Abstract
Whereas jasmonic acid (JA) and its amino acid conjugates, particularly JA-isoleucine (Ile), are known to play important roles in plant-herbivore interactions, whether other compounds also function as signals independently of JA-Ile and whether conjugates elicit systemic responses are unknown. To answer these questions, we simultaneously silenced JASMONATE-RESISTANT4 (JAR4) and JAR6, two functionally redundant enzymes in Nicotiana attenuata that conjugate JA to amino acids to produce plants (irjar4/6) with low levels of JA-Ile, JA-leucine (Leu), and JA-valine (Val; <16% of wild type). As expected, irjar4/6 plants are more vulnerable to herbivore attack, but only JA-Ile—not JA-Leu or JA-Val—applications restored the resistance of irjar4/6 plants, suggesting that JA-Leu and JA-Val do not mediate herbivore defense responses. Interestingly, the direct defense traits of irjar4/6 plants are significantly higher than those in LIPOXYGENASE3 (LOX3)-silenced (aslox3) plants, which are impaired in JA biosynthesis, and JA-Ile treatment could not fully restore the resistance of aslox3 plants. We thus conclude that JA, its precursors, or other metabolites complement the function of JA-Ile by eliciting a panoply of induced defenses. Similarly, transcriptional profiling of wild-type, irjar4/6, and aslox3 plants with microarrays demonstrated that JA-Ile and JA play overlapping yet distinct roles in herbivore defense. Analysis of transcripts in distal tissues demonstrated that JAR activity is essential in eliciting systemic responses. However, attempts to recover JA-13C6-Ile in systemic leaves and roots after feeding wounded leaves with 13C6-Ile were unsuccessful, suggesting that JA-Ile is not a long-distance signal, but is rather synthesized after the arrival of an unknown mobile signal to systemic tissues.
Plants defend themselves against herbivores with toxins or antinutritional compounds that directly affect herbivore physiology (direct defenses) as well as with the release of volatiles that recruit natural enemies, a form of indirect defense (Kessler and Baldwin, 2002). Because defenses are costly, plants usually express them only in response to herbivore attack (Baldwin, 1998; Zavala et al., 2004; Steppuhn and Baldwin, 2008). Interestingly, many induced defenses are found not only in damaged local leaves but also in undamaged systemic tissues located distal to the attack sites (van Dam et al., 2001; Schilmiller and Howe, 2005; Wasternack et al., 2006), thus allowing the plant also to cope with highly mobile herbivores. Many plant hormones are involved in these responses, including jasmonates, salicylates, and ethylene (Kahl et al., 2000; Ziegler et al., 2001; Peng et al., 2004; Halitschke and Baldwin, 2005; von Dahl et al., 2007). Among these, the jasmonate-signaling pathway plays a central role.
Jasmonates commonly refer to jasmonic acid (JA), as well as to its precursors, and to metabolites derived from the oxylipin biosynthesis pathway. According to the current model, oxylipin biosynthesis starts with the release of linolenic acid from chloroplast membranes. Following reactions catalyzed by 13-lipoxygenase (13-LOX), allene-oxide synthase (AOS), and allene-oxide cyclase, 12-oxo-phytodienoic acid (OPDA) is synthesized and transported to the peroxisome. Here, OPDA is converted to JA by OPDA reductase (OPR) and a β-oxidase complex (Delker et al., 2006). JA can either be methylated into methyl jasmonate (MeJA) by JA carboxyl methyltransferase, or conjugated with amino acids, particularly Ile (JA-Ile), by JASMONATE-RESISTANT1 (JAR1; Seo et al., 2001; Staswick and Tiryaki, 2004). Jasmonates are involved in the regulation of plant responses to environmental and developmental cues. Their functions in defending the plant against herbivores and some pathogens are well known (Liechti and Farmer, 2002). So far, the downstream responses of most jasmonates seem to be mediated by an F-box protein, CORONATINE-INSENSITIVE1 (COI1; Xie et al., 1998).
JA is an essential component of the herbivore-induced systemic signal. In grafting experiments conducted with tomato (Solanum lycopersicum), a JA biosynthetic mutant, suppressor of prosystemin-mediated responses2 (spr2), and a JA response mutant, jasmonic acid-insensitive1 (jai1), systemic signaling was observed to require both the biosynthesis of JA at the site of wounding and the ability to perceive a jasmonate signal in remote tissues (Li et al., 2002). Recently, a grafting experiment performed with an acyl-CoA oxidase1 (acx1) mutant, which is defective in an enzyme that catalyzes the first step in the β-oxidation stage of JA biosynthesis, further demonstrated that the mobile signal is JA or its derivatives (Li et al., 2005).
Nicotiana attenuata and its specialist herbivore Manduca sexta form a fruitful system in which to study plant-herbivore interactions (Baldwin, 1998). M. sexta attack elicits a number of well-described direct and indirect defense traits in N. attenuata (Baldwin, 2001). Direct defenses include nicotine, trypsin proteinase inhibitors (TPIs), Thr deaminase (TD), and several phenolic metabolites, which are synthesized by Phe ammonia lyase (PAL; Baldwin et al., 2001; Schittko et al., 2001; Kang et al., 2006). Indirect defenses include the emission of volatile organic compounds, such as green leaf volatiles—products of the hydroperoxide lyase (HPL) pathway—and some terpenes from the isoprenoid pathway (Kessler and Baldwin, 2002).
Applying M. sexta oral secretions (OS) to wounded leaves of N. attenuata dramatically amplifies the wound-induced JA burst within 90 min (Schittko et al., 2000). Exposing N. attenuata to volatile MeJA differentially regulates several growth- and defense-related genes (Schittko et al., 2001). Silencing a specific wound- and herbivore-induced 13-LOX isoform, LOX3, reduced the accumulation of OS-elicited JA and of two direct defenses, nicotine and TPI, as well as the levels of terpenes, which act as indirect defenses. LOX3-silenced plants are more susceptible to M. sexta attack, and MeJA treatment restores plant resistance. Moreover, expression analysis with a microarray enriched in herbivore-elicited genes revealed that oxylipins synthesized downstream of LOX3 are pivotal in up-regulating both defense genes (TPI, TD, HPL) as well as oxylipin synthesis genes (α-DIOXYGENASE [α-DOX]), and down-regulating genes involved in photosynthesis (RUBISCO and PSII; Halitschke and Baldwin, 2003). Similarly, silencing N. attenuata COI1 also impairs JA-elicited direct (nicotine levels, TPI activity) and indirect (cis-α-bergamotene emission) defenses. COI1-silenced plants are highly susceptible to M. sexta and to many herbivores not found on wild-type N. attenuata in its native habitats (Paschold et al., 2007).
