Abstract
RUNX1/AML1 is required for the development of definitive hematopoiesis, and its activity is altered by mutations, deletions, and chromosome translocations in human acute leukemia. RUNX1 function can be regulated by post-translational modifications and protein–protein interactions. We show that RUNX1 is arginine-methylated in vivo by the arginine methyltransferase PRMT1, and that PRMT1 serves as a transcriptional coactivator for RUNX1 function. Using mass spectrometry, and a methyl-arginine-specific antibody, we identified two arginine residues (R206 and R210) within the region of RUNX1 that interact with the corepressor SIN3A and are methylated by PRMT1. PRMT1- dependent methylation of RUNX1 at these arginine residues abrogates its association with SIN3A, whereas shRNA against PRMT1 (or use of a methyltransferase inhibitor) enhances this association. We find arginine-methylated RUNX1 on the promoters of two bona fide RUNX1 target genes, CD41 and PU.1 and show that shRNA against PRMT1 or RUNX1 down-regulates their expression. These arginine methylation sites and the dynamic regulation of corepressor binding are lost in the leukemia-associated RUNX1–ETO fusion protein, which likely contributes to its dominant inhibitory activity.
[Keywords: CD41, PU.1, arginine methylation, myeloid differentiation, AML1 target genes]
The RUNX1/CBFβ transcriptional regulatory complex is required for the development of murine definitive hematopoiesis (Okuda et al. 1996; Wang et al. 1996). RUNX1 (also known as AML1, CBFA2, and PEBP2αB) is the DNA-binding component of this complex, and its function is often compromised by mutations or chromosomal translocations in acute leukemia. The RUNX1–ETO fusion transcription factor, which is generated by the t(8;21) in acute myeloid leukemia (AML), has aberrant transactivating properties and can function as a dominant inhibitor of RUNX1 function and of other transcription factors as well (Mao et al. 1999; Nimer and Moore 2004; Zhang et al. 2004). RUNX1–ETO can promote the self-renewal of human and murine hematopoietic stem cells, which may increase the chance of developing secondary mutations, leading ultimately to the development of acute leukemia (Higuchi et al. 2002; Mulloy et al. 2003).
The human RUNX1 gene has three different isoforms, RUNX1a (which encodes a theoretical 250-amino-acid protein), RUNX1b (which contains 453 amino acids), and RUNX1c (which contains 480 amino acids). All contain the Runt DNA-binding domain, and both RUNX1b and RUNX1c contain transcriptional activating domains. There is no apparent functional difference between RUNX1b and RUNX1c despite their alternative N termini. (See Supplemental Fig. S1 for a sequence alignment. In this paper, we used amino acid number according to RUNX1b.) RUNX1 can act as either a transcriptional activator or a repressor, depending on the cellular and promoter context. RUNX1 often activates transcription weakly, but its interactions with other transcription factors such as GATA-1, ETS-1, and C/EBPα enhance its activating properties (Wotton et al. 1994; Zhang et al. 1996; Elagib et al. 2003). To achieve transcriptional activation, RUNX1 recruits coactivator molecules, such as ALY, p300, YAP, and MOZ (Bruhn et al. 1997; Kitabayashi et al. 1998, 2001; Yagi et al. 1999), which generally bind to the C terminus of RUNX1. However, RUNX1 can specifically silence CD4 gene expression during mature T-cell differentiation (Taniuchi et al. 2002); its repression of the p21 gene promoter in NIH3T3 cells has also been reported (Lutterbach et al. 2000). Although two distinct repressor complexes bind to RUNX1, the corepressor SIN3A complex (Lutterbach et al. 2000), and the Groucho/TLE repressor complex (Levanon et al. 1998), the precise mechanism through which specific genes are silenced by RUNX1 is not well understood.
RUNX1 is modified by phosphorylation (e.g., by the serine/threonine kinase ERK2) (Tanaka et al. 1996) and by acetylation (e.g., by the coactivator acetyltransferase p300) (Yamaguchi et al. 2004). Both modifications enhance transcriptional activation by RUNX1, thereby influencing its role in hematopoiesis. Recently, interactions of RUNX1 with the lysine methyltransferase SUV39H1 have been reported (Chakraborty et al. 2003; Reed-Inderbitzin et al. 2006), and a role for this methyltransferase in gene repression by RUNX1 has been suggested.
The arginine methylation of nonhistone proteins is being increasingly identified. PRMT1 is an arginine methyltransferase that functions to monomethylate or asymmetrically dimethylate arginine residues. PRMT1 is found in both the cytoplasm and the nucleus (Herrmann et al. 2005), and it accounts for most of the Type I arginine methyltransferase activity in cells (Tang et al. 2000). It is well established that arginine methylation by PRMT1 serves as a general marker for active transcription (Huang et al. 2005). PRMT1 targets histone H4R3 for arginine methylation, promoting the subsequent p300- and CARM1-mediated acetylation and methylation of histone tails (An et al. 2004). It also modifies coactivator molecules such as PGC-1 (Teyssier et al. 2005) and NIP45 (Mowen et al. 2004), and it is recruited by YY1 (Rezai-Zadeh et al. 2003), by nuclear hormone receptors (Wang et al. 2001), and by p53 for transcriptional activation (An et al. 2004).
RUNX proteins contain several potential arginine methylation sites, which prompted us to examine whether PRMT1 can methylate RUNX1 and alter its effect on gene expression. We show that PRMT1 directly interacts with RUNX1, methylates RUNX1, and functions as a coactivator for RUNX1-dependent transcriptional activation; and that reduction in the level of PRMT1 (or RUNX1) lowers the expression of several true RUNX1 target genes. While several sites within RUNX1 appear to be methylated, methylation of arginine residues within an RTAMR region of the RUNX1, just C-terminal to the Runt DNA-binding domain, abrogates the binding of SIN3A, thereby promoting RUNX1 transcriptional activity. The RTAMR region is not present in RUNX1–ETO, which binds SIN3A in a methylation-insensitive manner. We find variable levels of PRMT1, RUNX1, and arginine-methylated RUNX1 proteins in different hematopoietic cell lineages, which further suggests that this post-translational modification is involved in fine-tuning RUNX1 transcriptional regulatory activity.
Results
PRMT1 associates with RUNX1 in vivo
RUNX1 is a weak activator of several promoter-driven reporter constructs (Uchida et al. 1997; Elagib et al. 2003), but we found that PRMT1 enhances the effects of RUNX1 on both the IL-3 promoter (data not shown) and the CD41 promoter (Supplemental Fig. S2). Both MTA (a nonspecific methyltransferase inhibitor) and shRNA that knock down PRMT1 levels (Rezai-Zadeh et al. 2003) abrogate promoter activation by RUNX1.
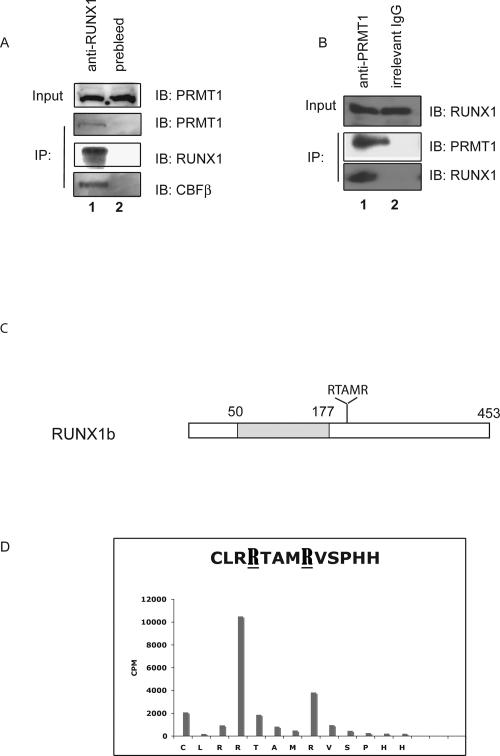
As these findings could reflect global effects of PRMT1 on transcription, we first examined whether PRMT1 physically interacts with RUNX1 in vivo. Using HEL cell extracts, we readily coimmunoprecipitated PRMT1 using an anti-RUNX1 antibody and immunoprecipitated RUNX1 protein using an anti-PRMT1 antibody, under stringent washing conditions (900 mM NaCl and 1% NP-40) (Fig. 1A,B). As PRMT1 is involved in RNA export (Yu et al. 2004) and can bind DNA via sequence-specific transcription factors (Rezai-Zadeh et al. 2003), we included RNase A and ethidium bromide in these immunoprecipitation reactions to exclude the possibility that its interactions with RUNX1 are mediated through RNA or DNA (Lai and Herr 1992). Neither treatment disrupted the RUNX1–PRMT1 immunoprecipitable complex, showing that PRMT1 and RUNX1 interact in a DNA- and RNA-independent manner.
