Abstract
Vaccination for human immunodeficiency virus type 1 (HIV-1) remains an elusive goal. Whether an unsuccessful vaccine might not only fail to provoke detectable immune responses but also could actually interfere with subsequent natural immunity upon HIV-1 infection is unknown. We performed detailed assessment of an HIV-1 gag DNA vaccine recipient (subject 00015) who was previously uninfected but sustained HIV-1 infection before completing a vaccination trial and another contemporaneously acutely infected individual (subject 00016) with the same strain of HIV-1. Subject 00015 received the vaccine at weeks 0, 4, and 8 and was found to have been acutely HIV-1 infected around the time of the third vaccination. Subject 00016 was a previously HIV-1-seronegative sexual contact who had symptoms of acute HIV-1 infection approximately 2 weeks earlier than subject 00015 and demonstrated subsequent seroconversion. Both individuals reached an unusually low level of chronic viremia (<1,000 copies/ml) without treatment. Subject 00015 had no detectable HIV-1-specific cytotoxic T-lymphocyte (CTL) responses until a borderline response was noted at the time of the third vaccination. The magnitude and breadth of Gag-specific CTL responses in subject 00015 were similar to those of subject 00016 during early chronic infection. Viral sequences from gag, pol, and nef confirmed the common source of HIV-1 between these individuals. The diversity and divergence of sequences in subjects 00015 and 00016 were similar, indicating similar immune pressure on these proteins (including Gag). As a whole, the data suggested that while the gag DNA vaccine did not prime detectable early CTL responses in subject 00015, vaccination did not appreciably impair his ability to contain viremia at levels similar to those in subject 00016.
Human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T lymphocytes (CTL) are a key arm of protective immunity in the immunopathogenesis of HIV-1 infection (25). Numerous studies of infected persons and experimental work in the macaque simian immunodeficiency virus model have shown that this immune response plays an antiviral role in early and chronic infection and wanes with later disease progression. Because CTL respond to epitopes presented by polymorphic human leukocyte antigen (HLA) class I molecules, the genetic backgrounds of both the infecting HIV-1 strain and the infected host individual affect the interaction of CTL and HIV-1 and determine the targeting of CTL responses against the virus (23).
The general goal of many HIV-1 vaccine approaches being developed is to produce CTL responses that will attenuate the disease course of natural infection. Although it is clear that HIV-1-specific CTL play an important role in delaying disease, a precise CTL measurement providing a “correlate of immunity” for protection from infection and/or disease remains elusive, despite recently developed assays such as the gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) that provide remarkably accurate measurements of CTL magnitude and targeting (21). At this stage, many human vaccine development efforts are focused on an initial benchmark of achieving CTL immunogenicity comparable to that of natural infection, with the goal of attenuating subsequent infection if it cannot be altogether prevented (13). To date, the most promising strategy for CTL immunogenicity has been vaccination with recombinant adenovirus type 5 (Ad5), while other strategies have been disappointingly inconsistent in generating CTL responses. Because all vaccine candidates that have advanced to human trials have shown immunogenic promise in animal models, the mechanism(s) of failure in humans is unclear.
It is yet unknown how vaccination prior to natural infection might affect the efficacy of CTL against HIV-1 in human vaccinees. The goal of preventive vaccines is priming of HIV-1-specific CTL responses that will attenuate or clear subsequent natural infection; however, it is possible that vaccination could be not only ineffective but actually deleterious. There are at least three major mechanisms whereby a vaccine could reduce the efficacy of the CTL response to subsequent HIV-1 infection. First, the vaccination could promote tolerance rather than immunoreactivity and blunt responsiveness to infection. Second, differences between the vaccine and challenge HIV-1 sequences could result in vaccine sequence-specific CTL responses that do not react against the infecting viral strain and even impair strain-specific CTL responses due to “original antigenic sin.” Third, the vaccine could subvert natural patterns of immunodominance and misdirect CTL to respond with less effective targeting; the response could be biased toward the protein(s) in the vaccine and not directed against other HIV-1 proteins not represented by the vaccine.
In this study, we perform a detailed evaluation of CTL responses and HIV-1 sequences of two persons who were infected at approximately the same time with the same strain of HIV-1. One of these individuals was infected during his vaccination course with a Gag DNA vaccine. The patterns of CTL reactivity against Gag and non-Gag proteins and the evolution of the HIV-1 genome in gag, pol, and nef are evaluated and compared for these two persons.
MATERIALS AND METHODS
Study participants.
Subject 00015 was a 43-year-old male participant in a vaccine trial of an HIV-1 gag DNA vaccine, who was documented to be HIV-1 seronegative at the time of enrollment but subsequently seroconverted during the study. Subject 00016 was a 44-year-old male sexual contact of subject 00015, who was initially HIV-1 seronegative but seroconverted near the time of his sexual contact with subject 00015. Both men reported other sexual partners in the months before seroconversion. This study was performed under an informed consent protocol approved by the UCLA institutional review board.
Vaccine protocol.
Subject 00015 was enrolled in a safety and immunogenicity trial of the Merck HIV-1 Gag DNA vaccine with aluminum phosphate adjuvant (HIV-1 gag DNA vaccine/AlPO4), which was previously tested and described for the macaque simian immunodeficiency virus model (6). He was randomized into the test (not control) group for the vaccine, which contained mammalian codon-optimized HIV-1 CAM-1 (19) gag sequence (closely resembling the clade B consensus). The study protocol consisted of vaccinations at weeks 0, 4, and 8 with 5 mg DNA with adjuvant, followed by a fourth booster vaccination with either the same DNA vaccine or a recombinant Ad5 HIV-1 gag vaccine. Subject 00015 completed the first three vaccinations but did not receive the fourth booster vaccination due to diagnosis with HIV-1 infection around the time of the third vaccination.