Previously, Kang et al. (2006) showed that silencing JAR4, the Arabidopsis (Arabidopsis thaliana) JAR1 homolog in N. attenuata, by virus-induced gene silencing (VIGS) impairs plant resistance to M. sexta and TPI activity and that the impaired phenotypes can be restored by exogenous JA-Ile application. These results indicate that JA-Ile is a very important oxylipin signal in plant-herbivore interaction. However, whether other signaling compounds also function as signals independently of JA-Ile and whether JA-Ile is the long-distance signal for the herbivore-induced systemic responses remain unclear.
Recently, we identified another JAR1 homolog in N. attenuata, JAR6. In independently silenced JAR4 and JAR6 transgenic plants, levels not only of JA-Ile/Leu, but also of JA-Val (Wang et al., 2007), were reduced. This result indicates JAR4 and JAR6 have a redundant function, synthesizing JA-amino acid conjugates in addition to JA-Ile. Additionally, it suggests that decreases in JA-Val and JA-Leu levels may impair plant resistance to herbivore attack.
In this study, we simultaneously silenced JAR4 and JAR6 (irjar4/6) by crossing independently silenced JAR4 and JAR6 plants. First, we examined whether JA-Leu and JA-Val are involved in plant-herbivore interactions. Second, we compared the phenotype of irjar4/6 and aslox3 plants to elucidate distinct roles of JA and other oxylipins versus JA-Ile in plant defense responses to herbivores. Finally, we investigated the role of JA-Ile in herbivore-induced systemic responses and examined whether JA-Ile is the mobile signal for these responses.
RESULTS
Silencing Both JAR4 and JAR6 Reduces JA-Amino Acid Conjugates without Influencing JA Levels
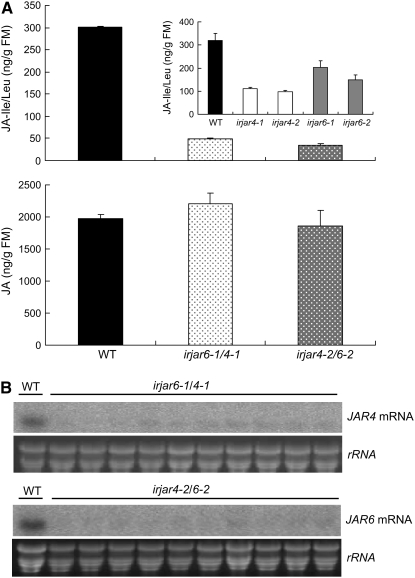
Previously, we reported that both NaJAR4 and NaJAR6 conjugate JA to Ile, Val, and Leu. Silencing JAR4 or JAR6 independently reduced levels of OS-elicited JA-Ile/Leu and JA-Val to 25.8% to 61.2% of levels found in wild-type plants (Wang et al., 2007). To obtain more completely silenced plants, we crossed homozygous T2 irjar4 plants with irjar6 plants to produce JAR4- and JAR6-silenced plants (irjar4/6). Two JAR4-silenced lines (irjar4-1 and irjar4-2) and two JAR6-silenced lines (irjar6-1 and irjar6-2) were used as both female and male parents and two F1 heterozygous lines (of eight) were selected for further study because of their low levels of JA-Ile/Leu 1 h after OS elicitation (Supplemental Fig. S1). The levels of OS-elicited JA-Ile/Leu were significantly lower in F1 heterozygotes than in independently silenced plants. Maximal elicited levels in irjar6-1/4-1and irjar4-2/6-2 were only 16% and 11.3% of those in wild-type plants. In contrast, the levels of OS-elicited JA in irjar6-1/4-1 and irjar4-2/6-2 did not significantly differ from those in wild-type plants (111.4% and 94.3% of wild type, respectively; Fig. 1A). JA-Val levels in irjar4/6 plants were below the level of detection of our liquid chromatography (LC)-mass spectrometry (MS). To measure JAR4/6 transcript levels in the heterozygotes, 10 randomly selected plants of each genotype were used for northern-blot analysis after OS elicitation. Transcript levels of all irjar4/6 plants were strongly reduced compared to wild-type plants (Fig. 1B).
Figure 1.
Silencing JAR4 and JAR6 in irjar4/6 plants. A, Mean (±se) of JA-Ile/Leu and JA concentrations in four replicate +1 leaves (one leaf position older than the source-sink transition leaf) of wild-type and irjar4/6 (irjar4-2/6-2, irjar6-1/4-1) plants, as well as of plants independently silenced for JAR4 or JAR6 (inset; irjar4-1, irjar4-2, irjar6-1, irjar6-2) 1 h after JA treatment. B, Accumulation of JAR4 or JAR6 transcripts in irjar4/6 and wild-type plants. RNA was isolated from 10 leaves growing at node +1 of separate irjar4/6 plants or five pooled +1 leaves of wild-type plants. Leaves were harvested 1 h (for irjar6-1/4-1 and wild-type control) or 3 h (for irjar4-2/6-2 and wild-type control) after OS treatment. A duplicate gel stained with ethidium bromide was used as an RNA-loading control.
Impaired Direct Defenses and M. sexta Resistance of irjar4/6 Plants Are Restored by Treatment with JA-Ile, But Not JA-Leu or JA-Val
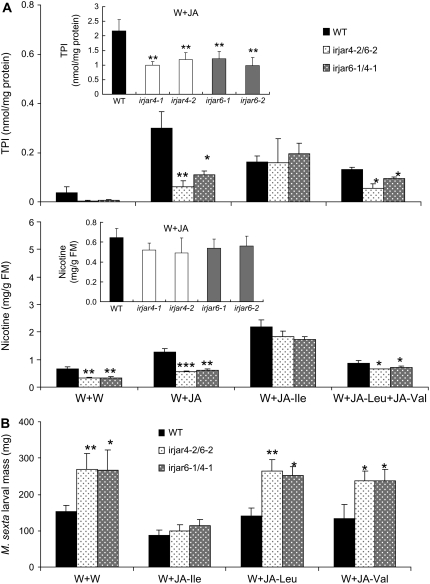
When JAR4 or JAR6 is independently silenced, levels of JA-elicited TPIs (<56.1% of those in wild-type plants; Student's t test; P ≤ 0.0061) but not nicotine are significantly reduced (>75.4% of those in wild-type plants; Student's t test; P ≥ 0.0646; Fig. 2A, insets). However, when JAR4 and JAR6 are simultaneously silenced, not only elicited TPI activity but also nicotine levels were significantly reduced. Compared to wounded controls, JA treatment amplified the differences in the two direct defenses between irjar4/6 (irjar4-2/6-2, irjar6-1/4-1) and wild-type plants. In irjar4/6 plants, TPI levels were <36.3% (Student's t test; P ≤ 0.0235) of those in wild-type plants; nicotine levels were <47.9% (Student's t test; P ≤ 0.0016) of those in wild-type plants (Fig. 2A). As expected, irjar4/6 plants are more susceptible than wild-type plants to herbivore attack. In glasshouse experiments, M. sexta gained significantly more mass (Student's t test; P ≤ 0.0428) on two lines of irjar4/6 plants than on wild-type plants when the plants were treated with wounding plus water (Fig. 2B).
