Abstract
We have previously found that the borders of evolutionarily conserved chromosomal regions often coincide with tumor-associated deletion breakpoints within human 3p12-p22. Moreover, a detailed analysis of a frequently deleted region at 3p21.3 (CER1) showed associations between tumor breaks and gene duplications. We now report on the analysis of 54 chromosome 3 breaks by multipoint FISH (mpFISH) in 10 carcinoma-derived cell lines. The centromeric region was broken in five lines. In lines with highly complex karyotypes, breaks were clustered near known fragile sites, FRA3B, FRA3C, and FRA3D (three lines), and in two other regions: 3p12.3-p13 (∼75 Mb position) and 3q21.3-q22.1 (∼130 Mb position) (six lines). All locations are shown based on NCBI Build 36.1 human genome sequence. The last two regions participated in three of four chromosome 3 inversions during primate evolution. Regions at 75, 127, and 131 Mb positions carry a large (∼250 kb) segmental duplication (tumor break-prone segmental duplication [TBSD]). TBSD homologous sequences were found at 15 sites on different chromosomes. They were located within bands frequently involved in carcinoma-associated breaks. Thirteen of them have been involved in inversions during primate evolution; 10 were reused by breaks during mammalian evolution; 14 showed copy number polymorphism in man. TBSD sites showed an increase in satellite repeats, retrotransposed sequences, and other segmental duplications. We propose that the instability of these sites stems from specific organization of the chromosomal region, associated with location at a boundary between different CG-content isochores and with the presence of TBSDs and “instability elements,” including satellite repeats and retroviral sequences.
The evolution of species and the development of cancer are both Darwinian processes based on variation and selection. In our earlier analysis of the human 3p12-p22 segment, we have found a certain concordance between human–mouse synteny breaks, and tumor-associated deletions (Kiss et al. 2002; Kost-Alimova et al. 2003). Later, based on comparative sequence analysis of one tumor-related deletion at 3p21.3 (named CER1), this association has been extended to other features of evolutionary plasticity, including gene duplications, retrotranspositions, and repeated chromosome rearrangements (Darai et al. 2005). Our cancer chromosome studies were focused on the analysis of deletions, detected by the “elimination test,” based on the transfer of human chromosome 3 (chr 3) into mouse fibrosarcoma (A9) cells, and the subsequent identification of eliminated versus retained chr 3 segments after in vivo tumor growth (Imreh et al. 1994; Yang et al. 1999; Kholodnyuk et al. 2002; Kost-Alimova and Imreh 2007). Therefore, the question remained open as to whether the association between tumor and evolutionary breaks observed in a model system could be found in human tumors, and if the answer is affirmative, do these break-prone regions have any structural characteristics?
Recently, it was shown that ∼5% of the human genome is composed of duplicated genomic segments, which emerged mostly during the past 35 million years of primate evolution. These segmental duplications (SDs) range from a few to hundreds of kilobases and share a high degree of sequence identity (>90%) (Eichler 2001; Samonte and Eichler 2002; Bailey and Eichler 2006). They have gone through extensive structural changes during a relatively short evolutionary time and were associated with chromosomal rearrangements in recent primate evolution (Samonte and Eichler 2002; Courseaux et al. 2003; Nahon 2003; Stankiewicz et al. 2003; Murphy et al. 2005a; Goidts et al. 2006; She et al. 2006). We decided to test whether these regions show signs of instability in human carcinoma cells, as judged by the analysis of tumor related breakpoints. Such analysis was not easy to perform earlier. Studies focused on specific sites like loss of heterozygosity (LOH) or locus-specific FISH were often biased by the choice of markers, guided by earlier studies and by the inevitable concentration on particular regions. Genome-wide studies, like karyotyping, metaphase CGH, multiplex FISH (M-FISH), or spectral karyotyping (SKY) have low resolution. In spite of these drawbacks, the earlier studies suggested a certain correspondence between evolutionary and cancer-related breakpoints. Our study showed a certain concordance between the positions of homozygous deletions at 3p12-p22 in human carcinoma lines and breaks on the mouse–human synteny maps (Kost-Alimova et al. 2003). Another human genomic region, 17p11.2-p12, is rich in SDs and is rearranged both in evolutionary and in cancer-related structural chromosome aberrations (Barbouti et al. 2004; Stankiewicz et al. 2004). Performing multispecies alignments, Murphy et al. (2005b) examined the relationship between the evolutionary and cancer-associated chromosome breakpoints using the Mitelman Database of Chromosome Aberrations in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman). They have found that frequent cancer-associated chromosome aberrations were close to evolutionary breakpoint regions three times as often as were the less frequent cancer-associated aberrations.
