Abstract
Germination of lettuce (Lactuca sativa) ‘Grand Rapids’ seeds is regulated by phytochrome. The action of phytochrome includes alterations in the levels of gibberellin (GA) and abscisic acid (ABA). To determine the molecular mechanism of phytochrome regulation of ABA metabolism, we isolated four lettuce cDNAs encoding 9-cis-epoxycarotenoid dioxygenase (biosynthesis; LsNCED1–LsNCED4) and four cDNAs for ABA 8′-hydroxylase (catabolism; LsABA8ox1–LsABA8ox4). Measurements of ABA and its catabolites showed that a decrease in ABA level coincided with a slight increase in the level of the ABA catabolite phaseic acid after red light treatment. Quantitative reverse transcription-polymerase chain reaction analysis indicated that ABA levels are controlled by phytochrome through down-regulation of LsNCED2 and LsNCED4 expression and up-regulation of LsABA8ox4 expression in lettuce seeds. Furthermore, the expression levels of LsNCED4 decreased after GA1 treatment, whereas the levels of expression of the other two genes were unaffected. The LsNCED4 expression was also down-regulated by red light in lettuce seeds in which GA biosynthesis was suppressed by AMO-1618, a specific GA biosynthesis inhibitor. These results indicate that phytochrome regulation of ABA metabolism is mediated by both GA-dependent and -independent mechanisms. Spatial analysis showed that after red light treatment, the ABA decrease on the hypocotyl side was greater than that on the cotyledon side of lettuce seeds. Moreover, phytochrome-regulated expression of ABA and GA biosynthesis genes was observed on the hypocotyl side, rather than the cotyledon side, suggesting that this regulation occurs near the photoperceptive site.
Plants undergo correct morphogenesis by responding to light and adapt to various forms of light. In addition to imbibition, some seeds require light to germinate and are called photoblastic seeds. Photoblastic seeds germinate after irradiation with red light (R; wavelength approximately 600–700 nm), and this effect of R is cancelled by successive irradiation with far-red light (FR; wavelength approximately 700–750 nm). The effects of R and FR are reversible. This type of germination, called photogermination, was discovered by experiments using lettuce (Lactuca sativa) ‘Grand Rapids’ seeds (Borthwick et al., 1952). The energy from wavelengths of R is mainly required for photosynthesis. This strategy allows survival of small seeds in which storage substances are low, such as the seeds of lettuce, tomato (Solanum lycopersicum), tobacco (Nicotiana tabacum), and Arabidopsis (Arabidopsis thaliana). The typical reversible photoreaction of lettuce seeds allowed Butler et al. (1959) to detect the pigment that acts as the R/FR receptor in extracts from etiolated cotyledons of Brassica rapa and etiolated shoots of Zea mays using differential spectrophotometry, and this photoreceptor was named phytochrome.
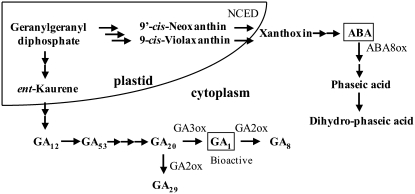
Many studies to elucidate the mechanisms of photoblastic lettuce seed germination have been carried out for over a half century, including biochemical, physical, and physiological studies, etc. (see many references that have been published). For example, the growth potential induced by R was measured by changing the osmotic pressure using mannitol (Takeba and Matsubara, 1979), and the induced growth potential was found to be due to degradation of storage protein (Takeba and Matsubara, 1977) and increased Glu and Gln levels in the hypocotyls of lettuce seeds (Takeba, 1980a, 1980b). The hypocotyl was found to be the most sensitive site in response to R in the lettuce seeds (Inoue and Nagashima, 1991). The role of the phytohormones GA and abscisic acid (ABA) on regulation of germination of the lettuce seeds is important. Application of bioactive GA mimics the effect of R (Kahn and Goss, 1957; Ikuma and Thimann, 1960), whereas application of ABA inhibits germination induced by R or application of bioactive GA (Kahn, 1968; Sankhla and Sankhla, 1968). Endogenous levels of the bioactive GA GA1 are up-regulated by phytochrome (Toyomasu et al., 1993), and those of ABA are down-regulated by phytochrome or application of GA1 in lettuce seeds (Toyomasu et al., 1994). These results suggest that control of the balance between endogenous levels of these two phytohormones is essential for regulation of germination of lettuce seeds by light. Furthermore, endogenous levels of GA1 are up-regulated by phytochrome through marked up-regulation of a GA 3-oxidase (Fig. 1) gene (Ls3h1, which is renamed LsGA3ox1 in this article) for the last step in GA1 biosynthesis (Toyomasu et al., 1998) and slight down-regulation of a GA 2-oxidase (Fig. 1) gene (LsGA2ox2) for inactivation of GA1 (Nakaminami et al., 2003). However, the molecular mechanisms of ABA metabolism in lettuce seeds have not been addressed.
Figure 1.
Metabolism of ABA and GA1 in lettuce seeds.
ABA is a sesquiterpenoid produced from C-40 carotenoids in plants (Milborrow and Lee, 1998; Hirai et al., 2000; Kasahara et al., 2004). The first committed step in ABA biosynthesis in plants is the oxidative cleavage of 9-cis-epoxycarotenoid to xanthoxin. The oxidative cleavage of cis-epoxycarotenoids catalyzed by 9-cis-epoxycarotenoid dioxygenase (NCED; Fig. 1) is the key regulatory step of ABA biosynthesis in higher plants (Nambara and Marion-Poll, 2005). ABA catabolism involves two pathways: hydroxylation and conjugation with Glc (Nambara and Marion-Poll, 2005). Hydroxylation pathways occur by irreversible hydroxylation, mainly at the 8′ position (Cutler and Krochko, 1999; Zhou et al., 2004). The 8′-hydroxyl ABA is spontaneously isomerized to phaseic acid (PA) and reduced to dihydrophaseic acid (DPA) or epi-DPA. 8′-Hydroxylation of ABA is catalyzed by ABA 8′-hydroxylase (ABA8ox; Fig. 1), which belongs to the P450 monooxygenase family (Nambara and Marion-Poll, 2005). We focused on NCED and ABA8ox as enzymes responsible for ABA biosynthesis and catabolism, respectively, to investigate ABA metabolism in lettuce seeds. The cDNAs encoding NCED and ABA8ox have been isolated from several plant species, including the model plant Arabidopsis (Schwartz et al., 1997; Tan et al., 1997; Neill et al., 1998; Qin and Zeevaart, 1999; Chermys and Zeevaart, 2000; Thompson et al., 2000; Iuchi et al., 2001; Kushiro et al., 2004; Saito et al., 2004; Chono et al., 2006; Millar et al., 2006; Yang and Choi, 2006; Yang and Zeevaart, 2006; Saika et al., 2007). Recently, it was reported that ABA levels are controlled by phytochrome through NCED and ABA8ox gene expression in Arabidopsis seeds and that germination is also regulated by phytochrome (Seo et al., 2006).
To elucidate the molecular mechanism regulating ABA levels, we isolated cDNAs encoding NCED and ABA8ox from lettuce and investigated the control of ABA levels and ABA metabolism gene expression by phytochrome. We also performed spatial analysis of endogenous ABA and its metabolic transcripts to compare the ABA reduction site with the photoperceptive site (Inoue and Nagashima, 1991) in lettuce seeds. We discuss ABA metabolism regulated by exogenously applied or endogenous bioactive GA.
