Abstract
The nuclear localization of genes is intimately tied to their transcriptional status in Saccharomyces cerevisiae, with populations of both active and silent genes interacting with components of the nuclear envelope. We investigated the relationship between the mammalian nuclear pore and the human genome by generating high-resolution, chromosome-wide binding maps of human nucleoporin 93 (Nup93) in the presence and absence of a potent histone deacetylase inhibitor (HDACI). Here, we report extensive genomic reorganization with respect to the nuclear pore following HDACI treatment, including the recruitment of promoter regions, euchromatin-rich domains, and differentially expressed genes. In addition to biochemical mapping, we visually demonstrate the physical relocalization of several genomic loci with respect to the nuclear periphery. Our studies show that inhibiting HDACs leads to significant changes in genomic organization, recruiting regions of transcriptional regulation to mammalian nuclear pore complexes.
[Keywords: NPC, nucleoporin, nuclear organization, Nup93, ChIP–chip]
The nucleus is a structurally and functionally complex organelle with a nonuniform interior consisting of distinct chromatin domains and several proteinaceous subcompartments. Chromosomes occupy nonrandom intranuclear positions with respect to each other and the nuclear periphery (Croft et al. 1999; Parada and Misteli 2002; Parada et al. 2002, 2004a, b; Tanabe et al. 2002a, b; Cremer et al. 2006). Chromosome positioning is believed to expose genomic loci to functionally distinct regions in the nucleus, generating transcriptional regulatory domains favoring either activation or repression. Distinct subnuclear regions also direct specific transcriptional programs by organizing genomic loci around specialized protein hubs. The nucleolus is one such region, mediating the localized transcription of ribosomal RNA genes encoded on multiple chromosomes. In the budding yeast, Saccharomyces cerevisiae, a nucleolar-proximal region has also been shown to cluster several tRNA genes (Thompson et al. 2003).
Components of the nuclear envelope have been shown to assert a repressive role in transcription. In yeast, the silent mating-type loci and telomeres are regulated by components of the nuclear periphery, including the nuclear pore complex (NPC) (Stavenhagen and Zakian 1994; Thompson et al. 1994; Maillet et al. 1996; Marcand et al. 1996; Andrulis et al. 1998; Feuerbach et al. 2002). However, several recent studies have reported NPC-proximal transcriptional activation, with the concomitant recruitment of induced genes from the nuclear interior to the periphery (Brickner and Walter 2004; Casolari et al. 2004, 2005; Menon et al. 2005; Cabal et al. 2006; Dieppois et al. 2006; Drubin et al. 2006; Schmid et al. 2006; Taddei et al. 2006; Brickner et al. 2007; Luthra et al. 2007; Sarma et al. 2007). NPC association can increase the efficiency of mRNA processing and export through associations with the SAGA and TREX complexes, regulate the absolute levels of gene expression, and establish an epigenetic state that confers transcriptional memory and rapid reactivation of genes (Cabal et al. 2006; Taddei et al. 2006; Brickner et al. 2007). Furthermore, several components of the nuclear transport machinery possess boundary activity, potentially facilitating the presence of both repressive and activating domains at the nuclear periphery (Ishii et al. 2002).
Elements of the nuclear periphery are also involved in transcriptional regulation in Drosophila melanogaster. In Drosophila, the dosage compensation complex (DCC), required for the twofold increase in gene expression on the peripherally localized male X chromosome, interacts with two nuclear pore components (Mendjan et al. 2006). The deletion of these pore proteins eliminates the hypertranscription of the male X chromosome, implicating Drosophila nuclear pores in transcriptional activation. In support of this, the Drosophila proteins E(y)2 and Xmas-2 were recently shown to regulate mRNA expression, export, and the subnuclear positioning of the hsp70 gene cluster (Kurshakova et al. 2007). E(y)2 and Xmas-2 are components of the Drosophila SAGA and TREX complexes, whose homologs in yeast play a role in NPC-associated transcriptional activation (Cabal et al. 2006; Dieppois et al. 2006; Taddei et al. 2006).
Interestingly, a recent genome-wide study in Drosophila probing interactions between the nuclear lamina and the genome uncovered a repressive role for the nuclear periphery (Pickersgill et al. 2006). Lamins are integral components of a protein network that lines the inner surface of the nuclear envelope between NPCs and have been shown to bind chromatin in vitro. These results suggest that distinct peripheral components, NPCs, and lamins, may have divergent roles in transcriptional activation and repression, respectively. In addition, a lamin-associated protein, LAP2β, binds to histone deacetylase 3 (HDAC3), a member of a large family of proteins that removes acetyl modifications from histones (Somech et al. 2005). Highly acetylated histones are found in the promoters of transcriptionally active genes, suggesting that the lamin-mediated enrichment of HDAC3 at the nuclear periphery could aid in the maintenance of a transcriptionally repressive environment (Fukuda et al. 2006).
Observations in both mouse and human cells indicate the presence of an equally diverse transcriptional regulatory domain at the nuclear periphery. For example, gene-poor chromosomes are reproducibly located near the nuclear periphery in human lymphocytes and fibroblasts (Croft et al. 1999; Boyle et al. 2001; Tanabe et al. 2002b). However, a study of the murine β-globin locus during erythroid maturation showed that significant transcriptional activity occurred at the nuclear periphery prior to the locus’s transit to the interior (Ragoczy et al. 2006). While NPCs have yet to be implicated in β-globin activation, evidence has emerged that a murine nucleoporin, Nup96, is involved in interferon gene regulation (Faria et al. 2006).
The mammalian NPC is thought to contain as many as 30 unique proteins that are present in multiple copies due to the eightfold symmetry of the complex (Cronshaw et al. 2002). Nucleoporin 93 (Nup93) is a mammalian nucleoporin centrally located in the nuclear pore (Rout et al. 2000; Krull et al. 2004). siRNA-mediated depletion of Nup93 in HeLa cells resulted in misshapen nuclei lacking some, but not all nucleoporins, while depletion of both Nup93 and the transmembrane nucleoporin Ndc1 led to a complete disruption of NPCs (Mansfeld et al. 2006). Nup93, as well as importin-α and CAS (an exportin), have been shown to interact with the histone acetyltransferase (HAT), CREB-binding protein (CBP) (Ryan et al. 2006). This interaction is similar to one seen in S. cerevisiae, where association of a HAT with the NPC is mediated by interactions with SAGA, a multimeric complex required for the expression of numerous yeast genes (Green et al. 2003; Rodriguez-Navarro et al. 2004). As mentioned previously, NPC–SAGA interactions are thought to play a major role in mediating gene recruitment to the NPC in S. cerevisiae (Cabal et al. 2006; Dieppois et al. 2006; Taddei et al. 2006).
HDAC inhibitors (HDACIs) globally elevate levels of histone acetylation in the nucleus by inhibiting Class I and II HDACs (Drummond et al. 2005). HDACI treatment also leads to the enrichment of acetylated histones at the nuclear periphery (Taddei et al. 2001; Gilchrist et al. 2004; Drummond et al. 2005). Local changes in genomic organization have also been reported following treatment with HDACIs (Taddei et al. 2001; Zink et al. 2004; Pickersgill et al. 2006). For example, repressed genes are no longer associated with lamins in Drosophila after treating cells with the HDACI trichostatin A (TSA) for 24 h (Pickersgill et al. 2006). In addition, the human CFTR gene moves away from the nuclear periphery upon treatment with TSA for 10 h (Zink et al. 2004). Importantly, several HDACIs are in clinical trials for various forms of cancer due to their ability to induce the expression of repressed genes that lead to growth arrest, differentiation, and apoptosis in transformed cells (Drummond et al. 2005; Glaser 2007).