Our multipoint FISH (mpFISH) method permits the detection of chr 3 rearrangements in tumor cell lines very efficiently (Darai-Ramqvist et al. 2006). We have chosen 10 carcinoma cell lines for the analysis of breakpoints. Chr 3 is one of the most rearranged chromosomes in different human carcinomas (Kost-Alimova and Imreh 2007; Kost-Alimova et al. 2007); renal cell carcinoma, which represents a majority of our cell line samples, is one of them (van den Berg and Buys 1997; Meloni-Ehrig 2002; see also Mitelman Database of Chromosome Aberrations in Cancer). The number of rearrangements varied from three up to 68 per karyotype as detected by M-FISH. Using mpFISH we found up to 20 chr 3 breaks per cell line. We detected a total of 54 different breakpoints on chr 3 in the 10 cell lines. In the lines with high karyotype complexity they clustered at three known fragile sites, FRA3B, FRA3C, and FRA3D, and at two other regions, 3p12.3-p13 and 3q21.3-q22.1. As we show, the characterization of the last two tumor break-prone regions sheds some light on five questions we were most interested in:
What is the main sequence feature of the tumor break-prone regions?
Do tumor break-prone regions colocalize with evolutionary break-prone regions?
What could be the mechanism of instability within the break-prone regions?
How is the instability maintained during long evolutionary time?
What is the selective value of the instability during evolution and in cancer?
Results
Tumor break-prone regions in carcinoma cell lines
We have previously analyzed several carcinoma cell lines by M-FISH and metaphase CGH. The average number of structural chromosomal changes varied in these cell lines and reflected the complexity of karyotype (Supplements 1 and 2). In this study we analyzed 10 carcinoma lines by painting three chromosomes (2, 3, and 12) and using mpFISH (Supplement 3). We detected 54 chr 3 breakpoints, of which 30 were involved in unbalanced translocations, 14 in interstitial deletions, seven in duplications, one in balanced translocations, and two in insertions (Supplement 4). We differentiated cell lines according to chromosome breakage frequency (Supplement 5). The first group (UOK125, KH39, and UOK115) had, on average, less than one break per chromosome, as shown by painting, and maximum one break per chr 3, as shown by mpFISH. The second group (TK-164, Caki-1, A498, and U2020) had one to three breaks per chromosome, as shown by painting, and two to four breaks per chr 3, as shown by mpFISH. The high karyotype complexity group, including Caki-2, UOK147, and HONE1, had more than three breaks per chromosome, as shown by painting, and 10–20 breaks per chr 3, as shown by mpFISH. Increased resolution (up to 1 Mb by mpFISH) did not improve the detection of rearrangements in low and moderate complexity lines, showing that the low complexity was not due to a lack of resolution. In contrast, the number of detected rearrangements increased with the improvement of the methods in the cell lines with high chromosome complexity Caki-2, UOK147, and HONE1. Totally, 41 breaks on chr 3 were detected in these three lines by mpFISH. These cell lines also had high ploidy: UOK147 and HONE1 were hypotetraploid, Caki-2 was hypopentaploid.
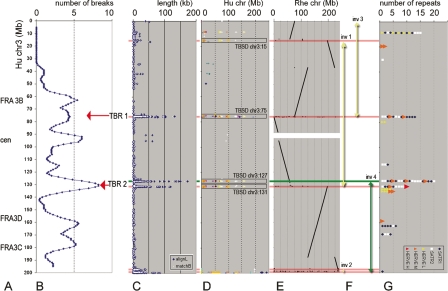
The mpFISH resolution was sufficient to conclude that the breakpoints were not identical, but clustered in particular regions spanning few megabases. To establish the criteria for the identification of tumor break-prone areas, we kept in mind that unbalanced rearrangements generated by end joining of fragments might involve a breakpoint distant to the fusion-point; and the break-prone region may span over several megabases, as exemplified by FRA3B (Becker et al. 2002). The probability of having a break-prone region near to a rearrangement breakpoint would be high within the adjacent region lost during a rearrangement and would decline at both ends of the region. The shape and size of single probability profiles were estimated empirically (Supplement 6), so that the superposition of such profiles gave the best break-prone area identification on the cumulative profile (Fig. 1A,B). We detected six major break-prone areas (more than four breaks in surroundings), of which four represented known instability regions (Fig. 1A). Two novel putative instability regions, designated as tumor break-prone regions 1 and 2 (TBR1 and TBR2), were identified within the 3p12-p13 (around the position of 75 Mb on chr 3) and 3q21.3-q22.1 bands (around the 130 Mb position), respectively. Totally, 16 breaks within the TBRs were detected in six cell lines with moderate and high complexity karyotypes (Supplements 4 and 5).
