Abstract
Embryonic stem (ES) cells offer insight into early developmental fate decisions, and their controlled differentiation may yield vast regenerative potential. The molecular determinants supporting ES cell self-renewal are incompletely understood. The homeodomain proteins Nanog and Oct4 are essential for mouse ES cell self-renewal. Using a high-throughput approach, we discovered DNaseI hypersensitive sites and potential regulatory elements along a 160-kb region of the genome that includes GDF3, Dppa3, and Nanog. We analyzed gene expression, chromatin occupancy, and higher-order chromatin structure throughout this gene locus and found that expression of the reprogramming factor Oct4 is required to maintain its integrity.
[Keywords: Nanog, Oct4, chromatin, reprogramming, self renewal]
Embryonic stem (ES) cells, which are derived from the inner cell mass (ICM) of the developing blastocyst, are endowed with unlimited self-renewal capacity, while they retain the potential to differentiate into all germ layers. There is considerable interest in revealing the genetic programs responsible for ES cell self-renewal, with the ultimate goal of directing differentiation to create a renewable tissue source for cell therapy. Currently, knowledge of the precise genetic determinants responsible for self-renewal is incomplete.
As components critical for ES cell self-renewal, several transcription factors, often referred to as core pluripotency factors, are subjects of intense study. One such factor, Nanog, was identified by virtue of its ability to promote self-renewal independent of the leukemia inhibitory factor (LIF)/gp130 signaling pathway (Chambers et al. 2003; Mitsui et al. 2003). Embryos depleted of Nanog fail to mature past 5 d of development (Mitsui et al. 2003). Nanog lies in a highly conserved genomic locus with a distinctive configuration. The Nanog gene is flanked by several other genes that are expressed early in development and/or involved in critical fate decisions. Colinearity of the genes encoding Aicda, Apobec1, GDF3, Dppa3 (also known as Stella and PGC7), and Nanog is conserved throughout orthologous regions in rodents and primates. The Aicda and Apobec1 genes encode DNA editing enzymes that may serve as epigenetic modifiers by selectively influencing the genomewide pool of methylatable targets during early development. Dppa3 is a maternal effect gene required for normal development. Recent work has shown that Dppa3 protects against inappropriate DNA demethylation in the maternal genome and at certain imprinted loci (Nakamura et al. 2007). GDF3 and Nanog are essential for ES cell self-renewal and contribute to early remodeling of the developing embryo. Interestingly, the region occupied by these early developmental modifiers comprises only 160 kb, or 0.004% of the entire genome. The organization of the cluster implies complex interactions that may provide a window into regulation of pluripotency-related genes.
Expression of the genes within the Nanog locus has been reported in several cancers. Amplification of the Nanog locus on chromosome 12 in humans has been found in essentially all germ cell tumors analyzed and coordinated overexpression of GDF3, Dppa3, and Nanog is observed in human seminomas and embryonal carcinomas, as revealed by expression profiling. Additionally, Nanog has been characterized as a marker for metastatic germ cell tumors (Korkola et al. 2006; Santagata et al. 2007).
Oct4 function is required during the earliest stages of development, and its depletion results in differentiation of the future embryo into extraembryonic trophectoderm. In concert with Nanog, Sox2, KLF4, and Myc, Oct4 can reprogram differentiated somatic cells to a pluripotent state (Okita et al. 2007; Wernig et al. 2007). Nanog participates in an autoregulatory circuit together with Oct4, Sox2 (Boyer et al. 2005; Loh et al. 2006), and likely other transcription factors (Ivanova et al. 2006; Matoba et al. 2006; Wang et al. 2006) to interact with genes in cis or trans that preserve self-renewal and enable differentiation into all three germ layers. Here, we describe initial efforts to dissect regulatory interactions and mechanisms operative in the Nanog locus. Using a high-throughput quantitative chromatin profiling (QCP) approach, we discovered DNaseI hypersensitive sites (HS) and potential regulatory elements along a 160-kb region of chromosome 6 that includes GDF3, Dppa3, and Nanog. We analyzed gene expression, chromatin occupancy, and higher-order chromatin structure throughout this locus. We demonstrate that genes within the Nanog locus are coregulated. Importantly, we show that several pluripotency-related nuclear factors co-occupy chromatin throughout the locus, and that Oct4 expression is required to maintain chromatin structure. Gene expression and chromatin structure within this locus may serve as a useful “molecular sensor” in cellular reprogramming studies.