Figure 1.
The endogenous RUNX1 and PRMT1 proteins physically interact in HEL cells. (A) An anti-RUNX1 antibody was used to immunoprecipitate PRMT1 from HEL cell extracts, using preimmune rabbit serum as control. As a positive control, CBFβ was detected among the anti-RUNX1 immunoprecipitated proteins. (B) An anti-PRMT1 murine antibody was used to immunoprecipitate RUNX1 from HEL cell extracts, using an irrelevant murine IgG antibody as negative control. (C) Schematic diagram of RUNX1b, showing the position of the Runt domain shaded and the RTAMR methylation site; the numbers on top correspond to the amino acid position. (D) Results of Edman degradation of an in vitro methylated synthetic peptide that contains amino acids 203–215 of RUNX1. Two sites in RUNX1 that are arginine-methylated by recombinant PRMT1 are identified.
To map the region in RUNX1 that interacts with PRMT1, we used various GST-RUNX1 fusion proteins (∼5 μg of protein per pull-down reaction) to pull down in vitro translated PRMT1 (Supplemental Fig. S3). Clearly, several different portions of the C terminus of RUNX1 can pull down PRMT1, which is similar to fibrillarin, a protein that interacts with PRMT1 through several regions (Yanagida et al. 2004). However, neither the Runt domain itself (GST-RUNX1 60–182) nor the N-terminal region of RUNX1 (GST-RUNX1 2–60) pulls down PRMT1. Thus, the binding of PRMT1 can occur via more than one domain within the RUNX1 C-terminal region. To further demonstrate the interaction of these proteins in vitro, we also used GST-PRMT1, which readily pulled down in vitro translated RUNX1 (Supplemental Fig. S3C).
Multiple sites in RUNX1 are arginine-methylated in vitro by PRMT1
PRMT1 does target nonhistone proteins for arginine methylation (Boisvert et al. 2003), so we determined if RUNX1 is arginine-methylated by PRMT1 in vitro and mapped the sites of methylation using mass spectrometry. By incubating in vitro translated RUNX1 with recombinant GST-PRMT1 and 3H-methyl-SAM, we readily detected arginine-methylated RUNX1 (using autoradiography) (data not shown). We also detected arginine methylation of RUNX1a at its C terminus (between amino acids 182 and 250) and within the Runt domain (within an “SGRGK” sequence that is also found at the N-terminal tail of histones H4 and H2A) (data not shown). We observed no methylation at the N terminus of RUNX1a (data not shown). Comparative mass spectrometric analysis of tryptic digests of the modified and unmodified RUNX1a protein identified these methylation sites. One m/z peak, at 1217.631 atomic mass units (amu), observed in the spectra of PRMT1-treated RUNX1 but not the untreated control, mapped to a predicted, monomethylated tryptic fragment of the sequence TAMRVSPHHPA (NCBI #557639) with a mass discrepancy of <12 ppm (0.015 Da) for the monoisotopic peak. This precursor ion was then selected for MALDI-TOF/TOF MS/MS analysis. The presence of unique fragment ions (b ions—originating at the N terminus) confirmed the identity of the peptide and allowed assignment of the methylation site to R210 in the published sequence (marked in bold and underlined in Fig. 1D), which is just C-terminal to the Runt domain, within a region shown to interact with the SIN3A repression complex (Lutterbach et al. 2000). This same region also interacts with PRMT1 (Supplemental Fig. S3). The R210 residue is present in RUNX1a, RUNX1b (R210), and RUNX1c (R237) (see Supplemental Fig. S1), but it is missing from RUNX1–ETO, which contains only 177 amino acids from RUNX1.
To further define arginine methylation within the C-terminal region of RUNX1, we performed in vitro methylation assays using a synthetic peptide that contains amino acids 203–215. The in vitro PRMT1-methylated peptide was sequenced using the Edman degradation method; not only was the arginine at position 210 (R210) methylated by PRMT1, but so was the arginine at position 206 (R206) (Fig. 1D), with the R206 site being the more dominant site within the small peptide. The R206 residue is present in RUNX1a, RUNX1b, and RUNX1c (position 233), but not in the RUNX2 or RUNX3 proteins.
RUNX1 is arginine-methylated in vivo
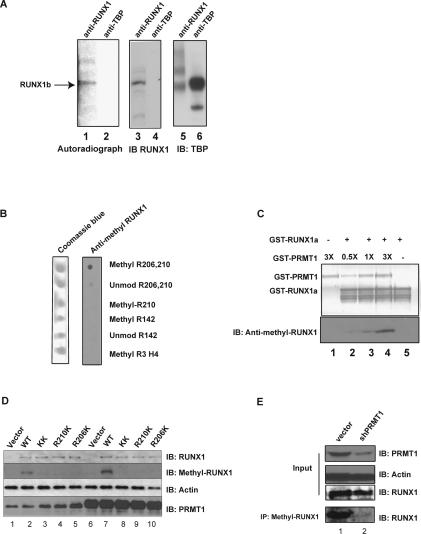
To determine whether RUNX1 exists as an arginine-methylated protein in vivo, we metabolically labeled several leukemia cell lines with [3H-methyl]-methionine in the presence of cycloheximide (to prevent methionine incorporation during protein synthesis). Using an anti-RUNX1 antibody to immunoprecipitate RUNX1 protein from the metabolically labeled cell extracts, we clearly detected 3H-methyl-RUNX1 in HEL cells (Fig. 2A, lane 1), and in Kasumi-1 and Meg 01 cells (data not shown). RUNX1 protein pulled down from the HEL cell extract by the anti-RUNX1 antibody runs at exactly the same position as radiolabeled RUNX1 (Fig. 2A, cf. lanes 3 and 1). The anti-TBP antibody pulled down neither methylated RUNX1 protein (Fig. 2A, lane 4) nor 3H-methyl-methionine labeled TBP (Fig. 2A, lane 2), demonstrating that the detected protein band is not due to de novo synthesis of RUNX1.
Figure 2.
RUNX1 is arginine-methylated in vivo. (A) HEL cells were treated with protein synthesis inhibitors before [3H-methyl]-methionine was added to the medium for labeling. The cell lysate was then immunoprecipitated with an anti-RUNX1 antibody (lanes 1,3) or an irrelevant (anti-TBP) antiserum (lanes 2,4). Methylated RUNX1 is shown in the left panel, and the middle and right panels are immunoprecipitation controls for the two antibodies used. (B) The anti-methyl-arginine-specific RUNX1 antibody efficiently recognizes the R206 and R210 diasymmetrically methylated peptide, but not the unmethylated peptide, the R210-only asymmetrically methylated peptide, the R142 asymmetrically methylated peptide, or the R3 asymmetrically methylated H4 histone tail peptide in a dot blot analysis. (C) The anti-methyl-arginine RUNX1 antibody recognizes recombinant RUNX1 only after in vitro methylation by recombinant GST-PRMT1. The top panel is the PVDF membrane stained for protein, and the bottom panel is a Western blot assay performed with the anti-methyl-arginine-RUNX1 antibody. (D) The wild-type RUNX1 protein, but not the R206K and R210K mutant proteins, is doubly arginine-methylated in HeLa cells, as detected using the anti-methyl-arginine RUNX1 antibody. Lane 1 is HeLa cells transfected with the empty pCDNA3 vector, whereas lanes 2–5 contain overexpressed wild-type RUNX1c or the various R-to-K RUNX1 mutants. Lane 6 contains HeLa cells that overexpress PRMT1 alone, and lanes 7–10 contain overexpressed RUNX1c and overexpressed PRMT1. The overexpression of PRMT1 leads to greater methylation of RUNX1 wild-type protein (cf. lanes 2 and 7), without changing the level of RUNX1 expression. Similarly, overexpression of RUNX1c did not change the level of PRMT1 (cf. lanes 1 and 2). (E) Knocking down PRMT1 in HEL cells with shRNA reduces the amount of arginine-methylated RUNX1 but not the total amount of RUNX1 protein. Lane 1 is the vector-integrated HEL cells. Lane 2 shows the decrease in PRMT1 and methyl RUNX1 in HEL cells that stably express PRMT1 shRNA. Actin levels serve as the loading control.