Figure 1.
Distribution of chr 3 features from 3-pter (top, 0 Mb) to 3-qter (bottom, 200 Mb). Megabase position is shown on vertical scale. (A) Approximate positions of known instability regions, including fragile sites FRA3B, FRA3C, and FRA3D (Schwartz et al. 2006) and pericentromeric region (cen). (B) Cumulative profile for 54 break regions detected by mpFISH in 10 carcinoma cell lines (for explanations, see Supplemental Fig. 6). Red arrows show newly identified tumor break-prone regions (TBRs). (C) SDs of different lengths. Blue rhomb (“alignL”), length of aligned duplicated sequence; white spot (“matchB”), number of match-pairs in aligned unit. (D) Dot-plot alignment of the duplicated (>90% homology) sequences longer than 10 kb, to different human chromosomes (shown with different colors) in relation to their megabase position on a chromosome (horizontal axis). (E) Dot-plot alignment of the rhesus orthologous sequences in relation to their megabase position on chromosome (horizontal axis). The human–rhesus synteny breaks, which correspond to human branch evolution, are shown by red horizontal lines. The human–mouse synteny break, which corresponds to a chromosome rearrangement during early primate evolution, is shown by a green horizontal line. (F) Four inversions in human chr 3 evolution: three occurred after divergence from rhesus branch (yellow arrows), the fourth after divergence from mouse (green arrow). (G) SATR1 and SATR2 satellite repeats, flanking the moderate score HERVE element (HERVE M) are associated with the TBRs, long SDs, and evolutionary chr 3 breaks. HERVE L and HERVE H: low and high score HERVE elements, respectively. Horizontal axis: number of the repeat elements within a particular site.
Tumor break-prone segmental duplication (TBSD)
We analyzed the distribution of SDs along chr 3 in relation to their size (Fig. 1C). The short SDs (<10 kb) were distributed relatively evenly along the chromosome, while the largest SDs (>100 kb) were located within three sites, at the 75, 127, and 131 Mb positions. These SDs were homologous to each other and to particular sites on the other chromosomes (Fig. 1D).
FISH analysis using BAC probes from TBR1 (Fig. 2), taken together with sequence analysis (Supplement 7) suggested the presence of two “SD-amplicons.” One of these (Fig. 2B) was largely similar to the 4q35 “SD-amplicon” reported by Ballarati et al. (2002). Homology with short arms of acrocentric chromosomes and the presence of BSR and LSau satellites suggest similarity to the low-copy sequences, characteristic for rDNA boundary, described by Bodega et al. (2006). On this basis, we call this “SD-amplicon” ribosomal-gene-cluster boundary SD (RBSD). The second “amplicon” was homologous to several chromosomal regions (Fig. 2A), which are similar to the described 3q21.3 “SD-amplicon” (Yue et al. 2005). Three of the homologous segments were at the 75 Mb position (chr3:75), at the 131 Mb position (chr3:131), and at the 127 Mb position (chr3:127). Use of FISH probes to identify the SDs and their borders showed that in the U2020 cell line, one breakpoint was exactly within the RBSD at chr3:75, and in HONE1 cell line, one breakpoint was within the chr3:131 SD (Supplement 3).
Figure 2.
