Abstract
Genetic variation within and among populations provides the raw material for evolution. Although many studies describe inter- and intraspecific variation of defensive metabolites, little is known about variation among plant populations within early signaling responses elicited by herbivory or by herbivore oral secretions (OS) introduced into wounds during feeding. In this study, we compare the OS-elicited early responses as well as the antiherbivore defensive metabolites in two accessions of the wild tobacco Nicotiana attenuata and show that, compared with an accession collected from Utah, an Arizona accession has lower herbivore-elicited activity of the salicylic acid-induced protein kinase, an important mitogen-activated protein kinase involved in herbivore resistance. These differences in salicylic acid-induced protein kinase activity were associated with substantially different levels of OS-elicited jasmonic acid, jasmonic acid-isoleucine conjugate, and ethylene bursts. Gene expression level polymorphism (ELP) determines phenotypic variation among populations, and we found the two accessions to have significantly different ELPs in the genes involved in early signaling responses to herbivory. In addition, we found differences between the Utah and the Arizona accessions in the concentrations of several secondary metabolites that contribute to N. attenuata's direct and indirect defenses. This study demonstrates significant natural variation in regulatory elements that mediate plant responses to herbivore attack, highlighting the role of ELP in producing a diversity of plant defense phenotypes.
How organisms adapt to their ever-changing environment remains a fundamental and challenging question in biology. Within a species, heritable phenotypic variation among populations, or even individuals within populations, reflects genetic differences, a central requirement for evolutionary change. In recent years, with the completion of several genome sequences, large-scale analyses have revealed substantial intraspecific genetic variation in humans (Morley et al., 2004), yeast (Saccharomyces cerevisiae; Brem et al., 2002), Drosophila (Nuzhdin et al., 2004), Caenorhabditis elegans (Denver et al., 2004), and Arabidopsis (Arabidopsis thaliana; Schmid et al., 2003, 2005; Koornneef et al., 2004; Nordborg et al., 2005; Clark et al., 2007), the model organism of choice for plant genetics.
Variations in the transcription of particular genes, called expression level polymorphism (ELP), is a hallmark (and perhaps often the cause) of diversified phenotypic traits (Doerge, 2002). ELPs in response to exogenously applied salicylic acid and jasmonic acid (JA) have been shown for many genes among different Arabidopsis accessions (Traw et al., 2003; Kliebenstein et al., 2006; van Leeuwen et al., 2007). In different populations, ELPs in particular genes have been shown to produce variation in pathogen resistance (Grant et al., 1995; Gassmann et al., 1999), flowering time (Johanson et al., 2000; Lempe et al., 2005; Werner et al., 2005), herbivore resistance, and secondary metabolite production (Kliebenstein et al., 2001, 2002; Lambrix et al., 2001). However, little is known about the influence of ELPs in diversifying herbivore-induced early signaling events.
Nicotiana attenuata, a wild tobacco plant whose range in western North America stretches from southern Canada to northern Mexico, has been intensively studied with regard to how it responds to the specialist herbivore Manduca sexta (Baldwin, 2001). The moment M. sexta begins to feed, a cascade of signaling events leads to increased defense levels (Fig. 1); N. attenuata plants recognize herbivory-specific elicitors derived from M. sexta oral secretions (OS), namely, fatty acid-amino acid conjugates (FACs), and quickly activate mitogen-activated protein kinase (MAPK) signaling and, subsequently, JA- and ethylene (ET)-mediated defense responses (Kahl et al., 2000; Halitschke and Baldwin, 2003; Wu et al., 2007a). These include: (1) a highly reconfigured transcriptome that encompasses changes in transcript levels of defense- and growth-related genes; (2) the release of volatile organic compounds (VOCs) that function as an indirect defense; and (3) the accumulation of herbivore-deterrent or -damaging secondary metabolites, for example, nicotine and trypsin proteinase inhibitors (TPIs; Reymond and Farmer, 1998; Hermsmeier et al., 2001; Kessler and Baldwin, 2001; Zavala et al., 2004b).
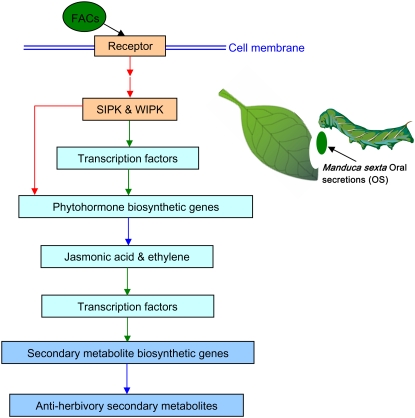
Figure 1.
Molecular events in N. attenuata after attack from M. sexta. Putative receptors in N. attenuata bind FACs present in the OS of M. sexta larvae. SIPK and WIPK, two MAPKs, are activated within minutes of the initiation of feeding. SIPK and WIPK regulate transcript levels of a number of transcription factors, which, in turn, mediate the transcript levels of genes encoding phytohormone biosynthetic enzymes or directly regulate the stability or activity of the enzymes by phosphorylation. The increased levels of phytohormones lead to changes in the transcript levels of transcription factors that regulate the transcript levels of genes involved in secondary metabolite biosyntheses. Red arrows, Direct interactions, e.g. phosphorylation; green arrows, transcriptional regulation; blue arrows, biosynthesis pathways.
TPIs are a class of important direct defense compounds in Nicotiana (as in many other plants); by inhibiting digestive proteinases in the M. sexta midgut, they slow growth and increase mortality of insect larvae (Glawe et al., 2003; Zavala et al., 2004b). N. attenuata accessions collected from southwestern Utah and Flagstaff, Arizona, have been previously found to produce distinct TPI activity levels: the Utah accession (UT) has herbivore-inducible TPI activity, whereas the Arizona accession (AZ) has no detectable TPI activity or mRNA expression, resulting from a deletion mutation in the TPI coding sequence that leads to the degradation of TPI mRNA by nonsense-mediated mRNA decay (Wu et al., 2007b). Nevertheless, a recent study by Steppuhn et al. (A. Steppuhn, M.C. Schuman, and I.T. Baldwin, unpublished data) has shown that AZ plants have an effective system of JA-mediated direct defenses against M. sexta, are fitter than UT plants when planted into the UT native environment, and suffer less attack from the most voracious native herbivores when both accessions are transformed to make them equivalent in nicotine and TPI production. This implies that AZ has reconfigured its system of direct defenses to compensate for the lack of TPI.
It is generally accepted that genetic changes in both cis- and trans-regulatory elements contribute remarkably more than changes in functional genes to phenotypic variation (Doebley and Lukens, 1998; Carroll, 2000; Gilad et al., 2006). Among the regulatory elements that account for herbivore resistance in plants, those involved in early responses are particularly interesting because many play key roles in transducing responses to various environmental stresses into changes in downstream gene expression and eventually into changes in phenotypic traits. Using the N. attenuata UT and AZ accessions, we examined phenotypic variation in the early responses to herbivory; furthermore, we analyzed a number of genes that are involved in these early responses to characterize how these genes' ELP may account for this phenotypic variation.
