Abstract
We describe a mutant of Zea mays isolated from a W22 inbred transposon population, widow's peak mutant1 (wpk1), with an altered pattern of anthocyanin synthesis and aleurone cell differentiation in endosperm. In addition, a failure of the developing mutant embryo to form leaf initials is associated with decreased expression of a subset of meristem regulatory genes that includes Abphyl1 and Td1. We show that the viviparous8 (vp8) mutant has a similar pleiotropic phenotype in the W22 inbred background in contrast to the viviparous embryo phenotype exhibited in the standard genetic background, and we confirmed that wpk1 is allelic to vp8. Further genetic analysis revealed that the standard vp8 stock contains an unlinked, partially dominant suppressor of the vp8 mutation that is not present in W22. Consistent with the early-onset viviparous phenotype of vp8, expression of several embryonic regulators, including LEC1/B3 domain transcription factors, was reduced in the mutant embryo. Moreover, reduced abscisic acid (ABA) content of vp8/wpk1 embryos was correlated with altered regulation of ABA biosynthesis, as well as ABA catabolic pathways. The ABA biosynthetic gene Vp14 was down-regulated in the nonsuppressed background, whereas the ZmABA8′oxA1a ABA 8′-hydroxylase gene was strongly up-regulated in both genetic backgrounds. Molecular analysis revealed that Vp8 encodes a putative peptidase closely related to Arabidopsis thaliana ALTERED MERISTEM PROGRAM1. Because the Vp8 regulates meristem development as well as seed maturation processes, including ABA accumulation, we propose that VP8 is required for synthesis of an unidentified signal that integrates meristem and embryo formation in seeds.
In flowering plants, seed development begins with double fertilization generating a diploid zygote that undergoes embryogenesis and a triploid central cell that develops as endosperm. As organogenesis nears completion, the embryo and endosperm enter a maturation phase characterized by developmental arrest and acquisition of dormancy.
Genetic studies in Arabidopsis (Arabidopsis thaliana) and maize (Zea mays) have identified two classes of transcription factors that are essential for seed maturation and dormancy processes. The first class of genes, exemplified by Arabidopsis LEC1 and L1L, encode HAP3-related transcription factors (Lotan et al., 1998; Kwong et al., 2003). The second class of genes, which includes Arabidopsis LEC2, FUS3, and ABI3, as well as maize Viviparous1 (Vp1), encodes B3 domain transcription factors (McCarty et al., 1991; Giraudat et al., 1992; Luerssen et al., 1998; Stone et al., 2001). Loss of function of these genes causes precocious germination of developing seeds (McCarty et al., 1989; Meinke, 1992, 1994; West et al., 1994). Although these genes have overlapping roles in regulation of downstream gene expression mediated by common cis-elements (Suzuki et al., 1997; Monke et al., 2004; Braybrook et al., 2006), differences in time of expression and spatial localization confer differential functions during seed development (Parcy et al., 1997; Nambara et al., 2000; Raz et al., 2001; Brocard-Gifford et al., 2003; Baumbusch et al., 2004; Santos Mendoza et al., 2005; To et al., 2006). Upon germination, expression of the embryonic regulators in seedlings is strictly repressed by the closely related VAL B3 factors (Suzuki et al., 2007).
Control of maturation by the B3 transcription factor network is further determined by interactions with hormone signaling pathways. For example, the ABI3/VP1 transcription factor has a unique capacity to interact with abscisic acid (ABA) signaling conferred by physical interaction with ABI5 (Hobo et al., 1999; Nakamura et al., 2001). This functionality enables integration of the LEC1/B3 network controlling embryogenesis with ABA signaling during the late phase of embryo maturation. FUS3, in turn, has been implicated in developmental regulation of ABA and GA biosynthesis in the seed (Nambara et al., 2000; Curaba et al., 2004; Gazzarrini et al., 2004) through regulation of key genes involved in hormone biosynthesis as well as turnover (for review, see Olszewski et al., 2002; Nambara and Marion-Poll, 2005). Finally, repression of embryonic development by the VAL B3 factors is promoted by an interaction with GA signaling (Suzuki et al., 2007).
Endosperm differentiation and maturation proceed in parallel with embryo development (for review, see Olsen, 2001). The mature endosperm of grass seeds consists of three principal cell types, starchy endosperm cells, aleurone cells, and basal endosperm transfer cells. Maize mutants that affect aleurone differentiation include Defective kernel1 (Dek1), Supernumerary aleurone layers1 (Sal1), and Crinkly4 (Cr4; Becraft et al., 1996, 2002; Lid et al., 2002; Shen et al., 2003). Whereas Dek1 and Sal1 genes are required during early endosperm development, Cr4 is required for aleurone differentiation late in endosperm formation (Becraft and Asuncion-Crabb, 2000). In addition, maize Vp1, an ortholog of Arabidopsis ABI3, is required for embryo maturation (McCarty et al., 1991) as well as activation of the anthocyanin biosynthesis pathway in aleurone cells (Hattori et al., 1992; Carson et al. 1997), indicating that the embryonic B3 genes also function in aleurone differentiation.
Here we describe genetic and molecular analysis of the maize vp8 mutant in embryo and endosperm development. We show that novel widow's peak1 (wpk1) mutations that alter the pattern of aleurone differentiation in the adgerminal region of the endosperm are allelic to vp8. Genetic analyses reveal that the vp8 phenotype is strongly conditioned by genetic background in maize due to action of an unlinked semidominant suppressor locus. Our results suggest that the pleiotropic effects of the vp8 mutation are mediated through regulation of specific meristem and embryonic regulatory genes and by regulation of genes controlling ABA biosynthesis and turnover in the developing seed. Finally, we cloned the Vp8 gene and show that it encodes a putative membrane peptidase closely related to Arabidopsis ALTERED MERISTEM PROGRAM1 (AMP1).
RESULTS
Isolation of wpk1 Mutants
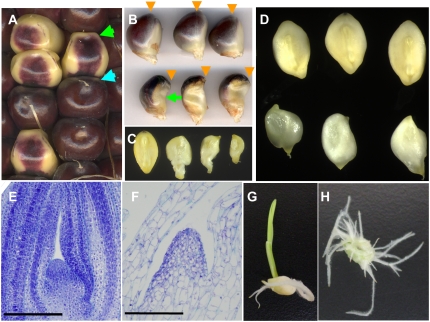
To search for new mutations that affect differentiation of aleurone in maize, we screened the UniformMu inbred transposon-tagging population (McCarty et al., 2005). We identified the wpk1 (wpk1-umu1) mutation that causes a distinctive pattern of pigmentation in the aleurone (Fig. 1A). A deficiency of anthocyanin accumulation in aleurone of wpk1 mutant seeds was most pronounced along the interface of the endosperm and embryo (Fig. 1, A and B) extending around the silk attachment site at the top of the kernel. In addition, the subaleurone endosperm of mature wpk1 kernels had a floury texture, resulting in less vitreous endosperm compared to wild-type kernels (Supplemental Fig. S1). Embryo development in wpk1 seeds was also severely affected. Compared to wild-type embryos, developing mutant embryos had irregular morphology, smaller size (Fig. 1C), and a slightly translucent appearance (Fig. 1D). Mutant embryos of dry seed were necrotic and nonviable (Supplemental Fig. S1). Although the dome structure of the shoot apical meristem (SAM) could be discerned in developing mutant embryos, leaf initials were frequently absent (Fig. 1, E and F). The cells in meristem as well as nonmeristem regions of the wpk1 embryo were strikingly enlarged. The cell enlargement phenotype was already apparent in the wpk1 developing embryo at 12 d after pollination (DAP; Supplemental Fig. S2). Consistent with the defect in the SAM structure of the embryo, the wpk1 mutants rarely formed a coleoptile and invariably failed to form leaves when developing embryos were rescued and grown on sterile culture medium (Fig. 1, G and H). Instead, the cultured mutant embryos formed prolific adventitious roots, suggesting that primary root meristem function was affected as well. We further noted that wpk1 mutant kernels were prone to abort early in development in growing seasons with high temperatures (Supplemental Table S1; i.e. compare the spring and fall seasons). In addition to visible seed abortion, we also detected significantly lower frequencies of wpk1 mutant seeds on self-pollinated heterozygous ears (Supplemental Table S1; e.g. 06S-e; P = 5 × 10−4). This result suggested that transmission of either male and/or female gametophytes carrying the wpk1 mutation was also affected.