Two-color FISH of TBR1 probes on normal human chromosomes. DAPI staining (blue) based karyotype. (A) BACs RP11-71K3 and RP11-1053M22 (red in Supplemental Fig. 7E) give red and green FISH signals, respectively, at 3p12-p13, 3q21-q22, 4-pter, 7-pter, 7q21, 8-pter, 11-pter, 11q13, 12-pter, and 16-pter, corresponding to large TBSD. (B) BACs RP11-666K17 and RP11-139H7 (green in Supplemental Fig. 7E), give red and green signals, respectively, at 3p12-p13 and on the short arms of acrocentric chromosomes, suggesting that this region represents an “SD-amplicon” homologous to ribosomal-gene-cluster boundary (RBSD), which was not identified by sequence analysis. Both probes were hybridizing also to the pericentromeric regions of chromosomes 1, 4, 20, and 22, corresponding to SDs 6, 7, and 8 in Supplemental Fig. 7A–D.
The other duplication partners were: chr3:15, chr4:4, chr4:9, chr7:7, chr7:97, chr8:7, chr8:12, chr11:4, chr11:67, chr11:71, chr12:8, and chr16:5 (Supplement 7). In the following, we will refer to these homologous sequences as tumor break-prone segmental duplication (TBSD) based on their involvement in chromosome breakages in carcinoma samples. Involvement of chr 3 TBSDs was shown using our mpFISH results; to analyze involvement of other TBSDs, we used the Mitelman Database of Chromosome Aberrations in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman). We found that pericentromeric and subtelomeric bands participated in carcinoma-associated chromosome rearrangements 2.5 times more often than interstitial bands. The majority of TBSD containing or adjacent bands (in the cases where a TBSD was located at the band boundary) were even more frequently involved (>3 times). The interstitial 3p13, 3q21, 7q22, and 11q13 bands were among the most frequent break-prone sites in carcinomas.
The TBSD expanded in Homininae
We compared the sequence of TBSD at chr3:75 with the 14 other TBSDs (chr3:15, chr3:127, chr3:131, chr4:4, etc.) using Dot Plot output of the PipMaker program. TBSD consisted of seven parts (Supplement 7), which were differently represented in different chromosomal locations as shown in Figure 3. Each part of a certain color is present at multiple human chromosomal locations, but has only one common orthologous segment in rhesus mouse, dog, opossum, and chicken genomes (Supplement 8A). This shows that the TBSD has originated from unique human–rhesus concestral sequences. In the chimpanzee, each of the multiple human TBSD locations has its own orthologous sequence, meaning that the expansion occurred after the separation of the rhesus and before the separation of the chimpanzee from the human branch.
Figure 3.
Schematic organization of the TBSDs from different human chromosomal sites as determined by comparison with the chr3:75 TBSD parts (see Supplemental Fig. 7G). All TBSDs, with the exception of those that are marked with a red arrow, are shown in pter–qter direction from bottom to top. Chromosomal segments, which have clearly identified conservation in rhesus genome, are shown in gray with corresponding rhesus chromosome number shown within the bar, and megabase position shown adjacent to the bar. Arrowhead on the bar indicates telomeric location of the rhesus orthologous sequence. Rounded end of the bar shows breakpoint of human–rhesus evolutionary inversions, indicated by black double-head arrows. The orange double-headed arrows show the inversions that occurred within this chromosomal region before human–rhesus, but after human–mouse divergence.
We have analyzed orangutan and gorilla chromosomes by FISH with three probes. The BAC RP11-266L17 from the region telomeric to chr3:75 TBSD gave unique signals on human, gorilla, and orangutan chromosomes, identifying the human chr 3 orthologs (red in Fig. 4). The chr3:75 TBSD probe, combined from RP11-71K3 and RP11-1053M22 (see Fig. 3A), gave multiple signals on gorilla chromosomes, including the human chr 3 orthologous chromosome. In contrast, the strong signal was missing from the human chr 3 ortholog on orangutan metaphases. It was present in only two other chromosome sites (green in Fig. 4A). According to Yue et al. (2005), this SD was missing on homologous chromosomes in siamang and Old World monkey as well, confirming that it expanded after the human–orangutan divergence. The RBSD probe, which was a combination of RP11-666K17 and RP11-139H7 (see Fig. 3B), gave multiple green signals at the short arms of acrocentric chromosomes in orangutan, like in human, suggesting that the RBSD expanded earlier than the TBSD. FISH analysis of the “4q35 SD,” which corresponds to our RBSD, in a number of apes showed that the duplication was present even in Old World monkeys, but suffered multiple changes during primate evolution (Ballarati et al. 2002). In contrast to human and gorilla, the orangutan had the BAC RP11-266L17 and RBSD probe signals separated on the chr 3 ortholog (Fig. 4B). This demonstrates that the chromosomal rearrangement at the TBSD site occurred in parallel with the TBSD expansion (in the homininae branch, after the divergence from other primates including orangutans).
