Abstract
Zinc finger domains are structures that mediate sequence recognition for a large number of DNA-binding proteins. These domains consist of sequences of amino acids containing cysteine and histidine residues tetrahedrally coordinated to a zinc ion. In this report, we present a means to selectively inhibit a zinc finger transcription factor with cobalt(III) Schiff-base complexes. 1H NMR spectroscopy confirmed that the structure of a zinc finger peptide is disrupted by axial ligation of the cobalt(III) complex to the nitrogen of the imidazole ring of a histidine residue. Fluorescence studies reveal that the zinc ion is displaced from the model zinc finger peptide in the presence of the cobalt complex. In addition, gel-shift and filter-binding assays reveal that cobalt complexes inhibit binding of a complete zinc finger protein, human transcription factor Sp1, to its consensus sequence. Finally, a DNA-coupled conjugate of the cobalt complexes selectively inhibited Sp1 in the presence of several other transcription factors.
Keywords: zinc fingers, transition metals, Sp1, nucleocapsid protein, HIV
Zinc finger domains consist of sequences of amino acids containing cysteine and histidine residues tetrahedrally coordinated about a zinc ion. Detailed structural information supports the existence of multiple classes of zinc fingers (1–3), and these include domains in which the zinc ion is coordinated by Cys2His2 (e.g., TFIIIA), Cys3His (e.g., retroviral nucleocapsid proteins), or Cys4 (e.g., glucocorticoid receptor). The zinc finger motif is crucial for specific DNA recognition by zinc finger transcription factors.
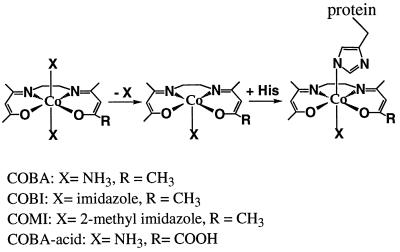
In this work, we describe a family of cobalt(III)-Schiff-base complexes [Co(III)-sb] that are effective zinc finger inhibitors. Related complexes were shown to have potent anti-inflammatory properties (4). The general structure of this class of complexes is shown in Fig. 1. We investigated the effect of a series of Co(III) complexes, cobalt(III) complex of bis(acetylacetone)ethylenediimine [acacen], reacted with zinc finger proteins including a synthetic peptide representing the first zinc finger region from HIV-1 nucleocapsid protein (NCp7) and Sp1, a human zinc finger transcription factor (5). CD and 1H NMR studies were performed to determine perturbations to zinc finger structure caused by reaction with the Co(III)-sb complexes. Gel-shift and filter-binding assays revealed that the Co(III)-sb effectively inhibited DNA binding by Sp1 and demonstrated that the Co(III)-sb could be specifically targeted by covalent attachment to an Sp1 recognition oligonucleotide.
Figure 1.
Cobalt(III) complex of acacen. The substitution of an axial ligand from cobalt(III) with a nitrogen from histidine results in a Co(III)-His complex. This histidine is from a peptide or protein containing a zinc finger domain. The complexes used in this work vary in the nature of the axial ligands, X, and the group at position R.
MATERIALS AND METHODS
Synthesis of Cobalt Complexes.
The syntheses of cobalt(III)-Sb complexes were performed as described (6).
Synthesis of Oligonucleotides.
Oligonucleotides were prepared on an Applied Biosystems 394 synthesizer, with addition of the phosporamidite of 2′-amino-2′-deoxyuridine as the terminal base for the sense strand and deprotected overnight at 55°C. DMT-2′-N-trifluoroacetyl-protected phosphoramidite was synthesized as reported (7). Oligonucleotides were purified over C18 SepPak columns (Waters). Then, 12-cc columns were washed twice with 20 ml of acetonitrile (EM Science), three times with 20 ml of H2O, and loaded with up to 1 mM DNA. Columns were washed three times with 20 ml of water, and the sample was eluted with 60:40 (H2O:acetonitrile). DNA was detected by absorbance at 260 nm. Samples were dried on a speed vac (Savant SC110a).
Synthesis of Oligonucleotides Conjugates (Co-Oligonucleotides).
Equimolar ratios of complement and sense strand in TE buffer (10 mM Tris⋅Cl, 1 mM EDTA, pH 8.0) were annealed by mixing, heating to 90°C, and cooling to room temperature. This sample was exchanged to H2O over a NAP-25 column and dried on a speed vac. One micromole of the double-stranded oligonucleotide, with 2′-aminouridine at the 5′ end, in 200 μl of 0.1 M Mes (Sigma), pH 4.5, was added to 7.6 mg of Co(acacen)(NH3)2 (COBA)-acid in 500 μl of the same buffer. This mixture was added to 38 mg of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC; Pierce) and reacted with gentle rocking overnight at room temperature. The product was purified over a NAP-25 (Pharmacia) column and dried on a speed vac. This sample then was purified further over hydroxyapatite (to isolate duplex DNA) in the batch mode (BioGel HTP DNA grade, Bio-Rad) with stepwise elution in sodium phosphate from 0.01–0.40 M, pH 6.8. Fractions containing double-stranded DNA, as determined by absorbance at 260 nm and thermal denaturation experiments, were desalted over C18 SepPak columns.
Synthesis of Peptides.
