Abstract
The activation of the ataxia telangiectasia mutated (ATM) and ATM/Rad3-related (ATR) kinases triggers a diverse cellular response including the initiation of DNA damage-induced cell cycle checkpoints. Mediator of DNA Damage Checkpoint protein, MDC1, and H2AX are chromatin remodeling factors required for the recruitment of DNA repair proteins to the DNA damage sites. We identified a novel mediator protein, Cep164 (KIAA1052), that interacts with both ATR and ATM. Cep164 is phosphorylated upon replication stress, ultraviolet radiation (UV), and ionizing radiation (IR). Ser186 of Cep164 is phosphorylated by ATR/ATM in vitro and in vivo. The phosphorylation of Ser186 is not affected by RPA knockdown but is severely hampered by MDC1 knockdown. siRNA-mediated silencing of Cep164 significantly reduces DNA damage-induced phosphorylation of RPA, H2AX, MDC1, CHK2, and CHK1, but not NBS1. Analyses of Cep164 knockdown cells demonstrate a critical role of Cep164 in G2/M checkpoint and nuclear divisions. These findings reveal that Cep164 is a key player in the DNA damage-activated signaling cascade.
[Keywords: DNA damage signal pathways, ATR, CEP164, RPA, MDC1, CHK1, H2AX]
The genome is subjected to continuous damage and repair. Free radicals generated during cellular metabolism, DNA replication errors, and exogenous carcinogens can all lead to DNA damage, which triggers multiple signal transduction pathways to slow down cell cycle progression, allowing for the repair of the damaged DNA. It has been proposed that these pathways sense DNA damage, activate cell cycle checkpoints, and recruit repair proteins to the damaged DNA (Sancar et al. 2004; Stucki and Jackson 2006). ATM (ataxia telangiectasia mutated) and ATR (ATM- and Rad3-related gene) are two evolutionarily conserved phosphatidylinositol kinase-related proteins that play critical roles in checkpoint activation (Shiloh 2003). Mutations in ATM lead to AT (ataxia telangiectasia) disorder (Savitsky et al. 1995), which is characterized by neuronal degeneration and cancer predisposition, while reduced ATR expression leads to Seckel syndrome (O’Driscoll et al. 2003), which is characterized by retarded development. Knockout of ATM and ATR in mice, respectively, demonstrates that ATR, but not ATM, is essential for embryogenesis and cell survival (Barlow et al. 1996; Brown and Baltimore 2000; Cortez et al. 2001).
ATM is activated by DNA double-strand break (DSB)-causing agents including ionizing radiation (IR), while ATR is activated by ultraviolet radiation (UV) as well as replication block (Abraham 2001). Interestingly, IR also activates ATR in an ATM- and Mre11–NBS1–Rad50 (MRN)-dependent manner (Jazayeri et al. 2005; Zhong et al. 2005; Myers and Cortez 2006). ATM and ATR share many substrates; i.e., Ser15 of p53 (Siliciano et al. 1997), and Ser1423 and Ser1524 of BRCA1 (Fabbro et al. 2004). In addition, phosphorylation of Ser139 of H2AX by ATM/ATR is well documented (Burma et al. 2001). Mediator of DNA damage checkpoint protein 1, MDC1, is a crucial component in the DNA damage response network (Goldberg et al. 2003; Lou et al. 2003; Shang et al. 2003; Stewart et al. 2003). For efficient phosphorylation of H2AX, its interacting protein, MDC1, is required (Stewart et al. 2003). MDC1 recruits ubiquitin ligase RNF8 to ubiquitinate H2AX and likely other substrates at the DNA damage site (Huen et al. 2007). The phosphorylation of MDC1 and H2AX operates upstream of the phosphorylation of replication protein A (RPA) in the ATM/ATR signal transduction pathway (Stewart et al. 2003). The identification of upstream players in DNA damage checkpoint pathways provides mechanistic insights into how genomic stability is maintained.
In this study, we report a novel mediator protein in DNA damage response, Cep164 (Graser et al. 2007), that interacts with ATR and ATM constitutively. DNA damage and replication stress induce phosphorylation of Cep164 and rapid relocalization of this protein into nuclear foci. We identify Ser186 of Cep164 as an in vitro and in vivo ATR/ATM phosphorylation site. DNA damage-induced phosphorylation of Cep164 at Ser186 does not require RPA, but instead depends on the presence of MDC1. Cep164 is essential for the phosphorylation of H2AX, MDC1, CHK2, and RPA upon UV and IR damage but is dispensable for the phosphorylation of NBS1 at Ser343. Furthermore, DNA damage-induced phosphorylation of CHK1 and activation of the G2/M checkpoint requires Cep164. These results highlight a critical role of Cep164 in ATM/ATR DNA damage signaling pathways.
Results
Identification of a novel ATR-associated protein, Cep164
ATR is evolutionarily conserved; its orthologs are Rad3 in Schizosaccharomyces pombe, Mec1 in Saccharomyces cerevisiae, and UVSB in Aspergillus nidulans, respectively. Amino acid sequence comparison of the ATR-interacting proteins: Rad26 (S. pombe), Pie-1 (S. cerevisiae), and UVSD (A. nidulans) revealed a weak homologous region within an ∼120-amino-acid coiled-coil domain (Wakayama et al. 2001). The ATR-interacting protein, ATRIP, a putative human homolog of Rad26, also contains this conserved region with limited overall amino acid sequence similarity (Cortez et al. 2001). We performed a BLAST sequence homology search of the human gene bank using amino acid sequences of UVSD and identified a putative Rad26 homologous region in the protein product of KIAA1052 (Supplemental Fig. S1). Further sequence analysis of KIAA1052 identified an open reading frame of 1455 amino acids. There are two amino acid sequences representing differentially spliced isoforms in the Gene Bank. The other is a recently reported novel centriole appendage protein named Cep164 that consists of 1460 amino acids (Graser et al. 2007). The 5-amino-acid difference is due to differential splicing of exons 9 and 26, respectively. At the N terminus, Cep164 consists of a WW domain, followed by a long predicted coiled-coil region (Berger et al. 1995), and 16 serine–glutamine/threonine–glutamine (SQ/TQ) sites that are potential ATM/ATR phosphorylation sites (Fig. 1A). Based on the presence of a putative Rad26 homologous region and the confirmed interaction with ATR (see below), we studied the role of Cep164/KIAA1052 in DNA damage response.
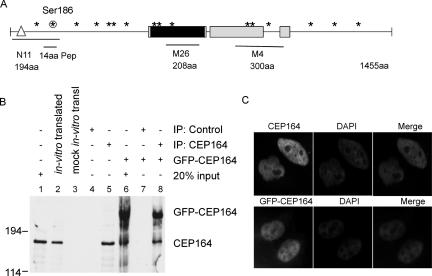
Figure 1.
Identification of an ATR-associated protein, Cep164, and its interaction with ATR and ATRIP. (A) A diagrammatic representation of domains and motifs in Cep164 sequence. Cep164 consists of 1455 amino acids. M26 and M4 are GST-Cep164 fusion peptides used as antigens for the production of antibodies. The corresponding antibodies are designated anti-M26 and anti-M4, respectively. N11 is a monoclonal antibody isolated using GST-Cep1641-194 fusion protein as antigen. The 14-amino-acid peptide and phospho-Ser186 peptide of Cep164 used to raise polyclonal anti-Cep164 and anti-phospho-Ser186 (anti-p Ser186) antibodies are shown. The asterisks indicate potential substrate sites (SQ/TQ) of ATR/ATM kinases; the encircled asterisk specifies Ser186 of Cep164. The triangle and black and gray rectangles depict the WW domain, Rad26 homologous domain, and coiled-coil region, respectively. (B) Specificity of Cep164 antibodies. Monoclonal anti-Cep164 antibody (M26) was used to immunoprecipitate endogenous or overexpressed GFP-Cep164. The same antibody was used for immunoblotting to detect Cep164 in HeLa cells lysate or in vitro translated product, or in the immunoprecipitates. (C) Nuclear localization of endogenous Cep164 or GFP-Cep164. HeLa cells were fixed and stained with anti-Cep164 (M4) and DAPI. HeLa cells were visualized under a fluorescence microscope 8 h after transfection.
To identify the protein product of Cep164, we prepared three GST-Cep164 fusion polypeptides consisting of amino acids 1–194, 665–873, and 887–1187 of Cep164, respectively, as shown in Figure 1A. The monoclonal antibody M26 recognized a protein of ∼180 kDa, a mass close to that of the predicted molecular mass of 164 kDa, in HeLa cells. The other monoclonal antibodies, M4 and N11, and polyclonal antibodies also detected a 180-kDa protein (Fig. 1B; data not shown). The mobility of in vitro translated Cep164 product was similar to that of the endogenous Cep164 (Fig. 1B, lanes 1,2). The monoclonal antibody M26 detected both the endogenous Cep164 protein as well as the GFP-Cep164 fusion protein in cells transfected with the GFP-Cep164 expression vector (Fig. 1B, lane 6). Taken together, we conclude that Cep164 encodes a 180-kDa protein.
Based on immunostaining, Cep164 was localized in the nuclei (Fig. 1C). Similarly, GFP-Cep164 fusion protein was also present in the nucleus (Fig. 1C, bottom panel). Results from cell fractionation were consistent with the immunostaining data (Supplemental Fig. S2).