To demonstrate that RUNX1 is indeed arginine-methylated at R206 and R210 in vivo, we generated an arginine methylation-specific anti-RUNX1 antibody that recognizes these methylated arginine residues. We used a dot blot assay to characterize the affinity-purified R206/R210 (RTAMR) directed antibody, and by using several different peptides, showed that the antibody specifically recognizes the doubly dimethylated R206/R210 peptide (Fig. 2B). It does not recognize the unmodified peptide, a R210 singly dimethylated peptide, a histone H4 tail R3-methylated peptide, or peptides containing the SGRGK motif in the Runt domain region. Using recombinant RUNX1a protein, which was arginine-methylated by PRMT1 in vitro, we showed that the anti-methyl RUNX1 antibody indeed recognizes PRMT1-methylated RUNX1 protein (Fig. 2C).
We also used this antibody to confirm that RUNX1 is arginine-methylated in the RTAMR region in vivo, first by overexpressing wild-type and different arginine mutant forms of RUNX1 protein in HeLa cells. Again, the antibody recognizes the methylated form of wild-type RUNX1, but not the R206/R210 double mutant form or the single R206K or R210K mutant forms of RUNX1 (Fig. 2D, cf. lanes 2 and 3,4,5), confirming that RUNX1 is doubly methylated in vivo. PRMT1 is clearly the relevant methyltransferase in vivo, as the amount of methylated RUNX1 increases following the overexpression of PRMT1 (Fig. 2D, cf. lanes 7 and 2) and is reduced when PRMT1 levels are lowered by shRNA in HEL cells (Fig. 2E), and in HeLa cells (Fig. 3A, cf. lanes 2 and 5).
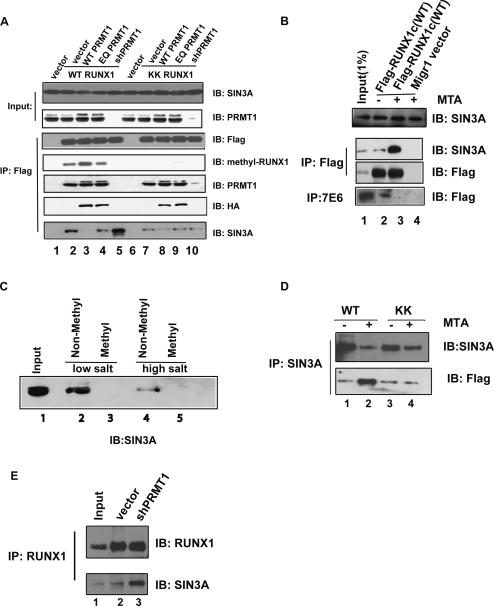
Figure 3.
PRMT1 regulates the association between RUNX1 and SIN3A by methylating RUNX1 at R206 and R210. (A) The association of SIN3A with wild-type RUNX1 but not the R206KR210K mutant is reciprocally affected by increasing or decreasing the level of PRMT1. Furthermore, PRMT1 enzymatic activity is required to decrease the association of RUNX1 with SIN3A. Anti-Flag antibody-coated agarose was used to immunoprecipitate Flag-tagged RUNX1 proteins from HeLa cells overexpressing wild-type or mutant forms of RUNX1 with wild-type PRMT1 (lanes 3,8), enzymatic dead PRMT1 (EQ) (lanes 4,9), or shRNA directed against PRMT1 (lanes 5,10). Equal loading in all lanes is shown by the SIN3A IB (in the top panel), the expression of RUNX1 protein is shown in the “IB: Flag” panel, the amount of RTAMR-methylated RUNX1 protein is shown in the “IB: methyl-RUNX1” panel, the level of PRMT1 in the anti-Flag immunoprecipitate is shown in the “IB: PRMT1” panel, the level of overexpressed PRMT1 in the anti-Flag immunoprecipitate is shown in the “IB: HA” panel, and the amount of SIN3A in the anti-Flag immunoprecipitate is shown in the bottom “IB: SIN3A” panel. (B) The methylation inhibitor MTA was added to HEL cells for 15 h, as indicated. More SIN3A is bound to Flag-RUNX1c (wild type) when MTA is present (cf. lanes 2 and 3 in A). (C) An arginine-methylated RUNX1 peptide does not pull down the SIN3A complex, whereas the unmethylated RUNX1 peptide does. (Lane 1, input) Ten percent nuclear extract, used for the peptide pulldown assay. Peptide-bound proteins were washed with 60 mM NaCl (lanes 2,3) or 100 mM NaCl (lanes 4,5). SIN3A protein is detected by an anti-SIN3A antibody. (D) In contrast, the association between SIN3A and the R206K R210K RUNX1 mutant protein (shown as KK in this figure) is not altered by MTA treatment. HEL cells expressing wild-type or mutant Flag-tagged RUNX1 were subjected to immunoprecipitation using an anti-SIN3A monoclonal antibody. The immunoprecipitated proteins were detected by anti-SIN3A and anti-Flag antibodies. (E) PRMT1 is primarily responsible for regulating the association of RUNX1 with SIN3A. Knocking down the level of PRMT1 by shRNA in a stably transfected HEL cell line increases the binding of RUNX1 with SIN3A. Input (lane 1) contains 10% of the amount of HEL cell extract used for the immunoprecipitation studies, whereas lanes 2 and 3 contain empty vector stably transduced and PRMT1-directed shRNA stably transduced cells, respectively. The immunoprecipitation was done using an anti-RUNX1 antibody; the coimmunoprecipitated protein was detected using the anti-SIN3A monoclonal antibody G11.
Treatment of HEL cells with MTA (a chemical that metabolically inhibits cellular [including PRMT catalyzed] methylation, at 1 mM) leads to substantially less methylated RUNX1 with little change in the amount of RUNX1 protein (Supplemental Fig. S4A,B). This effect is also observed using a generic methyl-arginine-specific antibody 7E6 (Fig. 3B). Similar results are obtained using Kasumi-1 cells (which also express RUNX1–ETO) (Supplemental Fig. S4C).
RUNX1 arginine methylation by PRMT1 regulates its association with SIN3A
It has been shown that SIN3A is recruited to RUNX1 through the RUNX1 amino acid 177–210 region (Lutterbach et al. 2000). Thus, the sites of in vivo arginine methylation in RUNX1 are within the SIN3A-interacting domain. To investigate whether methylation of the arginine residues in the SIN3A-interacting region of RUNX1 affects this interaction, we expressed the R-to-K mutant forms of RUNX1 and performed immunoprecipitation studies in HeLa cells. These studies show that both the wild-type and the mutant forms of RUNX1 bind to endogenous PRMT1 (Fig. 3A, lanes 2,7 next to bottom panel) and to SIN3A (Fig. 3A, lanes 2,7, bottom panel). However, while PRMT1 overexpression abrogates the binding of SIN3A to the wild-type RUNX1 protein (Fig. 3A, lane 3), it does not appreciably decrease SIN3A binding to the double R-to-K RUNX1 mutant (Fig. 3A, lane 8). Furthermore, knocking down PRMT1 expression using shRNA enhances the association of SIN3A with wild-type RUNX1 (Fig. 3A, lane 5) but does not change the association of SIN3A with the R-to-K mutant RUNX1 protein (Fig. 3A, lane 10). (Changing the amount of PRMT1 had no effect on the total amount of SIN3A or RUNX1 in the cell [Fig. 3A].) Since PRMT1 and SIN3A interact with RUNX1 through the same region, it is possible that PRMT1 and SIN3A compete for binding to RUNX1. To evaluate this possibility, we included the enzymatically dead form of PRMT1 (PRMT1 EQ, which interacts with RUNX1 as efficiently as does wild-type PRMT1 in the coimmunoprecipitation assay). (Note that the overexpressed PRMT1 proteins are HA-tagged and therefore run slightly larger than the endogenous PRMT1 in Fig. 3A; Western blot analysis using an anti-HA antibody shows that the overexpressed protein is detected as a single band.) SIN3A is still associated with RUNX1 in the presence of the overexpressed PRMT1 EQ mutant (Fig. 3A, lanes 4,9). Thus, arginine methylation of RUNX1 regulates its association with SIN3A, but the binding of PRMT1 to RUNX1 does not physically displace SIN3A.
To further demonstrate that methylation within this region directly disrupts the interaction between SIN3A and RUNX1, we performed a peptide pull-down assay. Methylated or nonmethylated synthetic peptides containing the RTAMR region of RUNX1 were used to pull down SIN3A from HEL cell nuclear extracts. While the nonmethylated peptide efficiently pulled down SIN3A, the methylated peptide did not (Fig. 3C). Thus, the short (14 amino acids) peptide that encompasses the RUNX1 methylation site contains the minimal region that can be recognized by the SIN3A complex.