Clinical monitoring.
During participation in the vaccine trial, subject 00015 was evaluated at day −30, at which time a screening HIV enzyme-linked immunosorbent assay (ELISA) was negative. Due to the fact that the study enrolled healthy, low-risk volunteers, HIV testing was not routinely performed during follow-up visits. Subject 00015 was seen in the clinic during each study visit. After his first vaccination at day 0, he had weekly clinic visits for 2 weeks. At the fourth week, he had his second vaccination, after which he had weekly clinic visits for 2 weeks, another clinic visit 2 weeks later, and then monthly clinic visits until week 26. At each visit, subject 00015 was questioned about high-risk sexual exposures for HIV-1 infection and denied any such exposures until he was found to have a positive HIV-1 serology at week 16. Safety laboratory monitoring (UCLA Medical Center clinical laboratories) was performed at weeks 1, 2, 5, and 6 and again at weeks 27 and 28.
Clinical testing of subject 00016 was obtained through his enrollment in E. S. Daar's acute infection cohort study, through the clinical laboratories at Cedars-Sinai Medical Center in Los Angeles.
Measurement of HIV-1 Gag-specific CTL responses by ELISPOT assay during the vaccine trial.
Initial perivaccination measurement of HIV-1-specific CTL responses was monitored at Merck using a previously described IFN-γ ELISPOT assay (7). In brief, peripheral blood mononuclear cells (PBMC) were screened using 20-mer peptides overlapping by 10 amino acids, representing the entire HIV-1 clade B Gag sequence of CAM-1. These peptides were screened in a single pool. The criteria for having a CTL response against Gag were previously established in validation assays (7) to be ≥55 spot-forming cells (SFC) per million PBMC and ≥4 times the negative control.
Measurement of total HIV-1-specific CTL responses by ELISPOT assay.
Subsequent IFN-γ ELISPOT monitoring was performed at UCLA. Whole-proteome HIV-1-specific CTL responses of both subjects were defined using CD8+ T lymphocytes as previously described (11, 22). Briefly, these lymphocytes were polyclonally expanded from PBMC using a CD3-CD4-bispecific monoclonal antibody (20) and these cells were screened using a standard IFN-γ ELISPOT assay, which has been demonstrated to produce results that are highly correlated to unexpanded cells. Synthetic peptides spanning all nine HIV-1 proteins were utilized for the screening (15-mers overlapping by 11 amino acids; NIH AIDS Research and Reference Reagent Program). These peptides were based on clade B consensus sequences, except for Gag (both clade B consensus and strain DU were tested) and Env (strain MN). The ELISPOT plates were scanned on an automated ELISPOT analyzer (Autoimmun Diagnostika GmbH, Germany). The initial screening utilized 53 peptide pools containing up to 16 peptides; individual peptide responses were subsequently deduced using a 4-by-4 matrix strategy and confirmed with individual peptides in further rounds of ELISPOT screening.
HIV-1 sequencing.
HIV-1 sequencing was performed as previously described (23, 24). Briefly, genomic DNA from the cryopreserved PBMC of both subjects (obtained between September 2002 and March 2003) was isolated using the Qiagen DNeasy kit (Qiagen, Valencia, CA) and the proviral HIV-1 genome sequences were amplified by nested PCR using specific primers as described by Altfeld et al. (2). The amplified DNA fragments were cloned into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA) and sequenced by using the BigDye sequence terminator kit, v3.1 (ABI, Foster City, CA) on an ABI 3130 genetic analyzer.
Phylogenetic analysis.
Subject 00015 and 00016 HIV-1 sequences were aligned against the Los Alamos HIV Database clade B consensus sequence and strain NL4-3 using CLUSTAL X, and manually edited. Neighbor-joining trees were constructed using the Tamura-Nei model of nucleotide substitution with a gamma distribution to accommodate rate variation using PHYLIP 3.64 software. The trees were rooted with Consensus B, and 1,000 bootstrap replications were performed. For each subject, the average pairwise nucleotide diversity and average nucleotide divergence between the samples were estimated using the Tamura-Nei model of nucleotide substitution. Standard errors were estimated using 1,000 bootstrap replications.
Nucleotide sequence accession number.
The sequences of the amplified DNA fragments obtained in this study have been submitted to GenBank (accession no. EU1080773 to -180846).
RESULTS
Subject 00015 received an HIV-1 Gag DNA vaccine, and was acutely infected with HIV-1 during the vaccination sequence.
