Abstract
Meiotic silencing of sex chromosomes may cause their depletion of meiosis-specific genes during evolution. Here, we challenge this hypothesis by reporting the identification of TEX11 as the first X-encoded meiosis-specific factor in mice. TEX11 forms discrete foci on synapsed regions of meiotic chromosomes and appears to be a novel constituent of meiotic nodules involved in recombination. Loss of TEX11 function causes chromosomal asynapsis and reduced crossover formation, leading to elimination of spermatocytes, respectively, at the pachytene and anaphase I stages. Specifically, TEX11-deficient spermatocytes with asynapsed autosomes undergo apoptosis at the pachytene stage, while those with only asynapsed sex chromosomes progress. However, cells that survive the pachytene stage display chromosome nondisjunction at the first meiotic division, resulting in cell death and male infertility. TEX11 interacts with SYCP2, which is an integral component of the synaptonemal complex lateral elements. Thus, TEX11 promotes initiation and/or maintenance of synapsis and formation of crossovers, and may provide a physical link between these two meiotic processes.
[Keywords: TEX11, male infertility, meiosis, synapsis, meiotic recombination, X chromosome]
In sexually reproducing organisms, meiosis reduces the chromosome complement by half to generate haploid gametes to allow for genome doubling at fertilization. During meiosis, homologous chromosomes undergo pair- ing, synapsis, recombination, and faithful segregation (Zickler and Kleckner 1999; Page and Hawley 2004). Syn- apsis is mediated by the assembly of the synaptonemal complex (SC), a multiprotein structure, and facilitates completion of homologous recombination that in turn allows exchange of genetic material between the paternal and maternal genomes. Homologous recombination, on the other hand, facilitates chromosome synapsis and ensures proper chromosome segregation. Defects in meiosis are a leading cause of both infertility and birth defects (trisomy and monosomy) in humans (Hassold and Hunt 2001).
In principle, meiosis-specific genes could be located anywhere in the genome. However, no mouse sex chromosome-linked mutants with meiosis-specific defects have been reported, leading to the perception that meiosis-specific factors are rarely if ever encoded by the sex chromosomes (Matzuk and Lamb 2002). This could result from an evolutionary situation set up by the process of meiotic sex chromosome inactivation (MSCI) that silences the transcriptional activity of the X and Y chromosomes during meiosis (Solari 1974; Handel et al. 1994). Our previous study that systematically identified germ cell-specific genes from mouse spermatogonia revealed that nearly one-third of these genes are located on the X chromosome (Wang et al. 2001). Enrichment of early spermatogenesis genes on the X chromosome was also substantiated by a separate genomic study (Khil et al. 2004). Although these X-linked germ cell-specific genes undergo MSCI during the pachytene stage of male meiosis, they are expressed in early meiosis, implicating a possible role in meiosis (Wang et al. 2005). To test this possibility, we characterized the function of one of the X-linked germ cell-specific genes, Tex11, and now demonstrate that TEX11 is a novel X-encoded meiosis factor required for male fertility.
Results
TEX11 forms discrete foci on meiotic chromosomes
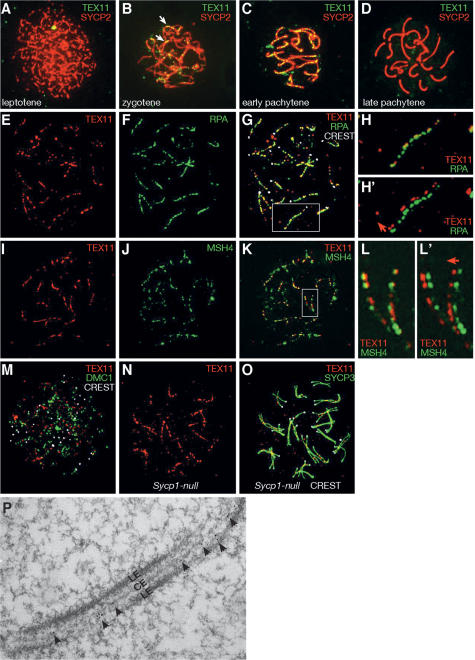
Tex11 is a germ cell-specific gene encoding a 947-amino-acid protein of unknown function (Wang et al. 2001). Protein sequence analysis revealed that TEX11 contains a tetratricopeptide repeat (TPR) protein–protein interaction domain but no other known protein motifs (Blatch and Lassle 1999). TEX11 protein was detected in testis but not in somatic tissues (data not shown). By immunostaining of spread nuclei, we found that TEX11 forms arrays of discrete foci along SCs in spermatocytes and fetal oocytes. In spermatocytes, TEX11 foci were not observed on asynapsed chromosomes in leptotene (Fig. 1A) but became apparent on synapsed regions in zygotene (Fig. 1B) and on the fully synapsed chromosomes in early pachytene (∼110 foci per cell) (Fig. 1C), suggesting that TEX11 may play a role in chromosomal synapsis. The TEX11 foci then disappeared in late pachytene (Fig. 1D) and were not observed in diplotene spermatocytes (data not shown). A similar spatiotemporal localization pattern of TEX11 was detected on female meiotic chromosomes (Supplemental Fig. S1), suggesting that TEX11 carries out a similar meiotic function in both sexes. The TEX11 foci were independently observed in wild-type meiocytes using two different antibodies, but they were absent in Tex11-deficient spermatocytes/oocytes (data not shown), validating the specificity of the TEX11 antibodies.
Figure 1.
TEX11 forms distinct foci on meiotic chromosomes. (A–D) Distribution of TEX11 foci in spermatocytes. TEX11 foci are absent in leptonema (A), present in zygonema (B) and early pachynema (C), but absent in late pachynema (D). (B) Note that TEX11 foci are present on synapsed regions (arrows) in zygonema. (E–G) TEX11 foci mostly (74%) colocalize with RPA foci in spermatocytes. The same spermatocyte is shown for TEX11 alone (E), RPA alone (F), and a composite image (G). CREST antiserum stains centromeres. Expanded view of the chromosome in the square in G is shown in H and H′ (with offset channels). (I–K) A majority of TEX11 foci (60%) overlap with MSH4 foci in spermatocytes. The same spermatocyte is shown for TEX11 alone (I), MSH4 alone (J), and a composite image (K). Expanded view of the two chromosomes in the square in K is shown in L and L′ (with offset channels). (H′,L′) The direction of deliberate shift of the red channel is indicated by an arrow. (M) TEX11 foci rarely overlap with DMC1 foci in spermatocytes. (N) TEX11 forms arrays of foci in Sycp1−/− pachynema. (O) TEX11 foci are located between the two aligned but separated lateral elements in Sycp1−/− pachynema. The same cell is shown in N. (P) Electron microscopy shows immunogold labeling of TEX11 between lateral elements (LEs) in spermatocytes. (CE) Central element.
