Abstract
We previously showed that the cysteines flanking the internal fusion peptide of the avian sarcoma/leukosis virus subtype A (ASLV-A) Env (EnvA) are important for infectivity and cell-cell fusion. Here we define the stage of fusion at which the cysteines are required. The flanking cysteines are dispensable for receptor-triggered membrane association but are required for the lipid mixing step of fusion, which, interestingly, displays a high pH onset and a biphasic profile. Second-site mutations that partially restore infection partially restore lipid mixing. These findings indicate that the cysteines flanking the internal fusion peptide of EnvA (and perhaps by analogy Ebola virus glycoprotein) are important for the foldback stage of the conformational changes that lead to membrane merger.
The avian sarcoma/leukosis virus (ASLV) Env protein is unusual among class I fusion proteins in that it contains an internal fusion peptide, a characteristic it shares with filovirus (Ebola virus and Marburg virus) glycoproteins. These fusion peptides are flanked by two Cys residues, which, based on structural and mutagenesis evidence, likely form a disulfide bond (9, 21). The ASLV fusion peptide contains one proline, and the filovirus fusion peptides contain two prolines, at their centers. Previous work has shown that these Pro residues are important for fusion (4, 8). The fusion peptide of EnvA is operationally defined as residues 22 to 37 of the transmembrane (TM) subunit. We previously provided evidence that the Pro in the middle of this peptide (P29) requires a β-turn structure (4) at some stage of fusion. We subsequently showed that the two Cys residues (C9 and C45) that define the likely disulfide-bonded loop encompassing the fusion peptide are required for infectivity and cell-cell fusion (5). In this study, we identified the specific stage of fusion that requires the Cys residues that flank the EnvA fusion peptide.
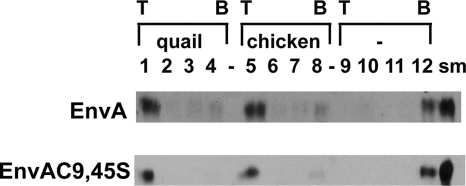
To begin our studies, we asked if the defect was in the first step of fusion: target membrane binding. Accordingly, we determined the ability of murine leukemia virus pseudotyped virions (10) bearing either wild-type EnvA or an EnvA harboring double Cys-to-Ser mutations at both positions 9 and 45 of the TM subunit (referred to below as EnvAC9,45S) to bind to target membranes in a receptor-dependent manner (6, 7, 15). As seen in Fig. 1, wild-type murine leukemia virus pseudovirions bound liposomes and floated to the top of the gradient when either the quail (lane 1) or the chicken (lane 5) form of the soluble Tva receptor (sTva) was used (7). In the absence of sTva, all of the pseudovirions remained at the bottom of the gradient (Fig. 1, lane 12). Similar receptor-dependent membrane association was observed for EnvAC9,45S (Fig. 1). These data show that the Cys residues (C9 and C45) that define the loop encompassing the fusion peptide are not required for receptor-triggered binding of EnvA-bearing virus particles to target membranes.
FIG. 1.
Receptor-triggered membrane association of EnvA and EnvAC9,45S. Virus-liposome association assays were modified from the work of Netter et al. (15). Briefly, 40 μl of virus was mixed with sTva (final concentration, 1 μM) and allowed to associate for 30 min on ice. After addition of 50 μl of liposomes (a 1:1:1:1.5 mixture of phosphatidylcholine, PE, sphingomyelin, and cholesterol; extruded through 100-nm-pore-size filters) and additional HM buffer (20 mM HEPES, 20 mM morpholineethanesulfonic acid, 130 mM NaCl [pH 7.5]) to bring the final volume to 100 μl, the samples were incubated at 37°C for 30 min and then returned to ice. The virus-liposome mixture was then diluted 1:1 with 80% (wt/vol) sucrose in HM buffer, transferred to a 700-μl centrifuge tube, and overlaid first with 400 μl of 25% sucrose and then with 50 μl of 5% sucrose. Samples were centrifuged at 150,000 × g in a 55 Ti rotor for 2 h. Four fractions, of 100, 150, and 150 μl and the remainder (approximately 200 μl), were collected from the top of the gradient. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose filters, probed with an anti-A tail antibody (7), and visualized with a horseradish peroxidase-conjugated anti-rabbit antibody. Either the quail or the chicken isoform of sTva or no receptor (-) was used to trigger membrane binding, as indicated. T, top of the gradient; B, bottom of the gradient; sm, a portion (1/10) of the starting sample used for the given triggering experiment.
