Abstract
Organ detachment requires cell separation within abscission zones (AZs). Physiological studies have established that ethylene and auxin contribute to cell separation control. Genetic analyses of abscission mutants have defined ethylene-independent detachment regulators. Functional genomic strategies leading to global understandings of abscission have awaited methods for isolating AZ cells of low abundance and very small size. Here, we couple laser capture microdissection of Arabidopsis thaliana stamen AZs and GeneChip profiling to reveal the AZ transcriptome responding to a developmental shedding cue. Analyses focus on 551 AZ genes (AZ551) regulated at the highest statistical significance (P ≤ 0.0001) over five floral stages linking prepollination to stamen shed. AZ551 includes mediators of ethylene and auxin signaling as well as receptor-like kinases and extracellular ligands thought to act independent of ethylene. We hypothesized that novel abscission regulators might reside in disproportionately represented Gene Ontology Consortium functional categories for cell wall modifying proteins, extracellular regulators, and nuclear-residing transcription factors. Promoter-β-glucuronidase expression of one transcription factor candidate, ZINC FINGER PROTEIN2 (AtZFP2), was elevated in stamen, petal, and sepal AZs. Flower parts of transgenic lines overexpressing AtZFP2 exhibited asynchronous and delayed abscission. Abscission defects were accompanied by altered floral morphology limiting pollination and fertility. Hand-pollination restored transgenic fruit development but not the rapid abscission seen in wild-type plants, demonstrating that pollination does not assure normal rates of detachment. In wild-type stamen AZs, AtZFP2 is significantly up-regulated postanthesis. Phenotype data from transgene overexpression studies suggest that AtZFP2 participates in processes that directly or indirectly influence organ shed.
Abscission zones (AZs) are tiers of small, densely cytoplasmic cells located at sites of organ detachment (Sexton et al., 1985). Within AZs reside one or more cell layers that separate in response to developmental or environmental cues. Shed is typically preceded by AZ cellular rounding coupled with differential enlargement on opposing sides of the future fracture plane (Morre, 1968; Sexton et al., 1985). Accompanying cell enlargement is the activation of multiple pectin and hemicellulose-modifying proteins that modify primary wall structure and reduce adhesion between AZ cells (del Campillo, 1999; Roberts et al., 2002). Accumulation of pathogenesis-related (PR) proteins limits invasion of pathogens into the proximal AZ face until wound sealing is complete (Roberts et al., 2000). Waxy protection layers finally form across the fracture plane.
Functional analyses of abscission-impaired mutants have identified genes that control abscission competence from the time AZ cells differentiate through the time they separate. LATERAL SUPPRESSOR (LS) encodes a VHIID regulatory gene (Schumacher et al., 1999) required for tomato (Solanum lycopersicum) pedicel AZ development (Malayer and Guard, 1964). JOINTLESS encodes a MADS-box gene (Mao et al., 2000) also required for AZ differentiation. Lesions in AZ formation in both ls and jointless mutants are restricted to flower and fruit pedicels; functional AZs are present in leaves and other structures (Butler, 1936; Malayer and Guard, 1964). Thus, AZ differentiation differs between organs. Differentiation must be complete before a plant is competent to respond to abscission signals (Osborne and Sargent, 1976). Although positional and biochemical differentiation may occur early in organogenesis (Kendall, 1918; Osborne and Sargent, 1976; McManus and Osborne, 1990, 1991), morphological features defining AZs may not be visible. In cotton (Gossypium hirsutum), clear AZ structure is not present until before shed, when further AZ cell division may occur (Leinweber and Hall, 1959; Bornman et al., 1967). In Arabidopsis (Arabidopsis thaliana) and tomato, clear morphological features define AZs long before abscission.
Genetic studies show abscission capacity to be influenced by disruptions in organ boundaries and other alterations in organ patterning. Partial fusion of sepals in the F-box gene mutant hawaiian skirt impairs shedding of Arabidopsis floral parts in which AZs appear to differentiate normally (González-Carranza et al., 2007b). Two BTB/POZ domain proteins, BLADE ON PETIOLE1 (BOP1) and BOP2, are expressed in regions overlapping the floral organ AZs (Ha et al., 2004; Hepworth et al., 2005; Norberg et al., 2005). A bop1bop2 double mutant exhibited abnormal organ patterning and loss of floral organ abscission (Hepworth et al., 2005; Norberg et al., 2005). Other transcription factors contributing to abscission competence include the structurally related MADS-box domain proteins AGL15 and AGL18; shedding of Arabidopsis floral parts is delayed in plants overaccumulating either protein (Fernandez et al., 2000; Adamczyk et al., 2007). AGL15 and AGL18 overexpressors exhibit concomitant slowing of other developmental transitions including flowering time and senescence (Fernandez et al., 2000; Adamczyk et al., 2007). Early flowering time and a slowing of floral senescence also accompany delayed abscission in plants with knocked-down levels of ACTIN-RELATED PROTEIN4 (ARP4; Kandasamy et al., 2005a). Similarly, knocking down ARP7 expression levels delays abscission of floral parts and alters flower development, impacting fertility (Kandasamy et al., 2005b). Pleiotropic phenotypes of ARP RNAi plants have been predicted to arise from aberrant gene transcription patterns caused by altered chromatin structure (Kandasamy et al., 2005b).
In wild-type plants, binding of ethylene to one or more ethylene receptors derepresses the ethylene signal transduction pathway leading to hormone-dependent responses including abscission. Once considered a fundamental abscission signal (Jackson and Osborne, 1970), ethylene is now viewed as an abscission rate regulator (Bleecker and Patterson, 1997; Patterson, 2001). Mutations in the etr1-1 ethylene receptor gene that render Arabidopsis plants insensitive to ethylene delay, rather than block, floral organ abscission (Patterson and Bleecker, 2004). Ethylene control of abscission is influenced in part via antagonism between ethylene and auxin. Abscission is delayed in plants with suppressed expression of the ARF2 member of the auxin response family (Ellis et al., 2005). ARF2 controls auxin signaling (Ellis et al., 2005) and ethylene synthesis is altered in arf2 mutants (Okushima et al., 2005). Nonhormone ligands controlling abscission include that encoded by INFLORESCENCE DEFICIENT IN ABSCISSION (IDA). Loss-of-function ida mutants fail to abscise Arabidopsis sepals, petals, and stamens (Butenko et al., 2003); transgenic lines overexpressing IDA rapidly shed all floral organs, as well as additional plant parts that do not normally abscise (Stenvik et al., 2006). The receptor for IDA is unknown but has been proposed to include receptor-like kinases (RLKs). Antisense inhibition of gene expression corresponding to the RLK termed HAESA (HAE) blocks abscission of Arabidopsis stamens, sepals, and petals (Jinn et al., 2000). Expression of HAE appears to be independent of ethylene synthesis or perception (Jinn et al., 2000). This is also the case with AGL15 (Fernandez et al., 2000) and ARP7 (Kandasamy et al., 2005b). At present, mechanisms by which putative ethylene-independent pathways contribute to abscission are poorly understood.
Many questions remain about abscission signaling. What is the primary abscission cue or cues? What regulators exert earliest control over signal perception and response? Why do some organs abscise in response to a given stimulus whereas others are retained? What genes control ethylene-dependent and independent pathways and how do pathways interact? Functional genomic approaches to addressing these questions have been hindered by an inability to obtain pure populations of AZ cells. Recently, we optimized methods for using laser-capture microdissection (LCM) to harvest highly enriched populations of specialized cells (Cai and Lashbrook, 2006). There, replum cell harvest was linked to ATH1 GeneChip studies of fruit maturation (Cai and Lashbrook, 2006). Here, we reveal dynamic changes in global gene expression taking place in AZs of Arabidopsis stamens progressing from prepollination to organ shed. Functional analyses of one AZ-up-regulated gene, ZINC FINGER PROTEIN2 (AtZFP2), provide evidence that this transcription factor participates in processes that directly or indirectly influence shedding of floral organs.
RESULTS AND DISCUSSION
LCM Facilitates AZ Transcriptome Profiling
Arabidopsis stamens, sepals, and petals abscise postpollination. Detachment is dependent upon separation of AZ cells residing at the bases of floral organs. Small AZ sizes and low numbers of AZ cells in Arabidopsis and other plants have historically confounded efforts to prepare enriched AZ populations for downstream analyses. We chose to isolate AZ cells using LCM, a method that has been successfully used to capture multiple specialized cell types from plants. Sources for laser-captured cells have included, but are not limited to embryos (Casson et al., 2005), the shoot apical meristem (Ohtsu et al., 2007a; Zhang et al., 2007), phloem (Asano et al., 2002; Vilaine et al., 2003; Yu et al., 2007), leaf epidermis (Nakazono et al., 2003; Murata and DeLuca, 2005), leaf mesophyll (Corpas et al., 2006), bundle sheath cells (Kerk et al., 2003), ovules and replums (Cai and Lashbrook, 2006), cell walls (Angeles et al., 2006), roots (Ramsay et al., 2004; Woll et al., 2005; Alkharouf et al., 2006; Dembinsky et al., 2007; Ithal et al., 2007; Klink et al., 2007), and floral organs (Nakada et al., 2006). LCM and its applications to biological studies in plants have been recently reviewed by Day et al. (2005, 2007), Nelson et al. (2006), and Ohtsu et al. (2007b).
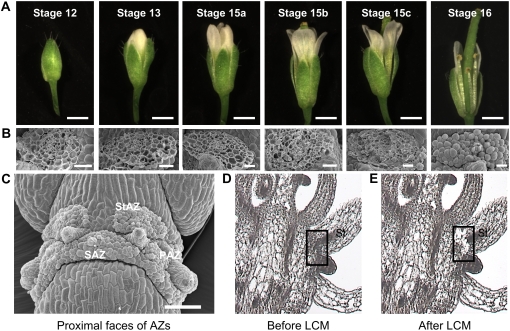
We chose stamens as a source of AZs for LCM studies because there are six stamens instead of four petals or sepals. This elevates the relative incidence of AZs in sectioned tissue and reduces the time required for cell capture. Whole flowers corresponding to five developmental stages linking prepollination to the onset of organ shed were fixed and paraffin-embedded and sectioned tissues were tape-transferred to slides (Cai and Lashbrook, 2006). Figure 1A depicts developmental stages of flowers from which stamen AZs were harvested. Stage numbers are those of Smyth et al. (1990). Anthesis occurs at Stage 13. Stage 15 was divided into substages a to c as defined in “Materials and Methods”. Stage 16 tissue was not prepared for LCM after it was determined that floral organs detached at their AZs during early fixation steps.
Figure 1.
Developmental stages of Arabidopsis flowers and scanning electron and light micrographs of floral organ AZs. A, Flower stages. Scale bar, 1 mm. B, SEM of stamen AZ fracture planes. Scale bar, 20 μm. C, SEM of proximal faces of AZs from stamens (StAZ), sepals (SAZ), and petals (PAZ). Scale bar, 100 μm. D and E, Sectioned floral tissue before (D) and after (E) LCM of stamen AZs (shown within boxes).