We sought to address whether TEX11 colocalizes with recombination-related proteins that also form foci on meiotic chromosomes. Mouse RAD51/DMC1, components of early meiotic nodules, form foci earlier during meiosis than RPA (replication protein A), a ssDNA-binding protein, which localizes to transition meiotic nodules (Plug et al. 1998; Moens et al. 2002). The protein composition of transition meiotic nodules changes during the course of meiotic recombination, and their other components include MSH4, MSH5, and BLM (Marcon and Moens 2005). A subset of transition nodules transforms into late meiotic recombination nodules (sites of crossovers) marked by MLH1.
Interestingly, a majority of TEX11 foci colocalized with RPA foci in pachynema (Fig. 1E–H, H′), whereas no overlap between TEX11 and DMC1 or MLH1 foci were observed (Fig. 1M; data not shown), suggesting that TEX11 might be a component of transition meiotic nodules. In further support of this notion, more than half of TEX11 foci overlapped with MSH4 foci in spermatocytes (Fig. 1I–L, L′). In addition, we found that TEX11 foci, like RPA foci, were located between the two aligned but separated lateral elements in synapsis-deficient Sycp1-null pachytene cells (Fig. 1N,O; de Vries et al. 2005). Consistent with the immunofluorescence results, electron microscopy showed that immunogold labeling of TEX11 was present as foci between lateral elements (Fig. 1P). Double immunoelectron microscopy revealed that TEX11 and RPA colocalize to electron-dense nodules on SCs (Supplemental Fig. S2). These data suggest that TEX11 might be a novel constituent of synapsis-independent transition meiotic recombination nodules and suggest that TEX11 might take part in the homologous recombination process.
TEX11 is required for meiosis and fertility in males
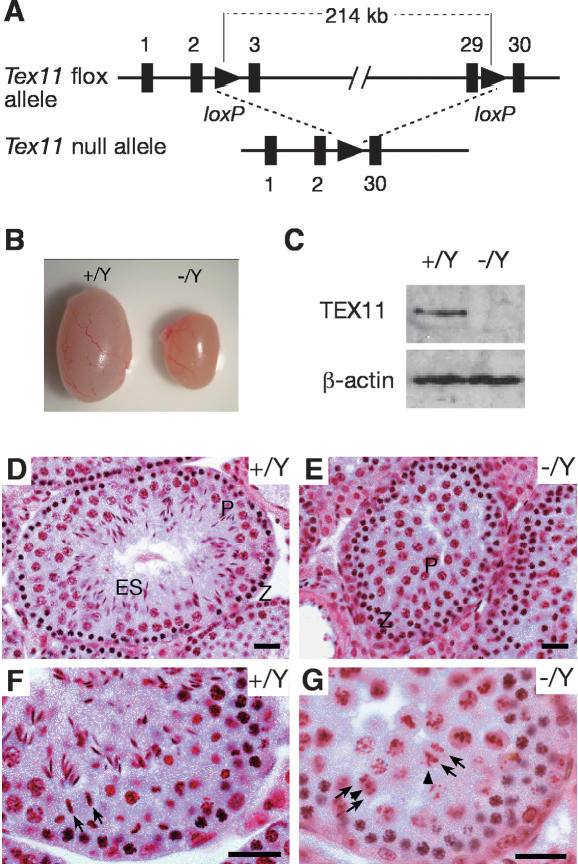
To uncover the functional role of Tex11 in meiosis, we generated a Tex11 conditional null mutant allele (Tex11fl) in mice using the Cre-loxP strategy (Fig. 2A). Sequence analysis revealed that the mouse Tex11 gene consists of 30 exons spanning a 224-kb genomic region on the X chromosome. We performed two consecutive targeting steps in embryonic stem (ES) cells to obtain a Tex11 conditional allele with loxP sites inserted in intron 2 and intron 29, which are 214 kb apart (Fig. 2A; Supplemental Fig. S3). Both Tex11fl/Y males and Tex11fl/fl females were fertile. To delete the Tex11 gene, Tex11fl mice were crossed with ACTB-Cre mice, in which Cre recombinase is expressed in all cells of the early embryo (Lewandoski et al. 1997). Tex11−/Y males and Tex11−/− females were viable and appeared to be healthy.
Figure 2.
Tex11 is essential for male meiosis. (A) Schematic diagram of Tex11fl and null alleles. Two loxP sites in the same orientation were introduced into introns 2 and 29. Exons are shown as black bars and the exon numbers are indicated. (B) Significant size reduction in 12-wk-old Tex11−/Y testis. (C) Western blot analysis shows the absence of TEX11 protein in Tex11−/Y testis. (D–G) Histological analysis of testes from 8-wk-old wild-type and Tex11−/Y mice. In contrast to wild-type tubules with full spermatogenesis (D), Tex11-deficient tubules (E) exhibit meiotic arrest. (F) Segregation of chromatin in a wild-type anaphase spermatocyte from a Stage XII tubule. Arrows indicate two chromatin masses (i.e., two sets of segregating chromosomes). (G) Abnormal chromosome segregation in Tex11-deficient Stage XII tubules. Arrows and arrowheads indicate abnormally segregating chromatin masses. (Z) Zygotene, (P) pachytene, (ES) elongated spermatids. Bars, 25 μm.
While Tex11−/− females were fertile with reduced litter size, Tex11−/Y males were sterile. Testes from 12-wk-old Tex11−/Y mice (84.5 ± 21.3 mg, n = 5 pairs) weighed 50% less than those from wild-type mice (180.5 ± 17.6 mg, n = 5 pairs) (Fig. 2B). In mutant mice, 98% of the Tex11 coding region was deleted, and absence of TEX11 protein in Tex11−/Y testes (Fig. 2C) confirmed that the mutant Tex11 allele was null. Sequence analysis did not reveal the presence of other transcripts or known noncoding RNAs in the Tex11 gene of mice, humans, and rats, strongly suggesting that the fertility defects observed in mutant mice were caused by loss of Tex11 function.