Figure 4.
Two-color FISH of TBR1 probes on orangutan chromosomes. BAC RP11-266L17 (red) identifies orangutan region, orthologous to unique human site telomeric to TBSD (see blue in Supplemental Fig. 7E). (A) Mixed BACs RP11-71K3 and RP11-1053M22 (green) identify TBSD orthologous sequences on two orangutan chromosomes, different from human chr 3 ortholog. (B) Mixed BACs, RP11-666K17 and RP11-139H7 (green), identify RBSD orthologous sequences as multiple signals on the short arms of acrocentric chromosomes. BAC RP11-266L17 signal (red) is not overlapping to green signal, showing that a chromosomal rearrangement occurred within the TBSD site during human–orangutan divergence.
TBRs are “hot spots” of chromosomal breaks in mammalian evolution
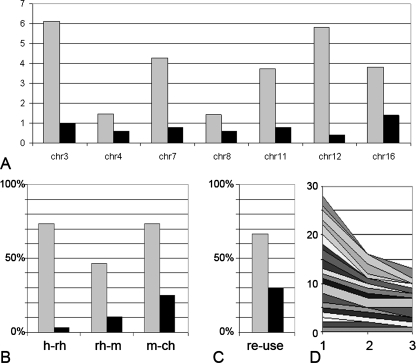
We analyzed the conservation of chr 3 segments in other sequenced species. Figure 1E shows a dot plot comparison of human chr 3 sequences against rhesus macaque (Macaca mulatta) chromosomal sequences. The presence of large conserved segments was in agreement with the results of FISH comparative analysis performed by Ventura et al. (2007). Five human-branch-specific breaks (see red horizontal lines in Fig. 1E) were identified using Mus musculus, Rattus norvegicus, Canis familiaris, Monodelphis domestica, and Gallus gallus chromosomal sequences as an out-group. These breaks, in contrast to the rhesus-branch-specific breaks, corresponded to the locations of long SDs (see Fig. 1C,D), three of which were within the TBSDs at the 15-, 75-, and 131-Mb positions. Comparing human chr 3 in the same way with mouse chromosomal sequences with the out-group of Canis familiaris, Monodelphis domestica, and Gallus gallus genomes, we found that the 127 Mb TBSD was colocalized with the only break that occurred in the primate-specific branch after divergence from other mammals (see green horizontal line in Fig. 1E). Recent human chr 3 rearrangement events could be traced on the basis of conserved fragments. Three inversions occurred during the evolution from the human–rhesus concestor (yellow vertical double-head arrows in Fig. 1F) and one inversion during the evolution from the human–rodent concestor (green vertical double-head arrow in Fig. 1F). This was also in agreement with comparative FISH studies in primates (Muller et al. 2000; Ventura et al. 2004), The reconstitution of ancestral chr 3 by these inversions (Supplement 8B) and comparison of orthologous regions between other species showed that the chr 3 TBRs were broken repeatedly during mammalian evolution. The random regions on chr 3 and on other chromosomes were broken and reused much less frequently than TBSD-containing sites (Fig. 5).
Figure 5.
Frequencies of breaks within regions surrounding the TBSDs during mammalian evolution. (A) Total number of human–rhesus, rhesus–mouse, and mouse–chicken synteny breaks per megabase within regions surrounding the TBSDs (gray) and within random 500-kb sites (black) taken from each 10 Mb of the particular chromosome (chr 3, chr 4, etc) sequence. (B) Percentage of regions, which had human–rhesus (h-rh), rhesus–mouse (rh-m), and mouse–chicken (m-ch) synteny breaks, counted from total number of analyzed 500-kb regions, which surrounded either TBSDs (gray) or random sites taken at each 10 Mb of TBSD containing chromosome sequences (black). (C) Frequency of reuse for synteny breakpoint region at TBSD (gray) and at random synteny break (black). Synteny breakpoint region is defined as a 500-kb region containing a minimum of one break in either human–rhesus or in rhesus–mouse or in mouse–chicken branch. Reuse of the synteny breakpoint region is defined as the occurrence of breaks in at least two of three mentioned branches during evolution. (D) Gradual decrease in frequency of rhesus–mouse and mouse–chicken synteny breaks with increase of distance from TBSD sites. Cumulative area shows the number of breaks within the 1st, 2nd, and 3rd sequence windows (point 1, 2, and 3, respectively) from different TBSD locations. For each TBSD location, windows 1, 2, and 3 represent successive sequence windows equal in size to TBSD, which flank TBSD (1), located next to both flanks (2), and most distant windows (3).