Disparate observations of both transcriptional activation and repression at the nuclear periphery in S. cerevisiae and Drosophila led us to investigate the presence of a similar regulatory domain in human nuclei. Using genomic location analysis (Ren et al. 2000), we report Nup93 interactions with human chromosomes 5, 7, and 16. To investigate the effects of global histone acetylation on NPC–chromatin interactions, we treated cells with TSA, a reversible HDACI that does not perturb gross nuclear structure (Taddei et al. 2001; Gilchrist et al. 2004; Pickersgill et al. 2006). Upon treatment with TSA, Nup93 distribution was significantly altered across all three chromosomes, indicative of a large-scale nuclear reorganization event with associated effects on gene expression. Analysis of Nup93-binding sites identified enrichments for several genomic features involved in transcriptional regulation. Together, these features define regions of functional interaction between the nuclear pore and the human genome.
Results
Identification of Nup93-binding sites
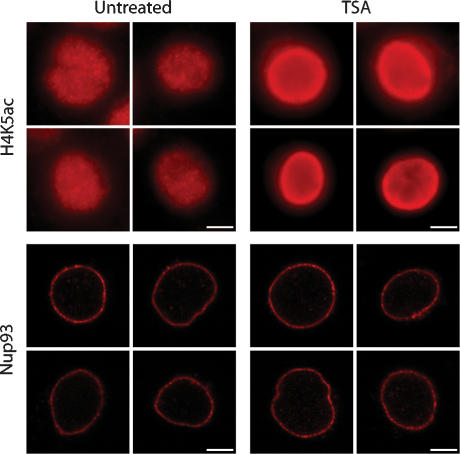
We investigated Nup93–genomic association in the presence and absence of TSA, a potent HDACI. We also performed chromatin immunoprecipitation (ChIP) on several other nucleoporins including Nup62, Nup107, and Nup205, isolating significant amounts of DNA (data not shown). Nup93 was chosen for further study due to the robustness of Nup93 ChIPs and the strictly maintained subnuclear localization of Nup93 at the nuclear envelope (Fig. 1). Immunofluorescence microscopy of HeLa S3 cells following a 12-h, 40-ng/mL TSA treatment revealed a marked increase in histone H4K5 acetylation at the nuclear periphery (Fig. 1), a finding supported by other studies (Taddei et al. 2001; Gilchrist et al. 2004). H3K9 acetylation was also enriched at the nuclear periphery (Supplemental Fig. 1), although to a lesser extent, consistent with previous observations (Gilchrist et al. 2004). Additionally, we found that Nup93 localization was unchanged following TSA treatment (Fig. 1). The subnuclear localization of two other nuclear envelope proteins, lamin B and LAP2, are also unaffected by TSA treatment (Taddei et al. 2001).
Figure 1.
TSA treatment of HeLa S3 cells. Histone acetylation was monitored with an anti-acetylated H4K5 antibody (H4K5ac) in untreated and TSA-treated HeLa S3 cells. Histone acetylation was enriched at the nuclear periphery after 12 h of exposure to 40 ng/mL TSA. H3K9 acetylation is also enriched, although to a lesser extent (Supplemental Fig. 1). The peripheral localization of Nup93 was unchanged following TSA treatment. Bars, 5 μm.
Next, we performed genomic location analysis by coupling ChIPs and microarray analysis (ChIP–chip) to map interactions between Nup93 and the human genome. This technique has been used extensively in S. cerevisiae and is emerging as a powerful tool in higher eukaryotes (Ren et al. 2002; Carroll et al. 2005, 2006; Kim et al. 2005). Asynchronously growing HeLa S3 cells were cross-linked, stabilizing both protein–protein and protein–DNA interactions. We then performed ChIPs of Nup93 and purified, amplified, and hybridized the associated DNA to Affymetrix tiled arrays (ChipE, also referred to as chip5) containing all nonrepetitive regions of human chromosomes 5, 7, and 16. This subset of chromosomes was examined based on preliminary data obtained from Nup62, Nup107, and Nup205 genomic association studies that showed significant interactions with chromosome 7 on ENCODE arrays, including binding at the HOXA locus and CFTR (data not shown). In addition, chromosome 7 has been shown to preferentially reside near the nuclear periphery in human cells (Boyle et al. 2001).
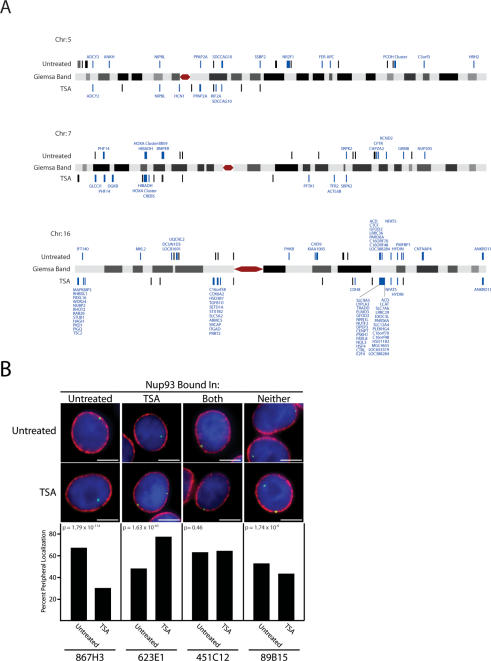
Nup93 was mapped to 207 sites on chromosomes 5, 7, and 16 in untreated cells and 170 sites in TSA-treated cells (Fig. 2A; Supplemental Table 1). The 18% reduction in total Nup93-binding sites was predominantly due to large-scale changes in binding to chromosome 5, where Nup93 association was reduced from 75 sites in untreated cells to 15 sites following TSA treatment (Fig. 2A; Supplemental Table 1). Significant binding sites were classified as either high confidence (143 untreated and 86 TSA sites, 10−12 < P ≤ 10−7) or very high confidence (64 untreated and 84 TSA sites, P < 10−12) based upon our statistical analysis (see the Supplemental Material).
Figure 2.
Nup93 binding map and subnuclear localization of associated genomic regions. (A) Nup93-binding sites identified by ChIP–chip are plotted on schematic representations of chromosomes 5, 7, and 16. Nup93-binding sites are denoted by black bars above or below each chromosome for untreated and TSA-treated cells, respectively. Nup93-binding sites directly overlapping coding regions are labeled blue with the associated gene displayed. Chromosomes are not to scale. (B) FISH analysis of Nup93 targets. Four genomic loci were visually mapped in untreated and TSA-treated HeLa S3 cells. From left to right, these loci interact with Nup93 in untreated cells, TSA-treated cells, both conditions, and neither condition. The top images are examples from untreated cells, while the bottom row are examples from TSA-treated cells. In each image, the FISH signal is green; DNA stained with DAPI is blue; and lamin B, a marker of the nuclear periphery, is red. The bar graphs report percent peripheral localization of each locus in untreated and TSA-treated cells. P-values comparing the difference in percent peripheral localization for each locus were calculated using the χ2 test. BAC probes used to study each genomic region are identified at the bottom of each column. Bars, 5 μm.