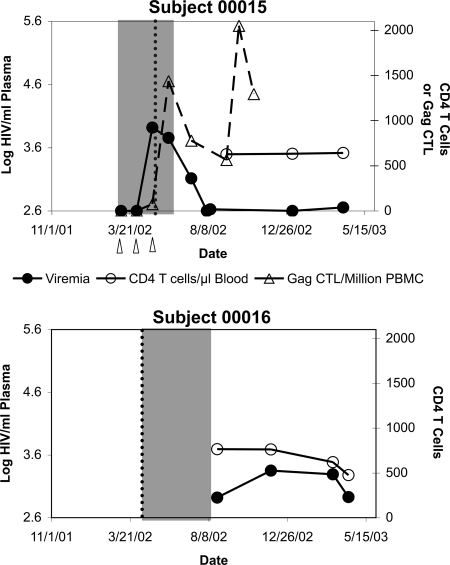
Subject 00015 was a healthy 43-year-old male enrolled in a clinical trial of the Merck HIV-1 gag DNA vaccine/AlPO4 vaccine (a DNA vector containing HIV-1 clade B CAM-1 isolate gag, administered with aluminum phosphate adjuvant) in January 2002 (Fig. 1, top panel). On 30 January 2002, he underwent prevaccination screening, including a negative ELISA for HIV antibodies. He subsequently received three Gag DNA vaccinations administered on 5 March 2002, 2 April 2002, and 30 April 2002. Repeat ELISA screening at his first vaccination visit on 5 March 2002 remained negative. On 5 May 2002, he presented with complaints of acute fever, malaise, headache, pharyngitis, and diffuse maculopapular rash. This syndrome was self-limited, but on 9 July 2002, he was found to have a positive HIV ELISA, confirmed by Western blotting. Retrospective plasma HIV-1 RNA screening revealed undetectable viremia at the first two vaccinations on 5 March 2002 and 2 April 2002, but a level of 8,300 HIV-1 genomes/ml at the third vaccination visit on 30 April 2002, followed by waning levels of 5,700 genomes/ml on 29 May 2002, 1,300 genomes/ml on 9 July 2002, and subsequently stable low-set-point viremia below 1,000 genomes/ml. Subject 00015 gave the additional history that he had had multiple high-risk sexual exposures before and during the vaccine trial. These data indicated that he was acutely infected in April 2002 during his vaccination series, with a peak of viremia occurring between 2 April 2002 and 29 May 2002, probably around 5 May 2002 (when he presented with symptoms of acute infection).
FIG. 1.
HIV-1 infection courses of subjects 00015 and 00016. The levels of plasma viremia and blood CD4+ T-lymphocyte levels are plotted for subject 00015 (top graph) and subject 00016 (bottom graph). For subject 00015, the vaccinations with Gag DNA are indicated by arrowheads, and the total Gag-specific IFN-γ ELISPOT response during the vaccine trial is plotted. The shaded regions indicate the intervals between the last negative and first positive HIV-1 ELISAs. The dotted lines indicate the time of onset of symptoms consistent with primary HIV-1 infection.
Subject 00016 was infected within a few weeks of subject 00015.
Subject 00016 was a 44-year-old male sexual contact of subject 00015. This individual had been previously tested by HIV ELISA and was negative on 15 November 2001 (Fig. 1, bottom panel). He presented for medical attention on 10 April 2002 with acute fever, pharyngitis, and myalgias, at which time an HIV ELISA was negative. However, a repeat ELISA and confirmatory Western blot on 20 August 2002 were positive, as well as detection of plasma HIV-1 RNA on 23 August 2002. Interestingly, subsequent set point viremia for subject 00016 also was low: below 1,000 RNA genomes/ml. While earlier viremia measurements were unavailable to narrow the timing of infection, the onset of the symptoms on 10 April 2002 suggested that subject 00016 was infected a few weeks before subject 00015 and was likely the source of HIV-1 transmission to subject 00015. These individuals therefore were both infected within a narrow window of time and likely infected with the same strain of HIV-1.
HIV-1 Gag-specific CTL responses to vaccination in subject 00015 were undetectable before infection.
During the vaccination trial, subject 00015 was screened for Gag-specific cellular immunity (CD8+ CTL) by IFN-γ ELISPOT assay using pooled Gag peptides. These assays demonstrated no detectable Gag-specific CTL during the first and second vaccinations on 5 March 2002 and 2 April 2002 and borderline responses at the third vaccination of 75 SFC per million PBMC on 30 April 2002 (Fig. 1, top panel). Subsequently, there was a rise in Gag-specific responses to 1,438 SFC/million PBMC on 29 May 2002, coincident with declining viremia. ELISPOT responses against Nef and Env peptide pools arose concurrently with the Gag-specific responses (data not shown). Thus, no Gag-specific responses were seen in the absence of viremia, and rising responses were seen during early viremia, indicating that detectable HIV-1-specific-CTL were not generated during the first two vaccinations, before the onset of viremia due to HIV-1 infection. The timing of rising CTL responses in relationship to declining viremia was consistent with the known temporal relationship of CTL to the drop of viremia at the end of acute infection (5, 15), suggesting that the Gag-specific responses were elicited by HIV-1 infection rather than by the vaccine.
The magnitude and breadth of Gag-specific CTL after HIV-1 infection were similar in subjects 00015 and 00016.
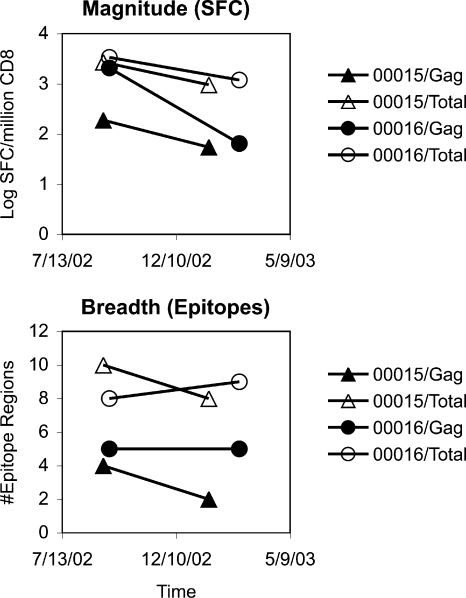
To examine and compare the global HIV-1-specific CTL responses of subjects 00015 and 00016 during early chronic infection, IFN-γ ELISPOT assays using pools of overlapping peptides spanning the entire HIV-1 proteome were performed. Two similar time points were analyzed for each individual in the immediate months after viremia reached the quasisteady-state set point after acute infection. The overall frequencies of CTL responses were assessed in terms of summed responses across all proteins (Fig. 2A). The magnitudes of total HIV-1 targeting were similar between subjects, with the subset of Gag-specific CTL being somewhat higher in subject 00016. The breadth of targeting was determined in terms of the total number of epitope regions (isolated 15-mer peptides or overlapping regions of consecutive peptides) recognized in each subject. The overall breadths also were similar between subjects, with the subset of Gag epitopes being slightly higher in subject 00016. One Gag (Gag 17-13) epitope region containing a previously described HLA-I A*03 epitope was targeted by both subjects (Table 1), who shared the A*03 haplotype in common. (Subjects 00015 and 00016 were A*02 A*03 B*15 B*56 C*01 C*03 and A*03 A*32 B*18 B*40 C*02 C*07, respectively.) Overall, these data demonstrated that subject 00015 was able to mount a Gag-specific CTL response after vaccination, which was not markedly dissimilar to that of subject 00016.