In Figure 1, B and C, scanning electron microscopy (SEM) visualizes surface features of AZ fracture planes at stages selected for abscission studies. Figure 1B depicts stamen AZ scars left on the parent plant after manual filament removal; Figure 1C shows parental sides of all floral organ fracture planes after organ detachment. The intact rounded AZ cells observed in Figure 1B (stages 15c and 16) and Figure 1C represent proximal AZ cells that have completely separated from contiguous distal AZ cells of the leaving organ (Patterson and Bleecker, 2004). In contrast, torn AZ cells observed in Figure 1B (stages 12–15c) testify that cell wall dissolution processes needed for separation from neighboring cells are not yet complete. In our studies, intact cells present on proximal fracture faces after organ removal are first evident on outermost AZ margins at stage 15c (Fig. 1B). Thus, final stages of separation occurring within a subset of stamen AZ cells have occurred by stage 15c. Cell separation is essentially complete between all stamen AZs of the proximal fracture plane by stage 16, when all AZ cells are rounded and intact (Fig. 1B) and organs detach when lightly touched.
Stamen AZ cells were laser-captured from flowers at stages 12 to 15c. Figure 1, D and E, shows representative flower sections before and after LCM of stamen AZs, respectively. Microdissected AZs included cells from the vascular bundle that passes through all abscission layers. Approximately 10,000 floral organ AZ cells could be captured in approximately 1 d and the amount of RNA subsequently isolated per laser-captured cell was approximately 10 to 15 pg (Cai and Lashbrook, 2006). Total RNA served as the template for preparing hybridization targets for ATH1 GeneChips (Cai and Lashbrook, 2006).
Probe Set Signals Regulated at the Highest Level of Statistical Significance Define a 551-Member Slice of the AZ Transcriptome
ATH1 GeneChips contain probe sets representing approximately 24,000 genes of the Arabidopsis genome. Replicated hybridizations of biotin-labeled aRNAs from AZs at stages 12 to 15c revealed statistically significant changes in probe set signal intensities corresponding to many AZ transcripts. We wished to restrict preliminary analyses to a manageable number of genes whose expression could provide a first glimpse of regulatory processes leading to cell separation. Restricting attention to the most significantly regulated probe sets (P ≤ 0.0001) generated a population of 551 transcripts (AZ551) representing the Arabidopsis stamen AZ transcriptome. An approximately 0.2% false discovery rate was determined using the method of Storey and Tibshirani (2003). A complete list of AZ551 genes is in Supplemental Table S1. Quantitative real-time PCR (Q-PCR) successfully validated microarray expression data for a subsample of 10 probe sets using stamen AZ RNA from laser-captured cells (data not shown). Supplemental Figure S1 depicts expression patterns of three validated transcripts related to GA metabolism or action. Both Q-PCR and microarray data showed similar trends in mRNA accumulation for the GA-related mRNAs shown in Supplemental Figure S1 and discussed later in the text.
Functional categorization of AZ551 using Gene Ontology Consortium (GO) tools (Berardini et al., 2004) on the Arabidopsis Information Resource Web site (http://www.arabidopsis.org) permitted comparisons between predicted functions of AZ transcripts and all Arabidopsis transcripts represented on the ATH1 GeneChip (Table I). Functional classification by the GO cellular component showed that the cell wall and extracellular matrix were represented in AZs at levels exceeding those in ATH1 by approximately 3- to 4-fold. The nucleus was represented in AZs at levels exceeding that of ATH1 by 1.6-fold (Table I). Consistent with AZ cellular component data was the increased representation of GO categories representing molecular function (Table I). Transcription factor activities within the AZ551 exceeded that of ATH1 by almost 2-fold. Collectively, the GO cellular component and molecular function data are consistent with known contributions of transcription factors and cell wall modifying proteins to abscission competence and cell wall disassembly, respectively (Mao et al., 2000; Roberts et al., 2002).
Table I.
Functional categorization of the 551 most significantly regulated transcripts in stamen AZs
Functional categories in which AZ transcript abundance exceeded that found in the ATH1 by at least 50% are represented. GO categorization data were determined on July 18, 2007. P-values were obtained using Fisher's exact test.
| GO Classification by Genes | Percent of Total Transcripts | AZ Overrepresentation | P-Value | |
|---|---|---|---|---|
| A. By cellular component | ATH1 | AZ551 | Fold Change | |
| Cell wall | 0.925 | 3.5 | 3.8 | 1.1 e-7 |
| Extracellular | 0.72 | 2.3 | 3.2 | 8.0 e-5 |
| Nucleus | 7.6 | 12.1 | 1.6 | 1.5 e-5 |
| Other membranes | 20.1 | 31.1 | 1.5 | 1.1 e-11 |
| B. By molecular function | ||||
| Transcription factor activity | 5.8 | 11.3 | 1.95 | 2.9 e-12 |
| C. By biological process | ||||
| Response to stress | 2.2 | 4.6 | 2.1 | 4.5 e-10 |
| Response to abiotic or biotic stimulus | 2.7 | 5.2 | 1.9 | 4.2 e-10 |
| Transcription | 3.7 | 5.4 | 1.5 | 2.2 e-6 |
| Signal transduction | 2.0 | 2.9 | 1.5 | 9.8 e-4 |
AZ Transcripts in the 551-Member Population Can Be Assigned to Eight Clusters with Both Distinct and Overlapping Functions
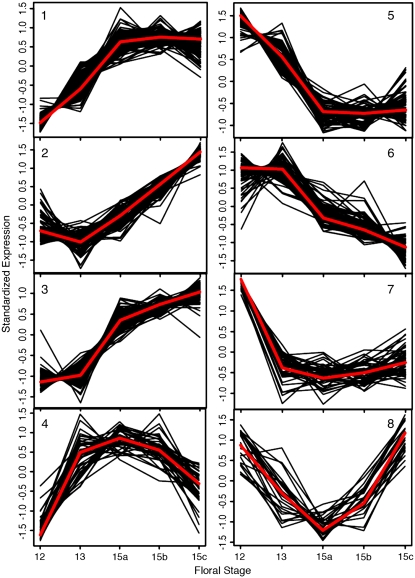
Cluster analysis is a statistical procedure that identifies and organizes patterns contained in complex data sets (Spellman, 2003). In genomic studies, clusters may represent common patterns of gene expression observed within a larger gene transcript population. Eisen et al. (1998) and others have shown that functionally related transcripts are often housed in the same cluster. Thus, knowing what biological functions are associated with a subset of transcripts within a cluster can provide important clues about potential roles for noncharacterized transcripts within the same population. As the first step toward establishing what discrete biological processes may collectively facilitate cell separation in AZs, we initiated cluster analyses of the AZ551 transcript set. The gap statistic method of Tibshirani et al. (2001) estimated that eight clusters could reasonably represent the gene expression patterns found within this transcript collection (Fig. 2). In brief, the gap statistic procedure assumes that a data set comprises a single component. This assumption serves as a null hypothesis that is rejected when statistical evidence supports the presence of additional clusters separated within expression space (Tibshirani et al., 2001). The K-medoid-based clustering algorithm of Kaufman and Rousseeuw (1990) used a distance metric to group AZ551 transcripts sharing similar distributions in expression space. In Figure 2, clusters 1 to 3 represent up-regulated genes within AZ551; clusters 5 to 7 contain down-regulated genes and the remaining two clusters represent genes that are both up- and down-regulated. Transcript identities are annotated in Supplemental Table S1.
Figure 2.
Classification of 551 AZ transcripts into eight clusters with similar patterns of gene expression. Clustering algorithms are described in “Materials and Methods”. [See online article for color version of this figure.]
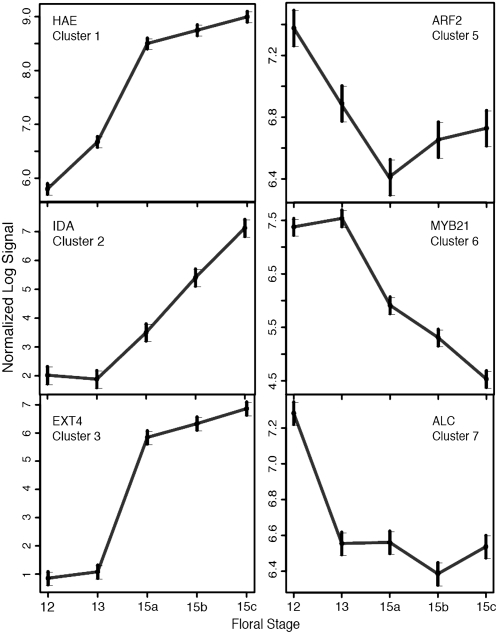
Figure 3 demonstrates that six clusters within the eight-member cluster collection contain transcripts shown by previous genetic studies to participate in cell separation processes associated with abscission or dehiscence. The effects on abscission of modulating gene expression corresponding to HAE (cluster 1), IDA (cluster 2), and ARF2 (cluster 5) were described earlier. mRNA for cluster 3's AtEXT4 (extensin 4) extensin, previously termed EXT1, was previously shown to accumulate in floral AZs and other tissues, where it is responsive to hormonal cues (Merkouropoulos and Shirsat, 2003). MYB21 gene expression (cluster 6) is required for male fertility; knockout mutants exhibit delayed anther dehiscence (Mandaokar et al., 2006). ALCATRAZ (cluster 7) is required for cell separation during silique dehiscence (Rajani and Sundaresan, 2001). Collectively, data of Figure 3 demonstrate that AZ551 contains genes previously defined as being expressed in separating plant cells. Their presence imparted confidence that AZ551 would contain functionally uncharacterized transcripts with potentially novel roles in abscission.
Figure 3.
Expression profiles of selected cell separation regulators represented in AZ551. Mean natural log signal intensities are plotted across developmental stages. Gene names are defined in the text.
In Table I, GO functional categories for AZ551 transcripts were compared with Arabidopsis genome transcripts on the ATH1 GeneChip. We similarly wished to establish if certain functions were overrepresented to statistically significant extents in one or more of the eight transcript clusters. In Table II, GO functional categorization of clusters coupled with an assessment of the statistical significance of relative category representation reveals general trends. First, processes occurring in cell walls are significantly overrepresented in clusters with up-regulated transcripts, consistent with known de novo synthesis of wall modifying proteins during abscission (Taylor and Whitelaw, 2001; Roberts et al., 2002). Statistically overrepresented functions within both up- and down-regulated clusters include nuclear functions involving transcription factors and nucleotide binding proteins, and transport functions (Table II). Supplemental Table S2 reveals transcript fold changes between floral stages for a subset of AZ551 genes. Annotation conventions within Supplemental Table S2 include those of Kende et al. (2004), Nakano et al. (2006), Palusa et al. (2007), and Rose et al. (2002).
Table II.