Histological analysis of testis from adult mice showed that disruption of Tex11 causes arrest of male meiosis and aberrant chromosome segregation in anaphase spermatocytes (Fig. 2D–G). Tubules of wild-type testis contained a full spectrum of spermatogenic cells (Fig. 2D,F). However, tubules of Tex11−/Y testis exhibited an arrest in late meiosis (Fig. 2E). We observed that anaphase spermatocytes in Tex11−/Y testis were defective (Fig. 2G). In wild-type anaphase cells, two sets of chromosomes segregated synchronously toward opposite spindle poles (Fig. 2F). In contrast, in Tex11-deficient anaphase cells, a few chromatin masses either migrated ahead or lagged behind the two major chromatin masses (Fig. 2G).
TEX11 promotes chromosomal synapsis in males
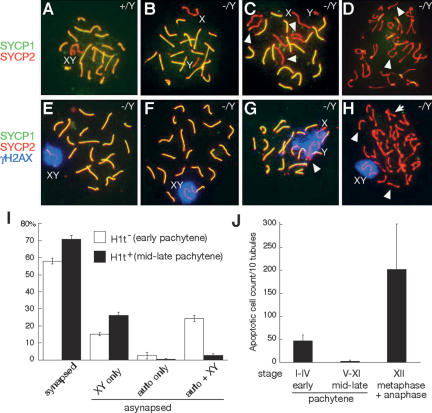
We next investigated whether Tex11−/Y pachytene spermatocytes exhibit defects in chromosomal synapsis by staining central and lateral elements of SCs with anti-SYCP1 and anti-SYCP2 antibodies, respectively (Fig. 3). In wild-type pachynema, 19 pairs of autosomes were fully synapsed, and the X–Y chromosomes were synapsed only in the pseudoautosomal region (Fig. 3A). Remarkably, we observed variable synaptic failure in Tex11−/Y pachynema; some chromosomes were not synapsed at all, whereas the majority of chromosomes were fully synapsed (Fig. 3B,C). Overall, 38% of pachytene spermatocytes from adult Tex11−/Y mice contained asynapsed chromosomes (two to 10 per nucleus, >900 pachytene nuclei scored). In contrast, only one of 150 wild-type pachytene nuclei analyzed had asynapsed sex chromosomes (<1%). Of the mutant pachytene spermatocytes with asynapsed chromosomes, 50% exhibited asynapsis of only the sex chromosomes (Fig. 3B), in 45% both sex chromosomes and autosomes were affected (Fig. 3C), and in 5% only the autosomes were affected. These observations strongly suggest that TEX11 contributes to initiation and/or maintenance of chromosomal synapsis in male meiosis.
Figure 3.
TEX11 promotes chromosome synapsis. (A) Wild-type pachynema with 19 pairs of fully synapsed autosomes (yellow) and partially synapsed X–Y chromosomes (red). (B) Tex11−/Y pachynema with asynapsed X–Y. (C) Tex11−/Y pachynema with eight asynapsed chromosomes: the X–Y and three pairs of autosomes (arrowheads). Notably, each pair of asynapsed homologs (as judged by equal SC length except for XY) is in close proximity and alignment, indicating a possible interaction. (D) Tex11−/Y diplonema with univalents (arrowheads). (E) An apparently normal Tex11−/Y pachynema with partially synapsed X–Y in the γH2AX-positive sex body. (F) Tex11−/Y pachynema. Asynapsed X–Y chromosomes are confined to a single γH2AX-positive domain. (G) Tex1−/Y pachynema with one pair of asynapsed autosomes (arrowhead) and asynapsed X–Y, all of which are γH2AX-positive. (H) Tex11−/Y diplonema with a large number of univalents (arrowheads). Some autosomes are bivalents (arrow). X–Y univalents are confined to a single γH2AX-positive domain. In contrast, all autosomal univalents are γH2AX-negative. (I) Analysis of asynapsed chromosomes in early (H1t-negative) and mid-to-late (H1t-positive) Tex11−/Y pachynema. Testes from four 7.5-wk-old Tex11−/Y mice were analyzed by triple immunostaining of spread nuclei with anti-SYCP1, anti-SYCP3, and anti-histone H1t antibodies. All pachynema in view were first divided into H1t− and H1t+ groups, and asynapsed chromosomes were then examined. More than 200 pachynema from each mouse were analyzed. (J) Two distinct rounds of apoptosis in adult Tex11−/Y testis. Seminiferous tubules were divided into three groups: H1t-negative (Stages I–IV), H1t-positive (Stages V–XI), and Stage XII.
BRCA1 recruits ATR to asynapsed chromatin in pachynema, which in turn phosphorylates H2AX (termed γH2AX) and leads to MSUC (meiotic silencing of unsynapsed chromatin) (Turner et al. 2005). To determine if MSUC occurs in Tex11−/Y spermatocytes, we examined the localization of γH2AX and ATR. The X–Y chromosomes were positively stained for γH2AX, regardless of whether they were synapsed or not (Fig. 3E,F). Asynapsed autosomes also resided in γH2AX-positive chromatin domains (Fig. 3G). The γH2AX staining was not observed on synapsed autosomes in late pachytene or on autosomes in diplotene spermatocytes from Tex11−/Y testis. As expected, asynapsed chromosomes in Tex11−/Y pachytene spermatocytes were ATR-positive (Supplemental Fig. S4). These data suggest that chromosomes asynapsed as a consequence of loss of TEX11 function are subject to MSUC.
