Abstract
We used pulsed-field gel electrophoresis and restriction fragment mapping to analyze the structure of Medicago truncatula chloroplast DNA (cpDNA). We find most cpDNA in genome-sized linear molecules, head-to-tail genomic concatemers, and complex branched forms with ends at defined sites rather than at random sites as expected from broken circles. Our data suggest that cpDNA replication is initiated predominantly on linear DNA molecules with one of five possible ends serving as putative origins of replication. We also used 4′,6-diamidino-2-phenylindole staining of isolated plastids to determine the DNA content per plastid for seedlings grown in the dark for 3 d and then transferred to light before being returned to the dark. The cpDNA content in cotyledons increased after 3 h of light, decreased with 9 h of light, and decreased sharply with 24 h of light. In addition, we used real-time quantitative polymerase chain reaction to determine cpDNA levels of cotyledons in dark- and light-grown (low white, high white, blue, and red light) seedlings, as well as in cotyledons and leaves from plants grown in a greenhouse. In white, blue, and red light, cpDNA increased initially and then declined, but cpDNA declined further in white and blue light while remaining constant in red light. The initial decline in cpDNA occurred more rapidly with increased white light intensity, but the final DNA level was similar to that in less intense light. The patterns of increase and then decrease in cpDNA level during development were similar for cotyledons and leaves. We conclude that the absence in M. truncatula of the prominent inverted repeat cpDNA sequence found in most plant species does not lead to unusual properties with respect to the structure of plastid DNA molecules, cpDNA replication, or the loss of cpDNA during light-stimulated chloroplast development.
For many years, the standard depiction of the chloroplast chromosome has been a genome-sized circular DNA molecule. For most plants, such as maize (Zea mays), restriction fragment mapping shows the chloroplast genome as divided into large and small single copy regions separated by inverted repeat sequences (IRA and IRB; Heinhorst and Cannon, 1993; Kunnimalaiyaan and Nielsen, 1997a). We previously found, however, most of the chloroplast DNA (cpDNA) of maize seedlings as linear and branched molecules with defined ends and proposed that the inversion isomers of IR-containing chloroplast genomes were produced during a recombination-dependent process of cpDNA replication (Oldenburg and Bendich, 2004a, 2004b) rather than the standard depiction of “flipping” recombination (without replication) within a circular molecule (Palmer, 1983, 1985). During plastid development, the form of maize cpDNA changes from multigenomic structures in the proplastids of meristematic tissue, where cpDNA replication occurs, to subgenomic molecules in expanding leaf cells after replication ceases, and finally to completely degraded DNA molecules in most chloroplasts of fully mature leaves (Oldenburg and Bendich, 2004a).
Some plants, such as Medicago truncatula, contain only one copy of the IR (Palmer, 1985; Saski et al., 2005). In this work, we ask whether the cpDNA in M. truncatula is similar to that in IR-containing plants with respect to structure and replication. Using in-gel procedures to protect large DNA molecules from breakage during DNA extraction, we find most M. truncatula cpDNA in linear and multigenomic branched forms rather than as circles. The ends of the linear molecules are not random, as expected from broken circles, and are near putative origins of cpDNA replication. Thus, we conclude that the absence of the prominent IR, which would preclude “flipping,” does not confer on the plastid genome any noticeable structural alterations when compared with the far more common IR-containing plastid genomes.
We previously found that a decline in the amount of cpDNA during chloroplast development in maize seedlings was triggered by light (Oldenburg et al., 2006). A second objective in this article is to determine whether the retention of cpDNA in M. truncatula is similarly affected by light. We find that after an initial stimulatory effect on cpDNA replication, light does indeed trigger a rapid decline in cpDNA, indicating that the absence of the IR does not prevent light-stimulated degradation of cpDNA. The rate of cpDNA degradation is dependent both on the intensity and the quality of the light.
RESULTS
Fragment Mapping of the Chloroplast Genome
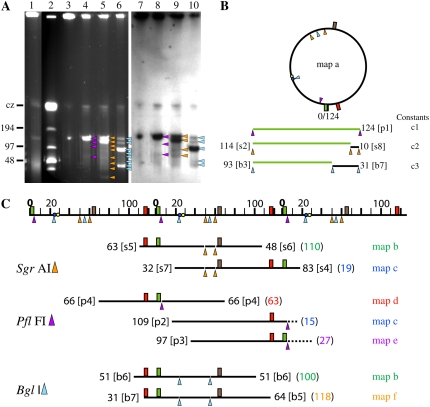
Pulsed-field gel electrophoresis (PFGE) reveals several forms of M. truncatula cpDNA (Fig. 1A, lane 1): linear molecules at the size of the genome (124 kb) with the dimer and higher concatemers bunched together in the compression zone, a well-bound fraction that did not migrate into the gel, and a smear of linear DNA molecules less than the size of the genome. As with light-grown maize (Oldenburg et al., 2006), no band of supercoiled circular DNA is observed with M. truncatula, but linear dimer and trimer bands generally lacking in maize are typically observed with M. truncatula at longer pulse times (Shaver et al., 2006).
Figure 1.
Maps of the M. truncatula chloroplast genome based on PflFI, SgrAI, and BglI digestions. A, PFGE and blot hybridization of undigested and enzyme-digested cpDNA. Lane 1, No pre-electrophoresis (see “Materials and Methods”); lane 2, size standards; lanes 3 to 10, after pre-electrophoresis. Lanes 1, 3, and 7, Undigested cpDNA; lanes 4 and 8, PflFI-digested cpDNA; lanes 5 and 9, SgrAI-digested cpDNA; lanes 6 and 10, BglI-digested cpDNA. Lanes 1 to 6, Ethidium fluorescence; lanes 7 to 10, ndhF hybridization. Fragment sizes for ethidium-stained PflFI- (p, purple triangles), SgrAI- (s, orange triangles), and BglI-digested (b, blue triangles) cpDNA in kilobase pairs and presence (+) or absence (−) of ndhF hybridization (Table I): p1, 124(+); p2, 109(−); p3, 97(+); p4, 66(+); s1, 124(+); s2, 114(+); s3, 101(−); s4, 83(+); s5, 63(+); s6, 48(−); s7, 32(−); s8, 10(−); b1, 124(+); b2, 112(+); b3, 93(+); b4, 77(−); b5, 64(−); b6, 51(+); b7, 31(+); and b8, 29(−). Lower exposures (not shown) show fragments s1 and s2 as discrete bands in lanes 5 and 9. Fragments s1, b1, and b2 probably result from incomplete digestion. cz, Compression zone. B, Map a, the circular map of the M. truncatula chloroplast genome. Symbols: purple triangle, PflFI; blue triangles, BglI; orange triangles, SgrAI; red rectangle, rbcL; green rectangle, ndhF; brown rectangle, petA; blue circle, homolog of Oenothera oriB; yellow circle, homolog of Oenothera oriA. Constants c1 to c3 are predicted fragments resulting from digestion of a circular or h-t multigenomic linear molecule with a one-cut (PflFI) or two-cut (SgrAI or BglI) restriction enzyme. C, Maps b to f, linear maps. A linear scale is shown at the top with units in kilobase pairs. The circular (B) and linear (C) maps are proportional but not drawn to the same scale. Linear maps of 124-kb monomers were constructed as follows: map c is constructed with an internal SgrAI constant fragment of 10 kb (s8) and two SgrAI end fragments (s4 and s7) of 83 and 32 kb, respectively, nearly equaling 124 kb and giving an end near 20 kb (near homologs of Oenothera oriA and oriB). Map f is constructed with an internal BglI constant fragment of 31 kb (b7) and two BglI end fragments (b5 and b7; 64 and 31 kb, respectively), nearly equaling 124 kb and giving an end near 120 kb (near rbcL, trnK-UUU, matK, and psbA genes). The dashed lines in PflFI maps c and e represent predicted fragments (15 and 27 kb, respectively) that were not visible by ethidium staining. Only maps supported by more than one enzyme digestion or probe are shown.