RESULTS
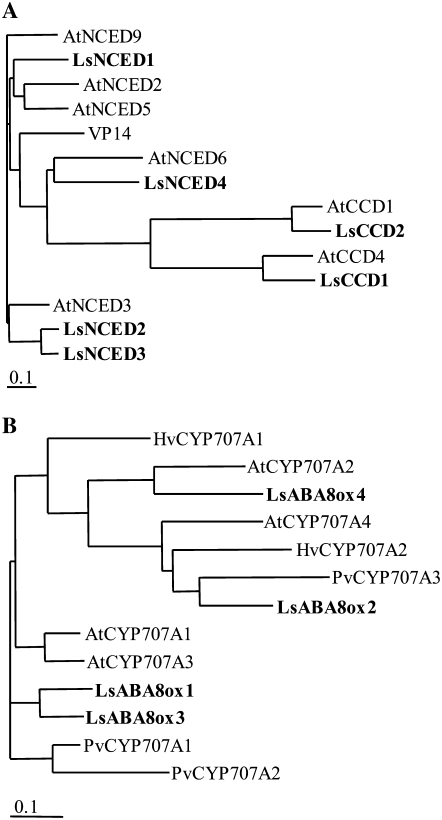
Cloning of cDNAs Encoding NCED in Lettuce
One set of degenerate primers (NCED-F and NCED-R) were designed on the basis of conserved amino acid sequences of plant NCED homologs, as described in “Materials and Methods.” cDNA fragments of the expected size of approximately 550 bp were amplified from the templates derived from seeds imbibed in the dark and from drought-treated leaves by reverse transcription (RT)-PCR with these degenerate primers. Sequence analysis of the subcloned fragments according to their sequence similarity with other plant NCEDs and carotenoid cleavage dioxygenase (CCD) indicated that four different NCED-like fragments and two CCD-like fragments were present. RACE was performed to determine each full-length cDNA sequence using gene-specific primers. End-to-end PCR was then performed using 5′- and 3′-end primers. Six fragments were named LsNCED1, NCED2, NCED3, NCED4, CCD1, and CCD2 (GenBank accession nos. AB120107, AB120108, AB120109, AB120110, AB120111, and AB120112, respectively; Fig. 2A). The predicted coding regions of LsNCED1, NCED2, NCED3, and NCED4 were 1,758, 1,815, 1,752, and 1,737 bp, encoding 585, 604, 583, and 578 amino acid residues, respectively. Those of LsCCD1 and CCD2 were 1,779 and 1,626 bp, encoding 592 and 541 amino acid residues, respectively. We could not find a lettuce NCED homolog in the database except for these six genes. Identities of amino acid sequences of six clones to the NCED in Arabidopsis (Iuchi et al., 2001) were as follows: LsNCED1/AtNCED5, 68%; LsNCED2/AtNCED3, 70%; LsNCED3/AtNCED3, 72%; LsNCED4/AtNCED6, 57%; LsCCD1/AtCCD4, 67%; and LsCCD2/AtCCD1, 80%. The degrees of identity of each LsNCED with other LsNCEDs and LsCCDs were approximately 50% to 80% and 30%, respectively. Genomic DNA fragments corresponding to these genes were isolated from lettuce and then sequenced. LsCCD2 included 13 introns, similar to AtCCD1, while LsCCD1 and LsNCEDs had no introns, similar to AtCCD4 and AtNCEDs (data not shown), suggesting that these lettuce genes closely resemble the CCD and NCED genes in Arabidopsis, not only in terms of the sequences but also the gene structures.
Figure 2.
Phylogenetic relationships among ABA metabolic enzymes. A, Phylogenetic tree of LsNCED and other plant NCEDs. Analyses were performed using the ClustalW program (http://clustalw.ddbj.nig.ac.jp/top-j.html). The accession numbers are as follows: AtCCD1, NM116217; AtNCED2, NM117945; AtNCED3, NM112304; AtCCD4, NM118036; AtNCED5, NM102749; AtNCED6, NM113327; AtNCED9, NM106486 (Arabidopsis); VP14, U95953 (Zea mays); and LsCCD1, LsCCD2, LsNCED1, LsNCED2, LsNCED3, and LsNCED4 (lettuce). B, Phylogenetic tree of LsABA8ox and other plant CYP707As. The accession numbers are as follows: AtCYP707A1, NM118643; AtCYP707A2, NM128466; AtCYP707A3, NM180805; AtCYP707A4, NM112814 (Arabidopsis); HvCYP707A1, DQ145932; HvCYP707A2, DQ145933 (Hordeum vulgare); PvCYP707A1, DQ352541; PvCYP707A2, DQ352542; PvCYP707A3, DQ352543 (Phaseolus vulgaris); and LsABA8ox1, LsABA8ox2, LsABA8ox3, and LsABA8ox4 (lettuce).
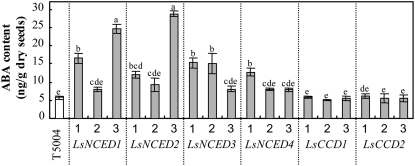
We examined whether the products of the four LsNCED genes are involved in ABA biosynthesis, while those of the two LsCCD genes are not. We confirmed that endogenous levels of ABA are accumulated in transgenic ABA-deficient Arabidopsis nced3 mutant T5004 (Iuchi et al., 2001) by overexpressing LsNCED genes. Transgenic T5004 lines in which each full-length cDNA was constitutively expressed under the control of the cauliflower mosaic virus 35S promoter were prepared, and homozygous T3 seeds 24 h after the start of imbibition were used to measure ABA content (Fig. 3). The endogenous ABA levels of the imbibed seeds of overexpressed lines of LsNCED1, NCED2, NCED3, and NCED4 were more abundant than those of T5004 seeds. On the other hand, ABA contents in LsCCD1 and LsCCD2 overexpressors were the same as those in the parent T5004. Therefore, overexpression of four LsNCEDs, but not LsCCDs, led to in vivo accumulation of ABA, indicating that these genes encode NCEDs responsible for ABA biosynthesis.
Figure 3.
Endogenous levels of ABA in the imbibed seeds of T5004 and transgenic lines. Endogenous levels of ABA in each seed incubated for 24 h under white light were measured by LC-MS/MS. Results are presented as means with ses from at least three independent seed batches. Lowercase letters indicate significant differences in a parameter at P < 0.05 (Tukey's honest significant difference test).
Cloning of cDNAs Encoding ABA8ox in Lettuce
To clone CYP707A homologs from lettuce, RT-PCR was performed with five degenerate primers (ABA8ox-F1, ABA8ox-F2, ABA8ox-R1, ABA8ox-R2, and ABA8ox-R3) designed on the basis of conserved amino acid sequences of CYP707A genes in Arabidopsis (Kushiro et al., 2004; Saito et al., 2004). Templates used here were cDNAs derived from R-treated seeds and drought-treated leaves. Three bands of approximately 740 bp (ABA8ox-F1 and ABA8ox-R1) from dehydrated leaves, 790 bp (ABA8ox-F1 and ABA8ox-R2) from R-treated seeds, and 980 bp (ABA8ox-F2 and ABA8ox-R3) from drought-treated leaves were amplified. The results of nucleotide sequence analyses indicated that the PCR products contained four different fragments. These four cDNAs were named LsABA8ox1, LsABA8ox2, LsABA8ox3, and LsABA8ox4 (GenBank accession nos. AB235917, AB235918, AB235919, and AB235920, respectively; Fig. 2B). The predicted coding regions of LsABA8ox1, ABA8ox2, ABA8ox3, and ABA8ox4 are 1,495, 1,455, 1,404, and 1,482 bp, encoding 464, 484, 467, and 493 amino acid residues, respectively. The deduced amino acid sequences of LsABA8ox showed 75% (LsABA8ox1/CYP707A1), 69% (LsABA8ox2/CYP707A4), 78% (LsABA8ox3/CYP707A1), and 67% (LsABA8ox4/CYP707A2) identity to CYP707A in Arabidopsis.