To confirm that the in vivo association of endogenous RUNX1 and SIN3A is regulated by arginine methylation in leukemia cells, we treated HEL cells with 1 mM MTA (for 15 h). MTA treatment abrogates arginine methylation of RUNX1 (Fig. 3B, bottom; Supplemental Fig. S4A) and enhances the in vivo association of SIN3A with wild-type RUNX1, without changing the levels of SIN3A (Fig. 3B), RUNX1, or PRMT1 (Supplemental Fig. S6A). We also generated stable cell lines expressing Flag-tagged wild-type or mutant (R206K and R210K) RUNX1 protein and immunoprecipitated SIN3A in the presence and absence of MTA. MTA treatment increases the amount of wild-type RUNX1 that coimmunoprecipitates with SIN3A, but not the amount of R206K/R210K mutant RUNX1 protein (Fig. 3D). We also checked the association of RUNX1 and SIN3A in HEL cells with reduced PRMT1 expression. The association of RUNX1 with SIN3A is clearly enhanced by the shRNA directed against PRMT1 (Fig. 3E). Thus, we conclude that the methyltransferase activity of PRMT1 regulates the association of SIN3A with RUNX1 through the R206 and R210 sites.
The R206K and R210K RUNX1 arginine mutant proteins do not activate transcription efficiently nor cooperate with PRMT1
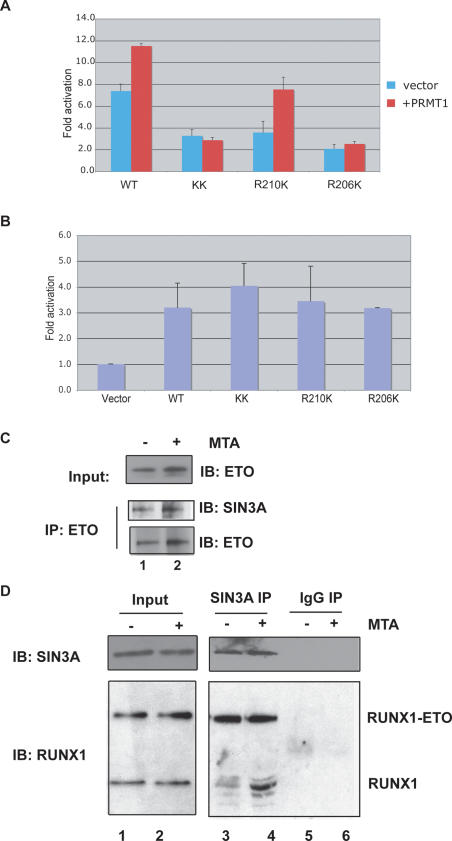
We mutated the arginine residues at positions R206 and R210 of RUNX1 to lysines, preserving their charge and best mimicking the side chain of arginine when it is not methylated, and then examined the transactivating properties of the mutant proteins. Despite being expressed similarly to the wild-type RUNX1 protein in HeLa cells (see Fig. 2D), all three RUNX1 mutants (R206K, R210K, and R206K/R210K) were less transcriptionally active on the IL-3 promoter (Fig. 4A, blue bars). Furthermore, while the activity of the wild-type RUNX1 protein and the R210K mutant was enhanced by coexpression of PRMT1 (Fig. 4A, red bars), neither the R206K mutant nor the double mutant R206K/R210K protein was more active when PRMT1was overexpressed. (The R206 site is the predominant site methylated by PRMT1 in vitro, and while the sequential order of methylation at these sites cannot be addressed by our experiments, perhaps R206 is the site that is first methylated in vivo, triggering partial dissociation of the SIN3A complex.)
Figure 4.
The in vivo association of RUNX1 with SIN3A (and presumably HDACs) regulates its transcriptional activity in HEL cells. (A) RUNX1 R-to-K mutants weakly activate the IL-3 promoter in HeLa cells compared with wild-type RUNX1, and the KK and R206K mutant forms do not respond to PRMT1 expression. The transfections were performed in the presence (in red) or absence (in blue) of PRMT1. (B) The addition of TSA restores transactivating properties to the RUNX1 R-to-K mutants on the IL-3 promoter. (C) The addition of MTA does not change the amount of SIN3A bound to ETO. Lanes 1 and 2 both contain HEL cell proteins immunoprecipitated with an anti-ETO antibody. (D) The addition of MTA for 12 h to Kasumi-1 cells enhances the binding of SIN3A to RUNX1, but has no effect on SIN3A binding to RUNX1–ETO. (Lanes 3,4) Following immunoprecipitation of SIN3A using an anti-SIN3A monoclonal antibody, the coimmunoprecipitated RUNX1 and RUNX1–ETO proteins were detected using an anti-RUNX1 antibody in a Western blot assay. (Lanes 5,6) Normal mouse IgG is used as the control.
The SIN3A complex recruits histone deacetylases (HDACs) to aid in gene repression and silencing, so to test whether the persistent interaction of the SIN3A complex with the R-to-K RUNX1 mutants is responsible for their reduced transcriptional activity, we examined whether the HDAC inhibitor trichostatin A (TSA) could rescue the transcriptional regulatory activity of the mutant proteins. In the presence of 100 ng/mL TSA, the mutants activated transcription of the IL-3 promoter plasmid to the same degree as wild-type RUNX1 (Fig. 4B). Thus, these mutants are not transcriptionally “dead,” although they may have lost the ability to shed corepressors and HDACs (or to recruit coactivators). Given its effects on the global level of histone acetylation, TSA did increase IL-3 promoter activity in the absence of RUNX1 or PRMT1 expression, as expected (data not shown).
Interaction of RUNX1–ETO with SIN3A is methylation-insensitive
It is known that ETO, the translocation partner of RUNX1 in the t(8;21), interacts with SIN3A (Gelmetti et al. 1998; Wang et al. 1998). After confirming this interaction (Fig. 4C), we examined whether it is also regulated by methylation. We did not find methylation-dependent regulation, as the anti-ETO antibody pulls down as much SIN3A from MTA-treated HEL cell extracts as from untreated HEL cell extracts. This led us to examine Kasumi-1 cells, a t(8;21) positive AML cell line that contains the RUNX1–ETO protein, RUNX1, and the RTAMR-methylated form of RUNX1 (Supplemental Fig. S4C). RUNX1–ETO lacks the RTAMR region (as shown in Supplemental Fig. S1) but contains nearly all of the ETO amino acid residues, and it functions as a potent repressor of RUNX1 function (Frank et al. 1995; Meyers et al. 1995; Yergeau et al. 1997). As predicted, MTA treatment of Kasumi-1 cells did not affect the amount of RUNX1–ETO associated with SIN3A, even though the amount of RUNX1 associated with SIN3A was increased several-fold (Fig. 4D, cf. lanes 3 and 4). Thus, the dynamic interaction between RUNX1 and SIN3A, which is regulated by arginine methylation at its C terminus (in the RTAMR site), is lost in the RUNX1–ETO fusion protein.
PRMT1 and RUNX1 regulate transcription of the CD41 gene in primary human CD34-positive hematopoietic cells
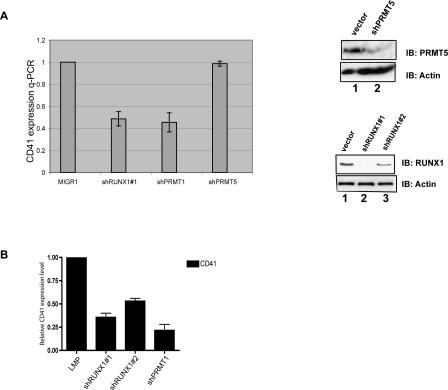
CD41 is an early marker of definitive hematopoiesis (Emambokus and Frampton 2003); it is also expressed in megakaryocytic cells and in primitive, multipotent progenitor cells (Mikkola et al. 2003). The CD41 promoter has been reported to be regulated by RUNX1 (Elagib et al. 2003), which we confirmed (Supplemental Fig. S2). We also determined that PRMT1 functions as a dose-dependent coactivator for RUNX1 on this promoter, using both PRMT1 overexpression and shRNA driven PRMT1 knockdown (data not shown). We find that CD41 is, indeed, a “PRMT1-sensitive” RUNX1 target gene, as expression of the CD41 endogenous gene is lowered 30%–50% in both primary human CD34+ cord blood cells (Fig. 5B) and in HEL cells (Fig. 5A) by reducing RUNX1 levels or PRMT1 (but not PRMT5) levels in the cell (as detected by real-time PCR) (Fig. 5A,B; Supplemental Fig. S5A). Furthermore, treating HEL cells with MTA significantly down-regulates CD41 expression (Supplemental Fig. S5B) without changing PRMT1 or RUNX1 levels, further implicating an endogenous methyltransferase activity (primarily PRMT1) in the in vivo regulation of CD41 gene expression by RUNX1.