We show that, in addition to the established difference in expression of the TPI gene, many other genes involved in antiherbivore defense display ELP in these two accessions. Compared with UT, AZ shows lower levels of herbivory-induced salicylic acid-induced protein kinase (SIPK) activity, an important MAPK related to herbivore defense in plants; furthermore, transcripts of SIPK and wound-induced protein kinase (WIPK), as well as two other MAPK genes, accumulate at different levels in AZ after wounding or herbivory. Compared with UT, transcripts of genes involved in phytohormone biosynthesis, as well as two transcription factors, have distinct levels of abundance in AZ; more importantly, the herbivore-induced production of JA, JA-Ile, and ET are partly impaired in AZ. Concentrations of several secondary metabolites involved in herbivore resistance also differed considerably between these two accessions. This work illustrates the high level of phenotypic and genetic variation in herbivore resistance traits among natural populations and provides insight into how variation in regulatory elements may contribute to phenotypic diversity and thus to plants' adaptation to their environments.
RESULTS
Herbivory Activates Lower Levels of SIPK Activity in AZ Than in UT Plants
After wounding or herbivore attack, N. attenuata rapidly activates SIPK and WIPK. Activation of SIPK and WIPK, upstream signal transduction components that modulate downstream phytohormonal and transcriptional changes in plants, is crucial for plants' ability to initiate herbivore resistance (Zhang and Klessig, 2001; Liu and Zhang, 2004; Wu et al., 2007a).
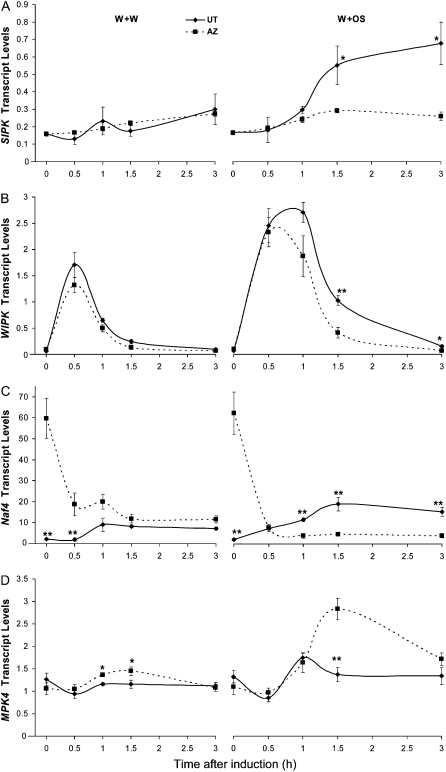
To investigate whether MAPKs are activated differently between these two accessions, we wounded both UT and AZ leaves with a pattern wheel (leaves at node +1) and immediately applied M. sexta OS (W+OS) or water (W+W) as a control. Samples were collected at different time points, and their MAPK activity was determined by an in-gel kinase assay (Fig. 2). After W+W treatment, UT and AZ plants showed the same levels of SIPK activity. W+OS induced much higher and longer-lasting SIPK activity than did W+W. Although 10 min after W+OS treatment, both accessions had the same levels of SIPK activity, W+OS-induced SIPK activity declined faster in AZ; after 30 and 60 min, SIPK in AZ showed less activity than in UT. Similar results were obtained in an independently repeated experiment (Supplemental Fig. S1), as well as in another experiment in which synthetic FACs, the components of M. sexta OS that elicit MAPKs (Wu et al., 2007a), were used (Supplemental Fig. S2). Activity levels of WIPK were much lower than those of SIPK (Wu et al., 2007a) and show a weak band in the in-gel kinase assay (Fig. 2); no difference between UT and AZ was detected.
Figure 2.
N. attenuata plants of the AZ accession have lower levels of M. sexta OS-elicited SIPK activity than do plants of the UT accession. N. attenuata UT and AZ plants were grown under identical conditions. Rosette-stage leaves (+1 position) were wounded with a pattern wheel; 20 μL of W+W or W+OS was applied to the wounds. Leaves from five replicate plants were harvested at the indicated times after elicitation and pooled. Kinase activity was analyzed by an in-gel kinase assay using myelin basic protein as the substrate.
Results of the in-gel kinase assay indicate that both AZ and UT plants can perceive FACs from M. sexta OS and, in response, activate SIPK and WIPK. However, a negative regulation system in AZ plants deactivates SIPK more quickly than it does in UT plants.
MAPK Genes Have Different Transcript Levels in UT and AZ Plants
Recent studies of MAPKs other than SIPK and WIPK have pointed to their critical roles in mediating plant stress responses (Tena et al., 2001; Zhang and Klessig, 2001; Pedley and Martin, 2005). We chose several MAPK genes that have been shown to be involved in early responses within the N. attenuata-M. sexta interaction and measured their transcript levels in both UT and AZ using quantitative real-time PCR (qRT-PCR).
After 1.5 h, W+OS treatment induced higher levels of SIPK transcript than did W+W treatment in UT. Neither treatment changed the levels of SIPK in AZ, and after 1.5 h, AZ had lower levels of W+OS-induced SIPK than did UT (Fig. 3A). Both treatments greatly induced WIPK in UT and AZ, with higher levels after W+OS treatment. Overall, WIPK transcript levels in UT and AZ did not differ from one another, except that WIPK levels in AZ declined more quickly 1.5 h after W+OS treatment (Fig. 3B).
Figure 3.
MAPK genes in N. attenuata UT and AZ accessions have different levels of transcript accumulation. N. attenuata UT and AZ plants were grown under identical conditions. Leaves at +1 position were wounded with a pattern wheel; 20 μL of either W+W or W+OS was applied to the wounds. Individual leaves from five replicate plants were harvested at the indicated times after elicitation. The mean (± se) expression levels of SIPK (A), WIPK (B), Naf4 (C), and MPK4 (D) were measured with qRT-PCR. Asterisks represent significantly different transcript levels between the UT and AZ accessions at the indicated times (unpaired t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; n = 5).
Naf4 is a close homolog of SIPK, which might be derived from a recent gene duplication event (Ren et al., 2006; Wu et al., 2007a). Remarkably, qRT-PCR showed that even when UT and AZ plants were not induced (Fig. 3C), AZ had approximately 25-fold higher levels of Naf4 transcript than did UT. After induction, a distinct transcript profile of Naf4 was also detected. In UT, treatment with either W+W or W+OS elevated the transcript levels of Naf4, with higher levels after W+OS treatment. Conversely, Naf4 transcript levels in AZ rapidly decreased severalfold after each treatment (Fig. 3C).