Overrepresented functions in transcript clusters relative to ATH1 probe sets
GO categorization data were determined on July 18, 2007. P-values were obtained using Fisher's exact test.
| Cluster No. | By GO Cellular Component | P-Value | By GO Molecular Function | P-Value |
|---|---|---|---|---|
| Up-regulated | ||||
| 1 | Nucleus | 0.015 | Transcription factor activity | 0.00002 |
| Cell wall | 0.0009 | Nucleotide binding | 0.0003 | |
| Other membranes | 0.029 | Transferase activity | 0.02 | |
| DNA or RNA binding | 0.03 | |||
| 2 | Cell wall | 0.0007 | Hydrolase activity | 0.002 |
| Extracellular | 0.017 | Other enzyme Activity | 0.0004 | |
| Endoplasmic reticulum | 0.007 | |||
| 3 | Nucleus | 0.03 | Transcription factor activity | 0.00001 |
| Cell wall | 0.003 | Other enzyme activity | 0.008 | |
| Other membranes | 0.0019 | |||
| Up-regulated early; down-regulated late | ||||
| 4 | Other membranes | 0.003 | Transporter activity | 0.037 |
| Mitochondria | 0.042 | |||
| Down-regulated | ||||
| 5 | Extracellular | 0.019 | Transferase activity | 0.008 |
| Other membranes | 0.00003 | Hydrolase activity | 0.0004 | |
| 6 | Nucleus | 0.0004 | Transcription factor activity | 1.5 e-7 |
| Cell wall | 0.03 | DNA or RNA binding | 0.001 | |
| Extracellular | 0.016 | |||
| Plasma membrane | 0.04 | |||
| 7 | Mitochondria | 0.03 | Transferase activity | 0.03 |
| Other membranes | 0.016 | Hydrolase activity | 0.04 | |
| Down-regulated early; up-regulated late | ||||
| 8 | – | Transporter activity | 0.025 | |
| Other enzyme activity | 0.027 |
Potential Functions of Up-Regulated Gene Clusters
Up-Regulated Cluster 1 Genes Encode Transcriptional Modulators, RLKs, Cell Wall Modifying Proteins, and Regulators of Hormone Biosynthesis, Action, and Transport
Cluster 1 transcripts increase in abundance between stages 12 and 15a and remain at high levels as abscission commences near stages 15b and 15c (Fig. 2). GO-annotated genes within cluster 1 are assigned to the nuclear compartment at rates surpassing those for other transcripts represented on the ATH1 GeneChip (Table II). Approximately 18% of cluster 1 mRNAs are assigned to a nuclear site versus 7.6% of ATH1 Arabidopsis genes and approximately 12% of AZ551 (data not shown). Myb transcription factors are well represented. Supplemental Table S2 reveals transcript fold changes between stages for a subset of AZ551 genes including Myb factors. Up-regulated genes include AtMYB4, 14, 45, 62, and 75 (Supplemental Table S2). All of these Mybs are modulated to some extent by Suc and nitrogen status with AtMYB14 also weakly regulated by auxin (Kranz et al., 1998). AtMYB4 and AtMYB75 gene products act as negative and positive transcriptional regulators, respectively, of phenylpropanoid biosynthesis (Borevitz et al., 2000; Jin et al., 2000; Rose et al., 2002). GO molecular function comparisons of cluster 1 transcripts show that transcription factor activities as well as activities for DNA, RNA, or nucleotide binding are overrepresented (Table II). Up-regulated genes in Supplemental Table S2 include RLKs that translate extracellular signals into cellular responses (Becraft, 2002). AZ551 RLKs present in annotations of Shiu and Bleecker (2001) include HAE (At4g28490), HAESA-LIKE2 (HSL2; At5g65710), FLAGELLIN SENSITIVE2 (At5g46330), and others (At1g09970, At3g14840, and At5g59010). HAE is noteworthy because Jinn et al. (2000) have reported that antisense inhibition of HAE prevented abscission of stamens, sepals, and petals. In our studies, developmental up-regulation of HAE at stage 12 (Supplemental Table S2) precedes first visible signs of cell separation at stage 15c (Fig. 1B) by several stages. We conclude that HAE must represent a very early contributor to abscission competence.
HAE appears to mediate steps within an ethylene independent abscission pathway as corresponding antisense mutants have the ethylene sensitivity to evoke a hormone-dependent triple response equivalent to that of wild-type controls (Jinn et al., 2000). Ethylene likely controls the rate of a separate abscission pathway that could communicate with components of the HAE route (Butenko et al., 2006). Our data suggest that both ethylene-dependent and -independent pathways are operational after stage 12 (Supplemental Table S2). Components of the former include ethylene response factors (ERFs) 003 and 072 (At5g25190 and At3g16770) and ethylene insensitive 3 (EIN3)-binding F-box protein 2 (EBF2; At5g25350) that contributes to SCF-dependent ubiquitination and degradation (Potuschak et al., 2003). Modulated ethylene action is accompanied by induction of multiple auxin responsive genes and accumulation of the PIN4 auxin efflux transcript (At2g01420). BR6OX2 (At3g30180) mediates the last step of brassinolide synthesis. It is up-regulated as is the brassinolide-enhanced target BEE3 (At1g73830) and GA-regulated GASA5 (At3g02885; Supplemental Fig. S1). Thus, expression of RLKs like HAE and HSL2 takes place in a complex backdrop of hormone signaling.
Abscission rate is partially controlled by cell wall modifying proteins (Lashbrook et al., 1998; Brummell et al., 1999). Almost 5% of transcripts in cluster 1 are assigned cell wall localization by GO analysis in contrast to almost 1% of all Arabidopsis genes (data not shown). Regulated wall transcripts in Supplemental Tables S1 and S2 include AtXTH28 (At1g14720), one member of the complex endotransglucosylase/hydrolase (XTH) gene family (Becnel et al., 2006). In current cell wall structural models, xyloglucans coat cellulosic microfibrils and tether microfibril neighbors (Hayashi, 1989; Carpita and Gibeaut, 1993). XTHs have one or both of two activities that can loosen cell walls during growth or strengthen them during assembly (Thompson and Fry, 1997, 2001; Rose et al., 2002). Similarly, cluster 1 transcripts include multiple peroxidases including PER21 (At2g37130), PER30 (At3g21770), and PER33 and/or PER34 (At3g49120 and/or At3g49110). Peroxidases, like XTHs, may loosen or stiffen cell walls in different cellular contexts (Passardi et al., 2005). In roots, PER34 increases root length (Passardi et al., 2005), presumably via cell wall loosening.
Up-Regulated Cluster 2 Transcripts Encode Proteins with Roles in Pectin Modification, Hormone Synthesis and Degradation, Receptor Binding, and Signal Transduction
Cluster 2 transcript levels rise throughout all stages assayed whereas cluster 1 mRNA abundance stabilizes after stage 15a (Fig. 2). It is possible that gene expression at later stages in cluster 2 contributes to processes necessary for final stages of organ detachment. Cluster 2 is enriched in transcripts with likely roles in cell wall structural modification (Table II). Regulated cell wall genes encode two glycosyl hydrolases with structural similarities to endo-β-1,3-glucanases (At4g18340) and endo-β-1,4-glucanases (At2g32990). However, most up-regulated cell wall genes encode pectolytic enzymes, including three pectin methylesterases (PMEs; At2g45220, At2g47550, and At4g02330), two polygalacturonases (At2g43890 and At3g07970), one pectate lyase-like protein (At3g27400), and a member of the invertase/PME inhibitor (PMEI) family (At3g47380). PMEs deesterify pectin to establish structurally and functionally distinct pectin classes (Ridley et al., 2001; Willats et al., 2001) and interact cooperatively with polygalacturonase (PG) to regulate homogalacturonan disassembly (Lashbrook, 2005). PMEIs regulate PMEs at the transcriptional and/or posttranslational levels. Pectate lyases, like PG, prefer homogalacturonan substrate de-esterified by PMEs, suggesting that cluster 2 transcripts have the potential to act cooperatively. The preponderance of pectin-modifying proteins is consistent with presumptive roles for pectin disassembly in reducing intercellular adhesion between AZ cells (Roberts et al., 2002). González-Carranza et al. (2002) reported that the promoter of abscission-related polygalacturonase PGAZAT (At2g41850) directed GUS expression to the bases of all floral organs after natural abscission was initiated. At2g41850 was not a member of our AZ551 population restricted by P-value. However, evaluation of its expression elsewhere in our data revealed low probe set signal intensities between stages 12 and 15b and increased expression at stage 15c (data not shown). These results may be consistent with the results of González-Carranza et al. (2002); PGAZAT gene expression initiated upon natural abscission would be expected to follow stage 15c, where the first signs of completed cell separation appear in Figure 1B.
IDA (At1g68765) encodes a 77-amino-acid ligand secreted into the extracellular matrix (Butenko et al., 2003). ida knockout mutants possess AZs of apparent normal structure but fail to abscise floral organs (Butenko et al., 2003). Like antisensed HAE plants (Jinn et al., 2000), ida mutants are ethylene sensitive (Butenko et al., 2003). Lines overexpressing IDA exhibit ectopic abscission of nonfloral organs along with normal organ shed (Stenvik et al., 2006). These data initially suggested that IDA was a component of an ethylene-independent pathway (Butenko et al., 2003), although subsequent studies showed ethylene might affect spatial distribution of IDA gene expression (Butenko et al., 2006). In fact, stimulation of IDA signaling in floral AZs may occur in a more complex hormonal background. Controllers of AZ hormone response may include ERF086 (At5g18560) and BRS1 (At4g30610), a suppressor of the brassinosteroid receptor BRI1. Up-regulated transcripts of hormone biosynthesis include anthranilate synthase (At5g05730), which encodes the enzyme catalyzing the first committed step of Trp biosynthesis and hence potential auxin production. GA 2-OXIDASE2 expression (At1g30040; Supplemental Fig. S1) leads to GA catabolism via 2-β-hydroxylation (Thomas et al., 1999). OPR1 (At1g76680) and/or OPR2 (At1g76690), homologs of the OPR3 jasmonate (JA) synthesis gene, are also up-regulated. The ATH1 GeneChip cannot distinguish between OPR1 and OPR2, but promoter-GUS studies suggest that up-regulated AZ activity is OPR1 (Biesgen and Weiler, 1999). OPR1 and OPR2 are considered unrelated to JA production (Schaller et al., 1998) because they act on steroisomeric forms of OPDA substrate unsuitable for JA biosynthesis (Schaller and Weiler, 1997; Schaller et al., 2000).
Likely Functions of Cluster 3 and 4 Genes Overlap with Those of Clusters 1 and 2
Multiple up-regulated Myb factors complement those of other clusters, including MYB2, 17, 37, 49, 68, 74, 116, and a Myb-like factor (Supplemental Table S2). MYB17 and MYB68 are regulated by nitrogen and sugar status (Kranz et al., 1998). Nutrient recycling precedes abscission, and transporter activity in AZ551 and cluster 4 exceeds that found in the whole genome (Tables I and II). Transporters that might remove nutrients from senescing or detaching organs are exemplified by cluster 3's sugar alcohol permease homolog, AtPLT5 (At3g18830). Strong up-regulation of AtPLT5 in floral AZs is consistent with previous reports suggesting a role in the reuptake of sugars released by polysaccharide degrading enzymes (Reinders et al., 2005).