We were interested how the human-branch-specific synteny breaks, which occurred before the human–rhesus divergence, are distributed around the TBSDs. We, therefore, studied the breaks, which were not associated with TBSD but with the surrounding region, since the TBSD appeared there after human–rhesus divergence. The sequence windows closest to TBSDs contained the highest number of the breaks (Fig. 5D), suggesting that the region that later accepted the TBSD represented a rearrangement “hot-spot” even earlier.
TBSDs are located at the transitions between CG-rich and CG-poor areas
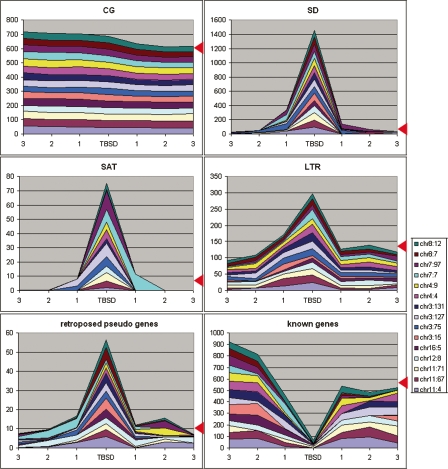
We analyzed different sequence characteristics of the 15 regions, corresponding to each TBSD and of three adjacent sequence windows of the same size (Fig. 6). Taking into account the observation that nine of these regions were at the transition between chromosomal bands, we oriented the analyzed sequences from R to G band (from left to right in Fig. 6). Seven regions (chr11:4; chr11:67, chr11:71, chr3:131, chr4:4, chr7:7, and chr8:12) appeared to have ascending megabase positions, while two (chr3:127 and chr8:7) were descending. These nine regions, aligned in the given orientation, showed a higher CG-content in the window flanking TBSD from the left compared with the right. We have therefore oriented the remaining six regions that did not show band transition, positioning the TBSD flanking window with the higher CG-content to the left. Then, four regions (chr12:8, chr16:5, chr3:15, and chr4:9) were oriented in increasing and two (chr3:75 and chr7:97) in decreasing megabase positions. The “CG” chart in Figure 6 shows that the TBSDs represent border regions between chromosomal areas with higher and lower CG-content. This prompted us to analyze isochores in the vicinity of TBSDs. We assessed the average GC levels over 500-kb DNA stretches distal and proximal to each TBSD, and estimated GC variation by measuring standard deviations of GC over these stretches using a 100-kb moving window, similarly to isochore analysis performed by Costantini et al. (2006). Results are shown in Figure 7. Eleven out of 15 TBSDs appeared at transitions between isochores; 10 of these transitions were statistically significant (difference exceeded standard deviations).
Figure 6.
Sequence features in the region surrounding TBSD. Cumulative area shows contents (in percentages) of particular sequence features, which were identified in different TBSD sites and in three successive sequence windows of the same size departing from the TBSD sites (points 1, 2, and 3, respectively). Red arrowheads show cumulative value for the same number of random regions, calculated based on average values for the human genome. Regions are pter–qter oriented, as described in the Results section, “TBSDs are located at the transitions between CG- rich and poor areas.”
Figure 7.
Isochore surrounding of TBSDs. Average GC levels were assessed over 500-kb DNA stretches distal and proximal to each TBSD. The colored boxes show correspondence of these stretches to different class isochores (see at right). (Ovals) TBSD; (black circles) telomeres; (double-headed arrows) recent human branch-specific evolutionary inversions. In the case of chr 3, three successive inversions changed the orientation of TBSD regions. In the case of chr 7, one additional inversion changed the orientation of the chr7:97 region as shown by the green arrow.