From the M. truncatula chloroplast genome sequence (GenBank accession no. AC093544), we identified restriction enzymes that cut only once (PflFI) or twice (BglI and SgrAI) per genome. We used these enzymes to determine whether the linear genomic concatemers have ends that are defined or random and contain head-to-tail (h-t), head-to-head (h-h), or tail-to-tail (t-t) genomic units. If there were defined ends, then discrete subgenomic fragments would be generated, whereas molecules with random ends would instead yield a broad range of subgenomic DNA sizes (Oldenburg and Bendich, 2001). Subgenomic restriction fragments can be obscured, however, if a smear of subgenomic DNA is present in uncut DNA (Fig. 1A, lane 1). Consequently, prior to restriction digestion, this smear as well as other mobile DNA were removed by pre-electrophoresis (Fig. 1A, lane 3), and the remaining well-bound forms were digested with restriction enzyme so that discrete subgenomic restriction fragments were detectable (Fig. 1A, lanes 4–6). Thus, there are defined, not random, ends for the M. truncatula cpDNA molecules, as we found for maize cpDNA (Oldenburg and Bendich, 2004b). For maize cpDNA, the complex forms in the well-bound fraction and linear monomers yield the same subgenomic restriction fragments (Oldenburg and Bendich, 2004b), and we assume that the same applies to M. truncatula.
For each type of concatemer (h-t, h-h, and t-t), digestion with PflFI would produce specific diagnostic fragments: monomer-sized for h-t concatemers and fragments larger and smaller than (but not equal to) the size of the genome for h-h, t-t, and h-h/t-t concatemers. The 124-kb genome-sized PflFI fragment that we found (Fig. 1A, lane 4) accounted for most of the mobile DNA and indicates that the linear concatemers are in an h-t arrangement. Although both circular forms (the monomer and the h-t dimer) and the h-t linear concatemer would generate genome-sized PflFI fragments, only linear forms would yield the subgenomic PflFI fragments that we also find in lane 4. The h-h/t-t circular and t-t linear arrangements are ruled out because they predict a larger than genome-sized PflFI fragment that was not observed. Because a linear trimer or higher oligomer containing an h-h linkage would also require a t-t arrangement, the h-h linear dimer is unlikely.
About 74% of the uncut cpDNA in lane 1 remained well bound, which represents branched linear and relaxed circular molecules. Of the well-bound DNA, about 35% remained in the well, 37% was in monomer fragments, and 14% was in subgenomic fragments following PflFI-digestion (Fig. 1A, lane 4). The residual well-bound DNA represents branched molecules that terminate at an enzyme recognition site (Oldenburg and Bendich, 2004b), and the genome-sized fragment represents the maximum amount in circular form (about 27% of uncut cpDNA), as determined by the fluorescence intensities of the PFGE fractions. The actual amount in circular form is probably much lower because this band also includes fragments that arise from branched h-t linear concatemers.
For a population of linear molecules (monomers and concatemers) that all have the same left and right ends, we would expect only two subgenomic fragments that sum to the size of the genome after digestion with a single-site restriction enzyme. These two fragments would also be produced from the ends of branched multigenomic forms. For the PflFI digest, however, there were three major subgenomic fragments (p2–p4 in Fig. 1A, lane 4, and C; Table I). Taken as pairs, the fragments can be summed to the approximate size of the genome (as done previously for maize [Oldenburg and Bendich, 2004b] and tobacco [Nicotiana tabacum; Scharff and Koop, 2006]) in three ways: p2 (109 kb) and a predicted fragment of 15 kb, p3 (97 kb) and a predicted fragment of 27 kb, and a pair of p4 fragments (66 kb; Fig. 1C; Table I). The 15- and 27-kb fragments are most likely not visible because their small size greatly reduces their ethidium stain visibility (Fig. 1A, lane 4). Similar reasoning can be applied independently for the BglI and SgrAI digests (Fig. 1, A and C; Table I). However, because these are two-cut restriction enzymes, two subgenomic fragments summing to the genome size are expected for circular molecules (instead of only one fragment of genome-size for PflFI) and three subgenomic fragments summing to the genome size (consisting of two end fragments and an internal fragment of 31 kb [BglI] or 10 kb [SgrAI]) are expected for linear molecules.
Table I.
Predicted and observed restriction fragments and chloroplast gene hybridization
| Enzymea | Fragmentb | Predicted Sizec | Measured Sized | Observed Fragment Intensitiese
|
|||
|---|---|---|---|---|---|---|---|
| EtBr | petA | ndhF | rbcL | ||||
| kb | |||||||
| PflFI | p1 | 124 | 124, 125 | +++ | +++ | +++ | |
| p2 | 109 | 109, 109 | + | + | |||
| p3 | 97 | 95, 99 | + | + | + | ||
| p4 | 66 | 64, 67 | + | + | + | ||
| SgrAI | s1 | 124 | 123, 124, 125 | +++ | +++ | +++ | +++ |
| s2 | 114 | 114, 114, 116 | +++ | +++ | +++ | +++ | |
| s3 | 101 | 102, 102, 103 | + | ||||
| s4 | 83 | 82, 83, 84 | + | ++ | ++ | ++ | |
| s5 | 63 | 63, 64, 66 | + | ++ | ++ | ++ | |
| s6 | 48 | 47, 48, 50 | + | ++ | |||
| s7 | 32 | 31, 32 | + | ||||
| s8 | 10 | 2, 5, 8 | + | ||||
| BglI | b1 | 124 | 120, 121, 124 | ++ | ++ | ++ | ++ |
| b2f | 112 | 105, 108, 111 | ++ | ++ | ++ | ++ | |
| b3 | 93 | 86, 87, 89 | +++ | +++ | +++ | +++ | |
| b4 | 77 | 76 | + | ||||
| b5 | 64 | 57, 63 | + | + | |||
| b6 | 51 | 41, 44, 50 | + | + | + | + | |
| b7 | 31 | 35, 37, 38 | +++ | + | + | ||
| b8 | 29 | 31 | + | ||||
Cut sites at 5,462 for PflFI; 50,410 and 59,726 for SgrAI; 23,921 and 55,251 for BglI.
Three pairs of PflFI subgenomic fragments (p2 + 15-kb, not detected; p3 + 27-kb, not detected; p4 + p4) sum to about genome size (Fig. 1C, c1). Three pairs of SgrAI subgenomic fragments (s3 + s8; s4 + s7; s5 + s6) sum to about 114 kb, the size of the genome (124 kb) minus the size of an internal fragment of 10 kb (s8; Fig. 1C, c2). Two pairs of BglI subgenomic fragments (b5 + b7; b6 + b6) sum to about 93 kb, the size of the genome minus the size of an internal restriction fragment of 31 kb (b7; Fig. 1C, c3). Additional internal fragments of 124 kb (p1), 114 kb (s2), and 93 kb (b3) can result from the digestion of circular or linear molecules as monomeric or concatameric units. The subgenomic end fragments (p2–p4; s3–s8; b4–b8) can only result from linear molecules, not circles (except for s8 and b7, which can be both end and internal fragments).