The peptides were synthesized at the Beckman Institute Biopolymer Synthesis Center at Caltech. The automated stepwise solid-phase syntheses were performed on an ABI 430A peptide synthesizer by using t-Boc chemistry (8). OCH2-phenylacetamidomethyl (PAM) or 4-methylbenzhydrilamine resin (Applied Biosystems) were used as a solid support. The side chain-protecting groups and peptide-resin bonds of the completed peptides were cleaved by using liquid anhydrous hydrofluonic acid in the presence of p-cresol and p-thiocresol, as scavengers for 60 min at 0°C (9). Peptides were purified by reverse-phase HPLC (HPLC liquid chromatography) on a semipreparative C18 column (10 × 1 cm; Brownlee Lab) by using a linear gradient of 0–50% aqueous acetonitrile/0.1% trifluoroacetic acid (TFA) over 120 min. The purity and the primary structure of the peptides were determined by capillary electrophoresis and mass assisted laser desorption ion time of flight (MALDI TOF) mass spectroscopy. Isolated peptides revealed the presence of the homogeneous target sequence by all analytical methods used.
Gel Shift.
3.5 pmol of Sp1 oligonucleotide (or OCT 1, CREB, CTF/NF1, AP1, GRE, and NF-kB, from Promega) were labeled at the 5′ end with ATP (Amersham) by T4 polynucleotide kinase using the manufacturer’s protocol (GIBCO/BRL). Samples of labeled oligonucleotide were removed and spotted to nitrocellulose before, and after purification through G-25 Sephadex spin columns pre-equilibrated with TE buffer (Boehringer Mannheim). Radioactivity bound to the filters was assayed on a Beckman LS 1801 scintillation counter. Labeling efficiency was calculated as percentage incorporation equals post spin counts times volume/prespin counts times volume.
Then, 0.263 pmols (1 μl of 25 ng/ml) of Sp1 transcription factor were incubated at room temperature for 1 h in 20 μl of gel-shift buffer A (25 mM Tris, pH 8.0/100 mM KCl/2 mM DTT/100 μM Zn Cl2/10% glycerol/50 μg/ml BSA); ref. 10) in the presence or absence of 28 pmol cobalt complex (1 μl of 0.01 mg/ml). Forty femptomoles of 32P-labeled oligonucleotide were added to each reaction tube and incubated for 30 min at room temperature. After 30 min, 2 μl of 0.1% bromophenol blue was added to all tubes. The reaction mixtures were run on 4% polyacrylamide gels (80:1 acrylamide:bisacrylamide) in 0.5 mM Tris-boric acid-EDTA buffer (45 mM Tris⋅borate/1 mM EDTA), which had been prerun for 30 min before loading. Gels were run at room temperature at 100 V and exposed to x-ray film (Kodak X-Omat AR), with an intensifying screen, overnight at −70°C and developed the following day.
Gel-shift assays by using cobalt–oligonucleotide conjugates were performed as above with minor modifications. Samples contained: 2 μl of 5x gel-shift buffer B [20% glycerol/5 mM MgCl2/2.5 mM EDTA/2.5 mM DTT/250 mM NaCl/50 mM Tris⋅HCl, pH 7.5/0.25 mg/ml poly(dI-dC)⋅poly(dI-dC) (Promega)]; positive controls contained 2 μl of HeLa nuclear extract from stock; experimental samples contained both HeLa extract and 4 pmol cobalt–oligonucleotide conjugates, and volume of all samples was adjusted to 9 μl with H2O. The mixtures were incubated at room temperature for 10 min. Two microliters of a 1:5 dilution from the appropriate radiolabeled oligonucleotide was added to each sample, and the samples were incubated for 20 min at room temperature. After 20 min, 1 μl of gel loading 10x buffer (250 mM Tris, pH 7.5/0.2% bromophenol blue/0.2% xylene cyanol/40% glycerol) was added to each reaction mixture, and the samples were loaded to 4% polyacrylamide native gels. Gels were run at 150 V, room temperature, then exposed to x-ray film, with an intensifying screen, for 16 h at −70°C.
Filter Binding.
Reaction mixtures were prepared as described above for a range of concentrations of cobalt complex. Samples were run in triplicate. After 30-min room temperature incubation with radiolabeled oligonucleotide, the reaction mixture was filtered through a nitrocellulose membrane (0.45 μm, Schleicher & Schull) under gentle suction; the tube was rinsed once with 50 μl wash buffer, and the filter was washed three times with 1 ml of wash buffer (10 mM Hepes, pH 7.5/1 mM EDTA, Promega protocol). Filters were dissolved in 9 ml of Filtron-X scintillation fluid (National Diagnostics), and radioactivity remaining on the filters was assayed on a Beckman LS 1801 scintillation counter.
To determine whether the binding of the conjugate was irreversible, it was first necessary to determine the binding saturation point for the amount of Sp1 used. One microliter of Sp1 (at 25 ng/ul from Promega) was incubated in 10 μl gel-shift buffer A with increasing amounts of 32P-labeled Sp1 oligonucleotide. Saturation of binding occurred with 2 μl of labeled oligonucleotide (0.6 pmol). The assays for irreversibility used this concentration of labeled Co-oligonucleotide or oligonucleotide. Resistance of Co-oligonucleotide binding to displacement by unlabeled oligonucleotide was assayed by incubating 1 μl of Sp1 in 10 μl of gel-shift buffer A with 2 μl of 32P-labeled Co-oligonucleotide or oligonucleotide. The samples were incubated at room temperature for 20 min; then excess unlabeled oligonucleotides (twofold) were introduced, and the samples incubated an additional 2 h. Samples were taken at various timepoints and analyzed by filter binding.
In a second assay to determine whether the conjugates bound irreversibly to transcription factor Sp1, samples containing 1 μl of Sp1, 10 μl of gel-shift buffer A, and 2 μl of 5′ [32P]Co-oligonucleotide were allowed to incubate for 1.5 h at room temperature. After incubation, one-half of the samples were diluted with 90 μl of gel-shift buffer A and all samples were left at room temperature an additional 1.5 h. Samples were then assayed by filter binding.
