Abstract
We investigated the functional and biochemical variability of Kunitz trypsin inhibitor (KTI) genes of Populus trichocarpa × Populus deltoides. Phylogenetic analysis, expressed sequence tag databases, and western-blot analysis confirmed that these genes belong to a large and diverse gene family with complex expression patterns. Five wound- and herbivore-induced genes representing the diversity of the KTI gene family were selected for functional analysis and shown to produce active KTI proteins in Escherichia coli. These recombinant KTI proteins were all biochemically distinct and showed clear differences in efficacy against trypsin-, chymotrypsin-, and elastase-type proteases, suggesting functional specialization of different members of this gene family. The in vitro stability of the KTIs in the presence of reducing agents and elevated temperature also varied widely, emphasizing the biochemical differences of these proteins. Significantly, the properties of the recombinant KTI proteins were not predictable from primary amino acid sequence data. Proteases in midgut extracts of Malacosoma disstria, a lepidopteran pest of Populus, were strongly inhibited by at least two of the KTI gene products. This study suggests that the large diversity in the poplar (Populus spp.) KTI family is important for biochemical and functional specialization, which may be important in the maintenance of pest resistance in long-lived plants such as poplar.
Plants actively respond to challenge by insect herbivores with rapid induction of a suite of biochemical defenses. These defenses can include both secondary metabolites and proteins, which may act as toxins, antifeedants, or antinutrients. One of the most common inducible herbivore defenses in plants is the rapid synthesis of proteinase inhibitors (PIs), small proteins that inhibit insect digestive proteases and lead to reduced insect growth rates or increased mortality (Ryan, 1990). Other induced antinutritive proteins of plants include oxidative enzymes that destroy or modify dietary amino acids and fatty acids, such as polyphenol oxidase, peroxidase, and lipoxygenase (Duffey and Felton, 1991), as well as the essential amino acid-degrading enzymes arginase and Thr deaminase (Chen et al., 2005, 2007). Defensive proteins that affect insect growth via other mechanisms include Cys proteases that permeabilize the insect peritrophic matrix (Pechan et al., 2002; Konno et al., 2004; Mohan et al., 2006). Some plants also enhance insect resistance by increasing the synthesis of alkaloids, terpenoids, glucosinolates, and phenolics in response to damage (Walling, 2000; Kessler and Baldwin, 2002). The types of secondary metabolites and proteins with potential antiherbivore activity are numerous and often taxon specific; consequently, plants can affect insects via many different mechanisms (for review, see Constabel, 1999).
PIs are widespread in the plant kingdom and have been described from many plant species and tissues. PIs comprise at least 10 distinct protein families classified by their amino acid sequence and the mechanistic class of proteinases they inhibit (Laskowski and Kato, 1980; De Leo et al., 2002; Rawlings et al., 2004). Most plant PIs, such as the well-characterized Kunitz and Bowman-Birk PI families, inhibit Ser proteinases, in particular trypsin. The Kunitz-type protease inhibitors, referred to hereafter as Kunitz trypsin inhibitors (KTIs), are proteins of approximately 20 kD with one or two disulfide bonds and a single reactive site. By comparison, Bowman-Birk inhibitors are smaller (approximately 8–10 kD), with high Cys content and two reactive sites (Richardson, 1991). Interaction of KTIs with their cognate proteases is well characterized; the reactive site of the KTI binds tightly to the active site of the protease in a substrate-like manner, and the KTI thus acts as a competitive inhibitor. Although the reaction is reversible, some KTIs have such strong affinities for their target protease that the reaction can be considered irreversible (Beynon and Bond, 1989). The structure of plant KTIs is a β-trefoil fold of 10 to 12 antiparallel β-strands connected by long loops (Song and Suh, 1998). The reactive site is located on a protruding reactive loop, which forms H-bonds with the active groove of the protease. Most KTI proteins have four conserved Cys residues that form two disulfide bridges, although examples with only one or no disulfide bridges are known (do Socorro et al., 2002; Araujo et al., 2005; Macedo et al., 2007). Evidence from several studies suggests that the first disulfide bridge, which surrounds the reactive loop, is necessary for the inhibitory activity of most KTIs (DiBella and Liener, 1969; do Socorro et al., 2002).
As a group, plant KTIs have extremely diverse protease targets and thus have negative effects on a broad range of phytophagous pests and pathogens. Individual KTIs typically have more specific activities. Whereas many plant KTIs inhibit trypsin or chymotrypsin, some can inhibit other Ser proteinases, such as elastase and subtilisin (Terada et al., 1994; Valueva et al., 2000; Revina et al., 2004; Sumikawa et al., 2006). KTIs that inhibit Cys or aspartic proteinases are also reported (Ritonja et al., 1990; Rowan et al., 1990; Heibges et al., 2003b). Moreover, some proteins belonging to the Kunitz family do not act as protease inhibitors (McCoy and Kortt, 1997) or may have lectin-like carbohydrate-binding or invertase inhibitor activity (Glaczinski et al., 2002; Macedo et al., 2004). Some plant KTIs are antimicrobial, presumably via inhibition of microbial proteinases (Kim et al., 2005; Park et al., 2005). Thus, determining a precise function of KTI-like proteins cannot be based only on primary sequence similarities, but requires in vitro assays for confirmation. Whereas the biochemical activity of many individual plant KTIs is known, only in potato (Solanum tuberosum) has extensive analysis of a large KTI family within the same species been reported (Heibges et al., 2003a, 2003b). In that study, the KTI family showed surprising sequence variation, including many nonsynonymous substitutions and indels, which appear to translate into functional diversity.
Poplar (Populus spp.) has been developed as a woody plant model in genomics and molecular biology and is especially useful as an experimental system for ecological genomics. As trees, poplars are long-lived organisms faced with many generations of pest insects and pathogens. The Populus trichocarpa genome is now fully sequenced (Tuskan et al., 2006) and large EST collections are available and can be used for digital analysis of gene expression and discovery (Sterky et al., 2004). Microarrays are also being used for global expression analysis. These resources all provide unprecedented access to genome-level information in a plant of significant ecological importance and worldwide distribution and make this an excellent system for study of plant defense proteins. The first KTI (win3) from hybrid poplar (Populus trichocarpa × Populus deltoides) was described by Gordon and coworkers (Bradshaw et al., 1990; Hollick and Gordon, 1993, 1995). Two wound-inducible win3 paralogs (PtTI1 and PtTI2) were later isolated from Populus tremuloides (Haruta et al., 2001). This study also described wound-inducible accumulation of the corresponding proteins and their in vitro inhibitor activity, consistent with a role for these KTIs in herbivore defense. Furthermore, heterologous expression of win3 in Nicotiana benthamiana leaves demonstrated modest, but significant, reduction in growth of Heliothis virescens (Lawrence and Novak, 2001). Recently, new wound-inducible KTIs from hybrid poplar were identified via EST gene discovery programs (Christopher et al., 2004; Ralph et al., 2006) and transcript-profiling studies have shown that several KTI genes are among the most strongly induced genes after wounding and herbivory (Major and Constabel, 2006; Ralph et al., 2006). This work points to a significant role of the KTI gene family in induced herbivore defense of poplar.
Here we characterize the protease inhibitory activities of five representative wound-inducible members of the KTI gene family as recombinant proteins from Escherichia coli. We determine that all the poplar KTI genes tested encode functional inhibitors, but with different inhibitor profiles, protease preferences, and stabilities. The combined expression patterns and inhibitory activities of the KTIs correlated with protease inhibitor activity of wounded poplar leaf extracts. We also demonstrate inhibitory activity of several KTIs against larval midgut proteases from bertha armyworm (BAW; Mamestra configurata) and forest tent caterpillar (FTC; Malacosoma disstria), pests of Populus and crucifers, respectively. Therefore, our study provides direct evidence of a role of the poplar KTI gene family in the inducible defense of poplar against insect herbivores.
RESULTS
The Poplar KTI Family Contains Many Diverse Members
As part of our ongoing analysis of the poplar defense response, we sought to characterize herbivore- and wound-inducible KTIs in detail. Wounding leaves induces transcript accumulation of several poplar KTIs, including win3 in hybrid poplar and the win3 orthologs PtTI1 and PtTI2 in P. tremuloides (Bradshaw et al., 1990; Haruta et al., 2001). Previously, we identified three novel KTIs among the most abundant and highly induced transcripts during the hybrid poplar wound response (Christopher et al., 2004; Major and Constabel, 2006). In addition, recent availability of the P. trichocarpa genome sequence facilitated comprehensive analysis of the KTI gene family. We determined that poplar has at least 22 KTIs (Fig. 1), but with sequence ambiguities the number of KTIs could be as high as 30. In this analysis, we included only gene models that were full length, contained the Kunitz motif ([L,I,V,M]-X-D-X2-G-X2-[L,I,V,M]-X5-Y-X-[L,I,V,M]), and at least one disulfide bond. KTI gene models with nucleotide sequences ≥99% identical were considered to be allelic; such sequences typically had synonymous-to-nonsynonymous substitution ratios of 1. For comparison, sequences ≤97% identical and with synonymous-to-nonsynonymous ratios of approximately 1.5 were not considered allelic. Phylogenetic analysis revealed that the KTI family is highly diverse; at the amino acid level, the similarity of the KTIs is as low as 25% (Supplemental Table S1). Inspection of the phylogenic tree revealed that the KTI family consists of several clades. KTIs within the same group are approximately 60% to 70% similar at the amino acid level, whereas KTIs from different groups share only approximately 30% of amino acid residues. Moreover, at least one of these homology groups (A) can be subdivided further into subgroups A1, A2, and A3; gwin3, the first poplar trypsin inhibitor (TI) to be studied (Bradshaw et al., 1990; Hollick and Gordon, 1993, 1995), and win3-like genes form subgroup A1. This subgroup also includes PtTI1 and PtTI2, very recently duplicated genes from P. tremuloides (Haruta et al., 2001; Talyzina and Ingvarsson, 2006). The gwin3 and win3.12 KTI genes were chosen for phylogenetic analysis because hypervariability of the win3 (TI2) locus prevented identification of a full-length gene model corresponding to gwin3 in the poplar genome (versions 1.0 and 1.1). The win3 locus has been shown to consist of several clustered KTI genes interspersed with repeats (Bradshaw et al., 1990; Hollick and Gordon, 1993), which is reflected by a region densely populated with repetitive DNA and truncated KTI gene models in the poplar genome sequence. Additional analysis will be required to resolve gene organization at this locus.