To determine the fate of pachynema with asynapsed chromosomes, we divided Tex11-deficient spermatocytes into two groups: histone H1t-negative (early pachynema) and H1t-positive (mid-to-late pachynema) (Fig. 3I; Supplemental Fig. S5; Cobb et al. 1999). The percentage of pachytene cells with asynapsed chromosomes involving both sex chromosomes and autosomes was drastically reduced as meiosis progressed to mid-to-late pachytene, whereas the relative percentage of cells with normal synapsis or with asynapsed sex chromosomes alone increased accordingly (Fig. 3I). We also found that apoptosis increased dramatically in Tex11−/Y testis in a stage-dependent manner. Using H1t as a marker along with DAPI staining of nuclear DNA to divide seminiferous tubules into three groups—H1t-negative (Stage I–IV), H1t-positive (Stages V–XI), and Stage XII—we observed two rounds of apoptosis in Tex11−/Y testis (Fig. 3J). The first round occurred in H1t-negative tubules and the second in Stage XII. Little apoptosis was observed in H1t-positive tubules (Stages V–XI) that contain mid-to-late pachytene spermatocytes. Notably, the second round of apoptosis (Stage XII) affected many more cells than the first round. Collectively, these data suggest that cells with asynapsed autosomes may be eliminated in Stages I–IV in response to a pachytene checkpoint that monitors completion of chromosome synapsis and/or by transcriptional silencing of autosomal genes that are crucial to meiosis due to MSUC (de Rooij and de Boer 2003; Turner et al. 2005). Cells having only asynapsed X–Y chromosomes might bypass a pachytene checkpoint and progress through the pachytene stage, since MSUC in these cells equals MSCI (Fig. 3I; Roeder and Bailis 2000; Eaker et al. 2002; Baarends et al. 2005; Turner et al. 2005). The question arises as to what could account for the second round of apoptosis (Stage XII) in Tex11−/Y testis (Fig. 3J). The majority (70%) of mid–late pachytene mutant spermatocytes had normal synapsis (Fig. 3I), but nearly all (96%) Tex11−/Y diplotene spermatocytes contained univalent chromosomes (Fig. 3D,H). Additionally, in these diplotene cells, sex chromosomal univalents remained γH2AX- and ATR-positive, whereas autosomal univalents were γH2AX- and ATR-negative (Fig. 3H; Supplemental Fig. S4). These results suggest de novo formation of autosomal univalents by desynapsis at the diplotene stage, possibly due to defective crossover formation at pachytene.
TEX11 promotes formation of meiotic crossovers in males
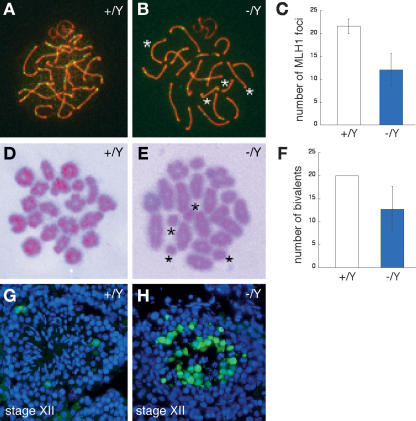
We monitored the formation of crossovers at pachytene. MLH1 is essential for crossover formation and marks sites of crossover (Baker et al. 1996; Edelmann et al. 1996). We detected a sharply reduced number of MLH1 foci (12.1 ± 3.5, n = 42 nuclei) in Tex11−/Y pachynema compared with wild-type (21.6 ± 1.6, n = 24 nuclei) (Fig. 4A–C), mainly due to a loss of foci from synapsed chromosomes, and only to a lesser degree caused by the presence of asynapsed X–Y chromosomes. Analysis of metaphase I chromosomes by Giemsa staining revealed a reduced number of bivalents (12.7 ± 5.0 bivalents per nucleus) in Tex11−/Y metaphase I spermatocytes compared with wild-type (20 bivalents per nucleus) (Fig. 4D–F). The presence of univalents could lead to chromosomal segregation defects at the subsequent anaphase I stage, and aberrant chromosome attachment to the spindle or chromosome nondisjunction could trigger the spindle checkpoint and induce apoptosis (Ashley et al. 1994; Li and Nicklas 1995; Odorisio et al. 1998; Eaker et al. 2002). We detected both defective chromosome segregation in Tex11−/Y anaphase spermatocytes (Fig. 2G) and massive apoptosis of germ cells in Stage XII tubules (containing the metaphase I and anaphase I spermatocytes) in Tex11−/Y testis (Figs. 3J, 4H), indicating that TEX11 promotes the formation of crossovers and thus is required for proper chromosome segregation during male meiosis.
Figure 4.
TEX11 modulates the formation of meiotic crossovers. (A–C) A dramatic reduction in the number of MLH1 foci (green) in Tex11−/Y pachynema (∼15 foci) relative to wild type (22 foci). (B) Note the absence of MLH1 foci on four synapsed chromosomes (asterisks) in the mutant. (D) Twenty bivalents (pairs of homologs) are connected via chiasmata in wild-type metaphase I spermatocytes. (E) Presence of univalents in Tex11-deficient metaphase I spermatocytes. Asterisks indicate four univalents. (F) A sharp decrease in the number of bivalents in Tex11-deficient metaphase I spermatocytes. (G) TUNEL analysis of wild-type tubules. A few apoptotic cells (green) are occasionally observed in Stage XII tubules. (H) Massive apoptosis in Tex11−/Y Stage XII tubules. Apoptotic cells presumably corresponded to anaphase I spermatocytes.
Chromosomal asynapsis and reduced crossover formation in Tex11-deficient females
In contrast to Tex11−/Y males, Tex11−/− females were fertile and produced offspring. However, Tex11−/− females displayed a reduction in litter size, generating on average 5.2 offspring per litter (5.2 ± 2.2). In comparison, heterozygous (Tex11+/−) littermates produced 9.6 offspring per litter (9.6 ± 2.0; P < 0.0001). Analysis of pachytene asynapsis and formation of MLH1 foci in fetal oocytes revealed a higher percentage (31.3%) of Tex11-deficient pachytene oocytes containing asynapsed chromosomes compared with wild-type/Tex11+/− oocytes (5.5%). The number of MLH1 foci (18.6 ± 2.6) in Tex11-deficient oocytes was slightly lower than that (23.5 ± 2.4) in wild-type/Tex11+/− oocytes. These data, together with studies of Tex11-deficient males, demonstrate that loss of Tex11 function causes the same effect on synapsis and crossover formation in both sexes, even though these defects in females are milder. The fertility in Tex11-deficient females could be explained by the fact that the number of MLH1 foci is close to 20 per cell in some oocytes. The reduced fertility in Tex11-deficient females could be caused by aneuploidy due to asynapsis and/or a reduced number of MLH1 foci in oocytes.