Figure 2.
Impaired direct defenses and resistance of irjar4/6 plant to M. sexta attack could be restored by treatment with JA-Ile but not JA-Leu or JA-Val. A, Mean (±se) levels of TPIs and nicotine in four replicate leaves growing at the +1 node of wild-type and two lines of irjar4/6 plants (irjar4-2/6-2 and irjar6-1/4-1), as well as plants independently silenced for JAR4 or JAR6 (inset; irjar4-1, irjar4-2, irjar6-1, irjar6-2). Rosette-stage plants were wounded and treated with 0.25 μmol JA (W + JA), JA-Ile (W + JA-Ile), JA-Val plus JA-Leu (W + JA-Val + JA-Leu), or water (W + W). Ninety-six hours after treatment, treated leaves were harvested for analysis. Asterisks represent significant difference between members of a pair (Student's t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001). B, Mean (±se) mass of 18 replicated M. sexta larvae after 9 d of feeding on wild-type and two lines of irjar4/6 plants (irjar4-2/6-2 and irjar6-1/4-1). Rosette-stage plants were wounded and treated with 0.25 μmol JA-Ile (W + JA-Ile), JA-Val (W + JA-Val), JA-Leu (W + JA-Leu), or water (W + W). One day later, one freshly hatched larva was placed on treated leaves of each treated plant. Asterisks represent significant difference between members of a pair (Student's t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
Because the OS-elicited bursts in JA-Leu/Ile and JA-Val in irjar4/6 plants are all significantly reduced (Supplemental Fig. S1A), which JA-amino acid conjugate was responsible for eliciting herbivore resistance was unclear. To determine whether JA-Val and JA-Leu are able to restore herbivore resistance in irjar4/6 plants, we treated plants with JA-amino acid conjugates and measured direct defense levels and herbivore resistance traits. Unlike JA-Ile treatment (Student's t test; TPI, P ≥ 0.6097; nicotine, P ≥ 0.1028), treatment with JA-Leu plus JA-Val restored neither TPI levels (Student's t test; P ≤ 0.0206) nor nicotine levels (Student's t test; P ≤ 0.0393; Fig. 2A). We also tested whether treatment with JA-Leu or JA-Val was able to rescue the herbivore resistance of irjar4/6 plants. Consistent with direct defense results, after 9 d of feeding, M. sexta larvae did not gain more mass on JA-Ile-treated plants (Student's t test; P ≥ 0.3072), but they did gain more mass on JA-Leu-treated (Student's t test; P ≤ 0.0459) and JA-Val-treated (Student's t test; P ≤ 0.0497) plants, demonstrating that the lack of JA-Ile, but not of JA-Leu or JA-Val, was responsible for the susceptibility of irjar4/6 plants (Fig. 2B).
Transcriptional Responses Elicited by Treatment with JA-Amino Acid Conjugates and MeJA in Wild-Type Plants
To further investigate the roles of JA-Ile, JA-Leu, and JA-Val in herbivore resistance, we used microarrays enriched in herbivore-elicited genes to compare the transcriptional responses of wild-type plants to treatments with the different JA conjugates. The analysis revealed that MeJA treatment up-regulated 114 genes and down-regulated 80 genes (expression levels at least 1.5-fold up or down; Supplemental Table S1). In response to treatments with JA-Ile, JA-Leu, and JA-Val, 43, 76, and 84 genes were up-regulated and 22, 41, and 64 genes were down-regulated, respectively (Supplemental Tables S2–S4). A Venn diagram reveals a high percentage of overlap between MeJA-regulated genes and JA-Leu- as well as JA-Val-regulated genes: 62% of JA-Leu-regulated genes and 44% of JA-Val-regulated genes are also regulated by MeJA. However, JA-Ile-elicited genes are different from MeJA-elicited genes: Only 22% were also regulated by MeJA (Supplemental Fig. S2A). Principal component analysis also revealed that JA-Leu-induced and JA-Val-induced transcriptional changes correlate significantly with changes induced by MeJA, but the changes induced by JA-Ile are distinct from the MeJA-elicited response (Supplemental Fig. S2B). These transcriptional responses are consistent with the conclusion from complementation experiments; although irjar4/6 plants show lower OS-elicited bursts of JA-Ile/Leu and JA-Val compared to wild-type plants, only the absence of JA-Ile can explain the impaired levels of TPI, nicotine, and herbivore resistance in irjar4/6 plants. The fact that the effects elicited by JA-Val and JA-Leu are highly similar to the effects elicited by MeJA, which suggests that these result in part from hydrolysis of the amino acid conjugate and deesterification of MeJA to form JA.
JA-Ile Treatment Restores OS-Elicited Transcriptional Responses Lacking in irjar4/6 Plants
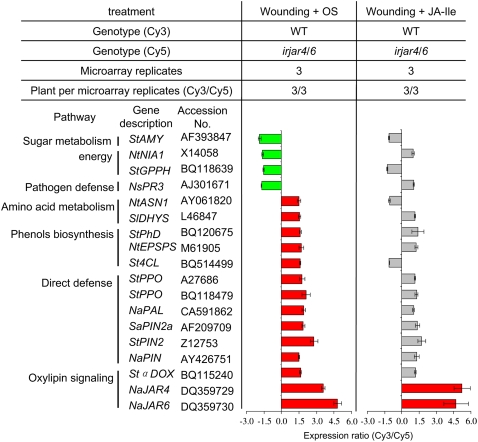
To further examine JAR4/6-dependent transcriptional responses, we used microarrays to compare transcript accumulation in response to OS elicitation in irjar4/6 (irjar4-2/6-2) and wild-type plants (Fig. 3). As expected, JAR4 and JAR6 transcripts are much lower, confirming that both JAR4 and JAR6 were thoroughly silenced in irjar4/6 plants. Genes up-regulated by JAR4/6 include several direct defense genes, such as PIN, PAL, α-DOX, and POLYPHENOL OXIDASE (PPO), as well as genes involved in biosynthesis of phenolics and amino acids. However, genes down-regulated by JAR4/6, which include some pathogen defense genes, such as PATHOGENESIS-RELATED3 (PR3) and several genes related to energy and heavy-metal homeostasis, are few (Fig. 3). After treatment with JA-Ile, all of the OS-elicited transcripts that were differentially regulated from wild type in irjar4/6 plants (with the exception of JAR4 and JAR6) were restored (Fig. 3), which is consistent with the interpretation that transcriptional changes in irjar4/6 plants result from the low amounts of JA-Ile in these plants.
Figure 3.
