Abstract
Although aquaporins (AQPs) have been shown to increase membrane water permeability in many cell types, the physiological role of this increase was not always obvious. In this report, we provide evidence that in the leafy stage of development (gametophore) of the moss Physcomitrella patens, AQPs help to replenish more rapidly the cell water that is lost by transpiration, at least if some water is in the direct vicinity of the moss plant. Three AQP genes were cloned in P. patens: PIP2;1, PIP2;2, and PIP2;3. The water permeability of the membrane was measured in protoplasts from leaves and protonema. A significant decrease was measured in protoplasts from leaves and protonema of PIP2;1 or PIP2;2 knockouts but not the PIP2;3 knockout. No phenotype was observed when knockout plants were grown in closed petri dishes with ample water supply. Gametophores isolated from the wild type and the pip2;3 mutant were not sensitive to moderate water stress, but pip2;1 or pip2;2 gametophores expressed a water stress phenotype. The knockout mutant leaves were more bent and twisted, apparently suffering from an important loss of cellular water. We propose a model to explain how the AQPs PIP2;1 and PIP2;2 delay leaf dessication in a drying atmosphere. We suggest that in ancestral land plants, some 400 million years ago, APQs were already used to facilitate the absorption of water.
Many aquaporins (AQPs) have been detected in animal cells (Agre et al., 2001; Zardoya and Villalba, 2001; Agre and Kozono, 2003) and plant cells (Chrispeels et al., 2001; Maurel et al., 2002; Luu and Maurel, 2005; Hachez et al., 2006). Although several studies reported that AQPs may increase membrane permeability to glycerol (Stroud et al., 2003), urea (Echevarria et al., 1994), and perhaps CO2 (Uehlein et al., 2003) or even ions (Yasui et al., 1999; Hazama et al., 2002), it is now well established that most of these transmembrane proteins increase the water permeability of cellular membranes. Thus, AQPs are potentially playing a role in many physiological processes (Maurel et al., 2002; Tyerman et al., 2002; Luu and Maurel, 2005) involving water flow through membranes. However, even in well-known processes such as transpiration, when plants are losing water due to environmental conditions, establishing a close correlation between AQPs and water transport appears complicated.
During the evolution of land plants two adaptation strategies emerged to cope with irregular water supply. A homoiohydric plant-like Arabidopsis (Arabidopsis thaliana), maintains its water status by controlling water potential, through turgor and osmotic pressure regulation. In this type of plant there are two parallel pathways for water transport known as the apoplastic and symplastic routes. The relative contribution of each pathway can vary, making it difficult to determine, with great accuracy, the magnitude of transmembrane water flow (Tyerman et al., 1999). Under most physiological conditions, water appeared to simply diffuse across the lipid bilayer (Finkelstein, 1987) and it has been difficult to show that water movement requires an increase in the water permeability (Pos) of the membrane bilayer by the action of AQPs. Hill et al. (2004) even suggested that the main function of AQPs is to act as sensors for osmotic and turgor pressure, and that their capacity to conduct water is consistent with the structure of a sensor.
We expected to find a closer connection between AQPs and transmembrane water flow in the moss Physcomitrella patens for several reasons. First, poikilohydric organisms such as mosses, whose water content varies with the external environment, do not regulate their water potential, and the need for turgor or osmotic sensors may not be as critical. Elucidation of the function of AQPs in bryophytes may hint at their function during the early stages of land plant evolution.
A second reason is that P. patens exhibits a simple, nonvascular structure in two different stages of its development. This moss, which lives in the open habitats of temperate regions, is subject to important variations in environmental parameters (light, humidity, and temperature). It is not a desiccation-tolerant plant and water exchanges depend on the developmental stage. In liquid medium, a single cell gives rise to filamentous protonema, i.e. branched files of single cells (Schaefer and Zryd, 2001). The protonema can only develop when fully surrounded by liquid water. Later, cell division continues to form the gametophore leaves, which develop in the air and form the aerial portion of the plant. The leaves have no cuticle or stomata and are one-cell thick. In the juvenile stage these do not possess vascular tissue but by the adult stage a single midrib develops (Sakakibara et al., 2003). Gametophore leaves gain or loose water depending on the relative humidity (RH) in the air measured far from the leaves (Nobel, 1999). Water is mainly drawn in by the leaves from external reservoirs through capillary action (Proctor and Tuba, 2002). It is lost through the aerial parts of the plant when RH decreases in the air near the cell wall. During this stage of development, water transpired from a gametophore must cross cellular membranes, so we hypothesized that AQPs could be involved in this process.
It has been known for some time that AQPs are present in P. patens, a well-established model system for various biological and evolutionary processes in higher plants (Schaefer, 2002; Quatrano et al., 2007; Shigyo et al., 2007). Twelve putative AQP sequences were first identified in a homology screen of a P. patens EST library (Borstlap, 2002), five of which belong to the PIP (plasma membrane intrinsic proteins) family. Since then, the first annotated P. patens ssp. patens genome sequence was released in 2007 (http://genome.jgi-psf.org). Twenty-five sequences are classified into a “carbohydrate transport and metabolism” family with the special annotation “aquaporin”. Nine AQPs (including the three genes described in this article) are considered as PIP, six others as TIP, another group of six as NIP, two as SIP, and two are without a specific subfamily name. A glycerol channel (Gustavsson et al., 2005) should probably be added to this list.
In this article, we report the cloning of three putative PIP in P. patens: PIP2;1, PIP2;2, and PIP2;3. Knocking out (Schaefer and Zryd, 1997) PIP2;3 did not lead to modifications in the Pos of protoplasts. On the contrary, the PIP2;1 or PIP2;2 knockouts resulted in a significant decrease in the Pos of protoplasts from mutant gametophores. Based on these results we conclude that both the PIP2;1 and PIP2;2 genes encode functional AQPs. Suppression of PIP2;1 and PIP2;2 reduced the tolerance of gametophores to water stress.
RESULTS
Cloning of Three AQP Genes
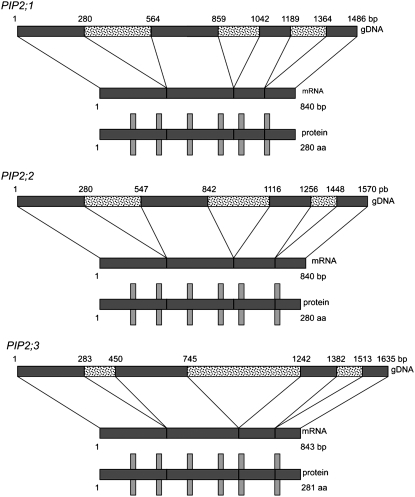
In the absence of the full P. patens genome sequence (this only became available at the beginning of 2007 at http://genome.jgi-psf.org/Physcomitrella), we searched EST databases for sequences similar to Arabidopsis AQPs (Nishiyama et al., 2003). This led to the isolation of three moss orthologs from the PIP2 family. Their gene structure was determined by comparing their coding and genomic sequences to characterize the number, position, and sequence of the introns. The three cloned homologs contain three introns, which is the case for all PIP genes (Johanson et al., 2001). The position of these introns is similar for the three genes (Fig. 1) as well as to the Arabidopsis PIP2 genes (Johanson et al., 2001).