FIG. 2.
Magnitude and breath of the HIV-1-specific CTL responses in subjects 00015 and 00016. (A) The magnitude of CTL targeting (as determined by IFN-γ ELISpot using 53 pools of 16 or fewer overlapping peptides spanning the entire HIV-1 proteome) is plotted for subjects 00015 and 00016. The screening peptides were based on the clade B consensus sequence for Pol, Nef, Tat, Rev, Vpu, Vif, and Vpr; strain MN sequence for Env; and strain DU sequence for Gag. Subsequent screening with clade B consensus Gag sequences revealed similar results to strain DU (not shown). (B) The number of epitope regions (as determined by subsequent analysis of individual 15-mer peptides) is plotted for subjects 00015 and 00016.
TABLE 1.
HIV-1-specific CTL targeting and epitope sequences in subjects 00015 and 00016a
| Protein | Location | Clade B consensus sequence | Subject 00015
|
Subject 00016
|
||
|---|---|---|---|---|---|---|
| CTL | Consensus sequence | CTL | Consensus sequence | |||
| Gag | 17-31 (p17) | EKIRLRPGGKKKYKL | Yes | -------------- (3) | Yes | -----------Q--- (2) |
| 25-39 (p17) | GKKKYKLKHIVWASR | Yes | --------------- (3) | ---Q----------- (2) | ||
| 213-222 (p24) | DRLHPVHAGP | --T------- (17) | --T------- (12) | |||
| 228-242 (p24) | MREPRGSDIAGTTST | --K-----------N (17) | --------------N (12) | |||
| 312-322 (p24) | EVKNWMTETLL | ----------- (17) | Yes | D---------- (12) | ||
| 397-411 (p7) | KEGHIAKNCRAPRKK | ----L-R-------- (3) | Yes | ----L-R-------- (2) | ||
| 425-435 (p7-p1) | DCTERQANFLG | ----------- (3) | Yes | ----------- (2) | ||
| 437-451 (p1-p6) | IWPSHKGRPGNFLQS | ND | Yes | ND | ||
| 461-475 (p6) | ESFRFGEETTTPSQK | ----L-----S--- (1) | Yes | ----------S--- (2) | ||
| 477-487 (p6) | EPIDKELYPLA | -T-G------- (1) | Yes | -A--------- (2) | ||
| Pol | 125-139 (protease) | HKAIGTVLVGPTPVN | Yes | Q-------------- (1) | Q-------------D (2) | |
| 141-155 (protease) | IGRNLLTQIGCTLNF | Yes | --------------- (18) | --------------- (12) | ||
| 421-435 (reverse transcriptase) | WASQIYAGIKVKQLC | Yes | --------------- (6) | --------------- (2) | ||
| 545-559 (reverse transcriptase) | KLPIQKETWEAWWTE | R------------M- (6) | Yes | R------------M- (1) | ||
| 625-639 (RNase) | TDTTNQKTELQAIHL | --------------- (6) | Yes | --------------- (3) | ||
| 933-947 (Int) | TKIQNFRVYYRDSRD | --------------- (3) | Yes | ------------H-- (2) | ||
| Env | 216-226 (gp120) | HYCAPAGFAIL | Yes | ND | ND | |
| 704-714 (gp41) | IVNRVRQGYSP | ND | Yes | ND | ||
| Vif | 121-135 | RNAILGHIVSPRCEY | Yes | -------V---I-D- (3) | ------R-------- (2) | |
| Nef | 81-91 | YKGALDLSHFL | Yes | ----V------ (23) | ----V------ (23) | |
| 105-115 | KRQDILDLWVY | ---E------- (23) | Yes | ---E------- (23) | ||
CTL responses (identified by IFN-γ ELISPOT with overlapping peptides) at the times shown in Fig. 2 are listed. The epitope region sequences correspond to isolated recognized 15-amino-acid peptides or the region of overlap between consecutive recognized peptides. Consensus sequences from each subject are given. The number of clonal sequences is given in parentheses. A dash indicates identity with the clade B consensus sequence. ND, not determined.
The infecting HIV-1 strains of subjects 00015 and 00016 were highly related and demonstrated similar degrees of diversity and divergence from clade B consensus sequences.