We examined whether there were interactions between endogenous Cep164 and ATR or ATRIP. Immunoprecipitation was carried out using Cep164, ATR, or ATRIP antibodies and analyzed by immunoblotting. The three proteins could be coimmunoprecipitated reciprocally (Fig. 2A, lanes 3–5). To further confirm the specific interaction among the three proteins, we performed immunoprecipitation in cells expressing Flag-tagged ATR. Flag-ATR and Cep164 also coimmunoprecipitate (data not shown). Previous studies indicated that ATR binds to chromatin during S phase (Hekmat-Nejad et al. 2000) and also upon DNA damage (Unsal-Kacmaz et al. 2002); hence the interaction seen between ATR and Cep164 could be mediated by DNA. Coimmunoprecipitation was carried out in the presence of ethidium bromide at the concentration of 50 μg/mL, which disrupts protein and DNA interaction (Lai and Herr 1992). Similar coimmunoprecipitation results were obtained in the presence of ethidium bromide (Fig. 2A, lanes 6–8). Taken together, we conclude that Cep164, ATR, and ATRIP interact physically in vivo.
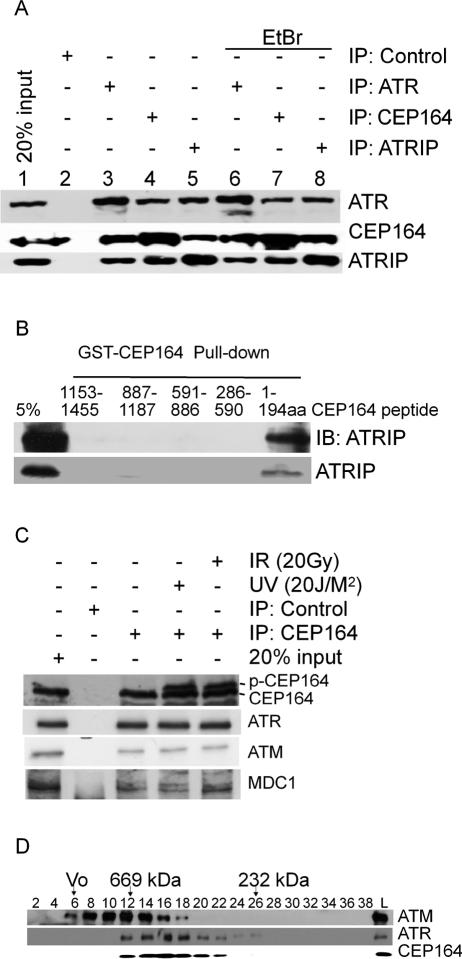
Figure 2.
Interaction of Cep164 with ATR and ATM complexes. (A) Interaction between Cep164 and ATR in vivo. HeLa cell lysates were incubated with anti-Cep164, anti-ATR, and anti-ATRIP antibodies, respectively, in the presence (lanes 2–5) or absence (lanes 6–8) of ethidium bromide. Immunoprecipitates were separated by SDS-PAGE followed by immunoblotting using monoclonal anti-Cep164 (M26) and anti-ATR (2B5) antibodies. (B) Direct interaction of Cep164 with ATRIP. (Top panel) The indicated GST fusion peptides were subjected to pull-down assay with HeLa cell lysate and blotted with ATRIP antibody. 35S-Methinonine-labeled ATRIP by in vitro transcription/translation was subjected to the indicated GST fusion peptide of Cep164 and the samples were electrophoresed; the gel was dried and exposed to X-ray film. (C) Effects of DNA damage on interaction between Cep164 and ATR or ATM. HeLa cells exposed to IR or UV were incubated for 1 h, and the Cep164 immunoprecipitates were prepared. The samples were analyzed by immunoblotting using ATM, ATR, and Cep164 antibodies. (D) Fractionation of HeLa cell lysate. HeLa cell lysates were fractionated using a Superdex-200 column, and samples were analyzed by immunoblotting.
To identify regions within Cep164 that mediate interaction with ATR and ATRIP, five different GST fusion peptides of Cep164 were made as indicated in Figure 2B. HeLa cell lysate was used for the pull-down assay with the GST fusion peptides. Immunoblotting of the pull-down samples revealed that the N-terminal peptide 1–194 amino acids of Cep164 interacts with ATRIP (Fig. 2B, top panel); this polypeptide also pulled down the in vitro translated ATRIP (Fig. 2B, bottom panel), confirming that Cep164 binds directly to ATRIP.
In order to determine whether Cep164 interacts with other proteins with a role in DNA damage-induced checkpoint activation pathway, samples of Cep164 immunoprecipitation were immunoblotted with ATM and MDC1 antibodies. Both MDC1 and ATM proteins associated with Cep164 (Fig. 2C, lane 3). ATM and MDC1 coimmunoprecipitated with Cep164 reciprocally (data not shown). It has been previously shown that the ATM-interacting protein Mre11 and NBS1 bind to ATRIP (Myers and Cortez 2006), which associates with Cep164 (Fig. 2B). On the other hand, MDC1 interacts physically with ATM through its FHA domain (Lou et al. 2006). Thus Cep164 is likely to be a component of both ATM and ATR kinase complexes.
To test whether DNA damage impacts the interaction of these proteins, HeLa cells were irradiated with IR and UV, and the lysates were analyzed using immunoprecipitation and immunoblotting. The interactions of Cep164 with ATR, ATM, and MDC1 were not altered by either IR- or UV-induced DNA damage (Fig. 2C, lanes 3–5). It has also been reported that the interaction of MDC1 with ATM is not altered by DNA damage (Stewart et al. 2003). Despite the fact that there was no change in the interactions, the Cep164 blot showed a slowly migrating band in the lysates of UV- and IR-irradiated cells.
HeLa cell lysates were fractionated by gel filtration chromatography, and the distribution of ATM, ATR, and Cep164 were determined. ATR was present in a 668-kDa complex (Fig. 2D), similar to a previous report (Unsal-Kacmaz and Sancar 2004); ATM was present in a 1.5-MDa complex (Lee and Paull 2004) as well as fractions containing smaller protein complexes. The pattern of Cep164 fractionation was similar to that of ATR protein and partially overlapped with ATM. When lysates prepared from HeLa cells were subjected to immunodepletion using anti-ATR antibody 2B6, a substantial quantity of Cep164 remained in the supernatant (Supplemental Fig. S3). Thus not all Cep164 is presented in ATR complexes.
Modification of Cep164 upon DNA damage and replication stress
We analyzed whether Cep164 was modified upon DNA damage. Cells were irradiated at 5, 10, 20, and 40 J/M2, and cell lysates were subjected to immunoblotting analysis using Cep164 antibody. As shown in Figure 3A, a slowly migrating band appeared in samples prepared from UV-irradiated cells. Intensities of the slowly migrating band increased upon increasing UV dosages, suggesting that the modification of Cep164 is UV dose-dependent. To determine the kinetics of the appearance of the slowly migrating Cep164, HeLa cells were irradiated with 20 J/M2 of UV, and the cells were harvested at 5, 15, 30, and 60 min post-UV. A slowly migrating band was detected 30 min post-irradiation. The intensity of the slowly migrating band increased with time (Fig. 3A, bottom panel) and persisted for >10 h (Supplemental Fig. S4). Immunoblotting with CHK1Ser317 and RPASer4/Ser8 phospho-specific antibodies showed that phosphorylation of CHK1 and RPA34 appeared at 5 and 60 min post-UV irradiation, respectively (Fig. 3A, bottom panel).
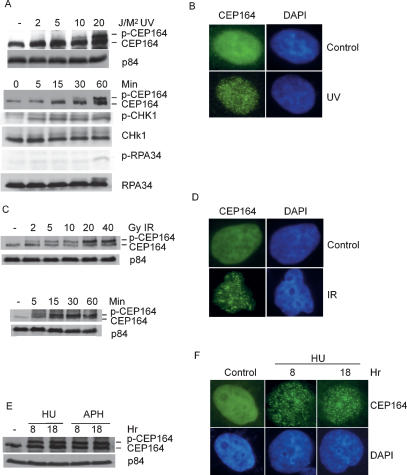
Figure 3.
Modification of Cep164 in response to DNA damage or replicative stress. (A) Modification of Cep164 upon UV irradiation. HeLa cells were treated with the indicated doses of UV, cells were lysed 1 h later, and lysates were analyzed by immunoblotting. (Top panel) Mitosin p84 is expressed at a constant level and serves as a loading control. UV-irradiated (20 J/M2) cells were lysed at the indicated time points post-UV irradiation. (Bottom panel) The samples were immunoblotted with the specified antibody. (B) UV-induced Cep164 foci formation. HeLa cells were exposed to UV at 20 J/M2 and incubated for 2 min. Cells were fixed and stained with anti-Cep164 (M4) antibodies. DNA was visualized by DAPI staining. (C) Modification of Cep164 upon IR. HeLa cells were treated with indicated doses of IR, and lysates were prepared 4 h later. Lysates were analyzed as described previously. IR-irradiated (20 Gy) cells were analyzed at different time points post-IR. (D) IR-induced Cep164 foci formation. HeLa cells were exposed to UV at 20 Gy and incubated for 2 min. Cells were fixed and stained with anti-Cep164 (M4) antibodies. DNA was visualized by DAPI staining. (E) Modification of Cep164 upon replication stress. HeLa cells were treated with 1 mM hydroxyurea or 2.5 μg/mL aphidicolin for the indicated period of time, and cell lysates were analyzed as described. (F) Replication stress-induced Cep164 foci formation. HeLa cells were exposed to 1 mM hydroxyurea for an indicated time, and cells were fixed and stained with anti-Cep164 (M4) antibodies and DAPI.
