Abstract
In bioassays with artificial diets, the 17-hydroxygeranyllinalool diterpenoid glycosides (HGL-DTGs) of Nicotiana attenuata function as antifeedants for the plant's adapted herbivore, tobacco hornworm (Manduca sexta). To determine whether HGL-DTGs have a defensive function in planta, we suppressed HGL-DTG production by silencing the source of the geranylgeranyl diphosphates (GGPPs) required for geranyllinalool biosynthesis, a key intermediate. We used virus-induced gene silencing to suppress transcript levels of GGPP synthase gene (Naggpps) and farnesyl diphosphate (FPP) synthase gene (Nafpps), northern blotting and real-time polymerase chain reaction to quantify transcript accumulations, and radio gas chromatography to analyze prenyltransferase specificity. Silencing Nafpps had no effect on the accumulation of HGL-DTGs but decreased leaf steroid content, demonstrating that DTG-synthesizing enzymes do not use GGPP derived from FPP and confirming FPP's role as a steroid precursor. Unlike plants silenced in the phytoene desaturase gene (Napds), which rapidly bleached, Naggpps-silenced plants had reduced HGL-DTG but not carotenoids or chlorophyll contents, demonstrating that Naggpps supplies substrates for GGPP biosynthesis for HGL-DTGs, but not for phytoene or phytol. Expression of Naggpps in Escherichia coli revealed that the recombinant protein catalyzes the GGPP synthesis from isopentenyl diphosphate and dimethylallyl diphosphate. When fed on silenced plants, hornworm larvae gained up to 3 times more mass than those that fed on empty vector control plants or plants silenced in Nafpps, the trypsin protease inhibitor gene, or the putrescine N-methyltransferase gene. We conclude that HGL-DTGs or other minor undetected diterpenoids derived from GGPP function as direct defenses for N. attenuata and are more potent than nicotine or trypsin protease inhibitors against attack by hornworm larvae.
Plants employ both direct and indirect defenses against herbivores. Direct defenses may provide physical barriers, act to poison herbivores or impede their digestion, or make plant tissues otherwise unappetizing. There are many ways of identifying the metabolites that function as defenses in plants. The effects on insect herbivores of secondary and primary metabolites such as polyphenols, quinones, alkaloids, some saponines, diterpenoid glycosides (DTGs), phytic acid, and trypsin protease inhibitors (TPIs) have been examined by supplementing artificial diets (De Boer and Hanson, 1987; Broadway and Duffey, 1988; Felton et al., 1989; Snook et al., 1997; Green et al., 2001; Pohlon and Baldwin, 2001; Jassbi et al., 2006). Unfortunately, some chemicals may be altered in the artificial diets (Usher et al., 1989) and the effectiveness of metabolites as defenses may depend on their chemical context (Felton et al., 1989).
Proteins are thought to be the most important nutrients for Lepidoptera herbivores, and some secondary metabolites, such as tannins, gossypol, and TPIs are thought to inhibit digestion of proteins (Damaty and Hudson, 1975; Feeny, 1976; Rhoades and Cates, 1976; Broadway and Duffey, 1988; Barbehenn and Martin, 1994); by binding to proteins, including digestive enzymes, tannins reduce the digestibility of proteins in larval midguts (Feeny, 1976; Rhoades and Cates, 1976; Downs et al., 2003). Many compounds have diverse modes of action. In addition to acting as an antifeedant, phytic acid may reduce the digestibility of foliage and fruits by forming complexes with dietary proteins and prevent the detoxification of toxins by inhibiting cytochrome P450 monooxygenases (Green et al., 2001). Moreover, the defensive function of some metabolites may depend on the herbivore's behavioral responses to other metabolites. In an example of a defensive synergism between metabolites, nicotine was found to be a particularly effective defense in Nicotiana attenuata against the larvae of Spodoptera exigua because the alkaloid prevented the larvae from compensating for the ingestion of TPIs by increasing consumption of leaf material (Steppuhn and Baldwin, 2007).
These complex interactions represent a principal reason why studies of metabolite defensive function in planta are thought to provide more reliable results compared to the use of artificial diet bioassays (Felton et al., 1989). Heterologous gene expression provides a means of examining the defensive role of metabolites in planta (Duan et al., 1996; Smigocki and Wilson, 2004; Zavala et al., 2004b). Introducing a Ser-type PI gene from potato (Solanum tuberosum) into rice (Oryza sativa) resulted in a new insect-resistant rice cultivar (Duan et al., 1996). Although the heterologous expression of a defensive gene in a plant can reduce herbivore performance, two complications can confound the extrapolation of the results to a natural host plant-herbivore interaction. First, specialist herbivores are often resistant to the metabolites of their host plant. Second, the newly introduced metabolite encounters a new chemical environment in the transgenic plant and may interact differently with these metabolites (Broadway, 1997; Winterer and Bergelson, 2001; Zavala et al., 2004b). Endogenous overexpression or silencing of genes that control the accumulation of metabolites provides the best tools for examining defensive function of a metabolite. Silencing a gland-specific P450 hydroxylase gene in the trichomes of Nicotiana tabacum resulted in suppression of biosynthesis of the predominant exudate component, cembratriene (CBT)-diol and increases in its precursor, CBT-ol. Transgenic plants with higher concentrations of the precursor diterpenoid CBT-ol showed greatly diminished aphid colonization, suggesting that the precursor was a more important defense against aphids than the (CBT)-diol (Wang et al., 2001).
Multiple direct defenses are known to function in the native tobacco N. attenuata; the alkaloid nicotine, a neurotoxin, functions as a potent direct defense, while antidigestive TPIs decrease the nutritional value of plants' tissues and slow insects' growth rates (Steppuhn et al., 2004; Zavala et al., 2004a, 2004b). N. attenuata emits volatile terpenoids, among other compounds, when attacked by herbivores; these terpenoids have been shown to attract the herbivores' natural predators and deter herbivore oviposition, serving as effective indirect defenses and reducing herbivore loads by up to 90% (Kessler and Baldwin, 2001). The defensive functions of nicotine and TPIs in N. attenuata against the specialist lepidopteran herbivore, tobacco hornworm (Manduca sexta), have been extensively examined by silencing the expression of key biosynthetic genes in planta (Steppuhn et al., 2004; Zavala et al., 2004b; Steppuhn and Baldwin, 2007). N. attenuata is a phytochemically diverse plant containing alkaloids (nicotine, anabasine, and anatabine), proteins (TPIs), phenolics (rutin, chlorogenic acids, and caffeoylputrescine), volatile terpenoids, and DTGs (Halitschke et al., 2000; Keinänen et al., 2001; van Dam et al., 2001; Lou and Baldwin, 2003; Steppuhn et al., 2004). Here, we examine the role of DTGs in the context of the other major secondary metabolites within the plant.
DTGs with geranyllinalool (GL) carbon skeletons (GL-DTGs) have been reported to be abundant secondary metabolites in several species of Nicotiana (Shinozaki et al., 1996; Snook et al., 1997; Keinänen et al., 2001; Jassbi et al., 2006). Two of the characterized 17-hydroxygeranyllinalool (HGL)-DTGs in N. tabacum showed antibiosis activity and acted as direct defenses inhibiting the growth of tobacco budworm (Heliothis virescens), a major pest on tobacco (Snook et al., 1997). The defensive role of HGL-DTGs against hornworm in Nicotiana was first proposed in a study that compared larval performance on three different species of Nicotiana, N. attenuata, N. bigelovii, and N. clevelandii, and found that larvae gained less mass when they fed on plants containing high HGL-DTG levels (Lou and Baldwin, 2003).
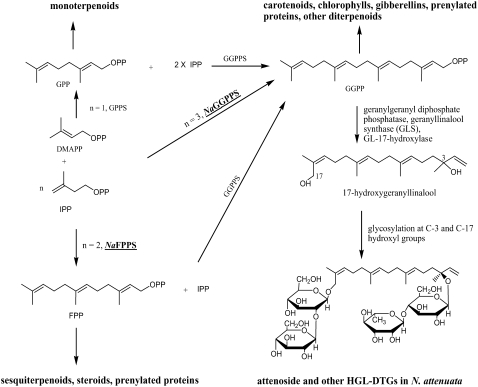
Prenyltransferases are key enzymes in the terpenoid biosynthesis pathway. They catalyze the alkylation of one or more (n = 1, 2, 3) molecules of isopentenyl diphosphate (IPP; C5), with an allylic diphosphate such as dimethylallyl diphosphate (DMAPP; C5) to produce geranyl diphosphate (GPP; C10), farnesyl diphosphate (FPP; C15), and geranylgeranyl diphosphate (GGPP; C20), respectively (Ohnuma et al., 1998; Dewick, 2002; Fig. 1). GGPP synthase may use DMAPP, GPP, or FPP in plants and bacteria, while in fungi and animals it uses FPP as its allylic substrate (Ohnuma et al., 1998; Dewick, 2002). GPP is the precursor of monoterpenoids, while FPP is involved in the biosynthesis of sesquiterpenoids, steroids, and triterpenoids. The acyclic all-trans GGPP is known to be the common precursor of all cyclic and acyclic diterpenoids (Guo et al., 1994). According to the proposed biosynthetic pathway (Fig. 1), geranylgeranyl (GG), the hydrolysis product of GGPP, is converted to GL and further hydroxylation and glycosylation yield HGL-DTGs.
Figure 1.
Biosynthetic pathway of terpenoids catalyzed by prenyltransferases and biogenesis of HGL-DTGs from GGPP.