CD Spectroscopy.
Peptides were solubilized at a concentration of 0.125 mg/ml in 25 mM phosphate buffer (pH 7.0) and placed in a 0.1-mm path length quartz cuvette. Spectra were averaged over eight scans from 260–195 nm on an Aviv 62DS spectropolarimeter (Aviv Associates, Lakewood, NJ). A twofold molar excess of ZnCl2 was added and the spectra was recorded again.
Inductively Coupled MS and Amino Acid Analysis.
One milligram of peptide in water was incubated with a molar equivalent of ZnCl2 at room temperature for 10 min, and this solution was added to 5 M equivalents of cobalt(III) complex with axial ligands X = 2-methylimidazole [Co(acacen)(2-methylimidazole)2; COMI] or NH3 (COBA). The peptide and cobalt complex mixture was incubated for 72 h at 4°C, placed in a 1,000-MW cutoff membrane (Spectrum) and dialysed against water at 4°C for 3 days. The amount of cobalt remaining in the sample was determined by inductively coupled plasma MS on a Perkin–Elmer/Sciex Elan 5000A. Peptide content for the same sample was determined by amino acid analysis on an Applied Biosystems 420a derivitizer/analyzer. This step was done for three different peptides: peptide 1, VKCFNCGKEGHIARNCRA; peptide 2, ITNDLRLKDWEHSQTLKNIT; and peptide 3, LQFSIETLNGRVQENAFKQQ.
NMR.
1H NMR data were acquired on a Bruker AM-500 or AMX-500 (T = 30°C, 5 mM D2O; D = 3H). Triplicate samples were prepared by dissolving 3.5 mg of peptide in 350 μl of D2O (Cambridge Isotope Laboratories, Cambridge, MA). One molar equivalent of ZnCl2 (3 μl of 0.5 M solution) was added to two of the samples; to one of these, 1 M equivalent of cobalt complex (17.2 μl of 40 mg/ml solution) also was added. The pD of all samples was adjusted to 7.0 by using NaOD (Cambridge Isotope Laboratories). COBA, in D2O (pD 7.19), was titrated into a 2.47 mM solution of unfolded peptide1 in D2O to a final cobalt complex:peptide ratio of 1:1. Spectra were acquired after each addition of cobalt complex.
HPLC.
Samples containing 1 mg of peptide in 1 ml of 0.001 M ZnCl2 (0.5 mM peptide) were prepared. Cobalt complexes were added to a final concentration of 0.5 mM and contained either axial ligand X = NH3 (COBA) or X = imidazole (Co(acacen)(imidazole)2; COBI) as shown in Fig. 1. Samples were passed through a cation exchange column (polyLC sulfoethyl A) in 5 mM potassium phosphate and eluted with a linear gradient of 0–500 mM KCl over 35 min.
4-(2-Pyridylazo)resorcinol (PAR) Assay for Zinc Release.
Peptides were solubilized in water at a concentration of 20 μM in the presence of 20 μM ZnCl2 (EM Science), and PAR (Sigma) was added to this solution to a final concentration of 0.1 mM. The absorbance at 500 nm of the peptide solution was monitored on a Hewlett–Packard 8452a UV-Vis Spectrophotometer as acacen or cobalt complex (COBA or COMI) was titrated into the solution (7.14 nmol/ml for each addition). Each sample was corrected against a blank containing 0.1 mM PAR (no peptide), which was titrated in parallel. The amount of zinc released by the peptide was determined by using ΔA500 = 6.6 × 104 M−1⋅cm−1.
RESULTS
Binding of Cobalt Complexes to a Model Zinc Finger Peptide.
The well known affinity of cobalt(III) for nitrogenous ligands lead us to suspect that histidine residues in zinc fingers could be suitable ligands. The peptide VKCFNCGKEGHIARNCRA and similar peptides have been extensively studied by numerous investigators as model zinc fingers (2, 11–17). This peptide has been shown to adopt zinc finger-like structure and bind double-stranded deoxyribonucleotides in solution (18). To demonstrate that the Co(III)-sb were indeed capable of binding to zinc finger peptides in a covalent manner, peptide (folded about zinc)-cobalt complex mixtures were subjected to HPLC separation on a cation exchange column under conditions that would disrupt any noncovalent interactions (data not shown). For these studies, we used COMI. COBI was used as a control because this complex is substitutionally inert. Free peptide eluted at 32 min, as did peptide in the presence of COBI. In the presence of the reactive COMI complex, however, the peptide elution shifts. Some amount of free peptide remains, but a large fraction of the peptide peak elutes ≈10 min earlier. These results indicate that only cobalt complex with substitutionally active ligand binds covalently to peptide and is consistent with axial ligand exchange of cobalt(III) as a mechanism for binding.
To determine the stoichiometry of cobalt complex binding to peptide, mixtures of peptide and COMI were subjected to extensive dialysis to remove noncovalently bound complexes and samples were removed for analysis of cobalt and peptide content. As shown in Table 1, histidine-containing peptides bound at least one cobalt complex whereas peptides without histidine residues did not. Furthermore, the presence of lysine, arginine, or cysteine residues did not influence the number of cobalt complexes bound.
Table 1.