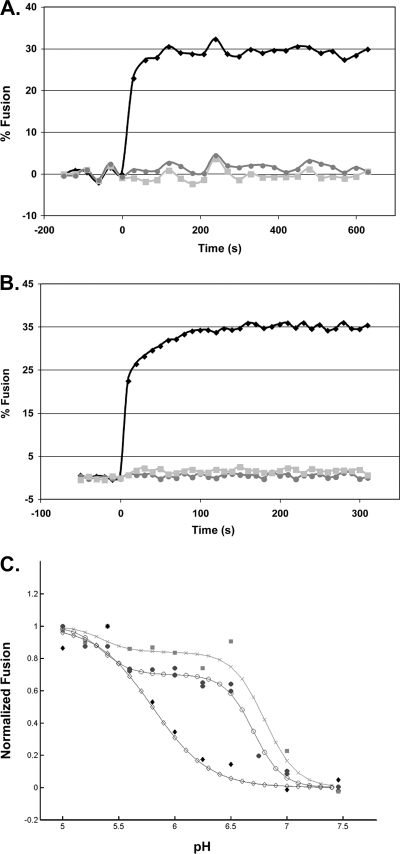
We then asked whether the cysteines are required for EnvA to reach the lipid mixing stage of fusion. To do this, we employed the fluorescence resonance energy transfer (FRET)-based virus-liposome fusion assay (18) adapted to a 384-well plate reader format. In this assay, the FRET pair consisting of (N-7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadecanoyl-sn-glycero-3-phosphatidylethanolamine (NBD-PE; Molecular Probes) and rhodamine-phosphatidylethanolamine (Rh-PE; Avanti) is added to liposomes in quantities that allow transfer of the NBD emission energy to Rh-PE in the liposome. Dilution of the probes upon fusion with a virus membrane can be measured by a loss of FRET. We first tested the validity of the assay by using the well- established infectious clone of ASLV subtype A (ASLV-A) [RCASBP(A)AP] (6a) and known triggers for and inhibitors of fusion. To this end, ASLV-A was incubated with sTva for 30 min at 4°C and pH 7.5. Liposomes containing the NBD-PE-Rh-PE FRET pair were then added and the mixture warmed to 37°C for 20 min to induce receptor-triggered target membrane association (6, 7). The samples were then adjusted to pH 5 (37°C), and the fluorescence was monitored. No fusion was observed if samples were incubated at pH 7.5. However, as seen in Fig. 2A, when the ASLV-A-sTva-liposome mixture was exposed to pH 5 at 37°C, fusion was rapid, reaching a maximum within 1 to 2 min. No fusion was observed when sTva was omitted from the virus-liposome mixture, and a peptide (R99) corresponding to the C-helix region of the EnvA TM subunit, which inhibits fusion and infectivity (6, 15), inhibited lipid mixing. Thus, the known conditions for ASLV-A Env (EnvA)-mediated fusion (dependence on preincubation with the receptor and subsequent exposure to a low pH [11, 12, 14], and inhibition by the C-helix peptide) are observed using this assay.
FIG. 2.