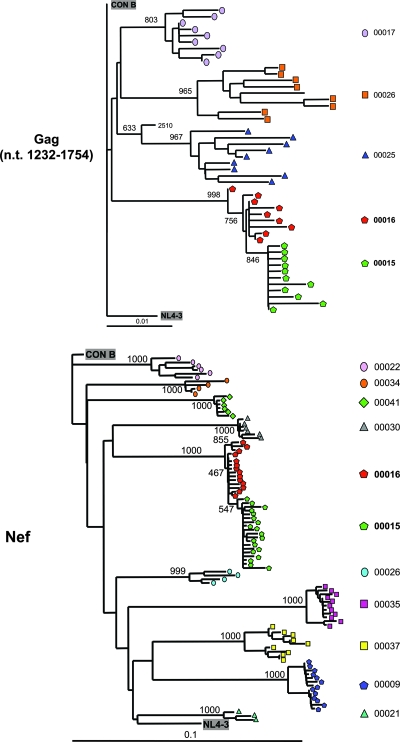
To confirm that subjects 00015 and 00016 were infected by the same strain of virus (likely through direct transmission), HIV-1 sequences were analyzed. Phylogenetic analyses confirmed that their infecting strains were highly related, indicating their common source and unrelatedness to the molecular clone (NL4-3) and other clinical sequences from our laboratory (Fig. 3). Notably, sequences from subject 00015 clustered as a distinct phylogenetic outgrouping from sequences of subject 00016 with a consistent branching pattern in 998/1,000 bootstrap replications in gag and 1,000/1,000 bootstrap replications in nef, thus suggesting their derivation from a common ancestor within subject 00016 sequences. These phylogenetic relationships suggested that subject 00015 was infected with HIV-1 from subject 00016, in agreement with the timing of clinical events.
FIG. 3.
Phylogenetic relationships of subjects 00015 and 00016 HIV-1 sequences to clade B consensus, NL4-3, and other primary isolates. Multiple cloned sequences from subjects 00015 and 00016 were compared to other contemporaneous primary isolate sequences, NL4-3, and Los Alamos National Laboratory HIV Sequence Database clade B consensus sequences. Neighbor-joining trees rooted on the clade B consensus sequence and evaluated with 1,000 bootstrap replicates are shown for gag (nucleotides 1232 to 1754 according to the HXB2 numbering system) and nef. Similar phylogenetic clustering relationships were observed for subject 00015 and 00016 pol and vif (not shown).
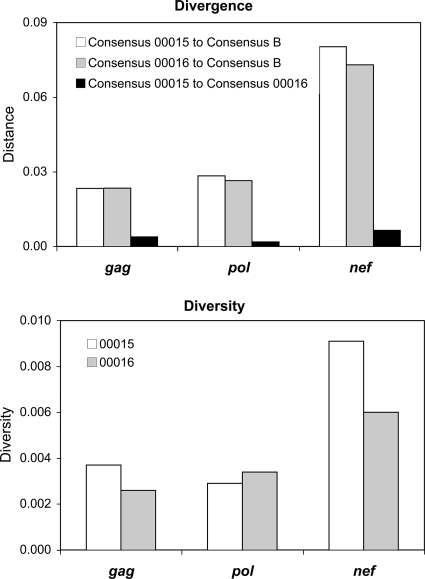
To explore and compare the evolutionary pressures on these viruses, the diversity and divergence of sequences were examined (Fig. 4). The diversity of HIV-1 sequences within each subject was comparable for regions in gag, pol, and nef, suggesting that the selective pressures driving viral diversification were similar between subjects for each of the encoded proteins. Comparison of the mean divergence of HIV-1 sequences of subjects 00015 and 00016 from the Los Alamos HIV Database clade B consensus sequence for gag, pol, and nef revealed similar degrees of divergence from the consensus for each genetic region. Given the concept that the clade B consensus sequence approximates the fittest wild-type sequence and that CTL immune pressure drives sequence evolution away from the consensus, these data suggested that CTL antiviral activities against HIV-1 in subjects 00015 and 00016 were similar between subjects for each of the encoded proteins. The overall similarity of genetic diversity and divergence between subjects for gag, pol, and nef implied that immune pressures on Gag, Pol, and Nef were similar between subjects (and not markedly different for Gag).
FIG. 4.
Phylogenetic divergence of gag, pol, and nef from subjects 00015 and 00016 versus clade B consensus sequences and intraindividual genetic diversity. Nucleotide sequences from subjects 00015 and 00016 and the Los Alamos HIV Database clade B consensus were assessed for divergence and diversity. The regions analyzed included nucleotides (HXB2 numbering) 1233 to 1755 (Gag amino acids 149 to 322), 2481 to 3019 (Pol amino acids 133 to 312), and 8797 to 9417 (Nef amino acids 1 to 207).
The CTL-targeted sequences showed little variation between the subjects.
HIV-1 escape from CTL through epitope mutation is thought to be a major mechanism of failure of immune containment, and thus the sequences targeted by CTL were examined for subjects 00015 and 00016 (Table 1). Of the seven epitope regions where at least three clonal sequences were available for each subject, five demonstrated the same consensus sequence for both subjects. For the two regions where the subject consensus sequences differed, the amino acid differences were near the ends of the sequences, and therefore did not necessarily affect the actual epitope within the region. While limited sequence data were available for the other epitope regions, most appeared to show similarity between subjects 00015 and 00016 (Table 1). In agreement with other studies demonstrating relative stability of CTL responses and epitope sequences during chronic infection, these data suggested that the CTL responses in subjects 00015 and 00016 during chronic infection targeted relatively conserved regions of HIV-1.
DISCUSSION
Given the observation that the HIV-1-specific CTL response is qualitatively associated with immune control of infection, the recent focus of many vaccine development efforts has been eliciting a CTL response that would protect from disease if not from infection with HIV-1. While attenuation of other viruses (such as poliovirus) has been a classical vaccine development strategy that would be expected to generate CTL responses, safety concerns have prompted a search for alternative strategies to deliver HIV-1 proteins to the HLA class I pathway. Because these strategies (such as DNA and nonreplicating recombinant virus vector vaccines) depart dramatically from attenuated-virus infections that would be expected to mimic natural CTL responses against the same virus, it is unknown how the immune outcome might differ. In theory, a novel HIV-1 vaccine could be not only unsuccessful at generating CTL responses but could elicit qualitatively disadvantageous responses or promote tolerance and thereby antagonize rather than promote immune containment in subsequent HIV-1 infection.