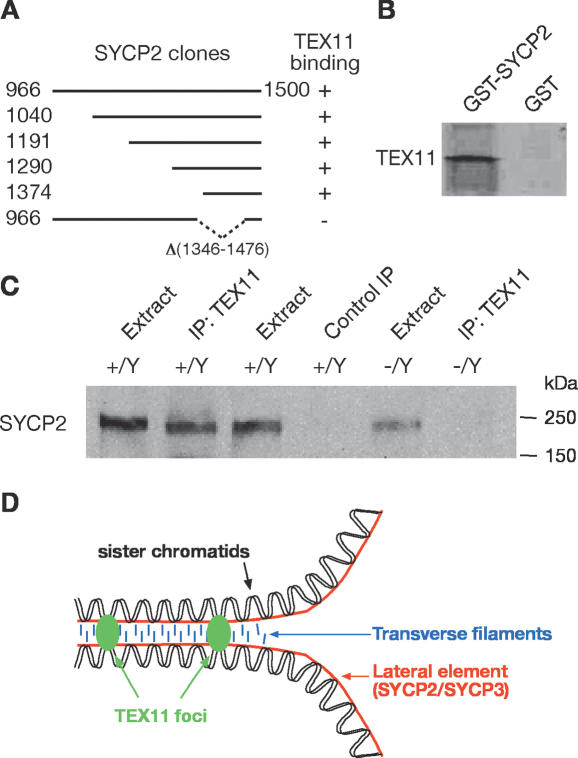
TEX11 interacts with SYCP2
To elucidate a potential molecular mechanism for the function of TEX11 in meiosis—i.e., to identify protein partners of TEX11—we performed a yeast two-hybrid screen of a mouse testis cDNA library using the full-length TEX11 as bait. The most frequently identified protein was SYCP2, an integral component of the lateral elements of the SC (Offenberg et al. 1998; Schalk et al. 1998). The SYCP2 C-terminal region (1374–1500 amino acids) was sufficient for interaction with TEX11 in both two-hybrid and GST pulldown assays (Fig. 5A,B). Furthermore, an internal deletion of SYCP2 (1346–1476 amino acids) abolished its interaction with TEX11 (Fig. 5A). This internal deletion (1346–1476 amino acids) was previously generated in Sycp2 mutant mice (Yang et al. 2006). We found that TEX11 foci were abundant on meiotic chromosomes in Sycp2 mutant pachytene oocytes (data not shown), suggesting that formation of TEX11 foci is independent of SYCP2. Coimmunoprecipitation from testicular protein extracts further demonstrated that TEX11 associates with SYCP2 in vivo (Fig. 5C). Thus, through its association with SYCP2, an SC component, TEX11 could provide a link between chromosomal synapsis and meiotic recombination (Fig. 5D).
Figure 5.
TEX11 interacts with SYCP2. (A) Five unique overlapping SYCP2 fragments identified in the two-hybrid screen using the full-length TEX11 protein as bait. Numbers indicate the position of terminal residues. The bottom clone with an internal deletion of SYCP2 (1346–1476 amino acids) does not interact with TEX11. (B) Binding of in vitro translated TEX11 to GST-SYCP2 (amino acids 1374–1500). (C) Coimmunoprecipitation of SYCP2 with TEX11 from testis. Immunoprecipitation was performed with rabbit anti-TEX11 antibodies or preimmune control serum and probed with guinea pig anti-SYCP2 antibodies on Western blot. (D) Model illustrating the association of TEX11 foci and the lateral elements at the zygotene stage. A pair of homologous chromosomes and transverse filaments of the SC are also shown.
Discussion
Our studies implicate TEX11 in initiation of chromosome synapsis. In support of such a function, loss of TEX11 results in an “all-or-none” synapsis defect on a per chromosome basis in early pachytene spermatocytes: A subset of chromosomes is completely asynapsed (presumably due to a failure in initiation), but other chromosomes are synapsed. The role in synapsis is further supported by colocalization of TEX11 with RPA foci, which are known to be present on synapsed regions (Plug et al. 1998; Moens et al. 2002). Importantly, TEX11 interacts with SYCP2, which is required for chromosomal synapsis (Yang et al. 2006). Additionally or alternatively, TEX11 could contribute to the elongation or stabilization of the SC. Thus, in the absence of TEX11, chromosome synapsis may be initiated but not propagated or maintained. Although little is known about the molecular underpinnings of synapsis initiation in mammals, this process has been extensively studied in yeast. A set of yeast proteins (Zip2, Zip3, Zip4, Msh4, Msh5, etc.), referred to as SIC (synapsis initiation complex) or ZMM proteins, are required for both chromosomal synapsis initiation and meiotic recombination (Borner et al. 2004; Tsubouchi et al. 2006). TEX11 exhibits some sequence homology with yeast Zip4 and Arabidopsis ZIP4. In Zip4 mutant yeast cells, Zip1, a component of yeast transverse filaments, localizes only in foci, indicating a failure of full synapsis (Tsubouchi et al. 2006). Furthermore, despite absence of sequence homology between Tex11 and Zip3, the yeast Zip3 mutant resembles the Tex11 mouse null mutant in one key aspect of synapsis. As in Tex11-deficient pachynema, Zip3-deficient pachytene cells exhibit an “all-or-none” synaptic defect on a per chromosome basis (Agarwal and Roeder 2000). These studies suggest that the TEX11-mediated synapsis pathways are functionally conserved among diverse organisms.
Several lines of evidence support the conclusion that TEX11 also plays a distinct role in meiotic recombination. Firstly, a majority of TEX11 foci overlap with RPA foci and MSH4 foci, which correspond to transition meiotic nodules. The formation of TEX11 foci between lateral elements in Sycp1-null pachytene spermatocytes further demonstrates that the synapsed structure is not essential for holding TEX11 to chromosomes; instead, it is likely that TEX11 is part of a recombination complex. Secondly, loss of TEX11 caused a subtle but distinct effect on initiation and/or maintenance of synapsis (a small subset of chromosomes are affected) but a more severe effect on crossover formation (almost a 50% loss of MLH1 foci). In particular, the number of MLH1 foci in Tex11−/Y pachytene spermatocytes with normal synapsis was greatly reduced, suggesting that the role of TEX11 in crossover formation is separate from its role in synapsis. Thirdly, yeast Zip4 and Arabidopsis Zip4 mutants exhibit defects in crossover formation, demonstrating the evolutionary conservation of the TEX11/ZIP4 functions in meiotic recombination (Tsubouchi et al. 2006; Chelysheva et al. 2007). Lastly, we observed two rounds of apoptosis in Tex11-deficient testis: in early pachytene spermatocytes and in anaphase I spermatocytes. These two rounds of apoptosis were distinct from each other, because increased apoptosis (the first round) was observed in H1t-negative tubules from juvenile Tex11-deficient testis, in which meiosis has yet to reach the anaphase of prophase I (Supplemental Fig. S6). Thus, in Tex11-deficient testis, asynapsis (if affecting autosomes) would result in cell elimination by the pachytene checkpoint and/or MSUC, whereas those cells that succeed in synapsis would fail to complete crossover formation and would be eliminated by the spindle checkpoint, resulting in male sterility (Roeder and Bailis 2000; Eaker et al. 2002; de Rooij and de Boer 2003; Turner et al. 2005).