To confirm that these four cDNAs encode ABA8ox, we carried out in vitro functional analysis. Each cDNA was expressed in yeast harboring an Arabidopsis P450 reductase to yield the recombinant protein (Pompon et al., 1996). PA was identified as a major product in each reaction mixture by liquid chromatography/tandem mass spectrometry (LC-MS/MS) analysis (Table I), indicating that each recombinant LsABA8ox catalyzed the 8′-hydroxylation of ABA. These results strongly suggested that LsABA8ox1 to 4 encode ABA8ox.
Table I.
Identification of the reaction product from ABA incubated with microsomes of yeast expressing fusion proteins derived from LsABA8ox
| Clone | Identified Metabolite | Retention Time on LC | Principal Ions/Relative Abundance |
|---|---|---|---|
| min | % of base peak | ||
| LsABA8ox1 | PA | 14.6 | 279 (M+, 12), 235 (6), 205 (46), 187 (20), 168 (29), 139 (100) |
| LsABA8ox2 | PA | 14.6 | 279 (M+, 12), 235 (6), 205 (46), 187 (20), 168 (29), 139 (100) |
| LsABA8ox3 | PA | 14.6 | 279 (M+, 12), 235 (6), 205 (46), 187 (20), 168 (29), 139 (100) |
| LsABA8ox4 | PA | 14.6 | 279 (M+, 12), 235 (6), 205 (46), 187 (20), 168 (29), 139 (100) |
| Authentic PA | 14.6 | 279 (M+, 12), 235 (6), 205 (46), 187 (20), 168 (29), 139 (100) |
Measurement of ABA and Its Catabolites in Germinating Lettuce Seeds
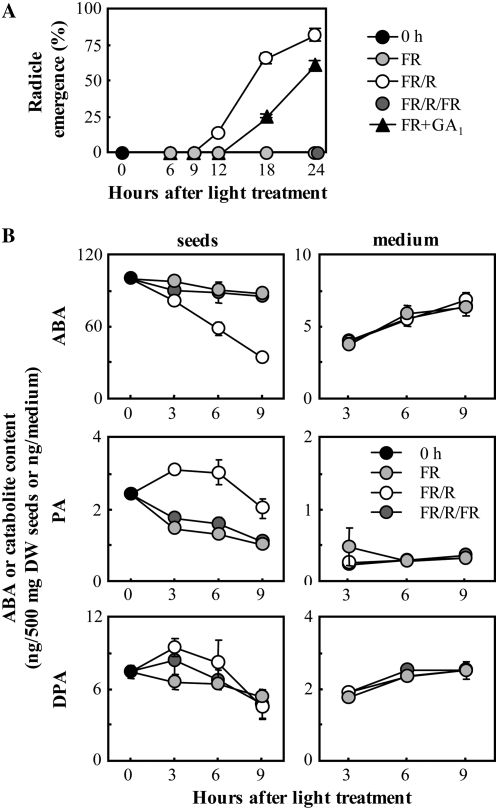
Endogenous levels of ABA decrease after R treatment during germination of lettuce seeds (Toyomasu et al., 1994). Here, we quantified ABA and its catabolites in FR-, FR/R-, and FR/R/FR-treated seeds, prepared as described in “Materials and Methods.” FR treatment was used as a control to suppress germination completely. FR/R/FR treatment was conducted to confirm photoreversibility of gene expression, which was expected if these genes are controlled by phytochrome. Radicle emergence was observed from 10 to 12 h after FR/R treatment under our experimental conditions (Fig. 4A). The endogenous levels of ABA decreased markedly after FR/R treatment, in contrast to the FR control (Fig. 4B). The level of PA was lower than that of ABA in the imbibed seeds, and increased slightly 3 to 9 h after FR/R treatment, compared to the FR control. The effect of R was cancelled out by successive FR treatment (see FR/R/FR). The levels of DPA were lower than those of ABA and were unaffected by light treatment. A peak of epi-DPA was not detected in seeds or medium (data not shown). The levels of ABA, PA, and DPA in each culture medium were also measured and were shown to be unaffected by light treatment, suggesting that these compounds in medium had diffused from the seed coats (Inoue, 1990). These results indicated that ABA reduction by R treatment in the seeds was due to metabolism and not to exudation, and catabolic intermediates, such as PA, were quickly catabolized in lettuce seeds.
Figure 4.
Quantification of endogenous ABA, PA, and DPA levels during incubation under different light conditions. A, Time course of germination of lettuce seeds under different conditions. 0 h indicates seeds imbibed for 3 h in the dark (just before light treatment). FR+GA1 indicates seeds treated with FR and 1 mm GA1. Experiments were performed in triplicate, and averages are shown with ses. B, The endogenous levels of ABA, PA, and DPA were analyzed by LC-MS/MS. DW, Dry weight. Experiments were performed in duplicate and averages are shown with ses.
Gene Expression during Germination
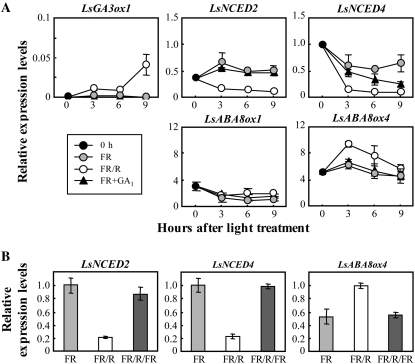
To investigate the regulatory mechanism of endogenous ABA, we performed expression analysis of LsNCED and LsABA8ox during germination by real-time quantitative RT-PCR (QRT-PCR). In a previous study, we showed marked accumulation of the transcripts of LsGA3ox1, encoding GA 3-oxidase, after FR/R treatment (Toyomasu et al., 1998). The LsGA3ox1 transcript was also quantified as a positive control (Fig. 5A). LsNCED2 and LsNCED4 transcripts were predominant in the imbibed lettuce seeds. Transcript levels of LsNCED1 and LsNCED3 were much lower than those of LsNCED2 and LsNCED4 and were negligible in the imbibed seeds (data not shown). LsNCED2 expression levels in FR seeds increased slightly 3 h after treatment. In contrast, transcript levels of LsNCED2 in FR/R seeds decreased 3 h after treatment and continued to decrease gradually until 9 h. A slight increment in LsNCED2 transcripts in FR seeds was not due to FR treatment, because the expression pattern in the FR seeds was almost the same as that of the imbibed seeds under conditions of continuous darkness (data not shown). The expression levels of LsNCED4 in the FR/R seeds decreased more drastically than those in the FR seeds 3 h after treatment and continued to decrease gradually until 9 h. On the other hand, LsABA8ox1 and LsABA8ox4 transcripts were abundant during seed germination among the LsABA8ox genes (Fig. 5A), and LsABA8ox2 and LsABA8ox3 transcript levels were negligible because of their much lower expression levels (data not shown). LsABA8ox4 transcript levels in FR/R seeds were higher than those in FR seeds at 3 h after treatment, while LsABA8ox1 transcripts were not markedly affected by light treatment. Successive FR treatment cancelled the effects of R on controlling the levels of LsNCED2, LsNCED4, and LsABA8ox4 expression (Fig. 5B). These results suggest that LsNCED2 and LsNCED4 were down-regulated and LsABA8ox4 was up-regulated by phytochrome in the imbibed lettuce seeds.