Nup93-binding sites were mapped to their nearest neighboring gene, resulting in the assignment of 86 untreated and 90 TSA-treated genes (some genes contained multiple Nup93-binding sites) (Supplemental Table 2). The untreated gene set did not contain any enriched gene ontology groups as reported by GOstat (Beissbarth and Speed 2004). However, the TSA-treated gene set contained two enriched ontology groups: DNA-binding activity (P = 0.002) and transcription factor activity (P = 0.02). These ontology groups include several genes that are members of the HOXA transcription factor cluster on chromosome 7, a gene-dense locus with extensive Nup93 association (Fig. 2A).
Mapping of Nup93 sites in untreated and TSA-treated cells across chromosomes 5, 7, and 16 revealed large changes in nuclear pore association (Fig. 2A). Significant reorganization of the q-arm of chromosome 5 occurred with the loss of statistically significant Nup93 binding over a 98-Mb region following TSA treatment (Fig. 2A). A similar change was seen on the q-arm of chromosome 7 where Nup93 association was lost over a 35-Mb region following TSA treatment (Fig. 2A). However, changes of this kind were not observed on chromosome 16, where many Nup93-binding sites, while mostly intergenic in untreated cells, overlapped with coding regions following TSA treatment (Fig. 2A). Interestingly, the subnuclear localization of chromosome territories has been shown to be unaffected by TSA, while local changes in chromatin condensation have been observed (Croft et al. 1999; Santos et al. 2002).
Visualization of Nup93-associated loci
To verify that Nup93-binding sites were located at the nuclear periphery, we performed FISH combined with confocal microscopy to visualize the intranuclear localization of several genomic loci. Four loci with distinct Nup93 association patterns were chosen (Fig. 2B). One locus, associated in untreated cells and absent in TSA-treated cells, was found at the nuclear periphery 66% of the time in untreated cells versus just 30% in TSA-treated cells (probe 867H3, P = 1.79 × 10−114) (Fig. 2B). Another locus, associated in TSA-treated cells and absent in untreated cells, was found at the nuclear periphery just 48% of the time in untreated cells versus 77% in TSA-treated cells (probe 623E1, P = 1.63 × 10−63) (Fig. 2B). A third locus, associated in both conditions, was found at the nuclear periphery 63% of the time in untreated cells and 64% of the time in TSA-treated cells (probe 451C12, P = 0.46) (Fig. 2B). Finally, a genomic region that was not bound in our ChIP–chip analysis was found at the nuclear periphery 52% and 43% of the time in untreated and TSA-treated cells, respectively (probe 89B15, P = 1.74 × 10−6) (Fig. 2B). The high percentage of peripheral localization observed for the control probe (89B15) in both conditions and probe 623E1 in untreated cells may reflect the peripheral positioning of chromosome 7 in mammalian cells (Boyle et al. 2001). Interestingly, probe 867H3, located on chromosome 5, shows only 30% peripheral association in TSA-treated cells, likely reflecting the preferentially internal positioning of chromosome 5. Associated P-values test the significance of the difference in the percent peripheral localization for each locus in untreated and TSA-treated cells. These results verify that ChIP–chip identifies Nup93-binding sites enriched at the nuclear periphery and is capable of distinguishing dynamic changes in localization that are influenced by TSA treatment.
Differential Nup93 binding was also observed at the CFTR locus on the q-arm of chromosome 7. Specifically, ChIP–chip showed that Nup93 was associated with the CFTR locus in untreated cells, but was not associated with the locus in TSA-treated cells (Fig. 2A). This result is consistent with previously reported visual evidence showing that CFTR is peripherally localized in mammalian cells and moves to the nuclear interior upon TSA treatment (Zink et al. 2004).
Comparison of Nup93 association in untreated and TSA-treated cells
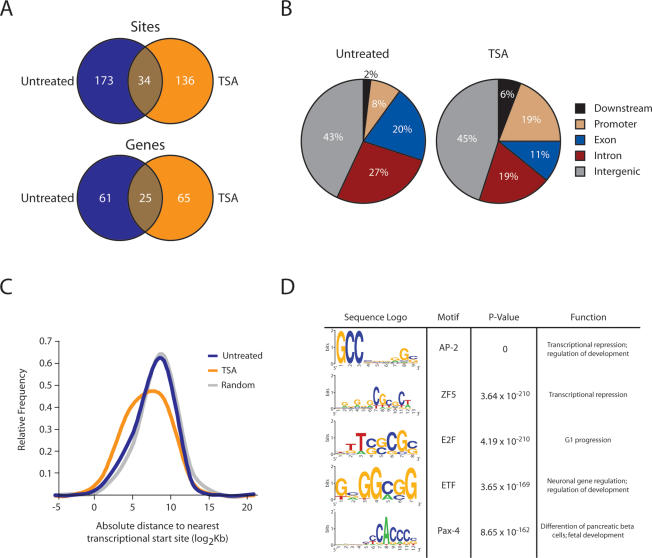
We investigated the magnitude of TSA-induced genomic reorganization and found that only 34 binding sites and 25 genes were shared between untreated and TSA-treated Nup93-binding sites, indicating a significant shift in peripheral association (Fig. 3A). Comparison of untreated and TSA-treated Nup93-binding sites with chip5 distributions reveals significant changes in association across five genomic regions: exons, introns, promoters, downstream elements, and intergenic regions (P = 0.001; χ2). Untreated binding sites are random relative to all categories except for introns when compared with chip5 (P = 0.002). However, when compared with chip5 distributions, TSA-binding sites are significantly enriched for introns and promoters (P = 0.005 and P = 1.2 × 10−14, respectively) and depleted for exons and intergenic regions (P = 5.7 × 10−4 and P = 5.4 × 10−5, respectively) (Fig. 3B). In agreement with this observation, we mapped the distance from the center of all untreated and TSA-treated Nup93-binding sites to the nearest transcription start site (Fig. 3C). Untreated binding sites show a slight bias toward the nearest transcription start site (P = 0.002), while TSA-binding sites are significantly closer to their nearest transcription start site (P = 6.8 × 10−6). Additionally, TSA-binding sites are significantly closer to the nearest transcription start site compared with untreated sites (P = 0.02).
Figure 3.
Nup93 association with the genome is significantly altered upon loss of histone deacetylation. (A) Venn diagrams showing the overlap between untreated and TSA-treated Nup93-binding sites and nearest genes. (B) Comparison of untreated and TSA-treated Nup93-binding sites with chip5 distributions reveals significant changes in association across five genomic regions: exons, introns, promoters, downstream elements, and intergenic regions (P = 0.001; χ2). Untreated sites are enriched for introns when compared with chip5 (P = 0.002). TSA-binding sites are significantly enriched for introns and promoters (P = 0.005 and P = 1.2 × 10−14, respectively) and depleted for exons and intergenic regions (P = 5.7 × 10−4 and P = 5.4 × 10−5, respectively) when compared with chip5 distributions. (C) Distribution of distances from the center of each untreated Nup93-binding site, TSA-treated Nup93-binding site, and random site to the nearest transcription start site. P-values for C were calculated by comparing untreated and TSA-treated Nup93-binding sites to randomly distributed sites using the two-tailed Mann-Whitney U-test. (D) Sequence logos for the five most significantly enriched transcription factor-binding motifs identified in TSA-treated Nup93-binding sites. One-hundred-six motifs were identified in TSA-treated binding sites by CEAS (Supplemental Table 3).