We propose that the dual function of TEX11 in synapsis and crossover fixation might be linked by the TEX11–SYCP2 interaction and could be explained by its focal localization on synapsed but not asynapsed chromosomes (Fig. 5D). Consistent with this model, the short length of the X–Y synapsed region might explain why asynapsis preferentially affects X–Y chromosomes. TEX11 forms distinct foci on the meiotic chromosomes, and these foci are located between the lateral elements of the SC. In contrast, SYCP2, an integral component of the lateral element, localizes continuously on the lateral elements. How could these two proteins interact while they appear to be spatially separate? Interestingly, a previous ultrastructural study reported that while both SYCP2 and SYCP3 localize continuously on the lateral elements, SYCP2 but not SYCP3 also localizes to “fuzzy connections” (presumably recombination nodules) between the lateral elements (Schalk et al. 1998). Thus, we postulate that TEX11 might interact with the pool of SYCP2 that is present in these “fuzzy connections.”
Many mutants of meiotic recombination nodule components (Dmc1, Msh4, Mlh1, etc.) abolish chromosomal synapsis and/or crossover formation in mice (Baker et al. 1996; Edelmann et al. 1996; Pittman et al. 1998; Yoshida et al. 1998; Kneitz et al. 2000). Remarkably, unlike these known meiosis factors, TEX11 promotes synapsis and crossover formation but is not essential for either process. Partial elimination of synapsis and crossover is not caused by any residual function of Tex11 in this mouse mutant, because the TEX11 protein is absent. One explanation could be that TEX11 is a regulatory but nonessential component of a multiprotein complex that modulates synapsis and crossover. Such a protein complex could be partially functional in the absence of TEX11.
Meiotic silencing of unpaired DNA or chromatin has emerged as a common process in diverse organisms such as filamentous fungi, nematodes, and mammals (Shiu et al. 2001; Fong et al. 2002; Baarends et al. 2005; Turner et al. 2005). Gene expression (microarray) studies show that meiotic silencing of sex chromosomes leads to depletion of male germline-intrinsic genes on the X chromosome in nematodes and mammals during evolution (Reinke et al. 2000; Khil et al. 2004). However, here we demonstrate that TEX11, encoded by the X chromosome, is a meiosis-specific protein, and that its loss causes meiotic failure in males. This study counters the hypothesis that the X chromosome may be depleted of meiosis-specific genes in mammals.
Our findings have important implications for male infertility in humans, which accounts for about half of the cases of infertility among couples. An estimated 15% of couples are affected by infertility worldwide (Matzuk and Lamb 2002). Given that disruption of Tex11 causes azoospermia in mice, mutations in the human TEX11 gene could cause infertility in men.
Materials and methods
Generation of anti-TEX11 polyclonal antibodies
The mouse Tex11 cDNA fragment encoding the C-terminal 100 amino acids (residues 848–947) was cloned into the pQE-30 expression vector (Qiagen). The 6xHis-TEX11 fusion protein was expressed in Escherichia coli, affinity-purified with Ni-NTA beads, and used to immunize two rabbits (Cocalico Biologicals, Inc.). The anti-TEX11 antiserum (serum 1966) was used for Western blot analysis (1:500). In addition, guinea pig polyclonal antibodies were raised against the C-terminal peptide of mouse TEX11 (EQLRALIVPPEDQGSVSSTNVAAQNHL) and affinity-purified.
Consecutive homologous gene targeting in ES cells
Two targeting constructs were generated. In the Tex11 targeting construct 1, we subcloned the floxed HyTK selection cassette into intron 2 (Supplemental Fig. S3). HyTK encodes a double selection marker for positive selection with hygromycin and negative selection with ganciclovir. Construct 1 consists of two homologous arms (2.1 kb and 2.0 kb). In targeting construct 2, the floxed HyTK selection cassette was inserted into intron 29 with two flanking homologous arms (2.3 kb and 2.8 kb). Homologous arms in both constructs were fully sequenced, and no mutations were found.
Four consecutive steps were executed to generate ES cells harboring a Tex11fl allele (Supplemental Fig. S3). Step 1, targeting ES cells with construct 1: V6.5 hybrid ES cells were electroporated with linearized targeting construct 1 and selected for integration with hygromycin B (120 μg/mL; Invitrogen). Hygromycin-resistant ES cell colonies (192) were screened by long-distance PCR. Twenty percent of clones were resistant due to homologous recombination, referred to as ES line A. Step 2, Cre-mediated recombination: Two independent line A clones were expanded, electroporated with the pOG231 plasmid that transiently expresses Cre recombinase, and subjected to negative selection with ganciclovir (2 μM; Sigma) for removal of the HyTK cassette. Viable colonies were screened by PCR. Cre-mediated recombination resulted in ES line B, which bears only one loxP site in intron 2. Step 3, targeting with construct 2: Two independent line B ES clones were electroporated with targeting construct 2 and screened as described in Step 1. The homologous targeting frequency was 17%. The resulting ES cells were referred to as ES line C. Step 4, second Cre-mediated recombination: Two independent line C ES cell clones were electroporated with the pOG231 plasmid and screened as described in Step 2. Recombination between the adjacent HyTK-flanking loxP sites resulted in the final Tex11fl allele, harboring two loxP sites 214 kb apart (Fig. 2A).
