Abstract
Colonization of Arabidopsis thaliana roots by nonpathogenic Pseudomonas fluorescens WCS417r bacteria triggers a jasmonate/ethylene-dependent induced systemic resistance (ISR) that is effective against a broad range of pathogens. Microarray analysis revealed that the R2R3-MYB-like transcription factor gene MYB72 is specifically activated in the roots upon colonization by WCS417r. Here, we show that T-DNA knockout mutants myb72-1 and myb72-2 are incapable of mounting ISR against the pathogens Pseudomonas syringae pv tomato, Hyaloperonospora parasitica, Alternaria brassicicola, and Botrytis cinerea, indicating that MYB72 is essential to establish broad-spectrum ISR. Overexpression of MYB72 did not result in enhanced resistance against any of the pathogens tested, demonstrating that MYB72 is not sufficient for the expression of ISR. Yeast two-hybrid analysis revealed that MYB72 physically interacts in vitro with the ETHYLENE INSENSITIVE3 (EIN3)-LIKE3 transcription factor EIL3, linking MYB72 function to the ethylene response pathway. However, WCS417r activated MYB72 in ISR-deficient, ethylene-insensitive ein2-1 plants. Moreover, exogenous application of the ethylene precursor 1-aminocyclopropane-1-carboxylate induced wild-type levels of resistance in myb72-1, suggesting that MYB72 acts upstream of ethylene in the ISR pathway. Collectively, this study identified the transcriptional regulator MYB72 as a novel ISR signaling component that is required in the roots during early signaling steps of rhizobacteria-mediated ISR.
The soil environment that is influenced by plant roots, the rhizosphere, is a nutrient-rich habitat providing niches for numerous microorganisms. Among these, many fungi and bacteria with properties beneficial to plants are present (Marx, 2004; Pozo et al., 2004). Some plant-beneficial bacteria (e.g. Bacillus and fluorescent Pseudomonas spp.; Kloepper et al., 2004; Weller, 2007), have been reported to protect plants against pathogenic microorganisms through different mechanisms, such as competition for nutrients, secretion of antibiotics and lytic enzymes, and stimulation of the plant's defensive capacity (Bakker et al., 2007). The latter phenomenon is commonly referred to as induced systemic resistance (ISR; Van Loon et al., 1998). ISR has been demonstrated in many plant species (e.g. bean [Phaseolus vulgaris], carnation [Dianthus caryophyllus], cucumber [Cucumis sativus], radish [Raphanus sativus], tobacco [Nicotiana tabacum], tomato [Solanum lycopersicum], and the model plant Arabidopsis [Arabidopsis thaliana]), and is effective against a broad spectrum of plant pathogens, including fungi, bacteria, viruses, and even insect herbivores (Van Loon et al., 1998).
The ability of plants to develop ISR in response to root colonization by Pseudomonas bacteria depends on the host-rhizobacterium combination (Van Loon et al., 1998; Pieterse et al., 2002). The nonpathogenic, rhizobacterial strain Pseudomonas fluorescens WCS417r has been shown to trigger ISR in several plant species and has served as a model strain to study ISR in Arabidopsis (Pieterse et al., 2002). Colonization of Arabidopsis roots by WCS417r triggers ISR against the bacterial leaf pathogens Xanthomonas campestris pv armoraciae and Pseudomonas syringae pv tomato DC3000 (Pst DC3000), the fungal leaf pathogen Alternaria brassicicola, the oomycete leaf pathogen Hyaloperonospora parasitica, and the fungal root pathogen Fusarium oxysporum f. sp. raphani (Pieterse et al., 1996; Van Wees et al., 1997; Ton et al., 2002b). Protection against these pathogens is characterized by a reduction in disease severity as well as an inhibition of pathogen growth.
Phenotypically, ISR resembles systemic acquired resistance (SAR) that develops upon primary infection with a necrotizing pathogen (Durrant and Dong, 2004). Although rhizobacteria-mediated ISR and pathogen-induced SAR are both effective against a broad spectrum of pathogens, their signal transduction pathways are distinct. The onset of SAR is accompanied by local and systemic increases in endogenous levels of salicylic acid (SA) and the transcriptional reprogramming of a large set of genes, including genes encoding pathogenesis-related (PR) proteins (Van Loon et al., 2006). Some PR proteins possess in vitro antimicrobial activity and are thought to contribute to the enhanced resistance state of SAR. Transduction of the SA signal requires functional NPR1, a regulatory protein that was identified in Arabidopsis through genetic screens for mutants impaired in their defense response to SA or its functional analogs (Dong, 2004). Plants that carry a mutation in the NPR1 gene accumulate normal or even higher levels of SA after pathogen infection, but are impaired in their ability to transcriptionally activate PR genes and to mount a SAR response. Although some rhizobacterial strains can activate the SA-dependent SAR pathway (De Meyer and Höfte, 1997), the large majority of the reported resistance-inducing fluorescent Pseudomonas spp. strains have been shown to trigger ISR in a SA-independent manner (Van Loon and Bakker, 2005). WCS417r-mediated ISR functions independently of SA as well, as demonstrated by observations that Arabidopsis genotypes impaired in SA accumulation or biosynthesis (i.e. NahG, eds5-1, sid2-1) were still able to develop wild-type levels of ISR upon colonization of the roots by WCS417r (Pieterse et al., 1996, 2002; Ton et al., 2002a). Analysis of the jasmonic acid (JA) response mutants jar1-1 and coi1-1, a range of ethylene (ET)-response mutants, and the SAR-compromised mutant npr1-1 revealed that components of the JA and the ET response are required for triggering ISR and that this induced resistance response, like SAR, requires NPR1 (Pieterse et al., 1998; Knoester et al., 1999; Van Wees et al., 2000; M.J. Pozo and C.M.J. Pieterse, unpublished data). However, the ISR and the SAR signaling pathways diverge downstream of NPR1 because, unlike SAR, ISR is not marked by the transcriptional activation of PR genes (Pieterse et al., 1996; Van Wees et al., 1997, 1999).
To identify genes that mark the onset of ISR, the transcriptome of Arabidopsis was surveyed in roots and leaves upon colonization of the roots by ISR-inducing WCS417r bacteria (Verhagen et al., 2004). Systemically in the leaves, no consistent changes in gene expression were observed in response to effective colonization of the roots by WCS417r, indicating that, in contrast to SAR, the onset of WCS417r-mediated ISR in the leaves is not associated with a major reprogramming of the transcriptome. However, after challenge inoculation of the induced plants with Pst DC3000, 81 genes showed a potentiated expression in the leaves, suggesting that these genes were primed to respond faster and/or more strongly upon pathogen attack. The majority of the primed genes appeared to be regulated by JA and/or ET signaling. Priming of pathogen-induced genes allows the plant to react more effectively to a subsequent invader, which might explain the broad-spectrum effectiveness of rhizobacteria-mediated ISR (Conrath et al., 2002, 2006). In contrast to constitutive activation of defense responses, priming does not require major metabolic changes when no pathogens are present. Therefore, it forms a low-cost defense strategy while acting against a broad spectrum of attackers (Van Hulten et al., 2006; Pieterse and Dicke, 2007).
Whereas in the leaves no changes in gene expression were evident before challenge inoculation, roots responded to colonization by ISR-inducing WCS417r bacteria with significant changes in the expression of 97 genes (Verhagen et al., 2004). To investigate the biological role of the root-specific, WCS417r-inducible genes in the onset of ISR, we systematically started to analyze T-DNA insertion mutants of these genes. In this study, we demonstrate that the WCS417r-responsive gene MYB72, encoding a R2R3-MYB-like transcription factor protein, functions as an essential component during the early steps of the ISR signaling cascade in Arabidopsis. MYB72 is a member of the large R2R3-MYB gene family of which 125 members have been identified in Arabidopsis (Kranz et al., 1998; Stracke et al., 2001; Yanhui et al., 2006). R2R3-MYB transcription factors are implicated in the regulation of various plant processes, although the function of most of them is still unknown (Stracke et al., 2001).