TBSDs and their close surroundings show enrichment in SDs, satellite, long terminal repeats, and retroposed pseudogenes
The “SD” chart in Figure 6 shows that the SD content was very high within the TBSDs, as expected. Interestingly, it was also significantly higher than the human genome average (red arrowhead) in the first flanking sequence window to left. The SD content gradually decreased with the distance from the TBSDs in both directions. In the third window it was below average. A similar tendency was recorded for satellite repeats (SAT chart in Fig. 6), for long terminal repeats that include retroposons (LTRs) and for retroposed sequences, including pseudogenes. The known gene content showed a mirror-like pattern with a minimal number of genes in the middle.
Analyzing the chr3:75 TBSD (Supplement 7), we found that it was flanked by satellite repeats SATR1 and SATR2, which contained an LTR repeat, HERVE, which belongs to the ERV1 endogenous retroviruses family. We studied the distribution of SATR1, SATR2, and HERVE along chr 3 (see Fig. 1G). Since the HERVE sequences varied considerably in the human genome, we classified the chr 3 HERVE sequences into three groups according to alignment scores. The intermediate score HERVE M with flanking SATR repeats were regularly associated with the chr3:75, chr3:127, and chr3:131 TBSDs. The majority of other TBSDs contained blocks of HERVE/SATR1, 2 at their boundaries and had increased sequence flexibility in the proximity of these blocks, suggesting the use of these elements for duplication and transposition (“putative instability element” in Fig. 3).
Discussion
Development of a new, high-resolution modification of FISH (mpFISH) allowed us to perform a thorough chr 3 analysis in 10 carcinoma cell lines and to identify tumor break-prone regions. Two of these regions (bands 3p12-13 and 3q21) were known to be involved in some cancer-associated chr 3 aberrations, but were not described previously as “instability sites.” The rearrangements within these novel “instability sites” designated TBRs were mostly unbalanced translocations. They were particularly frequent in hyperploid tumor lines with multiple chromosomal aberrations. Sixteen breaks within the TBRs and 21 breaks in fragile sites formed almost three-fourths of all breaks in the moderate (two to four chr 3 rearrangements in TK-164, Caki-1, A498, and U2020) and high complexity karyotype cell lines (10 to 20 chr 3 rearrangements in Caki-2, UOK147, and HONE1). Breaks around TBRs were found in all cell lines of these two categories, except one; while fragile sites were only broken in three cell lines, with high-complexity karyotype participating in interstitial deletions and amplifications. In the introduction we formulated five questions concerning tumor break-prone regions. We will attempt to answer these questions based on our results.
A TBR “epicentre” is characterized by specific sequence features
The chromosomal rearrangements had highest incidence in close proximity to “epicentres” of TBR1 and TBR2. We found one of the largest (∼250 kb in size) and evolutionarily youngest (spread after human–orangutan divergence) SDs (designated TBSD) there. These TBSDs were located at the 75, 127, and 131 Mb positions. The majority of the TBR breakpoints (10 of 16) were within 1-Mb areas surrounding the TBSDs; two were inside the SDs. The other six unbalanced translocation fusion points were within 5-Mb areas, surrounding the “epicenter,” and the TBSDs were lost due to these unbalanced translocations. This leaves us with two possible explanations. Breaks may occur within or close to the TBSD, followed by the digestion of free ends and the loss of the TBSD, before joining the other chromosomal fragment end. Alternatively, breaks occur within a large unstable region that surrounds the TBSD with a higher chance in its close surroundings.
The TBSD has a complex origin and is found at 15 locations in the human genome. The majority of them lie at the transition between higher and lower CG-content areas and are flanked by satellite (SATR1 and SATR2) and LTR (HERVE) repeats. Using the Mitelman Database of Chromosome Aberrations in Cancer, we found that these locations are also “hot spots” of carcinoma-related chromosomal rearrangements.
TBRs correspond to evolutionary break-prone regions
Structural instability provides an evolutionary opportunity for the recycling of certain chromosomal regions (Bourque et al. 2004; Zhao et al. 2004; Darai et al. 2005; Murphy et al. 2005b). We checked whether TBRs belong to these “recycling sites.”