Figure 1.
Phylogeny of KTI members from the genus Populus constructed by neighbor joining of p distance. KTI sequences were retrieved from the P. trichocarpa genome; only full-length TI sequences with a Kunitz motif sharing <96% amino acid identity with other sequences and having EST support were included in this analysis. Numbers at branches represent bootstrap support from 2,000 replicates (only values >80% are shown for clarity). Sequences are annotated with the protein ID from the P. trichocarpa version 1.0 genome sequence (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html). Sequences annotated as TI3, TI4, TI5, TI6, and TI7 are the putative P. trichocarpa orthologs of KTIs that have been previously identified (Christopher et al., 2004; Talyzina and Ingvarsson, 2006). Accessions of annotated TI sequences (GenBank gene index or protein ID from P. trichocarpa genome): GWIN3 (gi|20946), WIN3.12 (gi|169460), TI3 (739064), TI4 (555576), TI5 (574326), TI6 (725622), and TI7 (697808). Scale bar is p distance.
The three previously identified wound-inducible KTIs (PtdTI3, PtdTI4, and PtdTI5) each belong to separate homology subgroups (Christopher et al., 2004; Major and Constabel, 2006). TI3 forms subgroup A3 with 739063, whereas TI4 is a member of group B with another three genes. TI5 does not fit with any of the homology groups and is found on a separate branch. The two recently reported KTIs, TI6 and TI7, belong to subgroup A2, along with two other genes (Talyzina and Ingvarsson, 2006). Group C includes eight genes, none of which have been previously studied.
Inspection of sequence alignments of representative KTIs from each clade/subgroup further illustrates their sequence diversity (Fig. 2). This made aligning the sequences somewhat unreliable; to improve the quality of the alignment, secondary and tertiary structure predictions were made using JPred, SWISS-MODEL, CPHmodels, and ESyPred3D, which were used to manually edit and refine the alignment. We included the extensively studied soybean (Glycine max) KTI (GmKTI) and sporamin from Ipomoea batatas for comparison. Whereas all KTIs contain the Kunitz motif, they may otherwise share little overall amino acid similarity, including the position of gaps. However, it is interesting that the most conserved regions correspond to predicted β-sheets and, for poplar KTIs, the signal sequence. This is consistent with a recent analysis of molecular evolution of the poplar KTI group A, which showed that the loop regions connecting β-strands, in particular, are under positive selection (Talyzina and Ingvarsson, 2006). Furthermore, whereas some conserved residues are found within the reactive loop of poplar KTIs, this loop is highly variable, including the P1 residue of the reactive site (Fig. 2, boxed area and starred residues). The reactive loop of sporamin has atypical residues compared with GmKTI and other plant KTIs, but is similar to poplar KTIs. For the P1 site of the seven aligned poplar KTIs, six different residues are predicted, but none are Lys or Arg. These are most common among plant KTIs and considered specific for trypsin inhibition (Beynon and Bond, 1989). Interestingly, the entire predicted reactive loops are devoid of Lys or Arg, except for 75560. By contrast, many of the poplar KTIs have a Ser as the P1' residue. These conserved and variable regions are presumably indications of the selective pressures exerted on the KTI family (see “Discussion”).
Figure 2.
Alignment of representative KTI protein sequences from poplar. Soybean KTI (GmKTI) and sporamin of I. batatas (IbSPOA) are shown for comparison. Shading shows conserved (black) and similar (gray) amino acid residues, and dashes represent sequence gaps. A solid line above the alignment marks the signal peptides and inverted triangles denote the Kunitz motif (L,I,V,M)-X-D-(X2)-G-(X2)-(L,I,V,M)-(X5)-Y-X-(L,I,V,M). Below the alignment, two disulfide bridges formed by four conserved Cys residues are shown by brackets. Plus signs (+) denote free Cys residues present in poplar KTIs. The known structural features of GmKTI are indicated with arrows above the alignment (↔) delineating β-sheets, a boxed region indicating the reactive loop, and asterisks (*) denoting the P1 and P1′ reactive site residues. Sequences were retrieved from the National Center for Biotechnology Information with the following accession numbers: GmKTI (1AVU), IbSPOA (Q40084), WIN3 (CAA33539), TI2 (AAK32690), TI3 (AAQ84216), TI4 (AAQ84217), TI5 (AAQ84218), and TI6 (DT502517). Sequences for TIs 554072 and 75560 were retrieved from the P. trichocarpa version 1.0 genome (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html).
Poplar KTIs typically contain four Cys residues that form two conserved intramolecular disulfide bonds. An exception is TI5, which has only two Cys residues that form the first disulfide bond. All poplar KTIs, except TI5 and 554072, also have two additional free Cys residues located in a loop (Cys-165 and Cys-167, win3 numbering). Overall, poplar KTIs share very little similarity with GmKTI (<25% similarity), sporamin, and other well-characterized plant KTIs. This sequence diversity, especially of the reactive loop, predicted that poplar KTIs should have distinct inhibitory activity (see below).
Poplar KTIs Show Both Developmental and Wound-Inducible Expression Patterns
To determine in which tissues the various KTI genes are expressed, we queried PopulusDB for digital expression profiles derived from the abundance of ESTs in diverse cDNA libraries (Sterky et al., 2004). Expression patterns varied widely across the gene family (Fig. 3). We note that these digital northerns do not distinguish the expression patterns of highly similar sequences due to ambiguous EST clusters that correspond to several gene models. For example, KTIs TI3 and 739063, which have 87% nucleotide sequence identity (Supplemental Table S1), match EST sequences from the same cluster in PopulusDB and so their expression patterns are reported as identical. The highest levels of expression (EST abundance) were found for TI3/739063. These were expressed in essentially all tissues, whereas expression of other KTIs was more restricted. KTIs of the A1 and A2 subgroups (e.g. GWIN3, TI6, and TI7) were abundant in flowers, young leaves, and the apical shoot. None of the remaining KTIs appeared to be expressed at significant levels in healthy leaf tissue from which most cDNA libraries in PopulusDB are derived. Interestingly, many KTIs were expressed in floral tissues, most notably female catkins. Because the EST sets comprising this digital northern did not include any herbivore- or stress-induced tissues, we also included a summary of results from recent transcript-profiling studies (Christopher et al., 2004; Major and Constabel, 2006; Ralph et al., 2006; Fig. 3, right). This analysis indicated that inducible herbivore- and wound-induced expression also varies across the family; whereas most of the KTIs of group A are induced by wounding, the remaining KTIs, except TI4 and TI5, appeared not to have this pattern. However, for some genes, no wound-induced expression data are available.
Figure 3.
Digital northern of expression data for KTI transcripts. The PopulusDB database (http://poppel.fysbot.umu.se) was queried for each KTI and the number of ESTs from the corresponding cluster are shown (gray = no similar sequence in PopulusDB). For wound induction, data from Christopher et al. (2004), Ralph et al. (2006), and Major and Constabel (2006) are reported (+ for degree of induction, − symbols for repression, NC for no change of expression, and n.a. if no data were available). Sequences are annotated as in Figure 1 and the KTI phylogeny at left shows the sequence relationships among the different KTI members. Tissue abbreviations are: A + B, Cambial zone; UA, dormant cambium; G, tension wood; C, young leaves; I, senescing leaves; L, cold-stressed leaves; Q, dormant buds; P, petioles; F, flower buds; M, female catkins; V, male catkins; K, apical shoot; T, shoot meristem; N, bark; R, roots; S, imbibed seeds; Y, virus/fungus-infected leaves.
To confirm that the observed wound-induced changes in transcript levels translate into increased protein accumulation, we performed western-blot analysis using the two antibodies available for KTIs. The TI2 antibody is specific for KTIs in the win3/TI2 subgroup and detected several isoforms in leaves (Fig. 4). For simplicity, we collectively refer to these bands as TI2. The presence of multiple TI2 isoforms is consistent with the complexity and multiplicity of this subgroup (Bradshaw et al., 1990; Hollick and Gordon, 1993; Haruta et al., 2001). By contrast, on western blots the TI3 antibody recognized only a single band corresponding to TI3 protein (Fig. 4). A survey of hybrid poplar tissues determined that TI2-like proteins were found only in wounded leaves, whereas TI3 was present constitutively in most organs, including leaves, bark, wood, and roots (Fig. 4A). TI2 and TI3 were both constitutively present in female catkins of field-grown trees, with minor levels of both proteins in male catkins (Fig. 4B). TI2 protein accumulation in leaves was strongly induced by wounding and TI3 protein showed only modest wound induction. Furthermore, TI2 protein induction was clearly stronger in younger leaves (i.e. at leaf plastochron index [LPI] 3–5), whereas TI3 protein accumulation was relatively consistent independent of tissue age. These KTI proteins thus show very distinct patterns of protein accumulation, but both patterns were consistent with digital northern data (Fig. 3). After wounding, foliar levels of TI2 and TI3 proteins accumulated rapidly after wounding and with similar kinetics; no increase in protein was visible in the first 24 h, but both proteins accumulated to substantial levels 2 d after wounding (Fig. 4C). A similar pattern of TI2 protein accumulation was previously observed in P. tremuloides (Haruta et al., 2001).
Figure 4.
Western-blot analysis of TI2 and TI3 protein accumulation following wounding of poplar saplings. A, Leaves were wounded with pliers and 4 d later tissues were harvested from leaves at LPI 3 to 5 (L3), stem between internodes at LPI 3 to 5 (S3), leaves at LPI 9 to 11 (L9), bark at internodes LPI 9 to 11 (B9), wood at internodes LPI 9 to 11 (W9), and mature roots (R). Six paired control and wounded trees were separately analyzed, and representative blots are shown. B, Female and male catkins from field-grown P. trichocarpa trees were collected in May, 2007. C, Leaves were wounded and leaves 9 to 11 harvested at the indicated days. Two independent experiments were conducted and representative blots are shown.