Figure 1.
Phenotypes of the wpk1 mutant seeds and seedling. A, Ear segregating wpk1-umu1. Wild-type and wpk1 mutant seeds are indicated by blue and green arrows, respectively. B, Lateral views of mature seeds of wild type (top) and wpk1 (bottom) are shown. The positions of silk attachment are indicated by orange arrows. The green arrow highlights deficiency of anthocyanin accumulation in the germinal region of wpk1. C, A wild-type embryo (left) and three wpk1 embryos are shown. The embryos were excised from the seeds shown in B. D, Three developing embryos for wild type (top) and wpk1 (bottom) are shown. The embryos were excised from the seeds at 20 DAP. E and F, Embryonic SAMs of wild-type (E) and wpk1 (F) embryos at 16 DAP. Scale bars = 200 μm. The multiple embryos were sectioned and examined through histological analysis to confirm the altered development of SAM of the wpk1 mutant embryos. G and H, Seedlings from rescued wild-type control and wpk1 mutant embryos.
Aleurone Development in the wpk1 Mutant
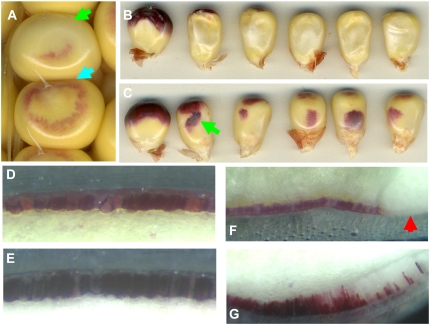
Taking advantage of the genetic background of the UniformMu population, we examined the pattern of aleurone differentiation marked by anthocyanin accumulation in the wpk1 mutant seeds. In wild-type kernels, pigmentation of aleurone was typically visible by 14 DAP in our spring field conditions, initiating in a ring of aleurone cells (Fig. 2A). In the wpk1 mutant, the onset of anthocyanin accumulation was delayed by up to several days and typically failed to occur at all in aleurone cells surrounding the embryo on the adgerminal face of the kernel. Within several days following the onset of pigment accumulation, the crown and adgerminal regions of wild-type kernels were completely pigmented, whereas the lower portion of the abgerminal aleurone remained colorless (Fig. 2B). In contrast to wild type, pigmentation of the abgerminal aleurone of wpk1 mutant seed was frequently enhanced relative to wild type, forming irregular patches of pigmented aleurone (Fig. 2C). This result suggested that Wpk1 regulation of anthocyanin accumulation is region specific.
Figure 2.
Aleurone development in wpk1 seeds. A, Developing wild-type (blue arrow) and wpk1 mutant (green arrow) seeds at 14 DAP are shown. B, Adgerminal views of developing wild-type embryo (left) and five wpk1 seeds (right) at 26 DAP. C, Abgerminal views of 26-DAP seeds shown in B. Patches of precocious anthocyanin accumulation are indicated by a light green arrow. D to G, The aleurone layer in abgerminal (D and E) and germinal (F and G) regions of wild-type (D and F) and wpk1 (E and G) kernels are shown. A red arrow indicates the position of the embryo in wild-type seed.
Maize Cr4, which encodes a plasma membrane receptor kinase, is a positive regulator of abgerminal aleurone differentiation in endosperm (Becraft et al., 1996). To test whether Cr4 and Wpk1 genetically interact in aleurone differentiation, we constructed cr4 wpk1 double-mutant seeds to examine pigmentation and patterning of the aleurone (Supplemental Fig. S3). The double-mutant seeds exhibited anthocyanin deficiencies in both adgerminal and abgerminal regions. The aleurone phenotype was consistent with an additive superposition of the cr4 and wpk1 single-mutant patterns of anthocyanin accumulation in the aleurone. This result implies that these two genes most likely function independently in aleurone development. The opposite polarities of the vp8 and cr4 phenotypes with respect to the adgerminal/abgerminal axis of the endosperm suggest that development of adgerminal and abgerminal domains may be separately regulated (Becraft and Asuncion-Crabb, 2000).
In addition to the distinctive pattern of anthocyanin pigmentation, the size and morphology of aleurone cells was strongly affected in wpk1 mutant seeds. The wpk1 mutant aleurone cells in the abgerminal, as well as adgerminal, region were markedly elongated in the anticlinal direction, but not in the periclinal plane (Fig. 2, D–G). Measurement of cell lengths of 15 cells shown in Figure 2E indicated an average length 1.6-fold greater than comparable aleurone cells in wild-type kernels. This result indicated that the Wpk1 gene suppresses anticlinal cell expansion of aleurone cells of the endosperm. Whereas cell elongation was most pronounced in the anthocyanin-deficient cells proximal to the embryo, slight elongation of aleurone cells was detected throughout the aleurone of wpk1 seeds, suggesting that cell size and anthocyanin accumulation patterns may be independently regulated by the Wpk1 gene.
Expression of Meristem-Related Genes in wpk1 Developing Embryo
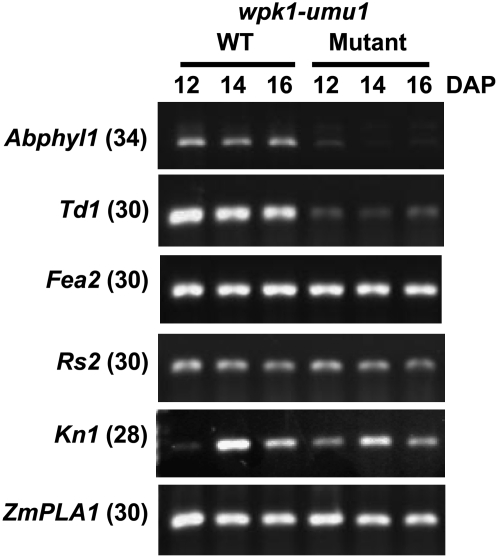
Because the wpk1 mutation significantly altered organization of the embryonic SAM, we determined whether expression of genes that are known to regulate SAM formation in grasses was affected during mid-to-late embryo development (12, 14, and 16 DAP) when wild-type W22 embryos continue to produce new leaf primordia (Supplemental Fig. S2). Interestingly, the wpk1 mutant affected expression of a subset, but not all, of SAM markers (Fig. 3). Expression of Abnormal phyllotaxy1 (Abphyl1; Asakura et al., 2003; Giulini et al., 2004) and Thick tassel dwarf1 (Td1; Bommert et al., 2005) genes was significantly reduced in the wpk1 developing embryo, whereas, on a total RNA basis, Fasciated ear2 (Fea2), Knotted1 (Kn1), Rough sheath2 (Rs2), and maize PLASTOCHRON1-like (ZmPLA1) genes (Hake et al., 1989; Timmermans et al., 1999; Tsiantis et al., 1999; Taguchi-Shiobara et al., 2001; Miyoshi et al., 2004) were expressed at comparable levels in wild-type and wpk1 embryos. In a replicate experiment using embryo samples harvested in a separate growing season (Supplemental Fig. S4), Abphyl1, Td1, Fea2, and ZmPLA1 genes showed similar patterns of expression, whereas expression of Kn1 and Rs2 genes was slightly reduced, suggesting an environmental effect on the gene expression. In any case, these results suggest that, in spite of having a strongly pleiotropic phenotype, the wpk1 mutation causes limited alterations in gene expression during meristem development.
Figure 3.
Expression of meristem-related genes in wpk1 embryos. RT-PCR analysis of meristem-related genes in developing wild-type and wpk1 mutant embryos at 12, 14, and 16 DAP. The numbers in parentheses indicate the numbers of cycles in the RT-PCR reactions. The cycle numbers were optimized to quantitatively examine expression for each gene.
