Abstract
Cytokinins regulate cell division and differentiation as well as a number of other processes implicated in plant development. The first step of cytokinin biosynthesis in Arabidopsis (Arabidopsis thaliana) is catalyzed by adenosine phosphate-isopentenyltransferases (AtIPT). The enzymes are localized in plastids or the cytoplasm where they utilize the intermediate dimethylallyl-diphosphate from the methylerythritolphosphate or mevalonic acid pathways. However, the regulatory mechanisms linking AtIPT activity and cytokinin biosynthesis with cytokinin homeostasis and isoprenoid synthesis are not well understood. Here, we demonstrate that expression of AtIPT3, one member of the adenosine AtIPT protein family in Arabidopsis, increased the production of specific isopentenyl-type cytokinins. Moreover, AtIPT3 is a substrate of the protein farnesyl transferase, and AtIPT3 farnesylation directed the localization of the protein in the nucleus/cytoplasm, whereas the nonfarnesylated protein was located in the plastids. AtIPT3 gain-of-function mutant analysis indicated that the different subcellular localization of the farnesylated protein and the nonfarnesylated protein was closely correlated with either isopentenyl-type or zeatin-type cytokinin biosynthesis. In addition, mutation of the farnesyl acceptor cysteine-333 of AtIPT3 abolishes cytokinin production, suggesting that cysteine-333 has a dual and essential role for AtIPT3 farnesylation and catalytic activity.
Cytokinins are involved in many aspects of plant growth and development. At the molecular level, cytokinins influence plant functions by stimulating cell division and differentiation in cooperation with other plant hormones (Haberer and Kieber, 2002; Howell et al., 2003; Kakimoto, 2003). Natural cytokinins are adenine derivatives carrying either an isoprene-derived molecule or an aromatic side chain at the N6 terminus (Sakakibara, 2006). In the first step of the isoprenoid cytokinin biosynthesis pathway, AMP, ADP, or ATP reacts with dimethylallyl-diphosphate (DMAPP; a central intermediate of the isoprenoid pathways) to the corresponding isopentenyl adenosine-5′-phosphates, which are subsequently metabolically converted to the monophosphate (iPRMP). Isopentenylation of adenosine phosphates is catalyzed by adenosine phosphate-isopentenyltransferase (IPT). In Arabidopsis (Arabidopsis thaliana), IPT proteins are encoded by a family of seven genes (AtIPT1 and AtIPT3 to AtIPT8; Kakimoto, 2001; Takei et al., 2001). The remaining two IPTs (IPT2 and IPT9) catalyze isopentenylation of adenine moiety of tRNAs (Golovko et al., 2002; Miyawaki et al., 2006). Subsequent conversion of iPRMP to isopentenyl adenosine (iPR) and isopentenyl adenine (iP) is catalyzed by 5′-nucleosidase and adenosine nucleosidase (Chen and Kristopeit, 1981a, 1981b). The iP nucleotides are transformed into the corresponding trans-zeatin nucleotides by CYP735A enzymes (Chen and Leisner, 1984; Takei et al., 2004a). Cytokinin level in plant tissues is regulated by the balance of de novo biosynthesis, interconversion between the different cytokinin species, inactivation, degradation, and transport. For example, irreversible inactivation of cytokinin results from glucosylation at the N7- or the N9-position of the adenine ring, whereas O-glucosylation at the N6 side chain terminal hydroxyl of zeatins is reversible and O-glucosylated zeatins are considered storage forms of the hormone (Brzobohaty et al., 1993; Mok and Mok, 2001). In contrast, degradation of cytokinin bases and nucleosides is catalyzed by cytokinin oxidase/dehydrogenase by cleavage of the side chain (Mok and Mok, 2001; Werner et al., 2003).
Among Arabidopsis IPTs, AtIPT3 is expressed in the vasculature throughout the plant, and its expression is increased by nitrate (Miyawaki et al., 2004; Takei et al., 2004b). AtIPT3 expression in Escherichia coli and Arabidopsis increases iP and iPRMP production, respectively, whereas the levels of iPRMP and iPR are decreased in the Atip3 mutant (Takei et al., 2001; Sakakibara et al., 2005; Miyawaki et al., 2006). Interestingly, AtIPT3 contains a typical C-terminal CaaX-box recognition motif for plant farnesyl transferase (PFT), which would suggest that this IPT protein is prenylated. If AtIPT3 is farnesylated, this would provide a unique situation where isopentenyl diphosphate and its isomer DMAPP would serve as precursors for farnesyl diphosphate (FPP) biosynthesis and IPT3 farnesylation as well as for AtIPT3-dependent cytokinin biosynthesis.
Modification of target proteins by farnesyl or geranylgeranyl is critical for the control of development, growth, and signaling (Yalovsky et al., 1999; Sinensky, 2000). Specific PFT or PGGT-I inhibitors block progression of the cell cycle in plant and animal cells, indicating that prenylation is required for the function of proteins involved in regulating cell division (Morehead et al., 1995; Qian et al., 1996; Tamanoi et al., 2001). For example, Ras, LKB1, CENP, and AtNAP1-1, which are involved in cell proliferation and differentiation or cytoskeletal functions, are prenylated proteins, and prenylation modulates their function by facilitating membrane association as well as protein-protein interaction or controlling protein function (Tamanoi et al., 2001; Hussein and Taylor, 2002; Martin and St Johnston, 2003; Galichet and Gruissem, 2006). Protein prenylation is conserved in animals and plants (Yalovsky et al., 1999), but unlike mice, in which PFT is essential for early embryonic proliferation, loss of Arabidopsis PFTβ (era1) or PFT/PGGT-Iα (plp) gene functions is not lethal, although the mutants are affected in their development (Yalovsky et al., 2000a; Running et al., 2004; Mijimolle et al., 2005). This could be due to the specific types of proteins that are prenylated in plants because most plant candidate protein prenyl transferase protein substrates differ from those identified in yeast (Saccharomyces cerevisiae) and animals (Galichet and Gruissem, 2003).
We investigated AtIPT3 to determine if the protein is farnesylated and to understand the potential role of AtIPT3 farnesylation during Arabidopsis development as well as for cytokinin biosynthesis. Here, we show that AtIPT3 is effectively farnesylated in vivo and that farnesylation regulates AtIPT3 subcellular localization, which affects cytokinin biosynthesis. In addition, AtIPT3 expression in E. coli, yeast, and Arabidopsis also demonstrates a role of Cys-333 for AtIPT3 catalytic properties. Characterization of AtIPT3 gain-of-function mutants demonstrated that increased cytokinin production affects plant development and contributes to the regulation of cell proliferation.