Characterization of lipid mixing induced by ASLV and VSV-EnvA. (A and B) Virus from infected cell culture medium was concentrated in 300,000-molecular-weight-cutoff Vivaspin columns and purified by banding on a step gradient of 25% and 60% sucrose (wt/vol). The visible band was collected, pelleted and concentrated by centrifuging through 20% sucrose, and resuspended in 150 μl HM buffer (20 mM HEPES, 20 mM morpholineethanesulfonic acid, 130 mM NaCl [pH 7.5]) overnight on ice at 4°C. To assay lipid mixing, 25 μg ASLV-A (A) or VSV-EnvA (B) was first incubated with sTva (solid black diamonds and solid light grey squares) or without sTva (solid dark grey circles) at pH 7.5 for 30 min at 4°C; liposomes (a 1:1:1:1.5:0.045:0.11 mixture of phosphatidylcholine, PE, sphingomyelin, cholesterol, Rh-PE, and NBD-PE) prepared by extrusion through 50-nm-pore-size filters (6) were added; and the mixture was warmed to 37°C for 15 min to trigger target membrane binding, after which baseline fluorescence at pH 7.5 was recorded for 5 min at 37°C. At time zero, the samples were adjusted to pH 5 and incubated at 37°C, and fluorescence (excitation at 460 nm; emission at 540 nm) was measured for 10 min. Where indicated, R99 was added to a final concentration of 100 μg/ml prior to acidification (solid light grey squares). Maximum possible NBD fluorescence was determined by adding NP-40 to a final concentration of 1% and measuring fluorescence at 37°C for 15 min. Percent fusion was calculated as (FpH − F0)/(FT − F0) × 100, where F0 is the baseline fluorescence (pH 7.5), FpH is the averaged fluorescence at the plateau at pH 5.0, and FT is the fluorescence at an infinite dilution (after disruption of the membranes with 1% NP-40). The data from triplicate samples were averaged. Results of a representative experiment are shown. Each experiment was repeated two or more times. (C) pH dependence of lipid mixing. Samples were treated as for panels A and B except that the pH was adjusted as indicated on the x axis. The results from triplicate samples of individual experiments were averaged, the value at pH 5 was set to 1, and fusion at each individual pH was reported as a fraction of the pH 5 value. The data from two independent ASLV-A experiments (solid dark grey circles), one VSV-EnvA experiment (solid light grey squares), and one influenza X:31 virus (Charles River Laboratories) experiment are plotted. The data for each virus were then fitted by a nonlinear least-squares method using the MatLab curve-fitting toolbox and the equation % fusion = c{1 − tanh[d(pH − pH0)]}, where pH0 is the pH at the inflection point of the curve, c is one-half the height between the initial and the maximal fusion for the curve, and the product(c·d) is the slope of the curve at the inflection point. The data from two independent ASLV experiments (filled circles) and one VSV-EnvA experiment (shaded squares) are plotted. For comparison, the pH dependence of influenza virus X:31 (Charles River Laboratories) was determined (filled diamonds). The fit curves are plotted using the corresponding open circles, X's, and open diamonds, respectively.
The experiments described above were performed using ASLV-A derived from chronically infected DF-1 cells. However, no mutant viruses bearing EnvA containing the double mutation C9,45S could be isolated from infected DF-1 cells (D. C. Melder, X. Q. Yin, S. E. Delos, and M. J. Federspiel, unpublished data). Thus, the fusion activity of EnvAC9,45S could not be assessed using native ASLV-A virions. Vesicular stomatitis virus (VSV) pseudotyped particles have been used extensively to study virus entry directed by foreign viral glycoproteins (17, 19), and EnvA is well incorporated into VSV pseudotyped particles (Z. Chen, unpublished data), We therefore assessed the fusion activity of EnvA on VSV pseudovirions. As seen with ASLV-A (Fig. 2A), the lipid-mixing activity of VSV-EnvA pseudovirions is rapid and complete within 1 to 2 min, requires preincubation with sTva and subsequent exposure to a low pH, and is inhibited by R99 (Fig. 2B). Therefore, the VSV-EnvA system appeared to be suitable for assessing the roles of C9 and C45 in EnvA-mediated fusion.