The wpk1 Mutant Is Allelic to vp8
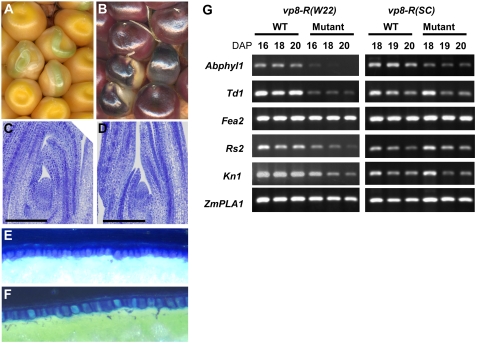
In a parallel study of the vp8 mutant (Robertson, 1955), we introgressed the reference vp8-R allele into the W22 inbred. In the original genetic background, vp8-R has a viviparous seed phenotype with a heterochronic effect on vegetative development (Evans and Poethig, 1997). We noticed that, in the W22 background, vp8-R conditioned a defective embryo rather than a viviparous phenotype and that pigmentation of the endosperm resembled the pattern observed in wpk1 kernels (Fig. 4, A and B). Genetic complementation tests performed by crossing mutant heterozygotes confirmed allelism of vp8-R and wpk1-umu1 (hereafter vp8-umu1) based on noncomplementation of the seed phenotype. Because the genetic background of the vp8-R stock distributed by the Maize Genetics Cooperation Stock Center is not documented, we use the designations vp8-R (SC) and vp8-R (W22) to distinguish the genotypes conferring viviparous and defective embryo phenotypes, respectively.
Figure 4.
Phenotypic analysis of vp8 seeds. A and B, Mature ears segregating vp8-R in SC (A) and W22 inbred (B) genetic backgrounds. C and D, Embryonic SAMs in developing wild-type and vp8-R (SC) embryos at 16 DAP. Scale bars = 200 μm. E and F, Abgerminal aleurone of wild-type and vp8-R mutant endosperms. G, RT-PCR analysis of meristem-related genes in vp8-R (SC) and vp8-R (W22) embryos. The RT-PCR condition for each gene was identical to that in Figure 3. The plants were grown simultaneously in a greenhouse.
To understand the basis for the altered expression of vp8-R in the SC and W22 backgrounds, we performed a characterization of embryo and aleurone development in vp8-R (SC) and vp8-R (W22) seeds and compared their phenotypes to vp8-umu1. In contrast to the vp8-R and vp8-umu1 mutations in the W22 background, vp8-R (SC) mutant embryos were viable if rescued prior to desiccation as previously reported (Evans and Poethig, 1997; Supplemental Fig. S3). The mutant was capable of forming a well-organized SAM and leaf initials with comparable size of the cells in the developing embryo (Fig. 4, C and D). The aleurone layer of vp8-R (SC) kernels did not show marked differences from wild type (Fig. 4, E and F). Whereas the absence of C1 and R1 alleles required for anthocyanin biosynthesis prevented evaluation of pigmentation patterns in the SC background, mutant aleurone cells were elongated only slightly compared to wild type (1.15-fold longer in the anticlinal direction compared to the wild-type control). Furthermore, unlike vp8-umu1, we did not detect increased early seed abortion or less than expected frequencies of viviparous seeds on the self-pollinated ears (Supplemental Table S2).
To compare meristem function in the SC and W22 backgrounds, expression profiles of meristem marker genes in developing vp8-R (SC) and vp8-R (W22) embryos were determined by reverse transcription (RT)-PCR (Fig. 4G). In the SC background, all of the meristem markers, except Abphyl1, showed comparable expression profiles in wild-type and mutant embryos. In marked contrast to vp8-R (W22) and vp8-umu1 embryos, vp8-R (SC) showed no evidence of altered expression of Td1. Whereas Abphyl1 expression was reduced in both the W22 and SC backgrounds, the effect was more pronounced in the W22 background. The comparatively subtle changes in gene expression of meristem-related genes in the SC background were consistent with the observation that the vp8-R (SC) mutant embryo was able to develop a functional SAM. Similar to vp8-umu1, Fea2 and ZmPLA1 genes were unaffected by vp8-R in both the W22 and SC backgrounds. Expression of Rs2 and Kn1 genes was slightly decreased in vp8-R (W22) embryos compared to wild type. These subtle differences are most likely due to environmental effects on embryo development because we detected similar differences in the vp8-umu1 between two seasons (Fig. 3; Supplemental Fig. S4). Overall, these results indicated that genetic background differences between W22 and SC had a much more profound effect on the vp8 phenotype than allele differences.
The SC Genetic Background Contains a Partially Dominant Suppressor of vp8
To identify genetic factors that interact with the vp8 mutation in W22 and SC genetic backgrounds, we analyzed the seed and seedling phenotypes of F1 plants generated by reciprocal crosses between vp8-R (SC) and vp8-umu1. All of the heteroallelic vp8 mutant seed from crosses made in either direction developed embryos comparable in size to wild type (Table I; Supplemental Fig. S5). The characteristic wpk1 phenotypes, including severe defective embryo and patterned anthocyanin deficiency, were not observed in the F1 seed. These results indicated that the SC background is able to suppress the wpk1 defective embryo sufficiently to produce a well-developed embryo that is in some cases viviparous. In spring (warm) as well as fall (cool) field environments, viviparous embryos were frequently observed in the F1 seeds derived from crosses between vp8-R (SC) females and vp8-umu1 male parents, whereas F1 seed from the reciprocal cross predominantly had an intermediate phenotype with near full-size embryos that were weakly viviparous. The qualitative difference in the phenotypes of reciprocal F1 hybrid seed suggested that either maternal factors or gametophytic transmission affected vp8 function in the developing seed. In this respect, it is noteworthy that, in some of the crosses involving vp8-R (W22) made in either direction, mutant seed was recovered at a lower than expected frequency consistent with Vp8 having a function in both male and female gametophytes (Table I).
Table I.
F1 seed phenotypes from heteroallelic crosses between vp8-R (SC) and vp8-umu1
06F and 06S represent the 2006 fall and 2006 spring seasons, respectively. Mutant embryos were classified as viviparous, intermediate, and wpk (aborted) based on the extent of shoot and scutellum development, as indicated in Supplemental Figure S3. χ2 tests and P values were calculated for a model that assumed that normal and all mutant classes (viviparous + intermediate + wpk) segregate 3:1.
| Female | Male | Normal | Mutant
|
P Value | ||
|---|---|---|---|---|---|---|
| Viviparous | Intermediate | wpk | ||||
| 06F +/vp8-R (SC) | 06F +/vp8-umu1 a | 130 | 32 | 0 | 0 | 0.12 |
| 06F +/vp8-R (SC) | 06F +/vp8-umu1 c | 65 | 11 | 0 | 0 | 0.03 |
| 06F +/vp8-R (SC) | 06F +/vp8-umu1 | 127 | 29 | 0 | 0 | 0.06 |
| 06F +/vp8-umu1 | 06F +/vp8-R (SC) | 153 | 0 | 29 | 0 | 4.7 × 10−3 |
| 06F +/vp8-umu1 | 06F +/vp8-R (SC) | 123 | 0 | 29 | 0 | 0.09 |
| 06F +/vp8-umu1 | 06F +/vp8-R (SC) | 139 | 1 | 22 | 0 | 1.5 × 10−3 |
| 06S +/vp8-R (SC) | 06S +/vp8-umu1 | 92 | 33 | 1 | 0 | 0.61 |
| 06S +/vp8-R (SC) | 06S +/vp8-umu1 | 83 | 5 | 0 | 0 | 2.9 × 10−5 |
| 06S +/vp8-R (SC) | 06S +/vp8-umu1 | 68 | 17 | 0 | 0 | 0.29 |
| 06S +/vp8-umu1 | 06S +/vp8-R (SC) | 99 | 3 | 3 | 0 | 5.0 × 10−6 |
| 06S +/vp8-umu1 | 06S +/vp8-R (SC) | 87 | 3 | 15 | 0 | 0.06 |
| 06S +/vp8-umu1 | 06S +/vp8-R (SC) | 150 | 1 | 10 | 0 | 1.0 × 10−7 |
To determine whether embryos with strong and intermediate viviparous phenotypes were viable and capable of growing into seedlings, we rescued these mutant embryos and placed them in sterile culture. Unlike vp8-umu1, the viviparous vp8-R (SC) mutants and heteroallelic F1 mutant embryos developed shoots at >90% frequency (Supplemental Fig. S6).