RESULTS
AtIPT3 Is a Farnesylated Protein
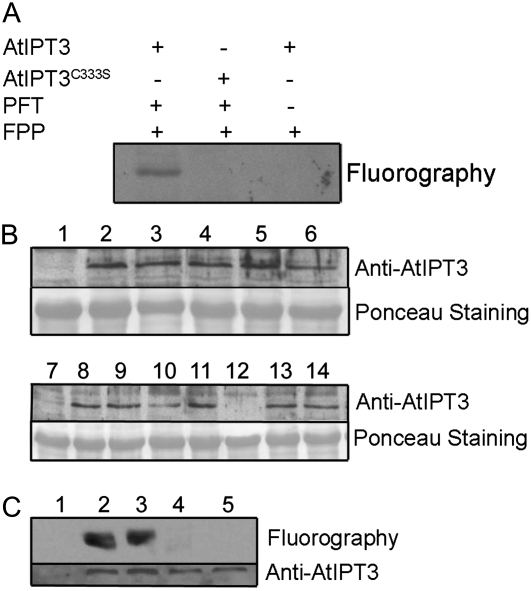
The presence of the CLVA motif at the C-terminal end of AtIPT3 suggested that the protein is a substrate of PFT. To test this hypothesis, we first incubated purified AtIPT3 with recombinant Arabidopsis PFT and [3H]FPP. AtIPT3 was labeled in the presence of both PFT and [3H]FPP (Fig. 1A). In addition, mutation of the conserved Cys farnesyl acceptor in the CLVA motif to Ser (AtIPT3C333S) confirmed that farnesylation required a functional farnesylation motif. In contrast, no prenylation was observed with PGGT-I using [3H]GGPP or [3H]FPP (data not shown).
Figure 1.
AtIPT3 is prenylated in vitro and in vivo. A, Wild-type (containing an intact CLVA CaaX box) and C333S (CLVA CaaX box mutated to SLVA) versions of AtIPT3 were used as substrates for plant protein prenyltransferases. Symbols + and − indicate the presence or absence of purified AtIPT3, purified AtPFT, and FPP. After electrophoresis and fluorography, exposure was carried out for 7 d. B, Western-blot analysis of 10-d-old wild-type (1 and 7), AtIPT3 (2–6), and AtIPT3C333S (8–14) T3 homozygous plant crude extracts. Eighty micrograms of total protein was loaded per well, and the membrane was probed with a polyclonal AtIPT3 antibody. C, In vivo prenylation assay. Fluorography and immunoblot of protein extracts from wild-type (1) and Arabidopsis plants expressing AtIPT3 (2 and 3) or AtIPT3C333S (4 and 5) proteins and labeled with farnesyl synthesized from 3H-MVA. Protein extracts were analyzed by SDS-PAGE in a 12% gel, followed by western blotting. Fluorography was carried out for 3 weeks.
To confirm that AtIPT3 is also farnesylated in vivo, we generated Arabidopsis transgenic plants in which the AtIPT3 and AtIPT3C333S cDNA were expressed under the control of the cauliflower mosaic virus 35S promoter. Seven AtIPT3 and six AtIPT3C333S T1 hygromycin-resistant plants with single T-DNA insertions were selected for further analysis. Five independent T3 homozygous AtIPT3 lines and seven AtIPT3C333S lines were analyzed by western blot using a polyclonal anti-AtIPT3 antibody. Except one AtIPT3C333S line, all transgenic lines expressed the transgene (Fig. 1B). Proteins from AtIPT3 and AtIPT3C333S plants labeled with [3H]mevalonic acid (MVA) were extracted and separated on SDS-polyacrylamide gels, which were then used either for immunoblot analysis with the anti-AtIPT3 antibody or for fluorography to detect labeled AtIPT3. AtIPT3 and AtIPT3C333S were expressed to a similar level in the transgenic plants (Fig. 1C). A labeled protein corresponding to the size of AtIPT3 was detected only in extracts from plants expressing AtIPT3 but not in plants expressing AtIPT3C333S or in wild-type plants (Fig. 1C). Together, these results establish that AtIPT3 is efficiently farnesylated in vivo as well.
AtIPT3 Subcellular Localization Is Regulated by Farnesylation
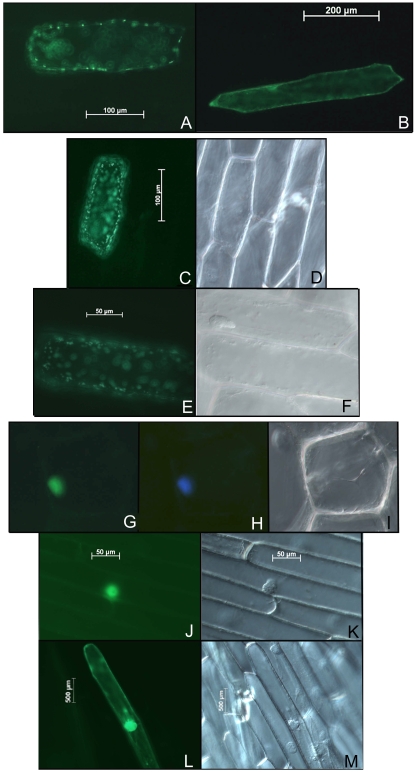
Most farnesylated proteins in yeast and animal cells are targeted to the plasma membrane (Sinensky, 2000), although in plants farnesylated APETALA1 has been found in the nucleus and AtNAP1;1 was found both in the cytoplasm and the nucleus depending on the farnesylation status of the protein (Yalovsky et al., 2000b; Galichet and Gruissem, 2006). AtIPT3 was reported to be located in plastids because the first 55 amino acids of the protein function as a chloroplast transit peptide (TP; Kasahara et al., 2004). Similarly, this TP was sufficient to direct GFP to plastids when expressed in onion (Allium cepa) epidermal cells (Fig. 2A). To investigate the function of farnesylation for AtIPT3 subcellular localization, TP-GFP-AtIPT3 and TP-GFP-AtIPT3C333S fusion proteins were constructed (see “Materials and Methods”) and transiently expressed in onion epidermal cells. TP-GFP-AtIPT3C333S was localized in the plastids (Fig. 2, C–F), whereas the absence of TP (GFP-AtIPT3C333S) resulted in the localization of the protein in the cytoplasm (Fig. 2B). In contrast, TP-GFP-AtIPT3 localization was primarily restricted to the nucleus 16 h after bombardment (Fig. 2, H–K). However, the protein was also present in the cytoplasm of some cells after 24 h (Fig. 2, L and M). Similarly, GFP-AtIPT3 was also found in the nucleus and in the cytoplasm of transformed cells (data not shown). Together, our data show that despite the presence of a chloroplast TP, farnesylation of AtIPT3 appears to direct most of the protein to the nucleus.
Figure 2.
Farnesylation affects subcellular localization of AtIPT3. The fusion constructs expressing TP-GFP (A), GFP-AtIPT3C333S (B), TP-GFP-AtIPT3C333S (C–F), and TP-GFP-AtIPT3 (G–M) proteins were introduced into onion epidermal cells by bombardment. GFP (A–C, E, G, J, and L) and 4′,6-diamino-phenylindole (H) fluorescence was observed using a fluorescence microscope.