Stoichiometry of cobalt complex binding to peptides
| Peptide | Cobalt:peptide |
|---|---|
| VKCFNCGHEGHIARNCRA | 1.44 ± 0.3:1 |
| ITNDLRLKDWEHSQTLKNIT | 1.57 ± 0.3:1 |
| LQFSSIETLNGRVQENAFKQQ | 1:469 ± 55 |
Inductively coupled mass spectrometry and amino acid analysis were used to determine the ratio of cobalt complexes bound to three different peptides. The presence of lysine, arginine, or cysteine residues does not influence the number of cobalt complexes bound.
Effect of Cobalt Complexes on Zinc Finger Peptide Structure.
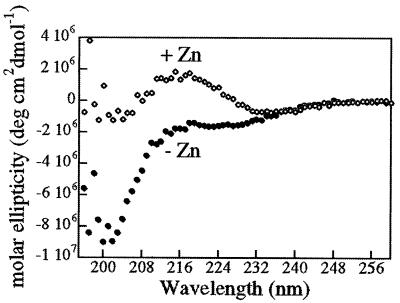
We examined the peptide by CD spectropolarimetry to verify that the peptide adopts a zinc finger structure in solution. In the absence of zinc, the peptide displays a spectrum characteristic of typical random coil structure with an intense negative band below 200 nm. Upon addition of zinc (Fig. 2), this spectrum reveals β type II turn content characteristic of zinc finger structures (17). The spectrum possesses features that are analogous to those displayed by the metalloprotein rubidoxin and the polypeptide ala2-gly2, both known to contain β type II turns (17), including minima at 185 nm and 227 nm and a maximum at 207 nm (19, 20). The ability of the peptides to form zinc fingers in solution and the effect of cobalt complexes on zinc finger structure were analyzed by NMR spectroscopy and fluorescence assays.
Figure 2.
Eighteen amino acid peptide adopts zinc finger conformation in the presence of zinc. Peptides were examined by CD spectropolarimetry (0.25 mM peptide in 25 mM phosphate buffer, pH 7.0) in the presence and absence of zinc. In the absence of zinc, the spectra is that of a typical random coil that changes upon the addition of zinc to a spectra characteristic of zinc fingers, with β type II turn content.
NMR Spectroscopy.
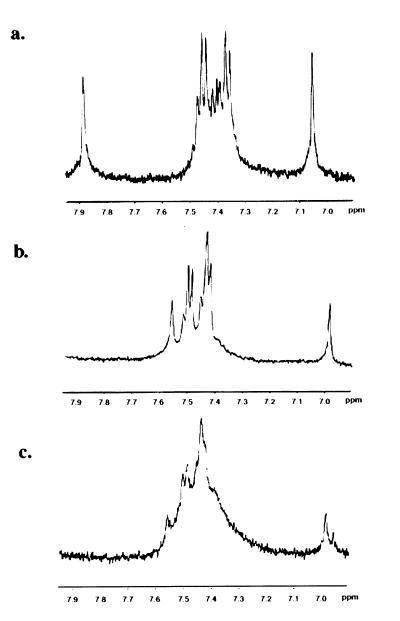
In the absence of zinc, the 1H NMR spectrum of the unfolded peptide contains peaks at 7.91 and 7.08 ppm, characteristic of free histidine (Fig. 3a) which shift in the presence of zinc to peaks at 7.58 and 7.01, which are representative of metal bound histidine (Fig. 3b) (5). In addition, the resolved multiplets indicate a stable tertiary structure (5). Upon addition of COBA, this multiplet structure is abolished. Over time, the spectrum in this region degrades to a broad peak (Fig. 3c), indicating that cobalt complex strongly perturbs the zinc finger structure.
Figure 3.
Cobalt(III) complex disrupts the structure of the zinc finger peptide. 1H NMR data were acquired on a Bruker AM-500 (T = 30°C, 5 mM D2O, pH 7.0); 3.5 mg of peptide were dissolved in 350 μl D2O (Cambridge Isotope Laboratories), for each of the samples. Approximately 1 M equivalent of ZnCl2 was added to the samples in a and b; 1 M equivalent of COBA also was added to c. In the absence of zinc as shown in a, histidine peaks appear at 7.08 and 7.91, which shift upon addition of zinc to those characteristic of metal bound histidine at 7.01 and 7.58 (b). Upon the addition of COBA, the spectrum degrades to a broad peak (c), suggesting a strong perturbation to the structure of the peptide.
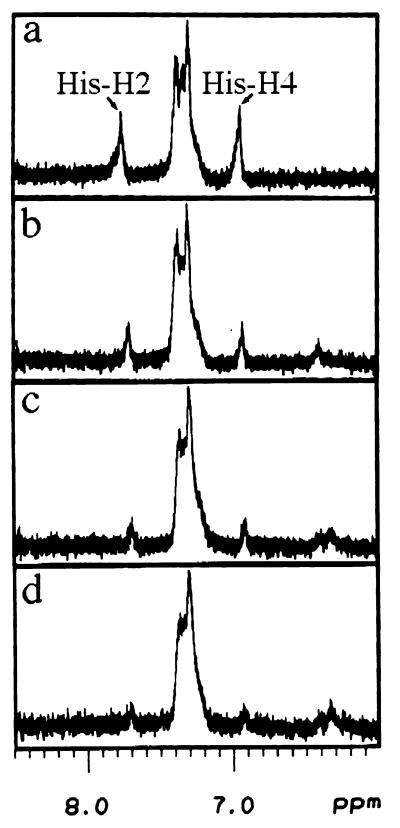
To determine the mechanism for the loss of zinc finger structure observed above, we performed titration studies. Titration of COBA to the unfolded peptide results in a disappearance of the imidazole proton peaks. As shown in Fig. 4, as cobalt complex is added to the peptide, the histidine peaks at 6.98 and 7.79 ppm gradually disappear. New peaks appear between 6.3 and 6.5 ppm. These results unambiguously demonstrate binding of the cobalt complex to the imidazole side chain.