We identified several enriched transcription factor-binding motifs in our TSA-treated data set by analyzing our sites with CEAS (Cis-regulatory Element Annotation System, http://ceas.cbi.pku.edu.cn), a Web-based ChIP–chip annotation system (Ji et al. 2006). TSA-treated Nup93-binding sites contained 106 enriched transcription factor-binding motifs; the top five are shown in Figure 3D with a complete list included in Supplemental Table 3. However, we identified only one enriched transcription factor-binding motif in untreated Nup93-binding sites, E2F (P = 8.71 × 10−7). Importantly, multiple iterations of randomly generated data sets failed to identify any significant motifs using CEAS. The increase in the number of transcription factor-binding motifs in the TSA-treated data set likely reflects the shift in Nup93 binding to promoter regions and transcription start sites. The identified motifs are recognized by a wide array of transcription factors whose diverse functions include transcriptional repression, transcriptional activation, G1 progression, and neuronal development.
Nup93 binding is proximal to differentially expressed genes
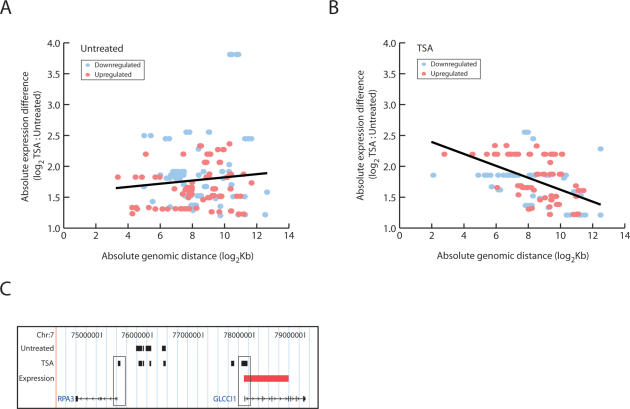
We performed RNA expression analysis that revealed patterns of expression upon TSA treatment: 753 genes were induced and 828 were repressed (Supplemental Table 4). Of these genes, 91 mapped to chromosome 5 (34 induced, 57 repressed), 70 to chromosome 7 (33 induced, 37 repressed), and 29 to chromosome 16 (17 induced, 12 repressed). However, there was no obvious overlap between Nup93-binding sites and transcriptionally induced or repressed genes. To probe the relationship further, we mapped all untreated and TSA-treated Nup93-binding sites to the nearest differentially expressed gene (induced and repressed). While no distance-to-gene relationship was uncovered for untreated Nup93-binding sites (Fig. 4A), we did uncover a strong correlation in TSA-treated Nup93-binding sites (Fig. 4B). One example of the proximal positioning of Nup93-binding sites to differentially expressed genes following TSA treatment is shown in Figure 4C. In untreated cells, Nup93-binding sites were identified in the intergenic region between two genes, RPA3 and GLCCI1, on chromosome 7. In TSA-treated cells, Nup93 sites were also found in both promoter regions, consistent with the promoter enrichment discussed earlier (Fig. 3B). Interestingly, while GLCCI1 expression was induced following TSA treatment, RPA3 expression was unaffected. Observations such as these show that differentially expressed genes may localize to the nuclear periphery, but that proximal positioning to the NPC is not always coupled with a transcriptional change.
Figure 4.
Nup93-binding sites are proximal to differentially expressed genes following TSA treatment. Untreated and TSA-treated Nup93-binding sites were mapped to the nearest differentially expressed gene. The log2-transformed distance-to-gene value (X-axis) was plotted against the magnitude of differential expression (Y-axis). Spearman rank order correlation was used to assess statistical significance. Down-regulated genes (blue) and up-regulated genes (red) are shown in each plot. The black trend lines were calculated by least squares best fit. (A) There was no correlation between distance-to-gene and magnitude of differential expression for untreated Nup93-binding sites (P = 0.0241; P = 0.7198). (B) Significant correlation was observed when comparing distance-to-gene and magnitude of differential expression for TSA-treated Nup93-binding sites (P = 0.5056; P = 6.28 × 10−11). (C) Nup93-binding sites near two genes on chromosome 7, RPA3 and GLCCI1. Untreated and TSA-treated sites overlap in the intergenic space while TSA treatment induces Nup93 binding to the promoter region of both genes, denoted by thin black rectangles. GLCCI1 expression is induced upon TSA treatment, illustrated by the red rectangle in the Expression track.
Nup93-binding sites are enriched for specific histone methylations
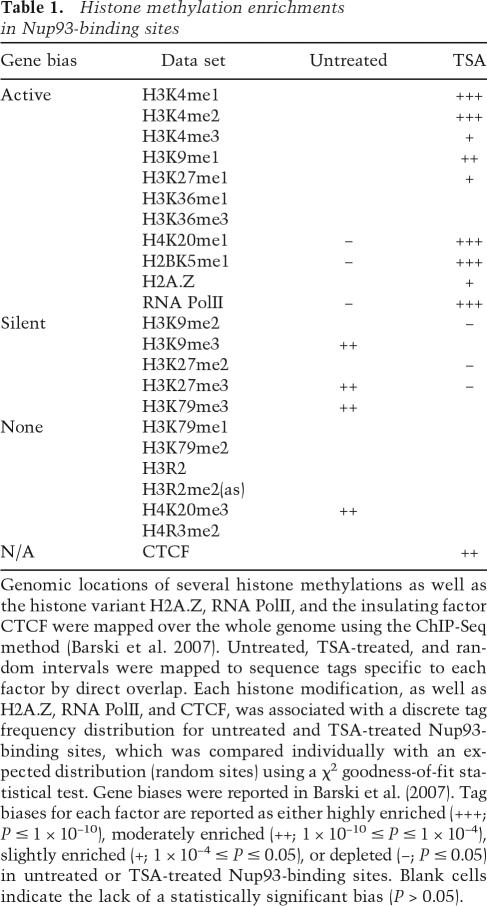
Untreated and TSA-treated Nup93-binding sites were enriched for histone methylations associated with transcriptionally silent and active regions, respectively (Table 1). A recent study mapped 20 unique histone methylations, the histone variant H2A.Z, the insulating factor CTCF, and RNA Polymerase II (PolII) over the whole human genome (Barski et al. 2007). We compared the reported distributions of histone methylations from Barski et al. (2007) with our untreated and TSA-treated Nup93-binding sites and found several significant overlaps (Table 1). Nine of the histone modifications are associated with active genes, five with silent genes, and six showed no bias. Untreated Nup93 sites were enriched for three specific histone modifications associated with silent genes, depleted for two modifications associated with active genes, and depleted for RNA PolII-binding sites. The opposite was found with TSA-treated Nup93 sites, which were enriched for seven histone modifications associated with active genes, depleted for three modifications associated with silent genes, and enriched in RNA PolII-, H2A.Z-, and CTCF-binding sites. These results indicate that regions associated with increased transcriptional activity become associated with Nup93 upon treatment with TSA.
Table 1.