Figure 1.
The putative P. patens PIP2 genes PIP2;1, PIP2;2, and PIP2;3. Schematic diagrams of the PIP2;1, PIP2;2, and PIP2;3 gene structures. The numbers on the genes correspond to the first and last bases of exons (dark boxes). The light boxes indicate introns. Vertical bars and boxes indicate the position of the splicing sites and transmembrane domains within the proteins.
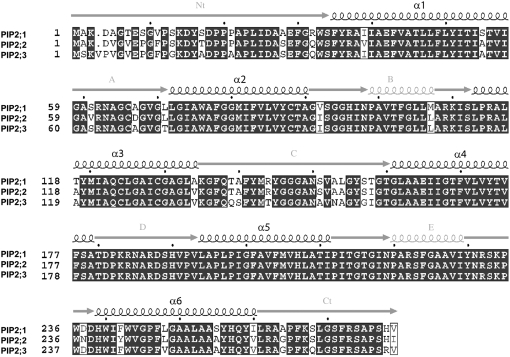
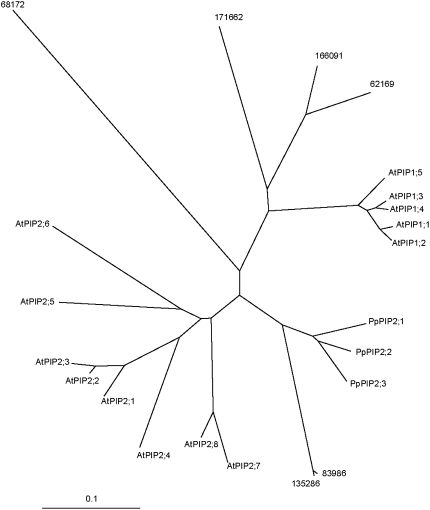
Open reading frames predict protein sequences of 280 or 281 amino acids. As expected for an AQP protein, six transmembrane domains (Stahlberg et al., 2001) are visible on the hydrophobicity profile of all three sequences (data not shown). The two NPA signature sequences usually detected in loops 2 and 5 of AQPs (Johanson et al., 2001) were also present in these homologs (Fig. 2). The alignment of the P. patens and Arabidopsis PIP protein sequences indicated that P. patens PIPs are more similar to PIP2 than to PIP1 AQPs (data not shown). Further molecular phylogenetic analysis (Fig. 3) confirmed the evolutionary relationship between these proteins. We named the three P. patens homologs: PIP2;1, PIP2;2, and PIP2;3.
Figure 2.
Alignment of the three encoded proteins PIP2;1, PIP2;2, and PIP2;3. This alignment was made using the programs Mutalin and ESPript (http://nosa-pbil.ibcp.fr). Predicted positions of the transmembrane domains (α1–α6) and external loops (A–E), from N terminus to C terminus, were identical for the three sequences and are shown by the sketches on top of the sequences.
Figure 3.
Molecular phylogenic relationship between Arabidopsis and P. patens PIPs. The evolutionary tree was generated with PROTDIST and NEIGHBOR of the PHYLIP package (http://bioweb.pasteur.fr/). The bar indicates a mean distance of 0.1 changes per amino acid residue. Numbers correspond to the protein identification from putative PpPIP (http://genome.jgi-psf.org). AtPIP, Arabidopsis PIP; PpPIP, P. patens PIP.
PIP2;1, PIP2;2, and PIP2;3 Expression Patterns
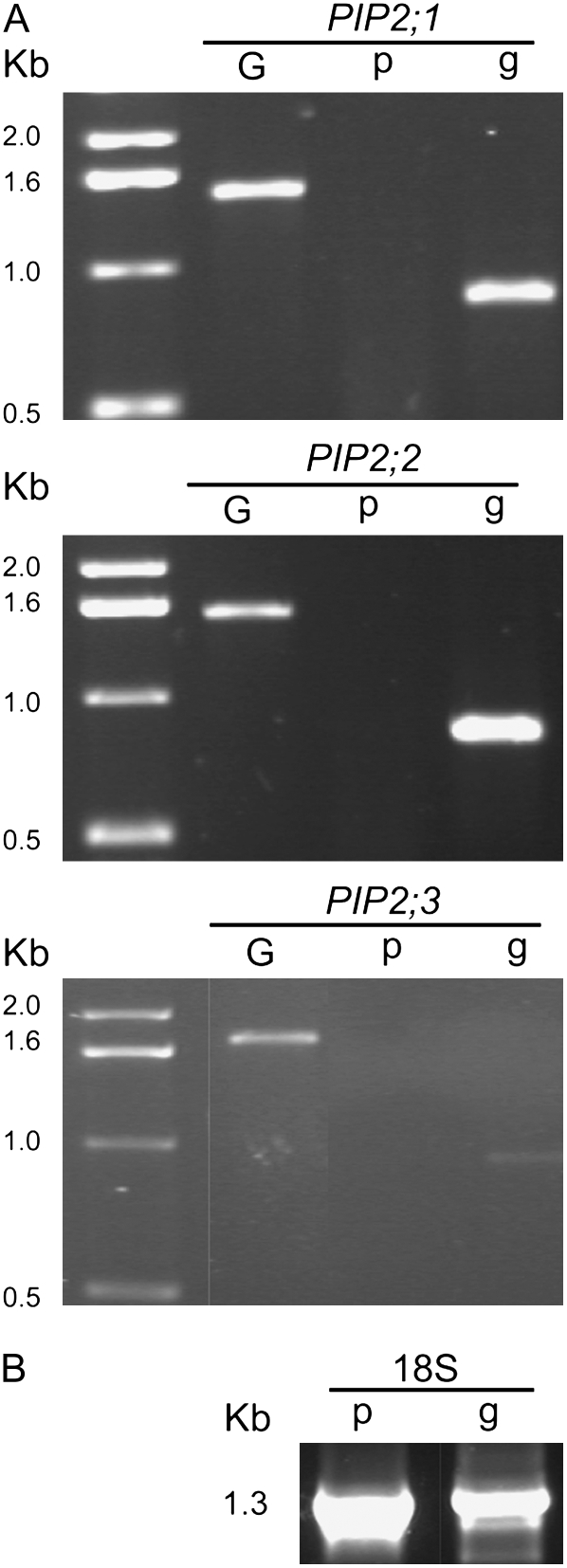
The expression patterns of the three genes were analyzed for two different stages of development (Fig. 4A). Total RNA was isolated from protonemata or gametophores, then reverse-transcribed. Control experiments were performed using constitutively expressed 18S RNA (Fig. 4B). We found that the transcripts were specifically accumulated in the gametophores (Fig. 4A, lanes g), but not detectable in protonema grown in liquid medium (Fig. 4A, lanes p). These results clearly showed that the transcripts are only expressed in the aerial parts of the plant.
Figure 4.