Silencing both JAR4 and JAR6 suppresses OS-elicited transcriptional response, which can be restored by JA-Ile supplementation. Leaves at node +1 of rosette-stage wild-type and irjar4/6 (irjar4-2/6-2) plants were wounded and treated with OS or 0.25 μmol JA-Ile. After 180 min, leaf material was harvested and RNA was extracted, transcribed into cDNA, and labeled with fluorescent dye (wild-type samples, Cy3; irjar4/6 samples, Cy5). For each treatment, three pools of wild-type samples and three pools of irjar4/6 samples were made, each consisting of three biological replicates. Each microarray was hybridized with one wild-type pool and one irjar4/6 pool of labeled cDNA. Three microarrays per treatment were hybridized and statistically analyzed. In the OS treatment (left bars), genes significantly (P < 0.05) higher (Cy3/Cy5 > 1.5) and lower (Cy3/Cy5 < −1.5) than those expressed in irjar4/6 plants are shown with red and green bars, respectively. In the JA-Ile treatment (right bars), genes not differently expressed (−1.5 < Cy3/Cy5 < 1.5) in two genotypes are shown with gray bars. Genes are identified by a two-letter genus/species designation followed by the gene name. St, Solanum tuberosum; Nt, Nicotiana tabacum; Ns, Nicotiana sylvestris; Sl, Solanum lycopersicum; Sa, Solanum americanum; AMY, β-amylase; NIA1, nitrate reductase; GPPH, α-glucan phosphorylase, H isozyme; ASN1, Asn synthase; DHYS, dehydroquinate synthase; PhD, prephenate dehydratase; EPSPS, 5-enolpyruvylshikimate-3-P synthase; 4CL, 4-coumarate-CoA ligase.
JA and Its Other Metabolites Are Also Important for Herbivore Resistance
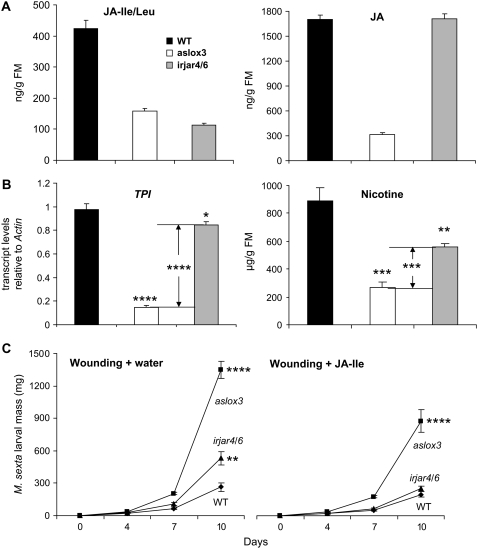
To determine whether JA must be modified to JA-Ile to elicit all the defenses that are effective against M. sexta attack, we compared resistance and defense responses of irjar4/6 plants with those of aslox3 plants, which are also known to have impaired direct and indirect defenses and be highly vulnerable to herbivore attack (Kessler and Baldwin, 2001; Halitschke and Baldwin, 2003). This comparison is particularly informative because, whereas both lines had suppressed JA-Ile levels, aslox3 plants also had reduced JA levels after OS elicitation. Compared to irjar4/6 (irjar4-2/6-2) plants, aslox3 plants showed largely reduced JA levels (18.6% of those in wild-type plants) and similarly reduced JA-Ile levels (37.2% of those in wild-type plants), whereas the JA levels of irjar4/6 did not differ from JA levels in wild-type plants (Fig. 4A). Although TPI transcripts were significantly reduced in both aslox3 (14.9% of those in wild-type plants; Student's t test; P < 0.0001) and irjar4/6 plants (86.6% of those in wild-type plants; Student's t test; P = 0.05), reductions were 6.4-fold larger in aslox3 plants (Student's t test; P < 0.0001). Similarly, although nicotine levels were significantly reduced in both aslox3 (29.9% of those in wild-type plants; Student's t test; P = 0.0004) and irjar4/6 plants (62.9% of those in wild-type plants; Student's t test; P = 0.0052), the reduction in aslox3 plants was 1.9-fold larger (Student's t test; P = 0.0003; Fig. 4B). Subsequently, we asked whether the herbivore resistance of aslox3 plants could be fully restored by treating wounds with JA-Ile. In control plants, after 10 d of feeding, larvae gained significantly more mass on both aslox3 and irjar4/6 plants (Student's t test; TPI; P < 0.0032); however, larvae on aslox3 plants gained 2.5-fold more than those on irjar4/6 plants. Moreover, after JA-Ile treatment, only the resistance of irjar4/6 plants was fully restored to levels of wild-type plants (Student's t test; P = 0.1874); larvae still gained significantly more mass on aslox3 plants treated with JA-Ile (Student's t test; P < 0.0001; Fig. 4C).
Figure 4.
JA-Ile is not the only active oxylipin signal eliciting antiherbivore defenses. A, Mean (±se) JA-Ile/Leu and JA concentrations of four replicate wild-type, aslox3, and irjar4/6 (irjar4-2/6-2) plants 1 h after OS treatment. B, Mean (±se) transcript levels of TPI and nicotine concentrations in four replicated wild-type, aslox3, or irjar4/6 (irjar4-2/6-2) plants 10 h (for TPI) or 72 h (for nicotine) after OS elicitation. C, Mean (±se) mass of 18 replicated M. sexta larvae feeding on wild-type, aslox3, and irjar4/6 plants (irjar4-2/6-2). Rosette-stage plants were wounded and treated with water (wounding + water) or 0.25 μmol JA-Ile (wounding + JA-Ile). One day later, one freshly hatched larva was placed on treated leaves of each treated plant. Asterisks represent significant differences between members of a pair (Student's t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
These results demonstrate that, whereas JA-Ile is clearly an important mediator of traits that confer resistance to M. sexta larvae, JA, its precursors, or the other JA conjugates also play significant roles in these responses.
Differences in OS-Elicited Transcriptional Responses between aslox3 and irjar4/6 Plants
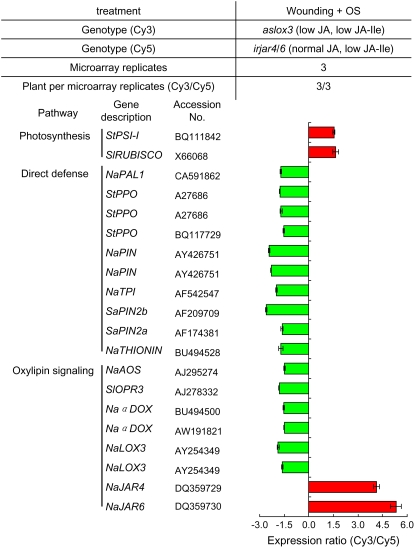
Because aslox3 plants are deficient in all the jasmonate derivatives, including JA-Ile, microarray analyses with aslox3 plants were expected to reveal the genes regulated by all LOX3-derived jasmonates, including JA-Ile. We labeled cDNA from OS-treated aslox3 plants with Cy3 and cDNA from irjar4/6 plants (irjar4-2/6-2) with Cy5. This hybridization scheme was expected to remove genes that are regulated by JA-Ile, leaving only genes specifically regulated by JA and other JA metabolites (Fig. 5). Up-regulation (red bars) or down-regulation (green bars) indicates that the expression was higher or lower in aslox3 plants than in irjar4/6 plants, respectively. Interestingly, some JA-Ile-regulated genes, such as PROTEINASE INHIBITOR (PIN), PAL, α-DOX, and PPO, were still significantly less expressed in aslox3 plants than in irjar4/6 plants, indicating that they are up-regulated by LOX3-derived jasmonates other than JA-Ile. The list of LOX3-regulated genes that could not be fully recovered by JA-Ile also contains many oxylipin-signaling genes such as AOS, OPR3, and many photosynthesis genes such as RUBISCO and PSI-I (Fig. 5). These results further support the hypothesis that JA-Ile is not the only active jasmonate signal: LOX3-derived jasmonates also play important roles in down-regulating genes involved in photosynthesis and up-regulating JA genes involved in biosynthesis.