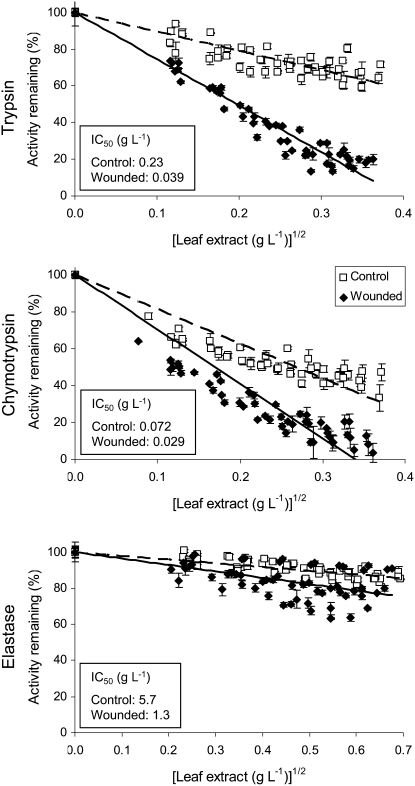
Because both transcript and protein levels of poplar KTIs increased in leaves in response to leaf damage, PI activity should also increase. We therefore assayed inhibition of Ser proteinases by crude leaf extracts using a kinetic assay (Worthington, 1988); such assays are more accurate than end point assays because activity measurements are effectively the means of continuous time points. Different amounts of leaf extract were tested with trypsin, chymotrypsin, and elastase, three common digestive enzymes found in lepidopteran pests. We first compared TI activity of leaf extracts from immature leaves at LPI 3 to 5 and mature leaves of LPI 9 to 11 and found that the activity was greatest for younger leaves of both untreated and wounded saplings (data not shown). We therefore focused on young leaves (LPI 3–5) for subsequent tests of inhibitory activity against other proteases, and calculated the 50% inhibitory concentrations (IC50) for control and wounded leaf extracts against the three proteases. For undamaged saplings, leaf chymotrypsin inhibition was higher than trypsin inhibition (IC50 of 0.072 g L−1 and 0.23 g L−1, respectively; see Fig. 5, insets), but wounded leaves contained similar levels of trypsin and chymotrypsin inhibitor activity (IC50 of 0.039 g L−1 and 0.029 g L−1, respectively). Elastase inhibitor activity represented only a small fraction of total inhibitory activity. All three proteinase inhibitory activities increased significantly following wounding (2.5- to 5.9-fold change in IC50; P ≤ 0.001). As observed in untreated leaf extracts, elastase inhibitory activity was a small proportion compared to trypsin and chymotrypsin after wound induction. Therefore, as already suggested by western analyses, these data confirm that KTI gene induction is followed by an increase in protease inhibitory activity in poplar leaves.
Figure 5.
Inhibitory activity of poplar crude leaf extracts against three commercial proteases. Increasing amounts of crude extracts from young leaves (LPI 3–5) before (control, squares) and 4 d after wounding (wounded, diamonds) were incubated with standard amounts of trypsin, chymotrypsin, and elastase, and the residual protease activity was measured. Six paired control and wounded trees (biological replicates) were used, and each sample analyzed in triplicate (technical replicates). Steeper slopes indicate more potent inhibition. Values are mean protease activity ±se (n = 3). Insets show the IC50 calculated from linear regression of all six independent biological replicates.
Recombinant Poplar KTI Proteins Exhibit Distinct Inhibitory Activity and Biochemical Properties
We predicted that the substantial sequence diversity of the poplar KTI genes should translate into diverse biochemical and biological properties of the corresponding proteins. To test this hypothesis, we characterized the functions of TI2, TI3, TI4, TI5, and TI6 proteins in detail because these represent the diversity of the KTI family. High levels of recombinant protein expressed in E. coli were observed for all KTI bacterial expression constructs, with the bulk of the recombinant proteins found in inclusion bodies, as illustrated for TI3 (Fig. 6A). Proteins were purified under denaturing conditions and dialyzed to renature the polypeptides and recover active protein. We initially tested the renaturation conditions used successfully for TI2 (Haruta et al., 2001), but found that, under these conditions, most of the other KTIs formed aggregates and precipitated. We therefore optimized the renaturation conditions for each KTI by altering the ionic strength, pH, and concentration of the reducing agent of the buffer. These tests showed that optimal refolding conditions varied considerably among the different KTIs (Table I). Thus, four of the KTIs were found to be soluble only in a buffer of high ionic strength, whereas TI3 was soluble in water or a buffered low-strength ionic solution. Moreover, whereas most KTIs were soluble near pH 8, their solubility differed greatly away from pH 8.
Figure 6.
Purification of recombinant TIs from E. coli. A, SDS-PAGE of recombinant TI3 protein preparations from different stages of purification. Lane 1, E. coli cells before induction with IPTG; lane 2, E. coli cells 24 h after induction with IPTG; lane 3, E. coli cell lysate without inclusion bodies; lane 4, lysed inclusion bodies containing TI3; lane 5, TI3 after purification on nickel nitrilotriacetic acid agarose resin column; lane 6, protein standard marker. B, SDS-PAGE of recombinant TI proteins. Lanes 1 to 5 are loaded with TI2 to TI6, respectively. Lane 6 is loaded with soybean KTI.
Table I.
Properties of recombinant poplar KTIsa
| KTI | Size (Amino Acid) | Predicted pI | Molecular Weight
|
Protein Solubilityb
|
||
|---|---|---|---|---|---|---|
| Predicted | Actual (SDS-PAGE) | Distilled, Deionized Water | 50 mm Tris (pH Range) | |||
| kD | ||||||
| TI2 | 213 | 4.42 | 23.1 | 24 | Insoluble | 2 and 5–12 |
| TI3 | 202 | 6.15 | 22.0 | 26 | Soluble | 2–4 and 9–12 |
| TI4 | 212 | 4.96 | 23.1 | 24 | Insoluble | 2–3 and 8–12 |
| TI5 | 214 | 3.98 | 22.9 | 30 | Insoluble | 7–12 |
| TI6 | 202 | 5.32 | 21.8 | 23 | Partially soluble | 2–3 and 7–12 |
Predicted Mr and pI were calculated by Vector NTI Advance 9 software; SDS-PAGE-determined Mr was extrapolated from the logarithmic-transformed migration distance of the proteins compared with protein standards (low range; Bio-Rad).
Recombinant protein solubility was assessed by dialysis of proteins in water and 50 mm Tris buffers of pH 2 to pH 12. For solubility in Tris buffer, the pH range for which the proteins were soluble is shown.
We verified the purity and concentrations of the successfully renatured KTIs by SDS-PAGE, which clearly showed a single major band for each KTI at an approximate predicted Mr (Fig. 6B). In addition, larger bands were apparent for TI3, TI4, and TI6; these may be protein dimers because the apparent Mr of these bands is twice that of the predominant bands. We previously reported putative dimers for TI2 (Haruta et al., 2001) and other plant KTIs are known to form dimers as well (Terada et al., 1994). Such dimers may be linked by an intermolecular disulfide bond formed between the free Cys residues of the KTIs (Fig. 2) because the predicted three-dimensional models suggest that residues are exposed on the surface of the proteins (data not shown). Whereas migration of TI2, TI4, and TI6 in gels corresponded well with their predicted Mr (Table I), TI3 and TI5 migrated slower than expected. This is likely the result of residual secondary protein structure because both recombinant proteins migrated according to their predicted Mr on denaturing urea-SDS-PAGE gels, which denature proteins more completely (data not shown).
Using in vitro assays with commercially available proteases, we confirmed that the recombinant and renatured poplar KTI proteins have protease inhibitory activity. Commercial GmKTI from soya was included for comparison because it has been extensively described in the literature. Because previous studies of plant KTIs have found a wide range of possible target proteases, we assayed inhibitor activity against several proteases: the Ser proteinases trypsin, chymotrypsin, elastase, and subtilisin, and the Cys proteinase papain. We again used kinetic assays at multiple TI concentrations to test inhibition of trypsin, chymotrypsin, and elastase. End point assays were used to test additional proteinases outside the chymotrypsin subfamily, the Ser protease subtilisin and Cys protease papain. These experiments confirmed that all KTIs tested are active protease inhibitors, but with remarkably different target preferences (Fig. 7). Thus, recombinant TI2 was a strong inhibitor of trypsin (Fig. 7, top), with activity levels similar to that of GmKTI, whereas TI3, TI5, and TI6 had intermediate levels of inhibitor activity. In contrast, TI4 did not inhibit trypsin at detectable levels. Against chymotrypsin, TI6 was the most potent of the recombinant poplar KTIs, although less potent than GmKTI, followed by TI3, TI5, and TI4 (Fig. 7, center). Although TI2 was very effective against trypsin, it failed to inhibit chymotrypsin significantly. The only poplar KTI with inhibitory activity against elastase was TI6 (Fig. 7, bottom). None of the poplar KTIs tested was active against subtilisin or papain.
Figure 7.
Inhibitory activity of purified recombinant poplar TIs against three commercial proteases. Titration of TI2 (blue diamond), TI3 (red square), TI4 (purple triangle), TI5 (orange closed circle), and TI6 (green x) against trypsin, chymotrypsin, and elastase. Soybean KTI (GmKTI; yellow dash) is shown for comparison and bovine serum albumin (indigo open circle) is a negative control. Data points represent the mean protease activity ±se (n = 3). Linear regression is used to calculate the IC50.