Before testing the mutant EnvA's, we characterized the pH dependence of lipid mixing. As seen in Fig. 2C, ASLV-A lipid mixing occurs in two stages, fit by two steps, with the first step occurring at a relatively high pH (pH 6.7; half-maximum fusion) and additional fusion occurring below pH 5.8. The pH curve for VSV-EnvA is similar to that for ASLV-A and is also fit as a two-step process. The similar biphasic nature of the two curves further suggests that the fusion characteristics of EnvA are retained in the VSV pseudotype system. As expected, and in contrast to EnvA, the lipid mixing activity of influenza virus displayed a low pH profile that rose as a single smooth curve. The unusually high pH threshold for lipid mixing observed for both ASLV-A and VSV-EnvA (onset at pH ∼7) may help explain our prior observation that ASLV-A labeled with pyrene can fuse with cells at a neutral pH (6); pyrene in a retrovirus membrane may promote lipid mixing. The second stage of EnvA-mediated lipid mixing at a lower pH could be due to fusion by a population of EnvA's that require a lower pH to convert them to a fusion-active form or may represent the transition from hemifusion to full fusion. The latter interpretation is consistent with the reported pH requirements for virus entry, infectivity, and cell-cell fusion (3, 13, 14; S. E. Delos, unpublished data), which are all posthemifusion events, and the reported delay between lipid mixing and fusion pore formation (12).
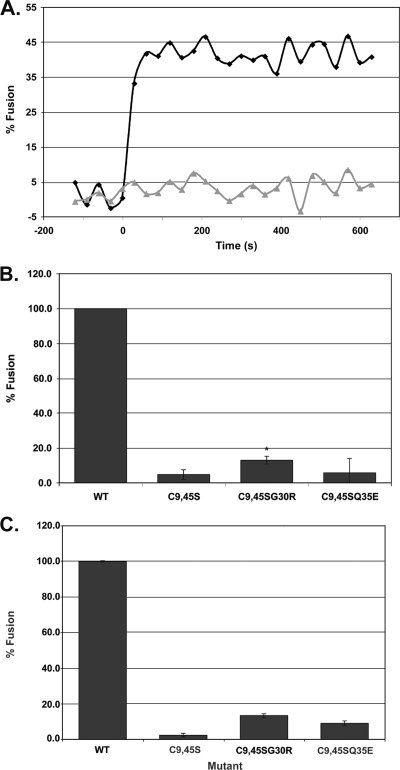
To address the question of whether EnvAC9,45S can mediate lipid mixing, we prepared VSV pseudovirions bearing wild-type EnvA or EnvAC9,45S, and used them to assess the ability of EnvAC9,45S to induce lipid mixing. EnvAC9,45S was well incorporated into VSV particles. As seen in Fig. 3A, no lipid mixing was observed for VSV-EnvAC9,45S. Thus, the cysteines flanking the fusion peptide are required for the lipid mixing step of fusion.
FIG. 3.
Characterization of lipid mixing induced by VSV-EnvAC9,45S and two second-site revertants. (A) Lipid mixing induced by VSV-EnvA (diamonds) and VSV-EnvAC9,45S (triangles). (B and C) Extent of lipid mixing for VSV-EnvA (wild type [WT]), VSV-EnvAC9,45S, VSV-EnvAC9,45SG30R, and VSV-EnvAC9,45SQ35E. Virus samples were prepared and lipid mixing assessed as described for Fig. 2, except that the relative amount of each virus used per assay was normalized for the relative incorporation of the various EnvA's (ranges, 0.63 to 1.42 for EnvAC9,45S, 0.63 to 0.86 for EnvAC9,45SG30R, and 0.71 to 1.02 for EnvAC9,45SQ35E, relative to wild-type EnvA) and the fusion observed for wild-type EnvA in the presence of R99 was used as pH0. Data in panel B are averages from four experiments, each carried out in triplicate. t tests were performed using the “two sample assuming unequal variances” function in Microsoft Excel [*, P(T ≤ t) = 0.0037]. Data in panel C are averages from duplicate samples from a single experiment. In panels B and C, FpH was calculated as for Fig. 2C.