Many DNA repair and checkpoint proteins form intranuclear foci at DNA damage/repair sites. The kinetics of Cep164 foci formation were analyzed using anti-Cep164 monoclonal antibody, M4. Cep164 foci were detected in HeLa cells within 2 min post-20 J/M2 of UV irradiation (Fig. 3B), and foci persisted for >10 h (data not shown). In accordance with previous report (Cortez et al. 2001), ATR foci were also observed in irradiated cells. The majority of Cep164 and ATR foci colocalized (Supplemental Fig. S5). Thus, as part of the early DNA damage response, Cep164 is modified and relocalized to DNA damage sites. To address whether Cep164 also became modified upon other assaults, HeLa cells were irradiated with 5, 10, 20, 30, and 40 Gy of IR, and protein lysates were prepared 4 h post-IR. Similar to UV irradiation, Cep164 was modified in a dose-dependent manner (Fig. 3C, top panel). A slowly migrating Cep164 band was detected within 5 min upon 20 Gy of IR, and the intensities of the slowly migrating band increased over time (Fig. 3C, bottom panel). IR-induced foci were readily detected within 5 min upon IR (Fig. 3D).
In addition to DNA damage induced by UV and IR, we tested the response of the Cep164 upon DNA replication stress. Hydroxyurea and amphidicolin are chemicals that lead to the replication forks’ stall by depletion of dNTPs or inhibition of DNA primase activity, respectively. A more slowly migrating band of Cep164 was detected after hydroxyurea and amphidicolin treatment (Fig. 3E); foci consisting of Cep164 were also detected upon hydroxyurea treatment (Fig. 3F). These data show that in addition to DNA damage, Cep164 is also modified in response to replication stress.
ATR- and ATM-mediated phosphorylation of Cep164
It was shown that caffeine inhibits ATM, ATR, and DNA-PK at 400 μM, 2 mM, and 10 mM, respectively (Sarkaria et al. 1999). Treating cells with 2 mM caffeine 1 h prior to UV irradiation inhibited the appearance of the slow-migration Cep164 band (Fig. 4A). This observation suggests that ATM or ATR might be responsible for the modification of Cep164. To confirm that the slowly migrating Cep164 band was indeed due to protein phosphorylation, lysates were treated with phosphatase. Phosphatase treatment reverted the slowly migrating band to a band with mobility similar to that of Cep164 in control cells (Fig. 4B), indicating that the slowly migrating band seen in UV-treated cells was indeed due to protein phosphorylation.
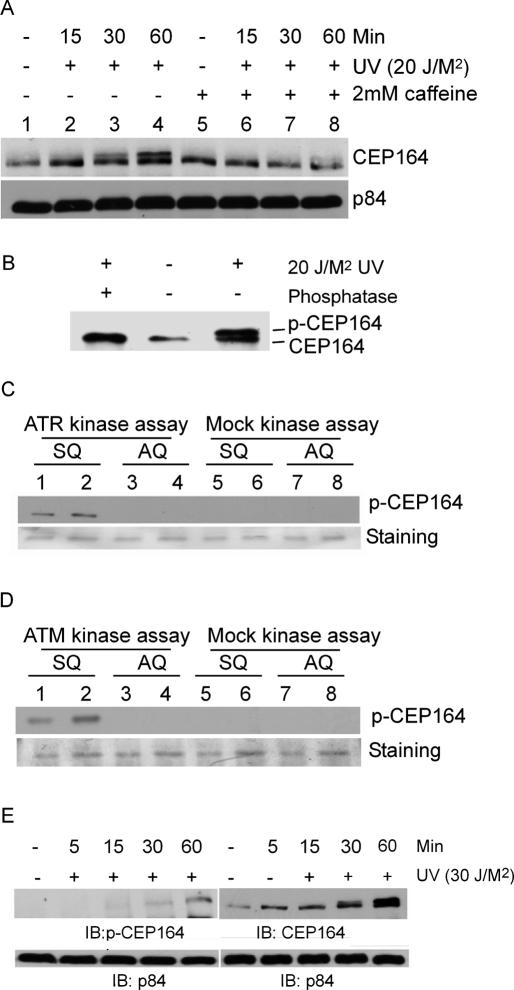
Figure 4.
Mapping of a phosphorylation site in Cep164. (A) The effect of caffeine on the phosphorylation of Cep164. HeLa cells were treated with 2 mM caffeine for 1 h and irradiated with UV at 20 J/M2. The treated and mock-treated cells were lysed at the indicated time points and analyzed as described. (B) Phosphatase treatment reverts the slowly migrating modified Cep164. HeLa cells were UV-irradiated with 20 J/M2 and incubated for 1 h. The nuclear lysates were prepared and treated with or without alkaline phosphatase and subjected to immunoblotting with anti-Cep164 antibody. (C) In vitro ATR kinase assays using GST-Cep164 substrate. The GST-Cep164wt (amino acids 174–194) and GST-Cep164S186A (Ser186 to Ala186) polypeptides were subjected to in vitro kinase assays using the ATR immunoprecipitates from lysates of UV-irradiated HeLa cells. Samples were gel-electrophoresed and immunoblotted with phospho-Ser186 antibody (top panel) or stained with Coomassie blue (bottom panel). (D) In vitro ATM kinase assays using GST-Cep164 substrate. Experiments were as described in C, except immunoprecipitated ATM was used in the assay. (E) Specificity of phospho-Ser186 antibody. HeLa cells were treated with 20 J/M2 of UV, and the cells were lysed at different time points as indicated. The samples were immunoblotted using phospho-Ser186, Cep164, and p84 antibodies, respectively.
The preferred SQ phosphorylation motif by ATM/ATR has been determined (Kim et al. 1999). To study phosphorylation of a preferred ATR phosphorylation site, Ser186 in the N-terminal 1–194 amino acids of the Cep164, GST fusion construct was made. Fusion protein was purified from Escherichia coli and subjected to in vitro ATR kinase assays. As expected, ATR phosphorylated the Cep164 N-terminal peptide (data not shown). To confirm that Ser186 is phosphorylated in vivo, phospho-specific antibodies were generated using a 14-amino-acid peptide (177–192) of Cep164. A GST-Cep164 fusion peptide (177–192) consisting of only one SQ site (Ser186Glu) and a mutant GST-Cep164 fusion peptide replacing Ser186 with Ala were generated. Purified fusion proteins were subjected to in vitro kinase assays using ATR; ATM or mock immunoprecipitates obtained from UV- or IR-irradiated HeLa cells, respectively. Immunoblotting analyses indicated that phosphorylated Ser186Glu, mediated by ATR or ATM, but not Ala186Glu peptide, was recognized by Cep164 phospho-specific antibodies (Fig. 4C,D, lanes 1–4). Anti-p-Cep164 antibodies did not recognize mock kinase phosphorylated peptide (Fig. 4C,D, lanes 5–8). Direct Western blotting analysis using lysates prepared from UV-irradiated HeLa cells also demonstrated that Ser186 was phosphorylated in vivo but not in control cells (Fig. 4E). The phospho-peptide antibodies reacted only with the slow-migration band and not the fast-migration band, suggesting that the antibodies react specifically with phosphorylated Ser186.
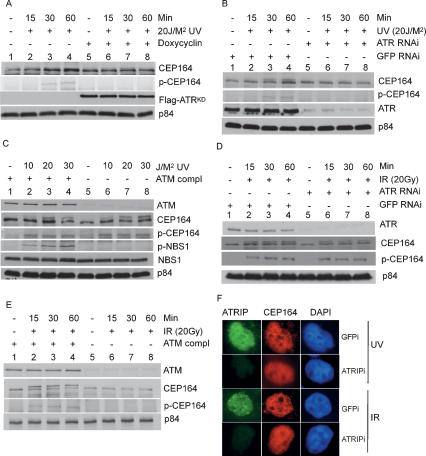
To ascertain the kinase-mediating phosphorylation of Ser186 of Cep164 in vivo, we analyzed a cell line expressing doxycyclin-induced ATRKD, which was shown previously to behave in a dominant-negative fashion (Cliby et al. 1998). In UV-irradiated and doxycyclin-induced cells, phosphorylated Cep164 was not detected (Fig. 5A); in contrast, the slow-migration band was found in cells without doxycyclin induction. Similar results were obtained when anti-p-Cep164 antibodies were used (Fig. 5A). In a complementary approach using siRNA-mediated knockdown of ATR, reduced phosphorylation of Cep164 was observed (Fig. 5B). Thus, ATR mediates phosphorylation of Cep164 upon UV irradiation.
Figure 5.
Cep164 is an in vivo substrate of ATR. (A) Phosphorylation of Cep164 in cells expressing ATRKD. Cells with tetracycline-regulated ATRKD expression vector were grown in the presence or absence of doxycycline for 72 h. Cells were harvested at different time points after UV irradiation. Lysates were analyzed using immunoblotting. (B) Phosphorylation of Cep164 in cells with ATR knockdown. HeLa cells were transfected with either ATRi or GFPi and were irradiated with 20 J/M2 of UV 72 h post-transfection. Lysates were prepared at the indicated time points and analyzed using Western blotting. (C) UV-induced phosphorylation of Cep164 in AT cells. AT lymphoblastoid cells and ATM-complemented cells were irradiated with 20 J/M2 and lysed at the indicated time points. Cell lysates were analyzed as described. (D) IR-induced phosphorylation of Cep164 in cells with ATR knockdown. HeLa cells were transfected with either ATRi or GFPi; cells were irradiated with 20 Gy of IR 72 h post-transfection. Cell lysates were analyzed using Western blotting. (E) IR-induced phosphorylation of Cep164 in AT cells. AT lymphoblastoid cells and ATM-complemented cells were irradiated with 20 Gy of IR. Cell lysates were analyzed using Western blotting. (F) ATR kinase activity and Cep164 foci. HeLa cells were knocked down with GFP RNAi or ATRIP RNAi, and the cells were irradiated with IR or UV. The cells were subjected to immunostaining with ATRIP and Cep164 antibody.