To help compare the inhibitory profiles of the KTIs, we calculated the IC50 for each KTI (Table II). Such IC50 values permit comparison of inhibitory activities relative to the various proteases tested and emphasize the very different activity profiles of the five poplar KTIs. We note that these are maximal IC50 values because we could not determine directly whether all protein in each preparation was fully refolded. However, we obtained similar inhibitory activity from independent recombinant protein preparations. The comparison of IC50 indicated that several KTIs have strong preferences for specific targets (Table II). TI2 appears to be a specific inhibitor of trypsin, whereas TI4 was specific to chymotrypsin. TI3 and TI5 both inhibited trypsin and chymotrypsin and TI6 inhibited chymotrypsin and elastase. Thus, all the recombinant KTIs tested were active as protease inhibitors and, as a group, inhibited the major digestive enzymes trypsin, chymotrypsin, and elastase.
Table II.
Summary of inhibitory activity (IC50a) of KTIs against different proteases
| KTI | Trypsin | Chymotrypsin | Elastase | Subtilisin | Papain |
|---|---|---|---|---|---|
| μm | |||||
| TI2 | 0.0280 | 2.95 | NA | NA | NA |
| TI3 | 0.152 | 0.152 | 74.1 | NA | NA |
| TI4 | 128 | 0.294 | NA | NA | NA |
| TI5 | 0.205 | 0.188 | 9.16 | NA | NA |
| TI6 | 0.643 | 0.111 | 0.731 | NA | NA |
| GmKTI | 0.0116 | 0.0567 | 3.65 | NA | NA |
ICs were calculated from the linear portion of the plot of residual proteinase activity against TI protein concentration (Fig. 7). NA, No activity.
Recent work has suggested that protein stability is a key function of plant defense proteins given the hostile environment of herbivore digestive systems (Chen et al., 2007). We found that KTI activity remained high even after dialysis at pH extremes (Table I). We therefore investigated KTI stability by assessing the denaturing effects of a reducing agent or increasing temperature. Purified recombinant KTIs were first incubated with increasing concentrations of dithiothreitol (DTT) for 30 min, after which remaining inhibitory activity was measured. These activities changed little up to 2 h and thus only data for 30-min incubations are shown (Fig. 8A). In general, each KTI protein was stable to a critical DTT concentration, beyond which it quickly lost activity. This critical concentration and the rate of activity loss differed for each KTI, giving rise to distinct stability profiles (Fig. 8A). TI4 and TI6 were both resistant to 0.1 mm DTT, but were differentially sensitive at higher concentrations. TI3 retained activity up to 1 mm DTT, but quickly lost activity above this level. TI5 was most resistant to reducing conditions because it retained 65% activity at 100 mm DTT, even after 2-h incubation. Interestingly, TI2 activity increased to an optimal 31.6 μm DTT, but quickly lost activity at higher concentrations and lost all activity beyond 3 mm DTT.
Figure 8.
Effect of denaturing conditions on activity of poplar TIs. A, KTIs were incubated in the presence of increasing concentrations of DTT for 30 min followed by incubation in iodoacetamide. B, KTIs were incubated at increasing temperatures for 30 min followed by incubation on ice. Data points represent mean TI activity ±se (n = 3). [See online article for color version of this figure.]
Similarly, we found that poplar KTIs had different stabilities to elevated temperatures. KTI proteins were incubated for 30 min at different temperatures and remaining inhibitory activities were compared with samples kept on ice (Fig. 8B). TI2 was most thermolabile, losing activity above 30°C. Similarly, TI3 was susceptible to temperatures above 40°C. We also repeated these experiments for TI2 and TI3 with gradual cooling after incubation at temperature and found similar loss of activity (data not shown). TI6 was more thermostable than both TI2 and TI3, remaining active after incubation at 60°C. Neither TI2, TI3, nor TI6 were boiling stable because they retained <20% activity after boiling. By contrast, TI5 was stable at all temperatures, retaining 80% to 100% activity at all temperatures assayed. TI4 was also stable at high temperatures, but exhibited enhanced activity of 125% to 130% when incubated at 60°C or greater.
In view of the apparent temperature stability of the KTIs, we also measured long-term stability by assaying recombinant KTI preparations at 4°C at 25-d intervals for extended time periods (Supplemental Fig. S1). We found no loss in activity within 150 d, although the measured activity of KTIs fluctuated between 80% and 130%. We extended this analysis to >1 year for TI2, again measuring activity periodically, and found no loss of activity (Supplemental Fig. S1). Overall, these tests demonstrated the remarkable stability of poplar KTI proteins.
Recombinant Poplar KTIs Inhibit Lepidopteran Midgut Proteases
The inhibitory activity of recombinant KTIs against commercially available proteases of mammalian origin prompted us to test these KTIs against digestive proteases from insects. We obtained midguts from BAW, a lepidopteran pest of crucifers. The midgut proteases of BAW have been well characterized and, like other lepidopteran larvae, consist primarily of Ser proteinases, particularly trypsin-, chymotrypsin-, and elastase-like enzymes (Hegedus et al., 2003). For these assays, different amounts of poplar KTIs were incubated with a standard quantity of BAW midgut extract and the remaining protease activity was measured. Assays were performed at two different pHs, which correspond to two pH optima of BAW midgut protease activity, using an end point assay with azocasein as a general protease substrate (Hegedus et al., 2003). In contrast to our results with commercial mammalian proteinases, TI3 was the only poplar KTI tested with significant activity against BAW midgut proteases (Fig. 9, top; data not shown). GmKTI was also an effective inhibitor, consistent with the results of Hegedus et al. (2003; Fig. 9, bottom). Both TI3 and GmKTI were more potent inhibitors of BAW gut protease activity at pH 11 than at pH 8. At pH 11, TI3 inhibited approximately 40% of BAW total midgut proteolytic activity, whereas GmKTI inhibited at least 50%. Extrapolation of the linear portions of these plots shows that only 0.47 μg of TI3 were needed to inhibit 40% of BAW protease activity, whereas 6.8 μg of GmKTI were required to inhibit the same activity. Thus, TI3 has a much greater affinity for at least some BAW midgut proteases. A similar pattern was observed for pH 8 assays, where TI3 was more potent, but had lower maximal inhibition of gut protease activity compared to GmKTI.
Figure 9.
Inhibition of midgut proteases from BAW (M. configurata) by poplar TI3 and soybean KTI (GmKTI). Increasing amounts of TI3 and GmKTI were preincubated with a constant amount of BAW midgut extract and the residual total proteolytic activity was assayed at pH 8 (cross) and pH 11 (diamond) with azocasein. Note the different x-axis scales for TI3 and GmKTI data. Values are mean protease activity ±se (n = 3).
We also tested poplar KTI inhibition of midgut extracts from FTC, a lepidopteran pest of poplars. We found that protease activity of FTC midguts is due primarily to Ser proteases with a pH optimum of 11.5 (data not shown). As with BAW protease assays, we measured residual protease activity after incubation of poplar KTIs or GmKTI with a standard quantity of FTC midgut extracts. All five KTIs inhibited FTC midgut proteolysis, although only TI2 and TI3 were strong inhibitors (Fig. 10). Comparison of inhibition curves revealed that TI3 inhibited FTC gut proteases with comparable apparent potency as GmKTI, and with greater potency than TI2 (Fig. 10). Furthermore, TI3 inhibited almost 70% of protease activity, whereas TI2 inhibited approximately 40%. These maximal inhibition values, which represent the proportion of the mixture of midgut proteases that is susceptible to inhibition, confirmed the potency of these KTIs against FTC and establishes their ecological relevance.
Figure 10.
Inhibition of midgut proteases from FTC (M. disstria) by two poplar TIs and soybean KTI (GmKTI). Increasing amounts of KTIs were preincubated with a constant amount of FTC midgut extract and the residual total proteolytic activity was assayed at pH 11.5 with azocasein. Values are mean protease activity ±se (n = 3). [See online article for color version of this figure.]
DISCUSSION
Kunitz TIs have been studied in many different plants and contexts, often with a focus on their potential for biotechnology-based pest control for agriculture; however, the diversity of KTIs within a plant species has not been extensively examined. The prevalence of KTIs in the poplar genome represents a unique opportunity to explore the variation and function of these proteins. Although the precise number of KTI genes in the P. trichocarpa genome has not yet been definitively determined, there could be as many as 30 genes. The region surrounding the win3/TI2 group, in particular, contains much repetitive DNA and appears to have evolved rapidly, making this clade of the gene family difficult to resolve. For functional analysis in the context of plant herbivore defense, we selected five herbivory-inducible genes that represent the diversity of the entire poplar KTI family. Our analysis indicates that each KTI gene product is a functional protease inhibitor with a distinct range of target protease preferences and shows differential stability to reducing agents and temperature.
Expression of KTI Genes Correlates with Inducible Protease Inhibitor Activity in Poplar Leaf Extracts
Based on our interest in herbivory, we chose to characterize representative poplar KTIs from homology groups that were wound or herbivore inducible. Results from published array and northern analyses suggested that group C KTIs generally do not respond to wounding (Fig. 3). Therefore, this group was not included in our analysis, although it is an important target for future study. The inducible nature of TI2 and TI3 was demonstrated at the protein level by western-blot analysis and these proteins showed rapid accumulation after wounding of leaves (Fig. 4). This suggests that, for the other KTIs, induced transcripts are also likely to result in increased levels of the gene products. Furthermore, we showed significant induction of total PI activity in wounded poplar leaves. Interestingly, wounded poplar leaf extracts inhibited chymotrypsin and trypsin almost equally, whereas control leaf extracts were more effective against chymotrypsin. This pattern likely reflects the strong inducibility of TI2 (Fig. 4) and its strong preference for trypsin (Table II). Only low levels of elastase inhibitory activity were found in leaf extracts, as predicted by our finding that only one KTI (TI6) had any significant antielastase activity (Fig. 7). Therefore, total PI activity in leaf extracts generally correlated with the activity of the expressed KTI proteins. Induction of protease inhibitory activity in crude extracts of wounded leaves is important because it demonstrates that up-regulation of KTI genes has a demonstrable impact on herbivore food quality.