Predicted fragment sizes were used to construct maps (Fig. 1C) and are expected restriction fragments (p1, s1, s2, s8, b1, b3, and b7) and averages for all other measured fragment sizes (p2–p4, s3–s7, and b2, b4, b5, b6, and b8).
Two determinations were made for PflFI and three were made for SgrAI and BglI.
Fragment intensities were judged visually as high (+++), medium (++), or low (+). Probes ndhF and rbcL were used with PflFI, and petA, ndhF, and rbcL with SgrAI and BglI.
Presence of this fragment is likely due to incomplete BglI digestion. Perhaps this fragment is due to the 77-kb (b4) fragment plus the internal, 31-kb (b7) fragment and pairs with a 16-kb fragment (too faint to be visible) to sum to the 124-kb genome.
We mapped the ends using probes to different regions of the genome: ndhF (Fig. 1, A–C; Table I), rbcL, and petA (Fig. 1, B and C; Table I). Map a (Fig. 1B) is circular and maps b to e (Fig. 1C) are linear, based on the hybridization of these probes to subgenomic fragments (Fig. 1A; Table I). The SgrAI and BglI fragment and hybridization data are consistent with maps generated by the PflFI digest with ndhF and rbcL hybridization (Fig. 1, A–C; Table I). Based on our physical mapping of M. truncatula cpDNA, the predominant ends of linear molecules lie near 20, 30, 60, 100, and 120 kb. Interestingly, two of the ends (Fig. 1C, maps c and e) lie near a sequence homologous to replication origins previously identified in Oenothera (Chiu and Sears, 1992); the other ends could be near previously unclassified origins. These results suggest that the ends may represent initiation sites for recombination-dependent DNA replication of linear molecules, as we inferred for maize (Oldenburg and Bendich, 2004b).
Genomes per Plastid after Transfer of Dark-Grown Seedlings to Light
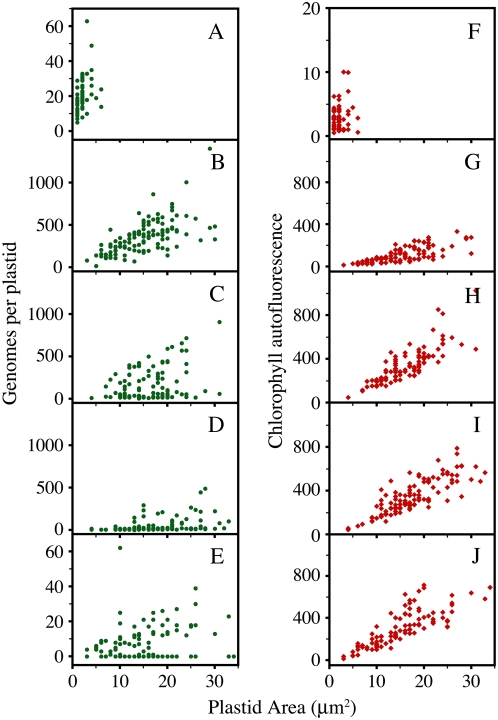
M. truncatula seedlings were harvested after 5 d of growth under one of the following light conditions: constant dark (Fig. 2, A and F); 3 d in the dark and then transferred to white light for 3 h (Fig. 2, B and G), 9 h (Fig. 2, C and H), or 24 h (Fig. 2, D and I) and then returned to the dark; and four 16-h-light/8-h-dark cycles followed by 1 d in the dark (Fig. 2, E and J). After the dark-grown cotyledons were transferred to the light, their appearance changed from a dull yellow color to pale green by 3 h, and fully green but not yet fully expanded by 24 h. Plastids were isolated from the entire cotyledon and analyzed for size, 4′,6-diamino-phenylindole (DAPI)-DNA fluorescence, and chlorophyll autofluorescence (Fig. 2, A–J; Table II). The etioplasts from the dark-grown seedlings (Fig. 2, A and F) were much smaller and had much less chlorophyll autofluorescence than the plastids from light-grown seedlings (Fig. 2, B–E and G–J). With 3 h of light exposure, plastids had much more cpDNA and reached nearly maximum size (Fig. 2B) and had increased chlorophyll content (Fig. 2G). With an additional 6 h of light, the plastids were fully developed (Fig. 2H) but experienced a 2-fold decrease in cpDNA (Fig. 2C). A 24-h white light exposure resulted in further reduction of DNA per plastid (Fig. 2D) with no change in chlorophyll autofluorescence (Fig. 2I). The seedlings grown under light/dark cycles had an average of only eight genomes per plastid, and 35 of the 102 plastids measured had no detectable DAPI-DNA fluorescence at all (Fig. 2E).
Figure 2.
The effect of white light on genome copy number and chlorophyll autofluorescence per plastid and plastid area. Plastids were isolated from cotyledons of 5-d-old seedlings grown under: constant dark; constant dark for 3 d, given a period of white light for 3, 9, or 24 h, and then returned to the dark; or 16-h-light/8-h-dark cycles. A to E, Genomes per plastid. F to J, Chlorophyll autofluorescence. Note that the scales in A, E, and F are changed. A and F, Constant dark; B and G, 3-h light period; C and H, 9-h light period; D and I, 24-h light period; E and J, 16-h-light/8-h-dark cycles. Each point represents one plastid. For statistical information, see Table II. [See online article for color version of this figure.]
Table II.
Plastid size, chlorophyll autofluorescence, and DNA content
| Light Conditionsa | No. of Plastids | Plastid Areab | Chlorophyll Autofluorescencec | Genomes per Plastidd | Plastids with DNA |
|---|---|---|---|---|---|
| % | |||||
| Dark | 57 | 2 ± 1 (1–6) | 3 ± 1 (0.5–10) | 21 ± 1 (5–63) | 100 |
| 3 h | 110 | 15 ± 1 (3–30) | 109 ± 7 (12–333) | 364 ± 19 (15–1400) | 100 |
| 9 h | 100 | 16 ± 1 (4–31) | 341 ± 16 (45–1023) | 180 ± 20 (7–908) | 100 |
| 24 h | 114 | 18 ± 1 (4–33) | 347 ± 15 (41–789) | 51 ± 8 (1–486) | 100 |
| Light/dark | 102 | 15 ± 1 (3–34) | 313 ± 20 (11–1297) | 8 ± 1 (0–62) | 66 |
Seedlings were grown for 5 d in constant dark (Fig. 2, A and F); constant dark with a 3- (Fig. 2, B and G), 9- (Fig. 2, C and H), or 24-h (Fig. 2, D and I) white light period after 3 d; or 16-h-light/8-hour dark cycles (Fig. 2, E and J).
Plastid area (μm2) was measured under white light. The average area ± se and (range) are given. The mean plastid area (μm2) for A and F is significantly lower than for B and G, C and H, D and I, or E and J (P < 0.0001).
The average chlorophyll autofluorescence per plastid ± se and (range) are given (as described in “Materials and Methods”). The mean autofluorescence for F is significantly lower than that for G to J (P < 0.0001), and G is significantly lower than H to J (P < 0.0001).