Results and Discussion
DNaseI hypersensitivity signature extends over the entire region containing Nanog and the other early-expressed genes
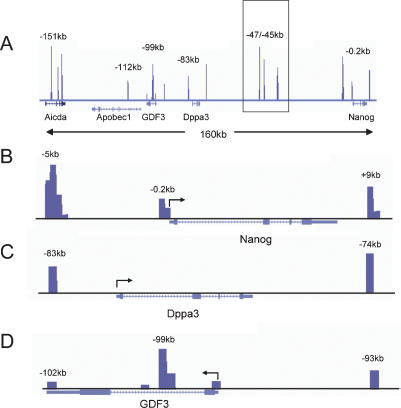
Little is known regarding the cis-acting elements responsible for Nanog regulation. A comprehensive survey of regulatory DNA within the Nanog locus has not been undertaken. DNaseI hypersensitivity is a sensitive indicator of regulatory elements in complex loci. As such, we mapped DNaseI hypersensitivity by QCP throughout the Nanog locus in mouse ES cells (Fig. 1; Dorschner et al. 2004). This high-throughput method permits assessment of HSs throughout the locus in an efficient manner and with great sensitivity. A strong HS was observed at −5 kb from the transcriptional start site of the Nanog gene, and a less prominent HS was seen at the proximal promoter. These regions have been reported previously to be bound by Sox2, Nanog, and other activators and repressors (Loh et al. 2006; Pereira et al. 2006). Interestingly, in addition to the predicted proximal HS features we observed at the Nanog proximal promoter, we identified a novel HS element downstream from the Nanog coding sequence positioned 9 kb from the translational start site. The Dppa3 gene also displayed robust DNase hypersensitivity 2 kb upstream of and 7 kb downstream from its translational start site. HSs are present at the proximal promoter and 3′ untranslated region of the GDF3 gene; however, two separate HSs form at the intron. The larger peak lying 1.1–1.5 kb from the translational start site is a strong candidate for a cis-control element of the gene.
Figure 1.
DNaseI HS determination by QCP. (A) Genomic structure of the Nanog locus. The intergenic cluster region is shown boxed. (B) Nanog regulatory elements are indicated with four peaks of HS located at the −5-kb region. A novel regulatory element is also seen at +9 kb. (C,D) QCP profiles for Dppa3 and GDF3 are shown. Note the large tandem HS peak in the GDF3 intron located −99 kb from the start of Nanog transcription. Arrows at the promoters indicate direction of transcription. Genomic positions relative to the Nanog start site are shown above each HS. Data were mapped to the March 2005 (mm6) genome assembly and are visualized within the University of California at Santa Cruz genome browser (Kent et al. 2002).
Many intergenic regulatory elements lie far upstream of the gene or gene loci that they control. A survey of the large intergenic regions separating Nanog from its neighboring genes revealed strong HSs located −47 kb, −45 kb, and −38 kb from the Nanog transcriptional start site and 26 kb downstream from Dppa3 (Fig. 1, boxed region). Consistent with the cell-specific nature of the chromatin structure of the entire Nanog locus, DNase hypersensitivity was not observed within 200 kb upstream of or downstream from the Nanog start site in a murine erythroleukemia (MEL) cell line (data not shown).