Analysis of transcript abundance from digital northerns showed highly diverse expression patterns across the KTI family. For example, based on EST abundance, win3/TI2 expression was highly specific, whereas TI3 showed a very broad pattern of expression, which was confirmed by western analysis (compare Figs. 3 and 4). The win3/TI2 protein accumulated to highest levels in young leaves, consistent with the corresponding transcripts being detected mostly in young leaves, the shoot apex, and floral organs (see below). Most of the other poplar KTIs also exhibited relatively tissue-specific patterns of expression, again with preferred expression in flowers (TI4) and shoot meristem (TI5; Fig. 3). By contrast, TI3 protein and transcripts accumulated in almost all tissues (Figs. 3 and 4). The apparently ubiquitous expression of TI3 is curious because PIs are thought to incur large fitness costs (Zavala et al., 2004). Presumably, TI3 has acquired new regulatory sequences during evolution, leading to its broad expression pattern, similar to the promoter of a KTI pseudogene (WCI-P1) from winged bean (Psophocarpus tetragonolobus), which confers strong constitutive expression in leaves, petioles, stems, roots, and seeds (Habu et al., 1997).
In general, analysis of EST databases predicted that the highest levels of KTI transcripts are present in poplar flowers, particularly female floral buds and catkins. For win3/TI2 and TI3, western analysis confirmed the accumulation of their corresponding proteins (Fig. 4). Hollick and Gordon (1995) had also previously shown that the win3.12 promoter drives developmentally regulated reporter gene expression in N. benthamiana reproductive organs, including flowers, pollen, and seeds. We did not observe win3/TI2 or TI3 protein accumulation in pollen or seeds (data not shown); this may be because win3 expression is temporally restricted during pollen and seed maturation (Hollick and Gordon, 1995). PIs are also known to be expressed in the flowers of other plant species, suggesting they play functional roles in plant reproductive organs (Botella et al., 1996; van Dam et al., 2001). Such roles could include regulation of proteolysis of storage proteins, especially in seeds, as well as defense against herbivore pests for these crucial organs. We note that other poplar defense-related proteins, including polyphenol oxidase, WIN4, and SP1/Pop3-like, also accumulate in female catkins (data not shown), suggesting that, at least in poplar, KTI gene expression may be part of a larger accumulation of defense proteins.
Biochemical Analysis Indicates Functional Diversification of Poplar KTI Genes
The poplar KTIs tested inhibited trypsin, chymotrypsin, and elastase with different profiles (Fig. 7). TI2 and TI4 had very specific inhibitory activities, being effective against only trypsin or chymotrypsin, respectively. By contrast, TI3 and TI5 inhibited both trypsin and chymotrypsin with similar efficiency. TI6 was the only elastase inhibitor, a rare activity among plant KTIs (Valueva et al., 2000; Araujo et al., 2005; Sumikawa et al., 2006). These functional differences presumably reflect the sequence diversity of the KTI family. Previous study of a KTI gene family, a biochemical analysis of potato KTIs, also revealed functional diversity both between and within the homology groups (Heibges et al., 2003a, 2003b). Thus, potato group A and B KTIs strongly inhibit trypsin, whereas potato group C KTIs do so only weakly. In contrast to poplar KTIs, potato KTIs also inhibit other enzymes, including subtilisin, the aspartic proteinases cathepsin D and aspergillopepsin, and Cys proteinases (Ritonja et al., 1990; Rowan et al., 1990; Valueva et al., 2000; Heibges et al., 2003b; Revina et al., 2004).
In poplar, the functional properties of individual KTIs could not be easily predicted based on primary sequence data. For example, although TI2 and TI6 protein sequences were most similar among the KTIs tested (Supplemental Table S1), their inhibitory activity was the most divergent. By contrast, TI3 and TI5 inhibited proteinases with similar potencies, but share little sequence similarity. Likewise, a comparison of reactive site and reactive loop residues revealed that these do not readily predict protease substrate preferences. This may relate to the atypical residues of poplar KTI reactive loops (Fig. 2). Our prediction of KTI reactive loops differs by a window of three amino acid residues from a previous study of Populus group A KTIs (Talyzina and Ingvarsson, 2006). We used secondary and tertiary structure predictions to predict reactive loop position, which provides a better estimate than sequence alignments with divergent plant KTIs. A similar approach has been used to predict the second reactive site of a winged-bean chymotrypsin inhibitor (Dattagupta et al., 1999). However, precise definition of the reactive loops and sites of poplar KTIs will require site-directed mutagenesis or structural determination.
By contrast, the reactive site residues of other types of plant KTIs may be useful indicators of inhibitor specificity. For example, Arg-Lys and Ile-Ser are often reactive site residues in inhibitors of trypsin, whereas Leu-Ser is often the reactive site for KTIs effective against chymotrypsin. Other plant KTIs may have more atypical reactive sites. Glu-Ser are the reactive site residues of DrTI from Delonix regia seeds and sporamin from Ipomoea batatas tubers (Pando et al., 2001; Yao et al., 2001), and these are also the predicted residues for the poplar TI2 and TI3 reactive sites (Fig. 2). The importance of Glu and Ser was shown by site-directed mutagenesis in sporamin. Here, changing reactive site residues from Glu to Arg or Ser to Ile completely abolish TI activity, despite the presence of Arg or Ile in reactive loops of other trypsin-inhibiting KTIs (Yao et al., 2001). Mutation of another negatively charged residue in the sporamin reactive loop also destroys inhibitory activity, indicating that the negatively charged residues in the sporamin reactive loop may contribute to inhibitory activity (Yao et al., 2001). Interestingly, 19 of 22 poplar KTIs have a conserved negatively charged residue in their reactive loops. Thus, molecular interactions between poplar KTIs and substrate proteinases may be similar to those in DrTI and sporamin.
We also observed that the biochemical stability of poplar KTIs differed considerably within the group. Similar to other protease inhibitors, several poplar KTIs retain activity in the presence of the reducing agent DTT and at high temperature (Garcia et al., 2004). However, these stability profiles varied greatly among the five poplar KTI proteins tested here. In our assays, TI5 was the most stable KTI, retaining activity at high levels of DTT and high temperature, whereas TI2 was very sensitive, losing one-half of its activity at 1 mm DTT and 50°C (Fig. 8). The presence of only a single disulfide bond in TI5 suggests it has evolved features that contribute to protein stability that are not affected by DTT. Although protein stability has not been measured for most plant KTIs that have one or no disulfide bonds, recent analysis of such a KTI from Inga laurina seeds did in fact show high stability in the presence of DTT (Macedo et al., 2007). Studies of other plant KTIs also reveal diverse stability profiles. Some KTIs are exceptionally resistant to reducing agents, boiling, and pH extremes, whereas others are sensitive (Garcia et al., 2004; Macedo et al., 2004). These data support the idea that antiherbivore proteins may be inherently more stable and resistant to proteolysis. Previous work from this laboratory found that the defensive enzyme polyphenol oxidase is stable in the gut of a lepidopteran herbivore and remained active in frass (Wang and Constabel, 2004). In a preliminary analysis with poplar-fed FTC, western blots showed clear bands corresponding to TI2 and TI3 in frass (data not shown). This suggests that poplar KTIs are stable and resist digestion within the guts of lepidopteran and perhaps other herbivores. Likewise, the tomato (Solanum lycopersicum) antiherbivore enzymes arginase and Thr deaminase were shown to be stable in midguts of Manduca sexta, where they destroy these nutritionally important amino acids, ultimately resulting in decreased larval growth (Chen et al., 2005, 2007).
The Role of KTIs in Poplar Herbivore Defense
In vitro tests with recombinant proteins showed that only TI3 is an active inhibitor of midgut proteases from BAW, a lepidopteran pest of canola (Brassica napus; Fig. 9). Comparison of the initial slopes of the TI3 and GmKTI inhibition curves indicated that TI3 has a stronger affinity for some BAW proteases than soybean GmKTI; however, neither protein completely abolishes proteolytic activity (Fig. 10). When we tested our recombinant KTIs against midgut proteases from FTC, which specialize on P. tremuloides in western Canada, all five KTIs inhibited proteolysis, although TI2 and TI3 were clearly most potent (Fig. 10). The effectiveness of herbivore-induced poplar KTIs on the digestive proteases of a poplar pest strongly supports an important role for KTIs in poplar defense. Furthermore, the differences in effectiveness that we measured against different pests, combined with the differences in target preferences against commercial proteases exhibited for the five KTIs, further support the idea of functional specialization within the poplar KTI gene family.
Compared with Arabidopsis (Arabidopsis thaliana), an annual species, the poplar genome has a larger suite of genes associated with insect resistance (Tuskan et al., 2006). Over its lifespan, a perennial, such as poplar, is exposed to more potential pest insects than an annual plant, and a broad array of protease inhibitors with activity against diverse proteases would seem advantageous. Lepidopteran pests contain complex mixtures of gut proteases, often differentially expressed, which can facilitate rapid adaptation of gut protease activity to ingested PIs in the herbivore diet (Jongsma et al., 1995; Mazumdar-Leighton and Broadway, 2001; Hegedus et al., 2003). Therefore, poplar KTIs should have many potential targets, both within any one pest species and among different poplar pests. Whereas we are aware that in vitro inhibition of gut proteases does not always correlate with a reduction of insect performance in vivo (Jongsma et al., 1995; Bown et al., 2004), we have used artificial diet bioassays with recombinant TI3 protein at biologically relevant concentrations to demonstrate that this inhibitor does in fact significantly reduce larval growth (I.T. Major, E. Despland, and C.P. Constabel, unpublished data). Previously, heterologous expression in N. benthamiana also demonstrated that win3 can reduce growth of H. virescens (Lawrence and Novak, 2001). Thus, some poplar KTI genes have already been shown to provide protection against insects, and it is likely that many of the others do the same. We therefore suggest that the diversity of genes in the KTI family is likely to provide ecological benefits to Populus.