RCAS viruses bearing EnvAC9,45S have been used to select for compensatory mutations (Melder et al., unpublished). Every recovered virus selected on DF-1 cells retained the original C9,45S mutations and had a second-site mutation in the fusion peptide at G30R, immediately adjacent to the central Pro (P29) (Melder et al., unpublished). In a second screen, on turkey embryo fibroblasts, 7 of 10 clones retained the original C9,45S mutations and contained the single additional mutation Q35E (Melder et al., unpublished). Thus, in each case, the loss of the flanking Cys pair was partially compensated for by a single second-site mutation to a charged residue within the fusion peptide. Viruses bearing G30R or Q35E on the wild-type backbone gave wild-type titers, but the same mutants in the context of EnvAC9,45S produced only ∼5 to 10% of wild-type titers on DF-1 cells (Melder et al., unpublished). Thus, when present alone, G30R and Q35E had no deleterious effect on fusion, indicating that the defects in the EnvAC9,45SG30R and EnvAC9,45SQ35E viruses must be due to the loss of C9 and C45, further demonstrating the importance of C9 and C45 for infectivity.
Since the defect of the parent mutation (EnvAC9,45S) is at the lipid mixing stage of fusion (Fig. 3A), we wondered whether either mutation (G30 to R or Q35 to E) could restore measurable lipid mixing activity to EnvAC9,45S commensurate with its partial rescue (∼5 to 10%) of infectivity. Accordingly, we prepared VSV pseudovirions bearing each of these triple mutations in EnvA and assessed their abilities to induce lipid mixing. As seen in Fig. 3B, we observed a small but statistically significant restoration of lipid mixing when G30 was replaced with R (in the C9,45S backbone). The level of restoration (to 13.0% ± 2.2% of the wild-type level) is similar to the level of recovery of infection for ASLV-A bearing EnvAC9,45SG30R. The results were more variable when Q35 was replaced with E (in the C9,45S backbone); we sometimes saw recovery to ∼10% of the wild-type level, but at other times we did not (Fig. 3B) (Z. Chen, unpublished data). These experiments were conducted with our standard amount of virus/reaction. When we doubled the amount of virus used, a small but noticeable amount of lipid mixing was observed for both mutant viruses (Fig. 3C). The poor rescue of lipid mixing for EnvAC9,45SQ35E versus EnvAC9,45SG30R is somewhat puzzling given their infectivity titers. One possibility is that the composition of our liposomes is not optimal for EnvAC9,45SQ35E.
In this study we identified the earliest step in fusion at which the Cys residues (C9 and C45) flanking the internal fusion peptide of ASLV EnvA are required. C9 and C45 are dispensable for receptor-triggered target membrane association, but they are stringently required for the lipid mixing stage of fusion. We envision two non-mutually exclusive roles for the cysteines flanking the EnvA fusion peptide: (i) to allow the EnvA TM subunit to fold back into a trimer of hairpins compact enough to mediate lipid mixing, perhaps involving interactions between the fusion loop and the pre-TM or TM regions (1, 2, 16, 20) of EnvA or (ii) to allow the fusion peptide to remain tightly associated with the target membrane during the foldback stage of fusion. Our findings are likely relevant to structural requirements for other internal fusion peptides flanked by Cys residues, notably those of the Ebola virus and Marburg virus glycoproteins.
Acknowledgments
We thank Shutoku Matsuyama for construction of the chicken sTva plasmid, Lukas Tamm for critical comments on the text, and John Delos for performing the best-fit analysis in Fig. 2C.
This work was supported by grants from the NIH to J.M.W. and S.E.D (AI22470) and to M.J.F. (AI48682). M.B.B. was supported in part by NIH award 5 T32 AI07076-27 to the Infectious Diseases Training Program at the University of Virginia.