Figure 5.
Changes in the transcript levels of LsNCED and LsABA8ox during incubation under different conditions. A, Expression levels of LsNCED and LsABA8ox after light and GA1 treatment. The expression levels of these genes were analyzed by QRT-PCR as described in the text. 0 h indicates seeds imbibed for 3 h in the dark (just before light treatment). FR+GA1 indicates seeds treated with FR and 1 mm GA1. The results were normalized to the expression of 18S rRNA (internal control), and the expression levels of all genes examined are given relative to a reference value, the transcript level of LsNCED4 at 0 h, set to 1. Two independent experiments were performed, and averages are shown with ses. B, Expression levels of LsNCED and LsABA8ox under photoreversible conditions 3 h after light treatment. The results were normalized to the expression of 18S rRNA, and then the highest value of each gene was set equal to 1. Two independent experiments were performed, and averages are shown with ses.
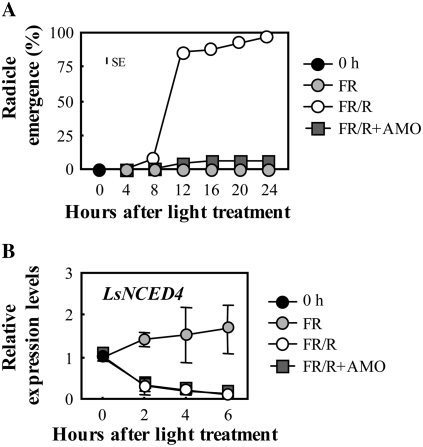
As reported previously, endogenous levels of ABA decreased after application of bioactive GA, which induced lettuce seed germination (Toyomasu et al., 1994). To examine whether LsNCED and LsABA8ox are regulated by exogenously applied GA, we performed QRT-PCR on GA1-treated seeds, prepared as described in “Materials and Methods.” The expression levels of LsNCED4 decreased in the GA1-treated seeds and were delayed relative to FR/R seeds, while there were no marked effects on the expression of other genes in comparison to the FR control (Fig. 5A). These patterns of decrease in the LsNCED4 transcript reflect the frequency of germination (Fig. 4A). The results strongly suggest that down-regulation of LsNCED4 was responsible for the ABA decrease after GA1 treatment. Therefore, we presume that LsNCED4 expression is down-regulated by endogenous GA1 accumulated in FR/R seeds. However, we could not entirely eliminate the possibility that LsNCED4 expression is also down-regulated by phytochrome. To examine whether the expression of LsNCED4 is also regulated by phytochrome, we carried out QRT-PCR using seeds in which GA biosynthesis was suppressed by treatment with GA biosynthetic inhibitor. A recent study showed that triazole-type GA biosynthetic inhibitors, such as uniconazole-P, inhibit ABA8ox activity (Saito et al., 2006) as well as ent-kaurene oxidase activity (Izumi et al., 1985). Both of these enzymes belong to the P450 monooxygenase family. Therefore, we used AMO-1618, which suppresses the early step (ent-copalyl diphosphate synthase, a member of the diterpene cyclase family) of GA biosynthesis (Kawaide et al., 1997). When we used 50 mg of seeds in the same volume of buffer and petri dish as in the above experiments, inhibition of GA biosynthesis by AMO-1618 suppressed germination up to approximately 5% (Fig. 6A). AMO-1618 treatment did not suppress down-regulation of LsNCED4 after FR/R (Fig. 6B), although the expression pattern in FR seeds using 50 mg of seeds was different from that using 500 mg of seeds (Figs. 5A and 6B). These results suggest that expression of LsNCED4 is down-regulated by phytochrome as well as GA.
Figure 6.
Changes in transcript levels of LsNCED4 during incubation with GA biosynthetic inhibitor. A, Time course of germination of lettuce seeds under different conditions. 0 h indicates seeds imbibed for 3 h in the dark (just before light treatment). Experiments were performed in triplicate and averages are shown with ses. The vertical bars represent the maximum size of the se bar within the figure. B, Expression levels of LsNCED4 in the seeds incubated in medium containing 50 mm AMO-1618. The expression levels of these genes were analyzed by QRT-PCR as described in the text. The results were normalized to the expression of 18S rRNA (internal control), and then the expression level at 0 h was set to 1. Two independent experiments were performed, and averages are shown with ses.
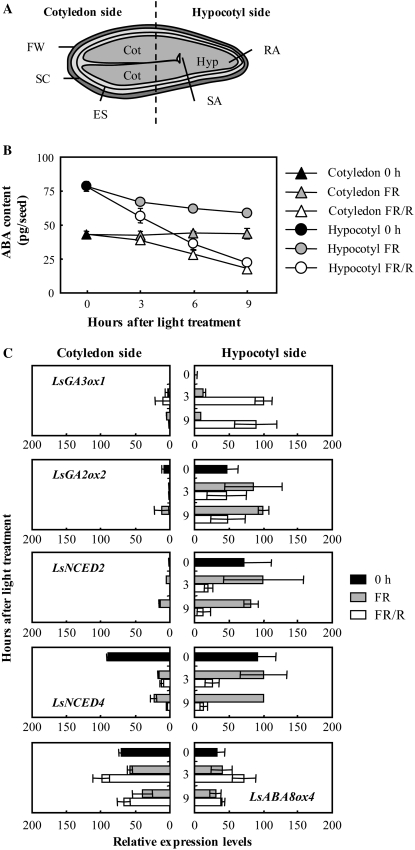
Spatial Analysis of Endogenous ABA and Transcripts of LsNCED and LsABA8ox
To examine the site of the ABA decrease in germinating seeds, we measured endogenous levels of ABA in seeds divided into two parts after freezing defined as the cotyledon side or the hypocotyl side. The cotyledon side was composed of the cotyledon, fruit wall, seed coat, and endosperm, while the hypocotyl side included the hypocotyl, root apex, shoot apex, part of the cotyledon, fruit wall, seed coat, and endosperm (Fig. 7A). Endogenous ABA levels in the imbibed seeds in the dark, just before light treatment, were higher on the hypocotyl side than on the cotyledon side (Fig. 7B; 0 h on the x axis). The ABA levels on both sides of the FR/R seeds decreased more markedly than those in the nongerminating FR seeds (Fig. 7B; compare white symbols and gray symbols). For example, ABA content in FR/R seeds minus those in FR seeds was −36 pg/seed on the hypocotyl side and −25 pg/seed on the cotyledon side 9 h after light treatment. The decrease by R of ABA content on the hypocotyl side was 1.4-fold greater than that on the cotyledon side.
Figure 7.
ABA and GA metabolism on the cotyledon side and the hypocotyl side of lettuce seeds. A, Frozen seeds were divided into two parts: cotyledon side, including the cotyledons (Cot), fruit wall (FW), seed coat (SC), and endosperm (ES); and the hypocotyl side, including the hypocotyl (Hyp), root apex (RA), shoot apex (SA), and part of the Cot, FW, SC, and ES. B, Quantification of endogenous ABA levels in the cotyledon side and the hypocotyl side by LC-MS/MS. 0 h indicates seeds imbibed for 3 h in the dark (just before light treatment). Experiments were performed in triplicate and averages are shown with ses. C, Changes in transcript levels of GA and ABA metabolism genes by QRT-PCR. The results were normalized to the expression of 18S rRNA (internal control), and then the highest value was set to 100. Two independent experiments were performed, and averages are shown with ses.