The interaction of poplar KTIs with a diversity of pest protease targets should be reflected in selective pressure on KTI genes. In evolutionary time, this would leave traces of positive selection and rapid molecular evolution in KTI genes in different poplar species or populations. We previously noted the high incidence of restriction fragment polymorphisms and high rates of nonsynonymous substitutions in aspen KTIs (Haruta et al., 2001; Miranda et al., 2004). More comprehensive analyses of molecular evolution have shown convincingly that the poplar KTI gene family is evolving rapidly, with some KTIs showing clear signs of positive selection (Ingvarsson, 2005; Talyzina and Ingvarsson, 2006). The origins of such strong selection pressures on the KTI family are difficult to demonstrate directly, but are predicted to relate to pressures from a diversity of herbivore pests and multiple digestive proteases. Therefore, gene duplication followed by strong positive selection from herbivores could lead to the biochemical diversification we observed in our experiments. Because poplars are long-lived trees, large and diverse families of defense genes may be crucial for survival against multiple pest species with short generation times capable of quick evolutionary changes.
MATERIALS AND METHODS
Plant Material and Wounding Treatments
Hybrid poplar (Populus trichocarpa × P. deltoids, clone H1-11) plants were propagated and maintained in the Bev Glover Greenhouse at the University of Victoria as described previously (Major and Constabel, 2006). Plants were typically 12 weeks old and 1 m tall, with approximately 30 leaves. Lateral shoots were removed periodically, but no less than 2 weeks prior to experiments, so that each plant consisted of a single main stem. Leaves were numbered using the LPI (Larson and Isebrands, 1971), with the index leaf (LPI 0) defined as the first developing leaf with a lamina length of >20 mm. For wound treatments, the margins of all leaves (LPI > 0) were crushed with pliers three times at 1-h intervals. Male and female catkins were sampled from a stand of black cottonwood trees (P. trichocarpa) growing on the campus of the University of Victoria. Immediately after harvesting, tissues were frozen in liquid nitrogen or on dry ice and stored at −80°C until analyzed.
Protein Extraction and Western-Blot Analysis
Protein was extracted from leaves of greenhouse-grown poplar by tissue homogenization in sodium phosphate buffer (100 mm, pH 7.0) containing 0.1% (v/v) Triton X-100, 5% (w/v) polyvinylpolypyrrolidone, and 0.1% (v/v) 2-mercaptoethanol. For catkins from field-grown trees, proteins were extracted with Tris buffer (50 mm, pH 7.5) containing 5% (w/v) SDS, 5% (w/v) polyvinylpolypyrrolidone, and 2.5% (v/v) 2-mercaptoethanol. Extracts were clarified by centrifugation, and soluble protein quantified using the Bradford (1976) method for sodium phosphate-buffer extracts or the RC DC protein assay (Bio-Rad) for Tris/SDS-buffer extracts. Proteins were separated by SDS-PAGE and electrotransferred onto polyvinylidene difluoride membranes (Pierce). Ponceau S (Sigma) staining was used to verify equal loading and transfer efficiency. Western-blot detection was carried out using polyclonal antibodies raised against TI2 (Haruta et al., 2001) and TI3 (see below). Immunocomplexes were detected using acid phosphatase- or horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) and blots were developed calorimetrically with 5-bromo-4-chloro-3-indoyl phosphate (Pierce) and nitroblue tetrazolium chloride (Pierce; acid phosphatase), or 3,3′-diaminobenzidine tetrahydrochloride (Sigma; horseradish peroxidase).
Identification of KTIs from the Poplar Genome and in Silico Analysis
To obtain the complete poplar KTI family, the P. trichocarpa version 1.0 genome sequence (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html) was queried for gene models annotated as Kunitz-type PIs. KTI gene models are based on the genome version 1.0 annotation because the more recently curated version 1.1 genome sequence (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html) was missing some KTI gene models that had EST support from GenBank and several models were truncated (data not shown); however, the genome version 1.1 annotations are shown for comparison (Supplemental Table S2). Gene models were combined with previously identified Populus KTI sequences (Bradshaw et al., 1990; Hollick and Gordon, 1993; Haruta et al., 2001; Christopher et al., 2004) and used to query the genome again for any Kunitz-like proteins not annotated as such. Gene models were removed that were not KTIs (i.e. lacking the Kunitz motif and having at least one disulfide bond). Many gene models encoded truncated KTIs that were flanked by regions of highly repetitive DNA and could not be resolved as separate TI genes (see “Results”; win3 locus). Truncated gene models were used to query the GenBank plant EST database to try to identify full-length sequences; in the absence of full-length ESTs in GenBank, these were not analyzed further. KTI gene models with nucleotide sequences ≥99% identical were considered allelic and only one gene model per locus was used for phylogenetic analysis.
Multiple amino acid alignments were made using ClustalW with default parameters, followed by manual adjustments in BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). To improve alignments, secondary structure predictions were made using Jpred (http://www.compbio.dundee.ac.uk/∼www-jpred) and tertiary structure predictions were made using SWISS-MODEL (http://swissmodel.expasy.org), CPHmodels (http://www.cbs.dtu.dk/services/CPHmodels), and ESyPred3D (http://www.fundp.ac.be/sciences/biologie/urbm/bioinfo/esypred). Predicted secondary and tertiary structures were compared and used to help align variable sites and indels. A neighbor-joining phylogenetic tree was constructed in MEGA 3.1 (http://www.megasoftware.net) using p distance (proportion of amino acid differences to the total number of sites compared) with pairwise deletion of gaps. MEGA recommends using p distance for pairwise deletion because it normalizes the number of differences based on the number of valid sites compared. Bootstrapping was used to test the robustness of the tree in MEGA 3.1. To determine preliminary KTI expression patterns via digital northern, we queried the PopulusDB EST database (http://poppel.fysbot.umu.se), which contains >100,000 EST sequences from 19 different tissue libraries (Sterky et al., 2004). An expression profile was constructed by determining the number of ESTs, according to their libraries, of each cluster.
Heterologous Expression and Purification of Recombinant KTI Proteins
TI2 was expressed and purified as described previously (Haruta et al., 2001). Signal peptides of each poplar KTI were predicted using SignalP (http://www.cbs.dtu.dk/services/SignalP). The coding sequence of poplar KTIs, minus the signal sequence, were PCR amplified from plasmid cDNA clones. TI3 (EST ID H1059; GenBank accession no. CN192549) was amplified with the primers TI-3s (5′-CGGGATCCGAAGCAGTGATCGATGCC-3′; BamHI site underlined) and TI-3a (5′-CCCAAGCTTCATCATTTTATACTCG-3′; HindIII double underlined). TI4 (H828; CN193330) was amplified with the primers TI-4s (5′-CGGGATCCTATACTGAGCCGGTGCTTG-3′) and TI-4a (5′-CCCAAGCTTTATGGATGAACTTAAAGG-3′). TI5 (H1685; CN192805) was amplified with the primers TI-5s (5′-CGGGATCCTCAGGGAATCCAGTGCTTG-3′) and TI-5a (5′-CCCAAGCTTTACAACAGCTTTTAATCC-3′). TI6 (WS0133_E08; DT502517) was amplified with the primers TI-6s (5′-CGGGATCCAAAGATGCTGCAGCAGTGCT-3′) and TI-6a (5′-CCCAAGCTTTTCATCTGGTTCAAACATAA-3′). PCR products were digested with BamHI and HindIII and cloned into the respective restriction sites of the pET21a (Novagen, EMD Biosciences) bacterial expression vector. The resulting pET-TI3, pET-TI4, pET-TI5, and pET-TI6 plasmids were sequenced and moved into Escherichia coli strain BL21(DE3) (Novagen) for expression.
Recombinant proteins were isolated from bacterial inclusion bodies as described previously (Haruta et al., 2001) and purified on a nickel nitrilotriacetic acid agarose affinity resin (Qiagen) and eluted along a pH gradient under denaturing conditions (8 m urea) as per the manufacturer's protocol. To renature the recombinant KTIs, we initially used our previous protocol (Haruta et al., 2001), but found that the proteins differentially aggregated and precipitated in buffers depending on ionic strength, concentration of the reducing agent, and pH (Table I). We therefore optimized the refolding conditions for each KTI and found a refolding buffer suitable for all the proteins such that subsequent in vitro assays for PI activity would not be influenced by different buffers. Recombinant KTIs were dialyzed against 50 mm Tris-HCl buffer (pH 8.0) at 4°C for 24 h with at least six changes of buffer. For comparison with in vitro assays of PI activity, soybean (Glycine max) KTI (GmKTI; Sigma) dissolved in distilled, deionized water was also dialyzed against 50 mm Tris-HCl buffer (pH 8.0) so that buffers were comparable. Protein concentrations were determined spectrophotometrically (A280) using molar extinction coefficients calculated by using Vector NTI Advance 9.0 software (Invitrogen) or reported by the supplier (GmKTI; Sigma), and were verified on Coomassie-stained SDS-PAGE gels.
TI3 antiserum was generated in rats using recombinant TI3 produced as above, but, in addition, we precipitated TI3 protein by dialysis against phosphate-buffered saline at 4°C. Rats were immunized with 100 μg TI3 protein in Freund's complete adjuvant (Sigma), with booster injections of 50 μg protein in Freund's incomplete adjuvant using standard procedures.
PI Assays
All PI activity assays were done in triplicate and the results shown as the means of three replicate assays. The PI activities of recombinant poplar KTIs against trypsin (EC 3.4.21.4; Sigma), chymotrypsin (EC 3.4.21.1; Sigma), and elastase (EC 3.4.21.36; Sigma) were determined by preincubating increasing concentrations of each KTI with a standard quantity of proteinase (6.6 × 10−4 g L−1 final assay concentration) in the appropriate assay buffer. PI activity was determined as described, by measuring residual proteinase activity as the rate of hydrolysis of specific chromogenic substrates (Worthington, 1988). Trypsin activity was assayed with p-toluene-sulfonyl-l-Arg methyl ester (Sigma) by monitoring the change in ΔA247 min−1; chymotrypsin activity was assayed with benzoyl-l-Tyr ethyl ester (Sigma) by monitoring the change in ΔA256 min−1; and elastase activity was assayed with N-succinyl-Ala-Ala-Ala-p-nitroanilide (Sigma) by monitoring the change in ΔA410 min−1. Trypsin and chymotrypsin activity assays were measured in 1-mL reactions using a UV-1601PC spectrophotometer (Shimadzu). Elastase activity assays were measured in 0.2-mL reactions in 96-well, nonstick microtiter plates (Corning) using a Sunrise microtiter plate reader (Tecan US). GmKTI (Sigma) and bovine serum albumin (Sigma) were assayed under identical conditions as positive and negative controls, respectively. The bovine serum albumin negative control ensured that small reductions in protease activity were due to actual inhibition and not substrate competition. KTI concentrations required to inhibit 50% of proteinase activity were calculated from the linear portion of the plot of residual proteinase activity against KTI protein (μm).