AtIPT3 Gain of Function Increases Cytokinin Production
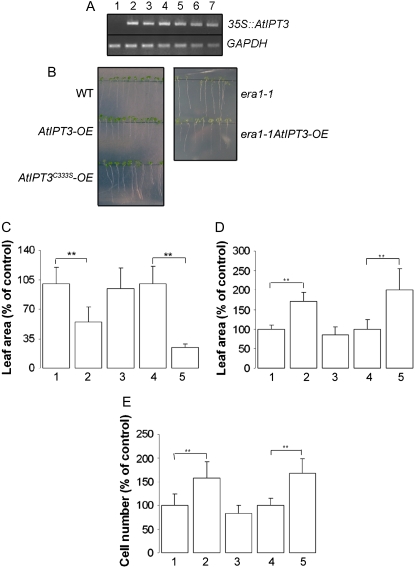
To examine the effects of AtIPT3 gain of function on cytokinin production and the potential role of its farnesylation in vivo, AtIPT3 was also expressed in era1-1 plants by crosses between six independent AtIPT3 homozygous plants that express a high level of the transgene and era1-1 (Fig. 3A). Cytokinin production was investigated in several independent AtIPT3 and era1-1AtIPT3 plants. All analyzed lines had elevated cytokinin accumulation in leaves compared to their respective wild type and era1-1 control lines. The lines AtIPT3 3.2.5 (AtIPT3-OE) and era1-1AtIPT3 3.7 (era1-1AtIPT3-OE) were therefore selected for full analysis. As shown in Table I, AtIPT3-OE plants had a 3.4-fold increase of total cytokinin. This accumulation was associated with a nearly 10-fold increase of iP ribotides [N6-(2-isopentenyl)adenosine 9-riboside 5′-phosphates; iPRPs], indicating that cytokinins are synthesized via the iPRP-dependent pathway. In addition, AtIPT3 overexpression resulted in a preferential production of iP-type cytokinins (11.2-fold), while the content of zeatin-type cytokinins was increased to a much lesser extent (1.5-fold). This accumulation of iP-type cytokinins was mainly due to the 12-fold increase of N6-(2-isopentenyl)adenine 7-glucoside (iP7G), which represented 53% of the pool of total cytokinin as compared to 15% in the wild type. In contrast, the content of trans-zeatin 7-glucoside (Z7G), the most abundant cytokinin present in wild-type plants, was only 1.5-fold increased in AtIPT3-OE plants, and its participation to the pool of total cytokinin decreased from 42% to 19% (Table I). Interestingly, era1-1 plants accumulated 2.2-fold more cytokinins than the wild type (Table I). Although iPRP content was 4-fold increased in era1-1 leaves, zeatin-type cytokinins predominantly accumulated in era1-1 with a 2-, 4-, 10-, and 15-fold increase of trans-zeatin 9-riboside (ZR), trans-zeatin O-glucoside (ZOG), trans-zeatin 9-riboside O-glucoside (ZROG), and dihydrozeatin 7-glucoside (DHZ7G), respectively. In contrast, the content of Z7G and trans-zeatin 9-glucoside decreased to 32% and 7% of the total pool of cytokinin compared to 42% and 13.6%, respectively, in the wild type. Similar to the situation in AtIPT3-OE plants, the increase in cytokinin production in era1-1AtIPT3-OE leaves was preferentially due to an accumulation of iP-type cytokinins (5.3-fold increase) rather than an increase of zeatin-type (1.7-fold increase) compared to era1-1 leaves. Cytokinin accumulation in era1-1AtIPT3-OE leaves was associated with a nearly 6-fold increase of iPRPs compared to era1-1, indicating that cytokinins are also synthesized via the iPRMP-dependent pathway in era1-1AtIPT3-OE plants. Furthermore, iP7G, like in AtIPT3-OE plants, was the predominant cytokinin (Table I). More importantly, ZR and ZROG content was 5.5- and 2-fold increased, respectively, in era1-1AtIPT3-OE leaves compared to era1-1. Overexpression of AtIPT3 in wild-type Arabidopsis had no effect on the content of cis-zeatin-type cytokinins, whereas a 2- and a 5-fold increase in cis-zeatin 9-riboside 5′-monophosphate (c-ZRMP) and cis-zeatin 9-riboside O-glucoside (c-ZROG), respectively, was measured in era1-1 leaves. Additionally, cis-zeatin riboside monophosphate content was 2-fold higher in era1-1AtIPT3-OE leaves than in era1-1. Surprisingly, cytokinin overproduction was almost completely abolished in AtIPT3C333S-OE plants. Replacement of the farnesyl Cys acceptor by Ser strongly reduced (2.7-fold) the accumulation of total cytokinins compared to AtIPT3-OE plants and only slightly increased cytokinin content (1.2-fold) compared to the wild type, indicating that the Cys might be important for AtIPT3 catalytic activity (Table I).
Figure 3.
Phenotypic analysis of AtIPT3, era1-1/AtIPT3, and AtIPT3C333S plants. A, Expression analysis of 35S∷AtIPT3 transgene was monitored in era1-1 (1) and era1-1AtIPT3 (2–7) 20-d-old plants using semiquantitative reverse transcriptase-mediated PCR. B, Root phenotype in 9-d-old wild-type, AtIPT3-OE, and AtIPT3C333S-OE seedlings and 11-d-old era1-1 and era1-1AtIPT3 seedlings grown on vertical plates. C, Average area of first leaves in 10-d-old wild-type (1), AtIPT3-OE (2), and AtIPT3C333S-OE (3) plants and 12-d-old era1-1 (4) and era1-1AtIPT3-OE (5) plants; **, P < 0.001. D and E, Average area of first leaves (D) and average abaxial epidermal cell number (E) in first leaves of 24-d-old wild-type (1), AtIPT3-OE (2), and AtIPT3C333S-OE (3) plants and in 26-d-old era1-1 (4) and era1-1AtIPT3-OE (5) plants; **, P < 0.001.
Table I.