The results of reciprocal crosses indicated that one or more dominant genetic factors in the SC background partially suppress the severe developmental defects caused by the vp8 mutation in W22. To estimate the number of genetic loci involved in suppression of vp8 in the SC background, we analyzed the phenotypes of F2 seeds generated from the heteroallelic F1 seeds as well as of F2 seeds from backcrosses of vp8-R (SC) with W22 (Table II). The mutant F2 seeds were grouped in three phenotype classes: strongly viviparous, intermediate, and wpk-like, respectively. Within the mutant class, the viviparous plus intermediate and wpk-like embryos occurred in a ratio that was consistent with segregation of a single, unlinked partially dominant suppressor. The SC and W22 inbreds are evidently homozygous for dominant and recessive alleles of the suppressor, respectively.
Table II.
F2 seed phenotypes from self-crosses of F1 backcrossed seeds and heteroallelic seeds
Fall season crosses (2005 [05F] and 2006 [06F]) were analyzed. The P values were determined from χ2 tests of a model assuming segregation of an incompletely dominant suppressor (i.e. the class of viviparous and intermediate phenotypes combined, and the wpk class segregate 3:1). The vp8 alleles segregating in F2 of heteroallelic hybrids were not distinguished.
| Female | Male | Normal | Mutant
|
P Value | ||
|---|---|---|---|---|---|---|
| Viviparous | Intermediate | wpk | ||||
| 05F +/+ (W22) | 05F +/vp8-R (SC) | 316 | 41 | 25 | 26 | 0.47 |
| 05F +/+ (W22) | 05F +/vp8-R (SC) | 326 | 40 | 45 | 18 | 0.08 |
| 05F +/+ (W22) | 05F +/vp8-R (SC) | 340 | 42 | 0 | 16 | 0.65 |
| 06F +/vp8-R (SC) | 06F +/vp8-umu1 | 350 | 17 | 28 | 15 | 1 |
| 06F +/vp8-R (SC) | 06F +/vp8-umu1 | 330 | 38 | 29 | 14 | 0.11 |
| 06F +/vp8-R (SC) | 06F +/vp8-umu1 | 232 | 35 | 19 | 18 | 1 |
Altered Regulation of ABA Synthesis and Turnover in the vp8 Mutant
The vp8-R (SC) genotype has been previously reported to have moderately reduced levels of ABA in the developing embryo (Neill et al., 1986), suggesting that ABA deficiency may contribute to the viviparous phenotype. We observed that viviparous vp8-R (SC) mutant embryos could be distinguished from wild-type embryos as early as 16 DAP consistent with the timing of ABA biosynthesis in maize embryos (Tan et al., 1997). However, the pleiotropic phenotypes of vp8-R (SC) exhibited during vegetative development are not readily attributed to ABA deficiency (Evans and Poethig, 1997).
To examine the potential role of hormone biosynthesis in the complex phenotype of vp8 mutants, we analyzed levels of three key plant hormones, ABA, auxin, and cytokinin, in developing embryo and endosperm tissues of wild-type and mutant seeds. Consistent with the previous report by Neill et al. (1986), the amount of ABA was significantly reduced in vp8-R (SC) developing embryo at 16 DAP (Fig. 5A), whereas auxin and cytokinin levels did not show clear differences between mutant and wild type (Supplemental Table S3). Moreover, consistent with the enhanced vp8 phenotype in the W22 background, accumulation of ABA in vp8-umu1 embryos was dramatically lower compared to the vp8-R (SC) and wild-type genotypes at 14 and 16 DAP (Fig. 5A; Supplemental Table S3). Whereas ABA levels measured on a fresh-weight basis in the SC and W22 backgrounds correlated with the severity of the mutant phenotype, we cannot rule out the possibility that this difference is due to indirect effect caused by the profound developmental defects of vp8 (W22) embryos. We did not detect significant differences in hormone levels in endosperms of wild-type and vp8 seed (Supplemental Table S3), indicating that Vp8 specifically affects ABA accumulation in the embryo.
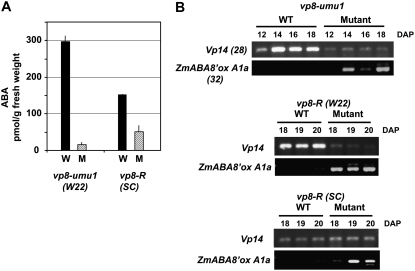
Figure 5.
ABA content and expression of Vp14 and ZmABA8′oxA1a in vp8 embryos. A, ABA content of developing wild-type (W) and vp8 mutant embryos (M) at 16 DAP. Error bars indicate the mean deviation of replicate experiments. B, RT-PCR analysis of Vp14 and ZmABA8′ox A1a expression in vp8 embryos in the W22 and SC genetic backgrounds. Total RNA was prepared from embryos of field-grown vp8-umu1 developing seeds and greenhouse-grown vp8-R (W22) and vp8-R (SC) seeds.
To gain insight into how ABA accumulation is regulated by Vp8, we analyzed expression of key genes, Vp14 and ZmABA8′oxA1a, that are implicated in control of ABA biosynthesis and degradation, respectively. The Vp14 gene encodes the major 9-cis-carotenoid dioxygenase expressed during maize embryo development (Tan et al., 1997), and ZmABA8′oxA1a is an ortholog of the ABA 8′-hydroxylases (Kushiro et al., 2004; Saito et al., 2004; Millar et al., 2006; Okamoto et al., 2006; Yang and Choi, 2006; Yang and Zeevaart, 2006; Saika et al., 2007) that catalyzes the first step in catabolism of ABA. As shown in Figure 5B, Vp14 expression was significantly lower in developing vp8-umu1 (W22) embryos, whereas expression of Vp14 in vp8-R (SC) embryos was similar to wild type. In contrast, ZmABA8′oxA1a expression was markedly elevated in vp8 mutant embryos in both the SC and W22 genetic backgrounds. These results indicate that the severe ABA deficiency evident in the W22 background correlates with simultaneous down-regulation of ABA biosynthesis and up-regulation of ABA catabolism pathways, whereas the moderate ABA deficiency conditioned by vp8 in the SC background is primarily due to elevated ABA catabolism. A key implication is that the dominant suppressor in the SC background restores regulation of ABA biosynthesis, but not repression of ABA catabolism. These results are consistent with the independent evidence that both ABA biosynthesis and turnover contribute to regulation of seed development and germination (for review, see Nambara and Marion-Poll, 2005).
Expression of Embryonic Regulators in the vp8 Mutant
Although the reduced ABA accumulation in vp8 mutant embryos is consistent with the viviparous phenotype, ABA deficiency alone seems unlikely to account for the pleiotropic phenotypes of vp8 in embryo, aleurone, and vegetative organs. To better understand the complex embryo phenotype, we analyzed expression of the LEC1-related factors and B3 domain transcription factors that regulate embryogenesis and maturation. In Arabidopsis, loss-of-function mutations at four loci, lec1, lec2, fus3, and abi3, prevent embryo maturation and induce a potential for viviparous seed development. The ABI3 gene and its maize ortholog, Vp1, are required for ABA-regulated gene expression late in seed development, whereas lec1, lec2, and fus3 genes affect earlier stages of seed development. Because aspects of the vp8 phenotype are manifest at early stages of seed development, we considered the possibility that Vp8 may also interact with the early-acting regulators of embryogenesis in maize.
To develop RT-PCR assays for expression of the maize embryo pathway, we searched available genome and EST databases to identify maize orthologs of the Arabidopsis HAP3 and B3 transcription factor genes. We identified three homologs of the Arabidopsis LEC1 and L1L HAP3-related genes (ZmLEC1, ZmL1La, and ZmL1Lb, respectively) based on the similarity of the HAP3 domains (M. Suzuki, unpublished data). A LEC1 ortholog identical to ZmL1La has been described previously (Zhang et al., 2002). Based on alignments of B3 domain sequences, we identified a single maize gene with roughly equal similarity to the Arabidopsis FUS3 and LEC2 genes, which we designated ZmFUS3.