PI activity against subtilisin A (EC 3.4.21.62; Sigma) and papain (EC 3.4.22.2; Sigma) was determined by preincubating increasing concentrations of each KTI with a standard quantity of proteinase in the appropriate assay buffer for 10 min (for subtilisin, 50 mm Tris-HCl, pH 7.5, at 37°C; for papain, 50 mm sodium acetate, pH 6.0, 2 mm DTT at 23°C). After azocasein (Sigma) was added to a final concentration of 1%, the reaction was followed for 1 h and stopped by the addition of TCA. Reactions were clarified by centrifugation, NaOH was added to the supernatant, and the A450 was measured and used to calculate residual proteinase activity.
PI activity of leaf extracts was determined against trypsin, chymotrypsin, and elastase by titrating leaf protein extracts with each proteinase and residual proteinase activity measured as described above. Percent proteinase inhibition was plotted against square root-transformed protein concentration (mg mL−1) of leaf extracts because the relationship between proteinase activity and protein extract was not linear. To directly compare levels of inhibitory activity in leaves, we calculated the total protein concentration of leaf extract, which inhibits 50% of proteinase activity (IC50). For statistical comparison of inhibitor activity in leaves of control and wounding treatments, we compared slopes [% inhibition/(protein extract)½] from linear regression analysis and calculated a P value (two-tailed) testing the null hypothesis that the slopes are identical (PI activity is equal from control and wound treatments).
For stability tests, recombinant KTIs were incubated under different conditions and, after the denaturing treatment, PI activity was compared with the activity of the untreated KTIs. Inhibition of trypsin was measured for TI2 and TI3, whereas chymotrypsin was used for TI4, TI5, and TI6. For thermostability assays, KTIs were incubated from 10°C to 100°C at 10°C intervals for 30 min, and then cooled on ice for approximately 30 min before measuring residual PI activity. Some thermostability experiments were also performed with gradual cooling after heating, as well as with different incubation times (ranging from 2.5–150 min), but results were the same for both assay variations. To test stability in the presence of a reducing agent, KTIs were incubated with DTT at concentrations increasing exponentially from 1 μm to 100 mm (final concentration) for 30 and 120 min. Reactions were terminated by adding iodoacetamide at twice the concentration of DTT before measuring residual PI activity. For long-term stability, KTIs were stored at 4°C and residual PI activity was measured at 25-d intervals for 100 d. For TI2, residual activity was further measured at 50-d intervals for an additional 200 d.
PI activity against insect proteases was determined from midgut extracts. BAW (Mamestra configurata) extracts were obtained from Dr. Dwayne Hegedus (Agriculture and Agri-Food Canada). Dr. Emma Despland (Concordia University) provided FTC (Malacosoma disstria) midguts dissected from fourth and fifth instar larvae that were reared on artificial diets (Despland and Noseworthy, 2006). FTC midguts were extracted into insect Ringer's solution and clarified by centrifugation, as described previously (Hegedus et al., 2003). A standard quantity of midgut extract was preincubated with increasing concentrations of KTIs. For BAW midgut extracts, protease activity was measured in two assay buffers of different pH (0.1 m Tris-HCl, pH 8.0; 0.1 m Gly-NaOH, pH 11.0). These correspond to the two pH optima of midgut protease activity (Hegedus et al., 2003). For FTC midgut extracts, protease activity was measured at pH 11.5 (0.1 m Gly-NaOH, pH 11.0), which corresponds to optimum protease activity (data not shown). Azocasein assays were carried out as above, except that the reaction was allowed to proceed for 6 h.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Long-term stability of poplar KTIs stored at 4°C.
Supplemental Table S1. Amino acid sequence similarities and identities among members of the poplar KTI family.
Supplemental Table S2. KTI gene models from P. trichocarpa genome versions 1.0 and 1.1.
Supplementary Material
Acknowledgments
We thank Dwayne Hegedus and Emma Despland for providing BAW midgut extracts and FTC midguts, respectively; Steven Ralph and Jörg Bohlmann for providing the TI6 clone (EST ID WS0133_E08; GenBank accession no. DT502517); Charles Melnyk for help with production of active recombinant TI proteins and protease inhibitor assays; Anna Isbister and Lan Tran for help with TI3 antibodies; Nicole Dafoe and Megan Towns for analysis of TI in tent caterpillar frass; and Brad Binges for help with maintaining trees at the Bev Glover Greenhouse.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (postgraduate scholarships to I.T.M. and grants to C.P.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: C. Peter Constabel (cpc@uvic.ca).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Araujo AP, Hansen D, Vieira DF, Oliveira C, Santana LA, Beltramini LM, Sampaio CA, Sampaio MU, Oliva ML (2005) Kunitz-type Bauhinia bauhinioides inhibitors devoid of disulfide bridges: isolation of the cDNAs, heterologous expression and structural studies. Biol Chem 386 561–568 [DOI] [PubMed] [Google Scholar]
- Beynon RJ, Bond JS (1989) Proteolytic Enzymes: A Practical Approach. IRL Press, Oxford University Press, New York
- Botella MA, Xu Y, Prabha TN, Zhao Y, Narasimhan ML, Wilson KA, Nielsen SS, Bressan RA, Hasegawa PM (1996) Differential expression of soybean cysteine proteinase inhibitor genes during development and in response to wounding and methyl jasmonate. Plant Physiol 112 1201–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown DP, Wilkinson HS, Gatehouse JA (2004) Regulation of expression of genes encoding digestive proteases in the gut of a polyphagous lepidopteran larva in response to dietary protease inhibitors. Physiol Entomol 29 278–290 [Google Scholar]
- Bradshaw HD Jr, Hollick JB, Parsons TJ, Clarke HR, Gordon MP (1990) Systemically wound-responsive genes in poplar trees encode proteins similar to sweet potato sporamins and legume Kunitz trypsin inhibitors. Plant Mol Biol 14 51–59 [DOI] [PubMed] [Google Scholar]
- Chen H, Gonzales-Vigil E, Wilkerson CG, Howe GA (2007) Stability of plant defense proteins in the gut of insect herbivores. Plant Physiol 143 1954–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA (2005) Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc Natl Acad Sci USA 102 19237–19242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher ME, Miranda M, Major IT, Constabel CP (2004) Gene expression profiling of systemically wound-induced defenses in hybrid poplar. Planta 219 936–947 [DOI] [PubMed] [Google Scholar]
- Constabel CP (1999) A survey of herbivore-inducible defensive proteins and phytochemicals. In AA Agrawal, S Tuzun, E Bent, eds, Induced Defenses against Pathogens and Herbivores. APS Press, St. Paul, pp 137–166
- Dattagupta JK, Podder A, Chakrabarti C, Sen U, Mukhopadhyay D, Dutta SK, Singh M (1999) Refined crystal structure (2.3 A) of a double-headed winged bean alpha-chymotrypsin inhibitor and location of its second reactive site. Proteins 35 321–331 [DOI] [PubMed] [Google Scholar]
- De Leo F, Volpicella M, Licciulli F, Liuni S, Gallerani R, Ceci LR (2002) PLANT-PIs: a database for plant protease inhibitors and their genes. Nucleic Acids Res 30 347–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despland E, Noseworthy M (2006) How well do specialist feeders regulate nutrient intake? Evidence from a gregarious tree-feeding caterpillar. J Exp Biol 209 1301–1309 [DOI] [PubMed] [Google Scholar]
- DiBella FP, Liener IE (1969) Soybean trypsin inhibitor: cleavage and identification of a disulfide bridge not essential for activity. J Biol Chem 244 2824–2829 [PubMed] [Google Scholar]
- do Socorro MCM, Oliva ML, Fritz H, Jochum M, Mentele R, Sampaio M, Coelho LC, Batista IF, Sampaio CA (2002) Characterization of a Kunitz trypsin inhibitor with one disulfide bridge purified from Swartzia pickellii. Biochem Biophys Res Commun 291 635–639 [DOI] [PubMed] [Google Scholar]
- Duffey SS, Felton GW (1991) Enzymatic antinutritive defenses of the tomato plant against insects. In P Hedin, ed, Naturally Occurring Pest Bioregulators. ACS Press, Washington, DC, pp 167–197
- Garcia VA, Freire Md GM, Novello JC, Marangoni S, Macedo MLR (2004) Trypsin inhibitor from Poecilanthe parviflora seeds: purification, characterization, and activity against pest proteases. Protein J 23 343–350 [DOI] [PubMed] [Google Scholar]
- Glaczinski H, Heibges A, Salamini R, Gebhardt C (2002) Members of the Kunitz-type protease inhibitor gene family of potato inhibit soluble tuber invertase in vitro. Potato Res 45 163–176 [Google Scholar]
- Habu Y, Peyachoknagul S, Sakata Y, Fukasawa K, Ohno T (1997) Evolution of a multigene family that encodes the Kunitz chymotrypsin inhibitor in winged bean: a possible intermediate in the generation of a new gene with a distinct pattern of expression. Mol Genet Genomics 254 73–80 [DOI] [PubMed] [Google Scholar]