Multiple regulated genes in clusters 3 and 4 control hormone signal transduction and cell wall structure. They include ERF043 (At4g32800), ERF092 (At3g23240), and ERF113 (At5g13330). Expression of ERF092, formerly termed ERF1, is induced by JA and ethylene in defense gene systems (Lorenzo et al., 2003). A potential hormone-regulated target during abscission is ethylene-responsive protein 33-like (At1g05710). Modulation of ethylene sensitivity is accompanied by activation of auxin-responsive proteins (At3g60690 and At5g35735). Up-regulation of a BRI1 receptor homolog (At3g13380) is consistent with the up-regulation in cluster 1 of the BRS1 component of early BR signaling (Li et al., 2001). Like HAE, BRI1 encodes a Leu-rich repeat RLK (Friedrichsen et al., 2000) and its induction in AZ551 suggests that RLK signaling is central to cell separation control. Expression data for two additional RLKs in cluster 4 are listed in Supplemental Table S2.
GO cellular component analysis suggests that the cell wall is an overrepresented site of cluster 3 gene expression (Table II). Cell wall modifying protein candidates (Supplemental Table S2) are AtXTH12 (At5g57530) and AtEXT4 (At1g76930). Another XTH, XTH7 (At4g37800) is up-regulated in cluster 4 (Supplemental Table S2). As noted earlier, XTHs can strengthen or loosen cell walls (Thompson and Fry, 2001; Rose et al., 2002). Extensins are insoluble proteins that increase cell wall strength. Merkouropoulos and Shirsat (2003) detected the expression of AtEXT4 within floral organ AZs. AtEXT4 overexpression increases the thickness of Arabidopsis inflorescences (Roberts and Shirsat, 2006). A plethora of regulated cluster 3 peroxidases (Supplemental Tables S1 and S2) may also contribute to cell wall modifications. The inclusion of potential cell wall strengthening proteins in cluster 3 could suggest a role for wall reinforcement during net disassembly.
Potential Functions of Down-Regulated Gene Clusters
Selected Myb Factors and Regulators of Hormone Response and Cell Wall Structure Are Down-Regulated in Clusters 5 and 6
Positive regulation of cell wall extensibility is counterbalanced by reduced expression of other genes. Several cellulose synthase (CESA) and synthase-like (CSLA) genes are down-regulated in cluster 5 (Supplemental Table S2). Some members of the CSLA gene family encode β-1,4-mannan and glucomannan synthases (Liepman et al., 2005). Hamann et al. (2004) postulated that individual CSLAs might exert tissue-specific roles given their low expression in whole organs; AtCSLA10, a cluster 5 gene, exhibited enhanced expression in flowers. Transcripts for expansin-like protein AtEXLA2 and expansin AtEXPA8 (At4g38400 and At2g40610) decline in AZs at stages 12 and 13, respectively. Expansins regulate cell wall extensibility via pH-dependent, nonenzymic means; this may involve interfering with hydrogen bonding of hemicelluloses and cellulose to convert wall tension to polymer creep (Cosgrove et al., 2002). Our data would not suggest an obvious role for AtEXPA8 or AtEXLA2 in regulating cell separation during stage 15. However, elevated EXP or EXP-like expression between stages 12 and 13 could facilitate subsequent entry of pectolytic enzymes into the cell wall matrix. Maximal EXP/EXP-like expression between stages 12 and 13 is accompanied by previously mentioned up-regulation of multiple genes encoding pectolytic enzymes. Reduced expression of expansin genes thereafter is accompanied by a reduction in mRNA abundance for a number of PG, PME, and PMEI genes (Supplemental Table S2, clusters 5 and 6), as well as XTH4 (At2g06850).
GO categorization assigns cluster 6 transcripts to the nucleus more often than it assigns all transcripts represented on the ATH1 GeneChip (Table II). Just as multiple Myb genes were up-regulated elsewhere, many Mybs are down-regulated in cluster 6 (Supplemental Table S2). These include AtMYB6, 21, 39, and 57. MYB21 is expressed in flower buds (Kranz et al., 1998) and in stamens where it is JA responsive (Mandaokar et al., 2006). MYB6, first characterized by Li and Parish (1995) is weakly responsive to ethylene, ABA, and IAA (Kranz et al., 1998). Other transcription factors in this cluster (Supplemental Table S1) include AGAMOUS (At4g18960) and an AGAMOUS-activated regulator of GA biosynthesis (At4g32980). Supporting a role for enhanced GA response postpollination is down-regulation of the RGL1 (Supplemental Fig. S1) and RGL2 negative regulators of GA response. Both of these DELLA proteins normally restrain cellular expansion. It would be interesting to determine if derepression of RGL targets potentiates AZ cell expansion.
Cluster 7 Contains Multiple Down-Regulated Genes of JA Biosynthesis
Cluster 7 transcripts exhibit steep declines in abundance after stage 12 (Fig. 2). A hallmark of such gene expression is the coordinate down-regulation of JA biosynthesis. JA synthesis requires sequential action of lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC), and OPR3 (Delker et al., 2006). JA can be converted to volatile methyl JA (MeJA) by JA carboxyl methyltransferase (At1g19640; Seo et al., 2001). Down-regulation of at least one enzyme from each of these steps occurs prior to abscission. Down-regulated genes in AZ551 include LOX3 and LOX3-like lipoxygenases (At1g17420 and At1g72520), AOS (At5g42650), and AOC1 (At3g25760). Transcripts for OPR3 (At2g06050) and JA carboxyl methyltransferase (At1g19640) were not represented in AZ551 restricted by P-value. High JA biosynthetic gene expression at stage 12 likely attests to requirements for JA in anther filament elongation, anther dehiscence, and pollen viability (Sanders et al., 2000; Stintzi and Browse, 2000).
Down-regulation of JA biosynthesis was unexpected in that both JA and MeJA promote abscission when applied externally (Ueda et al., 1996; Miyamoto et al., 1997; Hartmond et al., 2000). In citrus (Citrus sinensis), external application of MeJA or the structural and functional JA analog coronatine likely induces abscission by stimulating levels of ethylene (Hartmond et al., 2000; Burns et al., 2003). In bean (Phaseolus vulgaris), no ethylene production was evident following JA or MeJA-induced abscission (Ueda et al., 1996) although JA significantly promotes senescence (Ueda and Kato, 1982; Ueda et al., 1991). In Arabidopsis, exogenous JA also elicits premature senescence (He et al., 2002). Senescence may promote abscission by lowering auxin and/or inducing ethylene. Our results in Arabidopsis do not suggest an obvious direct role for JA or MeJA biosynthesis in controlling cell separation. Of note, reduction of JA levels after stage 13 is accompanied by apparent loss of JA responsiveness. Coronatine-responsive protein (At1g19670) is one down-regulated example. If JAs positively modulate abscission it is likely that they act indirectly or, like HAE, act very early in the abscission pathway. However, constitutive expression of the sole allene oxide synthase gene in Arabidopsis did not lead to precocious floral organ abscission in studies of Kubigsteltig and Weiler (2003). There, floral parts of the cas AOS overexpresser persisted along much of the inflorescence. Wild-type Arabidopsis plants typically retain flowers only at uppermost positions.
Cluster 8 Gene Expression Declines Postpollination But Is Restored to Near-Original Levels Prior to Cell Separation
Cluster 8 contains only 25 transcripts, and thus represents less than 5% of AZ551. mRNAs decline in abundance between stages 12 to 15a but are up-regulated to near-original levels by the time of cell separation (Fig. 2). GO analysis reveals cluster transporter activities are overrepresented relative to all ATH1 GeneChip transcripts (Table II). Electron transport proteins represent a subset of transporters; others shuttle one or more ions (e.g. H+, Na+, K+, and phosphate) or nucleotides (e.g. GTP), or nucleotide sugars (e.g. UDP-Gal). Resumption of cluster 8 transcript accumulation before cell separation may be necessary for energy generation, transmembrane trafficking, and/or maintenance of ionic homeostasis. The production of UDP-Xyl by the UXS3 isoform of UDP-GlcUA decarboxylase (At5g59290) could conceivably regulate cell wall synthesis late in abscission to temper cell wall disassembly. UDP-GlcUA decarboxylases generate substrate for making cell wall material and regulate biosynthetic enzymes through feedback mechanisms (Harper and Bar-Peled, 2002).
Functional Analyses Reveal Likely Participation of AtZFP2, a Zinc Finger Protein, in Processes Affecting Abscission Capacity
Figure 3 showed that AZ551 contains multiple expressed genes shown by previous reverse and forward genetic strategies to be associated with the abscission process. Their presence imparted confidence that AZ551 would contain other transcripts of unknown and potentially novel roles in cell separation. We hypothesized that likely sources of novel abscission genes would include the disproportionately represented GO functional categories for cell wall modifying proteins, extracellular regulators, and transcription factors (Table I). A subset of AZ551 transcripts was considered for potential functional analyses; one such target was AtZFP2 (At5g57520).
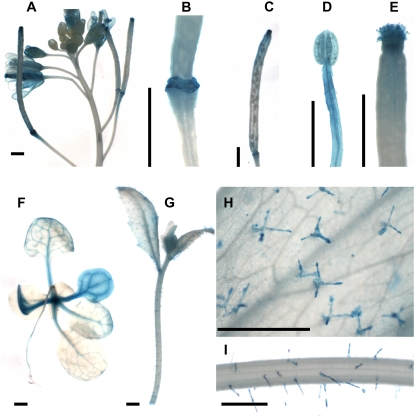
Zinc finger proteins regulate many developmental and stress responses (Takatsuji, 1998, 1999). At least 33 Arabidopsis ZFPs contain only one zinc finger domain (Englbrecht et al., 2004). Twenty-eight members of this so-called C1-l1 class of zinc finger proteins share a conserved QALGGH sequence within a putative DNA-contacting surface and a C-terminal Leu-rich sequence (Tague and Goodman, 1995; Takatsuji, 1998, 1999; Englbrecht et al., 2004). However, sequence alignments show low similarity elsewhere. Thus, ZFPs may carry out both distinct and overlapping functions. AtZFP2 is a single copy, single zinc finger domain gene (Tague and Goodman, 1995). Our transcriptional profiling revealed that AtZFP2 was up-regulated in cluster 3 of AZ551 (Supplemental Table S1). Promoter-GUS assays revealed enhanced AtZFP2∷GUS transgene expression in open flowers (Fig. 4A) and in floral AZ scars after organ shed (Fig. 4B). Reporter gene expression persisted in AZ scars late into silique maturation (Fig. 4C). The presence of strong expression in AZs validated AtZFP2 transcript accumulation patterns in microarrays and suggested a potential contribution of AtZFP2 to abscission. However, the AtZFP2 gene product appears to participate in non-AZ localized processes as well (Fig. 4, D–I). Promoter:GUS assays show that sites of AtZFP2∷GUS transgene expression include stamens and carpels (Fig. 4, D and E), cotyledons and major veins of rosette leaves (Fig. 4F), trichomes of inflorescence leaves (Fig. 4, G and H), and stems (Fig. 4, G and I).