Histone methylation enrichments in Nup93-binding sites
Genomic locations of several histone methylations as well as the histone variant H2A.Z, RNA PolII, and the insulating factor CTCF were mapped over the whole genome using the ChIP-Seq method (Barski et al. 2007). Untreated, TSA-treated, and random intervals were mapped to sequence tags specific to each factor by direct overlap. Each histone modification, as well as H2A.Z, RNA PolII, and CTCF, was associated with a discrete tag frequency distribution for untreated and TSA-treated Nup93-binding sites, which was compared individually with an expected distribution (random sites) using a χ2 goodness-of-fit statistical test. Gene biases were reported in Barski et al. (2007). Tag biases for each factor are reported as either highly enriched (+++; P ≤ 1 × 10−10), moderately enriched (++; 1 × 10−10 ≤ P ≤ 1 × 10−4), slightly enriched (+; 1 × 10−4 ≤ P ≤ 0.05), or depleted (−; P ≤ 0.05) in untreated or TSA-treated Nup93-binding sites. Blank cells indicate the lack of a statistically significant bias (P > 0.05).
Nup93 association with Giemsa (G) bands
To investigate chromosome-wide Nup93-binding patterns we compared the distribution of Nup93-binding sites with the G bands of chromosomes 5, 7, and 16. Staining with G dye reveals chromosome-wide banding patterns that are divided into five classes based on their staining intensity (Furey and Haussler 2003). The G-negative (Gneg) class consists of nonstaining bands, while the Gpos25, Gpos50, and Gpos75 classes contain progressively darker G-positive bands. The Gpos100 class consists of the darkest staining bands. Areas of heavy G staining represent condensed regions of the genome that have been shown to be gene poor, transcriptionally inactive, AT-rich, and replicated late in S phase. Of all the classes of G bands, Gneg bands have the highest density of genes, the largest average number of CpG islands, and are considered to represent euchromatin.
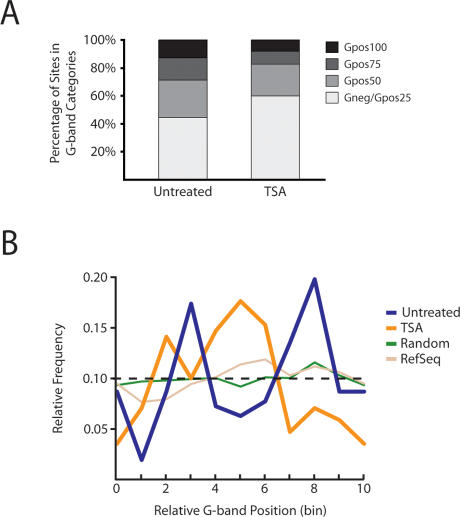
We identified nonrandom distributions of Nup93 across G bands in both untreated and TSA-treated data sets when compared with chip5 distributions (Fig. 5A). Untreated binding sites were enriched in Gpos50 bands (27% compared with 15% on chip5), while depleted in Gneg/Gpos25 bands (44% compared with 52% on chip5), suggesting a bias toward heterochromatin (P < 1.4 × 10−32; χ2). Conversely, TSA-induced binding sites were enriched in Gneg/Gpos25 bands (60% compared with 52% on chip5), while showing significant depletion in both Gpos75 and Gpos100 bands (9% and 8% compared with 16% and 13% on chip5, respectively), indicating a shift to regions of the genome enriched in euchromatin (P < 4.6 × 10−33; χ2) (Fig. 5A).
Figure 5.
Distribution of Nup93-binding sites within G bands. (A) Nup93 is enriched within Gpos50 bands and depleted in Gneg/Gpos25 bands in untreated conditions when compared with chip5 (P = 1.4 × 10−32; χ2). However, a similar comparison with chip5 distributions shows that TSA-treated Nup93-binding sites are enriched within Gneg/Gpos25 bands and depleted within Gpos75 and Gpos100 bands (P = 4.6 × 10−33; χ2). P-values were calculated by comparing the observed G-band distribution to chip5 G-band distributions using the χ2 test. (B) Binding distribution across G bands. All G bands were normalized by dividing each into 10 equal sections (bins). We assigned each Nup93 site a bin (0–9) based on its relative position between its nearest G-band boundaries. Each bin shown represents the relative position of several Nup93-binding sites (between 10 and 40 sites per bin). Intervals from randomized data sets were uniformly distributed across G bands, which reflected the probe distribution of chip5. Refseq genes failed to show significant positional enrichments (P = 0.97; χ2). However, both untreated and TSA-treated Nup93-binding sites were enriched in specific regions as shown by the peaks in blue and orange, respectively (P = 3.1 × 10−5 and P = 2.1 × 10−6, respectively; χ2). Furthermore, untreated and TSA-treated sites were positioned nonrandomly relative to each other, with significant TSA enrichment between two peaks of untreated enrichment (P = 5.4 × 10−23; χ2).
Next, we investigated the distribution of Nup93-binding sites within G bands. All G bands on chromosomes 5, 7, and 16 were normalized to the same length and then partitioned into 10 sections (bins). We assigned each Nup93 site a bin (0–9) based on its relative position between its nearest G-band boundaries. For example, a Nup93-binding site at the extreme 5′ G-band boundary would be placed in bin 0, while a binding site at the extreme 3′ G-band boundary would be placed in bin 9. As a result, each bin shown in Figure 5B represents the relative position of several Nup93-binding sites (between 10 and 40 sites per bin). A randomized data set failed to produce any enrichment across the normalized G bands, as did mapping of RefSeq genes (Fig. 5B). However, Nup93 had distinct enrichment patterns across G bands in both untreated and TSA-treated data sets, with two very strong peaks of enrichment in the untreated data set flanking a large region of enrichment found solely in the TSA-treated data set (Fig. 5B). These observations are indicative of a nuclear reorganization event induced by the inhibition of histone deacetylation.
Discussion
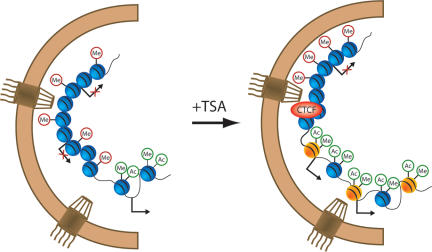
Our results show that the mammalian nuclear pore interacts with the human genome. To probe the functionality of this association we increased global levels of histone acetylation by treating cells with the HDAC inhibitor, TSA. Upon drug treatment, interactions between the nuclear pore and chromatin changed significantly. We observed changes in localized regions of transcriptional importance in addition to chromosome-wide changes in NPC association (Fig. 6).
Figure 6.
Model summarizing the factors involved in Nup93–genomic interactions. Untreated Nup93-binding sites contain a mix of active and inactive genes at the nuclear periphery, with enrichment for silent histone methylation marks (“Me” inside red circles). Following TSA treatment, Nup93-binding sites are enriched in several factors including the insulator protein CTCF, the histone variant H2A.Z (yellow spheres), and active histone methylation marks (“Me” inside green circles). Histone acetylation (“Ac” inside green circles) was enriched at the nuclear periphery following TSA treatment. One of the two NPCs on the right is proximal to a transcription start site, representing the promoter-binding and transcription factor-binding motif enrichment observed in Nup93-binding sites upon TSA treatment.