Expression pattern of PIP2;1, PIP2;2, and PIP2;3 transcripts in P. patens wild type. For each gene, amplification with 20-mer specific primers on genomic DNA was included as a control (A, lane G). RT-PCR analyses were performed on RNA from 4-d-old protonema (A, lane p) or 3- to 4-week-old gametophores (A, lane g). The integrity of the total RNA in the two preparations was assumed by checking the presence of constitutively expressed 18S RNA (B, lanes p and g). All experiments were repeated three times with independent plant samples.
Targeted Knockout of PIP2;1, PIP2;2, and PIP2;3
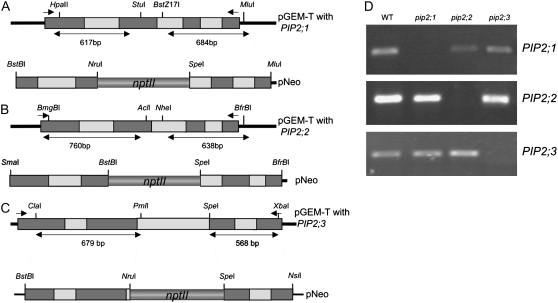
P. patens is especially amenable to gene knockout because of its high rate of homologous recombination (Schaefer and Zryd, 1997). Disruption constructs for the three genes were created by exchanging small fragments of the genes with an nptII selection cassette (Fig. 5). Knockout plants were screened by PCR amplification of the nptII cassette, and the 5′- and 3′-ends of the genes. The knockout of each of the three genes was confirmed by examining expression of the cDNA (Fig. 5D) in the three mutants, pip2;1, pip2;2, and pip2;3, using reverse transcription (RT)-PCR. As expected, the PIP2;1, PIP2;2, and PIP2;3 transcripts were detected in the wild type (Fig. 5D, lane WT) but not in the corresponding mutant (Fig. 5D). Despite similarities in the PIP2;1, PIP2;2, and PIP2;3, cDNA sequences, PCR demonstrated that each of three gene disruptions was specific.
Figure 5.
Schematic representation of the PIP2;1, PIP2;2, and PIP2;3 genes, corresponding knockout constructs, and molecular analysis of the knockout mutants. A, Gene structure and knockout construct of the PIP2;1 gene. B, Gene structure and knockout construct of the PIP2;2 gene. C, Gene structure and knockout construct of the PIP2;3 gene. Dotted lines and numbers of base pair indicate the regions of the genes used for insertion in pNeo. The position of the primers (little arrows) is detailed in “Materials and Methods”. D, Expression analysis of the wild-type and mutant plants. Total RNA from wild-type, pip2;1, pip2;2, and pip2;3 plants was reverse transcribed and used for PCR. The experiments were repeated three times with independent wild-type and mutant plant samples. The integrity of the total RNAs in the preparations was assumed by checking the presence of constitutively expressed 18S RNA.
Measurements of Pos on Isolated Protoplasts Suggest That PIP2;1 and PIP2;2 Function as AQPs
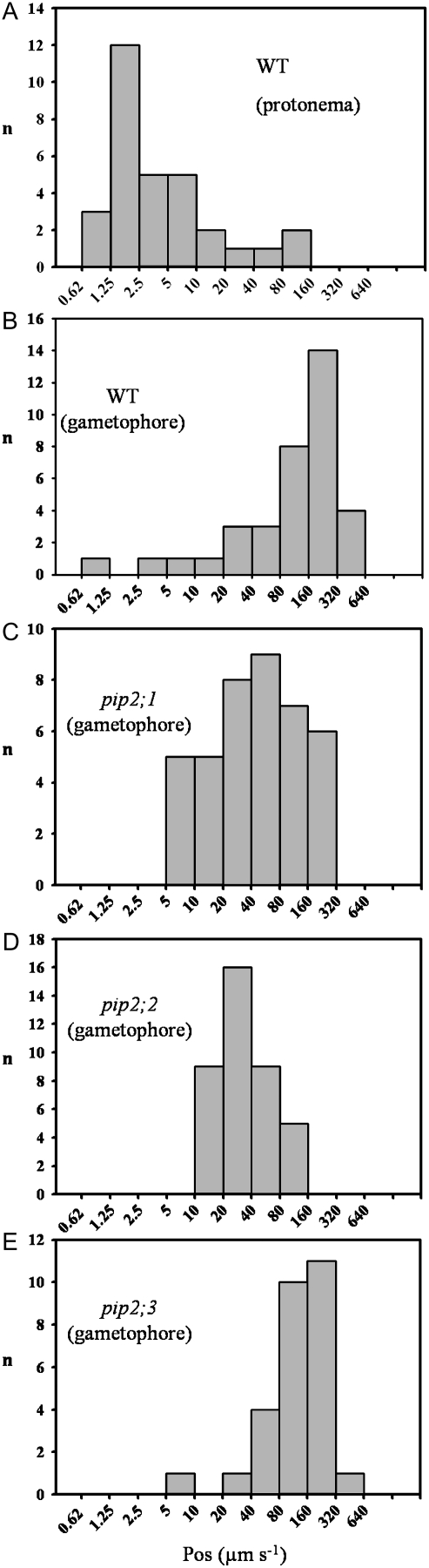
We measured the Pos, of protoplasts isolated from protonema and gametophore cells (Fig. 6), as previously described (Ramahaleo et al., 1999). The data are presented on a logarithmic scale because of the wide range of Pos values obtained. The protoplasts from wild-type protonema had a median value of Pos = 2.7 μm s−1 (Fig. 6A). This small value indicates simple diffusion across the lipid bilayer (Finkelstein, 1987). In contrast, the protoplasts from the wild-type gametophores had a median value of Pos = 159 μm s−1 (Fig. 6B). The large difference in Pos suggests that AQPs are present in the cellular membranes of the gametophore cells. These results are consistent with the expression pattern of our putative AQPs described in Figure 4.
Figure 6.
Histograms showing the Pos values for protoplasts from protonema and gametophores. The Pos values measured for 31 protoplasts from wild-type protonema (A) came from three different preparations. They had a diameter of: 30 ± 2.5 μm. The 36 protoplasts from wild-type gametophores (B) were from four different preparations. They had a diameter of 49.6 ± 7.8 μm. The 40 protoplasts from the pip2;1 gametophores (C) were from six different preparations. They had a diameter of 41.8 ± 4.4 μm. The 39 protoplasts from the pip2;2 gametophores (D) were from four different preparations. They had a diameter of 47.5 ± 8.5 μm. The 28 protoplasts from the pip2;3 gametophores (E) were from three different preparations. They had a diameter of 49.8 ± 5.6 μm. No correlation was found between Pos values and the diameter of the protoplasts from gametophore or protonema preparations. The diameter figures are given as mean ± sd; n is the number of protoplasts in each class.
Direct comparisons of wild-type and mutant gametophore protoplast Pos values were carried out (Fig. 6). The data did not pass the test for normality so the four samples were compared with a nonparametric ANOVA on ranks test followed by pairwise multiple comparison procedures (Dunn's method). There was no statistically significant difference (P < 0.05) in the median values between the wild type (159 μm s−1) and pip2;3 (134 μm s−1) or between pip2;1 (41 μm s−1) and pip2;2 (32 μm s−1), but the difference between wild type or pip2;3 versus pip2;1 or pip2;2 was significant.