This study examines two individuals who were infected with the same strain of HIV-1 approximately the same time, one of whom who was receiving a Gag DNA vaccine at the time of infection. This unfortunate and unusual situation allows a comparison of clinical outcome, immune responses, and viral evolution to assess for a possible impact of the vaccine. While it is clearly impossible to draw definitive conclusions from this limited study, the findings suggest that vaccination with this Gag DNA does not preclude effective immune containment of HIV-1. Both of these individuals have had unusually effective immune containment of HIV-1 with low levels of set point viremia in the absence of antiretroviral therapy (more than 4 years after infection at the time of this writing).
The explanation is unclear, but the similarity of clinical courses regardless of Gag DNA vaccination suggests that vaccination itself was not the cause of good immune containment in subject 00015 (although we have not excluded the possibility that there were non-IFN-γ-producing CTL responses elicited by vaccination). Neither of the HLA class I profiles of these men contain alleles known to be associated with retarded disease progression. They share only A*03, and their CTL responses also appear to be mostly nonoverlapping. Furthermore, subject 00015 does not have the CCR5 delta 32 mutation (not shown). The fact that both hosts have similarly atypical containment of infection suggests the possibility that a common virologic factor explains their relatively benign courses. Although defective Nef has been associated with decreased HIV-1 pathogenicity, the sequences of nef in these individuals reveal no such defects, and no obvious abnormalities are evident in the other sequenced regions of the viral genome to suggest a gross abnormality. Moreover, testing of reverse transcriptase/protease from subject 00016 demonstrates normal replicative capacity (data not shown).
Lack of detectable Gag-specific CTL responses in subject 00015 during his vaccination series before actual HIV-1 infection could have reflected either insufficient immunogenicity of the vaccine, or vaccine-induced tolerance. However, during subsequent HIV-1 infection, he displayed CTL responses against Gag with magnitude and breadth that were not markedly dissimilar to those of subject 00016, suggesting that the vaccine did not preclude CTL targeting of Gag through tolerance. Gag-specific CTL responses against four epitope regions were observed for subject 00015, and this likely underestimated his true response, due to limitations in CTL detection using peptides based on nonautologous HIV-1 sequences.
It is also possible that Gag-specific CTL below the limits of detection had already been primed in subject 00015 by the vaccine before actual HIV-1 infection. Among vaccine recipients in this trial, there were no Gag-specific CTL responses (as determined by IFN-γ ELISPOT) after the first and second vaccinations and about 26% responders after the third vaccination (data not shown). After an additional fourth booster vaccination with the same DNA vaccine or a recombinant Ad5 recombinant gag vaccine, 43% and 63% of vaccinees had responses, respectively (data not shown). Thus, it is unclear whether subject 00015 had primed CTL responses from his vaccinations before he sustained infection. Such preinfection priming by a vaccine could have several potential impacts on the immunopathogenesis of subsequent infection. The vaccine could skew the CTL response toward Gag, enhancing its immunodominance and secondarily reducing the response against other proteins. Another possibility is that the exposure to fixed Gag sequences (clade B consensus in nonreplicating Gag DNA), rather than the naturally evolving quasispecies of Gag during natural infection, could alter the pattern of CTL epitope targeting within Gag. The influence of CTL targeting is poorly understood, but appears to be important because the overall magnitude of the CTL response does not reliably correlate to immune control of viremia. Finally, vaccination could have contributed to better immune control in subject 00015 through priming of CD4+ T helper responses, given evidence that DNA vaccination can also prime such responses (9), although immune control in subject 00016 was similar despite lack of vaccination.
Although our data cannot provide definitive evidence regarding these issues, the clinical course of subject 00015 and the profile of his immunity do not suggest any vaccine-induced misdirection of Gag-specific CTL responses. Because Gag-specific CTL may be especially important for control of viremia (16, 26), the low set point viremia in subject 00015 suggests a lack of deficiency in his Gag-specific CTL response. The Gag-specific CTL response showed an epitopic breadth similar to that of subject 00016, and there was also a breadth of targeting of epitopes outside Gag, similar to subject 00016 and what has been seen typically in other studies. Although the breadth and magnitude of the HIV-1-specific CTL response are highly variable between different HIV-1-infected individuals (1, 3), the CTL responses of subject 00015 did not appear grossly unusual. While we did not perform more detailed CTL phenotyping for “polyfunctionality” of cytokine production, memory subset quality, or proliferative capacity, the similar levels of CTL between subjects and their parallel clinical courses were compatible with effective CTL responses regardless of vaccination status, in the context of our data regarding HIV-1 sequences and presumed viral fitness.
Besides using ELISPOT for an approximation of immune pressure, we also examined and compared HIV-1 sequence evolution in both subjects, because selective pressure by CTL is the major determinant of viral sequences in vivo (17). Examination of HIV-1 sequences in subjects 00015 and 00016 confirmed that they were infected with the same strain of virus, and the phylogenetic branching pattern further suggested that subject 00015 viruses are evolved from a subset of subject 00016 viruses, although we could not exclude that they were both infected from a third source. The sequence diversities within each subject were similar, suggesting similar immune pressures and constraints on viral evolution. Furthermore, levels of divergence from the clade B consensus sequence, which is considered an approximation of a “most fit” sequence, were also similar between subjects, suggesting comparable degrees of CTL pressure for viral evolution away from a fitter ancestor. HIV-1 sequence variation between the two subjects in CTL-targeted regions was limited, in agreement with observations that CTL escape mutations are rarely observed during the stable phase of chronic infection or occur very slowly (8, 12, 14). Overall, the similarity in phylogenetic parameters between the two subjects was compatible with their displaying similar degrees of CTL pressure against HIV-1, including earlier CTL responses that had already driven escape and since decreased to undetectable levels (12, 18).