Endogenous cytokinin level increases in AtIPT3-OE, era1-1, and era1-1AtIPT3-OE leaves but not in AtIPT3C333S-OE plants
The values show an average of three independent samples of 1 g. Each value represents the mean ± se. iP, N6-(2-isopentenyl)adenine; iPR, N6-(2-isopentenyl)adenosine; c-Z7G, cis-zeatin 7-glucoside; Z9G, trans-zeatin 9-glucoside.
| Cytokinin Metabolitea | Wild Type | AtIPT3-OE | era1-1 | era1-1AtIPT3-OE | AtIPT3C333S-OE |
|---|---|---|---|---|---|
| pmol g−1 FW | |||||
| iPRPs | 3.8 ± 0.5 | 32.8 ± 1.5 | 15.4 ± 4.0 | 90.9 ± 15.2 | 6.8 ± 0.6 |
| iPR | 1.7 ± 0.3 | 13.8 ± 4.3 | 2.9 ± 0.4 | 16.5 ± 3.9 | 3.4 ± 0.6 |
| iP | 0.7 ± 0.4 | 0.9 ± 0.5 | 3.8 ± 0.7 | 5.6 ± 1.5 | 2.1 ± 1.0 |
| iP7G | 22.6 ± 4.5 | 274 ± 39.0 | 32.0 ± 3.6 | 172.5 ± 5.8 | 32.3 ± 6.8 |
| iP9G | 1.1 ± 0.3 | 13.4 ± 2.6 | 0.6 ± 0.2 | 3.8 ± 0.6 | 1.4 ± 0.5 |
| ZR | 4.7 ± 1.4 | 7.2 ± 1.8 | 10.4 ± 1.7 | 57.7 ± 2.3 | 5.0 ± 1.3 |
| Z7G | 64.4 ± 5.6 | 96.9 ± 17.3 | 107.4 ± 10.2 | 163.6 ± 6.0 | 71.2 ± 3.7 |
| Z9G | 20.5 ± 0.3 | 35.4 ± 4.5 | 23.3 ± 1.2 | 44.0 ± 0.1 | 25.9 ± 2.2 |
| ZOG | 13.4 ± 0.1 | 18.5 ± 2.2 | 52 ± 4.0 | 69.6 ± 6.1 | 18.3 ± 2.1 |
| DHZ7G | 2.1 ± 0.1 | 3.1 ± 0.4 | 32.7 ± 2.3 | 46.3 ± 1.1 | 3.1 ± 0.5 |
| ZROG | 1.9 ± 0.2 | 2.1 ± 0.2 | 21.4 ± 1.3 | 42 ± 3.7 | 1.7 ± 0.4 |
| c-ZRMP | 2.1 ± 0.4 | 2.2 ± 0.3 | 4.2 ± 0.8 | 8.6 ± 1.6 | 3.3 ± 0.5 |
| c-Z7G | 5.9 ± 0.9 | 7.1 ± 1.1 | 8.0 ± 0.9 | 9.6 ± 1.2 | 7.6 ± 0.9 |
| c-ZOG | 4.0 ± 1.2 | 2.6 ± 0.0 | 3.4 ± 0.1 | 3.1 ± 0.4 | 3.8 ± 0.3 |
| c-ZROG | 2.6 ± 0.1 | 2.4 ± 0.3 | 12.6 ± 3.1 | 15.0 ± 1.1 | 2.0 ± 0.3 |
| Total | 151.5 ± 17.2 | 512.4 ± 76.0 | 330.1 ± 34.5 | 748.8 ± 50.6 | 187.9 ± 21.7 |
Leaf rosettes were harvested from plants grown for 5 weeks.
The high accumulation of cytokinin N-glucosides [IP7G and N6-(2-isopentenyl)adenine 9-glucoside (abbreviated IP9G)] in AtIPT3-OE and era1-1AtIPT3-OE plants indicated that the conversion of the primary products of the cytokinin biosynthesis to IP7G and IP9G could represent an important pathway involved in the maintenance of cytokinin homeostasis and induced in AtIPT3 plants. Degradation of cytokinins by cytokinin oxidase/dehydrogenase also occurs in Arabidopsis (Werner et al., 2003). However, cytokinin oxidase/dehydrogenase activity was altered neither in AtIPT3-OE nor in era1-1AtIPT3-OE plants compared to their corresponding controls (Table II). Together, these results show that the lack of protein farnesylation in era1-1 strongly affects cytokinin biosynthesis or/and metabolism. In addition, although the overall cytokinin content was very similar in AtIPT3-OE and era1-1AtIPT3-OE plants, the increased accumulation of zeatin-type cytokinins in era1-1AtIPT3-OE plants suggests that farnesylation affects IPT3-mediated cytokinin biosynthesis.
Table II.
Cytokinin oxidase/dehydrogenase activity is not altered in Arabidopsis plants expressing IPT3
The values show an average of four independent samples of 50 mg. Each value represents the mean ± se.
| Plant Genotype | CKX Activity |
|---|---|
| nmol adenine mg−1 protein h−1 | |
| Wild type | 3.26 ± 0.22 |
| AtIPT3-OE | 4.16 ± 0.17 |
| era1-1 | 4.59 ± 0.28 |
| era1-1AtIPT3-OE | 3.74 ± 0.36 |
| AtIPT3C333S-OE | 4.74 ± 0.23 |
Increased Cytokinin Accumulation Affects Plant Development and Cell Proliferation
The accumulation of cytokinins in plants expressing AtIPT3 has a significant effect on root development. Primary root length was reduced in AtIPT3-OE and era1-1AtIPT3-OE plants compared to the wild type and era1-1 (Fig. 3B). In addition, the transgenic plants also developed fewer lateral roots. Root development in AtIPT3C333S-OE plants, however, was not affected. To address the effect of AtIPT3 expression in leaves, we measured the area of the first leaves, which develop synchronously compared to other leaves (De Veylder et al., 2001; Beemster et al., 2005). Young AtIPT3-OE and era1-1AtIPT3-OE leaves were significantly smaller compared to wild-type and era1-1 leaves, respectively (Fig. 3C). However, with age, leaf expansion was increased in plants expressing AtIPT3, and, as shown in Figure 3D, 24-d-old AtIPT3-OE first leaves were 70% larger than wild-type leaves. Similarly, AtIPT3 expression in era1-1 also resulted in a 100% increase in size of mature 26-d-old leaves. Because a difference in leaf size can be caused by changes in cell number or cell size, we counted the number of epidermal cells on the abaxial surface of the first leaves in adult AtIPT3-OE and era1-1AtIPT3-OE plants. Cell number was 58% and 68% increased in AtIPT3-OE and era1-1AtIPT3-OE leaves, respectively, compared to the control plant (Fig. 3E), confirming that increased leaf size was primarily the result of an increased cell number. In contrast, leaf size and cell number were not changed in AtIPT3C333S-OE plants.