The decrease in Pos of more than 100 μm s−1 for pip2;1 or pip2;2, compared with wild type, suggests that AQP function is disrupted in these mutants, whereas the knockout of PIP2;3 has no effect on the permeability of isolated gametophores.
Effect of Water Stress on the Gametophores
All plants, including the wild type and the three mutants, exhibited variations in leaf size and number when gametophores were grown in closed petri dishes. No obvious phenotype could be identified, and within-group variation in morphology was as great as between-group variation. In a set of experiments (data not shown), petri dishes containing wild-type, pip2;1, pip2;2, and pip2;3 plants were opened at the same time and allowed to evaporate in a dry atmosphere (RH, 20%–30%; temperature, 20°C–22°C). The pip2;1 and pip2;2 mutant leaves appeared to become twisted more rapidly and in greater number than those of the wild type and pip2;3 plants.
The effect of water stress was also studied more directly on isolated gametophores. Three similar-sized gametophores from wild-type, pip2;1, and pip2;2 knockout plants, were deposited on a filter soaked with water (Fig. 7, A and B) or on a dry filter paper disc (Fig. 7C). The moisture was allowed to evaporate in the laboratory conditions. Gametophore leaves began slowly moving (bending and twisting) usually several minutes after the beginning of the experiment. These movements are assumed to indicate significant water loss from cells, i.e. sensitivity of the gametophore to the stress conditions. The times, t1 and t2, recorded at the start and finish of the movements were used as markers for water stress. The values of t1 and t2 are detailed in the legend to Figure 7.
Figure 7.
Effect of water stress on isolated gametophores. The top section of the plant gametophores was deposited at time t0 on dry or wet filter paper and exposed to a nonwater-saturated atmosphere. Times t1 and t2 correspond to initial and final leaf movements (bending or twisting) in gametophores. Photos are shown for: t0; t2; and tm = (t1 + t2)/2. A, Gametophores from wild-type, pip2;1, and pip2;2 plants on a wet filter. The variations in environmental parameters during the experiment were: RH, 60%; temperature, 24°C. For both mutants, t1 = 45 ± 5 min; t2 = 180 ± 5 min. No movement by the wild type was detected. B, Gametophores from wild-type, pip2;1, and pip2;2 plants on a wet filter. The variations in environmental parameters during the experiment were: RH, 55.5%; temperature, 28°C. For both mutants, t1 = 18 ± 3 min; t2 = 84 ± 3 min. No movement by the wild type was detected. In three repeat experiments similar to those shown in A or B, the values were, respectively: t1 = 42 ± 3 min and t2 = 120 ± 3 min (RH = 60%; 28°C); t1 = 21 ± 3 min and t2 = 100 ± 3 min (RH = 60%; 28°C); t1 = 36 ± 3 min and t2 = 105 ± 3 min (RH = 54%; 24°C). C, Gametophores from wild-type, pip2;1, and pip2;2 plants on a dry filter. The variations in environmental parameters during the experiment were: RH, 49%; temperature, 28°C. For both mutants and wild type, t1 = 5 ± 1 min and t2 = 28 ± 1 min. In three similar experiments, the values were, respectively: t1 = 6 ± 1 min and t2 = 36 ± 1 min (RH = 49%; 28°C); t1 = 20 ± 1 min and t2 = 52 ± 1 min (RH = 53.5%; 24°C); t1 = 15 ± 1 min and t2 = 46 ± 1 min (RH = 53.5%; 24°C). The accuracy of t1 and t2 (from ±1 to ±5 min, depending on the experiment) corresponded to the time interval between two photos. Scale bar, 10 mm.
When the gametophores were deposited on a dry filter, creating what we called “strong water stress” conditions, the filter sucked the liquid water still present on the leaves in a few seconds and the RH near the gametophores was probably rather low, close to that of the environment (<60%). The stress-induced movements started and ended at the same time in both the wild type and mutants in this experiment that is described in Figure 7C as well as in three other similar experiments.
We expected to create moderate water stress conditions by using a wet filter, assuming that the evaporation of water would increase humidity near the leaves. We found that when the gametophores were deposited on a wet filter, the wild-type or pip2;3 (data not shown) plants remained unaffected, whereas pip2;1 or pip2;2 knockout plants moved following the same time course in the experiment described in Figure 7, A and B, and in three other similar experiments.
Supplemental Videos S1 to S3 (see “Supplemental Data”) show wilting on dry and wet filters in a typical experiment. The strong water stress led all the gametophores to wilt at the same rate. The suppression of AQPs increased sensitivity to moderate stress conditions in two out of the three mutants.
DISCUSSION
PIP2;1, PIP2;2, and PIP2;3 Are Expressed in Wild-Type Gametophores and Were Specifically Knocked Out in Mutant Plants
We screened a whole plant P. patens EST library and identified three putative genes containing the two NPA repeated regions considered to be characteristic signatures of plant AQPs (Xie et al., 2003). The three sequences also show strong similarities with AQPs of the Arabidopsis PIP2 family (Fig. 3). We called them PIP2;1, PIP2;2, and PIP2;3 (Fig. 1).
Their expression pattern in wild type was examined using RT-PCR and is presented in Figure 4. The PIP2;1, PIP2;2, and PIP2;3 transcripts accumulated in the gametophores but could not be detected in the protonema stage. This expression pattern is consistent with a potential role as AQPs involved in transpiration. Under normal growth conditions the protonema does not experience water stress; it develops in dilute solutions with a water potential close to zero. We generated knockout mutants for PIP2;1, PIP2;2, and PIP2;3 (Fig. 5). The control experiments (Fig. 5D) indicated that, despite the close sequence similarity of the three proteins, the suppression of each gene was selective: pip2;1, pip2;2, and pip2;3 did not express their corresponding mRNA but expression levels of PIP2;1, PIP2;2, and PIP2;3 were normal in each of the two other mutants.
PIP2;1 and PIP2;2 Are Functional AQPs in Gametophore Cells
Several conclusions can be drawn from the Pos values measured on the various protoplasts populations (Fig. 6). Wild-type protonema protoplasts had low Pos values, suggesting an absence of active AQPs, whereas high Pos values for wild-type gametophore protoplasts indicate that active AQPs are functioning to transport water across the membrane in these cells. The decrease in Pos observed for the two knockout mutants (Fig. 6) strongly suggests that both PIP2;1 and PIP2;2 are functional AQPs.
PIP2;3 Has No Effect on Pos
Pos values were similar for wild-type and pip2;3 knockout gametophores. This suggests that, despite a close similarity between PIP2;1, PIP2;2, and PIP2;3 sequences, PIP2;3 does not code for an AQP with a significant role in water transport, at least in our growth culture conditions (RH = 100%). Thus, although like PIP2;1 and PIP2;2, PIP2;3 was only expressed in the gametophore, we have no indication concerning PIP2;3 function. We could, however, consider pip2;3 as a control, because it indicates that the recombination techniques used to obtain the mutants are not responsible for the decrease in Pos found with pip2;1 and pip2;2 plants.
pip2;1 and pip2;2 Plants Express a Phenotype under Moderate Water Stress Conditions
No phenotype was observed when the knockout mutants were grown in closed petri dishes with ample water supply. When dishes containing either wild type or mutants were opened simultaneously we noticed that pip2;1 and pip2;2 leaves dried more rapidly than wild-type or pip2;3 knockout leaves. Variations in transpiration rate due to fluctuations in environmental parameters (temperature, light, and RH) are assumed to be the same for all the plants so this did not appear to explain the difference.