The average genomes per plastid ± se and (range) are given (as described in “Materials and Methods”). The mean genomes per plastid for B is significantly higher than C, and C is higher than A, D, or E (P < 0.0001). All of the plastids for A, B, C, or D had DAPI-DNA fluorescence, whereas 35 (34%) of the 102 for E had no DAPI-DNA fluorescence.
The Effect of Tissue Age on the Abundance of cpDNA
We previously used real-time quantitative PCR (qPCR) with three primer sets (for petA, psbA, and ndhA) spaced widely across the chloroplast genome in order to quantify cpDNA levels in Arabidopsis (Arabidopsis thaliana; Rowan et al., 2007). The level of cpDNA determined with each of the primer sets was the same. As an alternative to DAPI-DNA fluorescence, we used qPCR with petA and psbA to study the effects of light on chloroplast genome copy number in M. truncatula. Table III shows that light affected the abundance of cpDNA in M. truncatula cotyledons and that petA and psbA gave indistinguishable values for cpDNA amount. We chose petA to conduct a more extensive qPCR analysis of the effects of tissue age and light quality, intensity, and duration on cpDNA abundance.
Table III.
Chloroplast per nuclear genome copies
| Light Conditionsa | Chloroplast/Nuclear Genomesb
|
|
|---|---|---|
| petA | psbA | |
| Dark | 1,047 ± 85 (781–1,314) | 1,052 ± 61 (929–1,113) |
| 2 h | 2,285 ± 181 (1,859–2,817) | 2,343 ± 256 (1,911–2,798) |
| 4 h | 2,109 ± 244 (1,607–2,937) | 1,852 ± 20 (1,833–1,872) |
| 16 h | 1,957 ± 113 (1,758–2,257) | 1,935 ± 457 (1,351–2,837) |
Seedlings were grown for 3 d in constant dark or 3 d constant dark followed by a 2-, 4-, or 16-h white light period.
The average chloroplast per nuclear genome copy number ± se and (range) are given (as described in “Materials and Methods”).
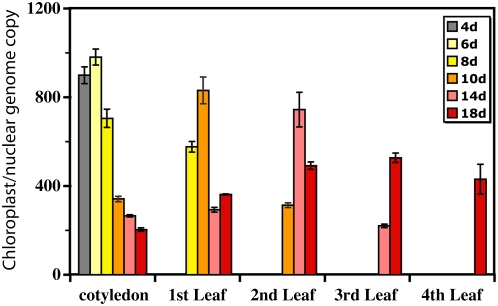
Total tissue DNA was extracted from cotyledons and from the first, second, third, and fourth leaves of 4-, 6-, 8-, 10-, 14-, and 18-d-old plants and analyzed by qPCR (Fig. 3; Table IV). In each case, there was an initial increase in cpDNA level from young to maturing tissues, followed by a decline with age relative to nuclear DNA. Note that cpDNA still remained in chloroplasts from the cotyledons and the first leaves 10 and 14 d after their initial measurements on days 4 and 8, respectively. The level of cpDNA continued to decline in the cotyledons well after full expansion (at about day 8) and chloroplasts presumably ceased dividing. Because M. truncatula does not exhibit endoploidy in the leaves (Kondorosi et al., 2000), the decline in cpDNA relative to nuclear DNA cannot be attributed to an increase in nuclear DNA with no change in cpDNA. We conclude that tissue age affects the decline in cpDNA and that the same pattern of cpDNA increase and decline occurs in the cotyledons and the leaves during the development of each tissue.
Figure 3.
The effect of tissue age on the abundance of cpDNA. Total tissue DNA was extracted from cotyledons and the first, second, third, and fourth leaves (if available) of 4-, 6-, 8-, 10-, 14-, and 18-d-old plants. The ratio of chloroplast to nuclear genome copies was determined by real-time qPCR with petA (chloroplast) and enod11 (nuclear) probes, respectively. Note that the absence of bars does not indicate readings of zero DNA but that these tissues were not present at those stages of plant development. For statistical information, see Table IV.
Table IV.
Chloroplast per nuclear genome copies
| Plant Tissuea Sample | Plant Age | Chloroplast/Nuclear Genomesb |
|---|---|---|
| d | ||
| Cotyledon | ||
| C-4 | 4 | 899 ± 38 (838–891) |
| C-6 | 6 | 981 ± 36 (955–1,053) |
| C-8 | 8 | 705 ± 41 (662–787) |
| C-10 | 10 | 342 ± 12 (330–353) |
| C-14 | 14 | 265 ± 5 (255–272) |
| C-18 | 18 | 204 ± 8 (187–212) |
| First Leaf | ||
| L1-8 | 8 | 577 ± 24 (553–600) |
| L1-10 | 10 | 831 ± 60 (771–891) |
| L1-14 | 14 | 293 ± 11 (272–308) |
| L1-18 | 18 | 362 ± 3 (360–365) |
| Second Leaf | ||
| L2-10 | 10 | 313 ± 11 (296–333) |
| L2-14 | 14 | 745 ± 78 (633–895) |
| L2-18 | 18 | 491 ± 17 (474–508) |
| Third Leaf | ||
| L3-14 | 14 | 221 ± 8 (205–229) |
| L3-18 | 18 | 527 ± 22 (493–567) |
| Fourth Leaf | ||
| L4-18 | 18 | 430 ± 67 (308–539) |
Total tissue DNA was extracted from cotyledons and leaves of 4- to 18-d-old plants grown in a greenhouse (Fig. 3).
The average chloroplast per nuclear genome copy number ± se and (range) are given for qPCR data using petA.
The Effects of Light Quality, Intensity, and Duration on the Abundance of cpDNA
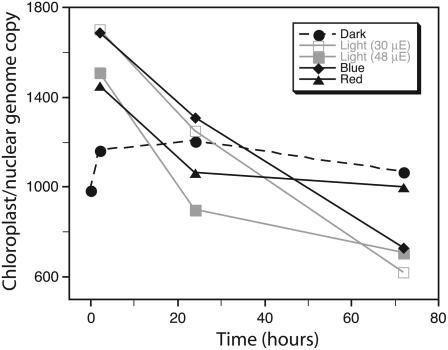
Seedlings were harvested after growth under each of the following light conditions: 3 d of constant dark with an additional 2, 24, or 72 h of dark; or 3 d in the dark followed by a period in the light (low white, 30 μE; high white, 48 μE; blue; or red) for 2, 24, or 72 h (Fig. 4; Table V). No dark treatment was given after illumination. After the dark-grown cotyledons were transferred to the light, their appearance changed from a dull yellow color to pale green by 2 h, fully green by 24 h, and still expanding up to 72 h. Total DNA was extracted from the entire cotyledon and analyzed by qPCR. After a slight increase, the level of cpDNA remained constant with time for the dark-grown seedlings; however, there was a sharp increase in cpDNA when the dark-grown seedlings were transferred to the light (low and high white, blue and red) for 2 h. At 72 h, cpDNA had declined to similar levels for the seedlings grown under low white, high white, and blue light. However, the rate of cpDNA decline was greater from 2 to 24 h for high white than low white or blue light illumination. For red light, cpDNA declined from 2 to 24 h, but unlike all the other light treatments, there was little additional decline at 72 h. In addition, the level of cpDNA increased when either the 72-h white light-grown seedlings were transferred to the dark for 24 h or the 72-h dark-grown seedlings were transferred to the light for 24 h (Table V). We conclude that illumination with white, red, or blue light is sufficient to trigger cpDNA replication, but only white and blue light promote cpDNA degradation with prolonged illumination. We also conclude that light can stimulate cpDNA replication regardless of the length of time the seedlings were kept in the dark and that the loss of cpDNA during light periods can be reversed during subsequent dark periods.