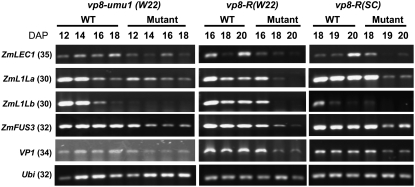
As shown in Figure 6, expression of ZmLEC1, ZmL1La, ZmL1Lb, ZmFUS3, and Vp1 was reduced in vp8 mutant embryos in both W22 and SC backgrounds. Whereas all five genes showed quantitatively lower expression in vp8 embryos late in development (18 and 20 DAP), ZmL1Lb expression, which was not detected in vp8 embryos after 18 DAP, showed the most striking qualitative difference between wild type and vp8 common to both backgrounds. Interestingly, the ZmL1La and ZmL1Lb genes were differentially expressed in the wild-type SC and W22 backgrounds. ZmL1La expression was relatively high compared to ZmL1Lb throughout development in SC embryos, whereas ZmL1Lb and ZmL1La were expressed at similar levels in W22 embryos. Hence, among the embryogenesis regulators tested, only ZmL1Lb and ZmL1La exhibited expression differences that correlated with partial suppression of the vp8 phenotype. In any case, because vp8 embryos in the suppressed background are fully formed, the qualitative differences in expression of the embryo pathway genes are likely to be caused by specific action of the Vp8 gene, but not by nonspecific effects due to gross morphological defects in embryo formation. This interpretation is further supported by the observation that expression of ZmL1Lb, in contrast to the other embryonic regulators, was clearly reduced in the vp8-R (SC) mutant at 18 DAP under greenhouse conditions. Although the viviparous phenotype of greenhouse-grown vp8-R (SC) embryos is barely discerned at 18 DAP, embryo genotypes were confirmed by subsequent RT-PCR analysis for the presence of detectable Vp8 mRNA (see Fig. 7B).
Figure 6.
Expression of LEC1/B3 embryonic genes in developing maize embryos. RT-PCR analysis of LEC1/B3 genes in vp8 developing embryos. Numbers in parentheses indicate the numbers of cycles in the RT-PCR reactions. The cycle numbers were optimized to quantitatively examine expression for each gene. The Ubiquitin (Ubi) gene was used as a control marker.
Figure 7.
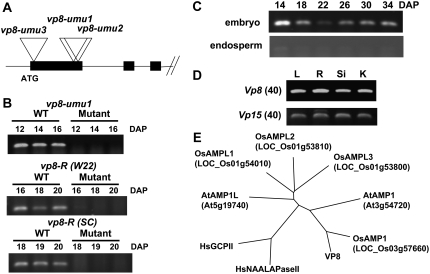
Structure and expression of the Vp8 gene. A, Three independent Mu-tagged alleles of vp8 mutant from the UniformMu population are shown. B, RT-PCR analysis of Vp8 expression in developing embryos of vp8 mutants. These results verified the genotype of embryos that were used for our RT-PCR analyses (Figs. 3–6) including embryo of vp8-R (SC) at 18 DAP. The 28 PCR cycles were run in the RT-PCR reactions for the Vp8 gene. C, RT-PCT analysis of Vp8 gene expression in embryo and endosperm of developing maize seeds. The 28 PCR cycles were run in the RT-PCR reactions for the Vp8 gene. D, RT-PCR analysis of expression of Vp8 gene in various plant tissues. The Vp15 gene (Suzuki et al., 2006) was used for control. The 40 cycles of PCR were run in the RT-PCR reactions, as indicated in the parentheses for both genes, to detect amplified products. L, Leaf from 10-d-old seedling; R, primary root from germinating seeds; Si, unpollinated silk; K, whole kernel (6 DAP). E, Unrooted ClustalW tree of AMP1/VP8-related proteins from maize (VP8), Arabidopsis (AtAMP1, AtAMP1L), rice (OsAMP1, OsAMPL1-3), and human glutamate carboxypeptidases (HsGCPII, HsNAALAPaseII).
Cloning of the Vp8 Gene by Transposon Tagging Performed in Silico
Our screen of the UniformMu population (McCarty et al., 2005) yielded three additional independent vp8 alleles that were confirmed by genetic complementation tests (designated vp8-umu2, vp8-umu3, and vp8-umu4, respectively). No consistent differences in the embryo and endosperm phenotypes could be discerned among a total of five vp8 alleles observed in the W22 background. Skewed F2 segregation ratios from self-pollinated heterozygotes were detected for multiple alleles (Supplemental Table S2). Moreover, RT-PCR analysis of meristem genes, ABA biosynthesis pathway genes, and LEC1/B3 embryo markers showed similar patterns as described for vp8-umu1 and vp8-R alleles (Supplemental Fig. S7).
To clone the vp8 locus, we performed MuTAIL-PCR (Settles et al., 2004), high-throughput sequencing, and bioinformatics cluster analysis (McCarty et al., 2005; Supplemental Information S1) to screen for allelic Mu insertions in the vp8-umu1, vp8-umu2, and vp8-umu3 mutants. Bioinformatics analysis detected overlapping MuTAIL sequences from the vp8-umu1 and vp8-umu2 lines that were derived from closely spaced insertions in a maize gene. Using gene-specific PCR, we confirmed the presence of a third nearby Mu insertion in the vp8-umu3 allele. Analysis of the flanking genomic sequences by BLASTX (Altschul et al., 1997) detected similarity to glutamate carboxypeptidases from various organisms, including homologs of the Arabidopsis AMP1 (Helliwell et al., 2001).
We performed RT-PCR to analyze expression of Vp8 in wild-type and vp8 mutant embryos (Fig. 7B). A Vp8 transcript was detected in the W22 inbred but not in vp8-umu1 and vp8-umu2 mutants, indicating that the transposon insertions disrupted transcription or mRNA stability. Although the molecular lesion in the vp8-R allele was not determined, the reference allele was also null for mRNA expression based on RT-PCR. In the developing seed of wild type, expression of the Vp8 gene was markedly higher in the embryo than in endosperm on a total RNA basis (Fig. 7C). Vp8 expression was detected as early as in 6-DAP developing seed, as well as in various vegetative tissues at lower levels (Fig. 7D), consistent with AMP1 gene expression in Arabidopsis (Helliwell et al., 2001; Vidaurre et al., 2007; Schmid et al., 2005).
Structure of the VP8 Protein
To determine the complete sequence of the VP8 protein, we isolated and sequenced a full-length cDNA of Vp8 mRNA by RT-PCR using RNA prepared from W22 developing embryos. We designed primers based on maize genome survey sequences that contained the predicted 5′ and 3′ untranslated regions of the gene. The cDNA sequence predicted a protein of 714 amino acids that aligns with two membrane-localized glutamate carboxypeptidases from human (Israeli et al., 1993; Pangalos et al., 1999). Comparison of amino acid sequences of the human glutamate carboxypeptidases and several plant homologs suggested that plants have two subfamilies of the peptidase-like proteins (Fig. 7D). In the tree, maize VP8, Arabidopsis AMP1, and rice (Oryza sativa) Os03g57660 form a distinct subfamily. In Arabidopsis, AMP1 has been shown to regulate shoot meristem development (Chaudhury et al., 1993; Conway and Poethig, 1997; Helliwell et al., 2001). To search for potential AMP/Vp8-related genes in maize, we analyzed all publicly available maize sequences, including a near-complete draft sequence of the whole maize genome (including 15,750 phase I bacterial artificial chromosome sequence assemblies; www.maizesequence.org). No other candidates for related maize paralogs were detected.
DISCUSSION
Our results show that maize Vp8 encodes a putative membrane-localized peptidase that is closely related to Arabidopsis AMP1. Loss of Vp8 function causes either lethality or precocious germination of the developing embryo, depending on the genetic background. The vp8 mutant is highly pleiotropic in the W22 inbred, indicating that the gene is essential for a wide range of developmental processes in maize. In the developing embryo, the Vp8 gene is required for expression of LEC1/B3 embryonic regulators, as well as for genes that regulate ABA synthesis and turnover. Vp8 effects on SAM organization are associated with reduced expression of a specific subset of meristem-related genes. Finally, we have identified a partially dominant suppressor that genetically interacts with vp8 in regulation of plant development in maize.