Our results complement two prior studies of CTL responses in vaccine recipients who had subsequent HIV-1 infection. Betts et al. examined an individual who received a recombinant canarypox vaccine (vCP205), who became HIV-1 infected approximately a year after finishing four vaccinations over 6 months (4). In contrast to our case, this individual developed detectable CTL (and helper) responses from vaccination, including CTL against a conserved B*27-restricted epitope that is frequently recognized in B*27+ slow progressors. Detailed analysis of these responses demonstrated a “polyfunctional profile” of multicytokine production upon stimulation, similar to that seen in slow progressors. After infection, however, these responses developed a hypofunctional profile typical for progressing HIV-1 infection, which the authors hypothesized could be due to vaccine-induced cellular immunity in the absence of humoral immunity. However, our subject had no detectable vaccine-induced CTL or antibody responses, and thus our data do not address this hypothesis. Another study by Horton et al. described 16 persons who also received recombinant canarypox vaccines and sustained HIV-1 infection subsequently, none of whom had developed detectable CTL responses from vaccination (10). Comparisons of these persons to unvaccinated controls demonstrated grossly similar patterns of CTL targeting and magnitude after HIV-1 infection, and one vaccinee developed a low viremia set point (without antiretroviral treatment) similar to our participant. Our data agree with the findings of Horton et al., demonstrating that apparently nonimmunogenic vaccination (by IFN-γ ELISPOT analysis) does not appear to interfere with subsequent CTL immunogenicity or immune containment of HIV-1, and provide further detail regarding the function of postinfection CTL responses and selective pressure on HIV-1 sequences compared to another person infected with the same viral strain.
In summary, HIV-1 infection of subject 00015 during HIV-1 Gag DNA vaccination appeared to have no observable detrimental effects on the ability of the immune response to contain HIV-1. Subject 00015 had similarly good immune control to that of subject 00016, who was infected with a closely related strain. Gag-specific CTL responses were absent in subject 00015 during vaccination but were mounted during infection and represented a typical proportion of a broad CTL response. HIV-1 genetic evolution in his HIV-1 gag, pol, and nef was similar to that in subject 00016, suggesting similar degrees of immune pressure between subjects. As a whole, these data provide evidence that lack of vaccine responsiveness does not necessarily preclude effective CTL responses in subsequent HIV-1 infection.
Acknowledgments
We are grateful to subjects 00015 and 00016 for their generous participation in this study.
This work was supported by PHS grants AI043203 (O.O.Y.), AI051970 (O.O.Y.), AI068449 (M.J.L.), and AI043638 (E.S.D.). Overlapping HIV-1 peptides and interleukin-2 were provided by the NIH AIDS Research and Reference Reagent Program.
Footnotes
Published ahead of print on 16 January 2008.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. R. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 772081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altfeld, M., T. M. Allen, X. G. Yu, M. N. Johnston, D. Agrawal, B. T. Korber, D. C. Montefiori, D. H. O'Connor, B. T. Davis, P. K. Lee, E. L. Maier, J. Harlow, P. J. Goulder, C. Brander, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420434-439. [DOI] [PubMed] [Google Scholar]
- 3.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 7511983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betts, M. R., B. Exley, D. A. Price, A. Bansal, Z. T. Camacho, V. Teaberry, S. M. West, D. R. Ambrozak, G. Tomaras, M. Roederer, J. M. Kilby, J. Tartaglia, R. Belshe, F. Gao, D. C. Douek, K. J. Weinhold, R. A. Koup, P. Goepfert, and G. Ferrari. 2005. Characterization of functional and phenotypic changes in anti-Gag vaccine-induced T cell responses and their role in protection after HIV-1 infection. Proc. Natl. Acad. Sci. USA 1024512-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. A. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 686103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casimiro, D. R., L. Chen, T.-M. Fu, R. K. Evans, M. J. Caulfield, M.-E. Davies, A. Tang, M. Chen, L. Huang, V. Harris, D. C. Freed, K. A. Wilson, S. Dubey, D. M. Zhu, D. Nawrocki, H. Mach, R. Troutman, L. Isopi, D. Williams, W. Hurni, Z. Xu, J. G. Smith, S. Wang, X. Liu, L. Guan, R. Long, W. Trigona, G. J. Heidecker, H. C. Perry, N. Persaud, T. J. Toner, Q. Su, X. Liang, R. Youil, M. Chastain, A. J. Bett, D. B. Volkin, E. A. Emini, and J. W. Shiver. 2003. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus, and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J. Virol. 776305-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubey, S., J. Clair, T. M. Fu, L. Guan, R. Long, R. Mogg, K. Anderson, K. B. Collins, C. Gaunt, V. R. Fernandez, L. Zhu, L. Kierstead, S. Thaler, S. B. Gupta, W. Straus, D. Mehrotra, T. W. Tobery, D. R. Casimiro, and J. W. Shiver. 2007. Detection of HIV vaccine-induced cell-mediated immunity in HIV-seronegative clinical trial participants using an optimized and validated enzyme-linked immunospot assay. J. Acquir. Immune Defic. Syndr. 4520-27. [DOI] [PubMed] [Google Scholar]