Cys-333 Has a Function in the Catalytic Activity of AtIPT3
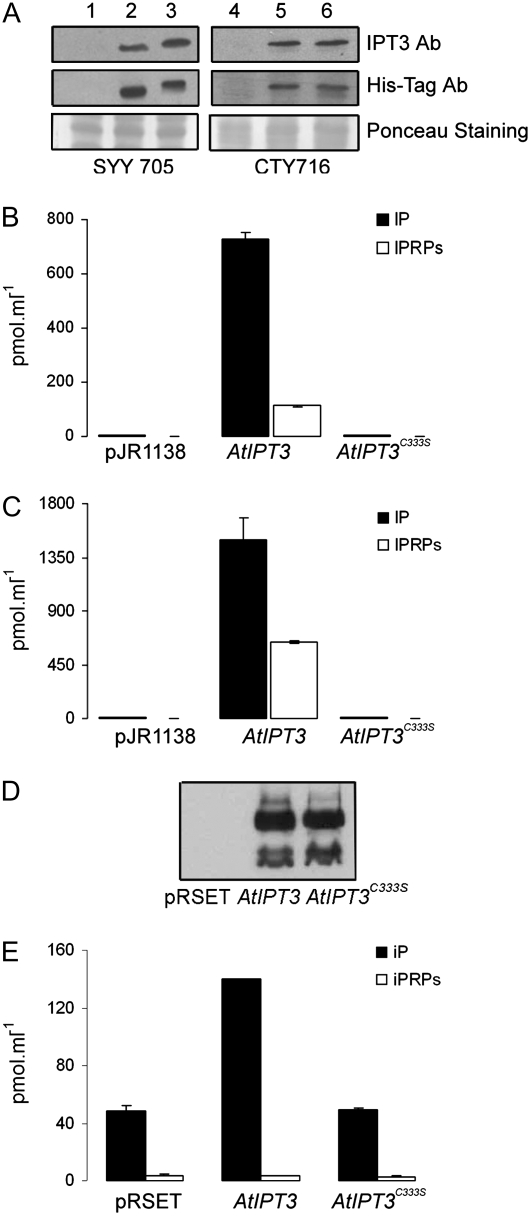
The decreased cytokinin production in AtIPT3C333S-OE plants and the absence of phenotypic alterations suggested that Cys-333 may have an important function for AtIPT3 catalytic activity. To investigate the function of Cys-333 for AtIPT3-mediated cytokinin biosynthesis, AtIPT3 and AtIPT3C333S were expressed in the yeast strains CTY716, which lacks PFT activity, and SYY705, which expresses the tomato (Solanum lycopersicum) PFT (Lavy et al., 2002). Western-blot analysis of yeast protein lysates using anti-AtIPT3 or His-tag antibody revealed that the two IPT3 proteins were expressed at similar levels in the two yeast strains (Fig. 4A). Attachment of farnesyl increases protein mobility in SDS-polyacrylamide gels (Zhu et al., 1993; Galichet and Gruissem, 2006). As expected, AtIPT3 migrated faster than AtIPT3C333S in SYY705 cell extracts (Fig. 4A, lines 2 and 3), whereas both proteins migrated at a similar position in CTY716 extracts (Fig. 4A, lines 5 and 6). The shift in AtIPT3 mobility in SYY705 protein extracts was reproducible and indicated that AtIPT3 was efficiently farnesylated in yeast. AtIPT3 expression resulted in the excretion of high level of iP and iPRPs in both yeast strains (Fig. 4, B and C). The level of accumulation of iP was similar in CTY716 and SYY705 strains as compared to the control (110- and 120-fold, respectively), whereas the level of iPRPs was somewhat higher in SYY705 cells compared to CTY716 cells (320- and 235-fold, respectively). In contrast, expression of AtIPT3C333S did not increase the level of any cytokinin in the two yeast strains (Fig. 4, B and C). To further substantiate our results, AtIPT3 and AtIPT3C333S were expressed in E. coli cells, which do not encode protein prenyl transferases. The two proteins were expressed at a similar level in bacteria (Fig. 4D), and whereas AtIPT3 caused a 3-fold accumulation of iP in the medium, AtIPT3C333S was inactive (Fig. 4E). Together, these data strongly suggest that Cys-333 is important for AtIPT3 catalytic activity and farnesylation of the protein.
Figure 4.
Excretion of cytokinin from yeast and E. coli cells expressing AtIPT3 and AtIPT3C333S. A, Immunoblot analysis of yeast SYY705 and CTY716 wild-type cells (1 and 3) and cells expressing AtIPT3 (2 and 5) or AtIPT3C333S (3 and 6). Protein extracts were analyzed by SDS-PAGE followed by western blotting using the polyclonal AtIPT3 and His-Tag antibodies. B and C, The iP and iPRP content in the culture medium of SYY105 (B) or CTY716 (C) yeast cells overexpressing AtIPT3 and AtIPT3C333S. D, Immunoblot analysis of E. coli cells expressing AtIPT3 and AtIPT3C333S in the presence of IPTG. Protein extracts were analyzed by SDS-PAGE followed by western blotting using the polyclonal His-Tag antibody. E, iP and iPRP content in the culture medium of E. coli cells expressing AtIPT3 and AtIPT3C333S in presence of IPTG.
DISCUSSION
The preferential formation of iP-type cytokinins in yeast, E. coli, and Arabidopsis plants expressing AtIPT3 indicates that AtIPT3-mediated cytokinin biosynthesis proceeds via the iPRP-dependent pathway that produces iPRPs as first products. These results confirmed previously reported data from Arabidopsis plants or E. coli cells expressing IPT3, which accumulated iPRPs and iP. The results are also consistent with Arabidopsis Atipt3 mutants that have decreased iPRP and iPR levels (Takei et al., 2001; Sakakibara et al., 2005; Miyawaki et al., 2006). Similar to other studies, our method of cytokinin nucleotide measurement did not allow us to determine if AtIPT3 uses AMP, ADP, or ATP as acceptor of the isopentenyl moiety for cytokinin biosynthesis (Kakimoto, 2001; Takei et al., 2001; Sun et al., 2003). Interestingly, both ZR and ZROG accumulated in era1-1AtIPT3-OE plants, and ZR, ZOG, DHZ7G, and ZROG levels were increased in era1-1 plants. In contrast, the content of zeatin-type cytokinins was not significantly altered in AtIPT3-OE plants, and only iP-type cytokinins accumulated in these plants. This reveals that the lack of protein farnesylation in era1-1 promotes trans-zeatin cytokinin production in Arabidopsis (Table III). Protein farnesylation controls IPT3 subcellular localization. The nonfarnesylated AtIPT3 is localized in the plastids, and the farnesylated AtIPT3 is located in the nucleus and the cytoplasm, at least in the onion epidermal cell assay. The observed dual subcellular localization of AtIPT3 could represent a regulatory mechanism of cytokinin production as a function of the activity of the cytoplasmic isoprenoid pathway (MVA pathway) and/or PFT. Depending on the efficiency of protein farnesylation and the availability of FPP for prenylation, AtIPT3 would be localized in the plastids or the nucleus. Consequently, and depending on the localization and the efficiency of cytokinin hydroxylase activities that catalyze hydroxylation of iP-type cytokinins, AtIPT3-mediated cytokinin production would then favor iP- or zeatin-type cytokinins by using DMAPP derived from the MVA pathway or the methylerythritolphosphate pathway, respectively (Table III). Whether the lack of protein farnesylation affects cytokinin biosynthesis only by modulating AtIPT3 subcellular localization or also influences the activity/localization of other proteins is currently not known, but the high accumulation of iPRPs in era1-1 plants would suggest a role of other farnesylated proteins in cytokinin production. Most of the small GTP-binding proteins are farnesylated (Tamanoi et al., 2001; Galichet and Gruissem, 2003), and cytokinin production is increased in tobacco (Nicotiana tabacum) plants expressing the rice (Oryza sativa) rgp1 gene (Sano et al., 1994), which would indicate that protein farnesylation could also influence cytokinin biosynthesis through modulation of small GTP-binding protein farnesylation. We attempted to further substantiate our analysis of AtIPT3 dual subcellular localization using biochemical fractionation. However, because of the low affinity of the IPT3 antibody, the results were not conclusive. Our results on IPT3 subcellular localization are also partially in contradiction with previous reports showing that AtIPT3 is only localized in plastids. These results were obtained from the expression of a fusion protein between the first 55 amino acids of AtIPT3 (which can function as a TP) and GFP (TP-GFP) or from stable expression of an AtIPT3-GFP fusion protein under the control of the AtIPT3 promoter (Kasahara et al., 2004; Miyawaki et al., 2006). However, a fusion of GFP at the C terminus of AtIPT3 would block farnesylation of the protein by masking the functional CAAX box.