Local chromatin changes alter NPC association
TSA treatment induces the global hyperacetylation of histones by reversibly inhibiting Class I and II HDACs. The resulting histone acetylation, which is enriched at the nuclear periphery in TSA-treated cells, decreases chromosome condensation through the recruitment of chromatin remodeling factors such as the SWI/SNF complex (Agalioti et al. 2002). Several other proteins, including CBP and the general transcription initiation factor, TFIID, are also recruited to histones through interactions with acetyl-lysine residues (Agalioti et al. 2002; Fukuda et al. 2006). Acetylated histones are typically enriched in the promoter regions of active genes and we found that Nup93 was highly enriched in these regions following TSA treatment. In addition, we identified >100 transcription factor-binding motifs in the TSA-treated Nup93-binding sites, a finding indicative of the functional repositioning of chromatin. Further evidence for a role of the mammalian nuclear transport machinery in transcriptional regulation comes from the transport protein CAS, the mammalian homolog of the yeast exportin, Cse1. A recent study has shown that CAS associates with p53 at the promoter regions of several proapoptotic genes and contributes to their transcriptional activation (Tanaka et al. 2007).
Histone methylation also plays an important role in transcriptional regulation. Several H3K4 methylations are associated with actively expressed genes, while the H3K27me3 modification is associated with silent genes (Barski et al. 2007). Accordingly, Nup93-binding sites in TSA-treated cells are significantly enriched in H3K4me1–3 and depleted in H3K27me3 (Table 1). The opposite histone methylation biases are observed for untreated Nup93-binding sites, which are random with respect to H3K4me1–3 and enriched in H3K27me3.
Transcriptional regulation at the NPC
TSA-treated Nup93-binding sites were enriched in promoter regions and were closer to transcription start sites than their untreated counterparts. We also show that following TSA treatment, Nup93-binding sites are enriched in RNA PolII-associated regions and are closer to genes exhibiting differential expression. These regions include genes that are both highly expressed and strongly repressed following drug treatment. The presence of both active and repressed genes at nuclear pores has been observed previously in S. cerevisiae (Casolari et al. 2004). The role of the NPC as a boundary element may help explain these observations (Ishii et al. 2002). Interestingly, TSA-treated Nup93-binding sites are enriched in CTCF-associated regions. CTCF, a protein that recognizes cis-acting transcriptional insulator elements and marks histone modification boundaries (Splinter et al. 2006; Barski et al. 2007), copurifies with the histone variant H2A.Z, as well as two components of the nuclear envelope, lamin A/C and importin α3/α1 (Yusufzai et al. 2004). These associations suggest that CTCF interacts with the nuclear periphery and that regions of CTCF-mediated regulation are repositioned proximal to nuclear pores following TSA treatment.
The balance of HAT and HDAC activity is essential for the maintenance of boundaries between active and repressed chromatin (Kimura et al. 2002; Suka et al. 2002). Indeed, boundary elements recruit both HATs and HDACs in order to prevent spreading of silent and active chromatin, respectively (West et al. 2004; Yusufzai et al. 2004). Mutations in HATs and HDACs can significantly shift the borders of functional regions in the genome. For example, deletion of a subunit of the Rpd3 HDAC complex in yeast leads to aberrant transcriptional initiation (Carrozza et al. 2005). The mammalian nuclear periphery contains at least one HAT and HDAC, CBP and HDAC3, respectively. CBP interacts with the NPC, while HDAC3 associates with the lamin-binding protein LAP2β. These proteins, along with CTCF, could be important components of boundary elements at the mammalian nuclear periphery.
Evidence from work in S. cerevisiae has shown that recently activated genes remain at the nuclear periphery for hours after they have been repressed (Brickner et al. 2007). This localization differs from their preactivated positioning, which appears to be random with respect to the nuclear periphery (Drubin et al. 2006). This novel form of transcriptional memory is functional; it was shown that the recently repressed GAL1 gene was activated faster than a long-term repressed form. In yeast, Nup2p and the histone variant H2A.Z were involved in this process. Interestingly, we found that TSA-treated Nup93-binding sites were enriched in H2A.Z-binding sites. The presence of H2A.Z at sites of NPC–chromatin interaction could mean a similar form of transcriptional memory is present in human nuclei. Alternatively, H2A.Z site enrichment could reflect the role of this histone variant in peripherally localized boundary activity as H2A.Z has been shown to copurify with CTCF in HeLa cells and block the spread of telomeric heterochromatin in yeast (Meneghini et al. 2003; Yusufzai et al. 2004).
Chromosome-wide changes in nuclear organization
Large-scale changes in NPC–chromosome interactions were observed after treatment with TSA. Stretches of chromosomes 5 and 7 were no longer associated, while chromosome 16 retained the majority of its associated regions (Fig. 2A). These changes appear to be part of an extensive nuclear reorganization event in response to the global increase in histone acetylation.
The NPC has been implicated in boundary activity in S. cerevisiae and chromosome looping is thought to occur at the nuclear periphery in Drosophila (Gerasimova et al. 2000; Ishii et al. 2002). In addition, evidence has emerged that two distally localized genes on murine chromosome 7 can be localized to the same nuclear compartment upon activation (Osborne et al. 2004). It is possible that CTCF-mediated insulator activity near mammalian NPCs could generate similar loops that functionally isolate regions of activation or repression at the nuclear periphery.
Chromosome G-band patterns represent regions of genomic enrichment in either heterochromatin or euchromatin. We show that Nup93-binding sites are more likely found in heterochomatin in untreated cells, while favoring euchromatin following TSA treatment. The significant and opposing histone modifications and G-band enrichments found in Nup93-binding sites reflect a global reorganization event that exchanges silent for active chromatin at NPCs.
In summary, we show that the human genome interacts with Nup93, a component of the mammalian nuclear pore. Nup93 is associated with regions of transcriptional repression and enriched heterochromatin content in HeLa cells. Upon global histone acetylation mediated by the HDAC inhibitor, TSA, Nup93 associates with regions important for transcriptional regulation. Our results hold significance for both the general mechanisms of gene expression in mammalian cells and the global effects of histone modifications on nuclear organization.
Materials and methods
Cell culture and drug treatment
HeLa S3 cells were grown in DMEM supplemented with 10% FBS. Exponentially growing cells were incubated in the presence of 40 ng/mL TSA (Sigma) for 12 h. Cells were grown to near confluency before fixation and harvesting for ChIP–chip, indirect immunofluorescence, FISH, and expression profiling.
Immunofluorescence
HeLa S3 cells were grown to near confluency and adhered to polylysine-coated coverslips. Cells were prepared for labeling as described previously (Brown et al. 1996). Primary antibodies used include anti-lamin B diluted 1:50 (Santa Cruz Biotechnology), anti-Nup93 diluted 1:200 (D. Forbes), anti-acetylated H4K5 diluted 1:100 (Abcam), and anti-acetylated H3K9 diluted 1:100 (Upstate Biotechnology). Other antibodies used for preliminary ENCODE studies include anti-Nup62 (BD Transduction Laboratories), anti-Nup107, and anti-Nup205 (generous gifts from V. Cordes). The secondary antibody used to detect polyclonal primaries was goat anti-rabbit Alexa594 diluted 1:1000 (Molecular Probes). Images were acquired on a Nikon E800 epifluorescence microscope equipped with a Radiance 2000 confocal laser scanning system (Bio-Rad).