Figure 4.
The cobalt(III) complex binds to a nitrogen of the imidazole ring. 1H NMR data were acquired on a Bruker AMX-500 (T = 30°C, pH 7.19). Aliquots of COBI were titrated to a 2.47 mM solution of unfolded peptide and incubated at room temperature for 1 h before measurement. The final cobalt complex:peptide ratios are as follows: 0:1 (a), 0.33:1 (b), 0.67:1 (c), and 1:1 (d).
PAR Assay.
PAR has an absorbance at 400 nm in the absence of zinc, which shifts to 500 nm when zinc is bound (Zn(II)PAR2) (21). By monitoring the change in absorbance at 500 nm, we were able to detect the release of zinc from a model zinc finger peptide upon interacting with cobalt complex. As zinc is released from the peptide, it is free to bind PAR as evidenced by an increase in the absorbance at 500 nm. Fig. 5 shows the change in absorbance at 500 nm as a function of increasing concentrations of either acacen, COBA, or COMI. It is evident that the addition of the equatorial ligand, acacen, alone does not induce a change in the absorbance at 500 nm, suggesting that no zinc is released by the peptide under these conditions.
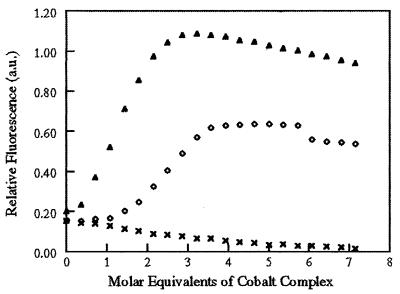
Figure 5.
Cobalt(III) complex causes the release of zinc from a zinc finger peptide. Increasing molar equivalents of acacen (x), COBA ([irco]), or COMI (▴) were added to a 20-μM solution of folded zinc finger peptide in the presence of the zinc-binding dye, PAR. PAR absorbs at 500 nm when zinc is bound and 400 nm in the absence of zinc. Binding of cobalt complexes displaces zinc as evidenced by an increase in the absorbance at 500 nm due to binding of free zinc by PAR. This increase in absorbance is complete after addition of 2.5 equivalents of COMI, corresponding to the release of 0.67 mol of zinc per mol of peptide. Titration with COBA is complete after 3.5 equivalents are added, corresponding to a release of 0.36 mol of zinc per mol of peptide. There is no increase in absorbance upon addition of acacen. All samples were corrected against a peptide-free blank containing PAR titrated in parallel.
In contrast, as COBA or COMI is titrated into the peptide solution, there is an increase in the absorbance at 500 nm, which is complete after the addition of 2.5 equivalents of COMI or 3.5 equivalents of COBA. The total increase in A500 of 0.886 after addition of COMI corresponds to the release of 0.67 mol of zinc per mol of peptide, using ΔA500 = 6.6 × 104 M−1⋅cm−1 (22). Addition of COBA results in a ΔA500 of 0.480, which corresponds to the release of 0.36 mol of zinc per mol of peptide. These results confirm that cobalt(III) complexes disrupt the structure of the model zinc finger peptide.
Effect of Cobalt Complexes on Zinc Finger Proteins.
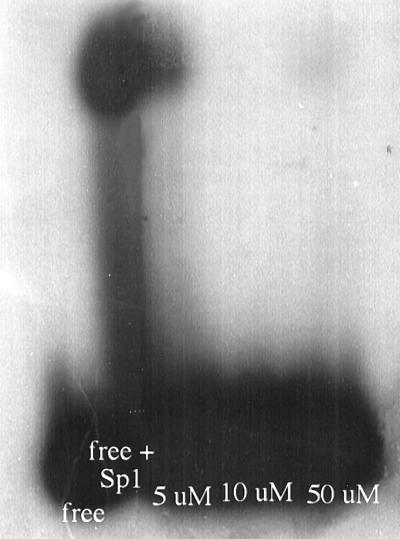
To determine whether the binding of cobalt(III) complexes to model peptides could be repeated with a full length zinc finger protein, we investigated the effects of cobalt complexes on the well characterized zinc finger protein Sp1 by using a gel-shift assay. The binding of cobalt complex to histidine residues that coordinate zinc should disrupt zinc finger structure and thus, inhibit binding of the protein to DNA. Sp1 contains three zinc finger-binding domains and recognizes the 6-bp consensus sequence 5′-GGGCGG. We used a commercially available, 22-bp oligonucleotide (Promega). Labeled oligonucleotide was incubated in the presence or absence of Sp1 and various amounts of cobalt complex, electrophoresed through a 4% polyacrylamide native gel, and the gel was exposed to x-ray film overnight. Free oligonucleotide migrates ahead of the larger complex of oligonucleotide bound by protein (shifted band, as shown in Fig. 6). It is evident that there is a loss of the shifted band as the amount of cobalt complex increases from a 2- to 25-fold molar excess over the amount of Sp1. This result indicates that cobalt chelate inhibits binding of Sp1 to its recognition oligonucleotide in a concentration-dependent manner.
Figure 6.
Cobalt(III) complex inhibits the binding of Sp1 to its recognition oligonucleotide. Sp1 was incubated with radiolabeled recognition oligonucleotide in the presence of increasing amounts of COBA as follows: lane 1, labeled oligonucleotide alone; lane 2, labeled oligonucleotide + Sp1; lane 3, labeled oligonucleotide + Sp1 +5 μM COBA; lane 4, labeled oligonucleotide + Sp1 +10 μM COBA; and lane 5, labeled oligonucleotide + Sp1 +50 μM COBA. The shifted band disappears with increasing amounts of COBA.