RESULTS
Knockout Mutant myb72-1 Is Blocked in Rhizobacteria-Mediated ISR
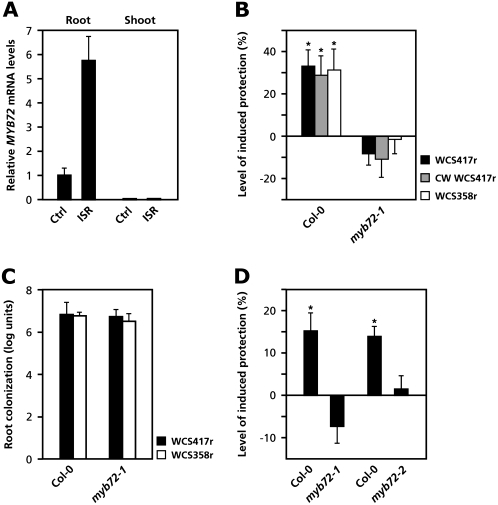
Previously, microarray analysis revealed a large set of genes that showed an altered expression pattern in the roots upon colonization by ISR-inducing WCS417r rhizobacteria (Verhagen et al., 2004). To investigate the role of these root-specific, WCS417r-induced genes in ISR signaling, we systematically analyzed knockout mutants of these genes for their ability to express WCS417r-ISR against Pst DC3000. A mutant with a T-DNA insertion in the MYB72 gene, which is specifically up-regulated in the roots upon colonization by WCS417r (Verhagen et al., 2004; Fig. 1A), was identified as being defective in the activation of ISR and was subjected to further detailed studies. Figure 1B shows that knockout mutant myb72-1 (SAIL_713G10) was unable to mount ISR against Pst DC3000 in response to colonization of the roots by WCS417r.
Figure 1.
ISR against Pst DC3000 is blocked in myb72 knockout mutants. A, Q-PCR analysis of MYB72 transcript levels in roots and shoots of Col-0 plants that were grown in soil for 2 weeks with or without ISR-inducing WCS417r bacteria. B, Levels of induced protection against Pst DC3000 in Col-0 and knockout mutant myb72-1. ISR was induced by growing the plants for 3 weeks in soil containing living ISR-inducing WCS417r or WCS358r bacteria, or crude cell wall material of WCS417r (CW WCS417r). Five-week-old plants were challenge inoculated with virulent Pst DC3000. Four days after challenge inoculation, the percentage of diseased leaves was assessed and the level of induced protection calculated on the basis of the reduction in disease symptoms relative to challenged, noninduced plants. The absolute proportions of diseased leaves in the control treatment were 53.9% (Col-0) and 50.7% (myb72-1). Asterisks indicate statistically significant differences compared to noninduced control plants (Student's t test, α < 0.05; n = 20). C, Numbers of rifampicin-resistant WCS417r or WCS358r bacteria (log10 of the number of cfu mL−1) in the rhizosphere of the plants at the end of the bioassay. In the rhizosphere of noninduced plants, no rifampicin-resistant bacteria were detected (detection limit 103 cfu g−1 root fresh weight). D, Quantification of WCS417r-induced protection against Pst DC3000 in wild-type Col-0 and knockout mutants myb72-1 and myb72-2. The level of induced protection was calculated as described above (B). The absolute proportions of diseased leaves in the control treatments were 78.0% (Col-0), 73.4% (myb72-1), 79.1% (Col-0), and 74.1% (myb72-2). Asterisks indicate statistically significant differences compared to noninduced control plants (Student's t test, α < 0.05; n = 20). All bioassays were repeated with similar results. Error bars represent ses.
Previously, rhizobacterial strain Pseudomonas putida WCS358r and a crude cell wall preparation of WCS417r were demonstrated to trigger the ISR signaling pathway in Arabidopsis, resulting in a similar level of induced protection against Pst DC3000 as ISR induced by live WCS417r bacteria (Van Wees et al., 1997). To find out whether ISR triggered by these inducers is also blocked in myb72-1, roots of Columbia-0 (Col-0) and myb72-1 plants were treated with killed WCS417r cells, or with living WCS358r bacteria, and tested for the expression of ISR. Col-0 plants treated with crude WCS417r cell wall material or living WCS358r bacteria both showed similar levels of protection against Pst DC3000 to that induced by live WCS417r cells (Fig. 1B). Knockout mutant myb72-1 was unable to mount ISR in response to any of the inducers, confirming that MYB72 is required for the onset of ISR by these activators.
To investigate whether the impaired ISR response of myb72-1 was caused by insufficient root colonization by the rhizobacterial strains, the number of rifampicin-resistant WCS417r and WCS358r bacteria per gram of root fresh weight was determined. No significant differences in the extent of root colonization between Col-0 and myb72-1 plants were observed (Fig. 1C). Thus, the inability of myb72-1 to express WCS417r-mediated ISR was not caused by reduced root colonization.
To confirm that the ISR-minus phenotype of knockout mutant myb72-1 was caused by disruption of the MYB72 gene, a second, independent T-DNA insertion mutant, designated myb72-2 (SALK_052993), was tested for its ability to express WCS417r-ISR. Figure 1D shows that myb72-2, like myb72-1, was unable to mount ISR against Pst DC3000, indicating that a functional MYB72 gene is required for the onset of WCS417r-ISR against this pathogen in Arabidopsis. Verification of the predicted T-DNA insertion sites in the myb72-1 and myb72-2 knockout mutants is described in Supplemental Figure S1. Because multiple experiments demonstrated that both mutants were impaired in their ability to mount WCS417r-ISR against Pst DC3000, mutant myb72-1 was used for further experiments.
Mutant myb72-1 Is Not Impaired in SAR or Resistance Induced by Methyl JA or ACC
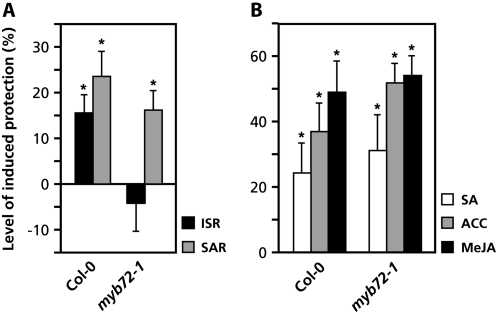
To investigate the effect of the myb72-1 mutation on pathogen-induced SAR, we compared the levels of rhizobacteria-mediated ISR and pathogen-induced SAR in this mutant. SAR was induced 3 d prior to challenge inoculation with virulent Pst DC3000 by infiltrating three lower leaves with avirulent Pst DC3000(avrRpt2). Wild-type Col-0 plants developed significant levels of protection against Pst DC3000 in response to induction of ISR and SAR (Fig. 2A). In contrast to ISR, SAR was expressed to wild-type levels in myb72-1, indicating that the ability to develop SAR was not altered in this mutant. Similarly, chemical induction of SAR by exogenous application of SA resulted in similar levels of protection against Pst DC3000 in Col-0 and myb72-1 (Fig. 2B), confirming that myb72-1 is not impaired in SAR.
Figure 2.
ISR, SAR, and chemically induced resistance against Pst DC3000 in Col-0 and myb72-1. A, Quantification of the level of ISR and SAR against Pst DC3000 in Col-0 and myb72-1 plants. ISR was induced by treatment of the roots with WCS417r bacteria. Induction of SAR was performed 3 d before challenge inoculation by pressure infiltrating three lower leaves with a suspension of avirulent Pst DC3000(avrRpt2) bacteria. The absolute proportions of diseased leaves in the control treatments were 72.8% (Col-0) and 75.1% (myb72-1). B, Level of induced resistance against Pst DC3000 in Col-0 and myb72-1 after exogenous application of either SA, ACC, or MeJA. Chemical inductions were performed by applying 1 mm SA, 1 mm ACC, or 0.1 mm MeJA as a soil drench 7 and 4 d prior to challenge inoculation with Pst DC3000. The absolute proportions of diseased leaves in the control treatments were 53.0% (Col-0) and 51.6% (myb72-1). For details on Pst DC3000 bioassays, see legend to Figure 1. Asterisks indicate statistically significant differences compared to noninduced control plants (Student's t test, α < 0.05; n = 20).
Like rhizobacteria-mediated ISR, exogenous application of methyl JA (MeJA) or the ET precursor 1-aminocyclopropane-1-carboxylate (ACC) triggers an enhanced level of resistance against Pst DC3000 (Van Wees et al., 1999). To examine the effect of the myb72-1 mutation on resistance induced by these chemicals, Col-0 plants were pretreated with MeJA or ACC at 7 and 4 d before challenge inoculation with virulent Pst DC3000. Figure 2B shows that myb72-1 developed wild-type levels of protection against Pst DC3000 in response to both chemicals, indicating the myb72-1 mutation has no effect on the ability to express enhanced resistance in response to ACC or MeJA. These findings suggest that MYB72 operates upstream of JA and ET in the ISR signaling pathway.