Figure 5.
Genes differently expressed in aslox3 and irajr4/6 plants. Leaves growing at +1 node of rosette-stage aslox3 and irjar4/6 (irjar4-2/6-2) plants were OS elicited. After 180 min, leaf material was harvested and RNA was extracted, transcribed into cDNA, and labeled with fluorescent dye (aslox3 samples, Cy3; irjar4/6 samples, Cy5). Three pools of aslox3 samples and three pools of irjar4/6 samples were made, each consisting of three biological replicates. Each microarray was hybridized with one aslox3 pool and one irjar4/6 pool of labeled cDNA. Three microarrays per treatment were hybridized and statistically analyzed. Genes significantly (P < 0.05) higher (Cy3/Cy5 > 1.5) and lower (Cy3/Cy5 < −1.5) expressed in aslox3 plants are shown with red and green bars, respectively. Genes are identified by two-letter genus/species designations followed by the gene name. St, Solanum tuberosum; Nt, Nicotiana tabacum; Sl, Solanum lycopersicum; Sa, Solanum americanum.
Simultaneously Silencing JAR4 and JAR6 Impairs Herbivore-Induced Systemic Responses
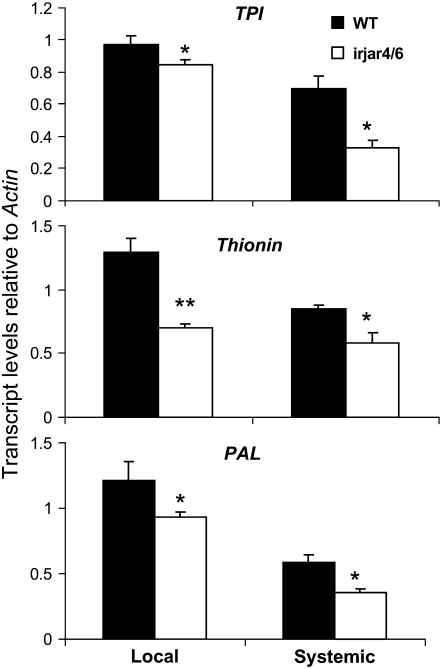
To investigate whether JA-Ile is involved in herbivore-induced systemic responses, we compared the transcript levels of TPI, PAL1, and THIONIN—well-known markers of systemic responses—in the local and systemic leaves of irjar4/6 (irjar4-2/6-2) and wild-type plants after OS elicitation. Leaves growing at node +1 (one leaf position older than the source-sink transition leaf) were OS elicited; orthostichous −4 leaves were harvested to measure systemic responses. As expected, transcript levels of all three genes were significantly lower in irjar4/6 local leaves (Student's t test; TPI, 86.6% of those in wild-type plants; P = 0.05; THIONIN, 54.5% of those in wild-type plants; P = 0.0062; PAL1, 76.5% of those in wild-type plants; P = 0.0422). Importantly, they were also significantly lower in systemic leaves (Student's t test; TPI, 47.5% of those in wild-type plants; P = 0.0133; THIONIN, 68.8% of those in wild-type plants; P = 0.02; PAL1, 60.0% of those in wild-type plants; P = 0.011; Fig. 6). These results suggest that JA-Ile plays an important role in transmitting herbivore-induced responses to undamaged leaves.
Figure 6.
Herbivore-induced systemic responses are impaired in irjar4/6 plants. Mean (±se) transcript levels of TPI, THIONIN, and PAL1 in five replicated local or systemic leaves 10 h (for TPI and THIONIN) or 2 h (for PAL1) after OS elicitation. Node +1 leaves were wounded and treated with OS. The untreated systemic leaf is growing at node −4. Asterisks represent significant difference between members of a pair (Student's t test; * P, < 0.05; ** P, < 0.01).
JA-Ile Itself May Not Be the Long-Distance Signal for Systemic Responses
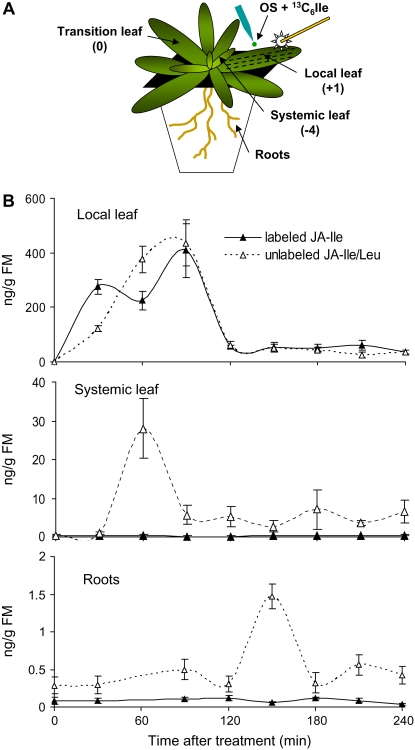
To investigate whether JA-Ile itself is the long-distance signal for herbivore-induced systemic responses or whether it has a different role in transmission of the systemic signal, 13C6-labeled Ile was mixed with M. sexta OS and supplied to the wounded leaves of hydroponic-cultured wild-type plants. Subsequently, unlabeled JA-Ile/Leu and 13C6-labeled JA-Ile were measured in treated local leaves, as well as in systemic leaves and roots (Fig. 7A). In the treated leaves, 90 min after treatment, similar levels of labeled JA-Ile and unlabeled JA-Ile/Leu were found (labeled JA-Ile, 408.4 ng/g fresh weight; unlabeled JA-Ile/Leu, 435.7 ng/g fresh weight). However, in untreated systemic leaves and roots, only unlabeled JA-Ile/Leu was detected. This reached peak levels at 60 (systemic leaf) and 150 min (roots; Fig. 7B). Because we could not distinguish JA-Ile and JA-Leu in our LC-MS-MS-MS analysis, it could be possible that the mobile signal is JA-Leu; however, we can exclude this because we show here that JA-Leu is not a functional jasmonate signal. In summary, these results indicate that de novo synthesis of JA-Ile is induced in distal tissue after wounding and OS elicitation of local leaves and suggest that, although JA-Ile is essential for systemic responses, it may not be the long-distance signal that triggers these responses.