To investigate the defensive role of DTGs in planta and to determine the biosynthetic pathway of HGL-DTGs in N. attenuata, we used virus-induced gene silencing (VIGS) to suppress the expression of the geranylgeranyl diphosphate synthase (GGPPS) gene (Naggpps) and the farnesyl diphosphate synthase (FPPS) gene (Nafpps). VIGS has been extensively used to investigate the function of genes (Liu et al., 2002; Lu et al., 2003; Benedito et al., 2004). It was successfully used to silence both the TPI gene (Natpi) in the leaves and the putrescine N-methyltransferase gene (Napmt) in the roots, suppressing the production of direct defenses TPI and nicotine, respectively (Saedler and Baldwin, 2004). We used Natpi- and Napmt-silenced plants as positive controls for our in planta feeding assays. Phytoene desaturase gene (Napds)-silenced plants bleach rapidly due to suppressed carotenoid production, and these were used as positive controls for the VIGS process (Liu et al., 2002). In addition to its role in the synthesis of diterpenoids, GGPP is also known to be the precursor of carotenoids and the phytol side chain of chlorophylls (Fray et al., 1995; Keller et al., 1998), and we consider the effects of silencing Naggpps on the total chlorophyll and carotenoid contents. Because FPP from Taxus baccata cell cultures is reported to be the substrate for GGPPS (Laskaris et al., 2000), we silenced Nafpps to clarify its role in the biosynthesis of HGL-DTGs.
RESULTS
VIGS of Naggpps and Nafpps
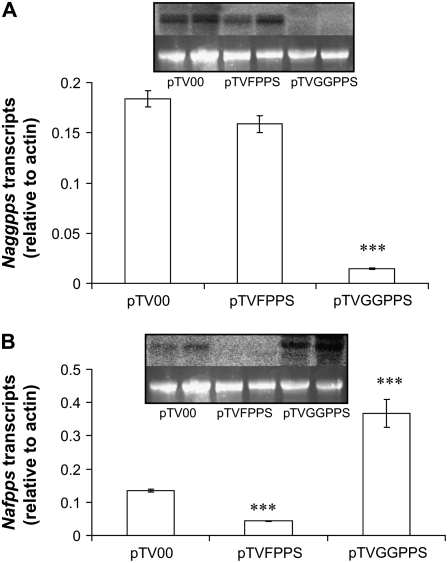
Approximately 20 d after inoculating early rosette-stage N. attenuata plants with Agrobacterium tumefaciens GV3101 harboring pTVFPPS, pTVGGPPS, pTV00 (empty vector control [EV]), and pTVPD plasmids, Napds-silenced plants became completely bleached, demonstrating successful gene silencing. To determine the silencing efficiency of Naggpps and Nafpps, total RNA was isolated from the leaves of the transformed plants and subjected to northern analysis (Fig. 2, insets); transcripts of both genes were substantially decreased in gene specific-silenced lines in comparison to EV controls.
Figure 2.
VIGS efficiency of Naggpps (A) and Nafpps (B). RT-PCR was run on cDNA from leaf samples of plants treated with pTV00, pTVFPPS, and pTVGGPPS. Transcript accumulation levels were determined relative to the N. attenuata actin gene and presented as the mean value (±se) of five biological replicates. ***, Significantly different from pTV00 at P < 0.0001. Insets: RNA was isolated and pooled from leaves of two biological replicates for each line pTV00, pTVFPPS, and pTVGGPPS, hybridized with radio-labeled probes for Naggpps (A) and Nafpps (B), washed and analyzed with a Fujifilm FLA-3000 Phosphoimager. Lower lanes: Ethidium bromide-stained agarose gel with rRNA as a control for equal RNA loading.
To quantify the silencing efficiency of Naggpps and Nafpps genes, the total RNA of the plants' leaf tissues was reverse transcribed and subjected to SYBR Green real-time (RT)-PCR, using the N. attenuata actin gene (EU273278) as an endogenous control. Using GGPPS-specific primers (Supplemental Table S1), we determined that the relative transcript levels in pTVGGPPS plants (n = 5; 0.015 ± 0.001) were an order of magnitude lower than those in pTVFPPS (n = 5; 0.16 ± 0.009) and EV plants (n = 5; 0.18 ± 0.008; Fig. 2A). When FPPS-specific primers were used, substantially higher levels were observed in EV (n = 5; 0.13 ± 0.01) and pTVGGPPS plants (n = 5; 0.37 ± 0.098) compared to pTVFPPS plants (n = 5; 0.04 ± 0.004; Fig. 2B). The VIGS efficiency (100 − [relative transcript accumulation of silenced line/relative transcript accumulation of EV] × 100) attained in Naggpps- and Nafpps-silenced lines was 92% and 69%, respectively.
Because the silencing efficiency of the Nafpps gene was only 69% and several homologs of fpps and ggpps have been reported in Artemisia tridentata and Arabidopsis (Arabidopsis thaliana; Hemmerlin et al., 2003; Lange and Ghassemian, 2003), we tested whether N. attenuata has more than one homologous copy of each gene. If so, this could influence the silencing, and if this gene has a redundant function, all other experiments might also be affected.
After construction of pTVFPPS and pUCFPPS, the Nafpps inserts of at least four clones of each plasmid were sequenced. There was complete identity in the amplified Nafpps sequence between the PCR primers among the clones of each plasmid. For primary cloning of three different Naggpps PCR fragments in pTVGGPPS, pTVGGPPS2, and pUCGGPPS and of the full-length Naggpps on pJETGGPPS, four independent PCRs with eight different primers on cDNA of N. attenuata as template were performed. After successful cloning, the Naggpps inserts of 14 clones (two of pTVGGPPS, two of pTVGGPPS2, six of pUCGGPPS, and four of pJETGGPPS) were sequenced. There were no differences in the amplified sequences between the respective PCR primers among all 14 clones. Because the primers were designed from sequence information from other Solanaceous plants (tomato [Solanum lycopersicum], Capsicum annuum, and N. tabacum), complete identity between primers and template could not be expected. For instance, a comparison of the full-length Solanaceous ggpps sequences available in GenBank reveals an 85% identity of the 3′ 900 bp of Naggpps (EF382626) to tomato ggpps (DQ267903) and a 79% identity to C. annuum ggpps (X80267). As a consequence, a few sequence differences between Naggpps and the clones in the primer regions occurred, showing that in our PCR system, primer/Naggpps template mismatches still allowed for efficient PCR amplification. Therefore, the amplification of only one Nafpps and only one Naggpps gene under conditions that allow primer/template mismatches represents strong evidence that there is only one copy of Nafpps and of Naggpps and no family of genes homologous to Nafpps and Naggpps in the genome of N. attenuata.
Southern-blot analysis confirmed that the N. attenuata genome harbored only one close homolog of fpps and ggpps (Supplemental Fig. S1, A and B); using a full-length Naggpps probe and a 350-bp Nafpps probe, all four digestions revealed only a single band. Together with the PCR fragment-sequencing results, these results lead us to conclude that there is no other close homolog of Nafpps and Naggpps in N. attenuata.
NaGGPPS and NaFPPS Protein Activity
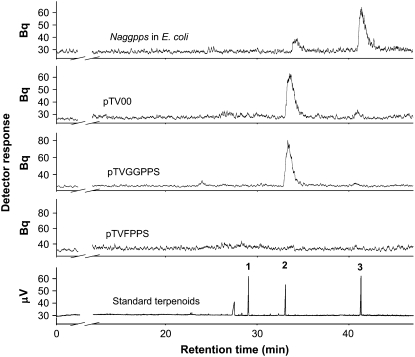
Radio gas chromatography (GC) analyses revealed that plant protein extracts of pTVGGPPS and pTVFPPS plants contained different levels of prenyltransferase activity. Whereas the extract of the pTVGGPPS line showed mainly FPPS activity (n = 5; 92% ± 2.7% farnesol, and 8% ± 1.9% GG of the total synthesized and hydrolyzed prenyldiphosphate products) and was comparable with the EV plants (n = 5; 98.5% ± 1.7% farnesol, and 1.5% ± 1.2% GG), the protein extract of pTVFPPS plants showed no measurable prenyltransferase activity (Fig. 3).
Figure 3.
Catalytic activity of recombinant NaGGPPS after heterologous Naggpps expression in E. coli and pTV00, pTVGGPPS, and pTVFPPS plant protein extracts. Purified recombinant NaGGPPS (top) and plant protein extract of the vector control pTV00, pTVGGPPS, and pTVFPPS (middle three segments) were assayed with DMAPP and [1-14C]IPP. The enzyme activity was measured by a radio-GC and identified by coinjecting unlabeled-terpene standards geraniol (1), farnesol (2), and geranylgeraniol (3) via a TCD measurement (bottom). The hydrolyzed main products are geranylgeraniol (3) and farnesol (2). The figure shows representative product activities of five assays.
Plant prenyltransferases are only highly similar in their amino acid sequences of the catalytic domain. In a phylogenetic tree of all prenyltransferases comparing the amino acid sequences of the complete open reading frame, FPPS cluster separately from the more related GGPPS or geranyl pyrophosphate synthase (GPPS; Burke and Croteau, 2002; Tholl et al., 2004; Bouvier et al., 2005; Schmidt and Gershenzon, 2007b). In contrast to FPPS, where it is possible to predict the putative enzyme activity, functional analyses of GGPPS are necessary to confirm enzyme activity. Because silencing Naggpps did not significantly change the levels of synthesized GGPP in the plants' total protein, we heterologously expressed full-length cDNA in Escherichia coli to determine if the cDNA of Naggpps codes for a functional GGPPS. After the putative chloroplast transit peptide was truncated, NaGGPPS functioned mainly as a GGPPS and showed only a minor amount of FPPS activity. The total synthesized and hydrolyzed prenyldiphosphate products were composed of 85% ± 3.1% GG and 15% ± 2.7% farnesol. The recombinant protein showed no GPPS activity (Fig. 3).