We tested whether Cep164 became phosphorylated upon UV irradiation in ATM-defective cells (AT cells). As shown in Figure 5C, UV-induced phosphorylation of Cep164 was similar in AT and ATM complemented cells. As expected, phosphorylation of NBS1 at Ser343 was aberrant in AT cells but not ATM complemented cells (Fig. 5C). Thus, ATR mediates phosphorylation of Cep164 upon UV irradiation.
ATR is activated by UV-induced DNA damage and replication stress, while ATM is activated by IR (Sancar et al. 2004). The phosphorylation of Cep164 upon IR was analyzed using HeLa cells with siRNA-mediated knockdown of ATR. In both control GFP and ATR knockdown cells, phosphorylation at Ser186 of Cep164 was detected at 15 min (Fig. 5D). However, in ATR knockdown cells, in contrast to GFP siRNA knocked down cells, there was no increase in Ser186 phosphorylation at 30 and 60 min post-IR. These data suggest that while ATR is dispensable for initial response, it is required for sustained Cep164 phosphorylation upon IR (Fig. 5E). Phosphorylation of Ser186 is compromised in AT cells (Fig. 5E, lanes 5–8), but complementation with ATM restored the IR-induced phosphorylation of Cep164 (Fig. 5E, lanes 1–4). Thus, IR-induced phosphorylation of Cep164 requires ATM.
Phosphorylation of Ser186 of Cep164 was observed 30 min post-UV radiation (Figs. 4E, 5A–C), but within 5 min upon IR radiation (Supplemental Fig. S6). In addition to IR- and UV-induced DNA damage, phosphorylation of Ser186 of Cep164 was observed in cells treated with alkylating agent (methyl methane sulfonate, MMS), intercalating agent (cisplatin), and inhibitor of topoisomerase II (doxorubicin) (Supplemental Fig. S7). The observation of Ser186 phosphorylation upon hydroxyurea and amphidicolin treatment (Supplemental Fig. S8) suggests that DNA damage as well as stalled replication forks lead to phosphorylation of Cep164 at Ser186.
To further confirm the roles of ATM/ATR in Cep164 foci formation, HeLa cells were treated with 2 mM caffeine to inactivate both kinases. No Cep164 foci were detected upon IR or UV (data not shown). Cep164 foci were diminished in ATRIP knocked down cells upon UV irradiation, but persisted upon IR irradiation (Fig. 5F). The data suggest that ATR/ATRIP is not crucial in the recruitment of Cep164 to DNA damage site upon IR-induced DNA damage.
Upstream molecules required for DNA damage-induced phosphorylation of Cep164
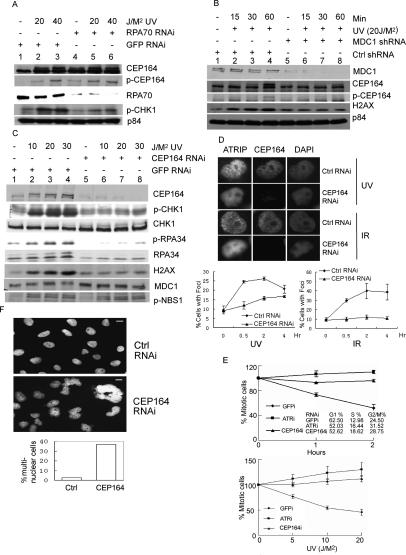
A critical role of RPA in DNA damage checkpoint activation has been reported in yeast, Xenopus, and mammalian cells. In S. cerevisiae, checkpoint activation is abolished in strains defective in ssDNA-binding protein RPA (Lydall and Weinert 1995). In Xenopus, checkpoint activation requires the loading of RPA onto chromatin (Shechter et al. 2004). RPA is a heterotrimer consisting of RPA70, RPA34, and RPA14 subunits. We studied whether UV-induced phosphorylation of Cep164 required RPA. RPA70 was knocked down by transient transfection of a specific siRNA. The cells were irradiated with 20 or 40 J/M2 of UV at 48 h post-transfection when RPA70 levels were greatly reduced (Fig. 6A). Consistent with the previous report, upon RPA70 knockdown, the phosphorylation of CHK1 at Ser317 was significantly reduced (Zou and Elledge 2003). Phosphorylation of Ser186 of Cep164 was detected in both the control and UV-irradiated RPA70 knockdown cells (Fig. 6A, lanes 4–6), while only irradiated GFP knockdown cells showed phosphorylation at Ser186 (Fig. 6A, lanes 2,3). Since replicative stress caused by RPA knockdown may lead to activation of ATR and phosphorylation of Cep164, whether RPA is required for augmentation of DNA damage-induced phosphorylation of Cep164 cannot be fully assessed.
Figure 6.
Cep164, MDC1, and RPA in ATR/ATM signaling pathways. (A) The effect of RPA70 knockdown on the phosphorylation of Cep164. HeLa cells were transfected with either RPA70i or GFPi; cells were irradiated with the indicated doses of UV 48 h post-transfection and lysed 1 h post-UV irradiation. Lysates were analyzed by Western blotting as indicated. (B) The effect of MDC1 knockdown on the phosphorylation of Cep164. HeLa cells were transfected with either MDC1i or GFPi; cells were irradiated with 20 J/M2 of UV 72 h post-transfection. Cell lysates were analyzed by immunoblotting as indicated. (C) The effects of Cep164 knockdown on the ATR signal pathway. HeLa cells were transfected with either Cep164i or GFPi; cells were irradiated with the indicated doses of UV 72 h post-transfection. Cells were harvested 1 h post-UV irradiation; samples were analyzed by immunoblotting. (D) HeLa cells were transfected with either control RNAi or Cep164 RNAi and irradiated with IR at 10 Gy or UV at 10 J/M2 48 h post-transfection, followed by immunostaining with ATRIP or Cep164 antibody at the time points as indicated in the bottom panel. Percentage of ATRIP foci was scored from 200 cells for each time point. Only cells with more than eight foci were counted. (E) G2/M checkpoint in cells with Cep164 knockdown. HeLa cells were transfected with Cep164i, ATRi, or GFPi and irradiated at 20 J/M2 48 h post-transfection. The cell cycle distributions are based on FACS analyses of GFPi- or Cep164i-transfected cells prior to radiation. (Top figure) Samples were collected at the indicated time points. (Bottom figure) The RNAi-transfected cells were irradiated with the indicated doses of UV, and the samples were collected 2 h after UV irradiation. These cells were fixed and stained with DAPI or costained with phospho-histone 3S10 antibody. The cells with condensed nuclear or positively stained with phospho-histone 3S10 antibody were considered as in G2/M phase. The percentage of cells in G2/M phase was determined. The G2/M percentage in control cells (without UV treatment) was artificially set as 100%, and all other percentages shown on both the top and bottom panels are relative ratios compared with the control. (F) Nuclear morphology of cells with Cep164 knockdown. HeLa-H2B-EGFP cells were transfected with Cep164 or Luciferase RNAi. Forty-eight hours after transfection, cells were fixed and the nuclear morphology was compared under the microscope. (Top) Nuclear morphology of cells transfected with Luciferase RNAi. (Middle) Nuclear morphology of cells transfected with Cep164 RNAi. (Bottom) The percentage of cells harboring multinuclei. Bars: top and middle figures, 10 μm.
H2AX functions in chromatin remodeling and is upstream of RPA in DNA damage-mediated signal transduction (Balajee and Geard 2004). H2AX is dispensable for IR-induced phosphorylation of MDC1; in contrast, Mdc1 is important for the phosphorylation of H2AX (Stewart et al. 2003; Stucki et al. 2005; Lou et al. 2006). To address whether chromatin remodeling is important for the phosphorylation of Cep164 at Ser186, MDC1 was knocked down by transient transfection using an MDC1-specific shRNA expression vector (Peng and Chen 2005). At 72 h post-transfection, expression of MDC1 was effectively reduced (Fig. 6B). In agreement with the previous report (Stewart et al. 2003), phosphorylation of H2AX was diminished in MDC1 knockdown cells, but not in control vector-transfected cells. Importantly, phosphorylation of Cep164 at Ser186 was significantly reduced (Fig. 6B). Taken together, MDC1-mediated chromatin remodeling is critical for DNA damage-induced phosphorylation of Cep164.
Cep164 is a crucial player in the early response of DNA damage-induced signal transduction
To explore the role of Cep164 in the DNA damage signal transduction pathway, Cep164 was knocked down using specific siRNAs. As shown in Figure 6C, the Cep164 protein level was substantially reduced in the Cep164 knockdown cells using either one of two Cep164 siRNAs (data not shown). Immunoblotting analyses were performed to address the impact of reduced expression of Cep164 in UV- or IR-induced cellular response. Phosphorylation of CHK1 at Ser317 was aberrant, while the total CHK1 protein levels were not changed; similarly, phosphorylation of RPA34 was reduced, and the total RPA levels did not alter during the time course studied. Phosphorylation of Ser139 of H2AX upon UV or IR was severely affected in Cep164 knocked down cells (Fig. 6C; Supplemental Fig. S9). Phosphorylation of MDC1 was also compromised. The phosphorylation of Chk2 at Thr68 was compromised in Cep164 knocked down cells upon IR irradiation, but phosphorylation of NBS1 at Ser343 was not affected by either UV or IR irradiation (Fig. 6C; Supplemental Fig. S9). Taken together, these data indicate that Cep164 is important for the proper phosphorylation of H2AX, RPA, CHK2, and CHK1 in DNA damage-induced checkpoint signaling cascade.