MPK4 is another MAPK involved at least in pathogen- and wounding-induced responses (Petersen et al., 2000; Gomi et al., 2005). We measured MPK4 transcript levels before and after W+W and W+OS treatments (Fig. 3D). Prior to treatment, AZ and UT showed similar levels of MPK4 transcript. In UT, neither treatment changed the levels of MPK4; in contrast, W+OS significantly increased MPK4 levels in AZ by 1.5 h (P = 0.0007, unpaired t test).
The ELP of these upstream kinases may result in different levels of their activity in UT and AZ, which may produce different downstream responses.
AZ Has Lower Herbivore-Induced Transcript Levels of WRKY Transcription Factors Than Does UT
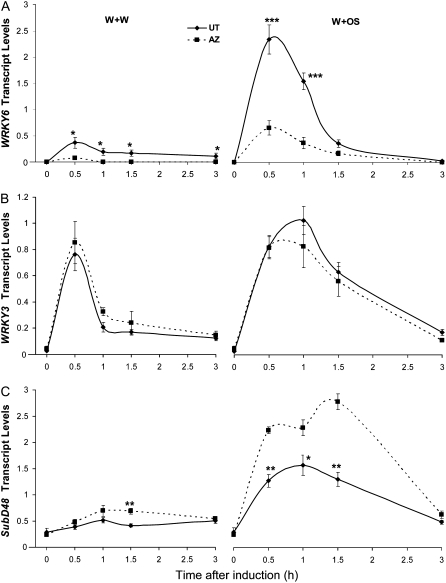
Transcription factors play critical roles in regulating gene transcript levels. WRKYs, which are plant-specific transcription factors, regulate various aspects of plant development and stress responses (Eulgem et al., 2000; Ulker and Somssich, 2004). It is known that transcription factors are the main targets of MAPKs (Hazzalin and Mahadevan, 2002; Yang et al., 2003). To investigate whether there is any ELP of WRKY genes between OS-elicited UT and AZ plants, which might result from the ELP of MAPKs, we measured the transcript abundance of three WRKY genes: WRKY3, WRKY6, and SubD48. WRKY6 has been shown to be an important signaling transduction component contributing to N. attenuata's responses to herbivory (M. Skibbe, N. Qu, I. Galis, and I.T. Baldwin, unpublished data) and is located downstream of SIPK and WIPK (Wu et al., 2007a). SubD48 is also implicated in herbivore resistance, as W+OS treatment induced higher levels of SubD48 transcript than did W+W (Wu et al., 2007a).
When not treated, both accessions had similar levels of WRKY6 transcript (P = 0.62, unpaired t test). Transcript abundance of WRKY6 reached maximum levels after only 0.5 h, increasing 450- and 3,250-fold after W+W and W+OS treatment, respectively, in UT (Fig. 4A). AZ had remarkably lower WRKY6 transcript levels after both treatments, although both accessions showed similar induction patterns, in that W+OS induced much higher levels of WRKY6 than did W+W. Although WRKY3 did not show greater induction by W+OS, W+W-induced WRKY3 transcript levels decreased faster than those induced by W+OS after reaching maximum levels in both accessions at 0.5 h. No statistical difference in WRKY3 transcript levels was detected between UT and AZ before or after either induction (Fig. 4B). Transcript levels of SubD48 were the same in UT and AZ when plants were not induced, and W+OS induced higher levels of SubD48 transcript than did W+W in both accessions (Fig. 4C). However, SubD48 levels were higher in W+OS-treated AZ than in UT plants.
Figure 4.
Transcript levels of different WRKY transcription factors in N. attenuata UT and AZ accessions. N. attenuata UT and AZ plants were grown under identical conditions. Rosette-stage leaves (+1 position) were wounded with a pattern wheel; 20 μL of either W+W or W+OS was applied to the wounds. Individual leaves from five replicate accessions were harvested at the indicated times after elicitation. The mean (± se) transcript levels of WRKY6 (A), WRKY3 (B), and SubD48 (C) were measured with qRT-PCR. Asterisks represent significantly different transcript levels between the UT and AZ accessions at the indicated times (unpaired t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; n = 5).
These data indicate that UT and AZ plants have different expression levels of transcription factors that are involved in herbivore resistance. The ELP of these trans-regulatory elements may also contribute to further variation in downstream gene transcript levels.
AZ Has Lower Levels of Herbivore-Induced JA, JA-Ile, and ET Than Does UT
JA, JA-Ile, and ET play critical roles in mediating plants' defense against herbivores (Reymond and Farmer, 1998; Halitschke and Baldwin, 2003; Staswick and Tiryaki, 2004; Kang et al., 2006; von Dahl et al., 2007). Evidence that SIPK and WIPK play central roles in mediating plants' JA and ET biosynthesis is increasing (Seo et al., 1995; Liu and Zhang, 2004; Wu et al., 2007a). To investigate whether the different levels of SIPK activity and ELP in many early signaling genes led to variation of phytohormone production in these two N. attenuata accessions, we measured both W+W- and W+OS-induced JA, JA-Ile, and ET levels.
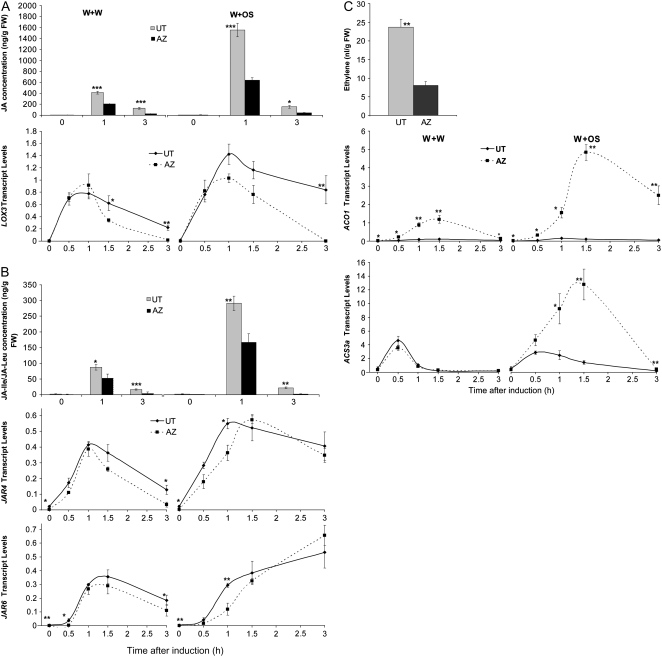
Without treatment, both accessions had similarly low levels of JA (approximately 6 ng/g fresh weight; P = 0.28, unpaired t test). In UT, W+OS treatment induced higher levels of JA than did W+W treatment (Fig. 5A, top), indicating that UT plants recognize herbivory and deploy herbivore-specific defenses. This “JA burst” was much smaller in AZ; W+OS induced only 40% of the JA levels in W+OS-treated UT (Fig. 5A, top). To examine whether the ELP of genes involved in JA biosynthesis contributed to this phenotype, we measured the transcript levels of LIPOXYGENASE3 (LOX3), which encodes an important enzyme in JA biosynthesis (Fig. 5A, bottom; Halitschke and Baldwin, 2003; Wasternack, 2007). When not induced, UT and AZ plants had the same levels of LOX3 transcript (P = 0.55, unpaired t test). Both W+W and W+OS greatly induced LOX3, which declined faster in AZ; at 3 h, W+W- and W+OS-treated AZ had as little as 7% and 0.2%, respectively, of the LOX3 levels found in UT.