Generation of Tex11fl and Tex11-deficient mice
Four Tex11fl/Y ES cell clones were injected into B6C3F1 blastocysts that were subsequently transferred to the uteri of pseudopregnant ICR females. One ES clone (C10-3) gave rise to seven chimeric mice. Two of the chimeric males transmitted the Tex11fl allele through the germline. Tex11fl mice were bred with ACTB-CreTg/Tg mice to delete Tex11 (Lewandoski et al. 1997).
Because Tex11−/Y males were sterile, generation of homozygous knockout (Tex11−/−) females required a different breeding strategy. We bred Tex11fl/Y males with Tex11+/− ACTB-CreTg/+ females. Twenty-five percent of the resulting female offspring were expected to be Tex11flox/− ACTB-CreTg/+. Because ACTB-Cre was strongly and ubiquitously expressed since the zygote stage, Tex11fl/− ACTB-CreTg/+ mice were equivalent to Tex11−/− ACTB-CreTg/+. The Tex11fl allele was not detected in genomic DNA prepared from multiple tissues (including ovary) from Tex11−/− ACTB-CreTg/+ mice by PCR. In addition, genotyping of >100 offspring sired by Tex11−/− ACTB-CreTg/+ females did not identify any pup that harbors the Tex11fl allele. Mice used were on a mixed genetic background (FVB, 129, and C57BL/6J). Animals were maintained and used for experimentation according to the guidelines of the Institutional Animal Care and Use Committee of the University of Pennsylvania.
TUNEL analysis and H1t immunofluorescence
Testes were fixed in 4% paraformaldehyde (PFA) for 3 h at 4°C, dehydrated in 30% sucrose overnight, prepared, and sectioned. TUNEL assays were performed with the ApopTag Fluorescein In Situ Apoptosis Detection Kit (catalog no. S7110, Serologicals Corporation/Chemicon) with or without concurrent immunostaining with anti-H1t antibodies. Cross-sections of tubules were divided into three categories: H1t-negative, H1t-positive, and Stage XII. TUNEL-positive cells were counted for each cross-section (Fig. 3J). More than 150 tubules were analyzed for each mouse.
Two-hybrid screen and immunoprecipitation
The full-length TEX11 coding region was cloned into the pAS2-1 vector through gap repair in yeast. A total of 2 × 106 clones from a mouse testis cDNA library (BD Biosciences) were screened according to standard protocols. After specificity tests, 53 clones were positive and sequenced. Sequence analysis revealed that eight clones encode SYCP2. For immunoprecipitation experiments, protein extracts were prepared by homogenizing testes from five postnatal day 20 mice per genotype. After sonication and centrifugation, extracts were incubated with protein A Sepharose and centrifuged prior to use for immunoprecipitation.
Histological, surface nuclei spread, immunofluorescent, and electron microscopy analyses
For histological analysis, testes were fixed in Bouin’s solution, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Metaphase spread cells were stained with 4% Gurr Giemsa (Invitrogen). The number of bivalents was derived using the following formula: the number of bivalents = 40 − n (40 is the theoretical maximum number of univalents; n is the total number of chromatin masses in each metaphase I nucleus). For immunofluorescent analysis of spread nuclei, testicular cell suspensions were prepared, immunostained, and analyzed. To obtain fetal oocytes, Tex11−/− ACTB-CreTg/+ females were caged with Tex11fl/Y males at 4 pm. Vaginal copulatory plugs were checked the next morning and recorded as 0.5 d post-coitum (dpc). Fetal oocytes were collected at 17.5 or 18.5 dpc. Primary antibodies used for immunofluorescence were as follows: rabbit anti-TEX11, guinea pig anti-TEX11 (1:200), rabbit anti-SYCP1 (1:100), rabbit and guinea pig anti-SYCP2 (sera 1918 and GP21, 1:100), rabbit anti-SYCP3 (1:500; a gift from S. Chuma) and our own (1:100), guinea pig anti-H1t (1:1000; a gift from M.A. Handel), rabbit anti-ATR (1:80; catalog no. PC538, EMD Biosciences), mouse anti-γH2AX (1:500; Upstate Biotechnology), human CREST antiserum (1:3000; a gift from N. Ringertz), rabbit anti-DMC1 (1:100; a gift from P. Moens), anti-RPA sera (1:400; a gift from P. Moens), anti-RPA monoclonal antibody (EMD Biosciences), and rabbit anti-hMSH4 antibodies (1:100) (Her et al. 2003). Various FITC-, Texas red-, AMCA-, Alexa fluor 488-, Cy5-, or TRITC-conjugated secondary antibodies were used. Slides were visualized under an Axioskop 40 fluorescence microscope. Images were captured with a digital camera (Evolution QEi, MediaCybernetics) and processed with ImagePro software (Phase 3 Imaging systems) and Photoshop (Adobe). Immunogold labeling was performed on 6-μm cryosections of shock-frozen mouse testis according to standard protocols with rabbit anti-TEX11 antibodies. Secondary antibodies were conjugated to 6-nm or 12-nm gold particles.
Acknowledgments
We thank E. Gleason for technical assistance, M.A. Handel for advice on the project and anti-H1t antibodies, P.E. Cohen for advice on phenotypic analysis, P. Moens for anti-RPA and anti-DMC1 antibodies, and S. Chuma for anti-SYCP3 antibodies. We thank M.A. Handel, S. Rozen, and P. de Boer for comments on the manuscript. This work was supported by an NIH grant GM076327 (to P.J.W.); by grants from the Swedish Cancer Society, the Swedish Research Council and the Karolinska Institutet (to C.H); by support from the Howard Hughes Medical Institute (to D.C.P); and by a grant from the Deutsche Forschungsgemeinschaft (to R.B.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1613608.