Figure 7.
Labeled JA-Ile was not transported to systemic tissues. A, Numbering of leaf positions in a rosette-stage N. attenuata plant growing in hydroponic cultures and illustration of treatments. The leaf undergoing the source-sink transition was designated as growing at node 0. The treated local leaf growing at node +1, which is older by one leaf position than the source-sink transition leaf, was wounded with a pattern wheel and treated with OS containing 0.625 μmol 13C6-labeled Ile. The untreated systemic leaf is growing at node −4. B, Mean (±se) of JA-Ile/Leu and 13C6-labeled JA-Ile in four replicated treated leaves, systemic leaves, and roots.
DISCUSSION
Here, we successfully silenced both JAR4 and JAR6 by crossing independently silenced JAR4 and JAR6 T2 homozygotes. F1 heterozygous irjar4/6 plants also showed extremely reduced JA-Ile/Leu levels (only about 15% of wild-type JA-Ile levels) and had impaired levels of nicotine, which are unaffected in the independently silenced lines. The irjar4/6 plants were therefore ideal for studying downstream responses regulated by JA-amino acid conjugates, and, because irjar4/6 plants did not show impaired JA levels, they were helpful for distinguishing the roles of JA and JA-amino acid conjugates in herbivore resistance. They were also particularly useful for elucidating the role of JA-amino acid conjugates in herbivore-induced systemic responses and in the mobile signaling that elicits these responses.
Deficiency of JA-Ile, But Not JA-Leu or JA-Val, Impairs the Elicited Direct Defenses and Herbivore Resistance of irjar4/6 Plants
Silencing JAR4 by VIGS significantly impaired the JA-Ile burst after OS elicitation and herbivore resistance, which could be restored by JA-Ile treatment, confirming that JA-Ile plays important roles in activating plant defenses (Kang et al., 2006). Simultaneously silencing JAR4 and JAR6 impaired levels of JA-Ile/Leu as well as JA-Val (Wang et al., 2007), suggesting that the impaired herbivore resistance of JAR4-silenced plants might also be caused by a deficiency of JA-Val or JA-Leu. However, two aspects of our results suggest JA-Val and JA-Leu are not involved. First, the compromised direct defenses, nicotine and TPI, in irjar4/6 plants could not be restored by application of JA-Leu plus JA-Val. Second, only JA-Ile, not JA-Val and JA-Leu, restored the impaired herbivore resistance of irjar4/6 plants. The fact that JA-Leu plus JA-Val treatment slightly increased levels of nicotine and TPIs can be explained by the results of microarray analysis of JA-Leu- and JA-Val-treated plants. The induced transcript profiles overlap highly among JA-Leu, JA-Val, and MeJA, suggesting that some of the exogenously applied JA-Leu and JA-Val was hydrolyzed to JA. Thus, a deficiency of JA-Ile, but not of JA-Leu or JA-Val, seems to be the real reason for the impaired herbivore susceptibility of JAR4/6-silenced plants. These results are consistent with findings in Arabidopsis, where JA-Leu and JA-Val are not effective root inhibitors and applying them to the medium could not restore the root phenotype of the jar1 mutant (Staswick and Tiryaki, 2004).
Both JA and JA-Ile Play Important But Distinct Roles
JA is well known to play a crucial role in plant-herbivore interactions. In N. attenuata, silencing the JA biosynthesis key gene, LOX3, or perception gene, COI1, greatly impairs herbivore-induced direct and indirect defenses, as well as plant resistance to M. sexta attack as well as to many other herbivores, including those not commonly found on these plants in nature (Kessler and Baldwin, 2001; Halitschke and Baldwin, 2003; Paschold et al., 2007). However, studies also support JA-Ile's importance in plant-herbivore interactions (Kang et al., 2006). More important, Thines et al. (2007) showed that JA-Ile is highly active in promoting interaction between the tomato COI1 and JAZ1 proteins, whereas JA and OPDA are inactive. Thus, a new question arises: How important is the role of JA-Ile in comparison to JA and its other known metabolites? Our results demonstrate that JA-Ile is not the only active oxylipin signal. Compared to aslox3 plants (which have reduced JA and JA-Ile levels), not only were the levels of direct defense traits of irjar4/6 plants much higher, but also M. sexta larvae gained much more mass on aslox3 plants than on irjar4/6 plants. More importantly, JA-Ile could not fully restore the herbivore resistance of aslox3 plants. These results demonstrate that JA and its precursor and/or its other metabolites also play important roles in plant-herbivore interaction. Although it has been shown that the JA precursor, OPDA, is an important signal for the defense against Bradysia impatiens and Alternaria brassicicola in the absence of JA (Stintzi et al., 2001), the tomato and Arabidopsis mutants deficient in ACX genes (encode key enzymes for β-oxidation) are susceptible to lepidopteran insects, suggesting that JA or its derivatives are more important in eliciting defenses against herbivores (Li et al., 2005; Schilmiller et al., 2007). The fact that Arabidopsis jar1 mutants are weak mutants of JA responses is consistent with this view. Unlike other JA biosynthesis and perception mutants in Arabidopsis (e.g. opr3 and coi1), the jar1 mutant is not sterile, suggesting that JAR1 is not necessary for the entire jasmonate response (Staswick et al., 1992; Xie et al., 1998; Stintzi and Browse, 2000).
Microarray analysis indicates some genes, such as PIN, PAL, PPO, and α-DOX, are simultaneously regulated by JA-Ile, as well as by JA or the other JA derivatives. Meanwhile, many oxylipin-signaling genes, including AOS and OPR3, and genes involved in photosynthesis, including RUBISCO and PSI-I, are more likely specifically regulated by JA or the other JA derivates. We propose that JA-Ile forms an activating complex with SCFCOI1, which degrades the JAZ1 repressor and thus activates the expression of direct defense genes PIN, PAL, PPO, and α-DOX. At the same time, JA and/or its other metabolites form different activating complexes with SCFCOI1, degrading one or more other JAZ proteins, JAZXs. JAZXs share similar functions with JAZ1, repressing the expression of direct defense genes PIN, PAL, PPO, and α-DOX. On the other hand, JAZXs have a specific function: repressing the expression of genes involved in JA biosynthesis, AOS and OPR3.