Figure 5.
Expression of CD41, a direct target gene of RUNX1, is regulated by RUNX1 and PRMT1 in primary cells. (A) Knocking down RUNX1 or PRMT1 levels (but not PRMT5 levels) using shRNA reduces CD41 expression in HEL cells, as shown by real-time PCR. The ability of the PRMT5- and RUNX1-directed shRNAs to lower the level of PRMT5 and RUNX1 protein is shown on the right. (B) CD41 expression is reduced in CD34+ cord blood cells grown in early cytokine mix by knocking down RUNX1 or PRMT1 using shRNA-expressing retroviral vectors.
Manipulation of PRMT1 levels by shRNA allows RUNX1 to repress CD41 gene expression
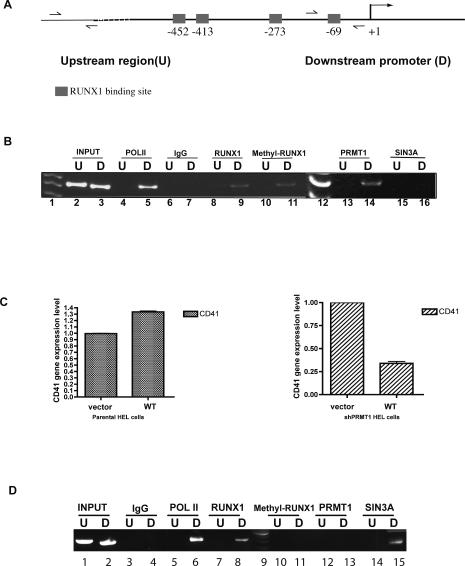
We used the RUNX1 RTAMR methyl-arginine-specific antibody in a chromatin immunoprecipitation (ChIP) assay to determine whether methylated RUNX1 is located on CD41 promoter sequences. RUNX1 and PRMT1 are found on the active CD41 promoter (and not on the upstream CD41 gene sequence), as is the RTAMR-methylated RUNX1 protein (see Fig. 6B, lane 11). Thus, the RTAMR arginine-methylated form of RUNX1 is found on an actively transcribed RUNX1-regulated target gene.
Figure 6.
Assessment of transcriptional regulators bound to the CD41 promoter. (A) A schematic diagram of the CD41 promoter is shown, indicating the upstream (U) and downstream (D) regions. (B) RUNX1 (lanes 8,9), PRMT1 (lanes 13,14), and methyl RUNX1 (lanes 10,11) are sitting on the actively transcribed CD41 promoter (∼70 bp upstream of the start site) in HEL cells, but not on an ∼5-kb upstream region of the gene (which we use as a control for the ChIP). An anti-Pol II antibody (lanes 4,5) served as a positive control and normal IgG (lanes 6,7) served as a negative control for the PCR reactions. Lanes 1 and 12 show the 50-bp DNA sizing ladder. Input lanes contain 0.01% of genomic input DNA. The PCR products were resolved using a 20% 1× TBE PAGE gel. (C) Comparison of the effects of overexpressing RUNX1 in HEL cells that stably express shPRMT1 (right panel) versus the parental HEL cells (left panel). RUNX1 slightly increases CD41 expression in HEL cells, but it reduces CD41 expression in cells with low PRMT1 levels. (D) ChIP assays show that in the shPRMT1-expressing HEL cells, overexpression of RUNX1 leads to the recruitment of SIN3A to the CD41 promoter (lane 15) but not PRMT1 or methyl-RUNX1 (lanes 11,13). (Lane 6) Pol II is present (and possibly stalled) on the CD41 promoter. Lane 9 is the DNA sizing ladder.
Theoretically, RUNX1 could function as a repressor of CD41 expression (by more strongly binding SIN3A) if PRMT1 levels were reduced. We examined this in HEL cells that we engineered to stably express shRNA against PRMT1 (these cells were also used in Fig. 5A). While RUNX1 modestly increased the endogenous level of CD41 expression in wild-type HEL cells (likely due to the high endogenous level of RUNX1), it decreased CD41 expression in the PRMT1 knockdown HEL cells (Fig. 6C, right panel). Examining the proteins bound to the CD41 promoter by ChIP analysis under these conditions showed that PRMT1 was no longer detected on the CD41 promoter in the knockdown cells; and although RUNX1 was detected on the CD41 promoter, arginine-methylated RUNX1 was not. However, SIN3A was now detected on the CD41 promoter (Fig. 6D). While not quantitative, these ChIP results are consistent with the role of PRMT1 in promoting RTAMR methylation of RUNX1, leading to its dissociation from SIN3A.
RUNX1 methylation is regulated during hematopoietic cell differentiation
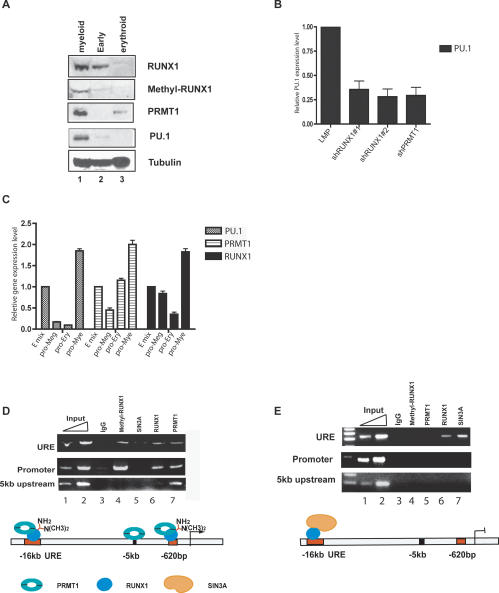
To determine if RUNX1 arginine methylation varies during normal hematopoietic cell differentiation, we first examined the levels of RUNX1-, PRMT1-, and RTAMR-methylated RUNX1 protein in human CD34+ cord blood cells grown for 4 d in one of four different cytokine cocktails (that either promote myeloid, erythroid, or megakaryocytic differentiation or that allow for retention of stem cell features [“early” cytokine mix]). PRMT1 is not detectable, and methyl RUNX1 is barely detectable in the cells grown in “early” mix, even though RUNX1 itself is present (Fig. 7A). With myeloid differentiation, RUNX1 increases, PRMT1 becomes detectable, and methyl RUNX1 protein is prominently seen. This coincides with the induction of PU.1 gene expression, an ETS protein that plays a key role in macrophage and B-cell development (Scott et al. 1994). In the erythroid mix, although PRMT1 is present, neither RUNX1 (nor, of course, methyl RUNX1) is detectable under these conditions, and PU.1 is not expressed (Fig. 7C).
Figure 7.
Regulation of RUNX1, PRMT1, and methyl RUNX1 levels during differentiation of human CD34+ cells in liquid culture. (A) Runx1 is arginine-methylated by PRMT1 during myeloid differentiation. CD34+ cells isolated from human cord blood cells were cultured under three different cytokine conditions: early (progenitor maintaining) mix, promyeloid differentiation mix, or proerythroid differentiation mix. PRMT1 protein is up-regulated under promyeloid and proerythroid differentiation conditions, RUNX1 and PU.1 protein are up-regulated with promyeloid differentiation mix and RUNX1 and PU.1 protein are both down-regulated in the proerythroid mix. (B) PU.1 expression is down-regulated in CD34+ cells grown in promyeloid mix following the reduction of RUNX1 or PRMT1 levels. (C) Real-time PCR results for the level of PRMT1, RUNX1, and PU.1 mRNA expression. (E) Early mix; (pro-Meg) megakaryocytic promoting mix; (pro-Ery) erythroid-promoting mix; (pro-Mye) myeloid-promoting mix. PU.1 expression is down-regulated in the pro-Ery and pro-Meg mix. RUNX1 is down-regulated in the pro-Ery mix. Both are up-regulated in the pro-Mye mix. (D) ChIP assays show that cells with higher PU.1 expression (grown in promyeloid mix) have both PRMT1 and methylated RUNX1 bound to the PU.1 promoter region and the URE. Only PRMT1 is bound to a region 5 kb upstream of the PU.1 transcriptional start site. (E) ChIP assays show that in the pro-Meg mix, SIN3A and RUNX1, but not PRMT1 or methyl RUNX1, are found on the URE. This accompanies the marked drop in PU.1 expression seen in these cells. Input lanes contain 0.01% and 0.05% of input genomic DNA used in panels D and E.