Nevertheless, to rule out the effect of local conditions, we then compared wilting of similar-sized isolated gametophores, which were deposited on the same dry filter paper disc. Because the entire gametophores were in contact with dry air, severe water stress was expected. In each of our experiments all the gametophores commenced and stopped wilting at the same time. We conclude that the AQPs do not modify sensitivity to dehydration of the gametophores in these conditions.
The situation was quite different when the gametophores were in contact with wet soil (water-soaked filter paper) through some of their leaves. Less stress was induced by these experimental conditions because only part of the leaves was transpiring in the air, whereas the other part could absorb liquid water. In contrast to the previous result, the water stress phenotype observed for the pip2;1 and pip2;2 knockout mutants supports an important role for the corresponding AQPs during transpiration.
Resistance to Transpiration by a Cell Surface in the Air: The Boundary Layer and Cell Membranes
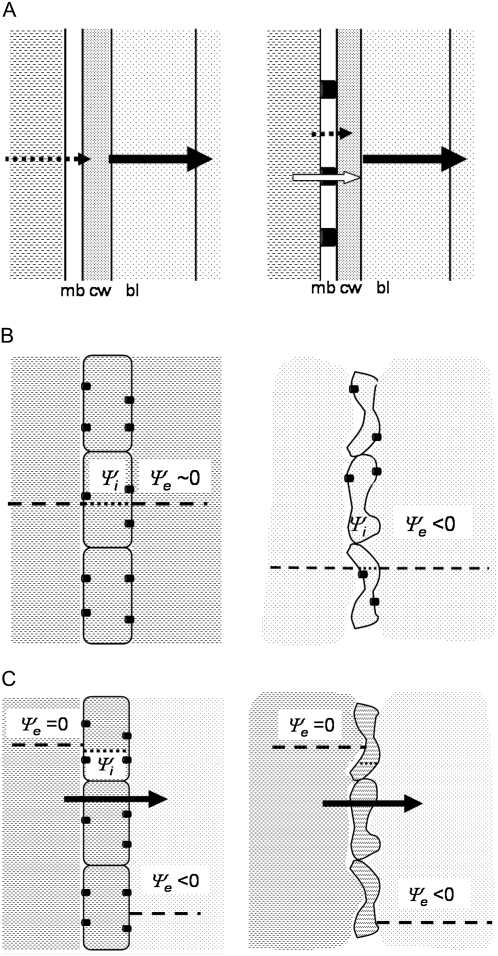
Hydrodynamic theory describes diffusion and convection currents in a gas phase. It shows that transverse resistance to the transfer of water molecules between a phase of liquid water and the ambient air is mainly located within a small layer of still air near the liquid surface, the so-called “boundary layer”. Tazawa and Okazaki (1997) suggested a dominant role for this layer during evaporation from a cell surface. Based on their experiments with Chara corallina cells, the authors concluded that, even without AQPs, the cell membranes were much more permeable than the boundary layer; the permeability of the lipid bilayer alone was at least 50 times higher than that of the boundary layer. Tazawa and Okazaki (1997) thus suggested that AQPs did not control transpiration, the resistance to water movement from the cell interior to the air being mainly located in the air (Fig. 8A). The same result was obtained when the experiments were performed with radish (Raphanus sativus) hypocotyls, suggesting that it was valid for all kinds of evaporating cell walls.
Figure 8.
Schematic drawing of transmembrane and transcellular transpiration flow (side view). A, Transpiration from a cell surface. The magnitude of the transmembrane flow (black arrow) is mainly determined by the resistance in the boundary layer (left drawing). Opening AQPs adds a pathway to water across the membrane, but does not modify the magnitude of the transpiration flow (right drawing). mb, Membrane; cw, cell wall; bl, boundary layer; dotted arrow, flow through the lipid bilayer; white arrow, flow through AQPs. B, When both sides of a cell are in contact with a water potential Ψe in the external medium, the cell water potential Ψi reaches an equilibrium value Ψi = Ψe. If Ψe ∼ 0 (left) the cell remains turgid; if Ψe is too negative (right) the cell becomes wilted. C, Transcellular transpiration flow from a cell with one side facing liquid water (Ψe ∼ 0), the other side facing the air. When the transpiration flow (black arrow) is not too high (medium stress), AQPs can keep Ψi close to zero by reducing the resistance of the membrane (left). For the same transpiration flow, if AQPs are removed from the membrane (right), the increase in resistance can produce a more dangerous drop in Ψi.
At first sight, our findings that cells lacking either one of two AQPs showed an increased rate of wilting may then seem counterintuitive and led us to analyze our experimental conditions more closely.
Environmental Conditions of a Transpiring Leaf
In the nonvascularized, one-cell-thick leaves of P. patens, each cell can exchange water with the external medium. If water potential values in the external medium, Ψe, are the same on each side of the leaf (Fig. 8B), the water potential within the cell, Ψi, will tend toward this value, corresponding to an equilibrium state without transpiration. There are two distinct cases: (1) the leaf is in contact with saturated air or dilute solutions (condensation or rain) are in contact with the leaf; Ψe is close to 0 and the cells remain fully turgid and sustain normal growth (Salisbury and Ross, 1992). (2) The leaf is transpiring in nonsaturated air (Ψe < 0); there is a loss of cellular water that can produce wilting. In both cases the cell water status is determined by the availability of water outside, not by the permeability of the cell membranes. This may explain the similar behavior observed between our mutants lacking AQPs and wild type; all the plants were turgid in closed petri dishes (no stress condition, Ψe ∼ 0) or wilted on dry filter paper (strong water stress conditions, too negative Ψe).
In the experiments on wet filters, our moderate stress conditions, we assume that parts of the leaves were either in contact with liquid water (Ψe = 0) or with nonsaturated air (Ψe ≤ 0). If a cell is in contact with liquid water on one side and with nonsaturated air on the other side a transcellular water flow is established (Fig. 8C). For the cell membrane in contact with liquid water on both sides, the resistance rm to water movement between cell interior and the liquid water outside is mainly due to the cell membranes (the resistance of the cell wall can be ignored because it is usually far smaller than that of the cell membranes; Nobel, 1999). This resistance is reduced if AQPs are present in the membranes. For the cell membrane in contact with the air, as already explained, the resistance between the cell interior and the air is mainly due to rbl, the resistance of the air (the boundary layer) near this surface; rbl does not depend on AQPs and, according to Tazawa and Okazaki (1997), rbl ≫ rm.