MYB72 Is Required for ISR against a Broad Spectrum of Pathogens
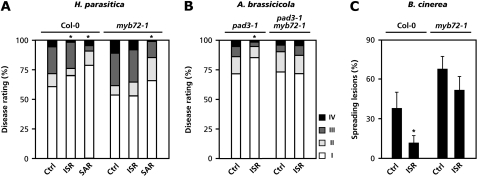
WCS417r-mediated ISR is effective against a broad spectrum of pathogens (Pieterse et al., 1996; Van Wees et al., 1997; Ton et al., 2002b). To examine whether MYB72 is required for the onset of broad-spectrum ISR, we tested the ability of myb72-1 to express ISR against the biotrophic oomycete H. parasitica and the necrotrophic fungi A. brassicicola and Botrytis cinerea. Figure 3A shows that WCS417r-ISR and benzothiadiazole (BTH)-induced SAR resulted in a relatively moderate, but statistically significant, level of protection of Col-0 plants against H. parasitica. Similar to the bioassays with Pst DC3000, mutant myb72-1 plants failed to develop ISR against this pathogen, whereas induction of SAR resulted in wild-type levels of induced resistance. To test the effectiveness of ISR against A. brassicicola, ISR bioassays were performed in the genetic background of the camalexin-deficient mutant pad3-1. In contrast to wild-type Col-0 plants, pad3-1 is susceptible to A. brassicicola infection and is routinely used in assays to test for induced resistance against this pathogen (Thomma et al., 1999; Ton et al., 2002b; Spoel et al., 2007). Figure 3B shows that induction of ISR in pad3-1 significantly reduced disease symptoms caused by A. brassicicola infection, whereas the pad3-1/myb72-1 double mutant failed to mount ISR against this pathogen. Similarly, Col-0 plants, but not myb72-1, expressed statistically significant levels of ISR against B. cinerea (Fig. 3C). Together, these results demonstrate that MYB72 is essential for ISR against different types of pathogens.
Figure 3.
ISR against H. parasitica, A. brassicicola, and B. cinerea is blocked in myb72-1. A, Quantification of ISR and SAR against H. parasitica. ISR was induced by growing the plants in soil containing ISR-inducing WCS417r bacteria. SAR was induced by applying 300 mm BTH as a soil drench 3 d before challenge. Plants were challenge inoculated with H. parasitica when 3 weeks old. Disease severity was determined 9 d after challenge. Disease ratings are expressed as the percentage of leaves (n = approximately 250) in disease-severity classes: I, no sporulation; II, trailing necrosis; III, <50% of the leaf area covered with sporangia; IV, >50% of the leaf area covered with sporangia, with additional chlorosis and leaf collapse. Asterisks indicate statistically significantly different distributions of the disease severity classes compared with the noninduced control treatments (χ2, α = 0.05). B, Quantification of ISR against A. brassicicola in pad3-1 and pad3-1/myb72-1. ISR was induced as described above. Plants were inoculated with A. brassicicola when 5 weeks old. Disease symptoms were determined 5 d after challenge. Disease ratings are expressed on the basis of symptom severity: I, no visible disease symptoms; II, nonspreading lesion; III, spreading lesion without tissue maceration; IV, spreading lesion with tissue maceration and sporulation of the pathogen. Asterisks indicate statistically significantly different distributions of the disease severity classes compared with the noninduced control treatments (χ2, α = 0.05; n = 120). C, Quantification of ISR against B. cinerea in Col-0 and myb72-1. ISR was induced as described above. Plants were inoculated with B. cinerea when 5 weeks old. Disease symptoms were determined 5 d after challenge. Disease ratings were expressed as percentage of leaves showing spreading lesions. Asterisks indicate statistically significant differences compared to noninduced control plants (Student's t test, α < 0.05; n = 20). Error bars represent ses. All experiments were repeated with similar results.
WCS417r-Induced Priming for Enhanced Callose Deposition Is Blocked in myb72-1
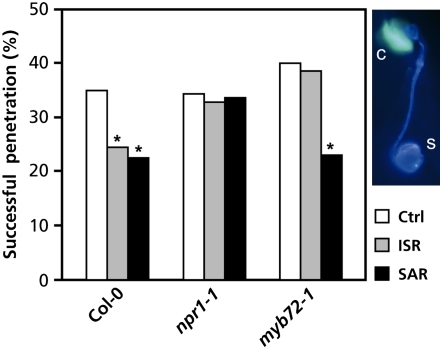
Induced resistance is often associated with priming for enhanced deposition of callose-containing papillae at sites of attempted pathogen attack or tissue injury (Kohler et al., 2002; Ton et al., 2005). Figure 4 shows that induction of ISR by WCS417r, or SAR by exogenous application of BTH, resulted in a significant decrease of the success rate of H. parasitica spores to penetrate the leaves of wild-type Col-0 plants due to enhanced callose depositions in the epidermal cell layer around the entry sites. Treated mutant npr1-1 plants that are blocked in their ability to express both ISR and SAR did not show this enhanced callose deposition at the sites of attempted pathogen attack and consequently did not display lower penetration success rates of the pathogen, confirming that priming for enhanced callose deposition is associated with ISR and SAR. Priming for enhanced callose deposition was normally expressed in myb72-1 upon induction of SAR with BTH, resulting in a significant decrease in successful spore penetration (Fig. 4). However, unlike BTH, WCS417r did not enhance callose deposition in myb72-1 in response to H. parasitica attack. Hence, although myb72-1 is not impaired in its ability to be primed for enhanced formation of callose-containing papillae, this defense-related trait is not expressed in myb72-1 in response to colonization of the roots by WCS417r bacteria. Together, these results indicate that MYB72 plays an important role in the onset of this primed defense response during ISR.
Figure 4.
Mutant myb72-1 is impaired in WCS417r-mediated priming for enhanced callose deposition at H. parasitica infection sites. Induced resistance against H. parasitica is associated with enhanced deposition of callose-containing papillae at sites of attempted penetration, resulting in a reduction of the number of spores that successfully penetrate into Arabidopsis leaves. Two days after challenge with H. parasitica, successful penetration of H. parasitica spores was quantified in leaves of Col-0, npr1-1, and myb72-1 plants. Leaves of plants of which the roots were pretreated with water (Ctrl), WCS417r (ISR), or BTH (SAR) were stained with calcofluor/aniline blue and analyzed by epifluorescence microscopy (UV). The inset shows a representative example of a germinating H. parasitica spore (s) triggering callose deposition (c) in the underlying epidermal cell. Callose-inducing spores were scored as unsuccessful penetrations, whereas those that did not trigger a callose response were scored as successful penetrations. The figure depicts the percentage of spores that successfully penetrated the host cell. Asterisks indicate statistically significant differences compared with the noninduced control treatments (χ2, α = 0.05; n = 150).
MYB72 Functions Upstream of or in Parallel with ET Signaling in the ISR Pathway
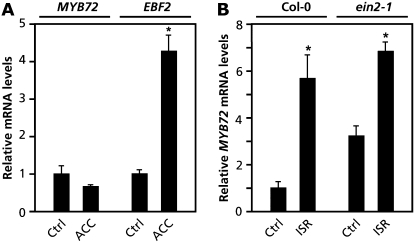
MYB72 transcripts accumulate in the roots upon colonization by WCS417r, whereas systemically in the leaves they are not detectable (Verhagen et al., 2004; Fig. 1A). Hence, MYB72 is likely to play a role in the early steps of the ISR signaling pathway. Previously, Knoester et al. (1999) demonstrated that, for the onset of ISR, ET signaling is required at the site of application of the ISR inducer. To investigate whether MYB72 gene expression is regulated by ET, MYB72 transcript levels were monitored in the roots by quantitative real-time PCR (Q-PCR) upon application of 0.1 mm ACC. Figure 5A shows that the ET-responsive gene EIN3 BINDING FACTOR2 (EBF2; Guo and Ecker 2003) is activated in ACC-treated roots. By contrast, MYB72 is not activated upon ACC treatment, indicating that MYB72 gene expression is not regulated by ET.
Figure 5.