Figure 4.
The effect of light quality, intensity, and duration on the abundance of cpDNA. Total cotyledon DNA was extracted from seedlings grown under: 3 d of constant dark with an additional 2, 24, or 72 h of dark; or 3 d constant dark followed by a period of light (low white, high white, blue, or red) for 2, 24, or 72 h. The ratio of chloroplast to nuclear genome copies was determined by real-time qPCR with petA (chloroplast) and enod11 (nuclear) probes, respectively. For statistical information, see Table V.
Table V.
Chloroplast per nuclear genome copies
| Light Conditionsa | Sample | Chloroplast/Nuclear Genomesb |
|---|---|---|
| Dark | ||
| 0 h | D1 | 982 ± 6 (976–989) |
| 2 h | D2 | 1,161 ± 40 (1,121–1,243) |
| 24 h | D3 | 1,202 ± 87 (1,075–1,370) |
| 72 h | D4 | 1,068 ± 22 (1,038–1,113) |
| 72 h + 24 h lightc | D5 | 1,502 ± 174 (1,218–1,820) |
| White light | ||
| 2 h | L1 | 1,700 ± 71 (1,629–1,771) |
| 24 h | L2 | 1,247 ± 146 (969–1,468) |
| 72 h | L3 | 620 ± 37 (545–662) |
| 72 h + 24 h darkd | L4 | 1,370 ± 226 (1,144–1,596) |
| Intense white light | ||
| 2 h | I1 | 1,510 ±10 (1,499–1,520) |
| 24 h | I2 | 898 ± 19 (879–917) |
| 72 h | I3 | 707 ± 27 (680–734) |
| Blue light | ||
| 2 h | B1 | 1,684 ± 174 (1,510–1,859) |
| 24 h | B2 | 1,306 ± 122 (1,184–1,428) |
| 72 h | B3 | 730 ± 149 (580–879) |
| Red light | ||
| 2 h | R1 | 1,448 ± 20 (1,428–1,468) |
| 24 h | R2 | 1,059 ± 147 (803–1,314) |
| 72 h | R3 | 996 ± 164 (832–1,160) |
Seedlings were grown for 3 d in constant dark with an additional 2, 24, or 72 h of dark; or 3 d constant dark followed by a 2-, 24-, or 72-h-light period with white, intense white, blue, or red light (Fig. 4).
The average chloroplast per nuclear genome copy number ± se and (range) are given for qPCR data using petA.
Seedlings were grown for 6 d in constant dark and then transferred to the light for 24 h.
Seedlings were grown for 3 d in constant dark, transferred to the light for 72 h, and then transferred back into the dark for 24 h.
DISCUSSION
We have characterized the DNA from M. truncatula chloroplasts with respect to the structure of DNA molecules and the persistence of cpDNA during light-stimulated chloroplast development. We wished to determine whether the absence of the prominent IR sequence in M. truncatula is associated with any obvious difference in these two parameters when compared with the IR-containing genomes of most plant species. We found no such differences. Our data do, however, indicate that the persistence of cpDNA may depend on the degree to which plastids develop in the dark.
Circular Maps and Linear Maps
Previous results from mapping and sequencing (sequence assembly is a mapping technique) for cpDNAs were interpreted to indicate that the cpDNA in vivo exists as a genome-sized circular DNA molecule (Heinhorst and Cannon, 1993; Kunnimalaiyaan and Nielsen, 1997a). Yet, our present results and those recently obtained for maize (Oldenburg and Bendich, 2004b) and tobacco (Scharff and Koop, 2006, 2007) show that most cpDNA molecules are actually in linear form. What is the basis for these conflicting conclusions? If cpDNA prepared in liquid is used for mapping, as was routinely done in the past, the concatemers will be broken because of hydrodynamic shear to yield fragments containing gene A linked to gene B as well as to gene Z, thereby generating a circular map. Such fragments produced within the chloroplast during light-stimulated degradation of cpDNA (and detected as the subgenomic smear of cpDNA sizes in lane 1 of Fig. 1A) will also contribute to the erroneous interpretation of circular chromosomes. Furthermore, the linear and circular forms of cpDNA may be readily distinguished with restriction enzymes that have only one or two recognition sites per genome rather than enzymes with many sites, as used in the past. Discrete subgenomic bands following digestion with a single-cutter is diagnostic for linear forms with defined ends. Starting with in-gel prepared DNA and focusing only on molecules of genomic length or larger (the well-bound, monomeric, and concatemeric PFGE fractions, all of which contain defined ends), linear maps are obtained.
Structure of cpDNA Molecules
The ends of linear cpDNA molecules have been mapped for the IR-containing maize (Oldenburg and Bendich, 2004b) and tobacco (Scharff and Koop, 2006) and now for M. truncatula that has no IR. For all three species, ends are located at positions near regions homologous to tobacco oriA (homologous to Oenothera oriB) and Oenothera oriA (Fig. 1C, maps c and e; Chiu and Sears, 1992; Kunnimalaiyaan and Nielsen, 1997b; Oldenburg and Bendich, 2004b), but for each species there are additional ends. For example, an end for tobacco, but not maize or M. truncatula, is located near tobacco oriB (Scharff and Koop, 2006, 2007). For M. truncatula, we mapped an additional end (Fig. 1C, map d) near gene psbK that corresponds to ends found in tobacco (Scharff and Koop, 2006) and two ends (Fig. 1C, maps b and f) that so far appear to be unique to M. truncatula. We conclude that for maize, tobacco, and M. truncatula, the mode of replication is likely the same and that some origins of replication are common among species and others may be species specific. As discussed for maize (Oldenburg and Bendich, 2004b), we propose that cpDNA replication does not begin with a D-loop-to-θ process. Instead, most cpDNA is produced in linear forms, and recombination-coupled replication creates the branched multi-genomic complexes that represent the segregating units in chloroplast inheritance.
With respect to the overall structure of M. truncatula cpDNA molecules, the presence or absence of the IR has no noticeable effect either when total cpDNA is fractionated by PFGE or when individual molecules are examined by fluorescence microscopy (Shaver et al., 2006). The PFGE profiles for uncut cpDNA from previously examined IR-containing plants include a prominent well-bound fraction (comprised mostly of branched linear forms larger than the size of the genome) and DNA migrating as simple linear molecules of genomic, oligomeric (collected at the compression zone, depending on the pulse time during electrophoresis), and subgenomic size (the smear of fragments near the bottom of the gel). For M. truncatula, we found this profile in lane 1 of Figure 1A and for cpDNA representing three other stages of leaf development (Shaver et al. 2006), as well as for pea (Pisum sativum), another species without an IR, at one stage of seedling development (Bendich and Smith, 1990). The subgenomic restriction fragments for M. truncatula and tobacco (Scharff and Koop, 2006) were less prominent than those for maize. This difference can be attributed to a larger size of the concatemers and branched forms giving a higher ratio of monomer-sized fragments to end fragments in M. truncatula and tobacco than in maize. Moving pictures of individual cpDNA molecules revealed multigenomic branched molecules with a structural complexity that is similar for pea (Bendich, 1991), M. truncatula (Shaver et al., 2006), and several IR-containing plants (Oldenburg and Bendich, 2004b; Rowan et al., 2004; Shaver et al., 2006), although the complexity of the molecules does depend on the age of the tissue. We therefore conclude that the IR has no apparent effect on the structure of cpDNA, although additional species without the IR need to be examined before this conclusion is firm.