Silencing Naggpps and Nafpps Decreases HGL-DTG Levels and Increases Larval Performance
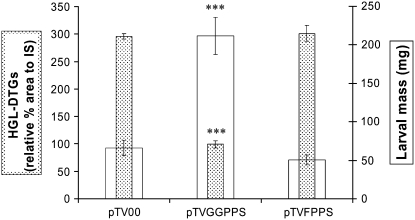
After 14 d of feeding on the VIGS plants, hornworm larvae gained 3- to 4-fold more mass on pTVGGPPS-silenced (n = 30; average mass [AM] = 211.3 ± 24.1 mg; t = 5.259; df = 54; P < 0.0001; 320.3% of the EV plants) than on pTVFPPS-silenced (n = 28; AM = 50.2 ± 6.4 mg; t = 1.337; df = 52; P = 0.187) or EV plants (n = 26; AM = 66.0 ± 10.1 mg; Fig. 4, white bars). In 40 biological replicates for each silenced line, the larvae fed for 18 d; the number of larvae that survived was as follows: pTVGGPPS, 30 (75%); pTVFPPS, 25 (62.5%); and pTV00, 19 (47.5%).
Figure 4.
Ingestion of HGL-DTGs is negatively correlated with hornworm larval performance. Dotted bars: RP-HPLC-UV (λ = 210 nm) relative area percentage of HGL-DTGs (means ± se, n = 9, area of the peaks at 24–28 min/area of thymols' peak × 100) of the methanolic extract of the leaves from plants not attacked by herbivores. White bars: hornworm larval mass (milligrams; means ± se; n = 40; in the beginning of the assay) after 14 d of feeding on N. attenuata plants inoculated with A. tumefaciens GV3101 carrying pTV00, pTVGGPPS, or pTVFPPS. ***, Significant differences from pTV00 at P < 0.0001.
To determine the effects of silencing Naggpps and Nafpps on secondary metabolite composition, a methanol extract of leaf material from silenced plants was analyzed by reverse-phase (RP)18 HPLC (Supplemental Fig. S2). Levels of HGL-DTGs were lower in pTVGGPPS plants than in pTVFPPS and pTV00 plants (Supplemental Fig. S2; Fig. 4, dotted bars). The average levels of HGL-DTGs in the herbivore-attacked leaves of pTVGGPPS and pTVFPPS plants were 33.6% (n = 10; mean relative area [MRA] % = 99.2 ± 12.3; t = 10.118; df = 18; P < 0.0001) and 101.9% (n = 10; MRA % = 300.7 ± 14.3; t = 0.271; df = 18; P = 0.789) of those measured in EV plants (n = 10; MRA % = 295.1 ± 14.9), respectively (Fig. 4, dotted bars).
Silencing Naggpps Decreases HGL-DTGs But Not Nicotine and TPI Levels, and Dramatically Increases Larval Performance; Silencing Natpi and Napmt Influences Neither HGL-DTGs Nor Caterpillar Performance
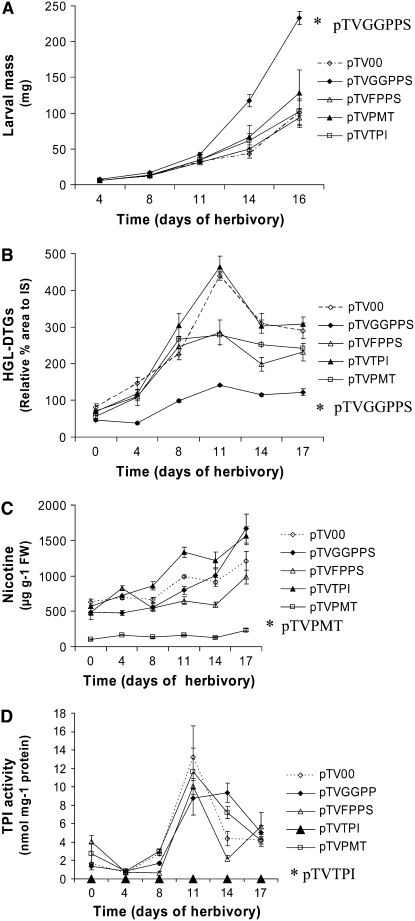
To compare the performance of larvae that fed on plants deficient in HGL-DTGs with those deficient in TPIs and nicotine, we repeated the experiments with the addition of Natpi- and Napmt-silenced plants. By day 16, the masses of larvae that fed on pTVGGPPS were significantly (n = 16; AM = 232.9 ± 9.4 mg; t = 7.003; df = 28; P < 0.0001) higher than the masses of those that fed on EV plants (n = 14; AM = 100.2 ± 17.2 mg). Masses of larvae that fed on pTVFPPS (n = 9; AM = 93.3 ± 13.4 mg), pTVPMT (n = 14; AM = 128.2 ± 32.0 mg), and pTVTPI (n = 14; AM = 102.0 ± 17.6 mg) were not significantly different from those that fed on EV plants (Fig. 5A). As in the first experiment, the number of larvae that survived to the end of the feeding bioassay was higher on pTVGGPPS plants (16, or 59%) than on the EV plants, where only 14 (52%) of 27 larvae survived.
Figure 5.
VIGS of Naggpps dramatically reduces HGL-DTG contents and increases hornworm larvae performance to a greater degree than the silencing of TPI and nicotine. A, hornworm larval mass (milligrams; mean ± se; n = 27 in the beginning of the assay) from N. attenuata plants inoculated with A. tumefaciens GV3101 carrying pTV00, pTVGGPPS, pTVFPPS, pTVTPI, and pTVPMT. B, Relative area percentage of HGL-DTGs (mean ± se; n = 6; RP-HPLC-UV peak area at 22–27 min/area of thymols' peak × 100, λ = 210 nm), nicotine (mean ± se; n = 6; μg g−1 leaf FW; C), and TPI activity (mean ± se; n = 5; nmol mg−1 protein; D). Nicotine and DTGs were measured in 40% methanolic extracts of the leaves of VIGS plants. *, Significantly different values from pTV00 with P < 0.05.
In the second VIGS experiment with additional Napmt- and Natpi-silenced plants, systemic leaf material was analyzed for HGL-DTGs, nicotine, and TPIs (Fig. 5, B–D). Leaves that had not been attacked by larvae were collected at different time points from locations above where the larvae were feeding. The levels of HGL-DTGs in the pTVGGPPS plants were the lowest compared to the other silenced and pTV00 plants at all time points (Fig. 5B). After 11 d of herbivore attack, HGL-DTGs reached their highest levels in all plants (up to 5.5-fold those of EV plants), but levels were still the lowest in pTVGGPPS plants, increasing 3-fold after herbivory. The levels of HGL-DTGs in pTVGGPPS, pTVFPPS, pTVPMT, and pTVTPI plants relative to those in pTV00 plants (n = 6; MRA % = 439.4 ± 11.4) were 32.1% (n = 6; MRA % = 140.9 ± 2.7; t = 25.465; df = 10; P < 0.0001), 64.9% (n = 6; MRA % = 285.3 ± 33.2; t = 4.394; df = 10; P = 0.0013), 63.3% (n = 6; MRA % = 278.1 ± 29.7; t = 5.15; df = 10; P = 0.0004), and 105.4% (n = 6; MRA % = 463.2 ± 29.9; t = 0.743; df = 10; P = 0.4746), respectively (Fig. 5B). By day 17, at the end of the feeding experiment, only levels of HGL-DTGs in pTVGGPPS plants (n = 6; MRA % = 122.4 ± 8.6; t = 7.277; df = 10; P < 0.0001) were significantly different (42.3%; P < 0.0001) from those of the EV plants.
Levels of nicotine increased 1.3- to 2.3-fold in the silenced lines and EV plants after 11 d of larval feeding, and silencing Natpi, Naggpps, and Nafpps changed the levels of nicotine to 134.5% (P = 0.001), 80.3% (P = 0.01), and 65.6% (P = 0.0002) of those of the EV. As expected, the nicotine content of the pTVPMT lines was only 16.7% (P < 0.0001) of that in EV controls (Fig. 5C). By day 17, HPLC analyses showed that the lowest levels of nicotine were in the leaves of pTVPMT plants (232.1 ± 21.6 μg g−1 leaf fresh weight [FW]; n = 6; t = 7.311; df = 10; P < 0.0001; 19.1% of EV plants) at the end of the feeding test, while nicotine levels in other silenced plants, pTVGGPPS (1,668.4 ± 205.9 μg g−1 FW; n = 6; 137.9%), pTVFPPS (985.0 μg g−1 FW of leaves; n = 6; 81.4%), and pTVTPI (1,564.7 ± 126.8 μg g−1 FW; n = 6; 129.4%), did not significantly differ from those of pTV00 plants (1,209.6 ± 131.9 μg g−1 FW; n = 6).
Analysis of TPI activity revealed similar levels and kinetics of inhibition among pTVGGPPS, pTVFPPS, pTVPMT, and pTV00 plants; as expected, TPI activity was negligible in the pTVTPI plants (Fig. 5D). Like levels of the other defensive substances, levels of TPIs increased 2.3- to 8.3-fold in the silenced and EV plants after 11 d of larval feeding.