Immunostaining with ATRIP antibody revealed that in Cep164 knocked down cells, the percentage of cells with ATRIP foci reduced at 0.5, 2, and 4 h post-IR or UV (Fig. 6D). At 4 h post-10 Gy of IR, ATRIP foci were detected in 38.7% of control knockdown cells, and only 10.8% in Cep164 knockdown cells. Similarly, there was a reduction of ATRIP foci upon UV. At 2 h post-10 J/M2 of UV, ATRIP foci was detected in 26.2% of control knockdown cells, and only 15.7% in Cep164 knockdown cells. While there were significantly fewer foci 0.5 and 2 h post-UV in Cep164 knockdown cells, percentages of foci-positive cells continued to increase over time, and by 4 h, there were 20.7% and 16.7% of ATRIP foci in control and Cep164 knockdown cells, respectively. It has been reported that RPA coated ssDNA recruits ATR/ATRIP to the site of DNA damage (Zou and Elledge 2003). The data presented here indicate that Cep164 is a new constituent in the recruitment and/or maintenance of ATRIP to the DNA damage site.
Reduced expression of Cep164 results in abrogation of G2/M checkpoint
ATR-mediated phosphorylation of CHK1 at Ser317 upon DNA damage is critical for the G2/M checkpoint (Zhao and Piwnica-Worms 2001). FACS sorting indicated that percentages of G1-phase cells were reduced in ATR (52.03%) and Cep164 (52.62%) knockdown cells, respectively, when compared with control GFP knockdown (62.50%) cells; and percentages of S- and G2/M-phase cells increased in ATR (16.44%; 31.52%) and Cep164 (18.62%; 28.75%) knockdown cells when compared with control GFP knockdown (12.98%; 24.50%) cells. To test whether Cep164 is required for the G2/M checkpoint, we scored the percentage of M-phase and G2-phase cells using phospho-histone 3 immunostaining in control GFP, ATR, and Cep164 siRNA knockdown cells, respectively, before and after UV irradiation. Comparing untreated, 1 h, and 2 h post-20 J/M2 of UV, no G2/M checkpoint activation was observed in Cep164 or ATR knockdown cells (Fig. 6E, top panel). Similar to the time course results, there was no G2/M checkpoint activation in either Cep164 or ATR knockdown cells upon treatment of different dosages of UV (Fig. 6E, bottom panel), thus reiterating the importance of Cep164 and ATR’s role in the G2/M checkpoint.
We analyzed the 4′,6′-diamidino-2-phenylindole (DAPI)-stained nuclear morphology of Cep164 knocked down cells. Approximately 37% of 600 scored cells were multinucleated (Fig. 6F). In contrast, only 2.7% of multinucleated cells were seen in the control GFP siRNA-transfected cells (Fig. 6F). In addition, there was an elevated number of giant cells (cell sizes doubled or greater than doubled) in Cep164 knockdown cells. A similar observation was made when Cep164 knockdown was performed in HeLa cells stably expressing H2B-EGFP (Fig. 6F). Multinucleated cells as well as giant cells are the hallmark of genomic instability, and this was observed in MDC1 knockdown cells (Lou et al. 2006).
Thus, Cep164, a novel player, has a functional link to both ATR and ATM signal transduction pathways in DNA damage-induced checkpoint activation. Both the presence of Cep164 and the kinase activities of ATM and ATR are important for checkpoint activation. The protein is upstream of RPA and CHK1/2 phosphorylation upon UV or IR irradiation. The abrogation of the G2/M checkpoint along with the formation of multinucleated and giant cells and the failure of ATRIP relocation to DNA damage sites upon Cep164 knockdown indicate that Cep164 is an important player required for genomic stability.
Discussion
The present study identified a novel player in DNA damage signal transduction. Cep164 was reported recently to be localized to the distal appendages of mature centrioles and was involved in the formation of primary cilia (Graser et al. 2007). We showed interaction between Cep164 and ATR/ATM, as well as the phosphorylation of Cep164 by ATR/ATM. By knocking down Cep164 and other key players in the DNA damage checkpoint pathways followed by the analyses of phosphorylation events, we place Cep164 upstream of RPA in the activation of CHK1 and CHK2.
The Cep164 protein
Based on amino acid sequence analyses, we identified several putative domains within Cep164: a WW domain, three coiled-coil regions, and a region with weak homology with the Rad26 domain. The WW domain has been shown to facilitate protein–protein interaction in both phosphorylation-dependent and -independent manners (Sudol et al. 2001). The specific function of the WW domain of Cep164, located in the N terminus of Cep164 (Fig. 1A), awaits future studies. The coiled-coil regions of Cep164 are similar to that of Rad50 and SMC family proteins (Anderson et al. 2001; Lowe et al. 2001); however, Cep164 lacks the conserved N- or C-terminal domains seen in the SMC family members. Recent studies demonstrate that the coiled-coil domain in ATRIP is involved in self-dimerization (Ball and Cortez 2005; Itakura et al. 2005), as well as augmenting the interaction between ATRIP and ATR (Ball and Cortez 2005). The tentative Rad26 homologous region of Cep164 is embedded in its first coiled-coil region (Fig. 1A), which also possesses self-dimerization functions (data not shown).
Using multiple approaches, we conclude that Cep164 encodes a 180-kDa protein. First, antibodies generated against different GST-Cep164 fusion proteins all recognized a 180-kDa protein (Fig. 1B). Second, reduction in Cep164 protein was seen in Cep164 knockdown cells using specific siRNA (Fig. 6C). Third, in vitro translated Cep164 migrated to a position similar to that of the endogenous protein was recognized by anti-Cep164 antibodies (Fig. 1B, lanes 1,2). Fourth, recombinant flag-Cep164 was recognized by an anti-Cep164 M26 antibody (Fig. 1B). Based on immunostaining experiments against the endogenous Cep164 as well as fluorescent microscopic observation of the recombinant GFP-Cep164 fusion protein, we conclude that Cep164 is localized to the nucleus (Fig. 1C). Chromatin fractionation has provided additional support for this conclusion (Supplemental Fig. S2). Consistent with previous reports (Andersen et al. 2003; Graser et al. 2007), we detected Cep164 in the centrosome (data not shown). The immunoprecipitation (Fig. 2A,C) and fractionation studies (Fig. 2D) suggest that Cep164 is associated mostly with the ATR/ATRIP complex; however, there is also overlapped distribution with ATM. Judging from the significant amount of Cep164 protein in the supernatant immuno-depleted of ATR (Supplemental Fig. S3), there is a certain population of Cep164 that does not interact with ATR/ATRIP. Activation of ATR by IR requires ATM (Jazayeri et al. 2005; Zhong et al. 2005; Myers and Cortez 2006); the ATM-interacting MER11 and NBS1 immunoprecipitate with ATRIP (Myers and Cortez 2006); and the phosphorylation of ATM at Ser1981 by UV and stalled replication forks required ATR but not the C-terminal NBS1 or Mre11 (Stiff et al. 2006). These studies demonstrate functional and biochemical links between ATM and ATR pathways. Cep164 protein may bridge ATM and ATR complexes; however, there is no change in the distribution of Cep164 molecules in ATR and ATM complex upon DNA damage (Fig. 2C).
ATR/ATM-mediated phosphorylation of Cep164
Both the in vitro and in vivo studies support that ATR and ATM phosphorylate Cep164. A wide range of DNA damage and replication stress triggers the phosphorylation of Ser186 with different kinetics (Supplemental Fig. S7). Upon IR irradiation, Ser186 phosphorylation was observed in 5 min (Fig. 3C; Supplemental Fig. S7), in contrast to the 30-min gap upon UV irradiation. The difference in the kinetics of Ser186 phosphorylation upon UV and IR might be due to difference in the nature of IR- and UV-induced DNA damage. IR causes single-strand breaks (SSBs), and if the breaks are localized closely to each other in opposite strands, DSBs also occur (Sato et al. 2005). The Mre11, NBS1, and Rad50 are critical for the processing of DNA ends damaged by IR and for the formation of RPA-coated ssDNA (Jazayeri et al. 2005; Zhong et al. 2005; Myers and Cortez 2006). In contrast, UV irradiation predominantly causes the formation of cyclobutane pyrimidine dimers (CPD) (Lippke et al. 1981) and pyrimidine pyrimidone (6-4PP) photo adducts (Mitchell and Nairn 1989) that are repaired by the nucleotide excision repair (NER) pathway. At higher UV doses, DSBs are induced due to the processing of clustered DNA damage. Recruitment of RPA to the DNA damage site upon UV is independent of the ATM and MRN complex (Jazayeri et al. 2005; Zhong et al. 2005; Myers and Cortez 2006). A key player in the NER pathway, XPA, is phosphorylated by ATR upon UV irradiation (Wu et al. 2006). In the absence of XPA, UV-induced phosphorylation of CHK1 at Ser317 mediated by ATR and the recruitment of RPA to chromatin are affected (Bomgarden et al. 2006). The kinetics of the phosphorylation of Ser186 may be influenced by proteins at the DNA damage sites that differ depending on the type of DNA damage.
UV-induced phosphorylation of Cep164 was similar in AT cells and AT cells complemented with ATM (Fig. 5C). On the other hand, IR-induced phosphorylation of Cep164 was not detected in AT cells but was readily detected in cells with ATR knockdown (Fig. 5D). However, the phosphorylation failed to sustain beyond 1 h in the latter case. ATM/ATR phosphorylates common SQ sites; it is unclear whether ATM/ATR works in a sequential manner to maintain the phosphorylation of specific sites; or alternatively, ATR prevents the dephosphorylation of the site through other mechanisms. It has been demonstrated that ATR phosphorylates H2AX at Ser139 upon UV irradiation in cells with ATM knockdown. Thus, it is likely ATR and ATM cooperate upon IR and UV as reported previously (Jazayeri et al. 2005; Myers and Cortez 2006).