Although it remains to be determined whether maize Vp8 is capable of complementing the Arabidopsis amp1 mutant, the similarities in the pleiotropic phenotypes of the amp1 and vp8-R (SC) mutants, as well as in the protein structures of AMP1 and VP8, suggest that these genes are likely orthologs. Whereas the precise mechanisms of SAM development and leaf differentiation are thought to be distinct in Arabidopsis and maize (Scanlon, 2000; Tsiantis and Hay, 2003; Champagne and Sinha, 2004), amp1 and vp8-R (SC) mutants, respectively, cause acceleration of leaf formation in both species (Chaudhury et al., 1993; Evans and Poethig, 1997). AMP1 suppression of lateral root formation (Vidaurre et al., 2007) is also consistent with Vp8 function (Fig. 1). In addition, the amp1 mutant gametophytes are less capable of producing seeds than wild type (Chaudhury et al., 1993), as observed in the vp8 mutant. Although amp1 does not consistently cause precocious germination, a possibly related phenotype, ectopic leaf initiation, has been described in the developing seeds (Conway and Poethig, 1997). Evans and Poethig (1997) noted observation of viviparous seeds from an amp1 allele. Furthermore, inspection of microscopy images of amp1 mutant seed (Mordhorst et al., 1998) indicates the presence of enlarged aleurone cells, consistent with the vp8 (W22) phenotype in maize. Structurally, VP8 is most similar to LOC_Os03g57660, the apparent rice candidate ortholog of Arabidopsis AMP1. Whereas our searches of maize sequence databases did not detect any additional members of the AMP1/VP8/LOC_Os03g57660 group in the maize genome, we detected evidence of other maize AMP1-like peptidases belonging to the AMP1-like family composed of At5g19740 (AMP1L) and three rice AMP1L proteins.
Although amp1 and vp8 have analogous phenotypes, the mutants differ in their reported effects on hormone synthesis. In Arabidopsis, amp1 mutant seedlings are reported to have elevated cytokinin levels compared to wild type (Chaudhury et al., 1993; Saibo et al., 2007), whereas we did not detect significant differences in cytokinin levels in wild-type and vp8 mutant embryos of maize. This discrepancy may well be due to the different tissues analyzed in the two species (seedling versus embryo). Cytokinin biosynthesis is evidently tightly regulated by spatial and temporal signals. In Arabidopsis and cereals, cytokinin turnover, as well as cytokinin responses, has been shown to be highly localized in plant tissues at various developmental stages (D'Agostino et al., 2000; Werner et al., 2003; Higuchi et al., 2004; Miyawaki et al., 2004; Takei et al., 2004; Hutchison et al., 2006; Riefler et al., 2006). For instance, in maize, expression of Abphyl1, which encodes a negative regulator of cytokinin signaling, is restricted to a subdomain of the embryonic SAM (Giulini et al., 2004). A cytokinin-activating enzyme encoded by rice LOG is expressed in a similar pattern in the vegetative SAM (Kurakawa et al., 2007). Whereas we show a predominant effect of vp8 on ABA synthesis and turnover in maize embryos, the hormone levels, including ABA content, have not been determined in the amp1 developing seeds. To precisely compare function of AMP1 and Vp8 in hormone accumulation, a more comprehensive and extensive analysis of hormone quantification at the equivalent stages of development, as well as at the cellular level, will be required.
Among the embryonic regulatory genes we analyzed, ZmL1Lb shows the earliest detectable difference in the expression in vp8 mutant embryos prior to discernible vivipary, thus suggesting that ZmL1Lb may be a primary target of unidentified factors derived from Vp8 function. In Arabidopsis, LEC1 has been proposed to be an upstream activator for FUS3 and ABI3 B3 domain genes (Kagaya et al., 2005). Therefore, decreased expression of ZmL1La and ZmL1Lb could account for down-regulation of ZmFUS3 and Vp1. Interestingly, lec1/fus3/lec2 and amp1/vp8 class mutants, respectively, cause heterochronic shifts in development, although apparently in opposite directions. In contrast to lec1, lec2, and fus3 mutants, amp1 does not cause ectopic development of trichomes on the cotyledons (Chaudhury et al., 1993; Keith et al., 1994; Meinke et al., 1994; West et al., 1994). Similarly, in maize, vp8 delays the expression of adult vegetative traits in leaves extending juvenile development (Evans and Poethig, 1997). Hence, there is not a simple relationship between heterochronic vegetative phenotypes and reduced expression of the LEC1/B3 genes in vp8 embryos. Similar pleiotropy is evident in regulation of Abphyl1 expression. Although loss of Abphyl1 function causes fasciation of the SAM in developing embryos as well as in plants (Jackson and Hake, 1999), vp8-R (SC) mutant plants do not show evidence of fasciation. We cannot completely rule out the possibility that the early decrease in ZmL1Lb and Abphyl1 expression may be caused indirectly by subtle morphological changes in the mutant embryo.
The down-regulation of LEC1/B3 genes in the vp8 may account for regulation of ABA accumulation as well as affect maturation-related gene expression. FUS3 has been shown to regulate ABA accumulation in developing seeds of Arabidopsis (Nambara et al., 2000; Gazzarrini et al., 2004). Hence, the up-regulation of the ZmABA8′oxA1a gene may be caused by reduced ZmFUS3 expression. Interestingly, the 5′ regions of maize and Arabidopsis ABA 8′-hydroxylase genes have multiple Sph/RY motifs (data not shown), which have been shown to mediate FUS3 binding (Reidt et al., 2000). The inference that ABA level is controlled indirectly through regulation of seed-specific factors is consistent with the fact that vp8 mutant plants lack phenotypes associated with ABA deficiency. The observation that vp8 (SC) embryos respond to ABA with normal sensitivity (Robichaud et al., 1980) suggests that seed-specific ABA deficiency accounts for the viviparous phenotype. Moreover, the floury endosperm and translucent embryo phenotypes of vp8, which are frequently associated with lower protein content in cereal seeds (for review, see Lopes and Larkins, 1993), are consistent with the broader role of LEC1/B3 regulators in gene expression for seed storage proteins and lipid accumulation (Keith et al., 1994; Meinke et al., 1994; Parcy et al., 1994; West et al., 1994; Nambara et al., 1995).
Whereas vp8 mutation alters development and gene expression in meristem as well as nonmeristem tissues of the embryo, it is not yet clear whether the dual effects are mediated by the same or distinct independent mechanisms. One possibility is that AMP1/Vp8 establishes a regulatory field that is interpreted locally to produce diverse responses in different tissues. In the meristem of Arabidopsis, AMP1 function is implicated in cytokinin signaling (Chaudhury et al., 1993; Helliwell et al., 2001). We have shown that Vp8 is required for normal expression of the Abphyl1 cytokinin response regulator during maize embryo development. This result suggests that Vp8 may at least indirectly regulate cytokinin signaling at a cellular level in the SAM of maize embryos. In another context, Vidaurre et al. (2007) have recently shown that the MP/ARF5 transcription factor interacts locally with AMP1 function in SAM development. Thus, a series of domain-specific transcription factors may interact with a ubiquitous AMP1/Vp8 function to precisely regulate meristem development. Likewise, LEC1/B3 genes, which are primarily expressed in nonmeristem tissues of the embryo, regulate diverse downstream targets, including seed storage protein genes under the apparent influence of AMP1/Vp8 function.