Transcription factors involved in ES cell self-renewal bind HS throughout the Nanog locus
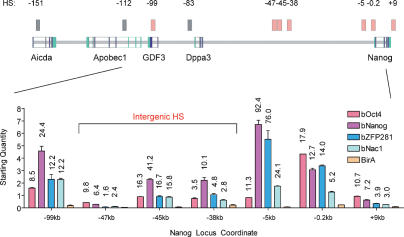
Transcription factors including Nanog, Oct4, Sox2, and others (such as ZFP281 and Nac1) collaborate to maintain ES cell pluripotency (Boyer et al. 2005; Loh et al. 2006; Wang et al. 2006). To determine the potential contribution of these factors toward transcriptional regulation, we examined chromatin occupancy of Oct4, Nanog, ZFP281, and Nac1 at HSs within the Nanog locus. These factors bind at shared positions at an unexpectedly high frequency throughout the locus (Fig. 2). Binding was most significant at the Nanog −5-kb HS, with near 100-fold enrichments over background in chromatin immunoprecipitation (ChIP) assays. Although Nanog and Sox2 binding were observed previously at both the −5-kb region and proximal promoter (Loh et al. 2006), our findings indicate that Oct4 also binds the upstream region. The Nanog proximal promoter and GDF3 intronic HS also exhibited high-level binding by Oct4, Nanog, ZFP281, and Nac1. Multispecies DNA alignments indicate conservation of potential Oct4-, Nanog-, and ZFP281-binding sites at the −99-kb GDF3 and −5-kb Nanog regulatory elements (Supplemental Fig. 1). Significant co-occupancy was also observed at the Nanog downstream HS. Perhaps the most striking finding is the presence of all four factors at intergenic HSs located at −47 kb, −45 kb, and −38 kb. It is of particular interest to note that Oct4, ZFP281, and Nac1 exhibit very similar enrichment ratios at the −99-kb and −45-kb HS, with Nanog binding being approximately twofold higher than the other three proteins. Oct4 binding was also evident at the −47-kb HS, coupled with a modest occupancy by Nanog. As expected, binding enrichment was significantly reduced when ES cells were induced to differentiate with retinoic acid and LIF withdrawal (data not shown). We also revealed that Oct4 may contribute to the regulation of Nac1. Depletion of Oct4 resulted in significant reduction of Nac1 mRNA, suggesting that uncoupling of the ES cell interactome network disrupts expression of multiple partners (Supplemental Figs. 2, 3).
Figure 2.
Transcription factors involved in ES cell self-renewal bind HS throughout the Nanog locus. ChIP was used to analyze enrichment of nuclear factors on chromatin within the Nanog locus in wild-type ES cells. A schematic of the Nanog locus located on murine chromosome 6 is shown at the top, with boxed lines depicting the exon structure of the genes and noncoding sequences in green. HSs are indicated as solid boxes at the positions noted relative to Nanog transcriptional initiation. Light-red boxes indicate binding at the genomic positions shown above. Gray boxes mark HSs where chromatin occupancy was not observed. Fold enrichment of chromatin occupancy by biotagged transcription factors over the BirA background control are indicated above each bar, and all values were normalized to an HPRT control.
The presence of HSs commonly demarcates cis-elements that function as enhancers in various test systems. We evaluated the ability of HSs throughout the Nanog locus to enhance transgene expression using two different experimental approaches (see Supplemental Fig. 4). Results from both assays indicated that HSs located at −99 kb, −45 kb, −5 kb, and +9 kb serve to enhance transcriptional activation. Therefore, they are likely to function as authentic regulatory elements in situ.
Our work implies that critical threshold levels of several proteins, including Oct4, Nanog, ZFP281, and Nac1 (among others), may be required to activate expression of genes within the Nanog locus. Differential enrichment of these factors at select promoter and intergenic regulatory elements suggests that precise factor stoichiometry might bridge cis-regulatory elements of multiple HSs together to maintain an open chromatin configuration and establish high-level transcriptional activation. How protein-bound promoter elements may form an active transcriptional complex through higher-order chromatin complexes is discussed below.
Oct4 ablation disrupts gene expression throughout the Nanog locus
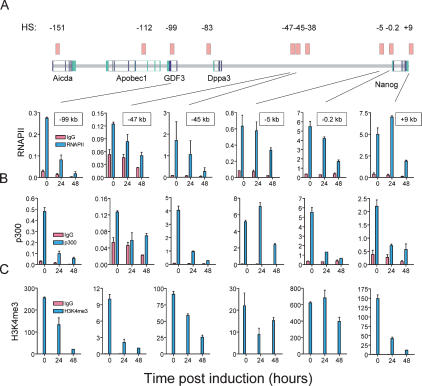
Next, we examined the consequences of Oct4 loss on the permissive chromatin state and gene expression within the Nanog locus. To study Oct4 repression in a controlled manner, we used ZHBTc4 ES cells. These cells lack functional endogenous Oct4 alleles and harbor a regulatable Oct4 transgene (Niwa et al. 2000). Cells are maintained in an undifferentiated state in the absence of doxycycline. Administration of doxycycline represses Oct4 expression and promotes trophectoderm differentiation. Doxycycline controlled repression of Oct4 results in >50% loss of Nanog mRNA within 12 h post-induction, well before morphological changes begin to appear in some cells at 48 h (Supplemental Fig. 2). In parallel, we also observed marked loss of Dppa3 (73%), GDF3 (70%), and Apobec1 (50%) transcripts within 12–24 h following Oct4 depletion. Such broad repression was not observed for the early, developmentally regulated Slc2a3/GLUT3 and FOXJ2 genes, which are located immediately downstream from Nanog; in contrast, their mRNA levels were significantly increased (Supplemental Figs. 2, 3, 5). As expected, loss of chromatin occupancy by Nanog was observed at HSs throughout the Nanog locus following controlled Oct4 depletion (Supplemental Fig. 6). It is interesting to note that the kinetics of Nanog occupancy loss at the −99-kb and −5-kb HSs differed from other HSs within the locus. This may be due in part to the presence of two Nanog-binding sites within these HSs (Supplemental Fig. 1). This possibility is supported by the observation that occupancy by Nanog is enriched to a greater degree at the −99-kb and −5-kb HSs in wild-type ES cells (Fig. 2).