FISH
Bacterial artificial chromosomes (BACs) were purified using a Large-Construct Kit (Qiagen). Digoxigenin-dUTP was incorporated into purified BACs using a DIG Nick Translation Kit (Roche). For each sample, 40–160 ng of DIG-dUTP-labeled probe were combined with 40 μg of unlabeled COT1 DNA (Roche) and 20 μg of salmon sperm DNA (Ambion), a portion of which was used for hybridization. Cells were prepared and probes hybridized as described previously (Solovei et al. 2002). The probe signal was detected using an antibody enhancer kit (Roche). Vectashield containing DAPI (Vector Laboratories) was added to the slides and the coverslips were sealed with nail polish. More than 200 cells were counted for each experimental condition. FISH signal was counted as peripheral if it either overlapped with or was immediately adjacent to the nuclear periphery signal.
ChIP–chip
ChIP was performed essentially as described (Brodsky et al. 2005) with the following differences. Chromatin was prepared from four independently grown batches of cells—two untreated and two treated with 40 ng/mL TSA for 12 h before fixation to generate replicates for each condition. Cleared chromatin was incubated overnight with polyclonal Nup93 antibody at 4°C. After incubation, a fresh batch of 50:50 protein A/G sepharose beads were added to the chromatin/antibody solution and incubated overnight. Beads were washed five times for 10 min each, followed by DNA elution overnight, proteinase K treatment, phenol extraction, and RNase treatment. Isolated DNA was amplified isothermally using random nonamers and Klenow polymerase (Invitrogen) for 2 h, yielding ∼4 μg of DNA per ChIP. DNA was prepared and hybridized to Affymetrix GeneChip Human Tiling 2.0R ChipE (chip5) arrays using fragmentation, hybridization, staining, and scanning procedures described previously (Kennedy et al. 2003). A sample of chromatin was set aside before immunoprecipitation and used to represent the input DNA.
Expression profiling
Total RNA was extracted with Trizol and purified with an RNeasy Mini Kit (Qiagen) with an added DNase step (RNase-free DNase; Qiagen). Fifteen micrograms of RNA from each sample were sent to the DFCI Microarray facility for labeling and hybridization to Affymetrix Human Genome U133 Plus2.0 expression arrays. Raw hybridization intensity (CEL) files from three biological replicates were processed using the RMA method (Bolstad et al. 2003; Irizarry et al. 2003a, b). Significant differentially expressed genes were considered those with average log2-transformed fold-difference ratios (TSA:Untreated) ≥1.2 or less than or equal to −1.2 and P < 0.01 (two-tailed Student’s t-test).
Analysis of genomic binding data
All genomic track data (sequence conservation scores, SNPs, Refseq genes, and Giemsa ideograms) were downloaded July 2006 from the University of Californai at Santa Cruz genome bioinformatics Web site (ftp://hgdownload.cse.ucsc.edu/goldenPath) for the May 2004 assembly. To maintain consistency in the track variables, these originally downloaded data were used for all subsequent analyses. Duplicate Nup93 ChIP samples were submitted to the Dana-Farber Cancer Institute Microarray Core facility for hybridization. Enrichments in ChIPs compared with input were calculated from raw intensity (CEL) files using a nonparametric statistical method implemented in Affymetrix Tiling Analysis Software (TAS) (Ghosh et al. 2006). Binding significances (expressed as probe P-values) were used to detect statistical dependencies between Nup93-binding enrichments and sequence conservation (see the Supplemental Material for a technical description of the method, including statistical significance assessment and false discovery).
Acknowledgments
We thank Alex Brodsky for help developing mammalian ChIP–chip; Beth Rasala for providing generous amounts of Nup93 antibody; Pamela Hollasch, Maura Berkeley, and Ed Fox at the Dana-Farber Cancer Institute’s Affymetrix Core Facility for microarray hybridization and scanning; Jennifer Waters and the Harvard Nikon Imaging Center at Harvard Medical School for help with confocal imaging; and J. Casolari, J. Hurt, and A. McKee for comments on the manuscript. This work was supported by a Ryan scholarship to C.R.B., a NIH training grant to C.J.K., and grants from the US National Institutes of Health to P.A.S. and D.J.F.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1632708.
References
- Agalioti T., Chen G., Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- Andrulis E.D., Neiman A.M., Zappulla D.C., Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beissbarth T., Speed T.P. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Boyle S., Gilchrist S., Bridger J.M., Mahy N.L., Ellis J.A., Bickmore W.A. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Brickner J.H., Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:E342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D.G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P.C., Widom J., Brickner J.H. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky A.S., Meyer C.A., Swinburne I.A., Hall G., Keenan B.J., Liu X.S., Fox E.A., Silver P.A. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 2005;6:R64. doi: 10.1186/gb-2005-6-8-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D., Lydon J., McLaughlin M., Stuart-Tilley A., Tyszkowski R., Alper S. Antigen retrieval in cryostat tissue sections and cultured cells by treatment with sodium dodecyl sulfate (SDS) Histochem. Cell Biol. 1996;105:261–267. doi: 10.1007/BF01463929. [DOI] [PubMed] [Google Scholar]
- Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.C., Hurt E.C., et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Carroll J.S., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll J.S., Meyer C.A., Song J., Li W., Geistlinger T.R., Eeckhoute J., Brodsky A.S., Keeton E.K., Fertuck K.C., Hall G.F., et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., Silver P.A. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Drubin D.A., Rando O.J., Silver P.A. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes & Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Cremer M., Dietzel S., Muller S., Solovei I., Fakan S. Chromosome territories—A functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Croft J.A., Bridger J.M., Boyle S., Perry P., Teague P., Bickmore W.A. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw J.M., Krutchinsky A.N., Zhang W., Chait B.T., Matunis M.J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G., Iglesias N., Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D.A., Garakani A.M., Silver P.A. Motion as a phenotype: The use of live-cell imaging and machine visual screening to characterize transcription-dependent chromosome dynamics. BMC Cell Biol. 2006;7:19. doi: 10.1186/1471-2121-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D.C., Noble C.O., Kirpotin D.B., Guo Z., Scott G.K., Benz C.C. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu. Rev. Pharmacol. Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- Faria A.M., Levay A., Wang Y., Kamphorst A.O., Rosa M.L., Nussenzveig D.R., Balkan W., Chook Y.M., Levy D.E., Fontoura B.M. The nucleoporin Nup96 is required for proper expression of interferon-regulated proteins and functions. Immunity. 2006;24:295–304. doi: 10.1016/j.immuni.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Feuerbach F., Galy V., Trelles-Sticken E., Fromont-Racine M., Jacquier A., Gilson E., Olivo-Marin J.C., Scherthan H., Nehrbass U. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 2002;4:214–221. doi: 10.1038/ncb756. [DOI] [PubMed] [Google Scholar]
- Fukuda H., Sano N., Muto S., Horikoshi M. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief Funct. Genomic Proteomic. 2006;5:190–208. doi: 10.1093/bfgp/ell032. [DOI] [PubMed] [Google Scholar]