Recently, PU.1 has been shown to be directly regulated by RUNX1 (Huang et al. 2008). We confirmed that PU.1 expression level is down-regulated in cells grown under myeloid culture conditions by knocking down RUNX1 and PRMT1, respectively (Fig. 7B). Thus RUNX1 and PRMT1 also cooperate to regulate gene expression in primary cells. This implies that RUNX1 repressor function could be more prominent in the absence of PRMT1 in early stem/progenitor cells, while the induction of PRMT1 during myeloid commitment might promote its activity functions; proving this theory would require much additional experimentation.
To accurately assess the importance of PRMT1 and RUNX1 in the regulation of the PU.1 gene in primary cells, we used Western blot analysis, ChIP studies, and quantitative PCR. The expression of PU.1 mRNA is significantly higher in cells grown in the promyeloid cytokine mix than in the “early” mix (Fig. 7C), and RUNX1, PRMT1, and methylated RUNX1 are readily detected on the PU.1 promoter region (which has a consensus RUNX1-binding site at base pair −620) as well as on the PU.1 upstream response element (URE) region (located 16 kb upstream of the promoter) (Fig. 7D). SIN3A is not present on the PU.1 promoter and is barely detectable on the PU.1 URE. We do detect PRMT1 on a region 5 kb upstream of the PU.1 transcription start site (that lacks an RUNX1-binding site), suggesting that PRMT1 may play a broader role in organizing chromatin structure independent of RUNX1.
When the cells differentiate along the erythroid or megakaryocytic lineage, reduced PU.1 expression is seen (Fig. 7C). Neither methyl RUNX1 or PRMT1 is detectable on the PU.1 promoter under the pro-Mega culture conditions (Fig. 7E). While RUNX1 and SIN3A can still be found on the URE, neither is found on the PU.1 promoter. Clearly, the lineage-specific occupancy of RUNX1 target gene promoters by PRMT1 is consistent with its proposed role as a positive regulator of RUNX1 transactivating function.
Discussion
Protein arginine methylation by PRMTs positively or negatively regulates protein–protein interactions, thereby regulating a variety of cellular processes that impact on gene transcription (Bedford and Richard 2005). We now show that PRMT1 is an important modulator of RUNX1-inducible gene expression, regulating the interaction of RUNX1 with the SIN3A corepressor complex, and functioning as a coactivator for RUNX1 (an effect that requires its methyltransferase activity). RUNX1 plays a critical role in the development of definitive hematopoiesis during fetal life (Okuda et al. 1996) and it regulates adult hematopoiesis as well. It is altered in acute myeloid and lymphoid leukemias by mutations, deletions, and translocations.
PRMT1 methylates arginine residues within several regions of RUNX1, including two arginine methylation sites at the C terminus of RUNX1 (R206 and R210) that we first identified using proteomic analysis. These methylation sites lack the “GRG” PRMT consensus methylation sequence (Gary and Clarke 1998). However, many proteins (e.g., PGC-1 and Stat-1) appear to be arginine-methylated by PRMT1 through nonconsensus sites (Mowen et al. 2001; Teyssier et al. 2005), and mass spectrometry data indicate that proteins contain many more methyl-arginine sites than predicted based on consensus arginine methylation sequences (Ong et al. 2004). We generated a methyl-arginine-specific anti-RUNX1 antibody that recognizes the asymmetrically dimethylated R206 and R210 residues in RUNX1 and show that these residues are methylated in vivo and that R206/R210-methylated RUNX1 protein binds target gene promoter sequences using ChIP assays. By modulating the level of PRMT1 in cells, using shRNA or overexpression, we show that PRMT1 regulates the level of arginine-methylated RUNX1 in the cell, and on two RUNX1 target gene promoters, CD41 and PU.1.
The effect of methylation of both arginines within the RTAMR region of RUNX1 is disruption of the interaction between RUNX1 and SIN3A. The binding of PRMT1 by RUNX1 does not physically displace SIN3A, as loss of SIN3A binding requires that the RTAMR motif in RUNX1 be methylated. Furthermore, mutating the arginine residues in the RTAMR region of RUNX1 to lysines eliminates the methylation-dependent dynamic nature of the RUNX1–SIN3A interaction. The PAH3 domain of SIN3A has been shown to interact with the region of RUNX1 that contains the RTAMR residues (Lutterbach et al. 2000); these interactions are quite hydrophilic. While PRMT1 can bind RUNX1 in this region, it binds other regions of RUNX1 as well, providing another reason why we did not detect competition between PRMT1 and SIN3A for binding to RUNX1. The physical interaction between RUNX1 and PRMT1 is not affected by methylation in this region, which would allow RUNX1 to recruit PRMT1 to chromatin, where it can further promote transcriptional activation by methylating Arg 3 on H4 (an activating mark for transcription) (Wang et al. 2001; Huang et al. 2005), thereby providing a better substrate for histone acetylation by p300, and subsequently for CARM1- mediated histone H3 arginine methylation (An et al. 2004).
The SIN3A complex functions as a corepressor by recruiting HDACs. Thus, although the R206 and R210 mutant forms of RUNX1 bind SIN3A in the presence of high levels of PRMT1, HDAC inhibitors (like TSA) increase transactivation by the R206 and R210 RUNX1 mutants to the wild-type RUNX1 level. This suggests that the inability of the R-to-K RUNX1 mutants to shed the SIN3A corepressor complex accounts for their low transcriptional activating properties. We find fine-tuning of the activating function of RUNX1 by arginine methylation and provide some evidence that the repressor properties of RUNX1 could be regulated by PRMTs. However, the ability of RUNX1 to repress CD41 expression in cells where PRMT1 is knocked down only implies that arginine methylation of RUNX1 can regulate this switch.
PRMT1 and methyl RUNX1 levels appear to be similarly regulated during hematopoietic cell differentiation. It may be that some target genes are activatable by unmethylated RUNX1, but expression of a target gene like PU.1 may require binding of (RTAMR) methylated RUNX1 to its promoter (and URE) sequences in order to be fully expressed. We showed that PRMT1-dependent methylation can regulate the binding of SIN3A to a RUNX1 target gene promoter; whether this is solely due to effects on RUNX1 or could also relate to effects on other PRMT1 target proteins, cannot be easily determined. The effects of RUNX1 on PU.1 expression clearly differs depending on cell type (Huang et al. 2008). However, the pattern seen in the human cells mimics that seen during mouse cell differentiation. For instance, PU.1 levels decrease as RUNX1 increases during megakaryocytic cell differentiation in the mouse, and loss of RUNX1 in this circumstance leads to PU.1 up-regulation (Huang et al. 2008). We also find that PU.1 is repressed during the megakaryocytic differentiation of human cells and also find down-regulation of PRMT1. Under these circumstances, we find RUNX1 associated with the SIN3 complex on the PU.1 URE region. Thus, RUNX1 seems to participate in a URE repressor complex involved in repressing PU.1 gene expression, even though it may not be the sole transcription factor involved. While PRMT1 expression is regulated during the process of hematopoietic cell differentiation, the signaling that controls PRMT1 expression is not known. Control of PRMT activity is another level of regulation that also appears to be regulated, as NGF, IFN-α, LPS, and T-cell activation have all been shown to increase the level of protein arginine methylation in the cell (Mowen et al. 2004; Bedford and Richard 2005).
We examined whether knockdown of RUNX1 or PRMT1 alters the colony-forming ability of human CD34+ cells in methylcellulose cultures. The numbers of CFU-GM, BFU-E, and CFU-GEMM were not altered by 60%–70% knockdown of RUNX1 (data not shown). Thus it may not be surprising that PRMT1 knockdown also did not affect CFU numbers. Given the need for evaluating methylation at the RTAMR site under circumstances in which RUNX1 activity is required (e.g., fetal liver hematopoiesis), studies of the ability of the KTAMK mutant to rescue RUNX1 function in the mouse will be undertaken.
RUNX1 is fused to ETO (a component of several corepressor complexes) by the t(8;21) that occurs in the FAB, M2 subtype of AML. The RUNX1–ETO fusion protein lacks the RUNX1 RTAMR sequence that dynamically interacts with the repressor complex SIN3A. Furthermore, the interaction of ETO with the SIN3A complex is constant despite varying levels of PRMT1 protein. Using Kasumi-1 cells, we showed that binding of SIN3A to RUNX1, but not to RUNX1–ETO, is altered when (arginine) methylation is blocked using MTA. The presence of ETO and the absence of RTAMR likely account for the inability of PRMT1-related arginine methylation to alter SIN3A binding to RUNX1–ETO. Thus, RUNX1–ETO can serve as a more persistent repressor for at least some of its target genes. Losing responsiveness to PRMT1-mediated signals may contribute more broadly to leukemogenesis as many leukemia-associated fusion transcription factors interact more tightly with corepressor complexes than their wild-type counterparts.