JA-Ile May Not Be the Mobile Signal for Herbivore-Induced Systemic Responses
The grafting experiment with the tomato acx1 mutant clearly demonstrated that JA, or a derivative of JA, is the transmissible signal for herbivore-induced systemic responses (Li et al., 2005). Because the lack of JA-Ile significantly reduces herbivore-induced TPI, PAL1, THIONIN, and nicotine in systemic tissues, JA-Ile is a good candidate for the long-distance signal. Importantly, nicotine can be restored to wild-type levels in irjar4/6 plants by applying JA-Ile to leaves, suggesting that exogenously supplied JA-Ile is transported to the roots, where nicotine is biosynthesized. However, our results suggest JA-Ile may not be the mobile signal. After 13C6-Ile was applied to wounded leaves, similar amounts of labeled JA-Ile and unlabeled JA-Ile/Leu were produced in treated local leaves; however, only unlabeled JA-Ile/Leu was detected in systemic leaves and roots. These results indicate that systemically occurring JA-Ile is synthesized de novo after the mobile signal arrives in systemic tissues. However, it is possible that the JA-Ile that was synthesized in planta from exogenously applied 13C6-Ile is not transported in the same way as JA-Ile synthesized from plant-derived Ile. For example, there is the possibility that the Ile in vascular bundles where a mobile signal is likely synthesized could not be labeled as efficiently as the Ile in mesophyll cells and therefore could not be detected in systemic tissues. Our negative results suggest an alternative hypothesis: that another mobile signal is transported to systemic tissues before JA-Ile synthesis. The mobile signal might be JA or other JA metabolites. According to this scenario, when the mobile signal arrives in systemic leaves, JA and de novo synthesized JA-Ile are perceived and activate responses just as they do in attacked leaves. The signal transported to the roots induces nicotine biosynthesis and is transported to the aboveground tissues where, together with the other direct defenses, it defends the plant against herbivores.
To summarize, we silenced both JAR4 and JAR6 in irjar4/6 plants, in which levels of JA-Ile/Leu and JA-Val were then greatly reduced. We demonstrated that the impaired herbivore resistance of irjar4/6 plants is caused by the lack of JA-Ile but not of JA-Leu or JA-Val. We also found that JA-Ile and other jasmonates play important but distinct roles in plant-herbivore interactions. Finally, we propose that JA-Ile, while important for herbivore-induced systemic responses, is not itself the mobile signal.
MATERIALS AND METHODS
Plant Growth and Manduca sexta Rearing
An inbred genotype of Nicotiana attenuata Torr. Ex Wats. (synonymous with Nicotiana torreyana; Solanaceae), originally collected from southwestern Utah in 1988, was transformed and used for all experiments. Seed germination and plant growth were conducted as described by Krügel et al. (2002). aslox3 plants, which are silenced in LOX3, a key enzyme of JA biosynthesis, were used (Halitschke and Baldwin, 2003). irjar4/6 plants are F1 heterozygous plants produced by crossing irjar4-2 and irjar6-1 T2 homozygous plants (female parents) with irjar6-2 and irjar4-1 T2 homozygous plants (male parents), respectively (Wang et al., 2007). Two lines, irjar4-2/6-2 and irjar6-1/4-1, were selected based on their low JA-Ile levels (Supplemental Fig. S1) and used for all experiments.
Eggs of Manduca sexta were obtained from North Carolina State University and kept in a growth chamber (Snijders Scientific; http://www.snijders-tilburg.nl) at 26°C/16 h light, 24°C/8 h darkness until the larvae hatched.
JA and JA-Amino Acid Conjugate Synthesis and Measurement, and Plant Treatments
JA-Ile, JA-Val, and JA-Leu were synthesized and measured as described previously (Wang et al., 2007). For MeJA, JA-Ile, JA-Val, JA-Leu treatment, and water control, leaves growing at nodes 0, +1, and +2 (the leaf undergoing the source-sink transition [T] was designated as growing at node 0) were wounded with a fabric pattern wheel, and the resulting wounds were immediately treated with 60 μL JA-Ile, MeJA, JA-Leu, JA-Val, or water (containing 12.5% ethanol; each leaf received 20 μL). The concentration of MeJA and JA-amino acid conjugate was 0.25 μmol/60 μL of 12.5% ethanol. For OS elicitation, the node +1 leaf was wounded as described above and immediately treated with 20 μL of OS. M. sexta larval OS were collected with Teflon tubing connected to a vacuum and stored under argon at −20°C.
Microarray Analysis
To analyze transcriptional changes in response to MeJA, JA-Ile, JA-Val, and JA-Leu, wild-type plants were treated as described above. Leaf material (leaf +1) from three plants per treatment was harvested 3 h after induction and replicates were pooled before RNA extraction. From each pool, mRNA was isolated, reverse transcribed, and labeled as previously described (Halitschke and Baldwin, 2003). Jasmonate-treated wild-type samples were Cy3 labeled and hybridized against water-treated wild-type samples labeled with Cy5.
To compare the OS- and JA-Ile-elicited transcriptional responses in irjar4/6 plants and to compare OS-elicited transcriptional responses of irjar4/6 plants and aslox3 plants, we elicited wild-type, irjar4/6, and aslox3 plants as described above. Leaf material (+1 leaf) from nine plants per genotype was harvested 3 h after treatment, and, for each microarray, leaves of three plants were pooled for RNA extraction. For all hybridizations, three replicate microarrays were performed.
We used a custom-made 1.4K microarray consisting of 50mer oligonucleotides. Sequences were selected from herbivore-induced genes identified in differential experiments and from public databases. All 1,405 clones were spotted 4-fold on each array. Spot intensities (SIs) for Cy3 and Cy5 were extracted from image files using AIDA software (Raytest; http://www.raytest.com). Raw signal intensities were local background (LBg) subtracted and LOWESS normalized using MIDAS (Saeed et al., 2003; also see http://www.tm4.org). Spots below 1.5× signal-to-noise ratio (=1.5× SI/LBg) were set to zero. For statistical analysis, all Bg-corrected SIs were 2Log transformed. Single slides were evaluated on the basis of an average treatment-to-control ratio >1.5 or <−1.5, and a P value (t test after exclusion of zero values) <0.05 as the criteria for significant regulation. To analyze the three replicate microarrays from the experiment, a nested ANOVA was performed on a total of 12 normalized Cy3 and Cy5 values for each clone. The obtained P values of the factor treatment for all clones were adjusted for multiple testing using Benjamini and Hochberg's (1995) step-up procedure for controlling the false discovery rate. To compare the overall transcriptional responses between the different jasmonate treatments and to determine the interarray distances, we performed principal component analysis using MeV (Saeed et al., 2003; see also http://www.tm4.org).
Real-Time PCR Assay
Total RNA was extracted with TRI reagent (Sigma; http://www.sigmaaldrich.com) according to the manufacturer's instructions and cDNA was prepared from 200 ng of total RNA with MultiScribe reverse transcriptase (Applied Biosystems; http://www.appliedbiosystems.com). Quantitative real-time PCR (ABI PRISM7000; Applied Biosystems; http://www.appliedbiosystems.com) was conducted using the quantitative PCR core reagent kit (Eurogentec; http://www.eurogentec.com) and gene-specific TaqMan primer pairs for the individual genes: TPIFP, 5′-TCAGGAGATAGTAAATATGGCTGTTCA-3′; RP, 5′-ATCTGCATGTTCCACATTGCTTA-3′; THIONIN FP, 5′-AACTATGGCTCGCTCCTTGTC-3′; RP, 5′-CTCATAGGCAACAAAAAGCATCA-3′; and PAL1 FP, 5′-TTTGCATACGCTGATGACGC-3′; RP, 5′-TGGAAGATAGAGCTGTTCGCG-3′.