Decline of cpDNA during Chloroplast Development
We previously showed that both the amount of DNA per chloroplast and the number of chloroplasts containing DNA decline during leaf development (Oldenburg and Bendich, 2004a; Rowan et al., 2004; Shaver et al., 2006). This decline is accompanied by changes in cpDNA structure from multigenomic forms to molecules of subgenomic size and eventually to chloroplasts containing no detectable DNA at all. Though our finding of Arabidopsis chloroplasts without DNA (Rowan et al., 2004) has been attributed to an artifact of DNase treatment during chloroplast isolation (Li et al., 2005; Zoschke et al., 2007), we found many or most chloroplasts without DNA in cytological sections, as well as in chloroplasts isolated without DNase for Arabidopsis and maize, but not tobacco (Oldenburg and Bendich, 2004a; Rowan et al., 2004; Shaver et al., 2006). We now show that cpDNA also declines in M. truncatula cotyledons and leaves. For the cotyledons, cpDNA decline would not be attributed to cell division because cell division does not occur during germination and cotyledon enlargement in other legumes (Lovell and Moore, 1970). Furthermore, during seed germination in Arabidopsis, the growth of cotyledons results from cell expansion and not cell division (Mansfield and Briarty, 1992, 1996).
In continuous light/dark cycles, M. truncatula cotyledon cpDNA is degraded because one-third of the chloroplasts lack detectable DNA (Fig. 2E). We also find that the DAPI-DNA fluorescence signal is not masked by chlorophyll or other components within the chloroplast because the 9-h, 24-h, and light/dark chloroplasts have similar chlorophyll autofluorescence (Fig. 2, H–J; Table II), but the number of genomes per plastid ranges from 900 to 0 (Fig. 2, C–E; Table II). The structure of individual DNA molecules changes from complex branched forms to subgenomic fragments in maize, Arabidopsis, tobacco, and M. truncatula as meristematic cells develop into expanded leaf cells (Oldenburg and Bendich, 2004a; Rowan et al., 2004; Shaver et al., 2006). The cpDNA loss during leaf development is most rapid in maize, less rapid in pea and M. truncatula, and slowest in tobacco (Shaver et al., 2006), which maintains cpDNA even in senescent leaves. Because M. truncatula and pea (which also lacks the IR) are intermediate in this hierarchy, the IR has no obvious effect on the rate of loss of cpDNA during leaf development. We conclude that there is a species-dependent effect on the rate of cpDNA loss, but cpDNA structure and stability are not dependent on the IR.
We previously showed that light can affect the loss or retention of cpDNA in maize seedlings (Oldenburg et al., 2006) and now show a similar result for cotyledons and leaves of M. truncatula. In dark-grown maize seedlings, proplastids develop into etioplasts, which might have an effect on the subsequent light-stimulated loss of cpDNA that differs from the loss for the direct proplastid-to-chloroplast transition. Maize seedlings grown in the dark for 14 d and transferred to the light experience massive cpDNA degradation within 3 d, leaving many chloroplasts with no detectable DNA (Oldenburg et al., 2006), as also found in light-grown plants (Oldenburg and Bendich, 2004a). For M. truncatula, cpDNA replication occurs within 2 h of transferring cotyledons from dark to light, and then the level of DNA per chloroplast rapidly declines over the next several hours (Fig. 5), although all chloroplasts retain some DNA after 24 h of light. Cellular levels of cpDNA, however, do not begin to decline until after 16 h of illumination, indicating that the initial decline in DNA per chloroplast is due to dilution by plastid division. This decline is then followed by a decrease in cpDNA per cell, which can only be attributed to degradation. Note that cpDNA degradation occurs before the cotyledons are fully expanded, indicating that the degradation is not related to senescence. The DNA degradation may result from light-induced damage to DNA that is not repaired. The degree of cpDNA retention seems to be species specific but does not depend on whether plants are grown under light/dark cycles (proplastids to chloroplasts) or if dark-grown plants are transferred to the light (etioplasts to chloroplasts).
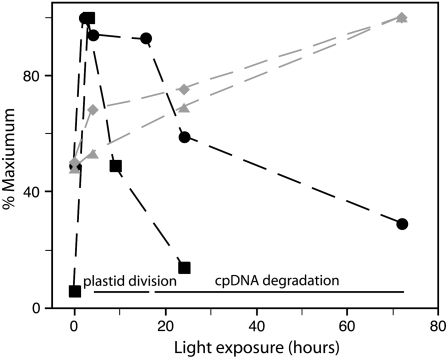
Figure 5.
Changes in cpDNA abundance during cotyledon development. Plastids were isolated from cotyledons of 5-d-old seedlings grown under constant dark, or constant dark for 3 d followed by a period of white light for 3, 9, or 24 h and then returned to the dark (Fig. 2; Table II). In a separate experiment, total cotyledon DNA was extracted from seedlings grown under 3 d of constant dark or 3 d constant dark followed by a period of white light for 2, 4, 16, 24, or 72 h (Fig. 4; Tables III and V). The ratio of chloroplast to nuclear genome copies was determined by real-time qPCR with petA (chloroplast) and enod11 (nuclear) probes, respectively. The lengths and widths of cotyledons were measured. Five cotyledon measurements were made after 0, 4, and 24 h of illumination, and three measurements were made after 72 h of illumination. The mean cotyledon lengths ± se after 0, 4, and 24 h were 4.2 ± 0.2, 4.6 ± 0.2, and 6 ± 0.3 mm, respectively. The mean cotyledon widths ± se after 0, 4, and 24 h were 1.4 ± 0.2, 1.9 ± 0.1, and 2.1 ± 0.1 mm, respectively. The cotyledon lengths after 72 h were 8, 8, and 10 mm, and the widths were 2.5, 3, and 3 mm. Black squares, Genomes per plastid; black circles, plastid genomes per nuclear genome copy; gray diamonds, mean cotyledon width; grey triangles, mean cotyledon length.
Factors That Influence the Retention or Loss of cpDNA
The level of plastid DNA depends on the balance between replication and degradation, processes likely to be influenced by communication between plastid and nucleus. Given that photoreceptors are involved in the regulation of numerous cellular events, we might expect that light signaling pathways are involved in setting plastid DNA levels. The persistence of cpDNA in illuminated seedlings may depend on the degree to which plastids develop in the dark during the onset of germination.