Silencing Nafpps Decreases Free Sterol Levels But Not Volatile Terpenoids
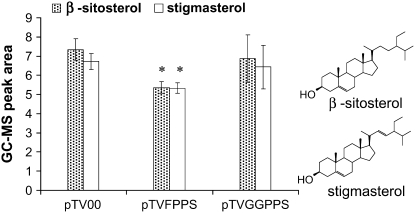
To investigate the effect of silencing Nafpps on terpenoid biosynthesis, the head spaces of whole plants were analyzed for volatile sesquiterpenoids. No significant differences (all P values > 0.1) were detected in their levels in comparison to other lines (data not shown). We assumed that Nafpps was involved in steroid biosynthesis, so the concentrations of the free steroids, β-sitosterol and stigmasterol, were measured in dichloromethane extracts of fresh leaves by GC-mass spectrometry (MS). Significant decreases were found in the levels of β-sitosterol (73% of the pTV00 plants; t = 3.099; df = 14; P = 0.0079) and stigmasterol (79% of the pTV00 plants; t = 2.721; df = 14; P = 0.0165) in pTVFPPS plants compared to their levels in EV plants (Fig. 6). Silencing Naggpps had no significant effect on the levels of free steroids in pTVGGPPS plants compared to those in EV plants.
Figure 6.
Levels of β-sitosterol (dotted bars) and stigmasterol (white bars) in free sterol extracts of N. attenuata plants are reduced in pTVFPPS plants but not in pTVGGPPS plants. Values are means ± se (n = 9) GC-MS peak areas. *, Significantly different values from pTV00 at P < 0.05.
Effects of Silencing Prenyltransferases on Photosynthetic Pigments
To evaluate the effect of silencing prenyltransferases on photosynthetic pigments, leaf chlorophyll contents were measured during the feeding bioassay with a portable chlorophyll meter (Supplemental Fig. S3). Total extractable chlorophylls (Chl.a + Chl.b) and carotenoids were measured in 80% aqueous acetone extracts of leaves with a UV/VIS spectrophotometer (Supplemental Fig. S4, A and B). Total chlorophyll and carotenoid content did not differ between silenced and pTV00 plants in the same treatment group (Supplemental Figs. S3 and S4). However, the total carotenoid levels of herbivore-attacked plants were significantly lower than in unattacked plants (Supplemental Fig. S4B).
DISCUSSION
Previous work with N. attenuata plants silenced in their ability to produce nicotine and TPIs suggested that the plants produce additional potent defense metabolites (Steppuhn and Baldwin, 2007). When inverted-repeat putrescine N-methyltransferase-silenced N. attenuata plants growing in a field plantation in the Great Basin Desert were elicited by methyl jasmonate (MeJA), the damage these plants received from native herbivores was as low as that of wild-type plants, suggesting the presence of other potent MeJA-inducible defenses (Steppuhn et al., 2004). The results from this study suggest that HGL-DTGs or other minor undetected diterpenoids derived from GGPP are these powerful defenses. Although artificial diet bioassays have indicated that HGL-DTGs have a defensive function (Snook et al., 1997; Jassbi et al., 2006), their defensive function had not been previously explored by manipulating in planta production, the gold standard of proof of defensive function. We silenced two prenyltransferase genes that were potentially involved in HGL-DTG biosynthesis, Naggpps and Nafpps, and compared the growth of hornworm larvae on the silenced lines with their growth on EV plants. In a second study, we used Natpi- and Napmt-silenced plants to compare the antifeedant potential of the well-studied defensive compounds, PIs and nicotine, with that of HGL-DTGs. Because silencing ggpps can reduce the levels of all diterpenoids, we examined the presence of other diterpenoids such as cembrane- and labdane-type diterpenoids, which are found in N. tabacum (Wang et al., 2001, 2004; Wang and Wagner, 2003) by means of GC-MS, HPLC, and NMR analyses of the chloroform extract of the plant, but we could not detect any; the different GL-DTGs were the major diterpenoid constituents of the plant that were extractable by methanol and water (Jassbi et al., 2006).
N. attenuata plants, which were inoculated with A. tumefaciens GV3101 carrying pTVFPPS, pTVGGPPS, pTVPMT, and pTVTPI plasmids, grew the same as EV plants. RT-PCR analyses of the total RNA extracted from leaves of the gene-silenced plants revealed that Naggpps and Nafpps are 92% and 69% silenced in their transcript levels, respectively (Fig. 2, A and B). However, the unexpected up-regulation of Nafpps after silencing Naggpps (Fig. 2B) can only be explained when our understanding of the pathways for the synthesis of different metabolites is more complete. Apparently, silencing prenyltransferase synthesis leads to the accumulation of certain metabolites and activates different pathways that increase the level of FPPS transcripts.
Radio-GC analyses of the radiolabeled synthesized and hydrolyzed prenyldiphosphates in plants' protein extracts confirmed the reduction of FPPS activity in pTVFPPS plants. Although the levels of synthesized GG were quite low and not significantly reduced after silencing Naggpps, heterologous expression of Naggpps in E. coli and analyses of its expressed product confirmed that NaGGPPS functions exclusively as a GGPPS (Fig. 3). The lower levels of synthesized GG in plant protein extracts indicated that the products of NaGGPPS do not accumulate and are instead rapidly used as a substrate for subsequent reactions such as the synthesis of HGL-DTGs.
We found that silencing Naggpps dramatically reduced HGL-DTG levels (Figs. 4, dotted bars, and 5B) but not levels of the other defensive metabolites, nicotine, and PIs (Fig. 5, C and D). Similarly, silencing Napmt or Natpi did not strongly affect the concentration of HGL-DTGs (Fig. 5B). Although the levels of induced HGL-DTGs in pTVFPPS and pTVPMT plants were less than those of EV plants after 11 and 14 d of herbivory (Fig. 5B), the differences in the levels of the suppressed metabolites (HGL-DTGs and nicotine) in the respective silenced plants (pTVGGPPS and pTVPMT) compared to those in EV plants were always significantly higher than those in the other silenced lines (Fig. 5, B and C; P < 0.0001). Silencing Natpi resulted in a line (pTVTPI) that was free of TPI activity. Overall, we conclude that transforming N. attenuata with pTVGGPPS, pTVPMT, and pTVTPI plasmids successfully silenced the production of HGL-DTGs, nicotine, and TPIs, respectively, without affecting the other plant metabolites, including carotenoids and chlorophylls. Therefore, these plants could be used to compare the defensive value of each metabolite in a plant-herbivore interaction. The dramatically increased performance of hornworm larvae feeding on pTVGGPPS plants argues that DTGs are a more effective defense against this herbivore than are TPIs and nicotine.
Although we did not detect significant effects of TPI and nicotine on larval growth in our experiment (Fig. 5A), the defensive function of nicotine and TPIs against hornworm herbivory has been well established by gene-silencing experiments with stably transformed lines of N. attenuata and Nicotiana sylvestris (Voelckel et al., 2001; Steppuhn et al., 2004; Zavala et al., 2004b). The experiments reported here use VIGS to reduce nicotine and TPI levels, and VIGS is known to elevate salicylic acid levels. Because both nicotine and TPIs utilize the jasmonic acid (JA)-signaling cascade for their induction after herbivore attack (Halitschke and Baldwin, 2003), it is possible that salicylic acid/JA antagonism and the resulting decrease in these defense metabolites could have reduced our ability to detect negative effects on larval growth. Moreover, hornworm tolerates doses of nicotine that unadapted insects find fatal (Snyder et al., 1994), and it is possible that other primary metabolites, such as proteins or carbohydrate levels altered by the VIGS system, may have minimized the influence of nicotine and TPIs on larval growth. Low nitrogen (N) supply attenuated JA-induced TPI and nicotine levels in N. attenuata (Lou and Baldwin, 2004), which points to the importance of plant growth conditions in influencing defensive function of particular metabolites.
N. attenuata germinates in the postfire environment and copes with large changes in soil N during postfire succession. As the N content of the soil in natural habitat of N. attenuata remains high for up to 3 years after a fire (Lynds and Baldwin, 1998), plants can take advantage of the high N levels to support the biosynthesis of N-intensive defenses, such as TPIs and nicotine; when the N content of soil decreases, plants can use another effective carbon-intensive defense, such as HGL-DTGs. Analyses of HGL-DTGs, nicotine, and TPIs (Fig. 5, B–D) revealed that after 11 d, all of these direct defenses were highly induced by larval feeding and reached their maximum concentration in the leaves of the plant. Increasing the levels of direct defenses encourages herbivores to move to new plants or leaves, helping the plant not to lose its entire canopy, and perhaps reduces competition of conspecific neighboring plants (Van Dam et al., 2000; Lou and Baldwin, 2003; Zavala and Baldwin, 2004; Paschold et al., 2007).
The lower levels of HGL-DTGs only in Naggpps-silenced plants suggest that GGPP is a precursor of HGL-DTG diterpenoids in N. attenuata (Fig. 1). In addition to its role in the biosynthesis of diterpenoids and carotenoids, GGPP is required to produce phytol, the side chain of chlorophylls. Carotenoids provide accessory pigments for photosynthesis; when they degrade, plants become photobleached (Fray et al., 1995; Keller et al., 1998). Silencing Naggpps, in contrast to silencing Napds, did not result in bleaching. The product of the silenced Naggpps gene is therefore likely involved in the synthesis of the precursor of diterpenoids rather than in the synthesis of carotenoids or chlorophylls' prenyl side chain. Levels of carotenoids were, however, decreased in all plants after herbivore attack (Supplemental Fig. S4B), consistent with the commonly observed down-regulation of photosynthesis-related genes in N. attenuata and other plants in response to herbivore attack (Bi and Felton, 1995; Hermsmeier et al., 2001; Heidel and Baldwin, 2004). Down-regulation of the carotenoids after herbivore attack is clearly not Naggpps dependent and may be controlled by other genes involved in carotenoid biosynthesis.