MYB72 expression is not regulated by ET. A, Q-PCR analysis of MYB72 transcript levels in the roots of 5-week-old Col-0 plants of which the roots were treated with a soil drench of water (Ctrl) or 0.1 mm ACC. To check the effectiveness of the ACC treatment, expression levels of the ET-responsive EBF2 gene were checked in the same samples. B, Q-PCR analysis of MYB72 transcript levels in the roots of Col-0 and ein2-1 plants 2 weeks after transfer of the seedlings to soil containing WCS417r bacteria (ISR) or not (Ctrl). Transcript levels in water-treated control plants were set at 1. Asterisks indicate statistically significant differences compared to noninduced control plants (Student's t test, α < 0.05).
To test whether WCS417r-induced expression of MYB72 in the roots requires ET sensitivity, MYB72 transcript accumulation was examined in the ET-insensitive, ISR-minus mutant ein2-1. Colonization of the roots by WCS417r bacteria activated MYB72 equally in both Col-0 and ein2-1 (Fig. 5B). These results indicate that MYB72 either acts upstream of ET signaling or is corequired with components from the ET signaling pathway during the onset of ISR.
MYB72 Is Required But Not Sufficient for ISR
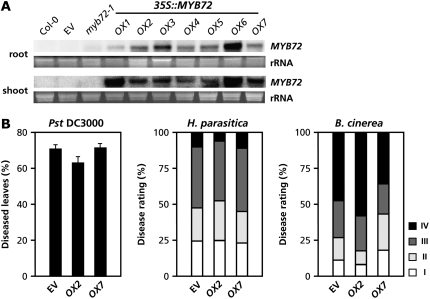
To investigate whether MYB72 is not only required but also sufficient for the onset of ISR, transgenic plants that constitutively express MYB72 (35S:MYB72) were generated and tested for enhanced disease resistance. Seven independent homozygous T3 lines (OX1–OX7) were phenotypically characterized. All transgenic lines displayed a phenotype that was similar to the parental Col-0 line and the empty vector (EV) control (Supplemental Fig. S2). RNA-blot analysis of roots and shoots of the 35S:MYB72 lines confirmed constitutive expression of MYB72 in all lines, albeit to varying levels (Fig. 6A). Bioassays for induced resistance assays were performed with Col-0, the EV control, and the seven 35S:MYB72 transgenic lines. Figure 6B shows that the level of basal resistance against Pst DC3000, H. parasitica, and B. cinerea in lines OX2 and OX7 was not significantly enhanced compared to that from the EV control line. Resistance assays with the other OX lines yielded similar results (data not shown), indicating that ectopic expression of MYB72 is not sufficient for the onset of ISR.
Figure 6.
35S:MYB72 overexpressors do not show enhanced levels of disease resistance. A, RNA gel-blot analysis of MYB72 transcript levels in roots and shoots of 5-week-old wild-type Col-0 plants and the mutant and transgenic lines EV (EV control), myb72-1, and 35S:MYB72-OX1 to -OX7. To check for equal loading, rRNA bands were stained with ethidium bromide. B, Levels of disease severity in transgenic EV, OX2, and OX7 upon inoculation with Pst DC3000, H. parasitica, and B. cinerea. 35S:MYB72 overexpressing lines OX1, OX3, OX4, OX5, and OX6 displayed similar levels of disease severity as OX2 and OX7 (data not shown). For details on pathogen bioassays, see legends to Figures 1 and 3.
MYB72 Physically Interacts with EIL3 in Vitro
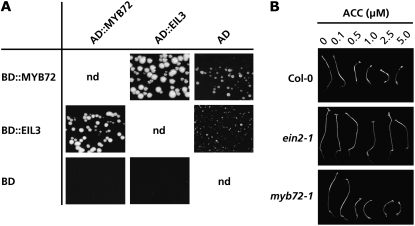
If MYB72 is essential but not sufficient for the onset of ISR, then additional components are likely to be corequired. Transcription factors usually exert their action in a complex with other proteins. Earlier, a systematic search for proteins that physically interact with MYB transcription factors was initiated by members of the EU-funded Regulatory Gene Initiative in Arabidopsis consortium. Using the ProQuest yeast two-hybrid system (Invitrogen), this screen revealed that MYB72 physically interacts with the ETHYLENE INSENSITIVE3 (EIN3)-like protein EIL3 (At1g73730; data not shown). To confirm the interaction of MYB72 with EIL3, the full-length coding regions of MYB72 and EIL3 were isolated, fused to the DNA binding domain (BD) and transcription activation domain (AD) of GAL4, and tested in a yeast two-hybrid assay. Figure 7A shows that cells containing the BD:MYB72 fusion with the AD:EIL3 fusion, and cells containing both the BD:EIL3 and the AD:MYB72 fusion were capable of growth on selective dropout (SD) medium to which 100 mm of the His biosynthesis inhibitor 3-amino-1,2,4-triazole (3-AT) was added. Cells containing either AD:MYB72 or AD:EIL3 in combination with GAL4 BD fused to the full-length Arabidopsis FRIGIDA protein (BD:FRI; Rutjens, 2007), which were used as negative controls, did not grow on the selective medium. Cells containing either the BD:MYB72 or the BD:EIL3 fusion with the empty GAL4 AD vector showed a low level of growth, indicating a low level of auto activation of the reporter gene in these combinations. Together, these results indicate that MYB72 and EIL3 interact in vitro. The EIL3 gene is expressed in the roots to similar levels as the constitutively transcribed ubiquitin gene UBI10 (Supplemental Fig. S3). Although EIL3 mRNA levels slightly rose in the roots upon colonization by WCS417r, this rise was not statistically significant. Coexpression of MYB72 and EIL3 makes the MYB72-EIL3 interaction in planta theoretically feasible. However, future studies on the interaction of MYB72 and EIL3 in planta should shed light on the occurrence and significance of this interaction in the onset of ISR.
Figure 7.
MYB72 interacts with EIL3 in a yeast two-hybrid assay. A, Yeast two-hybrid assay of interactions between the transcription factors MYB72 and EIL3 fused to the GAL4 DNA BD or transcriptional AD domain. Yeast cells (PJ69-4) containing either DB:MYB72 and AD:EIL3, or DB:EIL3 and AD:MYB72, or either of these fusions together with the vector control were grown on yeast SD medium. To suppress His formation that results from autoactivated HIS3-reporter gene activity, 100 mm of the His biosynthesis inhibitor 3-AT was added to the medium and yeast cells were grown for 3 d at 20°C. B, Triple response assay of etiolated Col-0, ein2-1, and myb72-1 seedlings grown for 7 d in the dark at 20°C on Murashige and Skoog agar containing 0, 0.1, 0.5, 1.0, 2.5, or 5.0 mm ACC.
The EIL3 paralogs EIN3, EIL1, and EIL2 have been demonstrated to function as key transcription factors of ET-regulated gene expression and to act as positive regulators of ET signaling (Stepanova and Ecker, 2000). Because MYB72 acts upstream of ET signaling or is corequired with components of the ET signaling pathway during the onset of ISR (Fig. 5), we investigated whether the ISR-minus phenotype of the myb72-1 knockout mutant is caused by a reduced sensitivity to ET. The triple response is a reaction of etiolated seedlings to ET and is commonly used as a reliable marker for ET sensitivity (Guzmán and Ecker, 1990). Etiolated Col-0, ET-insensitive ein2-1, and myb72-1 seedlings were grown in the dark on Murashige and Skoog agar plates with or without ACC. Ten days after germination, Col-0 seedlings grown on a concentration range of ACC showed a typical ET-induced growth inhibition of the hypocotyl and root, both characteristics of the triple response (Fig. 7B). As expected, the triple response was not apparent in the ET-insensitive ein2-1 seedlings. In contrast, in mutant myb72-1 seedlings, the triple response was indistinguishable from that in wild-type Col-0 plants. These results demonstrate that the absence of a functional MYB72 protein does not affect ET sensitivity. Hence, the inability of myb72-1 plants to mount ISR is not caused by an inability to react to ET in this mutant.
DISCUSSION
Colonization of the roots of Arabidopsis by nonpathogenic fluorescent Pseudomonas bacteria, such as WCS417r and WCS358r, leads to an enhanced level of resistance against a broad spectrum of pathogens in foliar tissues (Pieterse et al., 2002). Genes whose expression is changed in the roots upon colonization by ISR-inducing rhizobacteria are potentially involved in the onset of ISR. Here, we demonstrate the role of one of these genes, MYB72, in the onset of rhizobacteria-mediated ISR.