To assess the site of LsNCED and LsABA8ox expression in seed germination, in situ hybridization was performed, but no positive signals were detected because of the low expression levels. We then carried out expression analysis of LsNCED and LsABA8ox on the divided seeds described above by QRT-PCR. We also performed expression analysis of GA metabolic enzyme genes, LsGA3ox1 and LsGA2ox2, to compare the sites of GA1 accumulation and LsNCED4 expression, which may be down-regulated by endogenous GA1 in germinating seeds. The results of expression analysis of the GA and ABA metabolic enzyme genes are presented in Figure 7C. Transcripts of LsGA3ox1 and LsGA2ox2 were detected mainly on the hypocotyl side rather than the cotyledon side. The transcript levels of LsGA3ox1 and LsGA2ox2 on the hypocotyl side of the FR/R seeds were much higher and lower, respectively, than those of the FR seeds. Similarly, transcript levels of AtGA3ox1 and AtGA2ox2 were detected in the cortex and endodermis in the hypocotyl of Arabidopsis (Yamaguchi et al., 2001; Yamauchi et al., 2007). These results suggest that endogenous GA1 accumulated much more on the hypocotyl side than the cotyledon side in lettuce seeds. LsNCED2 transcripts were also detected predominantly on the hypocotyl side and decreased markedly after FR/R treatment. LsNCED4 transcripts were detected equally on both sides of the seed just before light treatment (0 h). The expression levels of LsNCED4 on the hypocotyl side decreased markedly after FR/R treatment, whereas expression on the cotyledon side showed almost equivalent decreases after FR and after FR/R treatment. The almost equivalent expression levels of LsNCED4 on the hypocotyl side of the FR seeds during incubation suggested that the reduction of LsNCED4 levels in the whole FR seeds (Fig. 5A) was due to their reduction on the cotyledon side. In contrast, LsABA8ox4 transcripts accumulated equally on both sides and increased after FR/R irradiation. These results indicated that LsNCED2 and LsNCED4 are mainly associated with a decrease in ABA on the hypocotyl side.
DISCUSSION
Lettuce seeds are excellent plant materials in which to investigate photogermination, and they have been used in many studies. The size (approximately 5 mm) and structure (straight embryo in a seed coat) of lettuce seeds are suitable for localization analysis. Inoue and Nagashima (1991) found the photoperceptive site in lettuce seeds by partial irradiation of the seeds with R through slit irradiation. The levels of Glu and Gln were also found to be increased in hypocotyls after R and GA (Takeba, 1980a, 1980b, 1980c, 1980d). Arabidopsis seeds, which are also photoblastic, are also suitable materials for genetic analysis and have been used in many studies of the function of various target genes for regulation of plant development since the 1990s. Using Arabidopsis seeds and their mutants or transgenic plants, characterization of the phytochrome family (Shinomura et al., 1996) and metabolic enzyme genes of GAs and ABA (Yamaguchi et al., 1998, 2001; Ogawa et al., 2003; Seo et al., 2006; Oh et al., 2007) provided important information on the mechanism of photogermination.
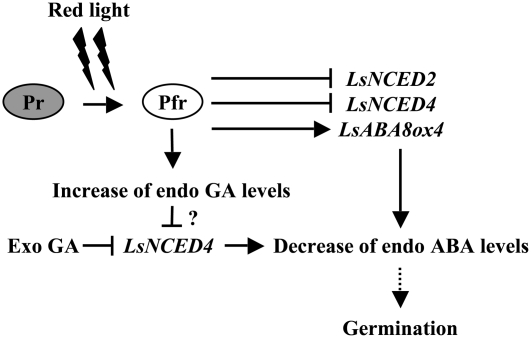
Our results indicated that down-regulation of LsNCED2 and LsNCED4 expression and up-regulation of LsABA8ox4 expression are involved in the decrease in ABA in lettuce seeds (Fig. 8). However, regulation of ABA metabolism by phytochrome in lettuce seeds was different from that in Arabidopsis seeds in terms of the involvement of GA. GA-independent signals appeared to contribute more prominently to phytochrome regulation of ABA levels in lettuce seeds than in Arabidopsis seeds. In lettuce, LsNCED2 and LsABA8ox4 were regulated by GA-independent signals, whereas LsNCED4 was regulated by both GA-dependent and GA-independent signals (Figs. 5A and 6B). This is in contrast to phytochrome regulation in Arabidopsis, in which AtNCED6 and AtCYP707A2 are regulated by both GA-dependent and GA-independent signals (Seo et al., 2006; Oh et al., 2007). These results indicate that the molecular mechanisms of regulation of ABA levels by phytochrome are partly different and that the detailed mechanisms vary according to plant species.
Figure 8.
Model of the regulatory mechanism of ABA levels on the germination of photoblastic lettuce seeds. Evidence for this model is discussed in the text. Arrows indicate activation, and T bars indicate negative regulation. Question mark indicates suggested relationships.
In previous studies using Arabidopsis seeds, in situ hybridization and reporter gene analysis were carried out to estimate localization of GAs and ABA (Yamaguchi et al., 2001; Okamoto et al., 2006; Seo et al., 2006; Yamauchi et al., 2007). However, these methods were not suitable for measurement of the precise content of the target mRNA, and, moreover, localization of the phytohormones could not be addressed because of the small size and curved embryos in Arabidopsis seeds. In this study, we measured ABA levels in dissected hypocotyl and cotyledon parts of lettuce by LC-MS/MS. This analysis demonstrated that the photoperceptive hypocotyl side showed more abundant reduction in ABA levels than the cotyledon parts (Fig. 7B). This is the first report, to our knowledge, of spatial information of the decrease in ABA by R in plant seeds. Consistent with these changes in ABA level, QRT-PCR expression analysis in dissected seeds showed that phytochrome regulation of LsNCED2 and LsNCED4 expression mainly occurred in the hypocotyl parts. We could not measure GA levels in each part, because the endogenous GA1 levels were approximately 1/100 those of ABA in lettuce seeds. However, the good agreement of changed spatial contents of ABA and expression patterns of ABA metabolism genes in lettuce seeds suggested that GA levels also increase in hypocotyls through up-regulation of LsGA3ox1 and down-regulation of LsGA2ox2 expression. As the hypocotyl was shown to be the most sensitive site in response to R in lettuce seeds (Inoue and Nagashima, 1991), it was suggested that levels of bioactive GA increase and those of ABA decrease near the photoperceptive site in the hypocotyls of lettuce seeds. Previous studies indicated that the growth potential of the hypocotyl in halved lettuce seeds is induced by R (Takeba, 1980d) or GA (Takeba, 1980c), and accumulation of Glu and Gln in the hypocotyls through up-regulation of Gln synthase gene expression by R or GA results in an increment in growth potential (Takeba, 1983, 1984; Sakamoto et al., 1990). These spatial analyses also suggest that biochemical events occurring in the hypocotyl of lettuce seeds are essential for germination in light. Recently, it was shown that phytochrome regulates GA and ABA metabolism genes in Arabidopsis seeds through PIL5, a negative regulator of phytochrome signals (Oh et al., 2007). Future studies should address whether photoperceptive cells are identical to the cells in which the levels of phytohormones change.
We were able to discriminate between universal and species-specific events by comparing the results obtained in lettuce and Arabidopsis. The information obtained by experiments using lettuce provides novel insights into the conserved and diverse mechanisms of photogermination among plant species. Further studies using Arabidopsis and/or lettuce seeds, such as investigation of the events in hypocotyls in seeds, will provide further important information to help us to understand the mechanisms of regulation of seed germination by light.