The isolation of vp8 alleles in the nonsuppressed W22 inbred background proved crucial to uncovering the unexpectedly broad and essential role of Vp8 in embryo and endosperm development. One of the notable findings is that Vp8 is likely required for normal cell division and expansion processes in the developing embryo as well as in the aleurone. Although Arabidopsis AMP1 is expressed throughout the tissues, expression of this gene is relatively higher in rapidly dividing tissues such as in shoot and root meristems (Schmid et al., 2005), implying involvement of AMP1/Vp8 gene function in cell division. Moreover, the nonsuppressed background effect enabled us to identify a semidominant suppressor of vp8 that rescues a discrete subset of phenotypes observed in the developing seed. The molecular basis for suppression is not known. Southern-blot analysis and searches of maize sequence databases failed to detect any closely related locus that correlated with the suppressed phenotype, suggesting that the suppressor is unlikely a partially redundant duplicate gene in the maize genome (data not shown). The evidence that the suppressor restores a subset of Vp8 functions (e.g. activation of Vp14 but not down-regulation of ZmABA8′oxA1a) suggests that the suppressor functions in the same pathway, but is not functionally redundant with Vp8. In addition, the differential effect on expression of Vp14 and ZmABA8′oxA1a by the suppressor suggests that activation of ABA synthesis and repression of ABA catabolism pathways are mediated by distinct Vp8-dependent mechanisms.
Our finding of a suppressor, together with the highly pleiotropic nature of the vp8 phenotype, suggests that a search for other interacting genes may be fruitful. Other genes that interact with Vp8 function in cereal seed development have so far not been identified. Whereas the maize terminal ear1 (te1), rice pla1, and pla2 mutants have accelerated leaf formation similar to that seen in vp8-R (SC) plants during vegetative development, no differences in seed development have been described in these mutants (Itoh et al., 1998; Veit et al., 1998; Miyoshi et al., 2004; Kawakatsu et al., 2006). Consistent with those findings, ZmPLA1 expression is unchanged in vp8 embryos. The recent significant finding that the MP/ARF5 auxin response factor genetically interacts with AMP1 (Vidaurre et al., 2007) suggests the possibility that the Vp8 and/or the suppressor might also interact with auxin signaling in developing seeds of maize.
The discovery that the Vp8 gene encodes a putative peptidase, together with the irregular pattern of aleurone pigmentation, is consistent with diffusion of a nonautonomous signal derived from Vp8 activity. Interestingly, Vp8 is expressed at significantly lower levels in endosperm than in embryo, suggesting the possibility that the abnormal endosperm development might be caused by embryo-derived diffusible signals. Several classes of plant peptides have been identified in signaling (for review, see Boller, 2005) and the VP8 may be involved in processing of one or more of these peptides. Vidaurre et al. (2007) has shown that AMP1-GFP fusion protein is localized to endomembranes in Arabidopsis, suggesting that the AMP1/VP8 peptidases may process peptides in intracellular compartments. The incompletely dominant nature of the suppressor found in the SC background indicates that dosage-sensitive factors influence Vp8 function in plant development.
MATERIALS AND METHODS
Plant Material
The vp8-R stock was obtained from the Maize Genetics Cooperation Stock Center. The vp8-R (W22) material used in this study was established by backcrossing the original vp8-R with the W22 inbred line five times. For the vp8-umu1, vp8-umu2, vp8-umu3, and vp8-umu4 alleles, the complementation tests were performed by generating at least five independent crosses between vp8-R and each of the vp8-umu heterozygous mutants.
Tissue Culture with Rescued Embryos
The vp8-umu1 and vp8-R (SC) heterozygous plants were self-pollinated and the resulting developing seeds were used for embryo rescue experiments. The heteroallelic F1 seeds were generated by crosses of vp8-umu1 (W22) and vp8-R (SC). Embryos excised from 20- or 22-DAP seeds were placed on culture medium, as previously described (Suzuki et al., 2006).
MuTAIL Library Construction and Sequence Assembly
The MuTAIL library construction and processing and assembly of the sequences were described previously (Settles et al., 2004; McCarty et al., 2005). A full description of these processes specifically with the vp8 mutants is available in the supplemental data.
RT-PCR Analysis
Total RNA was prepared from maize (Zea mays) embryos, DNaseI treated, and purified using the RNeasy kit (Qiagen). Total RNA from maize endosperm was extracted as previously described (McCarty, 1986). The RNA was further purified and DNaseI treated with the RNeasy kit. RT-PCR reactions were performed with 100 ng of total RNA in a total volume of 10 μL using the One-Step RT-PCR kit (Qiagen). The primers used for RT-PCR are listed in Supplemental Table S4.
Quantification of Hormones
Quantification of hormones was performed as described (Nakagawa et al., 2005; Naito et al., 2007) using a liquid chromatography-mass chromatography system (UPLC/Quattro Ultima Pt; Waters) with an ODS column (AQUITY-UPLC BEH-C18, 1.7 μm, 2.1 × 50 mm; Waters).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EU401893.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Hand sections of a dry mature kernel of W22 and wpk1-umu1.
Supplemental Figure S2. Thin sections of developing embryos at 12 DAP.
Supplemental Figure S3. Aleurone phenotype of cr4 vp8 double-mutant seed.
Supplemental Figure S4. RT-PCR analysis of meristem-related genes and embryo-expressed genes in wpk1-umu1 (vp8-umu1) developing embryos.
Supplemental Figure S5. Seedlings that were heteroallelic between vp8-umu1 and vp8-R (SC).
Supplemental Figure S6. F1 seeds between vp8-umu1 and vp8-R (SC) heteroallelic crosses.
Supplemental Figure S7. RT-PCR analysis of meristem-related genes and embryo-expressed genes in vp8-umu2 developing embryos.
Supplemental Table S1. Segregation of wpk1-umu1 seeds.
Supplemental Table S2. Segregation of vp8 seeds.
Supplemental Table S3. Hormone content in vp8 mutant seeds.
Supplemental Table S4. RT-PCR primers used in this study.
Supplemental Information S1. Full descriptions of MuTAIL construction and sequence assembly.
Supplementary Material
Acknowledgments
We thank the ICBR at the University of Florida for DNA sequencing. We also thank the Maize Genetics Cooperation Stock Center for providing vp8-R seed stock and Dr. Philip Stinard at the Center for providing pedigree information regarding this mutant allele.
This work was supported by the National Science Foundation (grant nos. 0077676 and 0322005).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Masaharu Suzuki (masaharu@ufl.edu).