MYB72 Belongs to a Transcription Factor Family of Which Several Members Are Involved in Stress Signaling
MYB72 is a member of a large class of genes that contain one or more MYB domains (Stracke et al., 2001). MYB genes were first identified as oncogenes derived from retroviruses in animal cells (Klempnauer et al., 1982). They encode transcription factor proteins that share the conserved MYB DNA BD (Jin and Martin, 1999). MYB proteins are categorized into subfamilies depending on the number of conserved repeats of the MYB domain. The ones from animals generally contain three MYB repeats, which are referred to as R1, R2, and R3. Most of the MYB-like genes in plants have only the R2 and R3 repeats (Kranz et al., 1998). According to an inventory of the Arabidopsis genome, MYB72 is one of approximately 125 genes that encode a putative R2R3-MYB protein in this plant species (Stracke et al., 2001). Gene expression analyses suggest a role for many R2R3-MYB proteins in a range of activities, such as plant secondary metabolism, development, regulation of cell death, stress tolerance, and pathogen resistance (Stracke et al., 2001; Yanhui et al., 2006). However, the biological functions of most of the MYB-like transcription factors have not been determined.
Within Arabidopsis, the MYB72 protein was found to possess highest homology with MYB10, MYB58, and MYB63 (Stracke et al., 2001), of which the functions are currently unknown. Alignment of the R2R3 domain of MYB72 with amino acid sequences of other plant species in the databases further revealed high homology with the MYB-like transcription factor protein OsLTR1 from rice (Oryza sativa; 75% identity; GenBank accession no. AAP92750), which has been implicated in JA-dependent defense responses (National Center for Biotechnology Information database locus information), and ZmMRP1 from maize (Zea mays; 75% identity; GenBank accession no. S04898; Supplemental Fig. S4). Besides MYB72, several other Arabidopsis MYB-like transcription factors have also been implicated to function in biotic or abiotic stress signaling. Mengiste et al. (2003) identified BOS1 (MYB108) and demonstrated a role for this protein in resistance against necrotrophic pathogens. Outside the conserved R2R3 domain, the amino acid sequence of MYB72 has no significant homology with that of BOS1, suggesting that both MYB transcription factors are not functionally related. This is confirmed by our observation that, in contrast to BOS1, overexpression of MYB72 does not result in enhanced resistance to necrotrophic pathogens (Fig. 6B). Other Arabidopsis MYB-like transcription factors have been demonstrated to function in drought stress responses (MYB2; Abe et al., 2003), hypersensitive cell death (MYB30; Vailleau et al., 2002), production of defense-related glucosinolates (MYB34/ATR1; Celenza et al., 2005), the wound response (MYB15; Cheong et al., 2002), and defense against insect herbivory (MYB102; De Vos et al., 2006). Also, in other plant species, MYB-like transcription factors play a role in the regulation of stress responses. For example, tobacco NtMYB2 was shown to positively regulate the expression of the PHE AMMONIA LYASE gene in response to wounding and elicitor treatment (Sugimoto et al., 2000), whereas rice OsMYB4 was shown to function as a key regulator in cold tolerance (Vannini et al., 2004).
Specificity of MYB72 Gene Expression
Previously, Kranz et al. (1998) analyzed the expression patterns of a large set of Arabidopsis MYB genes in different plant organs and under various conditions, such as treatment with various hormones, exposure to abiotic stress, and infection by Pst DC3000. In this study, MYB72 transcripts were not detected in any of the organs or conditions tested. Analysis of the Arabidopsis transcriptome using the Arabidopsis microarray database and analysis toolbox GENEVESTIGATOR (Zimmermann et al., 2004) confirmed that MYB72 is not activated in response to any of the hormones or biological agents tested (data not shown), suggesting that the induction of MYB72 in the roots upon colonization by nonpathogenic rhizobacteria is highly specific. However, low iron conditions or treatment of Arabidopsis roots with an excess amount of zinc do induce the expression of MYB72 in the roots (Colangelo and Guerinot, 2004; Van de Mortel et al., 2008). Studies by Thomine et al. (2003) and Van de Mortel et al. (2008) demonstrated that high zinc availability distorts iron uptake by the plant, thereby mimicking the iron-limiting conditions that activate MYB72. Interestingly, fluorescent Pseudomonas spp., such as WCS417r and WCS358r, produce large quantities of iron-chelating siderophores that facilitate the uptake of iron by the bacteria, thereby depriving their direct vicinity from iron (Bakker et al., 2007). Hence, it is not inconceivable that the induction of MYB72 is caused by enhanced iron stress that is inflicted in the roots upon colonization by the ISR-inducing rhizobacteria. This can be supported by the fact that both iron deprivation (Robinson et al., 1999) and colonization of the roots by WCS417r (Verhagen et al., 2004) lead to induction of the expression of FERRIC REDUCTION OXIDASE2 (FRO2; Robinson et al., 1999), a gene encoding a protein that plays a crucial role in the uptake of iron by the roots.
MYB72 Is Required in Early ISR Signaling
Previously, Knoester et al. (1999) demonstrated that ET signaling is required in the roots for the expression of ISR in the leaves. It was shown that mutant eir1-1, which is insensitive to ET in the roots only (Roman et al., 1995), develops no ISR when WCS417r bacteria were applied to the roots, but showed normal levels of ISR when WCS417r bacteria were infiltrated into the leaves. If ET signaling were required only for expression of ISR at the site of challenge inoculation, eir1-1 plants would develop normal levels of ISR in the leaves after application of WCS417r to the roots. However, this was not the case. Thus, ET signaling is required locally at the site of application of the inducer and may be involved in the generation or translocation of the systemically transported signal. Here, we demonstrated that WCS417r-induced expression of MYB72 is not regulated by ET because mutant ein2-1 plants accumulated normal levels of MYB72 transcripts in the roots upon treatment with WCS417r (Fig. 5B). In addition, the expression of MYB72 was not activated upon treatment of the roots with ACC (Fig. 5A). Together, these results demonstrate that MYB72 either acts upstream of ET or is corequired with components from the ET signaling pathway during the onset of ISR in the roots.
MYB72 Is Not Sufficient for the Onset of ISR
Although MYB72 is required for the onset of ISR, overexpression of the MYB72 gene did not result in enhanced disease resistance (Fig. 6). Hence, another signaling component is likely to be corequired for the expression of ISR. Yeast two-hybrid experiments revealed that MYB72 physically interacts with EIL3, a member of the EIN3 family of transcription factors (Fig. 7A). EIN3 and its closest paralogs, the EIN3-like proteins EIL1 and EIL2, are key transcription factors of ET-regulated gene expression and act as positive regulators of ET signaling (Stepanova and Ecker, 2000; Tieman et al., 2001; Guo and Ecker, 2004). They bind to promoters of ET-responsive genes, such as the regulatory gene ETHYLENE RESPONSE FACTOR1 (ERF1), and initiate a transcriptional cascade leading to the expression of ET-targeted genes (Chao et al., 1997; Solano et al., 1998). If EIL3 would function similarly as EIN3, EIL1, and EIL2 in ET signaling, then physical interaction with MYB72 may facilitate the as-yet-unidentified ET signaling event that is corequired with MYB72 in the roots for the onset of WCS417r-mediated ISR in the leaves. Besides a potential role in ET signaling, EIL3 (also called SLIM1 for SULFUR LIMITATION1) was recently shown to function as an important transcriptional regulator in the response of Arabidopsis to sulfur deprivation (Maruyama-Nakashita et al., 2006). Collectively, these data highlight that both MYB72 and EIL3 are part of the signaling network involved in the plant's response to biotic and abiotic stresses. Physical interaction between MYB72 and EIL3 in planta and its significance for the onset of ISR remains to be demonstrated and will be the subject of future study.