Figure 5.
N. attenuata UT and AZ accessions have different levels of wound- and OS-elicited phytohormones. N. attenuata UT and AZ plants were grown under identical conditions. Rosette-stage leaves (+1 position) were wounded with a pattern wheel; 20 μL of either W+W or W+OS was applied to the wounds. A, Mean (± se) JA concentrations in leaves harvested at indicated times were measured with HPLC-MS/MS (top); the mean (± se) transcript levels of LOX3 were measured with qRT-PCR (bottom). B, Mean (± se) JA-Ile/JA-Leu concentrations in leaves harvested at indicated times were measured with HPLC-MS/MS (top); the mean (± se) transcript levels of JAR4, JAR6 were measured with qRT-PCR (middle and bottom, respectively). C, Mean (± se) ET accumulated (top) from treated leaves enclosed in 250-mL flasks and measured after 5 h with a photoacoustic laser spectrometer (n = 3). The mean (± se) transcript levels of ACO1 and ACS3a in leaves harvested at indicated times were measured with qRT-PCR (middle and bottom, respectively). Asterisks represent significantly different transcript levels between the UT and AZ accessions at the indicated times (unpaired t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; n = 5).
Similar results were obtained for measurements of JA-Ile levels. W+OS treatment induced higher levels of JA-Ile than did W+W treatment in both accessions (Fig. 5B, top). At 1 h after either treatment, UT showed about 70% higher levels of JA-Ile than did AZ. JAR4 and JAR6 have been identified as the enzymes that conjugate JA and Ile to form JA-Ile (Wang et al., 2007); we measured the transcript levels of both genes (Fig. 5B, middle and bottom). AZ had basal transcript levels of both JAR4 and JAR6 that were almost 10-fold lower than those in UT (P = 0.038 and 0.005, respectively, unpaired t test). However, 1 to 1.5 h after induction, JAR4 and JAR6 transcript levels were similar in both accessions. The difference between JA-Ile levels in UT and AZ may have resulted from the different levels of both JA, a substrate for the conjugation reaction, and JAR activity because the basal levels of JAR protein might also be considerably lower in AZ.
W+W treatment did not induce a detectable amount of ET in UT or AZ (data not shown). After W+OS treatment, approximately 25 nL of ET was produced for every gram of UT leaf tissue (Fig. 5C, top); AZ produced only one-third as much. We measured the transcript profiles of two genes involved in ET biosynthesis, ACC OXIDASE1 (ACO1) and ACC SYNTHASE3a (ACS3a), in both accessions (Wang et al., 2002; von Dahl et al., 2007). Unexpectedly, levels of ACO1 transcript in untreated AZ plants were about 6-fold higher than in untreated UT plants (Fig. 5C, middle). In UT, W+OS-induced ACO1 transcript levels were no greater than those induced by W+W. In contrast, W+OS induced much higher levels of ACO1 than did W+W in AZ. Furthermore, AZ had much higher transcript levels of ACO1 than did UT under all conditions (Fig. 5C, middle). When untreated, UT and AZ had very similar ACS3a transcript levels (P = 0.48, unpaired t test), and W+W treatment induced similar ACS3a levels in both accessions (P > 0.064, unpaired t test); however, after W+OS treatment, ACS3a levels were severalfold higher in AZ than in UT (Fig. 5C, bottom).
These data show that UT and AZ have distinct herbivore-induced JA, JA-Ile, and ET responses. The ELP of phytohormone biosynthetic genes may contribute to the phenotypic variation between these accessions, but the differential regulation of ET production between the two accessions is clearly more complex.
UT and AZ Produce Different Levels of Antiherbivory Secondary Metabolites
Herbivore-induced kinase activation, and changes in phytohormones and the transcriptome, alter the accumulation of secondary metabolites to raise plant defenses against herbivore attackers. In N. attenuata, these herbivore-induced defenses include nicotine and TPI (Steppuhn et al., 2004; Zavala et al., 2004b). Because AZ plants have lost their TPI defense mechanism (Wu et al., 2007b), we analyzed the levels of nicotine as well as two phenolic compounds, chlorogenic acid and rutin, which are putative antiherbivore defenses (Isman and Duffey, 1982; Bi et al., 1997) and may constitute part of N. attenuata's direct defense.
Four days after plants were treated with W+W or W+OS, when levels of nicotine were at their highest (Baldwin et al., 1998), samples were harvested and analyzed by HPLC (Fig. 6A). Without treatment, AZ had approximately 50% more nicotine than did UT. Nicotine concentrations increased in both UT and AZ after treatment; nevertheless, AZ still contained 35% and 20% more nicotine compared to UT plants identically treated with W+OS and W+W, respectively.
Figure 6.
N. attenuata UT and AZ accessions produce different amounts of secondary metabolites that function as direct and indirect defenses. N. attenuata UT and AZ plants were grown under identical conditions. A to C, Rosette-stage leaves (+1 position) were wounded with a pattern wheel; 20 μL of either W+W or W+OS was applied to the wounds. Nontreated plants served as control. Four days after treatments, plant tissue samples were harvested; nicotine (A), chlorogenic acid (B), and rutin (C) concentrations were analyzed using HPLC. The emission of CAB was measured in treated leaves (+2 position) at 24 to 32 h posttreatment with either W+W or W+OS, with no treatment as control (D), or 150 μg MeJA in lanolin paste, with lanolin as control (E). Absolute emission is not comparable between D and E as these were two separate experiments (unpaired t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001, n = 5).
Compared with UT, more chlorogenic acid and rutin were detected in AZ (Fig. 6, B and C). Under normal conditions, AZ contained 1.3-fold more chlorogenic acid than did UT. After W+W and W+OS treatments, AZ still contained higher levels of chlorogenic acid. Rutin levels were not changed after either W+W or W+OS treatment; similar to concentrations of chlorogenic acid, concentrations of rutin in AZ were about 50% higher than in UT.