We next analyzed the presence of components of the basal transcriptional machinery at intragenic and intergenic HSs within the Nanog locus using RNA polymerase II-directed ChIP. We observed high level RNA polymerase II binding at the GDF3 intronic HS, as well as within the proximal promoter and distal Nanog regulatory elements located at the −5-kb and +9-kb HSs (Fig. 3A). A general loss of RNA polymerase II binding was observed throughout the locus upon repression of Oct4. Changes were evident by 24 h after the onset of Oct4 depletion, a time prior to overt differentiation. The reduction in RNA polymerase II enrichment was significant at the GDF3 intronic HS, the −45-kb HS and all Nanog promoter elements. The broad effects of Oct4 loss on RNA polymerase II binding are consistent with the consequences of Oct4 repression on expression of genes throughout the Nanog locus.
Figure 3.
Oct4 ablation disrupts gene expression throughout the Nanog locus. ChIP was used to analyze chromatin occupancy within HSs and at selected genomic positions in wild-type or Oct4-depleted ES cells during the time course indicated after doxycycline induction. Wild-type ES cells are indicated in blue and Oct4-depleted cells are in red. (A,B) Chromatin occupancy of RNA polymerase II and the transcriptional cofactor p300 are shown throughout the Nanog locus. (C) Enrichment of histone H3K4me3 is indicated. Values of antibody-bound and IgG control ChIPs are normalized to an HPRT control.
An established hallmark of enhancers is binding of the transcriptional coactivator p300. We surveyed the presence of p300 at HSs throughout the Nanog locus (Fig. 3B). High-level p300 enrichment overlapped RNA polymerase II occupancy observed at the Nanog proximal (−0.2-kb HS) and distal promoter (−5-kb HS), the −45-kb intergenic HS, and to a lesser extent at the GDF3 intron (−99-kb HS). Loss of p300 binding was significant at all HSs except −5 kb within 24 h following Oct4 depletion. Loss of enrichment at the −5-kb HS became evident by 48 h.
Significant enrichment of the transcriptionally active chromatin mark H3K4me3 was observed throughout the Nanog locus (Fig. 3C). The most prominent levels were again seen at the GDF3 intron, putative Nanog regulatory elements, and the postulated −45-kb enhancer. High levels were also observed at the Aicda −151-kb intronic HS and the −47-kb intergenic HS. Additionally, low-level enrichment was seen at the Dppa3 upstream HS and intergenic −47-kb and −38-kb HSs. Analysis of binding patterns for H3K9me3 further confirm that the locus is normally in a poised state for transcriptional activation, and that this state is altered following Oct4 depletion (Supplemental Fig. 7).
Our data demonstrate that chromatin within the identified HSs and, hence, proximal and distal cis-regulatory elements throughout the Nanog locus, is repressed following Oct4 depletion. Locus-wide alteration in chromatin structure and reduction of gene expression are observed as early as 12 h following Oct4 down-regulation and complete by 72 h (Fig. 3; Supplemental Figs. 2, 3). Importantly, changes in chromatin are evident well before morphological differentiation. Oct4 disruption appears to trigger remodeling of chromatin throughout the locus.