The different magnitude of rbl and rm has two consequences: (1) the total resistance to the transcellular flow, rbl + 2*rm, is nearly equal to rbl. It is not modified by AQPs and cells of approximately the same size, regardless of AQPs, should evaporate at the same rate; (2) for the same evaporation rate, the water potential inside the mutant and wild-type cells may be different: the larger the rm the larger the drop in cell water potential (Fig. 8). As a consequence, under moderate water stress, the wild type could maintain Ψi close to zero, but the suppression of an AQP in pip2;1 and pip2;2 mutants could produce a dangerous drop in water potential.
We conclude from our experiments that PIP2;1 and PIP2;2 code for functional AQPs. These transcripts were specifically expressed in the wild-type gametophores. The mutants lacking these genes wilted more easily, which means that their metabolism was suspended earlier than wild type under the same moderate stress conditions. The physiological advantage of a delay in wilting is obvious.
One interesting discussion point is that what we refer to as medium stress conditions may represent the situation of cells in the stomatal chamber of higher plant leaves, which are partly in contact with the intercellular air space. Therefore, even if AQPs are not required on the cell surface in contact with the air, AQPs could delay cellular water loss by facilitating water exchanges through the part of the cell surface in contact with the liquid phase.
The protonema, which normally grows in liquid medium, did not express PIP2;1 and PIP2;2. This is not surprising if the two AQPs carry out functions related to transpiration. Gametophores may have adapted various strategies to delay wilting, and increased cell membrane permeability to water could be one such strategy.
According to Proctor and Tuba (2002), poikilohydry is not the primitive starting point leading to homoiohydry, but an alternative strategy to cope with irregular water supply. When water is not easily available, rather than wasting energy maintaining a low water potential in their cells, poikilohydry plants suspend their metabolism during these periods of time. Our study suggests that more than 400 million years before the evolution of vascular plants, P. patens was already using AQPs to shorten these periods.
MATERIALS AND METHODS
Material
Plants of the moss Physcomitrella patens wild-type strain (Gransden) were provided by Martine Gonneau (Institut National de la Recherche Agronomique). Plants were grown axenically in petri dishes (90- or 45-mm diameter) containing a liquid culture medium called A′BCD-TES-NH4+ (Wang et al., 1980) solidified with 0.8% (w/v) agar. The petri dishes were wrapped with parafilm, so that plants developed in a water-saturated atmosphere. They were kept in a growth chamber at 23 to 25°C with a 16-h/8-h light/dark cycle. Light (60–75 μmol m−2 s−1) was provided by TLD 33 and chromasoleil TLD 83 (Philips) fluorescent tubes. The plants were subcultured every 7 d.
Standard molecular techniques were used (Sambrook et al., 1989). Restriction enzymes, Taq polymerase, pGEM-T vector, and T4 DNA ligase were obtained from Promega. Primers were custom-ordered from Proligos. PCR was performed in a thermocycler (Eppendorf). DNA sequencing was performed with an automated LI-COR 4000 sequencer.
Bacteria
The Escherichia coli strain DH5α used for plasmid DNA propagation was grown in 10% tryptone (DIFCO), 5% yeast extract (DIFCO), and 170 mmol L−1 NaCl.
Cloning of Three Putative AQP Coding Sequences
The 35 coding sequences of Arabidopsis (Arabidopsis thaliana) AQPs (Johanson et al., 2001) were used to screen the ESTs from the PHYSCObase (http://moss.nibb.ac.jp) database. One of these ESTs, referred to as 18342754 in GenBank, showed a strong similarity (E-value = 10−15) with the 5′ end of PIP2 AQPs. To amplify this partial sequence, two specific primers were designed: EST1For (5′-ggCTgTggAgCggCACTAgA-3′) and EST1Rev (5′-TggCAgAgAAgACggTgTAC-3′). PCR was carried out using total plant cDNA as the template. Total RNAs were isolated using the SV 96 total RNA isolation system kit (Promega) and reverse-transcribed with moloney murine leukemia virus reverse transcriptase RNase H Minus (Promega) according to the manufacturer's instructions. Two different sequences were amplified. To obtain the full-length coding sequence of each gene, 3′-RACE-PCR experiments were performed with the BD SMART mRNA amplification kit (BD Biosciences CLONTECH), according to the manufacturer's instructions. The primers used were: EST1Race (5′-ggCATATCAACCCAgCAgTCACgTTC-3′) and EST1BisRace (5′-ggCTTgCTgTTggCACgCAAAATCTC-3′). Once the 3′ cDNA ends were determined by RACE, gene-specific primers were designed to amplify the entire cDNA from the start-to-stop codons and then to amplify genomic sequences. PIP2;1 forward (5′-AgCAAACCATggCAAAAgAC-3′) and reverse (5′-gATggAggAACTCgTgCTTA-3′) primers and PIP2;2 forward (5′-AgCAAACCATggCAAAAgAC-3′) and reverse (5′-TgTCACgTTTAgATgTgACT-3′) primers were used.
The reversed sequence of a second EST (18334457 in GenBank) showed a strong similarity (E-value = 5 × 10−30) with the 3′ end of PIP2 AQPs. We obtained the full-length cDNA by following the same procedure as described above for EST 18342754. Two specific primers, EST2For (5′-CTggTgTACACCgTTTTCTC-3′) and EST2Rev (5′-TTgAACAggAgCAAATgCCC-3′), were designed to amplify a partial sequence and an EST2Race primer (5′-TgCCAgTTCCggTAATgggAATggTAg-3′) was used to determine the 5′ cDNA end. Two gene-specific primers, PIP2;3 forward (5′-ggTTTTgCgAggAAgAAgTT-3′) and reverse (5′-ggAATTTgTgAgggggCAAg-3′), allowed amplification of the entire cDNA.
The ClustalW program (Thompson et al., 1994) at http://www.infobiogen.fr/services/analyseq/cgi-bin/clustalw_in.pl. was used for sequence alignments.
Knockout Constructs for PIP2;1, PIP2;2, and PIP2;3
The pNeo plasmid was constructed as follows. The EcoRI fragment from pHP23b (Paszkowski et al., 1988), containing the neomycin phosphotransferase gene driven by the 35S cauliflower mosaic virus promoter, was removed and ligated into the EcoRI site of the pMCS5 vector (MoBiTec), generating pNeo with the neomycin phosphotransferase gene flanked by a multiple cloning site.
Knockout plasmids were constructed (Fig. 5) by enzymatic digestion of the three genes, and ligation into the pNeo plasmid. Each gene was cut with two different sets of enzymes to produce two fragments, which were then cloned into pNeo. PIP2;1 was digested with HpaII/StuI and BstZ17I/MluI to give two fragments: one of 617 bp and one of 684 bp (corresponding to 624 bp of the genomic sequence). These fragments were ligated in pNeo linearized by BstBI/NruI and SpeI/MluI for the first and second fragments, respectively. PIP2;2 was digested with BmgBI/AclI and NheI/BfrBI, to give two fragments: one of 760 bp and one of 638 bp (corresponding to 577 bp of the genomic sequence). These fragments were ligated in pNeo linearized by SmaI/BstBI and SpeI/BfrBI for the first and second fragments, respectively. For PIP2;3 two fragments (679 and 568 bp) were produced by digestion with ClaI/PmlI and SpeI/XbaI. These fragments were ligated in pNeo linearized by BstBI/NruI and SpeI/NsiI for the first and second fragments, respectively. These plasmids were amplified and linearized by ScaI for transformation. Linear DNA was purified by the Wizard SV Gel and PCR Clean-Up System from Promega, and then resuspended in sterile water at a concentration of 0.5 mg/mL.