MATERIALS AND METHODS
Light Sources and Plant Materials
R (7 Wm−2) was supplied by light-emitting diodes (MIL-R18; Sanyo Biomedical). FR (5.5 Wm−2) was also supplied by light-emitting diodes (MIL-R18) passed through a far-red acrylic filter (Deraglass A900, 2 mm thick; Asahikasei). Lettuce (Lactuca sativa) ‘Grand Rapids’ seeds were obtained by propagation of the same lot of seeds as used in our previous studies (Toyomasu et al., 1998; Nakaminami et al., 2003) obtained from South Pacific Seed in January 2005 and stored at 4°C with silica gel until use. Seeds (500 mg or 50 mg) were incubated according to the method described previously (Nakaminami et al., 2003). Three different light treatments were applied to seeds 3 h after the start of imbibition: irradiation with FR (FR treatment); FR followed by R (FR/R treatment); and FR, R, and FR, successively (FR/R/FR treatment). Each light irradiation lasted 5 min. Seeds imbibed in the dark for 3 h were also harvested (0 h) just before the light treatment. After light treatment, seeds were incubated in the dark at 25°C for 3, 6, and 9 h (500 mg) or 2, 4, and 6 h (50 mg). The seeds were then harvested and frozen in liquid nitrogen. Application of GA1 (1 mm) to the seeds was carried out as described previously (Nakaminami et al., 2003). Application of AMO-1618 (50 mm) to the seeds was performed at the start of imbibition. For dark incubation, seeds were handled under a dim-green safety light.
Lettuce seeds were sown on vermiculite and grown under continuous white light (6,000 lux) at 25°C for 1 month in a plant growth chamber (Biotron LPH 200; NK Systems). Plants were harvested and placed on filter paper under continuous white light at 25°C for 3 h. The dried leaves were collected and frozen. Total RNA was isolated and used for molecular cloning of ABA metabolic enzyme genes.
Molecular Cloning
The design of the degenerate primers for NCED was based on conserved amino acid regions of the NCED of Zea mays (Schwartz et al., 1997; Tan et al., 1997), Solanum lycopersicum (Thompson et al., 2000), Arabidopsis (Arabidopsis thaliana; Iuchi et al., 2001), Phaseolus vulgaris (Qin and Zeevaart, 1999), and Persea americana (Chermys and Zeevaart, 2000). The sequences of these oligonucleotides were 5′-TNTTYGAYGGNGAYGGNATG-3′ (NCED-F; forward primer, encoding LFDGDGM) and 5′-GTRCTRAARCGNTADTGNCT-3′ (NCED-R; reverse primer, encoding HDFAITE). The design of five degenerate primers for ABA8ox was based on conserved amino acid regions of the CYP707A family of Arabidopsis (Kushiro et al., 2004; Saito et al., 2004). The sequences of these oligonucleotides were 5′-GCNYTITTYTTYCAICARGG-3′ (ABA8ox-F1; forward primer, encoding AIFFHQG), 5′-TNGGNTGYCCNTGYGTNATG-3′ (ABA8ox-F2; forward primer, encoding LGCPCVN), 5′-CANGCYTCICKRAAIGTRAA-3′ (ABA8ox-R1; reverse primer, encoding VAERFTF), 5′-ACYTTCCANCCYTTIGGDAT-3′ (ABA8ox-R2; reverse primer, encoding VKWGKPI), and 5′-GGYTTNGGNGCNACYTCRAA-3′ (ABA8ox-R3; reverse primer, encoding PKPAVEF). The template cDNA for PCR was as described previously (Toyomasu et al., 1998) and was also applied to the dehydrated leaves. The reaction mixture (50 μL) contained 0.2 mm dNTPs, 1.5 mm MgCl2, 1 μm of each primer, and 2.5 units of Expand HF (Roche Diagnostics). The samples were heated to 94°C for 2 min, and then subjected to 40 cycles of 94°C for 1 min, 50°C (NCED) or 45°C (ABA8ox) for 1 min, and 72°C for 1 min, with a final extension of 72°C for 7 min. PCR fragments were cloned with the pGEM-T Easy vector (Promega). DNA sequencing was performed using a Big Dye terminator cycle sequencing kit (Applied Biosystems) and an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). RACE was performed using a Marathon cDNA amplification kit (CLONTECH) according to the methods described previously (Toyomasu et al., 1998).
Sequence Similarity Search and Alignment of Amino Acid Sequences
Homology searches of the databases were performed with BLAST (http://www.arabidopsis.org/Blast/index.jsp). Alignment of amino acid sequences was carried out using ClustalW, available from the DNA Data Bank of Japan home page (http://clustalw.ddbj.nig.ac.jp/top-j.html).
Functional Analysis of LsNCED by Heterologous Expression
Full-length cDNA was amplified by PCR using primers with restriction enzyme sites for XhoI at the 5′-end and KpnI at the 3′-end. The sequences of the primers used are shown in Table II. PCR fragments were subcloned into the pGEM-T Easy vector. The plasmids were digested with XhoI and KpnI and ligated into the Gateway entry vector pENTR1A (Invitrogen). The inserts were cloned into the pGWB2 vector (provided by Dr. Tsuyoshi Nakagawa, Shimane University) by the LR reaction according to the manufacturer's protocol. The plasmids were introduced into Agrobacterium tumefaciens by electroporation. T-DNA was transferred into the nced3 mutant T5004 (Iuchi et al., 2001) by the floral dipping method (Clough and Bent, 1998). T1 transgenic seeds were selected on half-strength Murashige and Skoog medium containing 0.8% (w/v) agar, kanamycin and hygromycin (each 25 μg/mL), and 100 μg/mL of chloramphenicol. T1 seedlings were transferred to soil and grown at 22°C under continuous white light. T2 plants were grown under the same conditions. Samples of 10 mg of seeds (T5004 and homozygous T3 transgenic lines) were placed on three filter papers (Filter Papers 3; Whatman) moistened with 2 mL of water in 60-mm tissue culture dishes (Iwaki). The seeds were incubated at 22°C under white light for 24 h and then harvested. Samples were extracted and subjected to LC-MS/MS analysis using 0.2 ng (±)-2-cis, 4-trans-ABA-d6 (Icon Services) as an internal standard, as described below in the section “Extraction and Mass Spectrometric Analysis of ABA, PA, and DPA.” Measurements of ABA in seeds of T5004 and each transformant line were taken from six and three independent seed batches, respectively.
Table II.
Specific primer sets used in construction of transgenic plants
Underlined text and underlined, bold text show the KpnI and the XhoI site, respectively.
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| LsCCD1 | GGTACCCAGACCACACAATGGATTCTC | CTCGAGACTTAAAACTGATGTTTCACTTAT |
| LsCCD2 | GGTACCCACAGATGGGCACCATGGAG | CTCGAGAATTGATAACACCATTGTATTAAT |
| LsNCED1 | GGTACCACCTCCAGCCCTTCATTCCT | CTCGAGGAAAAGCACATTCATGTTTCTTCA |
| LsNCED2 | GGTACCCGACATTAAAATAGCTTCAATCAC | CTCGAGAGATGAAATTTTACATAGTTATATA |
| LsNCED3 | GGTACCTCCACCAACTTTCAAATTGCTTAC | CTCGAGACTTAATACAACATAAAGTATATT |
| LsNCED4 | GGTACCCACCATTAAAGCTATCACGT | CTCGAGATTGAATTAAGCTTAATTAGCCAA |
Functional Expression of Lettuce ABA8ox
Functional analysis of LsABA8ox1 to LsABA8ox4 was performed using the method described by Kushiro et al. (2004). Full-length cDNA was cloned into a yeast (Saccharomyces cerevisiae) expression vector, pYeD60 (Pompon et al., 1996). The plasmid was transformed into yeast strain WAT11 (Pompon et al., 1996). Recombinant yeasts were grown in SGI medium for 24 to 36 and then transferred to SLI medium and induced by adding Gal for 12 h. The cell pellets were resuspended in 0.1 m potassium phosphate buffer, pH 7.6, and passed through a French press (20,000 psi). Microsomal fractions were suspended in 0.1 m potassium phosphate buffer, pH 7.6. To assay ABA 8′-hydroxylase, (+)-S-ABA was incubated with 2 μg of microsomal protein (in a volume of 100 μL) containing 0.5 mm NADPH, 0.5% (w/v) Triton X-100, and 1 mm (±)-dithiothreitol at 22°C for 16 h. The reaction was stopped by adding 1 n HCl, extracted with ethyl acetate, and dried by centrifugation under vacuum. The resulting sample mixture was dissolved in water and analyzed by LC-MS/MS. LC-MS/MS analysis was performed using the same method as described in the next section.