References
- Agarwal S., Roeder G.S. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- Ashley T., Ried T., Ward D.C. Detection of nondisjunction and recombination in meiotic and postmeiotic cells from XYSxr [XY,tp(Y)1Ct] mice using multicolor fluorescence in situ hybridization. Proc. Natl. Acad. Sci. 1994;91:524–528. doi: 10.1073/pnas.91.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarends W.M., Wassenaar E., van der Laan R., Hoogerbrugge J., Sleddens-Linkels E., Hoeijmakers J.H., de Boer P., Grootegoed J.A. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol. Cell. Biol. 2005;25:1041–1053. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.M., Plug A.W., Prolla T.A., Bronner C.E., Harris A.C., Yao X., Christie D.M., Monell C., Arnheim N., Bradley A., et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- Blatch G.L., Lassle M. The tetratricopeptide repeat: A structural motif mediating protein–protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Borner G.V., Kleckner N., Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Gendrot G., Vezon D., Doutriaux M.P., Mercier R., Grelon M. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 2007;3:e83. doi: 10.1371/journal.pgen.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J., Cargile B., Handel M.A. Acquisition of competence to condense metaphase I chromosomes during spermatogenesis. Dev. Biol. 1999;205:49–64. doi: 10.1006/dbio.1998.9101. [DOI] [PubMed] [Google Scholar]
- de Rooij D.G., de Boer P. Specific arrests of spermatogenesis in genetically modified and mutant mice. Cytogenet. Genome Res. 2003;103:267–276. doi: 10.1159/000076812. [DOI] [PubMed] [Google Scholar]
- de Vries F.A., de Boer E., van den Bosch M., Baarends W.M., Ooms M., Yuan L., Liu J.G., van Zeeland A.A., Heyting C., Pastink A. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes & Dev. 2005;19:1376–1389. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaker S., Cobb J., Pyle A., Handel M.A. Meiotic prophase abnormalities and metaphase cell death in MLH1-deficient mouse spermatocytes: Insights into regulation of spermatogenic progress. Dev. Biol. 2002;249:85–95. doi: 10.1006/dbio.2002.0708. [DOI] [PubMed] [Google Scholar]
- Edelmann W., Cohen P.E., Kane M., Lau K., Morrow B., Bennett S., Umar A., Kunkel T., Cattoretti G., Chaganti R., et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- Fong Y., Bender L., Wang W., Strome S. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science. 2002;296:2235–2238. doi: 10.1126/science.1070790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel M.A., Park C., Kot M. Genetic control of sex-chromosome inactivation during male meiosis. Cytogenet. Cell Genet. 1994;66:83–88. doi: 10.1159/000133672. [DOI] [PubMed] [Google Scholar]
- Hassold T., Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Her C., Wu X., Griswold M.D., Zhou F. Human MutS homologue MSH4 physically interacts with von Hippel-Lindau tumor suppressor-binding protein 1. Cancer Res. 2003;63:865–872. [PubMed] [Google Scholar]
- Khil P.P., Smirnova N.A., Romanienko P.J., Camerini-Otero R.D. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- Kneitz B., Cohen P.E., Avdievich E., Zhu L., Kane M.F., Hou H., Kolodner R.D., Kucherlapati R., Pollard J.W., Edelmann W. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes & Dev. 2000;14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M., Meyers E.N., Martin G.R. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- Li X., Nicklas R.B. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Marcon E., Moens P.B. The evolution of meiosis: Recruitment and modification of somatic DNA-repair proteins. Bioessays. 2005;27:795–808. doi: 10.1002/bies.20264. [DOI] [PubMed] [Google Scholar]
- Matzuk M.M., Lamb D.J. Genetic dissection of mammalian fertility pathways. Nat. Cell Biol. 2002;4 (Suppl.):s41–s49. doi: 10.1038/ncb-nm-fertilitys41. [DOI] [PubMed] [Google Scholar]
- Moens P.B., Kolas N.K., Tarsounas M., Marcon E., Cohen P.E., Spyropoulos B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA–DNA interactions without reciprocal recombination. J. Cell Sci. 2002;115:1611–1622. doi: 10.1242/jcs.115.8.1611. [DOI] [PubMed] [Google Scholar]
- Odorisio T., Rodriguez T.A., Evans E.P., Clarke A.R., Burgoyne P.S. The meiotic checkpoint monitoring synapsis eliminates spermatocytes via p53-independent apoptosis. Nat. Genet. 1998;18:257–261. doi: 10.1038/ng0398-257. [DOI] [PubMed] [Google Scholar]
- Offenberg H.H., Schalk J.A., Meuwissen R.L., van Aalderen M., Kester H.A., Dietrich A.J., Heyting C. SCP2: A major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res. 1998;26:2572–2579. doi: 10.1093/nar/26.11.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S.L., Hawley R.S. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Pittman D.L., Cobb J., Schimenti K.J., Wilson L.A., Cooper D.M., Brignull E., Handel M.A., Schimenti J.C. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- Plug A.W., Peters A.H., Keegan K.S., Hoekstra M.F., de Boer P., Ashley T. Changes in protein composition of meiotic nodules during mammalian meiosis. J. Cell Sci. 1998;111:413–423. doi: 10.1242/jcs.111.4.413. [DOI] [PubMed] [Google Scholar]
- Reinke V., Smith H.E., Nance J., Wang J., Van Doren C., Begley R., Jones S.J., Davis E.B., Scherer S., Ward S., et al. A global profile of germline gene expression in C. elegans. Mol. Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Roeder G.S., Bailis J.M. The pachytene checkpoint. Trends Genet. 2000;16:395–403. doi: 10.1016/s0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- Schalk J.A., Dietrich A.J., Vink A.C., Offenberg H.H., van Aalderen M., Heyting C. Localization of SCP2 and SCP3 protein molecules within synaptonemal complexes of the rat. Chromosoma. 1998;107:540–548. doi: 10.1007/s004120050340. [DOI] [PubMed] [Google Scholar]
- Shiu P.K., Raju N.B., Zickler D., Metzenberg R.L. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- Solari A.J. The behavior of the XY pair in mammals. Int. Rev. Cytol. 1974;38:273–317. doi: 10.1016/s0074-7696(08)60928-6. [DOI] [PubMed] [Google Scholar]
- Tsubouchi T., Zhao H., Roeder G.S. The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev. Cell. 2006;10:809–819. doi: 10.1016/j.devcel.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Turner J.M., Mahadevaiah S.K., Fernandez-Capetillo O., Nussenzweig A., Xu X., Deng C.X., Burgoyne P.S. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- Wang P.J., McCarrey J.R., Yang F., Page D.C. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- Wang P.J., Page D.C., McCarrey J.R. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum. Mol. Genet. 2005;14:2911–2918. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., De La Fuente R., Leu N.A., Baumann C., McLaughlin K.J., Wang P.J. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 2006;173:497–507. doi: 10.1083/jcb.200603063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Kondoh G., Matsuda Y., Habu T., Nishimune Y., Morita T. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]