A total of 16 potential ATR/ATM substrate sites (SQ/TQ) are present in Cep164 (Fig. 1B). Exactly how many of these sites are phosphorylated as well as which crucial sites are involved in the DNA damage response relating to the relocation of Cep164 to the DNA damage site and activation of downstream effectors are yet to be determined.
Cep164 is involved in the DNA damage-induced signal cascade
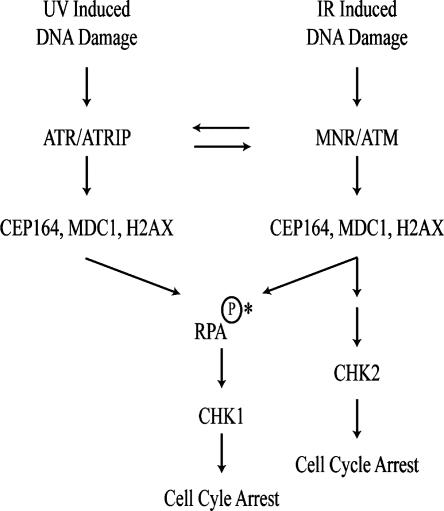
XPA, RPA, and XPC are all involved in the recognition of DNA lesions caused by UV; on the other hand, the MRN complex binds to the DNA DSBs. Studies have indicated that ATM/ATR and Rad17–RFC/Rad9/Hus1/Rad1 complexes are also critical sensors for DSBs (Sancar et al. 2004). It has been shown that ssDNA-coated RPA is critical for the recruitment of ATR/ATRIP to the DNA damage site and activation of ATR (Zou and Elledge 2003). A second RPA-independent ATR–ATRIP DNA-binding complex has also been identified (Bomgarden et al. 2004). Upon UV irradiation, increased phosphorylation of Ser186 of Cep164 was detected in RPA knockdown cells (Fig. 6A), suggesting that the previously identified RPA-independent complex may be involved in Cep164 phosphorylation. While phosphorylation of Ser186 of Cep164 remained the same, CHK1 phosphorylation was severely impaired, confirming a crucial role of RPA in the activation of CHK1. We place Cep164 upstream of RPA and parallel to MDC1 in the DNA damage signal cascade (Fig. 7) based on the following observations: (1) Kinetically, phosphorylation of RPA34Ser4/Ser8 occurs at 60 min post-UV (Fig. 3A, bottom panel), while phosphorylation of Ser186 of Cep164 appeared 30 min after UV irradiation. (2) Knockdown of hCep164 affects the phosphorylation of RPA34 (Fig. 6C), but RPA is not required for Cep164 phosphorylation (Fig. 6A). (3) Phosphorylation of MDC1 and H2AX, both upstream of RPA (Stewart et al. 2003), was diminished by knocking down Cep164 (Fig. 6C). (4) Knocking down Cep164 reduces ATRIP foci formation (Fig. 6D) that occurs at the time of RPA coating of ssDNA. While the kinetics of ATRIP foci formation were significantly slowed down upon UV radiation in Cep164 knockdown cells, no increase in the percentage of cells with ATRIP foci above the basal levels was seen in cells with Cep164 knockdown upon IR radiation. It is not clear whether this is due to the presence of compensative players in the UV response. Interestingly, persistent phosphorylation of Cep164 was detected in RPA70 knockdown cells in the absence of DNA damage (Fig. 6A). This may be due to replication stress induced by knocking down of RPA, leading to ATM/ATR-mediated phosphorylation of Cep164 (Araya et al. 2005).
Figure 7.
Cep164 and the ATM/ATR signal pathways. ATM and ATR serve as sensors responding to DNA damage induced by IR or UV, respectively. Mre11 nuclease of the MRN complex processes damaged DNA to generate ssDNA overhang. ATR phosphorylates CHK1 at Ser317 and activates the G2/M checkpoint. MDC1 and H2AX are upstream mediators whose phosphorylation is required for CHK1 phosphorylation and activation. The phosphorylation of Cep164 and MDC1 was compromised in cells with MDC1 or Cep164 knockdown, respectively, suggesting that these two proteins are mutually dependent for their proper regulation. RPA phosphorylation requires Cep164. See the text for details. (↓) Activation. (*) RPA is required for UV-induced Chk1 activation, but whether RPA phosphorylation is required has yet to be shown.
Phosphorylation of H2AX is a key step in the remodeling of chromatin upon DNA damage (Thiriet and Hayes 2005; Stucki and Jackson 2006). It is hypothesized that chromatin remodeling increases the accessibility of damaged DNA by repair proteins, allowing DNA resection, and formation of RPA-coated ssDNA (Morrison et al. 2004; van Attikum et al. 2004). H2AX molecules accumulate at the site of DNA damage and extend megabases away from the damage site to facilitate chromatin remodeling (Rogakou et al. 1999). The mediator protein, MDC1, coimmunoprecipitates with NBS1 (Goldberg et al. 2003; Stewart et al. 2003); MDC1 also brings ATM and H2AX together via a direct interaction with both proteins. MDC1 accumulates active ATM and H2AX at DNA damage sites, allowing the amplification of the DNA damage signals (Lou et al. 2006). MDC1 also recruits ubiquitin ligase RNF8 to ubiquitinate H2AX and likely other substrates at the DNA damage site, and both phosphorylation and ubiquitination modifications are crucial for DNA damage response (Huen et al. 2007; for review, see Petrini 2007). Thus, MDC1 is a central mediator in corralling sensors and chromatin remodeling factors to the DNA damage site. The reduced MDC1 and H2AX phosphorylation in Cep164 knockdown cells (Fig. 6C) and the MDC1-dependent Cep164 phosphorylation upon DNA damage (Fig. 6B) underlie the crucial roles of Cep164 as a mediator in the maintenance of genomic stability.
Multinucleated and giant cells, resulting from chromosome missegregation or abnormal cytokinesis, are frequently observed in cancer (Doussis et al. 1992); this phenomenon is likely related to dysfunctional checkpoint activation. For instance, inactivation of CHK1 results in polyploidy nuclei (Mihaylov et al. 2002). Similarly, polyploid nuclei and multiple centrosomes have been documented in cells with aberrant CDK1 (Itzhaki et al. 1997). Notably, accumulation of multinucleated cells with Cep164 knockdown (Fig. 6F) suggests a potential role for Cep164 in chromosome segregation, in addition to its functions in checkpoint signaling. Consistent with previous reports (Andersen et al. 2003; Graser et al. 2007), we also detected the presence of Cep164 at the centrosome (data not shown). Many checkpoint or DNA repair proteins including p53, CHK1, Chk2, BRCA1, and Rad51 have been shown to localize in the nucleus and the centrosomes (Kramer et al. 2004a). For example, human CHK1 localizes to centrosomes during interphase but not mitosis; the centrosome-associated CHK1 controls mitosis entry through regulating cyclin B–Cdk1 activity (Kramer et al. 2004b). In addition, the centrosomal fraction of CHK1 is phosphorylated upon DNA damage. Immobilization of kinase-inactive Chk1 to centrosomes leads to a defective G2/M checkpoint (Loffler et al. 2007). While a fraction of Cep164 is localized to the centrosome, specific contribution of the centrosomal versus the chromatin-associated Cep164 to checkpoint activation requires further studies. Taken together, our studies demonstrate that Cep164 is a novel mediator in the ATR/ATM signaling pathways and is also a centrosomal protein with a critical role in chromosome segregation.
Materials and methods
Cell culture
Cell lines were purchased from the American Type Culture Collection. Cells were cultured in Dulbecco’s Modified Eagle’s medium supplemented with 10% fetal bovine serum (Gibco). Cells were irradiated using a 137Cs γ-irradiator (Shepherd). Ultraviolet irradiation was performed using UV Stratalinker 2400 (Stratagene).
Plasmid construction
The DNA sequences encoding amino acids 1–194, 665–873, and 887–1187 (fragments consisting of 194, 208, and 300 amino acids, respectively) of Cep164 and amino acids 1–107 of ATRIP were obtained from cDNAs of kiaa1052 and ATRIP, respectively, using PCR, and were subcloned by ligating the PCR products into the Sma1 site of pGEX4-T3 vector. Fragments consisting of amino acids 177–192 (wild-type and mutant Cep164S186A) were cloned into a pGEX4-T vector at BamH1 and Xho1 sites.
Generation of antibodies
The GST-Cep164 fusion proteins consisting of amino acids 1–196, 665–873, and 887–1187 of Cep164 and amino acids 1–107 of ATRIP were overexpressed in E. coli and purified by affinity chromatography. Mouse polyclonal antisera specific for the fusion proteins and hybridoma cell lines, N11, M26, and M4 were generated according to standard procedures (Chen and Lee 1996).
Rabbit polyclonal anti-phospho-Ser186 antibody was raised against the KLH-conjugated peptide GELMLPpSQGLKTSA (Ser186). The phosphorylated peptide:unphosphorylated peptide reactivity ratio of affinity-purified Ser186-specific antibody was >99:1, as determined by ELISA (Bethyl Laboratories, Inc.).