In addition to direct defenses, N. attenuata deploys indirect defenses to attract predators of herbivores. One of the most important indirect defense compounds is cis-α-bergamotene (CAB), an herbivore-inducible VOC (Kessler and Baldwin, 2001). We found that W+OS treatment enhanced CAB emission in UT approximately 7-fold; however, neither W+W nor W+OS induced higher levels of CAB emission in AZ (Fig. 6D). Because CAB biosynthesis is regulated by JA (Halitschke et al., 2000), we examined whether the decreased JA levels in AZ after W+OS treatment were associated with diminished CAB levels. We applied 150 μg of methyl jasmonate (MeJA) dissolved in lanolin paste or pure lanolin paste (lanolin) as a control to both accessions and measured CAB emission. In both accessions, CAB emission was highly induced, but MeJA treatment did not recover the difference in CAB production between AZ and UT (Fig. 6E). This suggests that during evolution, AZ may have lost or reconfigured cis- or trans-elements involved in the regulation of CAB biosynthesis.
DISCUSSION
In this study, we examined the early herbivore-induced responses in two N. attenuata accessions collected from Utah and Arizona. We measured (1) the activity of two MAPKs, SIPK and WIPK; (2) the levels of the herbivore-induced phytohormones JA, JA-Ile, and ET; (3) differential transcript accumulation of several genes encoding MAPKs, transcription factors, and phytohormone biosynthesis enzymes, which we took to be indicative of ELPs in these genes; and (4) the associated variation in concentrations of direct and indirect defense-related secondary metabolites.
Natural Variation in Herbivore-Induced Signaling Events
MAPK signaling has been shown to play a critical role in mediating plant resistance to herbivores (Kandoth et al., 2007; Wu et al., 2007a). FACs from M. sexta OS are introduced into plant cells through wounds generated by herbivore feeding. These FACs bind to putative receptors in N. attenuata, enabling plants to perceive herbivory. This perception results in the activation of MAPKs, most importantly SIPK and WIPK (Wu et al., 2007a). We examined kinase activity and kinase gene transcript levels in both UT and AZ. Activity of SIPK and WIPK reached the same maximum levels in both accessions, indicating that both are equally able to perceive herbivory and initiate herbivory-induced signal cascades. Nevertheless, the speed with which SIPK activity levels decrease in AZ following simulated herbivory (W+OS) suggests that the negative regulation of SIPK is stronger in AZ than in UT. A recent study in Arabidopsis has shown that a phosphatase, AP2C1, might be responsible for this negative regulation (Schweighofer et al., 2007).
Both SIPK and WIPK modulate herbivory-induced JA biosynthesis (Wu et al., 2007a). AZ plants accumulate lower levels of JA after simulated herbivory, consistent with rapidly decreasing levels of herbivore-induced SIPK activity. Compared with JA biosynthesis, ET biosynthesis is simpler; only 2 enzymes, ACO and ACS, are responsible for converting S-adenosyl-Met to ET (Wang et al., 2002). SIPK directly phosphorylates certain (but not all) ACSs, dramatically increasing their stability (Liu and Zhang, 2004). Longer-lasting herbivore-induced SIPK activity levels may partly account for the greater amount of ET produced in UT plants than in AZ plants and could also explain why AZ produces less ET despite accumulating significantly higher levels of ACO and ACS transcripts than UT. However, the large differences in herbivore-induced JA and ET levels between UT and AZ plants are unlikely to have resulted solely from differences in SIPK activity. Variation of many genes involved in JA and ET biosynthesis probably also contributed to differences in the production of these two phytohormones in UT and AZ plants.
In addition to SIPK and WIPK, we found ELP in two other MAPK genes: Naf4 and MPK4. Naf4 shows high sequence homology to SIPK and thus may have a similar function (Ren et al., 2006). MPK4 is known to be involved in pathogen resistance and wound-induced responses but has as yet no known function in herbivore resistance (Petersen et al., 2000; Gomi et al., 2005). Because kinases are upstream regulatory elements that influence the transcription of a wide rage of genes, variation in their constitutive and/or induced levels is likely to produce large sets of ELPs.
Most MAPK targets are transcription factors (Hazzalin and Mahadevan, 2002). Two WRKY transcription factors, WRKY6 and SubD48, displayed ELP between UT and AZ; however, there was no ELP in a third WRKY (WRKY3). Transcript levels of WRKY6 have been shown to be positively regulated by both SIPK and WIPK (Wu et al., 2007a). Thus, we can assume that the different herbivory-induced SIPK activity in UT and AZ may have resulted in the observed ELP of WRKY6. MAPKs, JA, and ET are known to regulate expression of a wide array of genes. Therefore, many other transcription factors downstream of herbivory-induced MAPKs, JA, and ET are likely to also show ELP between UT and AZ.
The large and sometimes unpredictable variation between these two N. attenuata accessions in their herbivore-induced early responses is indicative not only of the complexity of the signaling networks but also of the cross-talk and feedback regulation that characterize the relationships among their components, which include receptors, kinases, phytohormones, and transcription factors, with each group of molecules potentially participating in multiple layers of regulation. Studying the molecular mechanisms that underlie the phenotypic changes in these early signaling events will eventually provide us with a detailed understanding of how plants fine-tune their signaling systems, as well as how evolutionary forces have shaped genetic variation among natural populations.
Natural Variation of Secondary Metabolites
Plants' reconfiguration of their transcriptomes and proteomes following herbivore attack results in the deployment of the last layer of the defense system, which comprises various antiherbivore secondary metabolites (Baldwin, 2001). We found that AZ produces greater amounts of both nicotine and phenolic compounds (chlorogenic acid and rutin) than does UT. Nicotine is a potent chemical defense that has a strong negative effect on M. sexta and on many other herbivores and has been previously shown to be a critical component of JA-mediated direct defense in AZ as well as in UT (Steppuhn et al., 2004). Nicotine biosynthesis is both up-regulated by JA and down-regulated by OS-induced ET (Baldwin, 1998; Voelckel et al., 2001; von Dahl et al., 2007). Thus, higher nicotine levels in AZ could indicate that decreased negative regulation by ET plays a greater role than decreased positive regulation by JA in nicotine accumulation after herbivory. Alternatively, they could reflect a greater availability of nitrogen for nicotine biosynthesis due to the lack of nitrogen-intensive TPI production in AZ. However, given that AZ produces constitutively higher amounts of nicotine, whereas ET and JA can only influence induced nicotine production, and that such high constitutive nicotine production was not seen in a line of the UT accession that was silenced for TPI (Zavala et al., 2004a), we assume that changes in certain trans- or cis-element(s) in the regulation of nicotine biosynthesis have enabled AZ to produce more nicotine than UT.