Model for ISR Signal Transduction
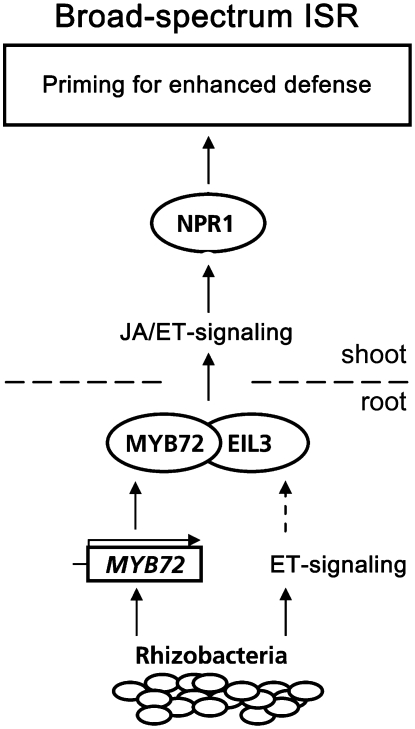
The identification of MYB72 as an important signaling component in the roots for the systemic onset of ISR adds a new factor to ISR signal transduction. Figure 8 summarizes our current understanding of the ISR signaling pathway. The local onset of WCS417r-ISR in the roots requires responsiveness to ET (Knoester et al., 1999) and is associated with an ET-independent activation of the MYB72 gene. MYB72 is required but not sufficient for the onset of ISR. Hence, MYB72 is assumed to act in concert with another signaling component. Because EIL3 interacts with MYB72 in vitro, EIL3 is a potential candidate in this respect. Systemically, in the leaves, expression of ISR requires responsiveness to both JA and ET and is dependent on NPR1 (Pieterse et al., 1998). The induced state of WCS417r-mediated ISR is not associated with major changes in defense-related gene expression (as opposed to SAR; Verhagen et al., 2004). Instead, ISR-expressing plants are primed to express a specific set of JA/ET-responsive genes faster and to a higher level upon pathogen infection (Van Wees et al., 1999; Hase et al., 2003; Verhagen et al., 2004). This enhanced defensive capacity allows the plants to respond faster and/or more strongly to attackers that trigger JA/ET-dependent defense responses without major metabolic changes in the absence of an intruder (Conrath et al., 2006). Therefore, ISR forms a low-cost defense strategy that is active against a broad spectrum of attackers (Van Hulten et al., 2006).
Figure 8.
Proposed model for the role of MYB72 in the signal transduction pathway controlling rhizobacteria-mediated ISR. Colonization of the roots by ISR-inducing P. fluorescens WCS417r leads to a local, ET-independent activation of the transcription factor gene MYB72. Downstream of, or in parallel with, MYB72, an as-yet-unidentified ET signaling component is required in the roots for the onset of broad-spectrum ISR in the leaves. Because EIL3 interacts with MYB72 in vitro, EIL3 is a potential candidate in this respect. Systemically, the ISR signal transduction cascade requires responsiveness to both JA and ET and is dependent on NPR1. Finally, induction of ISR is associated with priming for enhanced expression of a large set of JA- and ET-responsive genes that becomes apparent only after pathogen attack. This allows the plant to react more effectively to an invading pathogen, which may explain the broad-spectrum characteristic of rhizobacteria-mediated ISR.
MATERIALS AND METHODS
Cultivation of Rhizobacteria and Pathogens
Nonpathogenic, rifampicin-resistant Pseudomonas fluorescens WCS417r and Pseudomonas putida WCS358r bacteria were used for induction of ISR (Van Wees et al., 1997). Both strains were grown for 24 h at 28°C on King's medium B (KB) agar plates (King et al., 1954), as described previously (Pieterse et al., 1996). Crude cell wall material of WCS417r was prepared according to Van Wees et al. (1997). Colonization of the rhizosphere of wild-type and mutant plants by rifampicin-resistant WCS417r and WCS358r bacteria was examined at the end of each ISR bioassay as described (Pieterse et al., 1996).
An avirulent strain of Pseudomonas syringae pv tomato DC3000, carrying the avirulence gene avrRpt2 [Pst DC3000(avrRpt2); Kunkel et al., 1993], was used for SAR induction. Pst DC3000(avrRpt2) bacteria were grown overnight at 28°C in liquid KB medium supplemented with 25 mg mL−1 kanamycin. Virulent Pst DC3000 (Whalen et al., 1991) was used for challenge inoculations and cultivated in a similar manner in liquid KB medium without kanamycin. After centrifugation, bacterial cells were resuspended in 10 mm MgSO4, 0.015% (v/v) Silwet L-77 (Van Meeuwen Chemicals) to a final density of 2.5 × 107 cfu mL−1.
Hyaloperonospora parasitica strain WACO9 was maintained on susceptible Col-0 plants as described by Koch and Slusarenko (1990). Sporangia were obtained by washing leaves that were densely covered by sporangiophores in distilled water, collected by centrifugation, and resuspended in water to a final density of 5 × 104 cfu mL−1.
Alternaria brassicicola strain MUCL20297 was grown on potato dextrose agar (PDA; Difco Laboratories) plates containing penicillin (100 μg mL−1) and streptomycin (200 μg mL−1) for 2 weeks at 22°C. Conidia were harvested as described by Broekaert et al. (1990) and resuspended in water to a final density of 1 × 106 spores mL−1.
Botrytis cinerea strain B0510 was grown on one-half-strength PDA plates containing penicillin (100 μg mL−1) and streptomycin (200 μg mL−1) for 2 weeks at 22°C. Spores were collected and resuspended in one-half-strength potato dextrose broth (Difco Laboratories) to a final density of 5.5 × 105 spores mL−1. After a 3-h incubation period, the spores were used for inoculation of plants as described (Thomma et al., 1998).
Plant Growth Conditions
Seeds of wild-type Arabidopsis (Arabidopsis thaliana) Col-0, mutants ein2-1 (Guzmán and Ecker, 1990), npr1-1 (Cao et al., 1994), pad3-1 (Zhou et al., 1999), myb72-1 (SAIL_713G10), myb72-2 (SALK_052993), the double mutant pad3-1/myb72-1, the transgenic lines 35S:MYB72-OX1 to -OX7, and the corresponding EV control line EV were sown in quartz sand. Two-week-old seedlings were transferred to 60-mL pots containing a sand-potting soil mixture that had been autoclaved twice for 20 min with a 24-h interval. For ISR bioassays, a suspension of ISR-inducing WCS417r or WCS358r bacteria (109 cfu mL−1) had been mixed thoroughly through the soil to a final density of 5 × 107 cfu g−1, as described previously (Pieterse et al., 1996). Control soil was supplemented with an equal volume of 10 mm MgSO4. Plants were cultivated in a growth chamber with a 9-h day (200 μE m−2 s−1 at 24°C) and a 15-h night (20°C) cycle at 70% relative humidity. Plants were supplied with modified one-half-strength Hoagland nutrient solution (Hoagland and Arnon, 1938) once a week, as described (Pieterse et al., 1996).
Knockout Mutants
Homozygous knockout mutants SAIL_713G10 (Sessions et al., 2002), designated myb72-1, and SALK_052993 (Alonso et al., 2003), designated myb72-2, containing a T-DNA insertion in the MYB72 gene (At1g56160), were grown as described above. Confirmation of the T-DNA insert in SAIL_713G10 was obtained by PCR on genomic DNA using T-DNA left border primer T1 and MYB72-specific primers MYB72-F1 and MYB72-R1 following a procedure that was described previously (Sessions et al., 2002). Mutant SALK_052993 was checked for the presence of T-DNA using primer T2, and MYB72-specific primers MYB72-F2 and MYB72-R2. The primers used are listed in Supplemental Table S1. The exact insertion sites of the T-DNAs in myb72-1 and myb72-2 were determined by DNA sequencing of the PCR products.
Construction of Transgenic Plants
A fusion product of the cauliflower mosaic virus 35S promoter and the coding region of MYB72 was created by double-joint PCR as described by Yu et al. (2004). The resulting 35S:MYB72 fusion product was cloned into the binary vector pGreenII229 (Hellens et al., 2000). A derivative of the pGreenII229 vector was used as the EV control. Correct construction of the plasmids was verified by DNA sequencing. Subsequently, the binary vectors were transferred to Agrobacterium tumefaciens strain C58(pMP90) (Konzc and Schell, 1986), after which Arabidopsis Col-0 plants were transformed according to the floral-dip method (Clough and Bent, 1998). Transformants were selected by spraying T1 progeny with BASTA Finale SL14 (Bayer CropScience BV) according to the manufacturer's instructions. The resulting MYB72-overexpressing lines 35S:MYB72-OX1 to -OX7 were selfed and homozygous T3 lines were selected for use in disease resistance assays.
Induction Treatments
Induction of ISR with living rhizobacteria was performed by mixing ISR-inducing rhizobacteria through the soil as described above. For tests with killed cells, a crude cell wall preparation of WCS417r bacteria (in 10 mm MgSO4) was mixed through the soil in a similar manner, using the equivalent of the number of live bacteria introduced to the soil (5 × 107 cfu g−1). Seven days before challenge inoculation, a similar amount of the crude cell wall material was applied to each plant as a soil drench as described previously (Van Wees et al., 1997).