Designing of siRNAs
The siRNA duplexes were 21 base pairs with a 2-base deoxynucleotide overhang (Dharmacon Research). The sequences of Cep164 siRNA4 and siRNA5 oligonucleotides were GAAGA UACAGGAAGCUCAAdTdT and CUUCGCCAACGGGCAG UCUdTdT, respectively. While both siRNAs efficiently knock down Cep164, siRNA4 was used to perform all the knockdown experiments throughout this study. The control siRNAs used were UGGCUUUCUGUAGAGGACAUCdTdT and TTACGC TGAGTACTTCGAdTdT against GFP and luciferase, respectively. siRNAs for ATR, ATRIP (GGUCCACAGAUUAUUAG AUdTdT), and RPA70 were based on previous reports (Cortez et al. 2001; Zou and Elledge 2003). The plasmid harboring the siRNA sequence against MDC1 was a gift from Phang-Lang Chen (University of California at Irvine). Cells were transfected with siRNA duplexes by using Lipofectamine 2000 (Invitrogen) or RNAiFect (Qiagen), following the manufacturers’ instructions.
Chromatin fractionation
Whole-cell extracts were obtained by directly lysing cells in SDS sample buffer. To isolate chromatin, cells were lysed and fractionated as described (Mendez and Stillman 2000). Briefly, ∼2 × 106 cells were washed with PBS and resuspended in 200 μL of buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 5 μg/mL aprotinin, 5 μg/mL leupeptin, 0.5 μg/mL pepstatin A, 0.1 mM phenylmethylsulfonyl fluoride). Triton X-100 (0.1%) was added and mixed gently by inversion, followed by 5 min of incubation on ice. Nuclei were collected in pellet 1 (P1) by centrifugation at low speed (1300g, 5 min, 4°C). The supernatant (S1) was clarified by high-speed centrifugation (20,000g, 15 min, 4°C) to obtain the soluble fraction (S2). Nuclei were then washed once in buffer A, resuspended in 200 μL of buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 5 μg/mL aprotinin, 5 μg/mL leupeptin, 0.5 μg/mL pepstatin A, 0.1 mM phenylmethylsulfonyl fluoride), and incubated on ice for 30 min. Insoluble chromatin was collected by centrifugation (1700g, 5 min, 4°C), washed once in buffer B, and centrifuged at 1700g for 5 min. The final chromatin-enriched pellet (P3) was resuspended in SDS sample buffer and subjected to sonication. To release chromatin-bound proteins, nuclei (P1) were digested with 0.2 U of micrococcal nuclease (Sigma) in buffer A plus 1 mM CaCl2. After 1 min of incubation at 37°C, the nuclease reaction was stopped by addition of 1 mM EGTA. Nuclei were then lysed and fractionated as described above.
Gel filtration analysis
HeLa whole-cell extract was prepared in hypotonic buffer (20 mM HEPES at pH 7.9, 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT). A Superdex-200 PC 3.2/30 (2.4 mL) gel filtration column was equilibrated at 4°C in a solution containing 20 mM HEPES (pH 7.9), 250 mM NaCl, 1.5 mM MgCl2, and 2 mM DTT as reported previously (Unsal-Kacmaz and Sancar 2004).
Immunoblotting and immunoprecipitation
Cells were lysed either directly in SDS sample buffer containing 1 mM phenylmethylsulphonyl fluoride, 100 mM NaF, and 1 mM Na3VO4 or in EBC buffer supplemented with protease inhibitors (1 μg/mL aprotinin, 5 μg/mL leupeptin, 1 mM phenylmethylsulphonyl fluoride, 100 mM NaF, 1 mM Na3VO4). Protein concentration was determined by Bradford assay (Bio-Rad). Cell lysates obtained by lysis with EBC buffer were mixed with SDS sample buffer, and 30–60 μg of protein were subjected to SDS-PAGE. Proteins were transferred to Immobilon P (Millipore).
Monoclonal antibodies to NBS1, ATM, ATR, CHK1, and p84 were generated in the laboratory. Anti-Cep164 and anti-ATRIP monoclonal antibodies are available from GeneTex. The anti-CHK2 was purchased from Bethyl Laboratories, Inc. Anti-β-actin, anti-Flag antibodies were purchased from Sigma. The anti-phospho-H2AX (Ser139), anti-phospho-H3 (Ser10), and anti-MDC1 antibodies were obtained from Upstate Biotechnology. The polyclonal antibodies against ATR, Orc2, and Mek2, antibodies were purchased from Santa Cruz Biotechnolgy, Oncogene, and BD Biosciences, respectively. Anti-RPA34 and anti-RPA70 antibodies were obtained from LAB Vision Corporation, Inc., and Oncogene Research Products, respectively.
Cells were lysed in ice-cold EBC buffer containing 0.5% NonidetP-40 with protease inhibitors, and the lysates were spun at 14,000 rpm for 10 min. The supernatant was collected and subjected to protein estimation. One-milligram protein samples were precleared by incubation with Protein G/A Sepharose beads (1:1 ratio) for 1 h, and the samples were incubated with 4 μg of antibody for 2 h. The samples were incubated with Protein G/A Sepharose beads for 1 h and washed four times with EBC buffer containing 0.5% NonidetP-40. Bound proteins were boiled in SDS sample buffer. Independent immunoprecipitations were carried out by treating 0.3 mg/mL ethidium bromide during cell lysis as reported previously (Onclercq-Delic et al. 2003). The immunoprecipitates were then subjected to SDS-PAGE separation.
GST pull-down assay
Briefly, HeLa cell lysate or in vitro translated ATRIP labeled with 35S- methionine was mixed with the indicated GST fusion peptide and incubated for 90 min at 4°C, followed by incubation with Glutathione Sepharose 4B beads (Amersham Pharmacia) for 1 h. The samples were washed with TBS three times and boiled with equal amounts of 2× SDS loading buffer. The samples were subjected to gel electrophoresis.
Kinase assays
Endogenous ATR and ATM were immunoprecipitated from HeLa cells mock-treated or exposed to 10 Gy of IR using α-ATR (2B5) or α-ATM (3E8) IgGs. Recombinant Flag-ATRWt and ATRKD were immunoprecipitated using α-Flag-M2 antibodies. ATR and ATM kinase assays were performed as reported previously (Chen et al. 2001). Reaction products were separated by SDS-PAGE and analyzed by Coomassie staining and autoradiography.
Phosphatase treatment
HeLa cells were irradiated with UV at 20 J/M2 and incubated for 1 h. Approximately 2 × 106 cells were washed with PBS and resuspended in 200 μL of buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 5 μg/mL aprotinin, 5 μg/mL leupeptin, 0.5 μg/mL pepstatin A, 0.1 mM phenylmethylsulfonyl fluoride). Triton X-100 (0.1%) was added to the lysate and mixed gently by inversion, followed by 5 min of incubation on ice. Nuclei were collected in pellets by centrifugation at low speed (1300g, 5 min, 4°C) and were resuspended in phosphatase buffer (New England Biolab) with or without protein phosphatase (New England Biolabs) for 15 min at 35°C.
Immunofluorescence staining
For immunofluorescence staining, cells were fixed with 4% paraformaldehyde in PBS (pH 7.2) and permeabilized with 0.5% Triton X-100 in PBS. After 30 min of incubation in blocking buffer (10% FCS in PBS), cells were incubated with primary antibodies overnight at 4°C. After three washes with PBS, the cells were incubated with FITC- or Texas Red-conjugated secondary antibodies for 2 h at room temperature. The cells were then washed with PBS, counterstained with 0.05 μg/mL DAPI in PBS, and mounted in Immunon mountant. All antibodies were diluted in PBS supplemented with 5% FCS.
G2/M checkpoint assay
HeLa cells at 60%–70% confluence were transfected with siRNA duplexes against GFP, ATR, or Cep164, respectively. Forty-eight hours after transfection, cells were treated with UV at the indicated dosages and incubated for 2 h or as indicated and then fixed with 4% paraformaldehyde in PBS containing 0.1% Trion X-100. Cells were stained with DAPI and/or costained with anti-phospho-histone H3 followed by FITC-conjugated secondary antibody. The cells with condensed DNA and broken nuclear membrane or positive FITC fluorescence (in the case of anti-phospho-histone 3 immunostaining) were counted as mitotic cells. DAPI-positive cells were also counted. This experiment was repeated three times.
Acknowledgments
We thank the Kazusa Human cDNA project, Japan for the cDNA of KIAA1052, Dr. Eric A. Nigg (Max Planck Institute, Germany) for the cDNA of GFP-Cep164 and polyclonal antibodies, Dr. Stephen J. Elledge (Harvard Medical School) for ATRIP cDNA and antibody, Dr. Phang-Lang Chen (University of California at Irvine) for the shRNA construct of MDC1, and Yoon Kim and Sou-Ying Lee for generating reagents and reading the manuscript.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1627708.