The low level of CAB production in AZ illustrates the complex changes in its regulatory system; that AZ is able to produce CAB, but releases less than 10% as much as UT after OS-elicitation, indicates that the biosynthesis machinery is intact, but the regulatory system is constrained. That the decrease in production cannot be recovered with MeJA application shows that the bottleneck in regulation is downstream of the JA. Little is known about how CAB and other VOCs, which may also act as indirect defense compounds, are synthesized and regulated in N. attenuata. The synthesis of phenolic compounds in N. attenuata and their possible influence on M. sexta performance also remains to be characterized; some studies have revealed potential antiherbivore functions of phenolic compounds (Isman and Duffey, 1982; Bi et al., 1997). Further biochemical and genetic studies are needed to elucidate the ELPs in biosynthetic genes that may underlie the differential regulation of these compounds.
Ecological and Evolutionary Significance
Understanding the mechanisms responsible for the phenotypic variation in herbivore defense systems between these two N. attenuata accessions provides insights into their evolution. The TPI gene has been identified as a null allele in AZ (Zavala et al., 2004b; Wu et al., 2007b). In this study, we show that despite (or perhaps due to) the loss of its TPI defense mechanism, AZ produces greater amounts of nicotine and phenolics. The higher concentrations of these compounds in AZ may be a result of natural selection following the event of TPI mutation, enabling AZ to compensate for the loss of TPI. How the TPI null allele was fixed in AZ is unclear because this mutation could have considerably lowered the ability of AZ to survive under herbivory pressure. Using TPI-silenced transgenic plants, Zavala et al. (2004a) demonstrated that TPI production is costly under normal conditions because plants having lower TPI levels grow faster and have higher fitness due to the re-allocation of nitrogen; however, under selection pressure from herbivory, TPI-containing plants realize greater fitness due to the effectiveness of this defense (Zavala et al., 2004a, 2004b). One hypothesis is that after the TPI mutation event in AZ, there was very low selection pressure from herbivores; therefore, this null TPI allele was quickly fixed because it provided plants with higher fitness. An alternative scenario that could lead to the fixation of the null TPI allele would be genetic drift and small original population size. In either case, over time, (re-imposed) selection pressure from herbivory could have led to higher concentrations of other antiherbivore compounds, i.e. nicotine and phenolics. In nature, indirect defense compounds (e.g. CAB) are also highly effective in protecting plants from herbivory (Kessler and Baldwin, 2001). It is unclear why AZ has lost most of its CAB production. Steppuhn et al. (A. Steppuhn, M.C. Schuman, and I.T. Baldwin, unpublished data) have shown that AZ plants emit much higher levels of some monoterpenes than UT plants, both constitutively and inducibly. Whether in its natural habitat, AZ depends more on direct defense than indirect defense against herbivory or whether the predators in this habitat respond to other volatile signals remains to be studied.
Plants recollected in 2005 from the same region where the original AZ accession was collected in 1996 (near Flagstaff, Arizona) had the same null mutation in TPI (I.T. Baldwin and M.C. Schuman, unpublished data). This suggests that plants of the AZ accession have adapted to their native environment via as-yet unidentified defense or tolerance mechanisms. Coevolution has long been considered to have shaped the diversity of both plants and herbivores (Ehrlich and Raven, 1964; Mauricio, 2001; Zangerl and Berenbaum, 2005). Further ecological studies could provide the necessary data to connect diversity in herbivore-induced plant responses to differences in native herbivore population structures and their dynamics and to determine whether different selection forces imposed by diverse insect populations could have caused polymorphisms in defense compound production between native plant populations.
Recent studies of Arabidopsis populations have revealed considerable variation in both the genomic and the transcriptomic level (Kliebenstein et al., 2006; Clark et al., 2007; Kim et al., 2007). It is very unlikely that only one or a few genetic changes differentiate AZ and UT. We predict that thousands of genes show polymorphism in sequence and/or expression between these two accessions. This unknown pool of polymorphisms greatly restricts our ability to explain changes in many phenotypic traits between the UT and AZ accessions, especially because most of these traits are polygenic. Nevertheless, ever-increasing theoretical and practical advances have provided us with the ability to unravel the complex genetic basis that is responsible for the variations we have detected (Kliebenstein et al., 2001, 2002; Wentzell et al., 2007). Using biochemical, genetic, and population genetic tools, we expect to see variations in many loci that contribute to the phenotypic variation between UT and AZ; it will not be surprising if these loci turn out to be mostly regulatory elements: transcription factor-binding sites in promoters (cis-elements), genes coding for transcription factors (trans-elements), or even genes involved in modifying transcription factors, such as their binding activity, stability, and localization.
MATERIALS AND METHODS
Plant Material and Experimental Conditions
Seed collections of Nicotiana attenuata UT and AZ accessions are described by Glawe et al. (2003). Nearly homozygous UT and AZ accessions were obtained by inbreeding for 17 and seven generations in the glasshouse, respectively. In each generation, seeds were collected from one randomly selected plant for each accession. No artificial selection was conducted.
Seeds were germinated on agar plates containing Gamborg B5 media (Duchefa). Plants were grown in the glasshouse in 1-L individual pots at 26°C to 28°C under 16 h of light as in Krügel et al. (2002). All treatments were performed with plants in the rosette stage of growth. The second fully elongated (+1 position) leaves were used, unless otherwise noted. For W+W treatment, 20 μL of deionized water was immediately applied to wounds generated by a pattern wheel; similarly, for W+OS treatment, 20 μL of Manduca sexta OS (diluted 1:5 in water, v/v) was applied to wounds. For MeJA treatment, 20 μL of MeJA dissolved in lanolin (7.5 μg/μL) was rubbed on the base of the leaf with a small spatula; as a control, pure lanolin was used (lanolin). Samples from untreated plants were used as controls. Unless otherwise noted, five replicate plants were used. After specific time periods, leaves were excised, flash frozen in liquid nitrogen, and stored at −80°C until use.
qRT-PCR Analyses
Total RNA was extracted from approximately 100 mg leaf tissue using the Trizol reagent (Invitrogen). The qRT-PCR procedure is described in detail by Wu et al. (2007a). All RNA samples were reverse transcribed using oligo(dT) and the Superscript II enzyme (Invitrogen). qRT-PCR was performed on an ABI PRISM 7700 sequence detection system (Applied Biosystems) following conditions recommended by the manufacturer. In all analyses, a N. attenuata actin gene was used to normalize the concentrations of cDNA samples. The primers and probes used for the Taqman-based analysis, as well as the primers used for SYBR Green-based analyses, are listed in Supplemental Tables S1 and S2, respectively.
Phytohormone Analyses
JA and JA-Ile were analyzed using a HPLC-tandem mass spectrometry (MS/MS) method. Leaf tissue (approximately 200 mg) was ground in liquid nitrogen and extracted with ethyl acetate containing 200 ng of 13C2-JA and para-coumaric acid as internal standard for JA and JA-Ile/JA-Leu, respectively. After centrifuging, supernatants were dried in a concentrator (Eppendorf). The pellet from each sample was further extracted with 500 μL of 70% methanol (v/v). The supernatants were directly injected onto a 1200L HPLC-MS/MS (Varian), and the levels of JA and JA-Ile/JA-Leu were quantified by comparing their peak areas with those from internal standards (JA-Ile and JA-Leu could not be quantified separately due to their similar chemical properties and identical molecular weights).