Our results suggest that manipulating the level of PRMT1 activity could alter the balance between RUNX1–ETO and RUNX1 protein activity; such a strategy may favorably impact on the behavior of RUNX1–ETO-positive acute leukemia.
Materials and methods
Plasmids for expression of RUNX1 truncation and mutant proteins
A Flag tag and a 6-histidine tag were added to the N terminus and C terminus of the RUNX1c cDNA using PCR. The resulting cDNA was subcloned into the CMV promoter-driven pCDNA3 vector (Invitrogen) for use in transient transfection assays and for the preparation of in vitro translated protein. The site-specific RUNX1 mutant proteins, with R206 and R210 mutated to K individually or together, were prepared using PCR-based, oligonucleotide-directed mutagenesis.
The RUNX1 C-terminal truncation fragments were either generated using PCR, attaching BglII and XhoI restriction sites on the 5′ and 3′ ends, respectively, for subcloning into a GST Escherichia coli expression vector, pGEX-6p (Amersham), or were reported previously (Mao et al. 1999). The IL-3 promoter-driven reporter gene construct contains 5′-flanking sequences extending from 315 base pairs (bp) upstream of the start site to position +37, in the pGL3 basic vector (Uchida et al. 1997). The CD41 promoter-driven reporter gene construct was kindly provided by Dr. Adam Goldfarb (Elagib et al. 2003). The PRMT1 wild-type and mutant plasmids have been described (Strahl et al. 2001; Zhang and Cheng 2003).
Antibody generation
We generated a rabbit, anti-N-terminal RUNX1 polyclonal antibody using a peptide containing amino acids 2–17 from RUNX1b. We also generated a methyl-arginine-specific anti-RUNX1 antibody by immunizing rabbits with the C-terminal synthetic peptide (CLEQLRR [asymmetric-methylated] TAMR [asymmetric-methylated] VSPH) conjugated to KLH. The polyclonal antiserum was collected and purified using a peptide affinity column. The affinity of the antibody against the methyl-arginine RUNX1 was measured by ELISA and by dot blot assays (Sarma et al. 2004). For the dot blot assays, we spotted 5 pmol of peptide (measured by Ellman’s reagent) onto a nitrocellulose membrane. The control peptides were as follows: diasymmetric R210 peptide, CLRRTAMR (asymmetric methyl) VSPHH; methyl R142 Runt peptide, CRSGR (asymmetric methyl) GKSF; Runt peptide, CRSGRGKSF; H4 R3 methyl peptide SGR (asymmetric methyl) GKQGGKARAKAKTRSC.
Coimmunoprecipitation assays
Transiently transfected HEL cells, Kasumi-1 cells,and HeLa cells, were spun down at 200g for 10 min at 4°C, washed, and then resuspended at 4 × 107 cells per milliliter in EBC lysis buffer (20 mM Tris HCl at pH 8.0, 120 mM NaCl, 0.5% NP-40, 0.5% Empigen [Calbiotech, #324690], 10 mM NaF, 0.2 mM NaVO4,10 mM β-glycerophosphate) with freshly added DTT (1 mM), DNase I (1 μg/mL) (Worthington), and a proteinase inhibitor cocktail (Roche). The cells were incubated for 30 min on ice with sonication for 1 min at 20% amplitude with 10 sec on and 10 sec off. The supernatant was then collected after the mix was spun at 16,000g for 30 min. We mixed 200 μL of cell extract (∼2 mg), 800 μL of D100 buffer (20 mM HEPES at pH 7.9, 100 mM KCl, 0.2 mM EDTA, 0.5 mM PMSF, 20% glycerol) and ∼40 μL of anti-RUNX1 antibody cross-linked beads or anti-Flag agarose beads (Sigma) in the presence of both ethidium bromide (50 μg/mL) and RNase A (1 μg/mL). The reaction proceeded for 1–2 h at 4°C, then the beads were extensively washed and the RUNX1 protein was eluted using either the RUNX1 peptide or a Flag peptide at a final concentration of 0.5 mg/mL for 10 min at room temperature. Coimmunoprecipitation with other antibodies was done similarly, except that the beads were resuspended in SDS-PAGE sample buffer directly.
ChIP assays
HEL cells were maintained in RPMI medium supplemented with 5% fetal bovine serum (FBS) and 2 mM glutamine. Approximately 4 × 106 cells were used per ChIP reaction (per antibody), after cross-linking with 1% formaldehyde for 10 min at room temperature. ChIP was performed using the ChIP Assay Kit (UBI) according to a previously reported methodology (Zhao et al. 2001). The primer sequences used to amplify regions in the human CD41 promoter (GenBank accession no.: AC007722) containing the −69 RUNX1-binding sites are 5′-TCAGCCAT GAGCATCCACCCTCTG-3′ (forward) and 5′-TCCACAACC TCCCAGGCAGGAATG-3′ (reverse). A region ∼5 kb upstream of the CD41 transcription start site, which is devoid of RUNX1-binding sites, served as a control for each PCR. This region was amplified with the following primers: 5′-GGTGTGGAATTT CTGGCTTTGAAT-3′ (forward) and 5′- CTCTGTAACAATT AGGCCATATTTCCTT-3′ (reverse). For the PCR control reactions, 0.01% and 0.05% DNA inputs were used. The amplified DNA was analyzed after 35 cycles.
The ChIP primers for the human PU.1 promoter are as follows: 5′URE forward, GCACACATGCTTCCTGTGGTGACT; 5′ URE reverse, CCACGTGCCCTGACTCCCCTCCTAGC; proximal promoter forward, CTGGTCAGCAGGAAATTGGT; proximal promoter reverse, GGGAACTGGGCAGTTGTTTA; 5-kb upstream region forward, ATCTGTTCACATGGGCT TCC; 5-kb upstream region reverse, TGGTGGATAGGCA AGAAAGG.
Peptide pull-down assays
Methylated and nonmethylated peptides (used for antibody development) were synthesized, quantified, and conjugated to SulfoLink agarose (Pierce). For each pull-down reaction, 100 mg of nuclear extract were used with 10 μg of peptide-bound beads (50 μL) in D100 buffer plus 50 μg/mL ethidium bromide. After rotating overnight at 4°C, the beads were washed five times with 100 mM NaCl, 20 mM Tris (pH 8.0), and 1 mM DTT, plus protease inhibitors. The bound protein was then eluted with 1× SDS sample buffer and analyzed on 4%–12% NUPAGE gels.
CD34+ cell culture assays
CD34+ cells were isolated as reported (Mulloy et al. 2003) and cultured in one of four cytokine mixes. The cytokine mix used to maintain stem/progenitor cell status contained 20 ng/mL SCF, 20 ng/mL IL-6, 10 ng/mL FLT3 ligand, and 20 ng/mL TPO. The cytokine mix used to promote myeloid cell differentiation contained SCF (20 ng/mL), 10 ng/mL FLT3 ligand, 20 ng/mL IL-3, 20 ng/mL IL-6, 20 ng/mL G-SCF, and 20 ng/mL GM-CSF. The cytokine mix used to stimulate erythroid cell differentiation contained CSF (20 ng/mL) and EPO (6 μg/mL). The cytokine mix used to stimulate megakaryocytic cell differentiation contained 20 ng/mL SCF, 20 ng/mL TPO, and 50 ng/mL IL-11.
Generation of retroviral vectors, antibody reagents, and transient transfection of cell lines; GST pull-down assays; in vivo and in vitro arginine methylation assays; mapping of methylation sites; and real-time PCR assays
See the Supplemental Material.
Acknowledgments
We thank Arpi Nazarian for help with mass spectrometric analysis, Xiaomei Yan for technical assistance and critical advice, and Natalie Hua for assistance in preparing this manuscript. This work was supported by a Leukemia Lymphoma Society SCOR grant (to S.N., H.E.B., and P.T.), by the Herbert and Lee Friedman Memorial Fellowship (to X.Z.), by NCI Cancer Center Support Grant P30 CA08748, and by funding from the Mayo Foundation Scholar Program (to A.P.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1632608.