Because the amplification of full-length Naggpps and three different fragments of it from N. attenuata cDNA always yielded exactly the same Naggpps sequence and because a Southern blot with chromosomal N. attenuata DNA (Supplemental Fig. S1) suggested the presence of only one copy of Naggpps, we conclude that the N. attenuata genome harbors only one copy of Naggpps. This conclusion does not exclude the existence of additional nonhomologous ggpps genes in N. attenuata or the possibility of complex regulation or compartmentalization of the synthesis of GGPP, as is seen in Arabidopsis and tomato (Bartley and Scolnik, 1995; Keller et al., 1998; Lange and Ghassemian, 2003; Ament et al., 2006). This separation may be vital for plants that use different GGPPS to synthesize either primary metabolites, such as chlorophyll and carotenoids, necessary for plant growth, or induced defensive secondary metabolites. Some GGPPS may additionally catalyze the formation of GPP, as inferred from the high homology between the two enzymes (Gershenzon and Kreis, 1999; Burke and Croteau, 2002; Tholl et al., 2004; Bouvier et al., 2005). However, heterologous expression of plant GGPPS or GPPS in E. coli provides a means of clearly differentiating among product profiles (Engprasert et al., 2004; Schmidt and Gershenzon, 2007a, 2007b). The expression of Naggpps in E. coli and the analyses of its expressed product confirmed that Naggpps functions exclusively as a GGPPS (Fig. 3).
The ggpps gene in tomato is reportedly induced by both JA and methyl salicylate. However, the product of the gene, (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene, is induced mainly by JA (Ament et al., 2006). (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene is the dehydration product of GL, and because GL synthase levels in plants induced by JA are higher than in plants induced with methyl salicylate, GL synthase is also thought to be induced mainly by JA, especially because GL levels increase dramatically when spider mites attack the plant (Ament et al., 2006). As in tomato, the induction of HGL-DTGs in N. attenuata by MeJA and insect feeding indicates that the GL biosynthesis and induction pathways may be similar in both species. Like linalool and nerolidol, which are the allylically rearranged products of geraniol and farnesol, respectively, GL can be derived from an allylic rearrangement in GG. This conversion has been shown to be catalyzed by GL synthase, which was assayed in tomato (Ament et al., 2006). As reported for plaunotol (18-hydroxy geranylgeraniol), further hydroxylation at C-17 followed by glycosylation at C-3 and C-17 hydroxyls can produce HGL-DTGs in Nicotiana (Nualkaew et al., 2006; Fig. 1). From the above observations, we can conclude that at least four kinds of enzymes are involved in the biosynthesis of HGL-DTGs from GGPP in N. attenuata: GGPP phosphatase, which converts GGPP to GG; GL synthase, which converts GG to GL; GL-17-hydroxylase, which hydroxylates the Me-17 of the GL; and glycosylating enzymes, which convert HGL to different HGL-DTGs (Ament et al., 2006; Nualkaew et al., 2006).
Silencing Nafpps did not significantly affect the levels of HGL-DTGs, the performance of feeding larvae, or the chlorophyll and the carotenoid contents of the plants (Figs. 4 and 5; Supplemental Figs. S3 and S4), suggesting that NaGGPPS does not use FPP as a substrate and most probably catalyzes the condensation of three IPPs together with one DMAPP to produce GGPP (Fig. 1) and that the FPP produced by the Nafpps gene product is not involved in forming GGPP for carotenoid synthesis. To explore the effect of Nafpps and Naggpps silencing on volatile terpenoids, the headspaces of all of the silenced plants were analyzed, but no significant differences were observed between volatile sesquiterpenoids levels in any lines (data not shown).
The lower levels of free β-sitosterol and stigmasterol in dichloromethane extracts of leaves from pTVFPPS plants confirmed that Nafpps is involved in steroid synthesis in N. attenuata (Fig. 6; Brown, 1998). The presence of multiple fpps genes in A. tridentata, each with about 80% identity, suggested that the encoded isozymes function specifically. For example, one FPPS (FDS-1) has a major role in sesquiterpene production, while another (FDS-2) maintains a pool of FPP for other branches of the pathway (Hemmerlin et al., 2003). A Southern blot with chromosomal N. attenuata DNA (Supplemental Fig. S1) revealed the existence of only one copy of Nafpps. N. attenuata may contain additional but nonhomologous fpps genes that were not silenced by our VIGS construct. This may be one reason why we did not detect differences in volatile sesquiterpene production between pTV00 and pTVFPPS plants.
In summary, we found that silencing Naggpps had the greatest effect on larval performance and we conclude that it is N. attenuata's most effective defense gene against hornworm larvae. The gene product showed only GGPPS activity and synthesizes a precursor of HGL-DTGs. The agreement of these results with prior artificial diet feeding assays confirms the utility of a combination of in planta and bioassay approaches for identifying new defense metabolites. As Naggpps is not involved in primary metabolite synthesis and HGL-DTGs do not incorporate nutrients such as N that commonly limit growth, overexpressing Naggpps could increase the resistance of N. attenuata to hornworm herbivory without affecting the growth of the plant. Given the evidence from this and other studies for a strict separation of prenyltransferases involved in primary metabolism from those involved in secondary metabolism, and given the ubiquity of terpene synthesis, such an approach may also prove useful in agricultural plants. However, as has been shown in N. tabacum where the silencing of a P450 hydroxylase gene suppressed the accumulation of hydroxylated cembranoids (Wang et al., 2001, 2004; Wang and Wagner, 2003), silencing geranyllinalool synthase and GL-17-hydroxylase in N. attenuata may represent a more specific means of suppressing HGL-DTGs.
MATERIALS AND METHODS
Construction of Plasmids
The primer sequences used are listed in Supplemental Table S1. Cloning was done in Escherichia coli strains TOP10 and TOP10F' (Invitrogen, http://www.invitrogen.com). For the VIGS experiments, pTV00 derivatives were used to transform Agrobacterium tumefaciens strain GV3101.
The Naggpps and Nafpps gene fragments and the full-length Naggpps coding sequence were PCR amplified with proofreading DNA polymerases (VentR DNA Polymerase, http://www.neb.com, or Phusion High-Fidelity DNA Polymerase, http://www.finnzymes.fi) on cDNA from Nicotiana attenuata as template. Primers were designed using sequence information available in GenBank (http://www.ncbi.nlm.nih.gov) from other Solanaceous plants and correspond to the following sequences: GGPPS1-33 and GGPPS2-31 to Nicotiana tabacum GGPPS1 mRNA (AB041632), N. tabacum GGPPS2 mRNA (AB041633), and tomato (Solanum lycopersicum) GGPS2 mRNA (DQ267903); GGPPS3-33 and GGPPS4-31 to Capsicum annuum gene for GGPPS (X80267); GGPPS7-32, GGPPS8-33, and GGPPS11-31 to DQ267903; GGPPS10-26 to DQ267903 and X80267; FPPS1-33 and FPPS2-34 to C. annuum mRNA for FPPS (X84695) and tomato FPPS1 mRNA (AF048747); and FPPS3-34 and FPPS4-33 to N. tabacum mRNA for FPPS (U97330) and X84695. The PCR fragment obtained with primer pair GGPPS10-26 and GGPPS11-31 (1.1 kb) comprised full-length Naggpps (EF382626) and was cloned in pJET1 (3.1 kb; Fermentas, http://www.fermentas.com; DQ317600), yielding pJETGGPPS (4.2 kb). VIGS vector pTVGGPPS (5.9 kb) was obtained by cloning the 0.3-kb PCR fragment synthesized with primer pair GGPPS1-33/GGPPS2-31, comprising positions 491 to 807 of EF382626, in the BamHI and SalI sites of the tobacco rattle virus (TRV)-based plasmid pTV00 (Ratcliff et al., 2001). For sequence determination, the 0.3-kb PCR fragment amplified with primer pair GGPPS3-33/GGPPS4-31, comprising positions 100 to 320 of EF382626, was cloned in the same way, yielding pTVGGPPS2 (5.8 kb). The 0.3-kb PCR product obtained with primers GGPPS7-32 and GGPPS8-33 (positions 811–1093 of EF382626) was cloned as an NcoI/PstI fragment in pUCPDS5 (Bubner et al., 2006), resulting in pUCGGPPS (2.9 kb).
To silence Nafpps, the PCR fragment obtained with primer pair FPPS3-34 and FPPS4-33 was cloned as a BamHI/SalI fragment in pTV00, leading to silencing vector pTVFPPS (5.9 kb). Plasmid pUCFPPS was constructed by cloning the fragment synthesized with primers FPPS1-33 and FPPS2-34 in the NcoI and PstI sites of pUCPDS5. The GenBank accession numbers of the cloned Nafpps fragments are EF382631 and EF382632. The construction of the vectors pTVTPI (5.7 kb) and pTVPMT (6.2 kb) used for silencing the N. attenuata genes Napmt and Natpi was performed as described previously (pTVPMT2 and pTVPI2 in Saedler and Baldwin, 2004) by cloning the PCR fragments obtained with primer pairs TRV5-31/TRV6-31 (0.7-kb Napmt1 product) and TRV9-34/TRV10-36 (0.2-kb Natpi product) in the BamHI and HindIII sites of pTV00. pTVPD used for silencing Napds is a pTV00 derivative carrying a 206-bp fragment of the Nicotiana benthamiana phytoene desaturase (AJ571700) and was kindly provided by D. Baulcombe of the Sainsbury laboratory, Norwich, UK.