The insulator and boundary element protein CTCF is enriched at the −47-kb HS
CTCF is an insulator protein that serves to block enhancer function or selectively repress or enable gene activation in species as diverse as yeast and mammals (Bell and Felsenfeld 2000; Defossez and Gilson 2002). CTCF binds to the H19/Igf2 imprinting control region where it blocks enhancer activity in a methylation-controlled manner (Bell and Felsenfeld 2000; Hark et al. 2000). Recent whole genome analyses of in vivo binding and bioinformatic predictions further support a role for CTCF as a boundary element that may partition the genome (Kim et al. 2007; Xie et al. 2007). These studies revealed a consensus motif (CCGCGNGGNGGCAC) for CTCF binding. Using a refined consensus motif (CCD SNAGRKG GCRS), we scanned the Nanog locus and identified consensus CTCF candidate sites within HSs located at −147 kb, −112 kb, −78 kb, and −47 kb and within a highly conserved region at −55 kb (data not shown). We probed in vivo occupancy of CTCF at all these HSs and observed significant enrichment at the −47-kb intergenic HS (Supplemental Fig. 8). CTCF binding was diminished following Oct4 disruption, although this became evident only after the onset of overt differentiation. This suggests that loss of CTCF binding is a late step in locus-wide chromatin remodeling. The region of chromatin analyzed shows that CTCF binding is flanked by two nearby Oct4 sites (Supplemental Fig. 8). We speculate that this −47-kb HS might function as a boundary element to separate the discrete genic units within the locus and to allocate spatial and temporal control in the developing embryo. To corroborate the binding studies, we tested the −47-kb HS for in vivo insulator activity using a heterologous cell reporter assay. Two different variants of the −47-kb HS successfully shielded expression of the linked transgene from an adjacent enhancer, verifying that the −47-kb HS can function as an insulator element (Supplemental Fig. 9). Knockdown of CTCF within early embryos indicates this protein is essential for development between morula formation and blastocyst development (Fedoriw et al. 2004). An important function of CTCF may be to ensure proper regulation of genomic loci, such as Nanog, that participate in stem cell self-renewal.
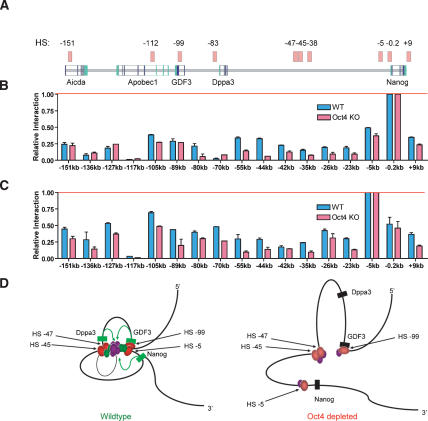
Long-range chromosomal contacts connect HS and create higher-order chromatin structure within the Nanog locus
DNA looping across distant chromosomal loci provides a mechanism for long-distance transcriptional control (Townes and Behringer 1990; Dekker et al. 2002; Drissen et al. 2004; Spilianakis and Flavell 2004; Vakoc et al. 2005). To ascertain whether chromosomal looping might join distant putative regulatory elements to strategic locations within the Nanog locus to form active transcriptional domains, we employed chromosome conformation capture (3C) (Dekker et al. 2002). Striking differences in chromatin conformation were observed between wild-type and Oct4-depleted cells (Fig. 4). At the Nanog proximal promoter, wild-type ES cells showed contact between HSs as far as 151 kb away, within the Aicda intronic HS (Fig. 4B). Significant interaction was also observed between the junction of the Apobec1 and GDF3 genes (−105 kb, located upstream of the −99-kb intronic HS), the Dppa3 proximal promoter (−80 kb), and all intergenic HSs between Dppa3 and Nanog (−55-kb, −44-kb, −42-kb, and −35-kb contacts). Upon Oct4 repression, contact between locus-wide HSs and the Nanog proximal promoter was greatly diminished (Fig. 4B). Loss of interaction with the Nanog proximal promoter was most striking at the Dppa3 promoter, the −47/−45/−38-kb intergenic HS cluster and a region of high-level conservation at −55 kb; however, significant loss at the Apobec1/GDF3 gene junction (marked by the −105-kb product) was also also observed. Diminished contact between the Nanog promoter and the −47/−45/−38-kb intergenic HS cluster or locus-wide gene promoters is consistent with the loss of Nanog and Oct4 gene expression in these cells, and collapse of a transcriptional complex at the Nanog promoter.