The online version of this article contains Web-only data.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Hagino T, Ohta Y, Aoki K, Yonekura-Sakakibara K, Deji A, Yamaya T, Sugiyama T, Sakakibara H (2003) Molecular characterization of His-Asp phosphorelay signaling factors in maize leaves: implications of the signal divergence by cytokinin-inducible response regulators in the cytosol and the nuclei. Plant Mol Biol 52 331–341 [DOI] [PubMed] [Google Scholar]
- Baumbusch LO, Hughes DW, Galau GA, Jakobsen KS (2004) LEC1, FUS3, ABI3 and Em expression reveals no correlation with dormancy in Arabidopsis. J Exp Bot 55 77–87 [DOI] [PubMed] [Google Scholar]
- Becraft PW, Asuncion-Crabb Y (2000) Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127 4039–4048 [DOI] [PubMed] [Google Scholar]
- Becraft PW, Li K, Dey N, Asuncion-Crabb Y (2002) The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129 5217–5225 [DOI] [PubMed] [Google Scholar]
- Becraft PW, Stinard PS, McCarty DR (1996) CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273 1406–1409 [DOI] [PubMed] [Google Scholar]
- Boller T (2005) Peptide signalling in plant development and self/non-self perception. Curr Opin Cell Biol 17 116–122 [DOI] [PubMed] [Google Scholar]
- Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S, Werr W (2005) thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132 1235–1245 [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR (2003) Regulatory networks in seeds integrating developmental, abscisic acid, sugar and light signaling. Plant Physiol 131 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson CB, Hattori T, Rosenkrans L, Vasil V, Vasil IK, Peterson PA, McCarty DR (1997) The quiescent/colorless alleles of viviparous1 show that the conserved B3 domain of VP1 is not essential for ABA-regulated gene expression in the seed. Plant J 12 1231–1240 [DOI] [PubMed] [Google Scholar]
- Champagne C, Sinha N (2004) Compound leaves: equal to the sum of their parts? Development 131 4401–4412 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Letham S, Craig S, Dennis ES (1993) amp1-a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J 4 907–916 [Google Scholar]
- Conway LJ, Poethig RS (1997) Mutations of Arabidopsis thaliana that transform leaves into cotyledons. Proc Natl Acad Sci USA 94 10209–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino IB, Deruere J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MM, Poethig RS (1997) The viviparous8 mutation delays vegetative phase change and accelerates the rate of seedling growth in maize. Plant J 12 769–779 [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7 373–385 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulini A, Wang J, Jackson D (2004) Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430 1031–1034 [DOI] [PubMed] [Google Scholar]
- Hake S, Vollbrecht E, Freeling M (1989) Cloning Knotted, the dominant morphological mutant in maize using Ds2 as a transposon tag. EMBO J 8 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Vasil V, Rosenkrans L, Hannah LC, McCarty DR, Vasil IK (1992) The Viviparous-1 gene and abscisic acid activate the C1 regulatory gene for anthocyanin biosynthesis during seed maturation in maize. Genes Dev 6 609–618 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Chin-Atkins AN, Wilson IW, Chapple R, Dennis ES, Chaudhury A (2001) The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13 2115–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mahonen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli RS, Powell CT, Fair WR, Heston WD (1993) Molecular cloning of a complementary DNA encoding a prostate-specific membrane antigen. Cancer Res 53 227–230 [PubMed] [Google Scholar]
- Itoh JI, Hasegawa A, Kitano H, Nagato Y (1998) A recessive heterochronic mutation, plastochron1, shortens the plastochron and elongates the vegetative phase in rice. Plant Cell 10 1511–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Hake S (1999) Control of phyllotaxy in maize by the abphyl1 gene. Development 126 315–323 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46 399–406 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Itoh J, Miyoshi K, Kurata N, Alvarez N, Veit B, Nagato Y (2006) PLASTOCHRON2 regulates leaf initiation and maturation in rice. Plant Cell 18 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P (1994) fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445 652–655 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols S, Olsen OA (2002) The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc Natl Acad Sci USA 99 5460–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes MA, Larkins BA (1993) Endosperm origin, development, and function. Plant Cell 5 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205 [DOI] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Misera S (1998) FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15 755–764 [DOI] [PubMed] [Google Scholar]
- McCarty DR (1986) A simple method for extraction of RNA from maize tissues. Maize Genet Coop News Lett 60 61 [Google Scholar]
- McCarty DR, Carson CB, Stinard PS, Robertson DS (1989) Molecular analysis of viviparous-1: an abscisic acid-insensitive mutant of maize. Plant Cell 1 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK (1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66 895–905 [DOI] [PubMed] [Google Scholar]
- McCarty DR, Settles AM, Suzuki M, Tan BC, Latshaw S, Porch T, Robin K, Baier J, Avigne W, Lai J, et al (2005) Steady-state transposon mutagenesis in inbred maize. Plant J 44 52–61 [DOI] [PubMed] [Google Scholar]
- Meinke DW (1992) A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258 1647–1650 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F (2006) Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J 45 942–954 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37 128–138 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Ahn BO, Kawakatsu T, Ito Y, Itoh J, Nagato Y, Kurata N (2004) PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc Natl Acad Sci USA 101 875–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monke G, Altschmied L, Tewes A, Reidt W, Mock HP, Baumlein H, Conrad U (2004) Seed-specific transcription factors ABI3 and FUS3: molecular interaction with DNA. Planta 219 158–166 [DOI] [PubMed] [Google Scholar]
- Mordhorst AP, Voerman KJ, Hartog MV, Meijer EA, van Went J, Koornneef M, de Vries SC (1998) Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Yamashino T, Kiba T, Koizumi N, Kojima M, Sakakibara H, Mizuno T (2007) A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci Biotechnol Biochem 71 1269–1278 [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Jiang CJ, Sakakibara H, Kojima M, Honda I, Ajisaka H, Nishijima T, Koshioka M, Homma T, Mander LN, et al (2005) Overexpression of a petunia zinc-finger gene alters cytokinin metabolism and plant forms. Plant J 41 512–523 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26 627–635 [DOI] [PubMed] [Google Scholar]
- Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S (2000) The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol 220 412–423 [DOI] [PubMed] [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S (1995) A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121 629–636 [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 5 165–185 [DOI] [PubMed] [Google Scholar]
- Neill SJ, Horgan R, Parry AD (1986) The carotenoid and abscisic acid content of viviparous kernels and seedlings of Zea mays L. Planta 169 87–96 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA (2001) ENDOSPERM DEVELOPMENT: cellularization and cell fate specification. Annu Rev Plant Physiol Plant Mol Biol 5 233–267 [DOI] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14 S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangalos MN, Neefs JM, Somers M, Verhasselt P, Bekkers M, van der Helm L, Fraiponts E, Ashton D, Gordon RD (1999) Isolation and expression of novel human glutamate carboxypeptidases with N-acetylated alpha-linked acidic dipeptidase and dipeptidyl peptidase IV activity. J Biol Chem 274 8470–8483 [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Misera S, Giraudat J (1997) The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V, Bergervoet JH, Koornneef M (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128 243–252 [DOI] [PubMed] [Google Scholar]
- Reidt W, Wohlfarth T, Ellerstrom M, Czihal A, Tewes A, Ezcurra I, Rask L, Baumlein H (2000) Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J 21 401–408 [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmulling T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DS (1955) The genetics of vivipary in maize. Genetics 40 745–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud C, Wong J, Sussex IM (1980) Control of in vitro growth of viviparous embryo mutants of maize by abscisic acid. Dev Genet 1 325–330 [Google Scholar]
- Saibo NJ, Vriezen WH, De Grauwe L, Azmi A, Prinsen E, Van der Straeten D (2007) A comparative analysis of the Arabidopsis mutant amp1-1 and a novel weak amp1 allele reveals new functions of the AMP1 protein. Planta 225 831–842 [DOI] [PubMed] [Google Scholar]
- Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S, Jikumaru Y, Fujimoto M, Arikawa T, Takahashi H, Ando M, et al (2007) Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol 48 287–298 [DOI] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol 134 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Mendoza M, Dubreucq B, Miquel M, Caboche M, Lepiniec L (2005) LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett 579 4666–4670 [DOI] [PubMed] [Google Scholar]
- Scanlon MJ (2000) Developmental complexities of simple leaves. Curr Opin Plant Biol 3 31–36 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Settles AM, Latshaw S, McCarty DR (2004) Molecular analysis of high-copy insertion sites in maize. Nucleic Acids Res 32 e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Li C, Min Z, Meeley RB, Tarczynski MC, Olsen OA (2003) sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein. Proc Natl Acad Sci USA 100 6552–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Settles AM, Tseung CW, Li QB, Latshaw S, Wu S, Porch TG, Schmelz EA, James MG, McCarty DR (2006) The maize viviparous15 locus encodes the molybdopterin synthase small subunit. Plant J 45 264–274 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Wang HH, McCarty DR (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 143 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H (2004) AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol 45 1053–1062 [DOI] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JA, McCarty DR (1997) Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA 94 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MC, Hudson A, Becraft PW, Nelson T (1999) ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284 151–153 [DOI] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiantis M, Hay A (2003) Comparative plant development: the time of the leaf? Nat Rev Genet 4 169–180 [DOI] [PubMed] [Google Scholar]
- Tsiantis M, Schneeberger R, Golz JF, Freeling M, Langdale JA (1999) The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284 154–156 [DOI] [PubMed] [Google Scholar]
- Veit B, Briggs SP, Schmidt RJ, Yanofsky MF, Hake S (1998) Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature 393 166–168 [DOI] [PubMed] [Google Scholar]
- Vidaurre DP, Ploense S, Krogan NT, Berleth T (2007) AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development 134 2561–2567 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ (1994) LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6 1731–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Choi D (2006) Characterization of genes encoding ABA 8′-hydroxylase in ethylene-induced stem growth of deepwater rice (Oryza sativa L.). Biochem Biophys Res Commun 350 685–690 [DOI] [PubMed] [Google Scholar]
- Yang SH, Zeevaart JA (2006) Expression of ABA 8′-hydroxylases in relation to leaf water relations and seed development in bean. Plant J 47 675–686 [DOI] [PubMed] [Google Scholar]
- Zhang S, Wong L, Meng L, Lemaux PG (2002) Similarity of expression patterns of knotted1 and ZmLEC1 during somatic and zygotic embryogenesis in maize (Zea mays L.). Planta 215 191–194 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.