- Haruta M, Major IT, Christopher ME, Patton JJ, Constabel CP (2001) A Kunitz trypsin inhibitor gene family from trembling aspen (Populus tremuloides Michx.): cloning, functional expression, and induction by wounding and herbivory. Plant Mol Biol 46 347–359 [DOI] [PubMed] [Google Scholar]
- Hegedus D, Baldwin D, O'Grady M, Braun L, Gleddie S, Sharpe A, Lydiate D, Erlandson M (2003) Midgut proteases from Mamestra configurata (Lepidoptera: Noctuidae) larvae: characterization, cDNA cloning, and expressed sequence tag analysis. Arch Insect Biochem Physiol 53 30–47 [DOI] [PubMed] [Google Scholar]
- Heibges A, Glaczinski H, Ballvora A, Salamini F, Gebhardt C (2003. a) Structural diversity and organization of three gene families for Kunitz-type enzyme inhibitors from potato tubers (Solanum tuberosum L.). Mol Genet Genomics 269 526–534 [DOI] [PubMed] [Google Scholar]
- Heibges A, Salamini F, Gebhardt C (2003. b) Functional comparison of homologous members of three groups of Kunitz-type enzyme inhibitors from potato tubers (Solanum tuberosum L.). Mol Genet Genomics 269 535–541 [DOI] [PubMed] [Google Scholar]
- Hollick JB, Gordon MP (1993) A poplar tree proteinase inhibitor-like gene promoter is responsive to wounding in transgenic tobacco. Plant Mol Biol 22 561–572 [DOI] [PubMed] [Google Scholar]
- Hollick JB, Gordon MP (1995) Transgenic analysis of a hybrid poplar wound-inducible promoter reveals developmental patterns of expression similar to that of storage protein genes. Plant Physiol 109 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK (2005) Molecular population genetics of herbivore-induced protease inhibitor genes in European aspen (Populus tremula L., Salicaceae). Mol Biol Evol 22 1802–1812 [DOI] [PubMed] [Google Scholar]
- Jongsma MA, Bakker PL, Peters J, Bosch D, Stiekema WJ (1995) Adaptation of Spodoptera exigua larvae to plant proteinase inhibitors by induction of gut proteinase activity insensitive to inhibition. Proc Natl Acad Sci USA 92 8041–8045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53 299–328 [DOI] [PubMed] [Google Scholar]
- Kim JY, Park SC, Kim MH, Lim HT, Park Y, Hahm KS (2005) Antimicrobial activity studies on a trypsin-chymotrypsin protease inhibitor obtained from potato. Biochem Biophys Res Commun 330 921–927 [DOI] [PubMed] [Google Scholar]
- Konno K, Hirayama C, Nakamura M, Tateishi K, Tamura Y, Hattori M, Kohno K (2004) Papain protects papaya trees from herbivorous insects: role of cysteine proteases in latex. Plant J 37 370–378 [DOI] [PubMed] [Google Scholar]
- Larson PR, Isebrands JG (1971) The plastochron index as applied to developmental studies of cottonwood. Can J For Res 1 1–11 [Google Scholar]
- Laskowski M Jr, Kato I (1980) Protein inhibitors of proteinases. Annu Rev Biochem 49 593–626 [DOI] [PubMed] [Google Scholar]
- Lawrence SD, Novak NG (2001) A rapid method for the production and characterization of recombinant insecticidal proteins in plants. Mol Breed 8 139–146 [Google Scholar]
- Macedo ML, Garcia VA, Freire MG, Richardson M (2007) Characterization of a Kunitz trypsin inhibitor with a single disulfide bridge from seeds of Inga laurina (SW.) Willd. Phytochemistry 68 1104–1111 [DOI] [PubMed] [Google Scholar]
- Macedo MLR, Freire MGM, Martins LTDM, Martinez DST, Gomes VM, Smolka MB, Toyama MH, Marangoni S, Coelho LCBB (2004) Novel protein from Labramia bojeri A. DC. seeds homologue to Kunitz-type trypsin inhibitor with lectin-like properties. J Agric Food Chem 52 7548–7554 [DOI] [PubMed] [Google Scholar]
- Major IT, Constabel CP (2006) Molecular analysis of poplar defense against herbivory: comparison of wound- and insect elicitor-induced gene expression. New Phytol 172 617–635 [DOI] [PubMed] [Google Scholar]
- Mazumdar-Leighton S, Broadway RM (2001) Identification of six chymotrypsin cDNAs from larval midguts of Helicoverpa zea and Agrotis ipsilon feeding on the soybean (Kunitz) trypsin inhibitor. Insect Biochem Mol Biol 31 633–644 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Kortt AA (1997) The 1.8 A crystal structure of winged bean albumin 1, the major albumin from Psophocarpus tetragonolobus (L.) DC. J Mol Biol 269 881–891 [DOI] [PubMed] [Google Scholar]
- Miranda M, Christopher ME, Constabel CP (2004) The variable nature of herbivore defense: evidence for a rapidly diverging Kunitz trypsin inhibitor gene in Populus. In QCB Cronk, J Whitton, RH Ree, IEP Taylor, eds, Plant Adaptation: Molecular Genetics and Ecology. NRC Press, Ottawa, pp 153–158
- Mohan S, Ma PWK, Pechan T, Bassford ER, Williams WP, Luthe DS (2006) Degradation of the S. frugiperda peritrophic matrix by an inducible maize cysteine protease. J Insect Physiol 52 21–28 [DOI] [PubMed] [Google Scholar]
- Pando SC, Oliva ML, Sampaio CA, Di Ciero L, Novello JC, Marangoni S (2001) Primary sequence determination of a Kunitz inhibitor isolated from Delonix regia seeds. Phytochemistry 57 625–631 [DOI] [PubMed] [Google Scholar]
- Park Y, Choi BH, Kwak JS, Kang CW, Lim HT, Cheong HS, Hahm KS (2005) Kunitz-type serine protease inhibitor from potato (Solanum tuberosum L. cv. Jopung). J Agric Food Chem 53 6491–6496 [DOI] [PubMed] [Google Scholar]
- Pechan T, Cohen A, Williams WP, Luthe DS (2002) Insect feeding mobilizes a unique plant defense protease that disrupts the peritrophic matrix of caterpillars. Proc Natl Acad Sci USA 99 13319–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph S, Oddy C, Cooper D, Yueh H, Jancsik S, Kolosova N, Philippe R, Aeschliman D, White R, Huber D, et al (2006) Genomics of hybrid poplar (Populus trichocarpa × deltoides) interacting with forest tent caterpillars (Malacosoma disstria): normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defences in poplar. Mol Ecol 15 1275–1297 [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Tolle DP, Barrett AJ (2004) Evolutionary families of peptidase inhibitors. Biochem J 378 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revina TA, Speranskaya AS, Kladnitskaya GV, Shevelev AB, Valueva TA (2004) Subtilisin protein inhibitor from potato tubers. Biochemistry (Mosc) 69 1092–1098 [DOI] [PubMed] [Google Scholar]
- Richardson M (1991) Seed storage proteins: the enzyme inhibitors. In LJ Rogers, ed, Amino Acids, Proteins and Nucleic Acids, Vol 5. Academic Press, Toronto, pp 259–305
- Ritonja A, Krizaj I, Mesko P, Kopitar M, Lucovnik P, Strukelj B, Pungercar J, Buttle DJ, Barrett AJ, Turk V (1990) The amino acid sequence of a novel inhibitor of cathepsin D from potato. FEBS Lett 267 13–15 [DOI] [PubMed] [Google Scholar]
- Rowan AD, Brzin J, Buttle DJ, Barrett AJ (1990) Inhibition of cysteine proteinases by a protein inhibitor from potato. FEBS Lett 269 328–330 [DOI] [PubMed] [Google Scholar]
- Ryan CA (1990) Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu Rev Phytopathol 28 425–449 [Google Scholar]
- Song HK, Suh SW (1998) Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator. J Mol Biol 275 347–363 [DOI] [PubMed] [Google Scholar]
- Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, Charbonnel-Campaa L, Lindvall JJ, Tandre K, Strauss SH, et al (2004) A Populus EST resource for plant functional genomics. Proc Natl Acad Sci USA 101 13951–13956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumikawa JT, Nakahata AM, Fritz H, Mentele R, Sampaio MU, Oliva ML (2006) A Kunitz-type glycosylated elastase inhibitor with one disulfide bridge. Planta Med 72 393–397 [DOI] [PubMed] [Google Scholar]
- Talyzina N, Ingvarsson P (2006) Molecular evolution of a small gene family of wound inducible Kunitz trypsin inhibitors in Populus. J Mol Evol 63 108–119 [DOI] [PubMed] [Google Scholar]
- Terada S, Fujimura S, Katayama H, Nagasawa M, Kimoto E (1994) Purification and characterization of two Kunitz family subtilisin inhibitors from seeds of Canavalia lineata. J Biochem (Tokyo) 115 392–396 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Valueva TA, Revina TA, Mosolov VV, Mentele R (2000) Primary structure of potato Kunitz-type serine proteinase inhibitor. Biol Chem 381 1215–1221 [DOI] [PubMed] [Google Scholar]
- van Dam NM, Horn M, Mares M, Baldwin IT (2001) Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J Chem Ecol 27 547–568 [DOI] [PubMed] [Google Scholar]
- Walling LL (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19 195–216 [DOI] [PubMed] [Google Scholar]
- Wang J, Constabel CP (2004) Polyphenol oxidase overexpression in transgenic Populus enhances resistance to herbivory by forest tent caterpillar (Malacosoma disstria). Planta 220 87–96 [DOI] [PubMed] [Google Scholar]
- Worthington CC (1988) Worthington Enzyme Manual. Worthington Biochemical Corporation, Freehold, NJ
- Yao PL, Hwang MJ, Chen YM, Yeh KW (2001) Site-directed mutagenesis evidence for a negatively charged trypsin inhibitory loop in sweet potato sporamin. FEBS Lett 496 134–138 [DOI] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Baldwin IT (2004) Constitutive and inducible trypsin proteinase inhibitor production incurs large fitness costs in Nicotiana attenuata. Proc Natl Acad Sci USA 101 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.