Plant Growth and VIGS
Smoke-treated N. attenuata seeds originating from glasshouse-grown generations of a collection originating from Utah were germinated on phytagel agar (Sigma, http://www.sigmaaldrich.com) as described previously and transferred to soil after about 10 d (Baldwin et al., 1994). All plants were grown under a 28°C/24°C 16-/8-h light/dark regime until the early rosette stage. The plants were transferred to a growth chamber under 20°C/17°C ± 1°C 16-/8-h light/dark at 70% ± 10% humidity for the VIGS inoculation. VIGS was used to silence the Naggpps gene, utilizing the TRV genome resident on the pTV00-derived plasmids and on plasmid pBINTRA6 (Ratcliff et al., 2001; Saedler and Baldwin, 2004). Approximately 6 d after plants were transferred to the growth chamber, they were coinoculated with A. tumefaciens GV3101 carrying pTVPD, pTVFPPS, pTVGGPPS, pTVPMT, or pTVTPI, and with the same Agrobacterium strain carrying pBINTRA6 according to the modified Ratcliff method (Ratcliff et al., 2001; Saedler and Baldwin, 2004). About 20 d after inoculation, the Napds-silenced plants bleached thoroughly, demonstrating successful silencing.
Larval Rearing and Feeding Bioassay
Larvae of the tobacco hornworm (Manduca sexta; Lepidoptera, Sphingidae) were hatched overnight at 28°C from eggs received from Carolina Biological Supply (http://www.carolina.com). One newly hatched larva of hornworm was released on a leaf of each silenced and nonsilenced (empty vector) plant when the corresponding leaf in the Napds-silenced plants (positive control) had completely bleached. A total of 40 and 27 replicate plants from each line were used in the bioassay in the first and second VIGS experiments, respectively (Figs. 4 and 5A). To measure larval performance, larvae were weighed after 4, 14, and 18 d in the first experiment (larval masses after 14 d of feeding are presented in Fig. 4) and after 4, 8, 11, 14, and 16 d of feeding in the second experiment (Fig. 5A). The insect-feeding bioassays were performed in the chambers and under the same conditions as the VIGS experiments. In the first VIGS experiment, Naggpps- and Nafpps-silenced plants were used; in the second experiment, Napmt-, Natpi-, Naggpps-, and Nafpps-silenced plants were used.
RNA Extraction, Northern Blotting, and RT-PCR
Samples were ground in liquid N with a mortar and pestle. Total RNA was extracted with TRI ReagentTM (Sigma) according to the manufacturer's instructions. RNA quality was checked on a denaturing 1% agarose gel (5% formaldehyde) and concentrations were measured spectrophotometrically at 260, 280, and 320 nm. RNA samples were stored at −80°C until use.
To determine the efficiency of silencing Naggpps and Nafpps genes, we conducted northern-blot analysis and used quantitative PCR analysis. RNA samples (10 μg, pooled from three individually grown plants) were size fractionated by 1.2% (w/v) agarose-formaldehyde gel electrophoresis and blotted onto a nylon membrane (GeneScreenPlus; PerkinElmer, http://www.perkinelmer.com) according to the manufacturer's protocol. Ethidium bromide fluorescence was used as the loading control. After blotting and UV cross-linking, blots were prehybridized for 1 h at 42°C in Ultrahyb hybridization buffer (Ambion, http://www.ambion.com) followed by overnight hybridization with the 32P-labeled probes. The blots were washed once, 15 min at 60°C with 2× SSC, 0.1% (w/v) SDS, followed by twice at 60°C in 0.1× SSC and 0.1% (w/v) SDS, 20 min each. Blots were exposed for 24 h on a phosphoimage film (Fujifilm, http://www.fujifilm.com). The signals were read by fluorescent image analyzer FLA-3000 (Fujifilm) and quantified with Aida Image Analyzer software (Raytest, http://www.raytest.com). The 313-bp PCR fragment obtained with primer pair GGPPS7-32/GGPPS8-33 and pUCGGPPS as template served as a probe for Naggpps (Supplemental Table S1). The level of Nafpps transcript was detected using probe PCR synthesized with primer pair FPPS1-33/FPPS2-34 and template pUCFPPS (Supplemental Table S1).
For RT-PCR analysis, five replicated biological samples were used. One microgram of total RNA obtained from each sample was reverse transcribed using oligo(dT) and Superscript II reverse transcriptase (Invitrogen) for a total volume of 20 μL. cDNA samples were further diluted with water to 40 μL, and 1 μL of the diluted cDNA was used for RT-PCR, carried out on a ABI PRISM 7700 Sequence Detection system (Applied Biosystems, http://www.appliedbiosystems.com) using qPCRTM Core kits (Eurogentec, http://www.eurogentec.com). For each analysis, a linear standard curve, threshold cycle number versus log (designated transcript level), was constructed using a series dilution of a specific cDNA standard; the transcript levels in all unknown samples were determined according to the sta ndard curve. The N. attenuata actin gene was used as an internal standard for normalizing cDNA concentration variations. The sequences of the primers used for SYBR Green-based RT-PCR (the N. attenuata actin gene, Actin-F1 and Actin-R1; Nafpps, FPPS FOR and FPPS REV; Naggpps, GGPPS FOR, GGPPS REV) are provided in Supplemental Table S1.
Southern Blotting
Genomic DNA was extracted from N. attenuata young leaves using the cetyl-trimethyl-ammonium bromide method (Doyle and Doyle, 1987). Quality and concentration were checked by agarose gel electrophoresis. Genomic DNA samples (5 μg) were digested with AseI, BamHI, BclI, or DraI (New England Biolabs, http://www.neb.com), size fractionated on a 0.8% (w/v) agarose gel, and blotted onto a nylon membrane (GeneScreenPlus; Perkin Elmer). Labeling, washing, and analytical procedures were done according to those used for northern blotting. The Naggpps probe was PCR synthesized with primer pair GGPPS10-26/GGPPS11-31 and pJETGGPPS as template and comprised the complete coding sequence of the gene. The Nafpps probe was the same used for northern blotting.
NaGGPPS and NaFPPS Assays in Plant Protein Extracts
Two grams frozen leaf tissue was ground in liquid N using a mortar and pestle and homogenized in extraction buffer containing 50 mm MOPSO, pH 6.8, 5 mm ascorbic acid, 5 mm sodium bisulfite, 5 mm dithiothreitol (DTT), 10 mm MgCl2, 1 mm EDTA, 10% (v/v) glycerol, 1% (w/v) polyvinylpyrrolidone (Mr = 10,000), 4% (w/v) polyvinylpolypyrrolidone, 4% (w/v) Amberlite XAD-4, and 0.1% (v/v) Tween 20. Extracts were shaken for 30 min at 4°C and centrifuged for 30 min at 10,000g. The filtered supernatant (Miracloth) was used for enzyme activity measurements that were carried out for the functional characterization of NaGGPPS.
Heterologous Expression of Naggpps in E. coli
To express Naggpps in E. coli, a fragment of this gene was amplified using the Expand High Fidelity PCR system (Roche Applied Science, http://www.roche-applied-science.com) with pJETGGPPS as template, thereby removing the coding sequence of the potential chloroplast transit peptide. The cleavage site of this peptide was predicted using the chloroP program (http://www.cbs.dtu.dk). With primer pair GGPPS14-25 and GGPPS13-24, a 0.9-kb fragment was synthesized; after an ATG start codon, the fragment contained codon 54 of EF382626. The fragment was cloned with T-overhangs in plasmid pCR-T7 CT TOPO (2.7 kb; Invitrogen) yielding Naggpps expression vector pGGPPS (3.6 kb). Expression strain E. coli BL 21(DE3) pLys S (Invitrogen) was transformed with pGGPPS. Bacterial cultures expressing Naggpps were grown using Overnight Express Autoinduction system 1 (Novagen, http://www.novagen.com) as described by the manufacturer's instructions but at 18°C. Bacterial pellets were resuspended in assay buffer without DTT and sonicated. The His-tagged recombinant protein was purified with nickel-nitrilotriacetic acid agarose columns (Qiagen, http://www.qiagen.com) according to the manufacturer's instructions. The recombinant protein was eluted with 250 mm imidazole in the assay buffer. After adding DTT, fractions were checked by SDS-PAGE and used to determine enzyme activity.
Functional Characterization of NaGGPPS
NaGGPPS assays were carried out in a final volume of 500 μL containing 20 mm 3-(N-Morpholino)-2-hydroxypropanesulfonic acid, pH 7.0, 10 mm of MgCl2, 10% (v/v) glycerol, and 2 mm DTT. Assays were carried out in triplicates of at least three biologically independent experiments with 40 μm [1-14C]IPP (2 MBq/μmol; Biotrend, http://www.biotrend.com) and 40 μm DMAPP (Echelon Biosciences, http://www.echelon-inc.com). After the reaction was initiated by adding recombinant protein, the assay mixture was covered with 1 mL pentane and incubated overnight at 30°C. To stop the assay and hydrolyze all diphosphate esters, a 1-mL solution with 2 units of calf intestine alkaline phosphatase (Sigma) and 2 units of potato (Solanum tuberosum) apyrase (Sigma) in 0.2 m Tris-HCl, pH 9.5 was added to each assay and incubated at 30°C overnight. After enzymatic hydrolysis, the resulting prenyl alcohols were extracted into 2 mL of diethyl ether and, after the addition of a standard terpene mixture (geraniol, farnesol, and geranylgeraniol), the organic extracts were evaporated under N2 and used for radio-GC measurements.
Radio-GC analysis was performed on a Hewlett-Packard (http://www.hp.com) HP6890 gas chromatograph equipped with a thermal conductivity detector (TCD) and Raga radioactivity detector (Raytest) and using a DB5-MS capillary column (J&W Scientific, http://www.jandw.com; 30 m × 0.25 mm i.d., 0.25-μm film thickness). The oven temperature was set 3 min at 70°C, followed by a gradient from 70°C to 240°C at 6°C min−1 and kept 3 min at the final temperature. The carrier gas was H2 with a flow rate of 2 mL min−1. Injector and TCD detector temperatures were maintained at 220°C and 250°C. The injection volume of the concentrated organic phase was 1 μL. The synthesized products, measured by radio-GC, were identified by comparing their retention times with those of coinjected authentic nonradioactive terpene standards, detected via the TCD. Protein concentrations in enzyme assays were measured according to Bradford (Bradford, 1976) using the Bio-Rad reagent with bovine serum albumin as standard. The protein concentration in each assay was equilibrated to a range of 50 to 100 μg mL−1.