To quantitate the degree of inhibition, samples were prepared in triplicate and analyzed by filter-binding experiments. Nitrocellulose filters bind protein but not DNA, so radioactivity remaining on the filters reflects protein-bound oligonucleotide. As the concentration of cobalt complex increases, the number of counts bound decreases, confirming the gel-shift results(data not shown). Addition of as little as 0.1 μg of cobalt complex (50% molar excess over amount of Sp1) reduced the counts bound to background levels, indicating a complete inhibition of Sp1 binding to the recognition oligonucleotide. Therefore, we conclude that cobalt complex effectively inhibits the function of Sp1.
Selective Inhibition of Zinc Finger Proteins.
To selectively inhibit the function of Sp1, we covalently attached Co(III)-sb to the recognition oligonucleotide for Sp1. The free Co(III)-sb complex can interact with any available histidines and has no specificity for Sp1. We predicted that the attached oligonucleotide would act to increase the local concentration of Co-sb in the zinc finger region of the protein. Sp1 would bind to oligonucleotide, and the proximity of Co-sb to histidine would allow it to bind avidly. Likewise, the bulk of the attached oligonucleotide should hinder interaction with other histidines outside of the DNA-binding region.
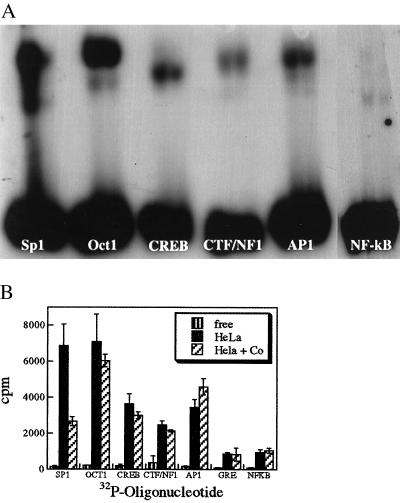
We covalently attached the cobalt complex to the Sp1 recognition oligonucleotide by placing a 2′-amino-2′-deoxyuridine residue at the 5′ end of the oligo. This oligo was reacted with a cobalt complex with carboxylic acid as the R group (COBA-acid in Fig. 1). Gel filtration purified oligonucleotide conjugates were then evaluated for specific inhibition of Sp1 out of a mixture of many other transcription factors in a HeLa nuclear extract system. Recognition oligonucleotides for the DNA-binding proteins Sp1, OCT 1, CREB, CTF/NF1, AP1, and NF-kB were labeled with 32P and individually incubated with HeLa extract in the presence or absence of cobalt-Sp1 oligonucleotide conjugates. The samples were separated by nondenaturing gel electrophoresis, and the autoradiograph is shown in Fig. 7a. The three lanes for each of the oligonucleotides represent: oligonucleotide alone, oligo plus HeLa extract, and oligo plus HeLa plus COBA-oligo conjugate. Only in the lane corresponding to Sp1 is there a loss of the shifted band in the presence of the COBA–oligonucleotide conjugates.
Figure 7.
COBA–oligonucleotide conjugates specifically inhibit Sp1. (a) Gel-shift assay, samples were prepared as described in Materials and Methods and run on 4% native polyacrylamide gels. Three lanes appear for each binding protein: lane 1, containing labeled recognition oligo alone; lane 2, labeled oligo + binding protein; and lane 3, labeled oligo + binding protein + COBA–oligonucleotide conjugate. Only lane 3 for Sp1 shows evidence of loss of shifted band in the presence of COBA-oligo conjugate. (b) Filter-binding assay, samples were prepared as in a and quantitated by filter-binding assays. Three lanes containing: free oligonucleotide ▥, oligo + binding protein ░⃞, and oligo + binding protein + COBA–oligonucleotide conjugate ▨ are shown for each of seven different DNA-binding proteins. Only Sp1 suffers a decrease of bound oligo in the presence of COBA–oligonucleotide conjugate.
Results of the gel-shift assays were confirmed, and the degree of inhibition was quantitated by filter-binding experiments (Fig. 7b). There was a 60% decrease in counts bound for Sp1 in the presence of 15-fold M excess of COBA–oligonucleotide conjugates. However, there is no significant change for any of the other DNA-binding proteins. This result demonstrates a selective inhibition of Sp1 when present in a mixture of six other transcription factors, with no nonspecific inhibition of other DNA-binding proteins.
Irreversible Inhibition of Zinc Finger Proteins.
The binding of cobalt complexes to the zinc finger protein Sp1 is irreversible. To determine which portion of the inhibition is caused by binding of the cobalt complex and not by competitive inhibition by the conjugated oligonucleotides, we assayed for irreversible binding of the cobalt conjugates to Sp1. Inhibition caused by cobalt complexes results from irreversible binding to histidine residues (23) and thus, should be resistant to treatments that would overcome a competitive inhibitor such as a free oligonucleotide. For example, one of the traits of competitive inhibition in enzymes is that inhibition can be overcome by sufficient amounts of substrate. Similarly, a competitively binding, labeled oligonucleotide should be displaced by sufficient amounts of unlabeled oligonucleotide. Because it was our intent to specifically evaluate the irreversibility of the binding by Sp1, we chose to work with the purified Sp1 system rather than with HeLa extract.