- 8.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3212-217. [DOI] [PubMed] [Google Scholar]
- 9.Graham, B. S., R. A. Koup, M. Roederer, R. T. Bailer, M. E. Enama, Z. Moodie, J. E. Martin, M. M. McCluskey, B. K. Chakrabarti, L. Lamoreaux, C. A. Andrews, P. L. Gomez, J. R. Mascola, and G. J. Nabel. 2006. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 DNA candidate vaccine. J. Infect. Dis. 1941650-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton, H., C. Havenar-Daughton, D. Lee, E. Moore, J. Cao, J. McNevin, T. Andrus, H. Zhu, A. Rubin, T. Zhu, C. Celum, and M. J. McElrath. 2006. Induction of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses in HIV vaccine trial participants who subsequently acquire HIV-1 infection. J. Virol. 809779-9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibarrondo, F. J., P. A. Anton, M. Fuerst, H. L. Ng, J. T. Wong, J. Matud, J. Elliott, R. Shih, M. A. Hausner, C. Price, L. E. Hultin, P. M. Hultin, B. D. Jamieson, and O. O. Yang. 2005. Parallel human immunodeficiency virus type 1-specific CD8+ T-lymphocyte responses in blood and mucosa during chronic infection. J. Virol. 794289-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamieson, B. D., O. O. Yang, L. Hultin, M. A. Hausner, P. Hultin, J. Matud, K. Kunstman, S. Killian, S. Altman, K. Kommander, B. T. M. Korber, J. V. Giorgi, and S. Wolinsky. 2003. Epitope escape mutation and decay of human immunodeficiency virus type 1 specific cytotoxic T lymphocyte responses. J. Immunol. 1715372-5379. [DOI] [PubMed] [Google Scholar]
- 13.Johnston, M. I., and A. S. Fauci. 2007. An HIV vaccine—evolving concepts. N. Engl. J. Med. 3562073-2081. [DOI] [PubMed] [Google Scholar]
- 14.Koibuchi, T., T. M. Allen, M. Lichterfeld, S. K. Mui, K. M. O'Sullivan, A. Trocha, S. A. Kalams, R. P. Johnson, and B. D. Walker. 2005. Limited sequence evolution within persistently targeted CD8 epitopes in chronic human immunodeficiency virus type 1 infection. J. Virol. 798171-8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 684650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masemola, A., T. Mashishi, G. Khoury, P. Mohube, P. Mokgotho, E. Vardas, M. Colvin, L. Zijenah, D. Katzenstein, R. Musonda, S. Allen, N. Kumwenda, T. Taha, G. Gray, J. McIntyre, S. A. Karim, H. W. Sheppard, and C. M. Gray. 2004. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J. Virol. 783233-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 2961439-1443. [DOI] [PubMed] [Google Scholar]
- 18.Oxenius, A., D. A. Price, A. Trkola, C. Edwards, E. Gostick, H. T. Zhang, P. J. Easterbrook, T. Tun, A. Johnson, A. Waters, E. C. Holmes, and R. E. Phillips. 2004. Loss of viral control in early HIV-1 infection is temporally associated with sequential escape from CD8+ T cell responses and decrease in HIV-1-specific CD4+ and CD8+ T cell frequencies. J. Infect. Dis. 190713-721. [DOI] [PubMed] [Google Scholar]
- 19.Saurya, S., Z. Lichtenstein, and A. Karpas. 2003. Characterization of gag gene of plasma HIV type 1 in combination therapy-treated AIDS patients with high viral load and stable CD4+ T cell counts. AIDS Res. Hum. Retrovir. 1973-76. [DOI] [PubMed] [Google Scholar]
- 20.Wong, J. T., and R. B. Colvin. 1987. Bi-specific monoclonal antibodies: selective binding and complement fixation to cells that express two different surface antigens. J. Immunol. 1391369-1374. [PubMed] [Google Scholar]
- 21.Yang, O. O. 2003. Will we be able to ‘spot’ an effective HIV-1 vaccine? Trends Immunol. 2467-72. [DOI] [PubMed] [Google Scholar]
- 22.Yang, O. O., W. J. Boscardin, J. Matud, M. A. Hausner, L. E. Hultin, P. M. Hultin, R. Shih, J. Ferbas, F. P. Siegal, M. Shodell, G. M. Shearer, E. Grene, M. Carrington, S. O'Brien, C. B. Price, R. Detels, B. D. Jamieson, and J. V. Giorgi. 2002. Immunologic profile of highly exposed yet HIV type 1-seronegative men. AIDS Res. Hum. Retrovir. 181051-1065. [DOI] [PubMed] [Google Scholar]
- 23.Yang, O. O., J. Church, C. M. R. Kitchen, R. Kilpatrick, A. Ali, Y. Geng, M. S. Killian, R. L. Sabado, H. Ng, J. Suen, Y. Bryson, B. D. Jamieson, and P. Krogstad. 2005. Genetic and stochastic influences on the interaction of human immunodeficiency virus type 1 and cytotoxic T lymphocytes in identical twins. J. Virol. 7915368-15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang, O. O., E. S. Daar, B. D. Jamieson, A. Balamurugan, D. M. Smith, J. A. Pitt, C. J. Petropoulos, D. D. Richman, S. J. Little, and A. J. L. Brown. 2005. Human immunodeficiency virus type 1 clade B superinfection: evidence for differential immune containment of distinct clade B strains. J. Virol. 79860-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, O. O., S. A. Kalams, A. Trocha, H. Cao, A. Luster, R. P. Johnson, and B. D. Walker. 1997. Suppression of human immunodeficiency virus type 1 replication by CD8+ cells: evidence for HLA class I-restricted triggering of cytolytic and noncytolytic mechanisms. J. Virol. 713120-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuniga, R., A. Lucchetti, P. Galvan, S. Sanchez, C. Sanchez, A. Hernandez, H. Sanchez, N. Frahm, C. H. Linde, H. S. Hewitt, W. Hildebrand, M. Altfeld, T. M. Allen, B. D. Walker, B. T. Korber, T. Leitner, J. Sanchez, and C. Brander. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 803122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]