ET was measured on a photoacoustic spectrometer (INVIVO). Three freshly detached leaves were treated either with W+W or W+OS, immediately sealed in a three-neck 250-mL flask with a round bottom, and kept in the glasshouse for 5 h. The headspace was flushed into a photoacoustic laser spectrometer with hydrocarbon-free clean air, and the ET concentration was quantified by comparing ET peak areas with peak areas generated by a standard ET gas.
Analysis of Nicotine, Chlorogenic Acid, and Rutin
Nicotine, chlorogenic acid, and rutin were analyzed using an HPLC-DAD method (Keinanen et al., 2001). About 100 mg of leaf tissue was extracted with 1 mL of extraction solution (40% methanol, 0.5% acetic acid). After centrifuging, the supernatants were collected and injected into an Agilent 1100 HPLC installed with an Inertsil ODS-3 column (Phenomenex). Nicotine, rutin, and chlorogenic acid in different concentrations were used as external standards.
Measurement of CAB
Volatiles were collected from plants following treatment to the +2 leaf with lanolin, MeJA in lanolin, W+W, or W+OS as described, or no treatment. Two separate sets of plants were used: one for lanolin or MeJA treatment, and one for control, W+W, or W+OS treatment. Plants were placed at a maximum distance from each other on a table in the glasshouse. At 24 h after treatment, treated leaves were enclosed in two 50-mL food-quality plastic containers secured with miniature claw-style hair clips. Ambient air flowed into the cage primarily through a trimmed P1000 pipette tip inserted into the bottom container, and was pulled out through a self-packed glass tube containing glass wool and 20 mg of SuperQ (Alltech), secured in a second trimmed P1000 pipette tip inserted into the top container. Airflow was powered by a manifold vacuum pump as described by Halitschke et al. (2000). Background volatiles present in ambient air were also collected using empty trapping containers; CAB was not present in quantifiable levels in background controls. Trapping was stopped after 8 h, and traps were either immediately eluted by spiking each trap with 400 ng of tetraline as an internal standard and pushing through 250 μL of dichloromethane into a gas chromatography vial, or stored at −20°C until elution. Flow rates through each trap were measured and included as correlates in statistical analyses.
Eluents were separated on a DB-5 column (Agilent) in a Varian CP-3800 gas chromatograph coupled with a Varian Saturn 4000 ion trap MS in EI mode. Relative emission of CAB was expressed in nanograms of tetraline per gram fresh weight per hour. Data were log-transformed prior to analysis to meet requirements of normality and homogeneity of variance; 1 was added to all values prior to transformation to accommodate 0 values.
Statistical Analysis
Statistics were done using the StatView software (SAS Institute).
In-Gel Kinase Assay
An in-gel kinase assay was performed following Zhang and Klessig (1997). Proteins were extracted from 100 mg of tissue with 250 μL of extraction buffer (100 mm HEPES, pH 7.5, 5 mm EDTA, 5 mm EGTA, 10 mm Na3VO4, 10 mm NaF, 50 mm β-glycerolphosphate, 1 mm phenylmethylsulfonyl fluoride, 10% glycerol, 10 mm dithiothreitol [DTT], and complete proteinase inhibitor cocktail tablets [Roche]). Protein samples containing 10 μg of total protein were separated on a 10% SDS-polyacrylamide gel embedded with 0.2 mg/mL myelin basic protein (Sigma). After electrophoresis, the gel was washed three times for 30 min each in a washing buffer (25 mm Tris, pH 7.5, 0.5 mm DTT, 0.1 mm Na3VO4, 5 mm NaF, 0.5 mg/mL bovine serum albumin, and 0.1% Triton X-100 [v/v]) at room temperature. Then, kinases in the gel were renatured in 25 mm Tris, pH 7.5, 0.5 mm DTT, 0.1 mm Na3VO4, and 5 mm NaF at 4°C overnight, with three changes of buffer. At room temperature, the gel was then incubated in a reaction solution (25 mm Tris, pH 7.5, 2 mm EGTA, 12 mm MgCl2, 1 mm DTT, and 0.1 mm Na3VO4) with 0.2 mm ATP plus 50 μCi of γ-32P-ATP (3,000 Ci/mmol) for 1 h. The reaction was stopped by a fixation solution (5% TCA and 1% sodium pyrophosphate [w/v]). After 6 h of washing with five changes, the gel was dried on a gel dryer (Bio-Rad), and image was obtained on an FLA-3000 phosphor imager system (FujiFilm Life Science).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. N. attenuata plants of the AZ accession have lower levels of M. sexta OS-elicited SIPK activity than do plants of the UT accession.
Supplemental Figure S2. N. attenuata plants of the AZ accession have lower levels of FAC-elicited SIPK activity than do plants of the UT accession.
Supplemental Table S1. Primers and probes used for Taqman-based qRT-PCR.
Supplemental Table S2. Primers used for SYBR Green-based qRT-PCR.
Supplementary Material
Acknowledgments
We thank Eva Rothe and Dr. Matthias Schöttner for their help with the phytohormone and HPLC analysis, Stefan Meldau for the in-gel kinase assay, and Emily Wheeler for editorial assistance.