HGL-DTGs and Nicotine Analysis
Six plants from each line were randomly selected and from each plant six systemic leaves that had not been herbivore attacked were harvested from locations above where the larvae fed. After being frozen in liquid N, leaves (150 mg) were extracted using 1 mL pure methanol or 0.5% acetic acid solution of 40% methanol in an ultrasonic bath for 35 min. Thymol (1 mg/mL, 50 + 800 μL of the methanolic extract) was used as an internal standard and RP18 HPLC analyses were performed as described previously for HGL-DTGs and nicotine (Keinänen et al., 2001; Jassbi et al., 2006). In analyses with an Inertsil ODS-3, 3 μm, 100 Å, 150- × 4.6-mm i.d. column (Ercatech AG, http://www.ercatech.ch), the HGL-DTGs' peaks eluted at 22 to 27 min and with a Luna 5 m C18 (2) 100 Å, 250- × 4.6-mm i.d. column, the HGL-DTGs' peaks eluted at 24 to 28 min (Phenomenex, http://www.phenomenex.com).
Analysis of Protease Inhibitors
A total of 150 mg of fresh leaf material of the plants was harvested, ground in liquid N, and extracted with 0.3 mL extracting buffer; subsequently, samples were subjected to radial diffusion assays as described previously (van Dam et al., 2001) and expressed as nmol mg−1 of leaf protein.
Analysis of Free Sterols
A total of 300 mg of the systemic leaf plant material, collected 14 d after herbivore treatment in the first VIGS experiment, was extracted as above but with 2 mL dichloromethane. The extracts (nine replicates for each silencing construct) were dried under an N2 stream, redissolved in 100 μL of dichloromethane, and subjected to GC-MS analyses. The GC-MS was carried out on a Varian 3800 gas chromatograph coupled to a Varian Saturn 2000 mass spectrometer operating in EI mode at 70 eV (http://www.varianinc.com). The GC equipped to a DB-5 MS (J&W Scientific column, 30-m × 0.25-mm i.d., 0.25-μm film thickness). The oven temperature was programmed from 285°C to 310°C at 5°C min−1 and kept for 10 min at 310°C. The carrier gas was helium with a flow rate of 1 mL min−1. The injector temperature was set at 310°C in splitless mode. The injection volume was 1 μL for all of the samples. Two major steroids, β-sitosterol and stigmasterol, were identified by comparing their retention times and mass spectrums with those of the authentic standards (from Fluka, http://www.sigmaaldrich.com, and Sigma).
Total Chlorophyll and Carotenoid Measurements
The chlorophyll contents of the plants' leaves were monitored by a portable SPAD-502 chlorophyll meter (Konica Minolta, http://www.konicaminolta.com) and expressed as a SPAD value (Netto et al., 2002). To measure the extractable chlorophylls and carotenoids, 100 mg of N2-frozen leaf tissues, collected 14 d after herbivore treatment, were extracted with 1 mL of 80% acetone for 30 min and diluted 10 times. Absorptions of the solutions at 664, 647, and 470 nm were measured using an Ultraspec 300 UV/VIS spectrophotometer. Concentrations of total carotenoids and chlorophylls were calculated according to the method described in Lichtenthaler (1987).
Statistical Analyses
The Student's t tests were performed using the algorithm embedded in StatView (http://www.imp.com).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF 382626, EF 382631, and EF 382632.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Southern-blot analysis of Naggpps and Nafpps genes in N. attenuata.
Supplemental Figure S2. RP-HPLC chromatograms of the methanolic extracts of the fresh leaves of N. attenuata plants silenced with A. tumefaciens GV3101 containing pTV00 (A), pTVGGPPS (B), and pTVFPPS (C), and structures of the HGL-DTGs (D) found in N. attenuata.
Supplemental Figure S3. Total chlorophyll contents expressed as SPAD (named after the company's Soil-Plant Analysis Division, Konica Minolta) mean values of three replicate leaves of nine plants (±se, n = 9) at different time intervals after seedlings were measured with a portable chlorophyll meter.
Supplemental Figure S4. Spectrophotometer-calculated concentration (mean ± se, n = 5, mg g−1 FW of leaves) of total chlorophyll (A), Chl b (λ = 647 nm), and Chl a (λ = 664 nm), and total carotenoids (B; λ = 470 nm) in 80% acetone extract of the leaves of VIGS plants after 14 d of herbivory (herbivore-attacked plants) and the plants of the same age but not attacked by herbivores.
Supplemental Table S1. Sequences of the primers used for the construction of plasmids, northern blotting, Southern blotting, and RT-PCR.
Supplementary Material
Acknowledgments
We thank David Baulcombe for the gift of pTV00, pTVPD, and pBINTRA; Jianqiang Wu for providing the N. attenuata actin gene primers; Emily Wheeler for editorial assistance; and the glasshouse team for growing the plants.
This work was supported by the Alexander von Humboldt Foundation (postdoctoral fellowship to A.R.J.) and by the Max Planck Society.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors is: Ian T. Baldwin (baldwin@ice.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ament K, Van Schie CC, Bouwmeester HJ, Haring MA, Schuurink RC (2006) Induction of a leaf specific geranylgeranyl pyrophosphate synthase and emission of (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene in tomato are dependent on both jasmonic acid and salicylic acid signaling pathways. Planta 224 1197–1208 [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Staszak-Kozinski L, Davidson R (1994) Up in smoke. I. Smoke-derived germination cues for post-fire annual, Nicotiana attenuata Torr. Ex. Watson. J Chem Ecol 20 2345–2371 [DOI] [PubMed] [Google Scholar]
- Barbehenn RV, Martin MM (1994) Tannin sensitivity in larvae of Malacosoma disstria (Lepidoptera): roles of the peritrophic envelope and midgut oxidation. J Chem Ecol 20 1985–2001 [DOI] [PubMed] [Google Scholar]
- Bartley GE, Scolnik PA (1995) Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Visser PB, Angenent GC, Krens FA (2004) The potential of virus-induced gene silencing for speeding up functional characterization of plant genes. Genet Mol Res 3 323–341 [PubMed] [Google Scholar]
- Bi JL, Felton GW (1995) Foliar oxidative stress and insect herbivory: primary compounds, secondary metabolites, and reactive oxygen species as components of induced resistance. J Chem Ecol 21 1511–1530 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Rahier A, Camara B (2005) Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res 44 357–429 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Broadway RM (1997) Dietary regulation of serine proteinases that are resistant to serine proteinase inhibitors. J Insect Physiol 43 855–874 [DOI] [PubMed] [Google Scholar]
- Broadway RM, Duffey SS (1988) The effect of plant protein quality on insect digestive physiology and the toxicity of plant proteinase inhibitors. J Insect Physiol 34 1111–1117 [Google Scholar]
- Brown GD (1998) The biosynthesis of steroids and triterpenoids. Nat Prod Rep 15 653–696 [Google Scholar]
- Bubner B, Gase K, Berger B, Link D, Baldwin IT (2006) Occurrence of tetraploidy in Nicotiana attenuata plants after Agrobacterium-mediated transformation is genotype specific but independent of polysomaty of explant tissue. Plant Cell Rep 25 668–675 [DOI] [PubMed] [Google Scholar]
- Burke C, Croteau R (2002) Geranyl diphosphate synthase from Abies grandis: cDNA isolation, functional expression, and characterization. Arch Biochem Biophys 405 130–136 [DOI] [PubMed] [Google Scholar]
- Damaty S, Hudson BJF (1975) Interaction of gossypol with cottonseed protein: potentiometric studies. J Sci Food Agric 26 1667–1672 [Google Scholar]
- De Boer G, Hanson FE (1987) Feeding responses to Solanaceous allelochemicals by larvae of the tobacco hornworm, Manduca sexta. Entomol Exp Appl 45 123–131 [Google Scholar]
- Dewick PM (2002) The biosynthesis of C5-C25 terpenoid compounds. Nat Prod Rep 19 181–222 [DOI] [PubMed] [Google Scholar]
- Downs CT, McDonald PM, Brown K, Ward D (2003) Effects of Acacia condensed tannins on urinary parameters, body mass, and diet choice of an Acacia specialist rodent, Thallomys nigricauda. J Chem Ecol 29 845–857 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19 11–15 [Google Scholar]
- Duan X, Li X, Xue Q, Abo-El-Saad M, Xu D, Wu R (1996) Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nat Biotechnol 14 494–498 [DOI] [PubMed] [Google Scholar]
- Engprasert S, Taura F, Kawamukai M, Shoyama Y (2004) Molecular cloning and functional expression of geranylgeranyl pyrophosphate synthase from Coleus forskohlii Briq. BMC Plant Biol 4 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeny P (1976) Plant apparency and chemical defense. In JW Wallace, RL Mansell, eds, Biochemical Interaction Between Plants and Insects, Recent Advances in Phytochemistry, Vol 10. Plenum Press, New York, pp 1–40
- Felton GW, Broadway RM, Duffey SS (1989) Inactivation of protease inhibitor activity by plant-derived quinones: complications for host-plant resistance against Noctuid herbivores. J Insect Physiol 35 981–990 [Google Scholar]
- Fray RG, Wallace A, Fraser PD, Valero D, Hedden P, Bramley PM, Grierson D (1995) Constitutive expression of a fruit phytoene gene in transgenic tomatoes causes dwarfism by redirecting metabolites from gibberellin pathway. Plant J 8 693–701 [Google Scholar]