Biological induction of SAR was performed 3 d before challenge inoculation by pressure infiltrating three lower leaves with a suspension of Pst DC3000(avrRpt2) bacteria at 107 cfu mL−1, as described (Pieterse et al., 1996).
For chemical induction of resistance, treatments were performed 7 and 4 d prior to challenge inoculation with Pst DC3000, or 3 d before challenge with H. parasitica WACO9. For the Pst DC3000 bioassays, the soil was drenched with 10 mL of either 100 μm MeJA, 1 mm ACC, or 1 mm SA. For SAR induction in the H. parasitica bioassays, the soil was drenched with 6 mL of 300 μm BTH. Control plants were treated with an equal volume of water. For induction of SAR in the H. parasitica bioassays, the functional SA analog BTH is routinely used instead of SA. The reason for this is the fact that, in our experimental setup, the young seedlings used in these assays are relatively sensitive to phytotoxic effects of SA. MeJA was purchased from Serva Brunschwig, ACC from Sigma-Aldrich Chemie BV, SA from Malinckrodt Baker BV, and BTH (BION) from CIBA-GEIGY GmbH.
Disease Resistance Assays
Pst DC3000 bioassays were performed essentially as described by Pieterse et al. (1996). Plants were challenged when 5 weeks old by dipping the leaves for 2 s in a solution of 10 mm MgSO4, 0.015% (v/v) Silwet L-77 containing 2.5 × 107 cfu mL−1 Pst DC3000 bacteria. Four days after challenge, disease severity was assessed by determining the percentage of diseased leaves per plant. Leaves were scored as diseased when showing necrotic or water-soaked lesions surrounded by chlorosis. The disease index was calculated by determining the proportion of leaves with disease symptoms per plant (n = 20).
H. parasitica bioassays were performed as described by Ton et al. (2002b). Three-week-old Arabidopsis Col-0, npr1-1, and myb72-1 plants were misted with a H. parasitica spore suspension containing 5 × 104 sporangiospores mL−1. Disease symptoms were scored at 9 d after inoculation for about 250 leaves per treatment. Disease ratings were expressed as severity of disease symptoms and pathogen sporulation on each leaf: I, no sporulation; II, trailing necrosis; III, <50% of the leaf area covered by sporangia; IV, >50% of the leaf area covered with sporangia, with additional chlorosis and leaf collapse. Quantification of the proportion of callose-inducing and noninducing spores was performed 2 d after inoculation as described by Ton et al. (2005). Inspection of the leaves was performed with an epifluorescence microscope containing a UV filter (bandpass 340–380 nm, longpath 425 nm). Callose-inducing spores were scored as unsuccessful penetrations, whereas those that did not trigger a callose response were scored as successful penetrations.
A. brassicicola bioassays were performed as described (Ton et al., 2002b). Because Arabidopsis Col-0 is resistant to A. brassicicola, whereas the camalexin-deficient mutant pad3-1 (Zhou et al., 1999) is susceptible (Thomma et al., 1999; Ton et al., 2002b), the role of MYB72 was investigated in the double mutant pad3-1/myb72-1, created through genetic crossing. When 5 weeks old, homozygous pad3-1 and pad3-1/myb72-1 plants (n = 20) were challenge inoculated with A. brassicicola by applying 3-μL droplets of water containing 1 × 106 spores mL−1 onto the second, third, and fourth true leaf pair of each plant. At 5 d after challenge, disease severity was determined. Disease rating was expressed on the basis of symptom severity: I, no visible disease symptoms; II, nonspreading lesion; III, spreading lesion without tissue maceration; IV, spreading lesion with tissue maceration and sporulation of the pathogen.
B. cinerea inoculations were performed with 5-week-old Col-0 and myb72-1 plants (n = 20) by applying 5-μL droplets of the spore suspension onto fresh needle-prick wounds on the second, third, and fourth true leaf pair of each plant as described (Thomma et al., 1998). At 5 d after challenge, disease severity was determined. Disease ratings were expressed as the percentage of leaves showing spreading lesions.
Gene Expression Analysis
Extraction of total RNA, RNA gel-blot analysis with a gene-specific probe for MYB72 (At1g56160), was performed as described (Van Wees et al., 2000; Verhagen et al., 2004).
Q-PCR was performed essentially as described by Czechowski et al. (2004). To check for genomic DNA contamination, a PCR with primers designed on EIL2 (At5g21120; EIL2-F and EIL2-R) was carried out. Efficiency of cDNA synthesis was assessed by Q-PCR, using primers of the constitutively expressed gene UBI10 (At4g05320; UBI10-F and UBI10-R). Primers for MYB72 (At1g56160; MYB72-F4 and MYB72-R4), EIL3 (At1g73730; EIL3-F and EIL3-R), and the ET-responsive gene EBF2 (At5g25350; EBF2-F and EBF2-R) were designed and checked as described by Czechowski et al. (2004). Nucleotide sequences of all primers are given in Supplemental Table S1.
Q-PCR analysis was performed in optical 96-well plates with a MyIQ Single Color Real-Time PCR detection system (Bio-Rad), using SYBR Green to monitor double-stranded DNA (dsDNA) synthesis. Each reaction consisted of 1 μL of cDNA, 0.5 μL of each of the two gene-specific primers (10 pmol μL−1), and 3.5 μL 2× IQ SYBR Green Supermix reagent (Bio-Rad) in a final volume of 15 μL. The following PCR program was used for all PCR reactions: 95°C for 3 min; 40 cycles of 95°C for 30 s, 59.5°C for 30 s, and 72°C for 30 s. Threshold cycle (CT) values were calculated using Optical System Software, version 1.0, for MyIQ (Bio-Rad). Subsequently, CT values were normalized for differences in dsDNA synthesis, using those of the constitutively expressed reference gene At1g13320 as described (Czechowski et al., 2005), after which the fold differences in transcript levels were calculated.
Yeast Two-Hybrid Assays
Constructs for yeast two-hybrid analyses were generated using vectors pDEST32 and pDEST22 (Invitrogen) for protein fusions to the GAL4 DNA BD or transcriptional AD, respectively. Full-length coding regions of MYB72 and EIL3 cDNA were introduced in both vectors using GATEWAY technology (Invitrogen), following the manufacturer's instructions. Clones containing the BD:MYB72, AD:MYB72, BD:EIL3, and AD:EIL3 fusions were checked by sequence analysis and subsequently used in the yeast two-hybrid assay. AD and BD plasmids were transformed into the a and α mating types of Saccharomyces cerevisiae strain PJ69-4, using a lithium acetate/polyethylene glycol protocol described by Gietz and Woods (2006). PJ69-4 carries ADE2, HIS3, URA, and LacZ reporters for reconstituted GAL4 activity (James et al., 1996). Transformants were selected on yeast-selective drop-out (SD) medium lacking either Leu (−leu) for selection of the BD vectors or Trp (−trp) for selection of the AD vectors. Opposite mating types were cocultured overnight on nutrient-rich YAPD medium (Sigma-Aldrich Chemie BV) at 30°C. Diploids harboring both plasmids were selected on SD medium lacking both Leu and Trp (−leu, −trp) and used in the yeast two-hybrid assay. Autoactivation levels of yeast transformants harboring either BD:MYB72 or BD:EIL3 were determined using SD medium lacking Leu, adenine, uracil, and His (−leu, −ade, −ura, −his), to which 0, 10, 25, 50, 75, or 100 mm of the His biosynthesis inhibitor 3-AT was added (Durfee et al., 1993). Because autoactivation could not be suppressed fully when using 100 mm 3-AT, only this concentration was used in the assay. The yeast two-hybrid assay of interactions between MYB72 and EIL3 was performed by growing the yeast strains with the different two-hybrid combinations on selective sd medium (−leu, −trp, −ade, −ura, −his, +3-AT) for 3 d at 20°C to reduce background growth levels.