Extraction and Mass Spectrometric Analysis of ABA, PA, and DPA
ABA and its metabolites in seed samples were extracted and quantified as described previously (Saika et al., 2007) using 50 ng of (±)-2-cis, 4-trans-ABA-d6, 2.5 ng of d3-PA, d3-DPA (provided by Dr. Suzanne R. Abrams, National Research Council of Canada), and d3-epi-DPA (provided by Dr. Nobuhiro Hirai, Kyoto University) as internal standards. The culture media were collected from dishes, and then 10 ng of (±)-2-cis, 4-trans-ABA-d6, 1 ng of d3-PA, d3-DPA, and d3-epi-DPA were added. The culture media were adjusted to pH 2 with 1 n HCl and extracted with ethyl acetate. The dehydrated ethyl acetate fractions were dried by centrifugation under vacuum. Extracts were dissolved in 300 μL of methanol and again dried by centrifugation under vacuum and subjected to LC (ACQUITY UPLC system; Waters)-MS/MS (Q-Tof premier; Micromass). The following fragments were used for quantification: 153 (ABA), 159 (d6-ABA), 139 (PA), 142 (d3-PA), 171 (DPA), 174 (d3-DPA), 171 (epi-DPA), and 174 (d3-epi-DPA). For ABA measurement from separated seed, frozen seeds (20 seeds) were separated into two parts on dry ice. Samples were extracted and subjected to LC-MS/MS analysis using 1 ng of (±)-2-cis, 4-trans-ABA-d6 as an internal standard, according to the method described previously (Saika et al., 2007).
Real-Time QRT-PCR
Total RNA was extracted from frozen seeds by the hot borate method (Wan and Wilkins, 1994) or on an RNAqueous column with Plant RNA Isolation Aid (Ambion). For expression analysis of divided seeds, samples of 50 mg of frozen seeds (fresh weight) were divided into two parts using a razor on dry ice. Total RNA was extracted from each portion using the RNAqueous column with Plant RNA Isolation Aid. cDNA was synthesized from aliquots of 1 μg total RNA using a QuantiTect Reverse Transcription kit (Qiagen). Real-time QRT-PCR using SYBR Green I was carried out on a sequence detector system (model 7000; Applied Biosystems) or a Thermal Cycler Dice real-time system (TP800; TaKaRa) as described previously (Kushiro et al., 2004). The sequences of the primers used are shown in Table III. The mean values of two or three replicates were normalized using 18S ribosomal RNA (rRNA) as an internal control.
Table III.
Specific primer sets used in the real-time QRT-PCR
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| 18SrRNA | GGAGCGATTTGTCTGGTTA | ATCTAAGGGCATCACAGACC |
| LsGA3ox1 | TGTTTGCACTTCCTATCCAACAG | TACGAGCATGTTCGTATGGAGAT |
| LsGA2ox2 | TGGAGGTCCACCATTGATG | CCTATTATCAGTCAACCTGGTCTTGTAG |
| LsNCED1 | AAACCCTACAATCCGACTATTCG | GGCCGCAGCTCTTTGTAAG |
| LsNCED2 | CTTCAGTTTCCTAAACAGTCTGTTGGTA | TGCTTTCAATCCATCTTCAACG |
| LsNCED3 | CAGTCGTCGGAGAAATTCCA | GCCTTTGTGTCTCCGTATGG |
| LsNCED4 | GGACACGGCTCATGGAATC | GCGAGATCACCGTCACGTT |
| LsABA8ox1 | GCTCGTGACCAAATCTCATCTC | TCAATGTGGGAAACCATGTG |
| LsABA8ox2 | TTGGGTTGTCCAAGTATCATGTTAG | CAAGCTTCGGAGATTATCAAGTGAT |
| LsABA8ox3 | CCTGCTAGCAAAGAGAGAATGC | CCCATCCTCGGAGTGATTG |
| LsABA8ox4 | GTGATGATATCGAGTCCTAAAGTAGCA | TTGAGATGCGAATGGTATGG |
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers LsNCED1: AB120107; LsNCED2: AB120108; LsNCED3: AB120109; LsNCED4; AB120110; LsCCD1: AB120111; LsCCD2: AB120112; LsABA8ox1: AB235917; LsABA8ox2: AB235918; LsABA8ox3: AB235919; LsABA8ox4: AB235920.
Acknowledgments
We thank Dr. Yusuke Jikumaru (RIKEN Plant Science Center) for support in quantification of ABA and its catabolites, and Mr. Atsushi Hanada, Ms. Shoko Shinoda, Ms. Sachiyo Harada, and Ms. Masayo Sekimoto for technical assistance. We are grateful to Dr. Suzanne R. Abrams (National Research Council of Canada), Dr. Nobuhiro Hirai (Kyoto University, Japan), and Dr. Tsuyoshi Nakagawa (Shimane University, Japan) for providing d3-PA and d3-DPA, d3-epi-DPA, and the vector pGWB2, respectively.
This work was supported in part by a Grant-in-Aid for Encouragement of Young Scientists (B) from the Japan Society for the Promotion of Science (JSPS; grant no. 15780081 to T.T.), and the JSPS 21st Century Centers of Excellence Program (Y.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Tomonobu Toyomasu (toyomasu@tds1.tr.yamagata-u.ac.jp).
Open Access articles can be viewed online without a subscription.