- Furey T.S., Haussler D. Integration of the cytogenetic map with the draft human genome sequence. Hum. Mol. Genet. 2003;12:1037–1044. doi: 10.1093/hmg/ddg113. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I., Byrd K., Corces V.G. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Hirsch H.A., Sekinger E., Struhl K., Gingeras T.R. Rank-statistics based enrichment-site prediction algorithm developed for chromatin immunoprecipitation on chip experiments. BMC Bioinformatics. 2006;7:434. doi: 10.1186/1471-2105-7-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist S., Gilbert N., Perry P., Bickmore W.A. Nuclear organization of centromeric domains is not perturbed by inhibition of histone deacetylases. Chromosome Res. 2004;12:505–516. doi: 10.1023/B:CHRO.0000034892.64739.ff. [DOI] [PubMed] [Google Scholar]
- Glaser K.B. HDAC inhibitors: Clinical update and mechanism-based potential. Biochem. Pharmacol. 2007;74:659–671. doi: 10.1016/j.bcp.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Green D.M., Johnson C.P., Hagan H., Corbett A.H. The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc. Natl. Acad. Sci. 2003;100:1010–1015. doi: 10.1073/pnas.0336594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003a;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003b;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Ishii K., Arib G., Lin C., Van Houwe G., Laemmli U.K. Chromatin boundaries in budding yeast: The nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- Ji X., Li W., Song J., Wei L., Liu X.S. CEAS: Cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–W554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy G.C., Matsuzaki H., Dong S., Liu W.M., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., et al. Large-scale genotyping of complex DNA. Nat. Biotechnol. 2003;21:1233–1237. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Umehara T., Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- Krull S., Thyberg J., Bjorkroth B., Rackwitz H.R., Cordes V.C. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell. 2004;15:4261–4277. doi: 10.1091/mbc.E04-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshakova M.M., Krasnov A.N., Kopytova D.V., Shidlovskii Y.V., Nikolenko J.V., Nabirochkina E.N., Spehner D., Schultz P., Tora L., Georgieva S.G. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. Embo J. 2007;26:4956–4965. doi: 10.1038/sj.emboj.7601901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R., Kerr S.C., Harreman M.T., Apponi L.H., Fasken M.B., Ramineni S., Chaurasia S., Valentini S.R., Corbett A.H. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- Maillet L., Boscheron C., Gotta M., Marcand S., Gilson E., Gasser S.M. Evidence for silencing compartments within the yeast nucleus: A role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes & Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- Mansfeld J., Guttinger S., Hawryluk-Gara L.A., Pante N., Mall M., Galy V., Haselmann U., Muhlhausser P., Wozniak R.W., Mattaj I.W., et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell. 2006;22:93–103. doi: 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Marcand S., Buck S.W., Moretti P., Gilson E., Shore D. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes & Dev. 1996;10:1297–1309. doi: 10.1101/gad.10.11.1297. [DOI] [PubMed] [Google Scholar]
- Mendjan S., Taipale M., Kind J., Holz H., Gebhardt P., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Meneghini M.D., Wu M., Madhani H.D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Menon B.B., Sarma N.J., Pasula S., Deminoff S.J., Willis K.A., Barbara K.E., Andrews B., Santangelo G.M. Reverse recruitment: The Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. 2005;102:5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Parada L., Misteli T. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 2002;12:425–432. doi: 10.1016/s0962-8924(02)02351-6. [DOI] [PubMed] [Google Scholar]
- Parada L.A., McQueen P.G., Munson P.J., Misteli T. Conservation of relative chromosome positioning in normal and cancer cells. Curr. Biol. 2002;12:1692–1697. doi: 10.1016/s0960-9822(02)01166-1. [DOI] [PubMed] [Google Scholar]
- Parada L.A., McQueen P.G., Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004a;5:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada L.A., Sotiriou S., Misteli T. Spatial genome organization. Exp. Cell Res. 2004b;296:64–70. doi: 10.1016/j.yexcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Pickersgill H., Kalverda B., de Wit E., Talhout W., Fornerod M., van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Ragoczy T., Bender M.A., Telling A., Byron R., Groudine M. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes & Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes & Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S., Fischer T., Luo M.J., Antunez O., Brettschneider S., Lechner J., Perez-Ortin J.E., Reed R., Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C.M., Harries J.C., Kindle K.B., Collins H.M., Heery D.M. Functional interaction of CREB binding protein (CBP) with nuclear transport proteins and modulation by HDAC inhibitors. Cell Cycle. 2006;5:2146–2152. doi: 10.4161/cc.5.18.3207. [DOI] [PubMed] [Google Scholar]
- Santos A.P., Abranches R., Stoger E., Beven A., Viegas W., Shaw P.J. The architecture of interphase chromosomes and gene positioning are altered by changes in DNA methylation and histone acetylation. J. Cell Sci. 2002;115:4597–4605. doi: 10.1242/jcs.00160. [DOI] [PubMed] [Google Scholar]
- Sarma N.J., Haley T.M., Barbara K.E., Buford T.D., Willis K.A., Santangelo G.M. Glucose-responsive regulators of gene expression in Saccharomyces cerevisiae function at the nuclear periphery via a reverse recruitment mechanism. Genetics. 2007;175:1127–1135. doi: 10.1534/genetics.106.068932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., Laemmli U.K. Nup–PI: The nucleopore–promoter interaction of genes in yeast. Mol. Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Solovei I., Walter J., Cremer M., Habermann F., Schermelleh L., Cremer T. FISH on three-dimensionally preserved nuclei. In: Beatty B., et al., editors. FISH, a practical approach. Oxford University Press; Oxford, UK: 2002. pp. 119–157. [Google Scholar]
- Somech R., Shaklai S., Geller O., Amariglio N., Simon A.J., Rechavi G., Gal-Yam E.N. The nuclear-envelope protein and transcriptional repressor LAP2β interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci. 2005;118:4017–4025. doi: 10.1242/jcs.02521. [DOI] [PubMed] [Google Scholar]
- Splinter E., Heath H., Kooren J., Palstra R.J., Klous P., Grosveld F., Galjart N., de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes & Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen J.B., Zakian V.A. Internal tracts of telomeric DNA act as silencers in Saccharomyces cerevisiae. Genes & Dev. 1994;8:1411–1422. doi: 10.1101/gad.8.12.1411. [DOI] [PubMed] [Google Scholar]
- Suka N., Luo K., Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- Taddei A., Maison C., Roche D., Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 2001;3:114–120. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Hediger F., Kalck V., Cubizolles F., Schober H., Gasser S.M. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Tanabe H., Habermann F.A., Solovei I., Cremer M., Cremer T. Non-random radial arrangements of interphase chromosome territories: Evolutionary considerations and functional implications. Mutat. Res. 2002a;504:37–45. doi: 10.1016/s0027-5107(02)00077-5. [DOI] [PubMed] [Google Scholar]
- Tanabe H., Muller S., Neusser M., von Hase J., Calcagno E., Cremer M., Solovei I., Cremer C., Cremer T. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. 2002b;99:4424–4429. doi: 10.1073/pnas.072618599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Ohkubo S., Tatsuno I., Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Thompson J.S., Johnson L.M., Grunstein M. Specific repression of the yeast silent mating locus HMR by an adjacent telomere. Mol. Cell. Biol. 1994;14:446–455. doi: 10.1128/mcb.14.1.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M., Haeusler R.A., Good P.D., Engelke D.R. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.G., Huang S., Gaszner M., Litt M.D., Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Yusufzai T.M., Tagami H., Nakatani Y., Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zink D., Amaral M.D., Englmann A., Lang S., Clarke L.A., Rudolph C., Alt F., Luther K., Braz C., Sadoni N., et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]