Grasses and legumes present an interesting contrast. For barley (Hordeum vulgare) and maize, seedling growth and plastid DNA synthesis occur in the dark, and maize plastids enlarge substantially in the dark (Baumgartner et al., 1989; Oldenburg et al., 2006). The cpDNA levels remain high until transfer to the light at which time cpDNA rapidly declines. Furthermore, plastid transcription increases and then decreases in either light- or dark-grown barley seedlings, but the decrease in transcription precedes cpDNA decline in the light (Baumgartner et al., 1989). Thus, when grasses are transferred to the light, their plastids are nearly fully developed and will almost immediately degrade their DNA. On the other hand, pea and M. truncatula do not exhibit plastid development in the dark, cpDNA levels do not increase or decrease until transfer to the light, and an increase in plastid transcription occurs in light-grown but not dark-grown pea seedlings (Sasaki et al., 1986; Dubell and Mullet, 1995a, 1995b). Thus, when transferred to the light, most steps of legume plastid development must still occur before cpDNA can be degraded. It appears that the decline in cpDNA after the transfer of dark-grown plants to the light is initially due to plastid division and then degradation in M. truncatula but mainly or entirely to degradation in maize.
For M. truncatula, short periods of light (white, red, or blue) stimulate plastid DNA replication in dark-grown plants, but further illumination results in plastid DNA reduction in white and blue but not red light (after an initial decrease due to plastid division). In red light, cpDNA levels remain high in the leaves of maize (Oldenburg et al., 2006) and the cotyledons of M. truncatula seedlings, probably because of a high rate of cpDNA replication and a low rate of degradation. The cpDNA retention in green chloroplasts under red light suggests that phytochrome does not promote cpDNA degradation. Instead, red-light signaling is likely involved in inhibiting degradation and promoting replication of cpDNA in maize and M. truncatula. In pea, Sasaki et al. (1986) concluded that the accumulation of cpDNA is mediated by phytochrome, and Dubell and Mullet (1995) showed that far-red light stimulated DNA synthesis.
In blue light, low levels of cpDNA are retained in the cotyledons of M. truncatula and the leaves of maize seedlings. The retention of DNA in chloroplasts in red light of relatively high intensity, but not lower intensity blue light, suggests that a blue light receptor (cryptochrome or phototropin) pathway triggers cpDNA degradation in both plants. For M. truncatula in white light, a higher fluence light initially results in a faster rate of cpDNA degradation than lower fluence, but after prolonged light exposure, the final level of cpDNA is the same. Therefore, higher fluence light may result in faster development of the chloroplast or the more rapid production of reactive oxygen species that damage DNA, which, if not repaired, is degraded, as in Escherichia coli (Skarstad and Boye, 1993). Nonetheless, some level of DNA is maintained in most chloroplasts of M. truncatula. The question still remains as to why different plants receiving the same signal (light) undergo high (maize) and low (pea, M. truncatula, and tobacco) rates of light-stimulated cpDNA degradation.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Plastid Isolation Procedure
Plastids were harvested from 5-d-old cotyledons, and DNA was extracted from 3- to 7-d-old cotyledons and 4- to 18-d-old plants. Seeds were sown in vermiculite and grown at 23°C to 25°C under fluorescent light or in the dark. Plastids were isolated with a high-salt buffer and no DNase as described previously (Shaver et al., 2006). For light quality experiments, seedlings were grown in boxes covered with clear, red, or blue cellophane. The light intensities were: 30 (low white), 48 (high white), 15 (red), and 2.5 (blue) μE m−2 s−1. The lengths and widths of cotyledons were measured at the longest and widest points, respectively, for dark-grown seedlings transferred to the light 3 d after sowing. Five cotyledon measurements were made after 0, 4, and 24 h of illumination, and three measurements were made after 72 h of illumination.
PFGE, Restriction Digestion, and Mapping of cpDNA
For PFGE and blot hybridization, plastids isolated from 7-d-old cotyledons were embedded in agarose gel plugs and lysed overnight at 48°C in 40 mm EDTA, pH 8, 1% sarkosyl with 200 μg/mL proteinase K. Phenyl-methyl-sulfonyl fluoride was used to inactivate the proteinase K, and samples were washed repeatedly in many gel-plug volumes of 10 mm Tris, 1 mm EDTA and stored at 4°C. PFGE, restriction digests, and hybridization with ndhF, petA, and rbcL probes were performed as described (Oldenburg and Bendich, 2004b). The contrast/brightness of images of the ethidium bromide (EtBr)-stained gel and hybridized membrane have been adjusted using Adobe Photoshop to highlight the less intense subgenomic bands. Pre-electrophoresis of DNA/agarose plugs was performed prior to restriction mapping to eliminate the smear of DNA of subgenomic size that would interfere with visualization of restriction fragments. For pre-electrophoresis, DNA plugs were placed into a conventional (not pulsed-field) 1.5% agarose gel, and electrophoresis was conducted for 1 h at 6 V/cm before the DNA plugs were removed from the gel and used for restriction digestion and mapping. For maps b to f in Figure 1C, the ends were determined by calculating the distance from the restriction site needed to generate a fragment size observed by EtBr-fluorescence and hybridization, as previously described (Oldenburg and Bendich, 2004b). For example, in map c, the end is located at approximately nucleotide 20,000. Fragment s4 is to the right of the SgrAI cut site and s7 is to the left, but only s4 hybridizes to ndhF, petA, and rbcL. Fluorescence intensities of the PFGE fractions were measured as described (Oldenburg and Bendich, 2004b).
Fluorescence Microscopy of cpDNA
For fluorescence microscopic imaging of the DAPI-stained plastids, the plastids were fixed in 0.8% gluteraldehyde and stored at 4°C, stained with 1 μg/mL DAPI, and 1% β-mercaptoethanol was added to reduce fading (Kuroiwa and Suzuki, 1980; Kuroiwa et al., 1981). Imaging was also performed with fixed plastids without the addition of DAPI. Imaging and quantification of DAPI-stained plastids were done using a 360ex460/50em filter, digital camera, and OpenLab software as previously described (Oldenburg and Bendich, 2004a; Oldenburg et al., 2006; Shaver et al., 2006). Imaging and quantification of plastid chlorophyll autofluorescence were done in the same way, except that a different filter (546ex540em) was used to capture the images, and the units for chlorophyll autofluorescence (Fig. 2, F–J; Table II) are reported in pixels as mean autofluorescence (pixels/μm2) × plastid area (μm2)/10.
The genome copy number per Medicago truncatula plastid was determined from the total DAPI fluorescence intensity (FI) using a method analogous to that previously described (Miyamura et al., 1986) and relative to vaccinia virus DNA. The total DAPI FI/virus was determined as 25 ± 11 pixels/μm2 (Oldenburg and Bendich, 2004a). This value was then used to calculate the relative FI of plastid to vaccinia particle, the vaccinia virus equivalents or “V” units (V = total DAPI FI/plastid divided by the average total DAPI FI per particle). Using vaccinia as the standard, the genome copy number was then calculated using the equation: copy number = 1.61 × V. The value 1.61 is a constant factor that accounts for the difference in DNA base composition and size between the vaccinia virus and the plastid genomes and was determined as (%A + T of virus genome/%A + T of plastid genome) × (bp vaccinia virus DNA/bp plant cpDNA), where %A + T for vaccinia (Copenhagen strain) is 66.6%, %A + T for M. truncatula is 66%, bp for vaccinia (strain vTF7.3) is 197,361, and bp for M. truncatula is 124,033 (accession no. AC093544). ANOVA was used to compare samples.