The PCR products were detected by gene-specific double-fluorescent dye-labeled TaqMan probes: TPI, TCCTTGCTCTCCTCCTCTTATTTGGAATGTCT; THIONIN, TTCATGGCATTTGCAGTCTTGGCAA; and PAL1, CAGAAACTGAGGCAAGTACTCGTCGACCAC.
Relative gene expression was calculated using a 2-fold dilution series of cDNAs, which had been transcribed from induced RNA samples of the same experiment and normalized by the N. attenuata Actin gene. Actin FP, 5′-GGTCGTACCACCGGTATTGTG-3′; Actin RP, 5′-GTCAAGACGGAGAATGGCATG-3′; FAM-labeled Actin probe, 5′-TCAGCCACACCGTCCCAATTTATGAGG-3′.
Analysis of Direct Defense Traits
Nicotine was analyzed by HPLC as described previously (Ziegler et al., 2001) with the following modification of the extraction procedure: Approximately 100 mg of frozen tissue was homogenized in 1 mL of extraction buffer utilizing the FastPrep extraction system (Savant Instruments; http://www.gmi-inc.com). Samples were homogenized in FastPrep tubes containing a 900-mg lysing matrix (BIO 101; http://www.qbiogene.com) by shaking at 6.0 m s−1 for 45 s. TPI activity was analyzed by radial diffusion activity assay as described previously (van Dam et al., 2001).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: NaTPI, AY426751; NaPAL1, CA591862; NaTHIONIN, BU494528; NaJAR4, DQ359729; NaJAR6, DQ359730.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phytohormone screening of irjar4/6 plants.
Supplemental Figure S2. JA-Ile elicits transcriptional responses of wild-type plants that are different from those elicited by MeJA, JA-Leu, and JA-Val as determined by microarray analyses.
Supplemental Table S1. List of selected genes differentially regulated by MeJA.
Supplemental Table S2. List of selected genes differentially regulated by JA-Ile.
Supplemental Table S3. List of selected genes differentially regulated by JA-Leu.
Supplemental Table S4. List of selected genes differentially regulated by JA-Val.
Supplementary Material
Acknowledgments
We thank Dr. Tamara Krügel for help in crossing; Eva Rothe and Dr. Matthias Schöttner for assistance in LC-MS analysis; Dr. Gustavo Bonaventure and Meredith Schuman for critical reading of the manuscript; Dr. Jin-Ho Kang for the comments on the experimental design; and Emily Wheeler for editorial assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ian T. Baldwin (baldwin@ice.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baldwin IT (1998) Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA 95 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT (2001) An ecologically motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiol 127 1449–1458 [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT, Halitschke R, Kessler A, Schittko U (2001) Merging molecular and ecological approaches in plant-insect interactions. Curr Opin Plant Biol 4 351–358 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57 289–300 [Google Scholar]
- Delker C, Stenzel I, Hause B, Miersch O, Feussner I, Wasternack C (2006) Jasmonate biosynthesis in Arabidopsis thaliana—enzymes, products, regulation. Plant Biol (Stuttg) 8 297–306 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Baldwin IT (2003) Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J 36 794–807 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Baldwin IT (2005) Jasmonates and related compounds in plant-herbivore interactions. J Plant Growth Regul 23 238–245 [Google Scholar]
- Kahl J, Siemens DH, Aerts RJ, Gabler R, Kuhnemann F, Preston CA, Baldwin IT (2000) Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore. Planta 210 336–342 [DOI] [PubMed] [Google Scholar]
- Kang JH, Wang L, Giri A, Baldwin IT (2006) Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid-isoleucine-mediated defenses against Manduca sexta. Plant Cell 18 3303–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291 2141–2144 [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53 299–328 [DOI] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT (2002) Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12 177–183 [Google Scholar]
- Li C, Schilmiller AL, Liu G, Lee GI, Jayanty S, Sageman C, Vrebalov J, Giovannoni JJ, Yagi K, Kobayashi Y, et al (2005) Role of β-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 17 971–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li C, Lee GI, Howe GA (2002) Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc Natl Acad Sci USA 99 6416–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti R, Farmer EE (2002) The jasmonate pathway. Science 296 1649–1650 [DOI] [PubMed] [Google Scholar]
- Paschold A, Halitschke R, Baldwin IT (2007) Co(i)-ordinating defenses: NaCOI1 mediates herbivore-induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant J 51 79–91 [DOI] [PubMed] [Google Scholar]
- Peng J, Deng X, Huang J, Jia S, Miao X, Huang Y (2004) Role of salicylic acid in tomato defense against cotton bollworm, Helicoverpa armigera Hubner. Z Naturforsch [C] 59 856–862 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34 374–378 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Howe GA (2005) Systemic signaling in the wound response. Curr Opin Plant Biol 8 369–377 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Koo AJ, Howe GA (2007) Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiol 143 812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko U, Hermsmeier D, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. II. Accumulation of plant mRNAs in response to insect-derived cues. Plant Physiol 125 701–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko U, Preston CA, Baldwin IT (2000) Eating the evidence? Manduca sexta larvae can not disrupt specific jasmonate induction in Nicotiana attenuata by rapid consumption. Planta 210 343–346 [DOI] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA 98 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of ro ot growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppuhn A, Baldwin IT (2008) Induced defenses and the cost-benefit-paradigm. In A Schaller, ed, Induced Plant Resistance to Herbivory. Springer, Berlin (in press)
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci USA 98 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448 661–665 [DOI] [PubMed] [Google Scholar]
- van Dam NM, Horn M, Mares M, Baldwin IT (2001) Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J Chem Ecol 27 547–568 [DOI] [PubMed] [Google Scholar]
- von Dahl CC, Winz RA, Halitschke R, Kuhnemann F, Gase K, Baldwin IT (2007) Tuning the herbivore-induced ethylene burst: the role of transcript accumulation and ethylene perception in Nicotiana attenuata. Plant J 51 293–307 [DOI] [PubMed] [Google Scholar]
- Wang L, Halitschke R, Kang JH, Berg A, Harnisch F, Baldwin IT (2007) Independently silencing two JAR family members impairs levels of trypsin proteinase inhibitors but not nicotine. Planta 226 159–167 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Stenzel I, Hause B, Hause G, Kutter C, Maucher H, Neumerkel J, Feussner I, Miersch O (2006) The wound response in tomato—role of jasmonic acid. J Plant Physiol 163 297–306 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094 [DOI] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Baldwin IT (2004) Constitutive and inducible trypsin proteinase inhibitor production incurs large fitness costs in Nicotiana attenuata. Proc Natl Acad Sci USA 101 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Keinanen M, Baldwin IT (2001) Herbivore-induced allene oxide synthase transcripts and jasmonic acid in Nicotiana attenuata. Phytochemistry 58 729–738 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.