Figure 4.
Reporter gene expression in plants harboring AtZFP2 promoter-GUS gene fusion. A, Promoter-GUS expression in transgenic inflorescences. B, AZ scars following floral organ detachment. C, AZ scars later in silique development. D, Stamen. E, Carpel. F, Two-week-old seedling. G, Inflorescence. H, Magnified view of leaf surface of G. I, Magnified view of stem surface of G. Scale bars, 1,000 μm.
As the first step toward testing AtZFP2's potential role in floral organ abscission, we constitutively expressed a 35S∷AtZFP2 transgene in Arabidopsis. Ninety-five hygromycin-resistant T1 plant lines were transplanted; almost half of these lines subsequently showed distinct phenotypes relative to wild-type controls. Q-PCR analysis of transgenic flowers was used to assess the combined level of 35S∷AtZFP2 and endogenous AtZFP2 (Supplemental Fig. S2A) and the 35S∷AtZFP2 transgene level alone (Supplemental Fig. S2B). Data in Supplemental Figure S2 show that transgenic flowers exhibit significant increases in total AtZFP2 transcript level relative to wild-type controls.
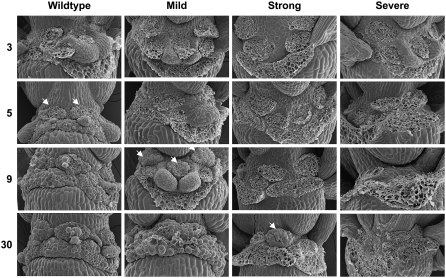
To analyze potential AtZFP2 functions, transgenic phenotypes were visually classified into three types: mild, strong, and severe. In all types, stamen filaments appeared shorter than in wild-type plants (data not shown) and plants were sterile. Transgenic lines were characterized in the T1 generation due to sterility. Other phenotypic changes observed in transgenic flowers are depicted in Supplemental Figure S3. Floral phenotypes included curving of carpels, greening of petals, and increases in trichome number (Fig. S3). Of special interest, transgenic lines were defective in abscission, with abscission delays accentuated in plants exhibiting strong and severe pleiotropic phenotypes. In Figure 5, floral organ retention is compared between wild-type plants (Fig. 5A) and transgenic lines with mild (Fig. 5B), strong (Fig. 5, C and E), and severe phenotypes (Fig. 5D). Because transgenic flowers do not open and have other defects, staging them using standard morphological features was not feasible. Instead, abscission was monitored as a function of inflorescence position. In wild-type plants, the flower stage at which white petals are first visible has been defined as position 1 and developmentally older flowers have increased position numbers (Patterson and Bleecker, 2004). In Figure 1A, position 1 in wild-type plants would correspond to stage 13: anthesis. AtZFP2 transgenic flowers that fail to open cannot, by definition, undergo anthesis. Therefore, we noted the number of floral buds present when abscission first occurred, not including the tight cluster of young flowers at the top of the inflorescence. Wild-type plants usually abscise parts 2 to 3 d after anthesis and, at that time, retain 4 to 6 intact flowers at uppermost positions (Fig. 5A). In contrast, abscission in transgenic 35S∷AtZFP2 plants with mild phenotypes is delayed by several positions (Fig. 5B). In strong phenotype transgenic plants, organ retention is very prolonged (Fig. 5, C and E). Over 40 closed buds are present on the strong inflorescence depicted in Figure 5E. The extremely slow inflorescence elongation rates of severe phenotype plants like that in Figure 4D limited the number of flowers on the inflorescence and all flower parts remained green for 3 to 4 weeks.
Figure 5.
Floral organ abscission in wild-type and 35S∷AtZFP2 plants. A, Inflorescence morphology in wild-type plants. B, Transgenic line with mild phenotype. C and E, Transgenic lines with strong phenotypes. D, Transgenic line with severe phenotype. F to I, Reproductive structures from plants in A to D, respectively. J to L, Floral organ abscission responses after cross-pollination with wild-type pollen grains. J, Mild transgenic line 4 d postpollination. K, Strong transgenic line 12 d postpollination. L, Severe transgenic line 32 d postpollination. Scale bars, 1 mm.
Figure 5, F to I, depict reproductive organs from the inflorescences shown in A to D, respectively. About 30 d postpollination, wild-type floral organs are long gone and the silique has elongated, browned, and initiated dehiscence (Fig. 5F). At this time, all floral organs have also detached from plants with mild 35S∷AtZFP2 phenotypes and mature siliques have initiated dehiscence (Fig. 5G). In contrast, retention of multiple floral organs is marked in plants with strong and severe transgene phenotypes (Fig. 5, H and I, respectively).
In wild-type Arabidopsis, developmental abscission follows pollination and fertilization. We wished to establish whether 35S∷AtZFP2 abscission delays could be due to impairment of these functions, particularly because the short stamen filaments of transgenic plants (data not shown) would be expected to limit effective pollination. Pollen grains from wild-type flowers were used to pollinate 35S∷AtZFP2 stigma; results are shown in Figure 5, J to L. Four days after pollination, the mild phenotype plant in Figure 5J has elongated siliques but still retains floral parts. Floral parts are retained even longer in strong phenotype plants (Fig. 5K; 12 d postpollination) and severe phenotype plants (Fig. 5L; 32 d postpollination). Collectively, these experiments show that pollination cannot rescue abscission delays or blockages in 35S∷AtZFP2 transgenic plants.
The increased floral organ retention seen in Figure 5 has several possible causes, including a failure to differentiate AZs or a failure to separate AZ cells once differentiated. In theory, such failures could occur in all AZs of the floral whorl or just a subset. For example, failure to differentiate or separate only the AZs of sepals would entrap petals and stamens and lead to unobservable abscission. To distinguish between these possibilities, SEM was used to visualize the parental faces of AZ fracture planes after manual removal of wild-type and transgenic floral parts (Fig. 6). When organs were removed from flowers at position 3 of wild-type plants or transgenic plants with mild, strong, or severe phenotypes, cells at bases of petals, sepals, and stamens were torn. Tearing is thought to reflect damage to cells that were tightly linked to their neighbors via cell wall connections at the time of organ removal, i.e. were nonabscised. At position 5, cells at the bases of all wild-type floral organs are intact, suggesting that middle lamellar connections to adjacent cells were at least partially dissolved due to initiation of stamen, sepal, and petal abscission. In Figure 6, arrowheads depict the position at which intact stamen AZ cells first appear. In 35S∷AtZFP2 transgenic lines with mild pleiotropic phenotypes, the first intact AZ cells for stamen AZs are observed at position 9. Thus, completion of stamen AZ cell separation in mild phenotype lines is delayed relative to wild-type stamen AZs by 2 to 3 d. Notably, sepal AZs in mutants with both strong and severe phenotypes do not exhibit features indicative of cell separation over all positions assayed—up to 30 positions in all. These data reveal that overexpression of AtZFP2 results in asynchronous abscission of different floral organs, behavior not seen in wild-type organs. It also suggests that some abscission-competent stamens and petals could be entrapped within a whorl of abscission-incompetent sepals at certain inflorescence positions in 35S∷AtZFP2 transgenic lines. We conclude that the native AtZFP2 gene product might participate in processes that initiate and/or coordinate cell separation in floral organ AZs.
Figure 6.
SEMs of wild-type and 35S∷AtZFP2 floral organ AZ fracture planes. Organs abscised naturally or were manually removed at designated positions along plant inflorescences. Arrowheads designate stamen AZs at the first position where intact cells were observed after organ removal.
Structural features of AtZFP2 suggest that this transcription factor may serve as an active repressor in processes affecting organ shed. AtZFP2 encodes a TFIIIA-type zinc finger protein containing the carboxy-terminal amino acid sequence DLSLRL. Hiratsu et al. (2004) showed that the DLSLRL domain of AtZFP2 had a moderate level of repression activity, similar to that observed in known repressors containing an ERF-associated amphiphilic repression (EAR) domain (Hiratsu et al., 2004). Adamczyk et al. (2007) discuss similar domains found in the MADS-box proteins AGL15 and AGL18 and observe their similarity to JOINTLESS-like MADS proteins (Johansen et al., 2002; Becker and Theißen, 2003). The mechanism whereby modulation of repressor activity could alter abscission capacity is uncertain but it is clear that pedicel abscission is blocked in jointless mutants (Butler, 1936) and floral organ abscission is delayed in plants overexpressing AGL15 (Fernandez et al., 2000), AGL18 (Adamczyk et al., 2007), and AtZFP2. It is possible that repression activity indirectly affects abscission competence via the slowing of certain developmental transitions seen in all of these abscission-impaired transgenic systems (Szymkowiak and Irish, 1999; Fernandez et al., 2000; Adamczyk et al., 2007).
CONCLUSION
We have characterized a 551-member slice of the AZ transcriptome whose expression is significantly regulated prior to developmental stamen shed. Supplemental Figure S4 summarizes floral stage intervals at which transcription of different gene classes is up- or down-regulated by at least 2-fold. Quantitative data corresponding to Supplemental Figure S4 is in Supplemental Table S2. Some expressed genes have been shown by others to be associated with the abscission and/or dehiscence processes, suggesting that the AZ551 population may also represent a valuable source of new cell separation determinants. This hypothesis was confirmed via functional analyses showing an influence of one up-regulated AZ551 gene, AtZFP2, on abscission. Abscission of 35S∷AtZFP2 floral organs was asynchronous and delayed; in plants with severe pleiotropic phenotypes abscission did not occur over time periods of 2 to 3 weeks. Interestingly, it appears that 35S∷AtZFP2 expression does not similarly inhibit cell separation during dehiscence because transgenic silique splitting was observed in Figure 5, G and I.
Comparison of AZ551 to the sequenced Arabidopsis genome reveals that cell wall proteins are disproportionately represented in AZs. Transcripts for PGs, pectate lyases, PMEs, and PMEIs are especially well represented, not surprising given the known roles of pectin-modifying enzymes in reducing adhesion between contiguous AZ cells. Our data set would actually be expected to underestimate the potential contributions of pectin modification to abscission control because the size of AZ551 was limited by F-testing to transcripts with P-values less than 0.0001. Indeed, additional pectin-related proteins exhibiting modulated accumulation prior to abscission are represented by signals lying just outside that window of statistical significance. These transcripts include PMEs and PGs not listed in Supplemental Tables S1 and S2. Interestingly, the significant up-regulation reported in Supplemental Table S2 for one PG (At3g07970) before abscission was at odds with prior reports that this transcript was not expressed in Arabidopsis AZs (González-Carranza et al., 2007a). We therefore investigated whether the At3g07970 probe set might be cross-hybridizing to another structurally similar PG present in the stamen AZ sample. BLAST analysis defined one structural relative of this PG as abscission-related PGAZAT/At2g41850 (González-Carranza et al., 2002). However, our expression profiles for At3g07970 are markedly different from those of PGAZAT (data not shown) and do not support the presence of substantial cross-hybridization. A second close structural relative of the At3g07970 PG is ADPG1/At3g57510 (Sander et al., 2001). The promoter of this Arabidopsis homolog of the oilseed rape (Brassica napus) polygalacturonase RDPG1 was previously shown to direct GUS expression to floral AZs of transgenic Arabidopsis (Sander et al., 2001). Our expression profiles for At3g07970 also significantly differ from those for RDPG1 (data not shown). At present, the reason for discrepancies between our data and that of González-Carranza et al. (2007a) is unclear.