References
- Borthwick HA, Hendricks SB, Parker MW, Toole EH, Toole VK (1952) A reversible photoreaction controlling seed germination. Proc Natl Acad Sci USA 38 662–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler WL, Norris KH, Siegelman HW, Hendricks SB (1959) Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proc Natl Acad Sci USA 45 1703–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chermys JT, Zeevaart JAD (2000) Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol 124 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chono M, Honda I, Shinoda S, Kushiro T, Kamiya Y, Nambara E, Kawakami N, Kaneko S, Watanabe Y (2006) Field studies on the regulation of abscisic acid content and germinability during grain development of barley: molecular and chemical analysis of pre-harvest sprouting. J Exp Bot 57 2421–2434 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip, a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cutler AJ, Krochko JE (1999) Formation and breakdown of ABA. Trends Plant Sci 4 472–478 [DOI] [PubMed] [Google Scholar]
- Hirai N, Yoshida R, Todoroki Y, Ohigashi H (2000) Biosynthesis of abscisic acid by the nonmevalonate pathway in plant, and by the mevalonate pathway in fungi. Biosci Biotechnol Biochem 64 1448–1458 [DOI] [PubMed] [Google Scholar]
- Ikuma H, Thimann KV (1960) Action of gibberellic acid on lettuce seed germination. Plant Physiol 35 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y (1990) Role of gibberellins in phytochrome-mediated lettuce seed germination. In N Takahashi, BO Phinney, J MacMillan, eds, Gibberellins. Springer-Verlag, New York, pp 289–295
- Inoue Y, Nagashima H (1991) Photoperceptive site in phytochrome-mediated lettuce (Lactuca sativa L. cv. Grand Rapids) seed germination. J Plant Physiol 137 669–673 [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27 325–333 [DOI] [PubMed] [Google Scholar]
- Izumi K, Kamiya Y, Sakurai A, Oshio H, Takahashi N (1985) Studies of sites of action of a new plant growth retardant (E)-1-(4-chlorophenyl)-4, 4-dimethyl-2-(1, 2, 4-trizol-1-yl)-penten-3-ol (S-3307) and comparative effects of its stereisomers in a cell free system of Cucurbits maxima. Plant Cell Physiol 29 821–827 [Google Scholar]
- Kahn A, Goss JA (1957) Effect of gibberellin on germination of lettuce seed. Science 125 645–646 [DOI] [PubMed] [Google Scholar]
- Kahn AA (1968) Inhibition of gibberellic acid-induced germination by abscisic acid and reversal by cytokinins. Science 125 645–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T (2004) Distinct isoprenoid origins of cis- and trans-zeatin biosynthesis in Arabidopsis. J Biol Chem 279 14049–14054 [DOI] [PubMed] [Google Scholar]
- Kawaide H, Imai R, Sassa T, Kamiya Y (1997) Ent-kaurene synthase from the fungus Phaeosphaeria sp. L487. cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. J Biol Chem 272 21706–21712 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encode ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milborrow BV, Lee HS (1998) Endogenous biosynthetic precursors of (+)-abscisic acid. IV. Carotenoids and ABA are formed by the ‘non-mevalonate’ triose-pyruvate pathway in chloroplasts. Aust J Plant Physiol 25 507–512 [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F (2006) Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J 45 942–954 [DOI] [PubMed] [Google Scholar]
- Nakaminami K, Sawada Y, Suzuki M, Kenmoku H, Kawaide H, Mitsuhashi W, Sassa T, Inoue Y, Kamiya Y, Toyomasu T (2003) Deactivation of gibberellin by 2-oxidation during germination of photoblastic lettuce seeds. Biosci Biotechnol Biochem 67 1551–1558 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56 165–185 [DOI] [PubMed] [Google Scholar]
- Neill SJ, Burnett EC, Desikan R, Hancock JT (1998) Cloning of a wilt-responsive cDNA from an Arabidopsis thaliana suspension culture cDNA library that encodes a putative 9-cis-epoxycarotenoid dioxygenase. J Exp Bot 49 1893–1894 [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Ymaguchi S, Hu J, Jikumaru Y, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272 51–64 [DOI] [PubMed] [Google Scholar]
- Qin X, Zeevaart JAD (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA 96 15354–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S, Jikumaru Y, Fujimoto M, Arikawa T, Takahashi H, Ando M, et al (2007) Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol 48 287–298 [DOI] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol 134 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Okamoto M, Shinoda S, Kushiro T, Koshiba T, Kamiya Y, Hirai N, Todoroki Y, Sakata K, Nambara E, et al (2006) A plant growth retardant, uniconazole, is a potent inhibitor of ABA catabolism in Arabidopsis. Biosci Biotechnol Biochem 70 1731–1739 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Takeba G, Shibata D, Tanaka K (1990) Phytochrome-mediated activation of the gene of for cytosolic glutamine-synthetase (GS1) during imbibition of photosensitive lettuce seeds. Plant Mol Biol 15 317–323 [DOI] [PubMed] [Google Scholar]
- Sankhla N, Sankhla D (1968) Reversal of (±)-abscisin II-induced inhibition of lettuce seed germination. Physiol Plant 21 190–195 [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JAD, McCarty DR (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276 1872–1874 [DOI] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun TP, Koshiba T, et al (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48 354–366 [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M (1996) Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 93 8129–8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeba G (1980. a) Changes revealed by a tracer technique in the amino acid metabolism of thermodormant and non-dormant New York lettuce seeds. Plant Cell Physiol 21 1627–1638 [DOI] [PubMed] [Google Scholar]
- Takeba G (1980. b) Accumulation of free amino acids in the tips of non-thermodormant embryonic axes accounts for the increase in the growth potential of New York lettuce seeds. Plant Cell Physiol 21 1639–1644 [DOI] [PubMed] [Google Scholar]
- Takeba G (1980. c) Effects of temperature, red light and hormones on the accumulation of free amino acids in osmotically growth-inhibited embryonic axes of New York lettuce seeds. Plant Cell Physiol 21 1645–1649 [DOI] [PubMed] [Google Scholar]
- Takeba G (1980. d) Phytochrome-mediated accumulation of free amino acids in embryonic axes of New York lettuce seeds. Plant Cell Physiol 21 1651–1656 [DOI] [PubMed] [Google Scholar]
- Takeba G (1983) Phytochrome-mediated increase in glutamine synthetase activity in photosensitive New York lettuce seeds. Plant Cell Physiol 24 1477–1483 [Google Scholar]
- Takeba G (1984) Effect of gibberellic acid on glutamine synthetase activity in two varieties of lettuce seeds, New York 515 and Grand Rapids. Plant Cell Physiol 25 239–247 [Google Scholar]
- Takeba G, Matsubara S (1977) Rapid disappearance of small fat bodies during the early stage of imbibition of lettuce seeds. Plant Cell Physiol 18 1067–1075 [Google Scholar]
- Takeba G, Matsubara S (1979) Measurement of growth potential of the embryo in New York lettuce seed under various combinations of temperature, red light and hormones. Plant Cell Physiol 20 51–61 [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JAD, McCarty DR (1997) Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA 94 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB (2000) Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol Biol 42 833–845 [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y (1998) Phytochrome regulation gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol 118 1517–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Tsuji H, Yamane H, Nakayama M, Yamaguchi I, Murofushi N, Takahashi N, Inoue Y (1993) Light effects on endogenous levels of gibberellins in photoblastic lettuce seeds. J Plant Growth Regul 12 85–90 [Google Scholar]
- Toyomasu T, Yamane H, Murofushi N, Inoue Y (1994) Effects of exogenously applied gibberellin and red light on the endogenous levels of abscisic acid in photoblastic lettuce seeds. Plant Cell Physiol 35 127–129 [Google Scholar]
- Yamaguchi S, Kamiya Y, Sun TP (2001) Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J 28 443–453 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun TP (1998) Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Takeda-Kamiya N, Hanada A, Ogawa M, Kuwahara A, Seo M, Kamiya Y, Yamaguchi S (2007) Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol 48 555–561 [DOI] [PubMed] [Google Scholar]
- Yang SH, Choi D (2006) Characterization of genes encoding ABA 8′-hydroxylase in ethylene-induced stem growth of deepwater rice (Oryza sativa L.). Biochem Biophys Res Commun 24 685–690 [DOI] [PubMed] [Google Scholar]
- Yang SH, Zeevaart JA (2006) Expression of ABA 8′-hydroxylases in relation to leaf water relations and seed development in bean. Plant J 47 675–686 [DOI] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223 7–12 [DOI] [PubMed] [Google Scholar]
- Zhou R, Cutler AJ, Ambrose SJ, Galka MM, Nelson KM, Squires TM, Loewen MK, Jadhav AS, Ross ARS, Taylor DC, et al (2004) A new abscisic acid catabolic pathway. Plant Physiol 134 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]