Table III.
Summary of the influence of protein farnesylation on AtIPT3 subcellular localization and cytokinin production
| Plant Genotype | AtIPT3 Farnesylation Status | AtIPT3 Subcellular Localization | Cytokinin Species Production |
|---|---|---|---|
| AtIPT3-OE | + | Nucleus/cytoplasm | iP-types (iPRPs, iPR, iP7G, and iP9G) |
| era1-1AtIPT3-OE | − | Plastids | iP-types (iPRPs, iPR, iP7G, and iP9G); zeatin-types (ZR and ZROG) |
The absence of an AtIPT3-mediated effect on cis-zeatin-type cytokinins is consistent with the model that tRNA degradation by AtIPT2 and AtIPT9 is the major source of the cis-zeatin cytokinins and that isoprenoid cytokinin biosynthesis does not influence cis-zeatin-type cytokinin formation (Kasahara et al., 2004; Miyawaki et al., 2006). However, the significant increase of c-ZRMP and c-ZROG in era1-1 and era1-1AtIPT3-OE plants suggests that the isomerization of trans-zeatin-type cytokinins could occur in Arabidopsis and that farnesylated proteins could play a role in this mechanism. A cis-trans-zeatin isomerase activity has been measured in Phaseolus vulgaris immature seeds (Bassil et al., 1993), but the existence of such an activity has not been demonstrated in Arabidopsis.
Cytokinin N-glucosides represented the main part of the pool of total cytokinins in AtIPT3-OE plants, and cytokinin oxidase/dehydrogenase activity was not altered in those plants. This indicates that N-glucosylation represents an important pathway of cytokinin inactivation and maintenance of cytokinin homeostasis in AtIPT3 Arabidopsis plants. Although cytokinin oxidases/dehydrogenases are involved in cytokinin degradation in many plants (Blagoeva et al., 2004), their overall activity was not altered in AtIPT3-OE plants. Seven genes for cytokinin oxidase/dehydrogenases have been identified in Arabidopsis, and their expression is increased by cytokinins (Werner et al., 2003). It is possible that the fast N-glucosylation of cytokinins produced in AtIPT3 Arabidopsis plants only allows accumulation of nonglucosylated cytokinins at levels that are too low for stimulation of the cytokinin oxidase/dehydrogenase activity. In addition, it is not clear if the accumulation of cytokinin N-glucosides is specific for AtIPT3 overexpression or if this might be a general response to overexpression of IPTs in Arabidopsis.
Mature wild-type and era1-1 Arabidopsis plants expressing AtIPT3 had enhanced leaf size with an increased number of cells, whereas root development was impaired. These phenotypes agree with the model that cytokinins modulate plant development through regulation of cell division (Sakakibara, 2006). Additionally, increased accumulation of cytokinins in era1-1 plants also provided an explanation for the enhanced leaf development and cell division observed in era1-1 leaves (A. Galichet, G.T.S. Beemster, and W. Gruissem, personal communication).
Based on the cytokinin measurement in AtIPT3C333S-OE Arabidopsis plants, as well as in yeast and E. coli cells expressing AtIPT3C333S, we postulate that Cys-333 is important for AtIPT3 catalytic activity. Although the attachment of a farnesyl group to Cys-333 does not appear to inhibit enzyme activity, this Cys by itself is of particular importance for AtIPT3 function. The carboxyl-terminal end of the different AtIPTs is not well conserved, and AtIPT3 is the only AtIPT possessing a CAAX box for modification by protein farnesylation. In contrast, the putative DMAPP-binding site, located at the amino-terminal end of the proteins, is well conserved among the different members of this protein family (Takei et al., 2001). Cys-333 might be important for the correct spatial conformation of the protein. It is also interesting to consider that Cys-333 has a dual function in directing the catalytic activity of AtIPT3 to different subcellular compartments as a function of the activity of the MVA pathway or PFT. Further experiments will be necessary to explore how cytoplasmic isoprenoid and cytokinin synthesis cooperate to maintain cytokinin homeostasis during plant development
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants used in our study were all derived from the Columbia accession line. Seeds were surface sterilized using 5% bleach and germinated on Murashige and Skoog medium. After 2 weeks, the seedlings were transferred to soil and grown in Conviron chambers with a 16-h-light/8-h-dark cycle at 23°C in 70% humidity.
Protein Production
AtIPT3 cDNA (At3g63110) was amplified by PCR using the primer IPT3-For (5′-ATGATCATGAAGATATCTATGGCT-3′) together with either IPT3-Rev1 (5′-GTCGACTCACGCCACTAGACACCGCGA-3′, for the wild-type version of the CaaX box, CLVA) or IPT3-Rev2 (5′-GTCGACTCACGCCACTAGACTCCGCGA-3′, for the C333S version, SLVA). Both fragments were cloned in the pCR 2.1-TOPO cloning vector (Stratagene), sequenced, and subsequently cloned in the pRSETa vector for protein expression in Escherichia coli. Recombinant proteins were purified on nickel-nitrilotriacetic acid agarose talon super-flow metal affinity resin (Clontech). For cytokinin analysis, E. coli colonies cultivated overnight on Luria-Bertani (LB) plates supplemented with ampicillin and chloramphenicol (both at 50 μg/mL) at 37°C were inoculated into 2 mL of LB media supplemented with ampicillin (50 μg/mL) and cultured for 4 h at 37°C. Suspension was further inoculated into 200 mL of LB for 2 h at 37°C and then 0.1 mm isopropylthio-β-galactoside (IPTG) was added. After culture for 2.5 h at 37°C, the optical density (A600) was measured, and bacterial suspension was frozen in liquid nitrogen and stored at −80°C.