In competition experiments, the labeled oligonucleotide was displaced more rapidly and completely than labeled cobalt–oligonucleotide and the decrease in counts bound plateaued 2 h after introduction of unlabeled competitor. Results show that 42.5 ± 5.4% of the COBA–oligonucleotide remains bound to Sp1 compared with 24.4 ± 7.8% unmodified oligonucleotide yielding a 90% confidence interval of a 14.6–20.7% difference in the two values.
In addition, we used a dilution assay as a means to test for irreversibility of the cobalt conjugate binding to Sp1. At equilibrium, reversibly bound oligonucleotide should dissociate from Sp1 in dilute solutions whereas irreversibly bound oligonucleotide will not. Sp1 oligonucleotide binding reduces to near background levels (4 ± 1%) after dilution, whereas 22.6 ± 4.8% of COBA–oligonucleotide remains bound. It is expected that some portion of the binding of COBA–oligonucleotide is mediated through the oligonucleotide and is reversible. These results produce a 90% confidence interval of a 16.3–20.9% difference, which correlates with the results of the competition studies.
DISCUSSION
Reactions of Co(III) Complexes with a Model Zinc Finger Peptide.
The results of our investigation clearly demonstrate that the cobalt(III) complexes react with a model peptide and disrupt zinc finger structure. Zinc finger peptide sequences have been extensively used to characterize the mechanism of DNA binding in glucocorticoid proteins (3), HIV nucleocapsid protein (13–17), and to study folding, metal-binding, and structure of zinc finger proteins (2, 14, 20, 24, 25). The first retroviral zinc finger region from HIV-1 nucleocapsid protein has been shown to form zinc finger-type structure in solution and is DNA-binding competent (12, 13, 17). In these investigations, CD and NMR verify that the peptide forms zinc finger-type structure in solution and that this structure is significantly perturbed by the binding of Co(III)-sb.
In NMR studies, the gradual loss of the imidazole proton peaks upon titration with the cobalt complex is clear evidence that the complex is binding to a nitrogen of the imidazole ring. This is supported by the results of the PAR assays, which demonstrate that zinc is released by the zinc fingers in the presence of cobalt complex, as would be expected if cobalt complex bound to the histidine imidazole and displaced zinc.
Stoichiometric studies using inductively coupled plasma MS and amino acid analysis demonstrate the covalent binding of cobalt complexes to peptides containing histidine residues; there is no significant binding to other peptides (see Table 1). The ratio of cobalt to peptide is independent of the number of lysine, arginine, or cysteine residues, indicating that these residues are not involved in binding cobalt complexes. It is important to note that the reduction potentials for the cobalt(III) complexes are too low to oxidize cysteines (6). Extensive dialysis of the Co(III)-sb and peptide reaction mixtures does not displace the complexes, suggesting that the binding of cobalt complexes to the peptides is irreversible.
Reactions of Co(III) Complexes with Zinc Finger Protein Sp1.
We investigated the ability of Co(III)-sb to inhibit the function of human transcription factor Sp1. Given the affinity of Co(III)-sb for histidines, we believed that the complexes would bind irreversibly to histidines in the zinc finger regions of Sp1, displacing the zinc and abolishing DNA-binding activity. The results of gel-shift and filter-binding assays indicate that Co(III)-sb effectively inhibits the binding of Sp1 to its recognition oligonucleotide. These data and the results of the peptide structural investigations demonstrate that the cobalt complex disrupts zinc finger structure and thus prevents binding of the protein to its recognition oligonucleotide.
Co-sb covalently attached to a recognition oligonucleotide for Sp1 was able to specifically inhibit Sp1 binding without inhibiting any of the other DNA-binding proteins examined. Both competition with unlabeled oligonucleotide and dilution were used to demonstrate covalent attachment of COBA to the Sp1 for a fraction of the conjugates. The majority of the binding appears to be reversible association with the oligonucleotide portion of the conjugate.
This report demonstrates that cobalt(III) complexes have significant effects on the structure and function of zinc finger-mediated DNA-binding. This site of inhibition represents a new target for antiviral agents. Antiviral agents have previously targeted the viral protein coat; however, the HIV virus has eluded such drugs by rapid mutation. Subsequently, attention was directed toward viral proteases and these treatments seem to be meeting with some success (19, 20, 24). The model peptide and protein used in our investigations are both key components in viral replication. The peptide comes from the zinc finger domain of the HIV nucleocapsid protein, a highly conserved region that is essential for proper packaging of HIV. The model protein selectively inhibited by Co(III) complexes in this investigation, Sp1, has been implicated in the activation of viral transcription. Cooperative interaction of Sp1 and NF-kB is required for stimulation of HIV-1 transcription, and this interaction is due to specific binding of the amino-terminal region of the RelA(p65) subunit to the zinc finger region of Sp1 (25).
The only other small molecules known to act on zinc fingers are a group of C-nitroso-substituted molecules that has been reported to remove zinc from the nucleocapsid protein, thereby disrupting the zinc finger domains, RNA packaging, and viral infectivity in vitro (12, 24, 26). The same researchers also report that a group of disulfide-substituted benzamides inhibits HIV nucleocapsid zinc fingers. These compounds are believed to oxidatively attack the cysteine thiolates of the zinc finger motif. It was proposed that the compounds selectively act on retroviral-type zinc fingers (CCHC) such as those on the nucleocapsid protein and do not act on the classical-type zinc fingers (CCHH) found in the other proteins. The mode of action for such discrimination is undetermined, and the effects of the drugs on eukaryotic proteins containing classical-type fingers have not been shown.
Our investigations demonstrate that cobalt(III) complexes bind to histidine residues of zinc finger domains and can inhibit selected transcription factors when conjugated to the recognition oligonucleotide for that protein. These complexes introduce a method for inhibition of zinc finger proteins which may be exploited for the treatment of various diseases.