- Gershenzon J, Kreis W (1999) Biochemistry of terpenoids: monoterpenes, sesquiterpenes, diterpenes, sterols, cardiac glycosides and steroid saponins. In M Wink, ed, Biochemistry of Plant Secondary Metabolism. Annual Plant Reviews, Vol 2. Sheffield Academic Press, Sheffield, UK, pp 222–299
- Green ES, Zangerl AR, Berenbaum MR (2001) Effects of phytic acid and xanthotoxin on growth and detoxification in caterpillars. J Chem Ecol 27 1763–1773 [DOI] [PubMed] [Google Scholar]
- Guo Z, Severson RF, Wagner GJ (1994) Biosynthesis of the diterpene cis-abienol in cell-free extracts of tobacco trichomes. Arch Biochem Biophys 308 103–108 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Baldwin IT (2003) Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J 36 794–807 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Keßler A, Kahl J, Lorenz A, Baldwin IT (2000) Ecophysiological comparison of direct and indirect defenses in Nicotiana attenuata. Oecologia 124 408–417 [DOI] [PubMed] [Google Scholar]
- Heidel AJ, Baldwin IT (2004) Microarray analysis of salicylic acid- and jasmonic acid-signaling in responses of Nicotiana attenuata to attack by insects from multiple feeding guilds. Plant Cell Environ 27 1362–1373 [Google Scholar]
- Hemmerlin A, Rivera SB, Erickson HK, Poulter CD (2003) Enzymes encoded by the farnesyl diphosphate synthase gene family in the big sagebrush Artemisia tridentata ssp. spiciformis. J Biol Chem 278 32132–32140 [DOI] [PubMed] [Google Scholar]
- Hermsmeier D, Schittko U, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. I. Large-scale changes in the accumulation of growth and defense-related plant mRNAs. Plant Physiol 125 683–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassbi AR, Zamanizadehnajari S, Kessler D, Baldwin IT (2006) A new acyclic diterpene glycoside from Nicotiana attenuata with a mild deterrent effect on feeding Manduca sexta larvae. Z Naturforsch 61b 1138–1142 [Google Scholar]
- Keinänen M, Oldham NJ, Baldwin IT (2001) Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuata. J Agric Food Chem 49 3553–3558 [DOI] [PubMed] [Google Scholar]
- Keller Y, Bouvier F, D'Harlingue A, Camara B (1998) Metabolic compartmentation of plastid prenyllipid biosynthesis, evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur J Biochem 251 413–417 [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291 2141–2144 [DOI] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51 925–948 [DOI] [PubMed] [Google Scholar]
- Laskaris G, Van der Heijden R, Verpoorte R (2000) Purification and partial characterisation of geranylgeranyl diphosphate synthase, from Taxus baccata cell cultures an enzyme that regulates taxane biosynthesis. Plant Sci 51 97–105 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148 350–382 [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus induced gene silencing in tomato. Plant J 31 777–786 [DOI] [PubMed] [Google Scholar]
- Lou Y, Baldwin IT (2003) Manduca sexta recognition and resistance among allopolyploid Nicotiana host plants. Proc Natl Acad Sci USA 100 14581–14586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Baldwin IT (2004) Nitrogen supply influences herbivore-induced direct and indirect defenses and transcriptional responses in Nicotiana attenuata. Plant Physiol 135 496–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC (2003) Virus-induced gene silencing in plants. Methods 30 296–303 [DOI] [PubMed] [Google Scholar]
- Lynds GY, Baldwin IT (1998) Fire, nitrogen, and defensive plasticity in Nicotiana attenuata. Oecologia 115 531–540 [DOI] [PubMed] [Google Scholar]
- Netto AT, Campostrini E, De Oliveira JG, Yamanishi OK (2002) Portable chlorophyll meter for the quantification of photosynthetic pigments, nitrogen and the possible use for assessment of the photochemical process in Carica papaya L. Braz J Plant Physiol 14 203–210 [Google Scholar]
- Nualkaew N, De-Eknamkul W, Kutchan TM, Zenk MH (2006) Membrane-bound geranylgeranyl diphosphate phosphatases: purification and characterization from Croton stellatopilosus leaves. Phytochemistry 67 1613–1620 [DOI] [PubMed] [Google Scholar]
- Ohnuma SI, Hirooka K, Tsuruoka N, Yano M, Ohto C, Nakane H, Nishino T (1998) A pathway where polyprenyl diphosphate elongates in prenyltransferases; insight into a common mechanism of chain length determination of prenyltransferases. Biol Chem 273 26705–26713 [DOI] [PubMed] [Google Scholar]
- Paschold A, Halitschke R, Baldwin IT (2007) Co(i) ordinating defense NaCOI1 mediates herbivore-induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant J 51 79–91 [DOI] [PubMed] [Google Scholar]
- Pohlon E, Baldwin IT (2001) Artificial diets ‘capture’ the dynamics of jasmonate-induced defenses in plants. Entomol Exp Appl 100 127–130 [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25 237–245 [DOI] [PubMed] [Google Scholar]
- Rhoades DF, Cates RG (1976) Toward a general theory of plant antiherbivore chemistry. In JW Wallace, RL Mansell, eds, Biochemical Interaction Between Plants and Insects, Recent Advances in Phytochemistry, Vol 10. Plenum Press, New York, pp 168–213
- Saedler R, Baldwin IT (2004) Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuata. J Exp Bot 55 151–157 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Gershenzon J (July 26, 2007. a) Cloning and characterization of isoprenyl diphosphate synthases with farnesyl diphosphate and geranylgeranyl diphosphate synthase activity from Norway spruce (Picea abies) and their relation to induced oleoresin formation. Phytochemistry 68 2649–2659 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Gershenzon J (2007. b) Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies). Phytochemistry 69 49–57 [DOI] [PubMed] [Google Scholar]
- Shinozaki Y, Tobita T, Mizutani M, Matsuzaki T (1996) Isolation and identification of two new diterpene glycosides from Nicotiana tabacum. Biosci Biotechnol Biochem 60 903–905 [DOI] [PubMed] [Google Scholar]
- Smigocki AC, Wilson D (2004) Pest and disease resistance enhanced by heterologous suppression of a Nicotiana plumbaginifolia cytochrome P450 gene CYP72A2. Biotechnol Lett 26 1809–1814 [DOI] [PubMed] [Google Scholar]
- Snook ME, Johnson AW, Severson RF, Teng Q, White RA Jr, Sisson VA, Jackson DM (1997) Hydroxygeranyllinalool glycosides from tobacco exhibit antibiosis activity in the tobacco budworm, Heliothis virescens (F.). J Agric Food Chem 45 2299–2308 [Google Scholar]
- Snyder MJ, Walding JK, Feyereisen R (1994) Metabolic fate of the allelochemicals in the tobacco hornworm Manduca sexta Biochem. Mol Biol 24 837–846 [Google Scholar]
- Steppuhn A, Baldwin IT (2007) Resistance management in a native plant: nicotine prevents herbivores from compensating for plant protease inhibitors. Ecol Lett 10 499–511 [DOI] [PubMed] [Google Scholar]
- Steppuhn A, Gase K, Kroch B, Halitschke R, Baldwin IT (2004) Nicotine's defensive function in nature. PLoS Biol 2 1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D, Kish CM, Orlova I, Sherman D, Gershenzon J, Pichersky E, Dudareva N (2004) Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell 16 977–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher BF, Bernays EA, Barbehenn RV, Wrubel RP (1989) Oral dosing of insects with feeding deterrent compounds. Entomol Exp Appl 52 119–133 [Google Scholar]
- van Dam NM, Hadwich K, Baldwin IT (2000) Induced responses in Nicotiana attenuata affect behavior and growth of the specialist herbivore Manduca sexta. Oecologia 122 371–379 [DOI] [PubMed] [Google Scholar]
- van Dam NM, Horn M, Mareš M, Baldwin IT (2001) Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J Chem Ecol 27 547–568 [DOI] [PubMed] [Google Scholar]
- Voelckel C, Kruegel T, Gase K, Heidrich N, Van Dam NM, Winz R, Baldwin IT (2001) Anti-sense expression of putrescine N-methyltransferase confirms defensive role of nicotine in Nicotiana sylvestris against Manduca sexta. Chemoecology 11 121–126 [Google Scholar]
- Wang E, Hall JT, Wagner GJ (2004) Transgenic Nicotiana tabacum L. with enhanced trichome exudate cembratrieneols has reduced aphid infestation in the field. Mol Breed 13 49–57 [Google Scholar]
- Wang E, Wagner GJ (2003) Elucidation of the functions of genes central to diterpene metabolism in tobacco trichomes using posttranscriptional gene silencing. Planta 216 686–691 [DOI] [PubMed] [Google Scholar]
- Wang E, Wang R, DeParasis J, Loughrin JH, Gan S, Wagner GJ (2001) Suppression of a P450 hydroxylase gene in plant trichome glands enhances natural-product-based aphid resistance. Nat Biotechnol 19 371–374 [DOI] [PubMed] [Google Scholar]
- Winterer J, Bergelson J (2001) Diamondback moth compensatory consumption of protease inhibitor-transformed plants. Mol Ecol 10 1069–1074 [DOI] [PubMed] [Google Scholar]
- Zavala JA, Baldwin IT (2004) Fitness benefits of trypsin protease inhibitor expression in Nicotiana attenuata are greater than their costs when plants are attacked. BMC Ecol 4 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Baldwin IT (2004. a) Constitutive and inducible trypsin proteinase inhibitor production incurs large fitness costs in Nicotiana attenuata. Proc Natl Acad Sci USA 101 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Hui D, Baldwin IT (2004. b) Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defenses. Plant Physiol 134 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.