Figure 4.
Long-range chromosomal contacts connect HSs throughout the Nanog locus. (A) A schematic of the Nanog locus, with HS indicated by light-red boxes. The region of chromosomal contact, relative to the Nanog start site, is shown below each graph. The red bar shows normalization against a nonchimeric amplification product at the specified region of analysis. HS up to 150 kb away make contact with the Nanog proximal promoter. (B,C) Chromosome conformation at the Nanog proximal promoter and −5-kb HS. Wild-type ES cells are indicated in blue and 72-h Oct4-depleted cells are in red. Higher-order structure is lost throughout the locus following Oct4 depletion. (D) A two-dimensional rendering of chromatin looping in three-dimensional space between distant regulatory elements (red ovals) and Nanog locus genes (green and black rectangles indicate active and repressed states, respectively). The model depicts DNA-bound factors within the proximal promoters of GDF3, Dppa3, and Nanog initiating contact (green arrows) with an active transcriptional node formed by RNA polymerase II (large purple oval); accessory DNA-binding or bridging transcription factors p300, ZFP281, Nac1, and CTCF (smaller purple ovals); and essential DNA-bound transcription factors Oct4 and Nanog (green ovals).
A similar pattern of chromosomal contact loss was observed when interactions at the Nanog −5-kb HS were analyzed (Fig. 4C). Again, following Oct4 depletion, contact loss was significant at the Apobec1/GDF3 gene junction, and to a lesser extent at the −151-kb HS with Aicda. Modest loss was observed at the Dppa3 proximal promoter, but was well-defined in the region containing the −47/−45/−38-kb intergenic HS cluster. Like the upstream −47/−45/−38-kb intergenic HS cluster, contacts were abolished following Oct4 removal. Analysis of contacts made with the GDF3 intronic HS corroborated results observed at the Nanog promoter and −5-kb HS (Supplemental Fig. 10).
A model for how distant cis-regulatory elements within the Nanog locus may interact is depicted in Figure 4D. In wild-type, self-renewing ES cells, DNA-bound transcription factors and cofactors occupying the proximal promoters of GDF3, Dppa3, and Nanog are in a transcriptionally active poised state. Shared essential DNA-binding factors, such as Oct4 and Nanog, may initiate contact with an active transcriptional node formed by RNA polymerase II and accessory DNA-binding or bridging transcription factors, such as p300, ZFP281, and Nac1. Initiation of Oct4 depletion results in down-regulation of Nanog. Loss of Nanog may be critical as it contacts DNA within the node at one or more sites within HS −5 and HS −99 that are proximal to Nanog and GDF3, as well as at intergenic HS −45 and HS −47. The resulting weakening of the two strongest links of the transcriptional complex may require only further subtle perturbations, supplied by loss of CTCF, p300, and Nac1, to complete collapse of the organized chromatin state. CTCF may act as a boundary element at HS −47 during a late step of commitment, together with an unidentified partner(s) further upstream between GDF3 and Dppa3, to partition the genes into a subcompartment of selective expression during development. This is implied by the pattern of gene expression observed in epiblast stem cells, in which GDF3 and Nanog are actively expressed, but Dppa3 is extinguished (Tesar et al. 2007).
Conclusion
We used high-throughput QCP to discover DNaseI HSs and potential regulatory elements along a 160-kb region of mouse chromosome 6 that is important for early fate decisions in the developing mouse embryo. The work reported here reveals a highly regulated, open chromatin domain surrounding the Nanog gene locus.
We also showed that Oct4 is essential for maintaining higher-order chromatin structure within the Nanog locus. Distant chromosomal contacts between regulatory elements within the Nanog locus are presumably mediated through DNA-bound protein complexes. A complex structure is suggested by a series of contacts between the Apobec1/GDF3 gene junction, Dppa3, Nanog, and the intergenic −47/−45/−38-kb HS cluster. We observed collapse of higher-order chromatin structure throughout the Nanog locus following depletion of Oct4. An altered chromatin conformation is concomitant with, and likely mediated by, loss of chromatin occupancy by Oct4 and Nanog, and other proteins (such as CTCF, Nac1, and ZFP281, among others). We suggest that evaluation of gene expression and chromatin structure within the Nanog locus gene cluster may serve as a useful sensor at the genomic level for changes during cellular reprogramming.