Real-Time qPCR
Total DNA for real-time qPCR was isolated from cotyledons and leaves using the GenElute Plant Genomic DNA Miniprep kit (Sigma). A 113-bp fragment of the chloroplast petA gene was amplified using the forward primer 5′-CTATTTCTGCATCCCCCTGA-3′ and the reverse primer 5′-TATACCGCCAGGACCAGAAC-3′. A 110-bp fragment of the chloroplast psbA gene was amplified using the forward primer 5′-ATCGCAGCTCCTCCAGTAGA-3′ and the reverse primer 5′-ATAGCCGCCGAAGTAGGAAT-3′. A 135-bp fragment of the nuclear enod11 gene was amplified using the forward primer 5′-AGGGTCAAGTTTTCGTTTCC-3′ and the reverse primer 5′-CGGTTTTACATTCATTATCCAC-3′. Amplification was performed using iQ SYBR Green Supermix or Bio-Rad iQ Supermix supplemented with SYBR Green I (Bio-Rad). Sample DNAs were denatured for 3 min at 94°C. Then 45 cycles of 15-s denaturation at 94°C, 15-s annealing at 55°C, and 2-s extension at 72°C were run using a Chromo 4 real-time PCR detection system. A melting curve from 65°C to 95°C was used to confirm the presence of single products. Data were analyzed using the Opticon Monitor 3 software. The copy number of cpDNA relative to nuclear DNA was calculated using the 2ΔΔCt method (Livak and Schmittgen, 2001; Pfaffl, 2001).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AC093544.
Acknowledgments
We thank Doug Ewing for assistance with growing plants.
This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education and Extension Service (grant no. 2002–35301–12021).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Arnold J. Bendich (bendich@u.washington.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Baumgartner BJ, Rapp JC, Mullet JE (1989) Plastid transcription activity and DNA copy number increase early in barley chloroplast development. Plant Physiol 89 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendich A (1991) Moving pictures of DNA released upon lysis from bacteria, chloroplasts, and mitochondria. Protoplasma 160 121–130 [Google Scholar]
- Bendich A, Smith S (1990) Moving pictures and pulsed-field gel electrophoresis show linear DNA molecules from chloroplasts and mitochondria. Curr Genet 17 421–425 [DOI] [PubMed] [Google Scholar]
- Chiu WL, Sears BB (1992) Electron microscopic localization of replication origins in Oenothera chloroplast DNA. Mol Gen Genet 232 33–39 [DOI] [PubMed] [Google Scholar]
- Dubell AN, Mullet JE (1995. a) Continuous far-red light activates plastid DNA synthesis in pea leaves but not full cell enlargement or an increase in plastid number per cell. Plant Physiol 109 95–103 [Google Scholar]
- Dubell AN, Mullet JE (1995. b) Differential transcription of pea chloroplast genes during light-induced leaf development. Plant Physiol 109 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinhorst S, Cannon GC (1993) DNA replication in chloroplasts. J Cell Sci 104 1–9 [Google Scholar]
- Kondorosi E, Roudier F, Gendreau E (2000) Plant cell-size control: growing by ploidy? Curr Opin Plant Biol 3 488–492 [DOI] [PubMed] [Google Scholar]
- Kunnimalaiyaan M, Nielsen BL (1997. a) Chloroplast DNA replication: mechanism, enzymes and replication origins. J Plant Biochem Biotechnol 6 1–7 [Google Scholar]
- Kunnimalaiyaan M, Nielsen BL (1997. b) Fine mapping of replication origin (oriA and oriB) in Nicotiana tabacum chloroplast DNA. Nucleic Acids Res 25 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T, Suzuki T (1980) An improved method for the demonstration of the in situ chloroplast nuclei in higher plants. Cell Struct Funct 5 195–197 [Google Scholar]
- Kuroiwa T, Suzuki T, Ogawa K, Kawano S (1981) The chloroplast nucleus: distribution, number, size, and shape, and a model for multiplication of the chloroplast genome during chloroplast development. Plant Cell Physiol 22 381–396 [Google Scholar]
- Li W, Ruf S, Bock R (2005) Constancy of organellar genome copy numbers during leaf development and senescence in higher plants. Mol Genet Genomics 275 185–192 [DOI] [PubMed] [Google Scholar]
- Livak K, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lovell P, Moore K (1970) A comparative study of cotyledons as assimilatory organs. J Exp Bot 21 1017–1030 [Google Scholar]
- Mansfield SG, Briarty LG (1992) Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can J Bot 70 151–164 [Google Scholar]
- Mansfield SG, Briarty LG (1996) The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana reserve mobilization. Int J Plant Sci 157 280–295 [Google Scholar]
- Miyamura S, Nagata T, Kuroiwa T (1986) Quantitative fluorescence microscopy on dynamic changes of plastid nucleoids during wheat development. Protoplasma 133 66–72 [Google Scholar]
- Oldenburg DJ, Bendich AJ (2001) Mitochondrial DNA from the liverwort Marchantia polymorpha: Circularly permuted linear molecules, head-to-tail concatemers, and a 5′ protein. J Mol Biol 310 549–562 [DOI] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ (2004. a) Changes in the structure of DNA molecules and the amount of DNA per plastid during chloroplast development in maize. J Mol Biol 344 1311–1330 [DOI] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ (2004. b) Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J Mol Biol 335 953–970 [DOI] [PubMed] [Google Scholar]
- Oldenburg DJ, Rowan BA, Zhao L, Walcher C, Schleh M, Bendich AJ (2006) Loss or retention of chloroplast DNA in maize seedlings is affected by both light and genotype. Planta 225 41–55 [DOI] [PubMed] [Google Scholar]
- Palmer JD (1983) Chloroplast DNA exists into two orientations. Nature 301 92–93 [Google Scholar]
- Palmer JD (1985) Comparative organization of chloroplast genomes. Annu Rev Genet 19 325–354 [DOI] [PubMed] [Google Scholar]
- Pfaffl M (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan B, Oldenburg D, Bendich A (2004) The demise of chloroplast DNA in Arabidopsis. Curr Genet 46 176–181 [DOI] [PubMed] [Google Scholar]
- Rowan BA, Oldenburg DJ, Bendich AJ (2007) A high-throughput method for detection of DNA in chloroplasts using flow cytometry. Plant Methods 3 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Nakamura Y, Matsuno R (1986) Phytochrome-mediated accumulation of chloroplast DNA in pea leaves. FEBS Lett 196 171–174 [Google Scholar]
- Saski C, Lee S, Daniell H, Wood T, Tompkins J, Kim H, Jansen R (2005) Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol Biol 59 309–322 [DOI] [PubMed] [Google Scholar]
- Scharff LB, Koop HU (2006) Linear molecules of tobacco ptDNA end at known replication origins and additional loci. Plant Mol Biol 62 611–621 [DOI] [PubMed] [Google Scholar]
- Scharff LB, Koop HU (2007) Targeted inactivation of the tobacco plastome origins of replication A and B. Plant J 50 782–794 [DOI] [PubMed] [Google Scholar]
- Shaver JM, Oldenburg DJ, Bendich AJ (2006) Changes in chloroplast DNA during development in tobacco, Medicago truncatula, pea, and maize. Planta 224 72–82 [DOI] [PubMed] [Google Scholar]
- Skarstad K, Boye E (1993) Degradation of individual chromosomes in recA mutants of Escherichia coli. J Bacteriol 175 5505–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Liere K, Börner T (2007) From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J 50 710–722 [DOI] [PubMed] [Google Scholar]