplex virus type-1 origin binding protein. DNA helicase activity. J. Biol. Chem. 2681220-1225. [PubMed] [Google Scholar]
- 9.Buermeyer, A. B., S. M. Deschenes, S. M. Baker, and R. M. Liskay. 1999. Mammalian DNA mismatch repair. Annu. Rev. Genet. 33533-564. [DOI] [PubMed] [Google Scholar]
- 10.Burkle, A. 2001. Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays 23795-806. [DOI] [PubMed] [Google Scholar]
- 11.Cesarman, E., E. A. Mesri, and M. C. Gershengorn. 2000. Viral G protein-coupled receptor and Kaposi's sarcoma: a model of paracrine neoplasia? J. Exp. Med. 191417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattopadhyay, S., Y. Chen, and S. K. Weller. 2006. The two helicases of herpes simplex virus type 1 (HSV-1). Front. Biosci. 112213-2223. [DOI] [PubMed] [Google Scholar]
- 13.Ebert, S. N., S. S. Shtrom, and M. T. Muller. 1990. Topoisomerase II cleavage of herpes simplex virus type 1 DNA in vivo is replication dependent. J. Virol. 644059-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewesuedo, R. B., and M. J. Ratain. 1997. Topoisomerase I inhibitors. Oncologist 2359-364. [PubMed] [Google Scholar]
- 15.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-Acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 665030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 692998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganem, D. 1996. Human herpesvirus 8 and the biology of Kaposi's sarcoma. Semin. Virol. 7325-332. [Google Scholar]
- 18.Gaspar, M., and T. Shenk. 2006. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc. Natl. Acad. Sci. USA 1032821-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grundhoff, A., and D. Ganem. 2004. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Investig. 113124-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gwack, Y., H. J. Baek, H. Nakamura, S. H. Lee, M. Meisterernst, R. G. Roeder, and J. U. Jung. 2003. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 232055-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenke, A. C., I. M. Stehle, F. Herrmann, T. Eisenberger, A. Baiker, J. Bode, F. O. Fackelmayer, and H. J. Lipps. 2004. Nuclear scaffold/matrix attached region modules linked to a transcription unit are sufficient for replication and maintenance of a mammalian episome. Proc. Natl. Acad. Sci. USA 10111322-11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawanishi, M. 1993. Topoisomerase I and II activities are required for Epstein-Barr virus replication. J. Gen. Virol. 742263-2268. [DOI] [PubMed] [Google Scholar]
- 23.Lieber, M. R., Y. Ma, U. Pannicke, and K. Schwarz. 2003. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4712-720. [DOI] [PubMed] [Google Scholar]
- 24.Lilley, C. E., C. T. Carson, A. R. Muotri, F. H. Gage, and M. D. Weitzman. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 1025844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, C. L., H. Li, Y. Wang, F. X. Zhu, S. Kudchodkar, and Y. Yuan. 2003. Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZIP protein with the origin. J. Virol. 775578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makhov, A. M., S. S. Lee, I. R. Lehman, and J. D. Griffith. 2003. Origin-specific unwinding of herpes simplex virus 1 DNA by the viral UL9 and ICP8 proteins: visualization of a specific preunwinding complex. Proc. Natl. Acad. Sci. USA 100898-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marintcheva, B., and S. K. Weller. 2001. Residues within the conserved helicase motifs of UL9, the origin-binding protein of herpes simplex virus-1, are essential for helicase activity but not for dimerization or origin binding activity. J. Biol. Chem. 2766605-6615. [DOI] [PubMed] [Google Scholar]
- 28.Marintcheva, B., and S. K. Weller. 2003. Helicase motif Ia is involved in single-strand DNA-binding and helicase activities of the herpes simplex virus type 1 origin-binding protein, UL9. J. Virol. 772477-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monier, K., J. C. Gonzalez Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2661-665. [DOI] [PubMed] [Google Scholar]
- 30.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2342-346. [DOI] [PubMed] [Google Scholar]
- 31.Richter, A., U. Strausfeld, and R. Knippers. 1987. Effects of VM26 (teniposide), a specific inhibitor of type II DNA topoisomerase, on SV40 DNA replication in vivo. Nucleic Acids Res. 153455-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sangrithi, M. N., J. A. Bernal, M. Madine, A. Philpott, J. Lee, W. G. Dunphy, and A. R. Venkitaraman. 2005. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 121887-898. [DOI] [PubMed] [Google Scholar]
- 33.Schulz, T. F. 1998. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J. Gen. Virol. 791573-1591. [DOI] [PubMed] [Google Scholar]
- 34.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun, R., S.-F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 732232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 785856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Y., H. Li, M. Y. Chan, F. X. Zhu, D. M. Lukac, and Y. Yuan. 2004. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: cis-acting requirements for replication and ori-Lyt-associated RNA transcription. J. Virol. 788615-8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, Y., Q. Tang, G. G. Maul, and Y. Yuan. 2006. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator. J. Virol. 8012171-12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West, S. C. 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4435-445. [DOI] [PubMed] [Google Scholar]
- 40.Wu, C. A., N. J. Nelson, D. J. McGeoch, and M. D. Challberg. 1988. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J. Gen. Virol. 62435-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, F. Y., J. Ahn, D. J. Alcendor, W. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 751487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, F. Y., S. E. Wang, Q.-Q. Tang, M. Fujimuro, C.-J. Chiou, Q. Zheng, H. Chen, S. D. Hayward, M. D. Lane, and G. S. Hayward. 2003. Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein and p21CIP-1. J. Virol. 778893-8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 936641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]