Footnotes
Published ahead of print on 9 January 2008.
REFERENCES
- 1.Armstrong, R. T., A. S. Kushnir, and J. M. White. 2000. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 151425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3309-319. [DOI] [PubMed] [Google Scholar]
- 3.Barnard, R. J., S. Narayan, G. Dornadula, M. D. Miller, and J. A. Young. 2004. Low pH is required for avian sarcoma and leukosis virus Env-dependent viral penetration into the cytosol and not for viral uncoating. J. Virol. 7810433-10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delos, S. E., J. M. Gilbert, and J. M. White. 2000. The central proline of an internal viral fusion peptide serves two important roles. J. Virol. 741686-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delos, S. E., and J. M. White. 2000. Critical role for the cysteines flanking the internal fusion peptide of avian sarcoma/leukosis virus envelope glycoprotein. J. Virol. 749738-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earp, L. J., S. E. Delos, R. C. Netter, P. Bates, and J. M. White. 2003. The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH. J. Virol. 773058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Federspiel, M. J., and S. H. Hughes. 1997. Retroviral gene delivery. Methods Cell Biol. 52179-214. [PubMed] [Google Scholar]
- 7.Hernandez, L. D., R. J. Peters, S. E. Delos, J. A. Young, D. A. Agard, and J. M. White. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 1391455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito, H., S. Watanabe, A. Sanchez, M. A. Whitt, and Y. Kawaoka. 1999. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J. Virol. 738907-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeffers, S. A., D. A. Sanders, and A. Sanchez. 2002. Covalent modifications of the Ebola virus glycoprotein. J. Virol. 7612463-12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landau, N. R., and D. R. Littman. 1992. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J. Virol. 665110-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama, S., S. E. Delos, and J. M. White. 2004. Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein. J. Virol. 788201-8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melikyan, G. B., R. J. Barnard, L. G. Abrahamyan, W. Mothes, and J. A. Young. 2005. Imaging individual retroviral fusion events: from hemifusion to pore formation and growth. Proc. Natl. Acad. Sci. USA 1028728-8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melikyan, G. B., R. J. Barnard, R. M. Markosyan, J. A. Young, and F. S. Cohen. 2004. Low pH is required for avian sarcoma and leukosis virus Env-induced hemifusion and fusion pore formation but not for pore growth. J. Virol. 783753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103679-689. [DOI] [PubMed] [Google Scholar]
- 15.Netter, R. C., S. M. Amberg, J. W. Balliet, M. J. Biscone, A. Vermeulen, L. J. Earp, J. M. White, and P. Bates. 2004. Heptad repeat 2-based peptides inhibit avian sarcoma and leukosis virus subgroup A infection and identify a fusion intermediate. J. Virol. 7813430-13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park, H. E., J. A. Gruenke, and J. M. White. 2003. Leash in the groove mechanism of membrane fusion. Nat. Struct. Biol. 101048-1053. [DOI] [PubMed] [Google Scholar]
- 17.Schornberg, K., S. Matsuyama, K. Kabsch, S. Delos, A. Bouton, and J. White. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 804174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stegmann, T., D. Hoekstra, G. Scherphof, and J. Wilschut. 1985. Kinetics of pH-dependent fusion between influenza virus and liposomes. Biochemistry 243107-3113. [DOI] [PubMed] [Google Scholar]
- 19.Takada, A., C. Robison, H. Goto, A. Sanchez, K. G. Murti, M. A. Whitt, and Y. Kawaoka. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 9414764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamm, L. K. 2003. Hypothesis: spring-loaded boomerang mechanism of influenza hemagglutinin-mediated membrane fusion. Biochim. Biophys. Acta 161414-23. [DOI] [PubMed] [Google Scholar]
- 21.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2605-616. [DOI] [PubMed] [Google Scholar]