Transformation Procedure
Protoplasts were isolated from a 7-d-old protonematal culture by incubation for 30 min in 1% driselase (D8037; Sigma) and dissolved in 0.48 m mannitol as previously described (Schaefer and Zryd, 1997). Transformation experiments were performed with 300 μL of protoplast suspension added to 20 μL of linearized DNA (prepared as described above), using the standard protocol (Schaefer and Zryd, 1997). After 7 d of incubation, regenerating colonies were transferred to NH4 medium supplemented with 50 mg/mL of paromomycin (P8692; Sigma). Stable antibiotic resistant clones were selected by a second round of growth of fragmented plants on a Pp-NH4 medium containing the antibiotic.
PCR Screening of Mutant Plants
For PCR analysis, genomic DNA was extracted using a genomic DNA quick preparation for PCR method. Fresh tissue (50 mg) was ground in 300 μL of extraction buffer (200 mm Tris-HCl, pH 7.5, 250 mm NaCl, 25 mm EDTA, and 0.5% SDS) and centrifuged at 12,000 rpm for 1 min. Isopropanol (200 μL) was added to the supernatant, left for 2 min at room temperature, and centrifuged at 12,000 rpm for 5 min. The pellet was resuspended in 100 μL of Tris EDTA buffer; 1 μL was used for the PCR. Three primers for the PIP2;1 genomic locus (Up21-2, 5′-TTTTCgTgTTggTgTACTgC-3′, PIP2;1 reverse, and Down21-8, 5′-CACTgCAAgTCACCAgAACT-3′), three primers for the PIP2;2 genomic locus (Up22-2, 5′-AgCAgTCATCgCTgAgTTTg-3′, PIP2;2 reverse, and Down22-8, 5′-gCATAgTCACCAAggCTggT-3′), and two primers for the PIP2;3 genomic locus (PIP2;3 forward and PIP2;3 reverse) were used in combination with two nptII gene-specific primers (neo4, 5′-ATgAACTgTTCgCCAgTCTT-3′ and neo6, 5′-CCTAAAACCAAAATCCAgTg-3′).
Protoplasts Used for Pos Measurements
Plants from 2- to 3-week-old subcultures, grown on solid medium, were used to prepare protoplasts for the permeability measurements. Protoplasts from the protonema or gametophore leaves were used in the experiments. A few strands of protonema were collected under a zoom microscope. They were almost completely digested after 90 min at 28°C in the following solution: driselase 1% (Sigma), mannitol 500 mmol/kg, macerozyme 0.1% (Yakult Honsha), polyvinylpyrrolidone 0.5%, cellulase RS 0.5% (Yakult Honsha), 5 mm MES buffered with Tris to pH 5.5. When gametophores (5–10) were taken from the same cultures, no protoplast was released after 90 min in this solution. The digestion had to be extended for 20 h at room temperature under sterile conditions to release enough protoplasts for our experiments.
Pos Measurements
Isolated protoplasts were resuspended in a storing solution (mannitol, 500 mmol/kg; Ca(No3)2, 1 mm; bovine serum albumin, 0.05% w/v; MES, 5 mm, buffered with Tris to pH 5.5), and Pos was measured at room temperature according to a technique previously described (Ramahaleo et al., 1999); the value of Pos was deduced from the initial rate of swelling of the protoplast, when transferred from the stock solution to a solution with lower osmoticum (mannitol, 100–300 mmol/kg; Ca(No3)2, 1 mm; bovine serum albumin, 0.05% w/v; MES, 5 mm, buffered with Tris to pH 5.5).
When the protonema from the wild type was digested for 90 min, the measured protoplasts had a median value of Pos = 2.7 μm s−1 (Fig. 6A). To detect the possible effect of a long incubation time on Pos, some protoplasts (10 from two different preparations) of the wild-type protonema were kept for 20 h in the digestion solution, before measurements. Under these conditions, the median value was 34.3 μm s−1. The long incubation time required to obtain protoplasts from the gametophores could therefore have led to an overestimation of their Pos (median value is 159 μm s−1). However, the ANOVA test on ranks indicated that the large increase in Pos from the protonema to the gametophore could not only be explained by longer digestion.
Water Stress
Isolated mutant and wild-type gametophores were grown in 100% RH and then subjected to water stress by exposure to lower RH from ambient air in the laboratory. The parameters of the laboratory environment (RH, 49% to 60%; temperature, 24°C to 28°C) were not regulated but fluctuations were ≤1% RH and ≤1°C within an experiment lasting 3 h or less. A surface mimicking wet ground evaporating in air was made by depositing a Whatmann filter paper disc (diameter 40 mm) on an open petri dish filled with a 1% agar solution (w/v) in water. When the dish was placed on the stage of an M3Z stereomicroscope (Leica), the paper remained wet for several hours under laboratory conditions. A dry filter paper disc (diameter 40 mm) was used to mimic dry ground. Gametophores of approximately the same size (see Fig. 7, A to C) were cut from 8-week-old wild-type and mutant plants. Immediately after depositing the gametophores on the filters, photos were taken every 1 to 5 min for 2 to 3 h, using a camera (EOS 300D; Canon) with an adapter for the stereomicroscope. Plants were exposed to a 1,300-lux cold light (KL 1500 LCD; Leica).
Software
The distribution of Pos values for all the groups of wild-type and mutant protoplasts did not always follow a normal distribution (the sd was about the same size as the mean) so we used nonparametric tests (Glantz, 1997) to compare the four groups: wild type, pip2;1, pip2;2, and pip2;3. Each Pos value was given a rank: the smallest was ranked 1. The next biggest ranked 2 and so on, up to the largest value. All the values were ranked without regard for the group from which they were derived. If wild type and mutant Pos values were the same, the average rank for each group should be similar to the average rank computed without regard for the grouping.
We used the nonparametric Dunn's test and one-way ANOVA on ranks test from the program SigmaStat (Systat Software GmbH) to identify significant differences between average ranks, i.e. between the groups. The graphs were drawn with SigmaPlot (Systat Software GmbH).
Sequence data from this article have been deposited with the EMBL/GenBank database/ libraries under accession numbers AY494191 (PIP2;1), AY494192 (PIP2;2), and DQ018113 (PIP2;3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Video S1. Wilting of a gametophore deposited on a wet filter (associated with Figure 7A; 1 s on the video corresponds to 5 min in real time).
Supplemental Video S2. Wilting of a gametophore deposited on a wet filter (associated with Figure 7B; 1 s on the video corresponds to 3 min in real time).
Supplemental Video S3. Wilting of a gametophore deposited on a dry filter (associated with Figure 7C; 1 s on the video corresponds to 1 min in real time).