Pectin modification is most prevalent at stage 13 whereas multiple regulators of the hemicellulosic-cellulosic structure are turned on at stage 12 (Supplemental Fig. S4). It is possible that modifications of the hemicellulose-cellulose structure by XTHs, expansins, peroxidases, and/or extensins before or at stage 13 facilitate entry of pectolytic proteins to their substrates as occurs in ripening fruit (Lashbrook, 2005). Alternatively, cell wall changes prior to stage 13 could include reinforcement activities of some XTHs, peroxidases, or extensins. Although net cell wall modification during abscission results in disassembly, wall stiffening early in the abscission pathway would be a way to temper the rate of pectin and hemicellulose disassembly. Given the large size of gene families for cell wall modifying proteins (The Arabidopsis Genome Initiative, 2000), our data should help prioritize gene family members for future functional analyses to answer such questions.
The HAE mRNA component of the ethylene-independent abscission route accumulates significantly between stages 12 and 13, whereas the earliest signs of cell separation occur late in stage 15. Ethylene signaling components with the same pattern of mRNA accumulation include two ERFs and an EIN3-binding protein component of the SCF complex used for ubiquitination-mediated degradation (Potuschak et al., 2003). Thus, ethylene-dependent and -independent pathways seem to be initiated at similar times. Potential convergence of ethylene-dependent and -independent routes may link cell separation regulators (Butenko et al., 2006). The activation of both receptor kinase and ethylene-regulated pathways so close to the time of floral pollination emphasizes that developmental competence to respond to abscission cues is realized before cellular rounding or other visible manifestations of cell separation.
Early induction of abscission pathways presents both opportunities and challenges to researchers interested in understanding abscission control. Opportunities include the potential to identify novel, early regulators suitable for use in plant improvement. Crops including soybean and cotton exhibit high rates of precocious shed prior to fruit set, limiting yield potential (Wiebold et al., 1981; Guinn, 1982). Others like citrus resist detachment at harvest and require chemical harvesting aids that may bring undesirable side effects (Kender et al., 2001). Our studies should help identify gene targets whose modification may reduce or promote shedding of commercially important organs. However, a key challenge will be devising methods to select targets that preferentially control abscission rather than multiple plant processes such as were observed with AtZFP2. For example, distinguishing genes controlling abscission from those coregulating unrelated processes like anther dehiscence, pollination, and/or floral senescence will be important. Of course, it is possible that the primary abscission cue that initiates ethylene-independent and/or -dependent abscission pathways may be derived from these or other events.
A second challenge will be to determine if current abscission models should incorporate hormones beyond ethylene and auxin. Our studies reveal significant changes in the synthesis, perception, and transport of many hormones in AZs prior to organ shed. These include enhancement of GA catabolism, derepression of GA signal transduction, down-regulation of JA biosynthesis, enhanced auxin conjugation and efflux, and increased expression of BR-related genes including the BRI1 receptor (Supplemental Fig. S4; Supplemental Table S2). Some hormonal changes are likely to contribute to processes other than abscission or may influence abscission via ethylene and/or auxin. Certainly, complex modulation of hormone status in stamen AZs suggests the potential for significant cross-talk between abscission pathways. We are presently assessing the scope of possible hormone communication in detaching organs via the analysis of an expanded population of highly regulated AZ genes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia grown in a growth chamber under daily cycles of 16-h light and 8-h darkness at 21°C were fertilized with Peters EXCEL (Scotts). Light intensity was approximately 100 to 130 μmol m−2 s−1. Flowers were harvested when plants were approximately 6 weeks old and bore a total of 12 to 14 open flowers and siliques. Flowers were collected at the same daily time (4–5 pm) to reduce circadian effects. Floral stages were assigned according to descriptions of Smyth et al. (1990). Flowers at stage 12 (“petals level with long stamens”), 13 (“petal visible, anthesis”), and early stage 15 (“stigma extends above long anthers”; S15a) were collected from the primary inflorescence of the same plant. Flowers of the same stage from the same plant batch were pooled as one independent sample. Three separately grown plant batches formed three biological replications. Flowers at stage S15c were approximately 24 h older than those at stage S15a. Flowers one position above S15c flowers were termed stage S15b. Flowers at stages 15b and 15c were harvested from different plant batches 6 months later. Flowers were fixed, processed, and paraffin-embedded according to Cai and Lashbrook (2006).
LCM, RNA Extraction, aRNA Preparation, and Microarray Hybridization
Protocols of Cai and Lashbrook (2006) were used for longitudinal paraffin sectioning, LCM, and RNA isolation, amplification, and biotin labeling. Paraffin sections were mounted onto adhesive-coated slides (Cai and Lashbrook, 2006) with a paraffin tape transfer system (Instrumedics). Prior to LCM, tape was carefully removed from slides submerged in xylenes using forceps. AZ microdissection using the Pix-Cell II LCM system employed CapSure Macro LCM caps (Arcturus Engineering). RNA was isolated from microdissected cells with the PicoPure RNA isolation kit (Arcturus Engineering). Biotin-labeled aRNA was generated with the BioArray RNA amplification and labeling system (Enzo Life Sciences). ATH1 GeneChip (Affymetrix) hybridizations and scanning at the Affymetrix GeneChip Facility of Iowa Sate University employed Affymetrix protocols.
Data Analysis
Genes that changed significantly over the five developmental stages were selected by applying linear mixed model analysis (Wolfinger et al., 2001; Nettleton, 2006) with the SAS mixed model procedure. Stage was considered as a fixed effect and plant batch as a random effect. Natural log transformed MAS 5.0 signal intensities were used as response variables. False discovery rate was calculated as per Storey and Tibshirani (2003); 551 genes with P ≤ 0.0001 are listed in Supplemental Table S1. Annotation of each microarray element was accomplished with the “microarray elements search and download” program on The Arabidopsis Information Resource Web site (www.arabidopsis.org). Functional categorization of AZ551 using GO analytical tools (Berardini et al., 2004) permitted comparisons between predicted functions of AZ transcripts and transcripts within the Arabidopsis genome. Fisher's exact test established statistical significance of overrepresented functions (Fisher, 1922).
Cluster Analysis
AZ551 transcripts were clustered using R software (http://www.R-project.org). The mean normalized signal intensity for each probe set was calculated from three biological replications. A data matrix was constructed with rows corresponding to genes and columns representing means of each gene at each stage. Signal intensities of each gene were standardized so that each gene (row) would have a mean of zero and an sd of 1. Eight clusters were chosen by the gap statistic (Tibshirani et al., 2001). The K-medoid clustering method was used to assign membership of each probe set (Kaufman and Rousseeuw, 1990).
AtZFP2 Cloning and Transformation
Full-length AtZFP2 cDNA was reverse transcribed (RETROscript; Ambion) from flower RNA and amplified with Platinum Taq DNA Polymerase High Fidelity (Invitrogen). Primers used were: forward 5′-CACCATGGACTACCAGCCAAACACATC-3′; reverse 5′-TTAGAGCCTTAAGGATAAGTCAAG-3′.
For promoter-GUS construction, a 5′ upstream region of 3,000 bp preceding the 5′ untranslated region of AtZFP2 was amplified from genomic DNA. Primers used were: forward 5′CACCCATTTTCCCTATTGGTTGACGTC-3′; reverse 5′-AGAGAAGTGTGTTTGAAGAGTTTGG-3′.
Transformation constructs were obtained with Gateway technology. Cloning blunt-ended PCR product into pENTR/D-TOPO vector (Invitrogen) generated an entry clone. Pvu1 enzyme digestion destroyed kanamycin resistance of the entry construct. An LR recombination reaction was performed using Gateway LR clonase II enzyme mix (Invitrogen). This transferred AtZFP2 and the promoter from the entry construct into Gateway Destination vectors PMDC32 and PMDC164, respectively (Curtis and Grossniklaus, 2003). Vectors were introduced into Agrobacterium tumefaciens (strain GV3101) by freeze and thaw (Holsters et al., 1978). Arabidopsis was transformed by floral dip (Clough and Bent, 1998) and transgenic plants selected on Murashige and Skoog-hygromycin plates (50 mg/L).
Q-PCR
For validation of gene expression pattern obtained from microarray studies, Q-PCR employed gene-specific primers provided by the QuantiTect primer assay (QIAGEN). The QuantiTect Reverse transcription kit (QIAGEN) generated cDNA using RNA isolated from laser-captured stamen AZs. Reactions in a Mx3000P instrument (Stratagene) used the QuantiTect SYBRGreen PCR kit (QIAGEN); 18S rRNA was amplified in parallel with each target gene as an internal control. A relative amount of Q-PCR product was represented by −ΔΔCt according to described methods (Livak and Schmittgen, 2001; Ehlting et al., 2005). Basically, threshold detection cycles (Ct) were normalized using corresponding 18S rRNA Ct values to generate ΔCt values. ΔCt values for each gene were compared to the highest ΔCt value among 15 samples obtained for that gene to generate −ΔΔCt values. Q-PCR was replicated three times and results were shown in plots parallel with normalized signal intensities from microarray data.
For examinations of the transcript level of 35S∷AtZFP2 and the combined level of 35S∷AtZFP2 and endogenous AtZFP2 in transgenic plants, flower RNA was reverse transcribed. The primer set for amplifying the transgene only is: forward 5′-TGGTGGTCATCAAAACGCTCAT-3′ and reverse 5′-ATTGCCAAATGTTTGAACGA-3′. The reverse primer recognizes the nopaline synthase terminator of the transgene. The QuantiTect primer assay, At_ZFP2-1-SG (QIAGEN), was then used to collectively PCR amplify 35S∷AtZP2 and endogenous AtZFP2. Relative expression was calculated according to Livak and Schmittgen (2001) with 18S rRNA as the internal control and wild-type expression normalized to 1. Three biological replications were conducted for wild-type flowers; eight biological replications were conducted for 35S∷AtZFP2 flowers.
Histochemical Analysis of GUS Activity
Gus staining used 5-bromo-4-chloro-3-indolyl-b-d-GlcUA substrate (Jefferson et al., 1987). Tissues from T2 plants were incubated at 37°C overnight in 0.1 m phosphate buffer containing 10 mm EDTA, 0.5 mm K-ferricyanide, 0.5 mm K-ferrocyanide, 1.0 mm X-glucuronide, and 0.1% Triton X-100. Tissues were cleared in 70% ethanol and viewed under a stereomicroscope.