Immunoblots
Nitrocellulose membranes were first blocked for 2 h at room temperature with 4% nonfat milk and subsequently incubated overnight at 4°C with the polyclonal anti-AtIPT3 antibody (diluted 1:100), washed with Tris-buffered saline containing Tween 20, and incubated 1 h with 10,000-fold diluted secondary antibody conjugated with horseradish peroxidase for detection with an ECL kit (Amersham Pharmacia Biotech).
In Vitro and in Vivo Prenylation Assays
Both in vitro and in vivo prenylation assays were performed as previously described (Yalovsky et al., 1997, 2000a).
GFP Constructs and Fluorescence Microscopy
Part of the AtIPT3 gene (Met-1-Ser-55, TP) was fused in frame to the amino terminus of the GFP gene in pGFP-MRC (Rodriguez-Concepcion et al., 1999). AtIPT3 and AtIPT3C333S (Arg-56-Ala-336) were cloned in frame to the carboxy terminus of the GFP gene, containing the TP or not. All constructs were sequenced. Onion (Allium cepa) epidermal cells were transformed by plasmid bombardment as described (Scott et al., 1999). Samples were examined for GFP fluorescence 16 or 24 h after transformation using a Zeiss Axioplan 2 fluorescence microscope at 520 nm.
Construction of AtIPT3 Plants
AtIPT3 and AtIPT3C333S cDNA were cloned in the modified vector pCAMBIA 1380 containing a cauliflower mosaic virus 35S promoter (kindly provided by L. Gomez-Gomez). The constructs were introduced into Agrobacterium tumefaciens strain LBA4404. These strains were used to transform Arabidopsis Columbia plants by floral dip (Clough and Bent, 1998), and transgenic plants were selected on hygromycin. The 35S∷AtIPT3 transgene was introduced by crossing into era1-1 plants to construct era1-1AtIPT3 plants.
RNA Isolation and RT-PCR
RNA was extracted using Qiagen (Chatsworth) RNeasy columns according to the manufacturer's instructions. For RT-PCR analysis, 5 μg of total RNA was treated with DNase I, and DNA-free RNA was transcribed using an oligo(dT) primer and moloney murine leukemia virus reverse transcriptase (Clontech). Aliquots of the generated cDNA, which equaled 50 ng of total RNA, were used as a template for PCR with gene-specific primers.
Yeast Expression
His-AtIPT3 and His-AtIPT3C333S fusion genes were cut from pRSETa constructs and were directionally cloned in pJR1138 vector containing the LEU2 marker (Yalovsky et al., 1997). The resulting plasmids were used to transform the yeast (Saccharomyces cerevisiae) strains CTY716 (MATa ade2 lys2 his3 trp1 leu2 ura3 ram1 AD2) and SYY705 (MATa ade2 lys2 his3 trp1 leu2 ura3 ram1 AD2, pSYYFTA1 [high copy number URA3 LeFTA], pSYFTB2 [high copy number TRP1, LeFTB]; Lavy et al., 2002). Yeast cells were cultivated on solid YNB media (6.7 g/L yeast nitrogen base, 20% Glc, 50 μg/mL His, 50 μg/mL Lys for SYY105 strain or plus 50 μg/mL Trp and 50 μg/mL uracil for CTY716, 20% bacto agar) for 2 d at 30°C. Some colonies were inoculated into YNB liquid medium and cultured under agitation overnight at 30°C. The optical density (A600) was measured, and the suspension was then frozen in liquid nitrogen and stored at −80°C.
Cytokinin Determination
Endogenous cytokinins were extracted by methanol/formic acid/water (15/1/4, v/v/v) from 55-d-old Arabidopsis rosette leaves, homogenized in liquid nitrogen, and purified using dual-mode solid phase extraction method (Dobrev and Kaminek, 2002). Cytokinin ribotides were determined as corresponding ribosides following their dephosphorylation by alkaline phosphatase. Yeast and E. coli suspensions were filtrated, and 1 mL of culture medium was acidified with HCOOH (pH 3 or less) and purified directly on cation-exchange reversed phase MCX Oasis column (Waters). Detection and quantification were carried out using HPLC/mass spectrometry (MS; Finnigan) operated in the positive ion full-scan MS/MS mode using a multilevel calibration graph with [2H]-labeled cytokinins as internal standards. Detection limits of different cytokinins were between 0.5 and 1.0 pmol per sample. Results represent averages of analyses of three independent samples and of two HPLC MS/MS injections for each sample.
Detection of Cytokinin Oxidase/Dehydrogenase Activity
Cytokinin oxidase/dehydrogenase activity was measured as previously described (Motyka and Kaminek, 1994). Arabidopsis leaf rosettes were powdered in liquid nitrogen and further homogenized in 0.1 m Tris-HCl buffer, pH 7.5. After the purification on polyvinylpolypyrrolidone (Sigma) column and removal of nucleic acids by polyethyleneimine (Polymin P; Serva Feinbiochemica) proteins were precipitated with ammonium sulfate, and protein concentrations were determined according to the method of Bradford using bovine serum albumin as a standard. The standard radioisotope assay based on the conversion of [3H]N6-(2-isopentenyl)adenine to adenine was used for determination of cytokinin oxidase/dehydrogenase activity (Motyka et al., 2003).
Acknowledgments
We thank Dr. Hitoshi Sakakibara for the AtIPT3 antibody, Dr. Shaul Yalovsky for the CTY7166 and SYY705 yeast strains, Dr. Václav Motyka for determination of cytokinin oxidase/dehydrogenase activity in Arabidopsis, and Jiří Malbeck and Alena Trávníčková for HPLC/MS/MS.
This work was supported by ETH Zurich (to W.G.), by the Grant Agency of the Academy of Sciences of the Czech Republic (grant no. A600380507 to M.K.), and by the Ministry of Education, Youth and Sports of the Czech Republic (grant no. LCO6034 to M.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Wilhelm Gruissem (wgruissem@ethz.ch).
Open Access articles can be viewed online without a subscription.