Supplementary Material
Acknowledgments
We thank Martine Gonneau for introducing us to P. patens. We thank Professor Hervé Dupont, Dr. Loïc Faye, and Professor Susannah Gal for their critical comments on the manuscript.
This work was supported by a grant from the Conseil Régional de Haute Normandie (to D.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jean-Paul Lassalles (jp.lassalles@univ-rouen.fr).
The online version of this article contains Web-only data.
References
- Agre P, Borgnia MJ, Yasui M, Neely JD, Carbrey J, Kozono D, Beitz E, Hoffert J, Leitch V, King LS (2001) Discovery of the aquaporins and their impact on basic and clinical physiology. In Aquaporins, Vol 51. Academic Press, London, pp 1–38
- Agre P, Kozono D (2003) Aquaporin water channels: molecular mechanisms for human diseases. FEBS Lett 555 72–78 [DOI] [PubMed] [Google Scholar]
- Borstlap AC (2002) Early diversification of plant aquaporins. Trends Plant Sci 12 529–530 [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ, Morillon R, Maurel C, Gerbeau P, Kjellbom P, Johansson I (2001) Aquaporins of plants: structure, function, regulation, and role in plant water relations. In Aquaporins, Vol 51. Academic Press, London, pp 277–334
- Echevarria M, Windhager EE, Tate SS, Frindt G (1994) Cloning and expression of AQP3, a water channel from the medullary collecting duct of rat kidney. Proc Natl Acad Sci USA 91 10997–11001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein A (1987) Water Movement through Lipid Bilayers, Pores, and Plasma Membranes: Theory and Reality, Vol 4. Wiley-Interscience, New York
- Glantz SA (1997) Primer of Biostatistics. McGraw-Hill, New York
- Gustavsson S, Lebrun AS, Norden K, Chaumont F, Johanson U (2005) A novel plant major intrinsic protein in Physcomitrella patens most similar to bacterial glycerol channels. Plant Physiol 139 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez C, Zelazny E, Chaumont F (2006) Modulating the expression of aquaporin genes in planta: a key to understand their physiological functions? Biochim Biophys Acta 1758 1142–1156 [DOI] [PubMed] [Google Scholar]
- Hazama A, Kozono D, Guggino WB, Agre P, Yasui M (2002) Ion permeation of AQP6 water channel protein—single-channel recordings after Hg2+ activation. J Biol Chem 277 29224–29230 [DOI] [PubMed] [Google Scholar]
- Hill AE, Shachar HB, Shachar HY (2004) What are aquaporins for? J Membr Biol 197 1–32 [DOI] [PubMed] [Google Scholar]
- Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjovall S, Fraysse L, Weig AR, Kjellbom P (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol 126 1358–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu DT, Maurel C (2005) Aquaporins in a challenging environment: molecular gears for adjusting plant water status. Plant Cell Environ 28 85–96 [Google Scholar]
- Maurel C, Javot H, Lauvergeat V, Gerbeau P, Tournaire C, Santoni V, Heyes J (2002) Molecular physiology of aquaporins in plants. Int Rev Cytol 215 105–148 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Fujita T, Shin IT, Seki M, Nishide H, Uchiyama I, Kamiya A, Carninci P, Hayashizaki Y, Shinozaki K, et al (2003) Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implication for land plant evolution. Proc Natl Acad Sci USA 100 8007–8012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobel PS (1999) Physicochemical and Environmental Plant Physiology. Academic Press, San Diego
- Paszkowski J, Baur M, Bogucki A, Potrykus I (1988) Gene targeting in plants. EMBO J 7 4021–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor MCF, Tuba Z (2002) Poikilohydry and homoihydry: antithesis or spectrum of possibilities? New Phytol 156 327–349 [DOI] [PubMed] [Google Scholar]
- Quatrano RS, McDaniel SF, Khandelwal A, Perroud PF, Cove DJ (2007) Physcomitrella patens: mosses enter the genomic age. Curr Opin Plant Biol 10 182–189 [DOI] [PubMed] [Google Scholar]
- Ramahaleo T, Morillon R, Alexandre J, Lassalles J (1999) Osmotic water permeability of isolated protoplasts. Modifications during development. Plant Physiol 119 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara K, Nishiyama T, Sumikawa N, Kofuji R, Murata T, Hasebe M (2003) Involvement of auxin and a homeodomain-leucine zipper I gene in rhizoid development of the moss Physcomitrella patens. Development 130 4835–4846 [DOI] [PubMed] [Google Scholar]
- Salisbury FB, Ross CW (1992) Plant Physiology. Wadsworth Publishing, Belmont, CA
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Woodbury, New York
- Schaefer DG (2002) A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annu Rev Plant Biol 53 477–501 [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zryd JP (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11 1195–1206 [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zryd JP (2001) The moss Physcomitrella patens, now and then. Plant Physiol 127 1430–1438 [PMC free article] [PubMed] [Google Scholar]
- Shigyo M, Tabei N, Yoneyama T, Yanagisawa S (2007) Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol 48 179–185 [DOI] [PubMed] [Google Scholar]
- Stahlberg H, Heymann B, Mitsuoka K, Fuyijoshi Y, Engel A (2001) The aquaporin superfamily: structure and function. In Aquaporins, Vol 51, pp 39–119
- Stroud R, Savage D, Miercke L, Lee J, Khademi S, Harries W (2003) Selectivity and conductance among the glycerol and water conducting aquaporin family of channels. FEBS Lett 555 79–84 [DOI] [PubMed] [Google Scholar]
- Tazawa M, Okazaki Y (1997) Water channel does not limit evaporation of water from plant cells. J Plant Res 110 317–320 [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyerman SD, Bonhert HJ, Maurel C, Steudle E, Smith JAC (1999) Plant aquaporins: their molecular biology, biophysics and significance for plant water relations. J Exp Biol 50 1055–1071 [Google Scholar]
- Tyerman SD, Niemietz CM, Bramley H (2002) Plant aquaporins: multifunctional water and solute channels with expanding roles. Plant Cell Environ 25 173–194 [DOI] [PubMed] [Google Scholar]
- Uehlein N, Lovisolo C, Siefritz F, Kaldenhoff R (2003) The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 425 734–737 [DOI] [PubMed] [Google Scholar]
- Wang TL, Cove DJ, Beutelmann P, Hartmann E (1980) Isopentenyladenine from mutants of the moss, Physcomitrella Patens. Phytochemistry 19 1103–1105 [Google Scholar]
- Xie J, Wehner T, Wollenberg K, Purugganan MD, Conkling MA (2003) Intron and polypeptide evolution of conserved NPA motif regions in plant aquaporins. J Am Soc Hortic Sci 128 591–597 [Google Scholar]
- Yasui M, Hazama A, Kwon TH, Nielsen S, Guggino WB, Agre P (1999) Rapid gating and anion permeability of an intracellular aquaporin. Nature 402 184–187 [DOI] [PubMed] [Google Scholar]
- Zardoya R, Villalba S (2001) A phylogenetic framework for the Aquaporin family in eukaryotes. J Mol Evol 52 391–404 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.