Materials and methods
Cell culture
ES cell lines (CJ7, J1, or ZHBTc4; a kind gift from A. Smith) were maintained in standard ES media. ZHBTc4 ES cells were cultured in the above medium and induced to differentiate with 100 ng/mL doxycycline. Tagged ES cell lines were described previously (Wang et al. 2006). MEL cells were cultured in DMEM supplemented with 10% fetal calf serum, 2 mM L-glutamine, and 50 U/mL penicillin/streptomycin.
mRNA expression analysis
cDNA was obtained using the Qiagen RNeasy kit. Reverse transcription was performed with SuperScript III RT from Invitrogen according to the manufacturer’s recommendations. Quantitative real-time PCR was performed on cDNA in a 25-μL SYBR Green reaction using a Bio-Rad iCycler. Expression was normalized to HPRT.
Array analysis and bioinformatics
Affymetrix array data was obtained from the Novartis GeneAtlas Web site (Su et al. 2004). Gene and intergenic DNA alignments were done with CLUSTALW using the AlignX program from the VectorNTI suite.
DNaseI digestion and DNA purification
DNaseI digestions on CJ7 nuclei were done according to a standard protocol (Dorschner et al. 2004).
Quantitative ChIP (Q-ChIP)
Murine CJ7 ES cells were used for ChIP of RNA polymerase II, TATA-binding protein (TBP), p300, H3K4me3, H3K27me3, H3K9me3, and CTCF using standard antibody pull-down methods following cross-linking in media containing 1% formaldehyde for 10 min and sonication to nucleosomal-sized fragments averaging 400–500 base pairs. J1 ES cells expressing BirA and biotin-tagged proteins (Oct4, Nanog, Zfp281, and Nac1) were processed similarly.
3C analysis
Chimeric PCR products produced from 3C in wild-type or doxycycline-treated ZHBTc4 ES cells were distinguished using primers that are available upon request. Briefly, ES cells were treated with 4% formaldehyde for 10 min at room temperature, quenched with 0.125 M glycine, trypsinized, pelleted, and washed with cold PBS. Cells were then sequentially treated with 0.3% SDS, then 1.8% Triton X-100 to sequester the SDS, before overnight digestions with HindIII, T4 ligation for 4 h at 16 degrees, cross-link reversal, and DNA cleanup. Control samples without cross-linking or without T4 ligase were treated in parallel. Each reaction was quantified within the exponential phase of amplification using a SynGene GeneSnap imaging system and the GeneTools analysis program.
Acknowledgments
We thank Austin Smith for the ZHBTc4 ES cells and members of the Orkin laboratory for comments. S.H.O. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1606308.
References
- Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Defossez P.A., Gilson E. The vertebrate protein CTCF functions as an insulator in Saccharomyces cerevisiae. Nucleic Acids Res. 2002;30:5136–5141. doi: 10.1093/nar/gkf629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dorschner M.O., Hawrylycz M., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nat. Methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- Drissen R., Palstra R.J., Gillemans N., Splinter E., Grosveld F., Philipsen S., de Laat W. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes & Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoriw A.M., Stein P., Svoboda P., Schultz R.M., Bartolomei M.S. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkola J.E., Houldsworth J., Chadalavada R.S., Olshen A.B., Dobrzynski D., Reuter V.E., Bosl G.J., Chaganti R.S. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006;66:820–827. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Matoba R., Niwa H., Masui S., Ohtsuka S., Carter M.G., Sharov A.A., Ko M.S. Dissecting oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS ONE. 2006;1:e26. doi: 10.1371/journal.pone.0000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Pereira L., Yi F., Merrill B.J. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell. Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S., Ligon K.L., Hornick J.L. Embryonic stem cell transcription factor signatures in the diagnosis of primary and metastatic germ cell tumors. Am. J. Surg. Pathol. 2007;31:836–845. doi: 10.1097/PAS.0b013e31802e708a. [DOI] [PubMed] [Google Scholar]
- Spilianakis C.G., Flavell R.A. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Townes T.M., Behringer R.R. Human globin locus activation region (LAR): Role in temporal control. Trends Genet. 1990;6:219–223. doi: 10.1016/0168-9525(90)90182-6. [DOI] [PubMed] [Google Scholar]
- Vakoc C.R., Letting D.L., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B.E., Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Xie X., Mikkelsen T.S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E.S. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]