SEM
Flowers were fixed in formaldehyde-acetic acid (45% ethanol, 2.5% acetic acid, and 2.5% formalin) overnight, dehydrated in an ethanol series, and critical-point dried (DCP-1; Denton Vacuum). Samples were mounted on aluminum stubs, silver painted, sputter-coated (Denton Vacuum) with Au/Pd, and viewed under a SEM (JSM-5800LV; JEOL) at 10 kV.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Validation of microarray results using Q-PCR.
Supplemental Figure S2. Q-PCR measurement of transcripts representing combined 35S∷AtZFP2/endogenous AtZFP2 and 35S∷AtZFP2 only in flowers of wild-type and 35S∷AtZFP2 plants.
Supplemental Figure S3. Phenotypes of wild-type and 35S∷AtZFP2 floral organs.
Supplemental Figure S4. Developmental timing of accumulation of selected AZ551 transcripts.
Supplemental Table S1. AZ551, the 551 most significantly regulated genes of the stamen AZ transcriptome.
Supplemental Table S2. Fold changes of selected AZ551 transcripts within floral stage intervals.
Note Added in Proof
Up-regulated expression of the At3g07970 polygalacturonase in floral organ AZs prior to abscission was previously reported by Kim and Patterson (Kim J, Patterson SE [2006] Expression divergence and functional redundancy of polygalacturonases in floral organ abscission. Plant Signal Behav 1: 281–283).
Supplementary Material
Acknowledgments
We thank Dr. Jiqing Peng for performing microarray hybridizations and Drs. Madan Bhattacharyya, Harry Horner, Patrick Schnable, and Steve Whitham for critical review of this manuscript. We are grateful to Dr. Dan Nettleton for helpful comments on the manuscript and for providing SAS and R-code modified for use in this study. The authors gratefully acknowledge two anonymous reviewers for providing constructive comments on the manuscript.
This work was supported by Iowa State University (ISU) Agricultural Experiment Station, ISU Plant Sciences Institute, ISU College of Agriculture, and the U.S. Environmental Protection Agency (Cooperative Agreement no. CR–83281101).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Coralie C. Lashbrook (ccl@iastate.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50 1007–1019 [DOI] [PubMed] [Google Scholar]
- Alkharouf NW, Klink VP, Chouikha IB, Beard HS, MacDonald MH, Meyer S, Knap HT, Khan R, Matthews BF (2006) Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224 838–852 [DOI] [PubMed] [Google Scholar]
- Angeles G, Berrio-Sierra J, Joseleau JP, Lorimier P, Lefebvre A, Ruel K (2006) Preparative laser capture microdissection and single-pot cell wall material preparation: a novel method for tissue-specific analysis. Planta 224 228–232 [DOI] [PubMed] [Google Scholar]
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H, Kadowaki K (2002) Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J 32 401–408 [DOI] [PubMed] [Google Scholar]
- Becker A, Theißen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29 464–489 [DOI] [PubMed] [Google Scholar]
- Becnel J, Natarajan M, Kipp A, Braam J (2006) Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol 61 451–467 [DOI] [PubMed] [Google Scholar]
- Becraft PW (2002) Receptor kinase signaling in plant development. Annu Rev Cell Dev Biol 18 163–192 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesgen C, Weiler EW (1999) Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10, 11-reductases from Arabidopsis thaliana. Planta 208 155–165 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Patterson SE (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornman CH, Spurr AR, Addicott FT (1967) Abscisin, auxin, and gibberellin effects on the developmental aspects of abscission in cotton (Gossypium hirsutum). Am J Bot 54 125–135 [Google Scholar]
- Brummell DA, Hall BD, Bennett AB (1999) Antisense suppression of tomato endo-1,4-β-glucanase Cel2 mRNA accumulation increases the force required to break fruit abscission zones but does not affect fruit softening. Plant Mol Biol 40 615–622 [DOI] [PubMed] [Google Scholar]
- Burns J, Pozo L, Arias C, Hockema B, Rangaswamy V, Bender C (2003) Coronatine and abscission in citrus. J Am Soc Hortic Sci 128 309–315 [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB (2003) INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Stenvik GE, Alm V, Saether B, Patterson SE, Aalen RB (2006) Ethylene-dependent and -independent pathways controlling floral abscission are revealed to converge using promoter:reporter gene constructs in the ida abscission mutant. J Exp Bot 57 3627–3637 [DOI] [PubMed] [Google Scholar]
- Butler L (1936) Inherited characters in the tomato. II. Jointless pedicel. J Hered 37 25–26 [Google Scholar]
- Cai S, Lashbrook CC (2006) Laser capture microdissection of plant cells from tape-transferred paraffin sections promotes recovery of structurally intact RNA for global gene profiling. Plant J 48 628–637 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3 1–30 [DOI] [PubMed] [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K (2005) Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J 42 111–123 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Fernandez-Ocana A, Carreras A, Valderrama R, Luque F, Esteban FJ, Rodriguez-Serrano M, Chaki M, Pedrajas JR, Sandalio LM, et al (2006) The expression of different superoxide dismutase forms is cell-type dependent in olive (Olea europaea L.) leaves. Plant Cell Physiol 47 984–994 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D (2002) The growing world of expansins. Plant Cell Physiol 43 1436–1444 [DOI] [PubMed] [Google Scholar]
- Curtis M, Grossniklaus U (2003) A gateway TM cloning vector set for high-throughput functional analysis of genes in plants. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RC, Grossniklaus U, Macknight RC (2005) Be more specific! Laser-assisted microdissection of plant cells. Trends Plant Sci 10 397–406 [DOI] [PubMed] [Google Scholar]
- Day RC, McNoe LA, Macknight RC (2007) Transcript analysis of laser microdissected plant cells. Physiol Plant 129 267–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo E (1999) Multiple endo-1,4-β-D-glucanase (cellulase) genes in Arabidopsis. Curr Top Dev Biol 46 39–61 [DOI] [PubMed] [Google Scholar]
- Delker C, Stenzel I, Hause B, Miersch O, Feussner I, Wasternack C (2006) Jasmonate biosynthesis in Arabidopsis thaliana: enzymes, products, regulation. Plant Biol 8 297–306 [DOI] [PubMed] [Google Scholar]
- Dembinsky D, Woll K, Saleem M, Liu Y, Fu Y, Borsuk LA, Lamkemeyer T, Fladerer C, Madlung J, Barbazuk B, et al (2007) Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol 145 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, et al (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42 618–640 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman P, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132 4563–4574 [DOI] [PubMed] [Google Scholar]
- Englbrecht CC, Schoof H, Bohm S (2004) Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DE, Heck GR, Perry SE, Patterson SE, Bleecker AB, Fang SC (2000) The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12 183–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA (1922) On the interpretation of χ2 from contingency tables, and the calculation of P. J R Statist Soc Ser A 85 87–94 [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza ZH, Elliott KA, Roberts JA (2007. a) Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J Exp Bot 58 3719–3730 [DOI] [PubMed] [Google Scholar]
- González-Carranza ZH, Rompa U, Peters JL, Bhatt AM, Wagstaff C, Stead AD, Roberts JA (2007. b) HAWAIIAN SKIRT: an F-box gene that regulates organ fusion and growth in Arabidopsis. Plant Physiol 144 1370–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza ZH, Whitelaw CA, Swarup R, Roberts JA (2002) Temporal and spatial expression of a polygalacturonase during leaf and flower abscission in oilseed rape and Arabidopsis. Plant Physiol 128 534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinn G (1982) Fruit age and changes in abscisic acid content, ethylene production, and abscission rate of cotton fruits. Plant Physiol 69 349–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CM, Jun JH, Nam HG, Fletcher JC (2004) BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol 45 1361–1370 [DOI] [PubMed] [Google Scholar]
- Hamann T, Osborne E, Youngs H, Misson J, Nussaume L, Somerville C (2004) Global expression analysis of CESA and CSL genes in Arabidopsis. Cellulose 11 279–286 [Google Scholar]
- Harper AD, Bar-Peled M (2002) Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol 130 2188–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmond U, Yuan R, Burns JK, Grant A, Kender W (2000) Citrus fruit abscission induced by methyl-jasmonate. J Am Soc Hortic Sci 125 547–552 [Google Scholar]
- Hayashi T (1989) Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol 40 139–168 [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW (2005) BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17 1434–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M (2004) Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem Biophys Res Commun 321 172–178 [DOI] [PubMed] [Google Scholar]
- Holsters M, De Waele D, Depicker A, Messens E, Van Montagu M, Schell J (1978) Transfection and transformation of A. tumefaciens. Mol Gen Genet 163 181–187 [DOI] [PubMed] [Google Scholar]
- Ithal N, Recknor J, Nettleton D, Maier T, Baum TJ, Mitchum MG (2007) Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol Plant Microbe Interact 20 510–525 [DOI] [PubMed] [Google Scholar]
- Jackson MB, Osborne DJ (1970) Ethylene, the natural regulator of leaf abscission. Nature 225 1019–1022 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC (2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 14 108–117 [PMC free article] [PubMed] [Google Scholar]
- Johansen B, Pedersen LB, Skipper M, Frederiksen S (2002) MADS-box gene evolution-structure and transcription patterns. Mol Phylogenet Evol 23 458–480 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Deal RB, McKinney EC, Meagher RB (2005. a) Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J 41 845–858 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Deal RB, Meagher RB (2005. b) Arabidopsis ARP7 is an essential actin-related protein required for normal embryogenesis, plant architecture, and floral organ abscission. Plant Physiol 138 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw P (1990) Finding Groups in Data: An Introduction to Cluster Analysis. Wiley, New York
- Kendall J (1918) Abscission of flowers and fruits in the Solanaceae, with special reference to Nicotiana. University of California Publications in Botany 5 347–428 [Google Scholar]
- Kende H, Bradford KJ, Brummell DA, Cho HT, Cosgrove DJ, Fleming AJ, Gehring C, Lee W, McQueen-Mason S, Rose JKC, et al (2004) Nomenclature for member of the expansin superfamily of genes and proteins. Plant Mol Biol 55 311–314 [DOI] [PubMed] [Google Scholar]
- Kender W, Hartmond U, Burns J, Yuan R, Pozo L (2001) Methyl jasmonate and CMN-pyrazole applied alone and in combination can cause mature orange abscission. Sci Hortic (Amsterdam) 88 107–120 [Google Scholar]
- Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM (2003) Laser capture microdissection of cells from plant tissues. Plant Physiol 132 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Overall CC, Alkharouf NW, MacDonald MH, Matthews BF (2007) Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean (Glycine max) roots infected by the soybean cyst nematode (Heterodera glycines). Planta 226 1389–1409 [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16 263–276 [DOI] [PubMed] [Google Scholar]