References
- An W., Kim J., Roeder R.G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Bedford M.T., Richard S. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Boisvert F.M., Cote J., Boulanger M.C., Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell. Proteomics. 2003;2:1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- Bruhn L., Munnerlyn A., Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes & Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Sinha K.K., Senyuk V., Nucifora G. SUV39H1 interacts with AML1 and abrogates AML1 transactivity. AML1 is methylated in vivo. Oncogene. 2003;22:5229–5237. doi: 10.1038/sj.onc.1206600. [DOI] [PubMed] [Google Scholar]
- Elagib K.E., Racke F.K., Mogass M., Khetawat R., Delehanty L.L., Goldfarb A.N. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- Emambokus N.R., Frampton J. The glycoprotein IIb molecule is expressed on early murine hematopoietic progenitors and regulates their numbers in sites of hematopoiesis. Immunity. 2003;19:33–45. doi: 10.1016/s1074-7613(03)00173-0. [DOI] [PubMed] [Google Scholar]
- Frank R., Zhang J., Uchida H., Meyers S., Hiebert S.W., Nimer S.D. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- Gary J.D., Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- Gelmetti V., Zhang J., Fanelli M., Minucci S., Pelicci P.G., Lazar M.A. Aberrant recruitment of the nuclear receptor corepressor–histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann F., Lee J., Bedford M.T., Fackelmayer F.O. Dynamics of human protein arginine methyltransferase 1(PRMT1) in vivo. J. Biol. Chem. 2005;280:38005–38010. doi: 10.1074/jbc.M502458200. [DOI] [PubMed] [Google Scholar]
- Higuchi M., O’Brien D., Kumaravelu P., Lenny N., Yeo E.J., Downing J.R. Expression of a conditional AML1–ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- Huang S., Litt M., Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes & Dev. 2005;19:1885–1893. doi: 10.1101/gad.1333905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Zhang P., Hirai H., Elf S., Yan X., Chen Z., Koschmieder S., Okuno Y., Dayarm T., Growney J.D., et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat. Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- Kitabayashi I., Yokoyama A., Shimizu K., Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17:2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi I., Aikawa Y., Nguyen L.A., Yokoyama A., Ohki M. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J. 2001;20:7184–7196. doi: 10.1093/emboj/20.24.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J.S., Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D., Goldstein R.E., Bernstein Y., Tang H., Goldenberg D., Stifani S., Paroush Z., Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B., Westendorf J.J., Linggi B., Isaac S., Seto E., Hiebert S.W. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 2000;275:651–656. doi: 10.1074/jbc.275.1.651. [DOI] [PubMed] [Google Scholar]
- Mao S., Frank R.C., Zhang J., Miyazaki Y., Nimer S.D. Functional and physical interactions between AML1 proteins and an ETS protein, MEF: Implications for the pathogenesis of t(8;21)-positive leukemias. Mol. Cell. Biol. 1999;19:3635–3644. doi: 10.1128/mcb.19.5.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers S., Lenny N., Hiebert S.W. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol. Cell. Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola H.K., Fujiwara Y., Schlaeger T.M., Traver D., Orkin S.H. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101:508–516. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- Mowen K.A., Tang J., Zhu W., Schurter B.T., Shuai K., Herschman H.R., David M. Arginine methylation of STAT1 modulates IFNα/β-induced transcription. Cell. 2001;10:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- Mowen K.A., Schurter B.T., Fathman J.W., David M., Glimcher L.H. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol. Cell. 2004;15:559–571. doi: 10.1016/j.molcel.2004.06.042. [DOI] [PubMed] [Google Scholar]
- Mulloy J.C., Cammenga J., Berguido F.J., Wu K., Zhou P., Comenzo R.L., Jhanwar S., Moore M.A., Nimer S.D. Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood. 2003;102:4369–4376. doi: 10.1182/blood-2003-05-1762. [DOI] [PubMed] [Google Scholar]
- Nimer S.D., Moore M.A. Effects of the leukemia-associated AML1–ETO protein on hematopoietic stem and progenitor cells. Oncogene. 2004;23:4249–4254. doi: 10.1038/sj.onc.1207673. [DOI] [PubMed] [Google Scholar]
- Okuda T., van Deursen J., Hiebert S.W., Grosveld G., Downing J.R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Ong S.E., Mittler G., Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- Reed-Inderbitzin E., Moreno-Miralles I., Vanden-Eynden S.K., Xie J., Lutterbach B., Durst-Goodwin K.L., Luce K.S., Irvin B.J., Cleary M.L., Brandt S.J., et al. RUNX1 associates with histone deacetylases and SUV39H1 to repress transcription. Oncogene. 2006;25:5777–5786. doi: 10.1038/sj.onc.1209591. [DOI] [PubMed] [Google Scholar]
- Rezai-Zadeh N., Zhang X., Namour F., Fejer G., Wen Y.D., Yao Y.L., Gyory I., Wright K., Seto E. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes & Dev. 2003;17:1019–1029. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K., Nishioka K., Reinberg D. Tips in analyzing antibodies directed against specific histone tail modifications. Methods Enzymol. 2004;376:255–269. doi: 10.1016/S0076-6879(03)76017-0. [DOI] [PubMed] [Google Scholar]
- Scott E.W., Simon M.C., Anastasi J., Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Briggs S.D., Brame C.J., Caldwell J.A., Koh S.S., Ma H., Cook R.G., Shabanowitz J., Hunt D.F., Stallcup M.R., et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Kurokawa M., Ueki K., Tanaka K., Imai Y., Mitani K., Okazaki K., Sagata N., Yazaki Y., Shibata Y., et al. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol. Cell. Biol. 1996;16:3967–3979. doi: 10.1128/mcb.16.7.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Frankel A., Cook R.J., Kim S., Paik W.S., Williams K.R., Clarke S., Herschman H.R. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 2000;275:7723–7730. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Osato M., Egawa T., Sunshine M.J., Bae S.C., Komori T., Ito Y., Littman D.R. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;11:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- Teyssier C., Ma H., Emter R., Kralli A., Stallcup M.R. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes & Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida H., Zhang J., Nimer S.D. AML1A and AML1B can transactivate the human IL-3 promoter. J. Immunol. 1997;158:2251–2258. [PubMed] [Google Scholar]
- Wang Q., Stacy T., Miller J.D., Lewis A.F., Gu T.L., Huang X., Buhweller J.H., Bories J.C., Alt F.W., Ryan G., et al. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- Wang J., Hoshino T., Redner R.L., Kajigaya S., Liu J.M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Huang Z.Q., Xia L., Feng Q., Erdjument-Bromage H., Strahl B.D., Briggs S.D., Allis C.D., Wong J., Tempst P., et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Wotton D., Ghysdael J., Wang S., Speck N.A., Owen M.J. Cooperative binding of Ets-1 and core binding factor to DNA. Mol. Cell. Biol. 1994;14:840–850. doi: 10.1128/mcb.14.1.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R., Chen L.F., Shigesada K., Murakami Y., Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Kurokawa M., Imai Y., Izutsu K., Asai T., Ichikawa M., Yamamoto G., Nitta E., Yamagata T., Sasaki K., et al. AML1 is functionally regulated through p300-mediated acetylation on specific lysine residues. J. Biol. Chem. 2004;279:15630–15638. doi: 10.1074/jbc.M400355200. [DOI] [PubMed] [Google Scholar]
- Yanagida M., Hayano T., Yamauchi Y., Shinkawa T., Natsume T., Isobe T., Takahashi N. Human fibrillarin forms a sub-complex with splicing factor 2-associated p32, protein arginine methyltransferases, and tubulins α 3 and β 1 that is independent of its association with preribosomal ribonucleoprotein complexes. J. Biol. Chem. 2004;279:1607–1614. doi: 10.1074/jbc.M305604200. [DOI] [PubMed] [Google Scholar]
- Yergeau D.A., Hetherington C.J., Wang Q., Zhang P., Sharpe A.H., Binder M., Marin-Padilla M., Tenen D.G., Speck N.A., Zhang D.E. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1–ETO fusion gene. Nat. Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- Yu M.C., Bachand F., McBride A.E., Komili S., Casolari J.M., Silver P.A. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes & Dev. 2004;18:2024–2035. doi: 10.1101/gad.1223204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.E., Hetherington C.J., Meyers S., Rhoades K.L., Larson C.J., Chen H.M., Hiebert S.W., Tenen D.G. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF α2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol. Cell. Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kalkum M., Yamamura S., Chait B.T., Roeder R.G. E protein silencing by the leukemogenic AML1–ETO fusion protein. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
- Zhao X., Pendergrast P.S., Hernandez N. A positioned nucleosome on the human U6 promoter allows recruitment of SNAPc by the Oct-1 POU domain. Mol. Cell. 2001;7:539–549. doi: 10.1016/s1097-2765(01)00201-5. [DOI] [PubMed] [Google Scholar]