References
- Bassil NV, Mok D, Mok MC (1993) Partial purification of a cis-trans-isomerase of zeatin from immature seed of Phaseolus vulgaris L. Plant Physiol 102 867–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GT, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inze D, Vuylsteke M (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoeva E, Dobrev PI, Malbeck J, Motyka V, Strnad M, Hanus J, Vankova R (2004) Cytokinin N-glucosylation inhibitors suppress deactivation of exogenous cytokinins in radish, but their effect on active endogenous cytokinins is counteracted by other regulatory mechanisms. Physiol Plant 121 215–222 [DOI] [PubMed] [Google Scholar]
- Brzobohaty B, Moore I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K (1993) Release of active cytokinin by a beta-glucosidase localized to the maize root meristem. Science 262 1051–1054 [DOI] [PubMed] [Google Scholar]
- Chen CM, Kristopeit SM (1981. a) Metabolism of cytokinin: deribosylation of cytokinin ribonucleoside by adenosine nucleosidase from wheat germ cells. Plant Physiol 68 1020–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Kristopeit SM (1981. b) Metabolism of cytokinin: dephosphorylation of cytokinin ribonucleotide by 5′-nucleotidases from cytosol. Plant Physiol 67 494–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Leisner SM (1984) Modification of cytokinins by cauliflower microsomal enzymes. Plant Physiol 75 442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inze D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrev PI, Kaminek M (2002) Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J Chromatogr A 950 21–29 [DOI] [PubMed] [Google Scholar]
- Galichet A, Gruissem W (2003) Protein farnesylation in plants: conserved mechanisms but different targets. Curr Opin Plant Biol 6 530–535 [DOI] [PubMed] [Google Scholar]
- Galichet A, Gruissem W (2006) Developmentally controlled farnesylation modulates AtNAP1;1 function in cell proliferation and cell expansion during Arabidopsis leaf development. Plant Physiol 142 1412–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovko A, Sitbon F, Tillberg E, Nicander B (2002) Identification of a tRNA isopentenyltransferase gene from Arabidopsis thaliana. Plant Mol Biol 49 161–169 [DOI] [PubMed] [Google Scholar]
- Haberer G, Kieber JJ (2002) Cytokinins. New insights into a classic phytohormone. Plant Physiol 128 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SH, Lall S, Che P (2003) Cytokinins and shoot development. Trends Plant Sci 8 453–459 [DOI] [PubMed] [Google Scholar]
- Hussein D, Taylor SS (2002) Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J Cell Sci 115 3403–3414 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (2001) Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol 42 677–685 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (2003) Perception and signal transduction of cytokinins. Annu Rev Plant Biol 54 605–627 [DOI] [PubMed] [Google Scholar]
- Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T, Kamiya Y, Yamaguchi S, Sakakibara H (2004) Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J Biol Chem 279 14049–14054 [DOI] [PubMed] [Google Scholar]
- Lavy M, Bracha-Drori K, Sternberg H, Yalovsky S (2002) A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14 2431–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, St Johnston D (2003) A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature 421 379–384 [DOI] [PubMed] [Google Scholar]
- Mijimolle N, Velasco J, Dubus P, Guerra C, Weinbaum CA, Casey PJ, Campuzano V, Barbacid M (2005) Protein farnesyltransferase in embryogenesis, adult homeostasis, and tumor development. Cancer Cell 7 313–324 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37 128–138 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowska D, Tabata S, Sandberg G, Kakimoto T (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA 103 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DW, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52 89–118 [DOI] [PubMed] [Google Scholar]
- Morehead TA, Biermann BJ, Crowell DN, Randall SK (1995) Changes in protein isoprenylation during the growth of suspension-cultured tobacco cells. Plant Physiol 109 277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motyka V, Kaminek M (1994) Cytokinin oxidase from auxin- and cytokinin-dependent callus cultures of tobacco (Nicotiana tabacum L.). J Plant Growth Regul 13 1–9 [Google Scholar]
- Motyka V, Vankova R, Capkova V, Petrasek J, Kaminek M, Schmulling T (2003) Cytokinin-induced upregulation of cytokinin oxidase activity in tobacco includes changes in enzyme glycosylation and secretion. Physiol Plant 117 11–21 [Google Scholar]
- Qian D, Zhou D, Ju R, Cramer CL, Yang Z (1996) Protein farnesyltransferase in plants: molecular characterization and involvement in cell cycle control. Plant Cell 8 2381–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Yalovsky S, Zik M, Fromm H, Gruissem W (1999) The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J 18 1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running MP, Lavy M, Sternberg H, Galichet A, Gruissem W, Hake S, Ori N, Yalovsky S (2004) Enlarged meristems and delayed growth in plp mutants result from lack of CaaX prenyltransferases. Proc Natl Acad Sci USA 101 7815–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57 431–449 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kasahara H, Ueda N, Kojima M, Takei K, Hishiyama S, Asami T, Okada K, Kamiya Y, Yamaya T, et al (2005) Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc Natl Acad Sci USA 102 9972–9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Seo S, Orudgev E, Youssefian S, Ishizuka K (1994) Expression of the gene for a small GTP binding protein in transgenic tobacco elevates endogenous cytokinin levels, abnormally induces salicylic acid in response to wounding, and increases resistance to tobacco mosaic virus infection. Proc Natl Acad Sci USA 91 10556–10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Wyatt S, Tsou PL, Robertson D, Allen NS (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26: 1125, 1128–1132 [DOI] [PubMed]
- Sinensky M (2000) Recent advances in the study of prenylated proteins. Biochim Biophys Acta 1484 93–106 [DOI] [PubMed] [Google Scholar]
- Sun J, Niu QW, Tarkowski P, Zheng B, Tarkowska D, Sandberg G, Chua NH, Zuo J (2003) The Arabidopsis AtIPT8/PGA22 gene encodes an isopentenyl transferase that is involved in de novo cytokinin biosynthesis. Plant Physiol 131 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Sugiyama T (2001) Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J Biol Chem 276 26405–26410 [DOI] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H (2004. b) AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol 45 1053–1062 [DOI] [PubMed] [Google Scholar]
- Takei K, Yamaya T, Sakakibara H (2004. a) Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J Biol Chem 279 41866–41872 [DOI] [PubMed] [Google Scholar]
- Tamanoi F, Kato-Stankiewicz J, Jiang C, Machado I, Thapar N (2001) Farnesylated proteins and cell cycle progression. J Cell Biochem Suppl 37 64–70 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Trueblood CE, Callan KL, Narita JO, Jenkins SM, Rine J, Gruissem W (1997) Plant farnesyltransferase can restore yeast Ras signaling and mating. Mol Cell Biol 17 1986–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Kulukian A, Rodriguez-Concepcion M, Young CA, Gruissem W (2000. a) Functional requirement of plant farnesyltransferase during development in Arabidopsis. Plant Cell 12 1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Rodriguez-Concepcion M, Bracha K, Toledo-Ortiz G, Gruissem W (2000. b) Prenylation of the floral transcription factor APETALA1 modulates its function. Plant Cell 12 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Rodriguez-Concepcion M, Gruissem W (1999) Lipid modifications of proteins: slipping in and out of membranes. Trends Plant Sci 4 439–445 [DOI] [PubMed] [Google Scholar]
- Zhu JK, Bressan RA, Hasegawa PM (1993) Isoprenylation of the plant molecular chaperone ANJ1 facilitates membrane association and function at high temperature. Proc Natl Acad Sci USA 90 8557–8561 [DOI] [PMC free article] [PubMed] [Google Scholar]