References
- Abraham R.T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Anderson D.E., Trujillo K.M., Sung P., Erickson H.P. Structure of the Rad50 x Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J. Biol. Chem. 2001;276:37027–37033. doi: 10.1074/jbc.M106179200. [DOI] [PubMed] [Google Scholar]
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Araya R., Hirai I., Meyerkord C.L., Wang H.G. Loss of RPA1 induces Chk2 phosphorylation through a caffeine-sensitive pathway. FEBS Lett. 2005;579:157–161. doi: 10.1016/j.febslet.2004.11.066. [DOI] [PubMed] [Google Scholar]
- Balajee A.S., Geard C.R. Replication protein A and γ-H2AX foci assembly is triggered by cellular response to DNA double-strand breaks. Exp. Cell Res. 2004;300:320–334. doi: 10.1016/j.yexcr.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Ball H.L., Cortez D. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J. Biol. Chem. 2005;280:31390–31396. doi: 10.1074/jbc.M504961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., et al. Atm-deficient mice: A paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Berger B., Wilson D.B., Wolf E., Tonchev T., Milla M., Kim P.S. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomgarden R.D., Yean D., Yee M.C., Cimprich K.A. A novel protein activity mediates DNA binding of an ATR–ATRIP complex. J. Biol Chem. 2004;279:13346–13353. doi: 10.1074/jbc.M311098200. [DOI] [PubMed] [Google Scholar]
- Bomgarden R.D., Lupardus P.J., Soni D.V., Yee M.C., Ford J.M., Cimprich K.A. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase η on ATR checkpoint signaling. EMBO J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.J., Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- Chen G., Lee E. The product of the ATM gene is a 370-kDa nuclear phosphoprotein. J. Biol. Chem. 1996;271:33693–33697. doi: 10.1074/jbc.271.52.33693. [DOI] [PubMed] [Google Scholar]
- Chen M.J., Lin Y.T., Lieberman H.B., Chen G., Lee E.Y. ATM-dependent phosphorylation of human Rad9 is required for ionizing radiation-induced checkpoint activation. J. Biol. Chem. 2001;276:16580–16586. doi: 10.1074/jbc.M008871200. [DOI] [PubMed] [Google Scholar]
- Cliby W.A., Roberts C.J., Cimprich K.A., Stringer C.M., Lamb J.R., Schreiber S.L., Friend S.H. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Guntuku S., Qin J., Elledge S.J. ATR and ATRIP: Partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Doussis I.A., Puddle B., Athanasou N.A. Immunophenotype of multinucleated and mononuclear cells in giant cell lesions of bone and soft tissue. J. Clin. Pathol. 1992;45:398–404. doi: 10.1136/jcp.45.5.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbro M., Savage K., Hobson K., Deans A.J., Powell S.N., McArthur G.A., Khanna K.K. BRCA1–BARD1 complexes are required for p53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage. J. Biol. Chem. 2004;279:31251–31258. doi: 10.1074/jbc.M405372200. [DOI] [PubMed] [Google Scholar]
- Goldberg M., Stucki M., Falck J., D’Amours D., Rahman D., Pappin D., Bartek J., Jackson S.P. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., Nigg E.A. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Nejad M., You Z., Yee M.C., Newport J.W., Cimprich K.A. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Sawada I., Matsuura A. Dimerization of the ATRIP protein through the coiled-coil motif and its implication to the maintenance of stalled replication forks. Mol. Biol. Cell. 2005;16:5551–5562. doi: 10.1091/mbc.E05-05-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki J.E., Gilbert C.S., Porter A.C. Construction by gene targeting in human cells of a ‘conditional’ CDC2 mutant that rereplicates its DNA. Nat. Genet. 1997;15:258–265. doi: 10.1038/ng0397-258. [DOI] [PubMed] [Google Scholar]
- Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C., Lukas J., Jackson S.P. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2005;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Kim S.T., Lim D.S., Canman C.E., Kastan M.B. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Kramer A., Lukas J., Bartek J. Checking out the centrosome. Cell Cycle. 2004a;3:1390–1393. doi: 10.4161/cc.3.11.1252. [DOI] [PubMed] [Google Scholar]
- Kramer A., Mailand N., Lukas C., Syljuasen R.G., Wilkinson C.J., Nigg E.A., Bartek J., Lukas J. Centrosome-associated Chk1 prevents premature activation of cyclin-B–Cdk1 kinase. Nat. Cell Biol. 2004b;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- Lai J.S., Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Paull T.T. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lippke J.A., Gordon L.K., Brash D.E., Haseltine W.A. Distribution of UV light-induced damage in a defined sequence of human DNA: Detection of alkaline-sensitive lesions at pyrimidine nucleoside–cytidine sequences. Proc. Natl. Acad. Sci. 1981;78:3388–3392. doi: 10.1073/pnas.78.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler H., Bochtler T., Fritz B., Tews B., Ho A.D., Lukas J., Bartek J., Kramer A.2007DNA damage-induced accumulation of centrosomal Chk1 contributes to its checkpoint function Cell Cycle 6 25412548 [DOI] [PubMed] [Google Scholar]
- Lou Z., Chini C.C., Minter-Dykhouse K., Chen J. Mediator of DNA damage checkpoint protein 1 regulates BRCA1 localization and phosphorylation in DNA damage checkpoint control. J. Biol. Chem. 2003;278:13599–13602. doi: 10.1074/jbc.C300060200. [DOI] [PubMed] [Google Scholar]
- Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M.A., Celeste A., Manis J.P., van Deursen J., Nussenzweig A., Paull T.T., et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Lowe J., Cordell S.C., van den Ent F. Crystal structure of the SMC head domain: An ABC ATPase with 900 residues antiparallel coiled-coil inserted. J. Mol. Biol. 2001;306:25–35. doi: 10.1006/jmbi.2000.4379. [DOI] [PubMed] [Google Scholar]
- Lydall D., Weinert T. Yeast checkpoint genes in DNA damage processing: Implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- Mendez J., Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylov I.S., Kondo T., Jones L., Ryzhikov S., Tanaka J., Zheng J., Higa L.A., Minamino N., Cooley L., Zhang H. Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol. Cell. Biol. 2002;22:1868–1880. doi: 10.1128/MCB.22.6.1868-1880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D.L., Nairn R.S. The biology of the (6-4) photoproduct. Photochem. Photobiol. 1989;49:805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- Morrison A.J., Highland J., Krogan N.J., Arbel-Eden A., Greenblatt J.F., Haber J.E., Shen X. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- Myers J.S., Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M., Ruiz-Perez V.L., Woods C.G., Jeggo P.A., Goodship J.A. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- Onclercq-Delic R., Calsou P., Delteil C., Salles B., Papadopoulo D., Amor-Gueret M. Possible anti-recombinogenic role of Bloom’s syndrome helicase in double-strand break processing. Nucleic Acids Res. 2003;31:6272–6282. doi: 10.1093/nar/gkg834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A.M., Chen P.L. NFBD1/Mdc1 mediates ATR-dependent DNA damage response. Cancer Res. 2005;65:1158–1163. doi: 10.1158/0008-5472.CAN-04-2508. [DOI] [PubMed] [Google Scholar]
- Petrini J.H.J. A touching response to damage. Science. 2007;316:1138–1139. doi: 10.1126/science.1143700. [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Boon C., Redon C., Bonner W.M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L.A., Unsal-Kacmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sarkaria J.N., Busby E.C., Tibbetts R.S., Roos P., Taya Y., Karnitz L.M., Abraham R.T. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- Sato M., Sasaki H., Kazui T., Yokota J., Kohno T. Probing the chromosome 9p21 region susceptible to DNA double-strand breaks in human cells in vivo by restriction enzyme transfer. Oncogene. 2005;24:6108–6118. doi: 10.1038/sj.onc.1208750. [DOI] [PubMed] [Google Scholar]
- Savitsky K., Sfez S., Tagle D.A., Ziv Y., Sartiel A., Collins F.S., Shiloh Y., Rotman G. The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. Hum. Mol. Genet. 1995;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- Shang Y.L., Bodero A.J., Chen P.L. NFBD1, a novel nuclear protein with signature motifs of FHA and BRCT, and an internal 41-amino acid repeat sequence, is an early participant in DNA damage response. J. Biol. Chem. 2003;278:6323–6329. doi: 10.1074/jbc.M210749200. [DOI] [PubMed] [Google Scholar]
- Shechter D., Costanzo V., Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 2004;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Siliciano J.D., Canman C.E., Taya Y., Sakaguchi K., Appella E., Kastan M.B. DNA damage induces phosphorylation of the amino terminus of p53. Genes & Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S., Wang B., Bignell C.R., Taylor A.M., Elledge S.J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- Stiff T., Walker S.A., Cerosaletti K., Goodarzi A.A., Petermann E., Concannon P., O’Driscoll M., Jeggo P.A. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M., Jackson S.P. γH2AX and MDC1: Anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst). 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Sudol M., Sliwa K., Russo T. Functions of WW domains in the nucleus. FEBS Lett. 2001;490:190–195. doi: 10.1016/s0014-5793(01)02122-6. [DOI] [PubMed] [Google Scholar]
- Thiriet C., Hayes J.J. Chromatin in need of a fix: Phosphorylation of H2AX connects chromatin to DNA repair. Mol. Cell. 2005;18:617–622. doi: 10.1016/j.molcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K., Sancar A. Quaternary structure of ATR and effects of ATRIP and replication protein A on its DNA binding and kinase activities. Mol. Cell. Biol. 2004;24:1292–1300. doi: 10.1128/MCB.24.3.1292-1300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsal-Kacmaz K., Makhov A.M., Griffith J.D., Sancar A. Preferential binding of ATR protein to UV-damaged DNA. Proc. Natl. Acad. Sci. 2002;99:6673–6678. doi: 10.1073/pnas.102167799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Fritsch O., Hohn B., Gasser S.M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- Wakayama T., Kondo T., Ando S., Matsumoto K., Sugimoto K. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:755–764. doi: 10.1128/MCB.21.3.755-764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Shell S.M., Yang Z., Zou Y. Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group A by ataxia telangiectasia mutated and Rad3-related-dependent checkpoint pathway promotes cell survival in response to UV irradiation. Cancer Res. 2006;66:2997–3005. doi: 10.1158/0008-5472.CAN-05-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H., Bryson A., Eckersdorff M., Ferguson D.O. Rad50 depletion impacts upon ATR-dependent DNA damage responses. Hum. Mol. Genet. 2005;14:2685–2693. doi: 10.1093/hmg/ddi302. [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]