Acknowledgments
We thank Drs. Keiko Kato, Scott Fraser, and Harry Gray for helpful discussion and Bassil Dahiyat for assistance with the CD measurements. This work was supported by the Biological Imaging Center of the Beckman Institute and the Redox Pharmaceutical Corporation.
ABBREVIATIONS
- Co(III)-sb
cobalt(III) Schiff-base complex
- COBA
Co(acacen)(NH3)2
- COBI
Co(acacen)(imidazole)2
- COMI
Co(acacen)(2-methylimidazole)2
- COBA-acid
Co(aciden)(NH3)2
- PAR
4-(2-pyridylazo)resorcinol
- acacen
bis(acetylacetone)ethylenediimine
References
- 1.Berg J M. Curr Opin Struct Biol. 1993;3:11–16. [Google Scholar]
- 2.Berg J M. Annu Rev Biophys Biophys Chem. 1990;19:405–421. doi: 10.1146/annurev.bb.19.060190.002201. [DOI] [PubMed] [Google Scholar]
- 3.Archer T K, Hager G L, Ominchinski J G. Proc Natl Acad Sci USA. 1990;87:7560–7564. doi: 10.1073/pnas.87.19.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wooley P H, Whalen J D. Agents Actions. 1992;35:273–279. doi: 10.1007/BF01997511. [DOI] [PubMed] [Google Scholar]
- 5.South T L, Kim B, Summers M F. J Am Chem Soc. 1989;111:395–396. [Google Scholar]
- 6.Bottcher A, Takeuchi T, Hardcastle K I, Meade T J, Gray H B, Cwikel D, Kapon M, Dori Z. Inorg Chem. 1997;36:2498–2504. [Google Scholar]
- 7.Meade T J, Kayyem J F. Angew Chem. 1995;34:352–354. [Google Scholar]
- 8.Sluka J P, Horvath S J, Glasgow A C, Simon M I, Dervan P B. Biochemistry. 1990;29:6551–6561. doi: 10.1021/bi00480a002. [DOI] [PubMed] [Google Scholar]
- 9.Stewart J M, Young J. Solid Phase Peptide Synthesis. 2nd. Ed. Rockford, IL: Pierce Chemical Co.; 1984. [Google Scholar]
- 10.Shi Y G, Berg J M. Science. 1995;269:282–284. doi: 10.1126/science.7536342. [DOI] [PubMed] [Google Scholar]
- 11.Lam W C, Maki A H, Casas-Finet J R, Erickson J W, Kane B P, Souder R C, II, Henderson L E. Biochemistry. 1994;33:10693–10700. doi: 10.1021/bi00201a017. [DOI] [PubMed] [Google Scholar]
- 12.Rice W G, Schaeffer C A, Harten B, Villinger F, South T L, Summers M F, Henderson L E, Bess J W, Jr, Arthur L O, McDougal J S, et al. Nature (London) 1993;361:473–475. doi: 10.1038/361473a0. [DOI] [PubMed] [Google Scholar]
- 13.South T L, Summers M F. Protein Sci. 1993;2:3–19. doi: 10.1002/pro.5560020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surovoy A, Dannull J, Moelling K, Jung G. J Mol Biol. 1993;229:94–104. doi: 10.1006/jmbi.1993.1011. [DOI] [PubMed] [Google Scholar]
- 15.Dannull J, Surovoy A, Jung G, Moelling K. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aldovini A, Young R. Virology. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laussac J P, Cung M T, Érard M, Mazarguil H. C R Acad Sci Paris Ser III. 1991;313:183–186. [PubMed] [Google Scholar]
- 18.Summers M F, Henderson L E, Chance M R, Bess J W, Jr, South T L, Blake P R, Sagi I, Perez-Alvarado G, Sowder R C, III, Hare D R, et al. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brahms S, Brahms J. J Mol Biol. 1980;138:149–178. doi: 10.1016/0022-2836(80)90282-x. [DOI] [PubMed] [Google Scholar]
- 20.Smith J A, Pease L G. CRC Crit Rev Biochem. 1980;8:315–399. doi: 10.3109/10409238009105470. [DOI] [PubMed] [Google Scholar]
- 21.Kuwahara J, Coleman J E. Biochemistry. 1990;29:8627–8631. doi: 10.1021/bi00489a019. [DOI] [PubMed] [Google Scholar]
- 22.Hunt J B, Neece S H, Schachman H K, Ginsburg A. J Biol Chem. 1984;259:14793–14803. [PubMed] [Google Scholar]
- 23.Takeuchi T. The Electronic Structure of Distorted Porphyrins and Cobalt Schiff Base Derivatives as Novel Enzyme Inhibitors. Pasadena, CA: California Institute of Technology; 1996. [Google Scholar]
- 24.Rice W G, Schaeffer C A, Graham L, Bu M, McDougal J S, Orloff S L, Villinger F, Young M, Oroszlan S, Fesen M R, et al. Proc Natl Acad Sci USA. 1993;90:9721–9724. doi: 10.1073/pnas.90.20.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perkins N D, Agranoff A B, Pascal E, Nabel G J. Mol Cell Biol. 1994;14:6570–6583. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice W G, Supko J G, Malspels L, Bess R W, Jr, Clanton D, Bu M, Graham L, Schaeffer C A, Turpin J A, Domamgala J, et al. Science. 1995;270:1194–1197. doi: 10.1126/science.270.5239.1194. [DOI] [PubMed] [Google Scholar]