ACC-Induced Triple Response
Seeds of Arabidopsis were surface sterilized for 5 min in 5% (v/v) sodium hypochlorite, washed in 70% (v/v) ethanol, and air dried. Seeds were subsequently distributed evenly on 1.0% (w/v) agar medium (pH 5.7) containing 0.5% (w/v) Murashige and Skoog salts (Duchefa BV), 0.5% (w/v) Suc, and different concentrations of filter-sterilized ACC, which was added from a 10 mm stock solution. The effect of ACC-derived ET on hypocotyl and primary root length in etiolated seedlings was determined essentially according to Guzmán and Ecker (1990). After pregermination in the dark for 2 d at 4°C, seedlings were grown for an additional 3 to 7 d at 20°C in darkness after which the triple response was monitored.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization of T-DNA knockout mutants myb72-1 and myb72-2.
Supplemental Figure S2. Morphological phenotype of transgenic 35S:MYB72 lines.
Supplemental Figure S3. Q-PCR analysis of EIL3 mRNA levels in roots of Arabidopsis Col-0 plants grown in soil with or without ISR-inducing P. fluorescens WCS417r bacteria.
Supplemental Figure S4. Alignment of AtMYB72 with other R2R3-MYB transcription factor proteins.
Supplemental Table S1. Nucleotide sequences of the primers used in this study.
Supplementary Material
Acknowledgments
We thank Hans van Pelt, Hans van Aken, and Dr. Hannes Lans for technical assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Corné M.J. Pieterse (c.m.j.pieterse@uu.nl).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Bakker PAHM, Pieterse CMJ, Van Loon LC (2007) Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology 97 239–243 [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Terras FRG, Cammue BPA, Vanderleyden J (1990) An automated quantitative assay for fungal growth. FEMS Microbiol Lett 69 55–60 [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J (2005) The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol 137 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89 1133–1144 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Chang H-S, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U, Beckers GJM, Flors V, García-Agustín P, Jakab G, Mauch F, Newman M-A, Pieterse CMJ, Poinssot B, Pozo MJ, et al (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19 1062–1071 [DOI] [PubMed] [Google Scholar]
- Conrath U, Pieterse CMJ, Mauch-Mani B (2002) Priming in plant-pathogen interactions. Trends Plant Sci 7 210–216 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38 366–379 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer G, Höfte M (1997) Salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 induces resistance to leaf infection by Botrytis cinerea on bean. Phytopathology 87 588–593 [DOI] [PubMed] [Google Scholar]
- De Vos M, Denekamp M, Vuylsteke M, Van Loon LC, Smeekens SCM, Pieterse CMJ (2006) The Arabidopsis thaliana transcription factor AtMYB102 functions in defense against the insect herbivore Pieris rapae. Plant Signal Behav 1 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X (2004) NPR1, all things considered. Curr Opin Plant Biol 7 547–552 [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev 7 555–569 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (2006) Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol Biol 313 107–120 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7 40–49 [DOI] [PubMed] [Google Scholar]
- Guo HW, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF (EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115 667–677 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase S, Van Pelt JA, Van Loon LC, Pieterse CMJ (2003) Colonization of Arabidopsis roots by Pseudomonas fluorescens primes the plant to produce higher levels of ethylene upon pathogen infection. Physiol Mol Plant Pathol 62 219–226 [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42 819–832 [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI (1938) The water culture method for growing plants without soil. Calif Agric Exp Stn Bull 347 36–39 [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41 577–585 [DOI] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of phycocyanin and fluorescin. J Lab Clin Med 44 301–307 [PubMed] [Google Scholar]
- Klempnauer KH, Gonda JJ, Bishop JM (1982) Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular pregenitor c-myb: the architecture of a transduced oncogene. Cell 31 453–463 [DOI] [PubMed] [Google Scholar]
- Kloepper JW, Ryu C-M, Zhang SA (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94 1259–1266 [DOI] [PubMed] [Google Scholar]
- Knoester M, Pieterse CMJ, Bol JF, Van Loon LC (1999) Systemic resistance in Arabidopsis induced by rhizobacteria requires ethylene-dependent signaling at the site of application. Mol Plant Microbe Interact 12 720–727 [DOI] [PubMed] [Google Scholar]
- Koch E, Slusarenko A (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schwindling S, Conrath U (2002) Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol 128 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzc C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204 383–396 [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16 263–276 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ (1993) RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene AvrRpt2. Plant Cell 5 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18 3235–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx J (2004) The roots of plant-microbe collaborations. Science 304 234–236 [DOI] [PubMed] [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15 2551–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Dicke M (2007) Plant interactions with microbes and insects: from molecular mechanisms to ecology. Trends Plant Sci 12 564–569 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van Wees SCM, Hoffland E, Van Pelt JA, Van Loon LC (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8 1225–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Van Wees SCM, Ton J, Van Pelt JA, Van Loon LC (2002) Signalling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biol 4 535–544 [Google Scholar]
- Pieterse CMJ, Van Wees SCM, Van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, Van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo MJ, Van Loon LC, Pieterse CMJ (2004) Jasmonates—signals in plant-microbe interactions. J Plant Growth Regul 23 211–222 [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397 694–697 [DOI] [PubMed] [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR (1995) Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics 139 1393–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjens BPW (2007) Control of plant architecture by the TALE homeobox genes ATH1 and PENNYWISE. PhD thesis. Utrecht University, Utrecht, The Netherlands
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao QM, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Ecker JR (2000) Ethylene signaling: from mutants to molecules. Curr Opin Plant Biol 3 353–360 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4 447–456 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Takeda S, Hirochika H (2000) MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 12 2511–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Lelièvre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H (2003) AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J 34 685–695 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Nelissen I, Eggermont K, Broekaert WF (1999) Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J 19 163–171 [DOI] [PubMed] [Google Scholar]
- Tieman DM, Ciardi JA, Taylor MG, Klee HJ (2001) Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J 26 47–58 [DOI] [PubMed] [Google Scholar]
- Ton J, De Vos M, Robben C, Buchala AJ, Métraux J-P, Van Loon LC, Pieterse CMJ (2002. a) Characterisation of Arabidopsis enhanced disease susceptibility mutants that are affected in systemically induced resistance. Plant J 29 11–21 [DOI] [PubMed] [Google Scholar]
- Ton J, Jakab G, Toquin V, Flors V, Iavicoli A, Maeder MN, Métraux J-P, Mauch-Mani B (2005) Dissecting the β-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell 17 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Van Pelt JA, Van Loon LC, Pieterse CMJ (2002. b) Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol Plant Microbe Interact 15 27–34 [DOI] [PubMed] [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylidès C, Roby D (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Mortel JE, Schat H, Moerland PD, Ver Loren van Themaat E, Van der Ent S, Blankestijn H, Ghandilyan A, Tsiatsiani S, Aarts MGM (2008) Expression differences for genes involved in lignin, glutathione and sulfate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ 31 301–324 [DOI] [PubMed] [Google Scholar]
- Van Hulten M, Pelser M, Van Loon LC, Pieterse CMJ, Ton J (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA 103 5602–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon LC, Bakker PAHM (2005) Induced systemic resistance as a mechanism of disease suppression by rhizobacteria. In ZA Siddiqui, ed, PGPR: Biocontrol and Biofertilization. Springer, Dordrecht, The Netherlands, pp 39–66
- Van Loon LC, Bakker PAHM, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36 453–483 [DOI] [PubMed] [Google Scholar]
- Van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44 135–162 [DOI] [PubMed] [Google Scholar]
- Van Wees SCM, De Swart EAM, Van Pelt JA, Van Loon LC, Pieterse CMJ (2000) Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc Natl Acad Sci USA 97 8711–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wees SCM, Luijendijk M, Smoorenburg I, Van Loon LC, Pieterse CMJ (1999) Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol Biol 41 537–549 [DOI] [PubMed] [Google Scholar]
- Van Wees SCM, Pieterse CMJ, Trijssenaar A, Van't Westende YAM, Hartog F, Van Loon LC (1997) Differential induction of systemic resistance in Arabidopsis by biocontrol bacteria. Mol Plant Microbe Interact 10 716–724 [DOI] [PubMed] [Google Scholar]
- Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 37 115–127 [DOI] [PubMed] [Google Scholar]
- Verhagen BWM, Glazebrook J, Zhu T, Chang H-S, Van Loon LC, Pieterse CMJ (2004) The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol Plant Microbe Interact 17 895–908 [DOI] [PubMed] [Google Scholar]
- Weller DM (2007) Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97 250–256 [DOI] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60 107–124 [DOI] [PubMed] [Google Scholar]
- Yu J-H, Hamari Z, Han K-H, Seo J-A, Reyes-Dominguez Y, Scazzocchio C (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41 973–981 [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Glazebrook J (1999) Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.