- Kubigsteltig I, Weiler E (2003) Arabidopsis mutants affected in the transcriptional control of allene oxide synthase, the enzyme catalyzing the entrance step in octadecanoid biosynthesis. Planta 217 748–757 [DOI] [PubMed] [Google Scholar]
- Lashbrook C (2005) New insights into cell wall disassembly during fruit ripening. Stewart Postharvest Review 3. http://www.stewartpostharvest.com/archives/archives_issue3_october2005.htm (January 30, 2008)
- Lashbrook CC, Giovannoni JJ, Hall BD, Fischer RL, Bennett AB (1998) Transgenic analysis of tomato endo-β-1,4-glucanase gene function. Role of Cel1 in floral abscission. Plant J 13 303–310 [Google Scholar]
- Leinweber CL, Hall WC (1959) Foliar abscission in cotton. I. Effect of age and defoliants on the respiratory rate of blade, petiole, and tissues of the abscission zone. Bot Gaz 120 144–151 [Google Scholar]
- Li J, Lease KA, Tax FE, Walker JC (2001) BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 98 5916–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SF, Parish RW (1995) Isolation of two novel myb-like genes from Arabidopsis and studies on the DNA-binding properties of their products. Plant J 8 963–972 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malayer JC, Guard AT (1964) A comparative developmental study of the mutant side-shootless and normal tomato plants. Am J Bot 51 140–143 [Google Scholar]
- Mandaokar A, Thines B, Shin B, Lange BM, Choi G, Koo YJ, Yoo YJ, Choi YD, Browse J (2006) Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J 46 984–1008 [DOI] [PubMed] [Google Scholar]
- Mao L, Begum D, Chuang HW, Budiman MA, Szymkowiak EJ, Irish EE, Wing RA (2000) JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406 910–913 [DOI] [PubMed] [Google Scholar]
- McManus MT, Osborne DJ (1990) Identification of polypeptides specific to rachis abscission zone cells of Sambucus nigra. Physiol Plant 79 471–478 [Google Scholar]
- McManus MT, Osborne DJ (1991) Identification and characterization of an ionically bound cell wall glycoprotein expressed preferentially in the leaf rachis abscission zone of Sambucus nigra L. J Plant Physiol 38 63–67 [Google Scholar]
- Merkouropoulos G, Shirsat AH (2003) The unusual Arabidopsis extensin gene atExt1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta 217 356–366 [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Oka M, Ueda J (1997) Update on the possible mode of action of the jasmonates: focus on the metabolism of cell wall polysaccharides in relation to growth and development. Physiol Plant 100 631–638 [Google Scholar]
- Morre DJ (1968) Cell wall dissolution and enzyme secretion during leaf abscission. Plant Physiol 43 1545–1559 [PMC free article] [PubMed] [Google Scholar]
- Murata J, De Luca V (2005) Localization of tabersonine 16-hydroxylase and 16-OH tabersonine-16-O-methyltransferase to leaf epidermal cells defines them as a major site of precursor biosynthesis in the vindoline pathway in Catharanthus roseus. Plant J 44 581–594 [DOI] [PubMed] [Google Scholar]
- Nakada M, Komatsu M, Ochiai T, Ohtsu K, Nakazono M, Nishizawa NK, Nitta K, Nishiyama R, Kameya T, Kanno A (2006) Isolation of MaDEF from Muscari armeniacum and analysis of its expression using laser microdissection. Plant Sci 170 143–150 [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS (2003) Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 3 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T, Tausta S, Gandotra N, Liu T (2006) Laser microdissection of plant tissue: what you see is what you get. Annu Rev Cell Biol 57 181–201 [DOI] [PubMed] [Google Scholar]
- Nettleton D (2006) A discussion of statistical methods for design and analysis of microarray experiments for plant scientists. Plant Cell 18 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg M, Holmlund M, Nilsson O (2005) The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132 2203–2213 [DOI] [PubMed] [Google Scholar]
- Ohtsu K, Smith MB, Emrich SJ, Borsuk LA, Zhou R, Chen T, Zhang X, Timmermans MCP, Beck J, Buckner B, et al (2007. a) Global gene expression analysis of the shoot apical meristem of maize (Zea mays L.). Plant J 52 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsu K, Takahashi H, Schnable PS, Nakazono M (2007. b) Cell type-specific gene expression profiling in plants by using a combination of laser microdissection and high-throughput technologies. Plant Cell Physiol 48 3–7 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Mitina I, Quach HL, Theologis A (2005) AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J 43 29–46 [DOI] [PubMed] [Google Scholar]
- Osborne DJ, Sargent JA (1976) The positional differentiation of abscission zones during the development of leaves of Sambucus nigra and the response of the cells to auxin and ethylene. Planta 132 197–204 [DOI] [PubMed] [Google Scholar]
- Palusa SG, Golovkin M, Shin SB, Richardson DN, Reddy ASN (2007) Organ-specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in Arabidopsis. New Phytol 174 537–550 [DOI] [PubMed] [Google Scholar]
- Passardi F, Cosio C, Penel C, Dunand C (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24 255–265 [DOI] [PubMed] [Google Scholar]
- Patterson SE (2001) Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol 126 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SE, Bleecker AB (2004) Ethylene-dependent and -independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol 134 194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115 679–689 [DOI] [PubMed] [Google Scholar]
- Rajani S, Sundaresan V (2001) The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol 11 1914–1922 [DOI] [PubMed] [Google Scholar]
- Ramsay K, Wang ZH, Jones MGK (2004) Using laser capture microdissection to study gene expression in early stages of giant cells induced by root-knot nematodes. Mol Plant Pathol 5 587–592 [DOI] [PubMed] [Google Scholar]
- Reinders A, Panshyshyn JA, Ward JM (2005) Analysis of transport activity of Arabidopsis sugar alcohol permease homolog AtPLT5. J Biol Chem 280 1594–1602 [DOI] [PubMed] [Google Scholar]
- Ridley BL, O'Neill MA, Mohnen D (2001) Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57 929–967 [DOI] [PubMed] [Google Scholar]
- Roberts J, Whitelaw C, Gonzalez-Carranza Z, McManus M (2000) Cell separation processes in plants-models, mechanisms and manipulation. Ann Bot (Lond) 86 223–235 [Google Scholar]
- Roberts JA, Elliott KA, Gonzalez-Carranza ZH (2002) Abscission, dehiscence, and other cell separation processes. Annu Rev Plant Biol 53 131–158 [DOI] [PubMed] [Google Scholar]
- Roberts K, Shirsat AH (2006) Increased extensin levels in Arabidopsis affect inflorescence stem thickening and height. J Exp Bot 57 537–545 [DOI] [PubMed] [Google Scholar]
- Rose J, Braam J, Fry S, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43 1421–1435 [DOI] [PubMed] [Google Scholar]
- Sander L, Child R, Ulvskov P, Albrechtsen M, Borkhardt B (2001) Analysis of a dehiscence zone endo-polygalacturonase in oilseed rape (Brassica napus) and Arabidopsis thaliana: evidence for roles in cell separation in dehiscence and abscission zones, and in stylar tissues during pollen tube growth. Plant Mol Biol 46 469–79 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Biesgen C, Mussig C, Altmann T, Weiler EW (2000) 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210 979–984 [DOI] [PubMed] [Google Scholar]
- Schaller F, Hennig P, Weiler EW (1998) 12-Oxophytodienoate-10,11-reductase: occurrence of two isoenzymes of different specificity against stereoisomers of 12-oxophytodienoic acid. Plant Physiol 118 1345–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Weiler EW (1997) Enzymes of octadecanoid biosynthesis in plants—12-oxo-phytodienoate 10,11-reductase. Eur J Biochem 245 294–299 [DOI] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci USA 96 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Seto H, Yamakawa H, Ohashi Y (2001) Transient accumulation of jasmonic acid during the synchronized hypersensitive cell death in tobacco mosaic virus-infected tobacco leaves. Mol Plant Microbe Interact 14 261–264 [DOI] [PubMed] [Google Scholar]
- Sexton R, Lewis L, Trewavas A, Kelly P (1985) Ethylene and abscission. In JA Roberts, GA Tucker, eds, Ethylene and Plant Development. Butterworths, London, pp 173–196
- Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT (2003) Cluster analysis and display. In D Bowtell, J Sambrook, eds, DNA Microarrays: A Molecular Cloning Manual. Cold Spring Harbor Laboratory Press, Woodbury, NY, pp 569–581
- Stenvik GE, Butenko MA, Urbanowicz BR, Rose JK, Aalen RB (2006) Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 18 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymkowiak EJ, Irish EE (1999) Interactions between Jointless and wild-type tomato tissues during development of the pedicel abscission zone and the inflorescence meristem. Plant Cell 11 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tague BW, Goodman HM (1995) Characterization of a family of Arabidopsis zinc finger protein cDNAs. Plant Mol Biol 28 267–279 [DOI] [PubMed] [Google Scholar]
- Takatsuji H (1998) Zinc-finger transcription factors in plants. Cell Mol Life Sci 54 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H (1999) Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol Biol 39 1073–1078 [DOI] [PubMed] [Google Scholar]
- Taylor J, Whitelaw C (2001) Signals in abscission. New Phytol 151 323–339 [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Thomas S, Phillips A, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Fry SC (1997) Trimming and solubilization of xyloglucan after deposition in the walls of cultured rose cells. J Exp Bot 48 297–305 [Google Scholar]
- Thompson JE, Fry SC (2001) Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. Plant J 26 23–34 [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Walther G, Hastie T (2001) Estimating the number of clusters in a data set via the gap statistic. J R Statist Soc B 63 411–423 [Google Scholar]
- Ueda J, Kato J (1982) Inhibition of cytokinin-induced plant growth by jasmonic acid and its methyl ester. Physiol Plant 54 249–252 [Google Scholar]
- Ueda J, Miyamoto K, Hashimoto M (1996) Jasmonates promote abscission in bean petiole explants: its relationship to the metabolism of cell wall polysaccharides and cellulase activity. J Plant Growth Regul 15 189–195 [Google Scholar]
- Ueda J, Miyamoto K, Sato T, Momotani Y (1991) Identification of jasmonic acid from Euglena gracilis Z as a plant growth regulator. Agric Biol Chem 55 275–276 [Google Scholar]
- Vilaine F, Palauqui JC, Amselem J, Kusiak C, Lemoine R, Dinant S (2003) Toward phloem deciphering: a transcriptome analysis of the phloem of Apium graveolens. Plant J 36 67–81 [DOI] [PubMed] [Google Scholar]
- Wiebold W, Ashley D, Boerma HR (1981) Reproductive abscission levels and patterns for eleven determinate soybean cultivars. Agron J 73 43–46 [Google Scholar]
- Willats W, McCartney L, Mackey W, Knox J (2001) Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 47 9–27 [PubMed] [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8 625–637 [DOI] [PubMed] [Google Scholar]
- Woll K, Borsuk L, Stransky H, Nettleton D, Schnable PS, Hochholdinger F (2005) Isolation, characterization and pericycle specific transcriptome analyses of the novel maize (Zea mays L.) lateral and seminal root initiation mutant rum1. Plant Physiol 139 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YY, Lashbrook CC, Hannapel DJ (2007) Tissue integrity and RNA quality of laser microdissected phloem of potato. Planta 226 797–803 [DOI] [PubMed] [Google Scholar]
- Zhang X, Madi S, Borsuk L, Nettleton D, Elshire RJ, Buckner B, Janick-Buckner D, Beck J, Timmermans M, Schnable PS, et al (2007) Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet 3 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.