This work was supported by the Max-Planck-Society.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ian T. Baldwin (baldwin@ice.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baldwin IT (1998) Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA 95 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT (2001) An ecologically motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiol 127 1449–1458 [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT, Gorham D, Schmelz EA, Lewandowski CA, Lynds GY (1998) Allocation of nitrogen to an inducible defense and seed production in Nicotiana attenuata. Oecologia 115 541–552 [DOI] [PubMed] [Google Scholar]
- Bi JL, Murphy JB, Felton GW (1997) Antinutritive and oxidative components as mechanisms of induced resistance in cotton to Helicoverpa zea. J Chem Ecol 23 97–117 [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L (2002) Genetic dissection of transcriptional regulation in budding yeast. Science 296 752–755 [DOI] [PubMed] [Google Scholar]
- Carroll SB (2000) Endless forms: the evolution of gene regulation and morphological diversity. Cell 101 577–580 [DOI] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 338–342 [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Thomas WK (2004) High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430 679–682 [DOI] [PubMed] [Google Scholar]
- Doebley J, Lukens L (1998) Transcriptional regulators and the evolution of plant form. Plant Cell 10 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge RW (2002) Mapping and analysis of quantitative trait loci in experimental populations. Nat Rev Genet 3 43–52 [DOI] [PubMed] [Google Scholar]
- Ehrlich PR, Raven PH (1964) Butterflies and plants: a study in coevolution. Evolution Int J Org Evolution 18 586–608 [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 199–206 [DOI] [PubMed] [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 20 265–277 [DOI] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP (2006) Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440 242–245 [DOI] [PubMed] [Google Scholar]
- Glawe GA, Zavala JA, Kessler A, Van Dam NM, Baldwin IT (2003) Ecological costs and benefits correlated with trypsin protease inhibitor production in Nicotiana attenuata. Ecology 84 79–90 [Google Scholar]
- Gomi K, Ogawa D, Katou S, Kamada H, Nakajima N, Saji H, Soyano T, Sasabe M, Machida Y, Mitsuhara I, et al (2005) A mitogen-activated protein kinase NtMPK4 activated by SIPKK is required for jasmonic acid signaling and involved in ozone tolerance via stomatal movement in tobacco. Plant Cell Physiol 46 1902–1914 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269 843–846 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Baldwin IT (2003) Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J 36 794–807 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Kessler A, Kahl J, Lorenz A, Baldwin IT (2000) Ecophysiological comparison of direct and indirect defenses in Nicotiana attenuata. Oecologia 124 408–417 [DOI] [PubMed] [Google Scholar]
- Hazzalin CA, Mahadevan LC (2002) MAPK-regulated transcription: a continuously variable gene switch? Nat Rev Mol Cell Biol 3 30–40 [DOI] [PubMed] [Google Scholar]
- Hermsmeier D, Schittko U, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. I. Large-scale changes in the accumulation of growth- and defense-related plant mRNAs. Plant Physiol 125 683–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isman MB, Duffey SS (1982) Toxicity of tomato phenolic-compounds to the fruitworm, Heliothis-Zea. Entomol Exp Appl 31 370–376 [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290 344–347 [DOI] [PubMed] [Google Scholar]
- Kahl J, Siemens DH, Aerts RJ, Gabler R, Kuhnemann F, Preston CA, Baldwin IT (2000) Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore. Planta 210 336–342 [DOI] [PubMed] [Google Scholar]
- Kandoth PK, Ranf S, Pancholi SS, Jayanty S, Walla MD, Miller W, Howe GA, Lincoln DE, Stratmann JW (2007) Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc Natl Acad Sci USA 104 12205–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Wang L, Giri A, Baldwin IT (2006) Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid-isoleucine-mediated defenses against Manduca sexta. Plant Cell 18 3303–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinanen M, Oldham NJ, Baldwin IT (2001) Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuata. J Agric Food Chem 49 3553–3558 [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291 2141–2144 [DOI] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M (2007) Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet 39 1151–1155 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T (2002) Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics 161 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T (2001) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, West MA, van Leeuwen H, Kim K, Doerge RW, Michelmore RW, St Clair DA (2006) Genomic survey of gene expression diversity in Arabidopsis thaliana. Genetics 172 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55 141–172 [DOI] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT (2002) Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12 177–183 [Google Scholar]
- Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, Gershenzon J (2001) The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia ni herbivory. Plant Cell 13 2793–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D (2005) Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet 1 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio R (2001) An ecological genetic approach to the study of coevolution. Am Zool 41 916–927 [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG (2004) Genetic analysis of genome-wide variation in human gene expression. Nature 430 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3 e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM (2004) Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol Biol Evol 21 1308–1317 [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2005) Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol 8 541–547 [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120 [DOI] [PubMed] [Google Scholar]
- Ren D, Yang KY, Li GJ, Liu Y, Zhang S (2006) Activation of Ntf4, a tobacco MAPK, during plant defense response and its involvement in hypersensitive response-like cell death. Plant Physiol 141 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1 404–411 [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, Weisshaar B, Mitchell-Olds T (2005) A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169 1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Sorensen TR, Stracke R, Torjek O, Altmann T, Mitchell-Olds T, Weisshaar B (2003) Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res 13 1250–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, et al (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Okamoto M, Seto H, Ishizuka K, Sano H, Ohashi Y (1995) Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science 270 1988–1992 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppuhn A, Gase K, Krock B, Halitschke R, Baldwin IT (2004) Nicotine's defensive function in nature. PLoS Biol 2 E217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Asai T, Chiu WL, Sheen J (2001) Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 4 392–400 [DOI] [PubMed] [Google Scholar]
- Traw MB, Kim J, Enright S, Cipollini DF, Bergelson J (2003) Negative cross-talk between salicylate- and jasmonate-mediated pathways in the Wassilewskija ecotype of Arabidopsis thaliana. Mol Ecol 12 1125–1135 [DOI] [PubMed] [Google Scholar]
- Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7 491–498 [DOI] [PubMed] [Google Scholar]
- van Leeuwen H, Kliebenstein DJ, West MA, Kim K, van Poecke R, Katagiri F, Michelmore RW, Doerge RW, St Clair DA (2007) Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid. Plant Cell 19 2099–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelckel C, Schittko U, Baldwin IT (2001) Herbivore-induced ethylene burst reduces fitness costs of jasmonate- and oral secretion-induced defenses in Nicotiana attenuata. Oecologia 127 274–280 [DOI] [PubMed] [Google Scholar]
- von Dahl CC, Winz RA, Halitschke R, Kuhnemann F, Gase K, Baldwin IT (2007) Tuning the herbivore-induced ethylene burst: the role of transcript accumulation and ethylene perception in Nicotiana attenuata. Plant J 51 293–307 [DOI] [PubMed] [Google Scholar]
- Wang KL, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell (Suppl) 14 S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Halitschke R, Kang JH, Berg A, Harnisch F, Baldwin IT (2007) Independently silencing two JAR family members impairs levels of trypsin proteinase inhibitors but not nicotine. Planta 226 159–167 [DOI] [PubMed] [Google Scholar]
- Wasternack C (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell AM, Rowe HC, Hansen BG, Ticconi C, Halkier BA, Kliebenstein DJ (2007) Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet 3 1687–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Warthmann N, Trainer GT, Ecker JR, Chory J, Weigel D (2005) Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc Natl Acad Sci USA 102 2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT (2007. a) Herbivory rapidly activates mapk signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19 1096–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Kang JH, Hettenhausen C, Baldwin IT (2007. b) Nonsense-mediated mRNA decay (NMD) silences the accumulation of aberrant trypsin proteinase inhibitor mRNA in Nicotiana attenuata. Plant J 51 693–706 [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD, Whitmarsh AJ (2003) Transcriptional regulation by the MAP kinase signaling cascades. Gene 320 3–21 [DOI] [PubMed] [Google Scholar]
- Zangerl AR, Berenbaum MR (2005) Increase in toxicity of an invasive weed after reassociation with its coevolved herbivore. Proc Natl Acad Sci USA 102 15529–15532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Baldwin IT (2004. a) Constitutive and inducible trypsin proteinase inhibitor production incurs large fitness costs in Nicotiana attenuata. Proc Natl Acad Sci USA 101 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Hui D, Baldwin IT (2004. b) Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defenses. Plant Physiol 134 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (1997) Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6 520–527 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.