Human–rhesus sequence comparison identified breaks between homologous segments on chr 3 (see Fig. 1E). Remarkably, only the human-branch-specific breakpoints (see red and green horizontal lines in Fig. 1E) colocalize with the large SDs, confirming association between evolutionary breaks and SDs. Three large segment inversions, which occurred during the evolution of human chr 3 from common primate ancestor (see Fig. 1F), have TBSDs at the breakpoints. Analyzing all human chromosomes, we concluded that 10 out of 15 TBSDs were involved in chromosomal inversions during recent human evolution, and the surrounding regions were rearranged not only recently (see Fig. 3), when the TBSDs were inserted, but also earlier in mammalian evolution (see Fig. 5). TBSD regions were reused during mammalian evolution in 70% of cases, positioning the TBRs among the “hottest spots” of evolution.
Organization of TBSDs and their surroundings (see Figs. 3, 6) suggest that multiple rearrangement events, including high-transposition activity, duplications, and deletions occurred during evolution. Fourteen out of 15 (93%) TBSDs showed copy-number polymorphism in the human population, as reported by Redon et al. (2006).
Our findings confirm and add more evidence to Haaf and colleagues’ hypothesis about evolutionary plasticity of the primate-specific chr 3 breakpoint regions (Tsend-Ayush et al. 2004; Yue et al. 2005, 2006; Yue and Haaf 2006).
Possible mechanisms of instability at the TBSDs
As we mentioned, the breakpoints detected by us on chr 3 in carcinoma cell lines were clustered around, but not necessarily within the TBSD; the majority of rearrangements were unbalanced translocations leading to loss of the TBSD. This speaks against nonallelic homologous recombination (NAHR), which was suggested as a main mechanism of chromosomal rearrangements that involve SDs (Samonte and Eichler 2002; Stankiewicz and Lupski 2002; Locke et al. 2003; Dennehey et al. 2004; Muller et al. 2004; Shaw and Lupski 2004; Murphy et al. 2005a). NAHR does not explain all evolutionary rearrangements that involve the TBSDs as well (see Fig. 3). Inversions between chr3:75 and chr3-pter; chr3:15 and chr3:131 show no homology between the flanks. The organization of chr3:15 and chr3:131 regions suggests that the inversion has split the ancestral TBSD. A similar “split-mechanism” is also likely for the rearrangement at the chr 7 TBSD sites.
TBSDs were inserted into TBRs after human–rhesus divergence. In the majority of cases, they flanked homininae-specific inversions (see black double-headed arrows in Fig. 3). However, two inversions that involved the chr3:127 and chr12:8 sites (see orange double-headed arrows in Fig. 3), occurred before human–rhesus divergence, and the majority of the TBSD sites were reused for chromosome breakage even earlier in mammalian evolution. This shows that the instability is not restricted to the TBSDs themselves. In this respect, it should be mentioned that the content of other SDs, other satellite and LTR repeats, is high in close proximity of TBSD and decreases gradually with increasing distance from it (see Fig. 6). It is possible that these features reflect the instability of the chromosomal region organization that favors the acceptance of transposable elements, retroposed genes, and duplications. It is known that such unstable organization characterizes the telomere and centromere (Murphy et al. 2005b; Bailey and Eichler 2006). We have found that the TBSDs often occur at former or potential telomere sites (data not shown). An intriguing feature of the TBSD regions is their location at the border between R- and G-bands, signaling the transition between higher and lower CG-content zones or isochores. The higher CG-content parts of four regions, chr3:75, chr3:127, chr4:4, and chr4:9, correspond to the ancestral terminal sites mentioned. The high CG-content parts also show higher levels of SDs, LTR repeats, and retroposed pseudogenes in the TBSD flanking region, in contrast to more distant regions (see Fig. 6).
Interestingly, three out of seven primate-specific inversions (see chr3:75-pter; chr7:7-97, and chr11:4-71 in Fig. 7), seem to split pre-existing homogeneous isochores; the other three inversions (see chr3:127-qter; chr4:4-9, and chr8:7-12 in Fig. 7) split the ancestral regions with closer CG-content. In the putative ancestral state, all inversion neighborhoods were different in CG-content by not more than 5%, while in the derived human state, the difference at nine of 15 transitions was >5%. The majority of isochore transitions in the human genome are <5% (Costantini et al. 2006); therefore, we suggest that formation of high-contrast isochore boundaries at the recent chromosomal inversion breakpoints in the derived state may explain their high instability. Less contrast transitions at the breakpoints in the ancestral chromosomal state is probably related to their reuse by breakage earlier in evolution.
Isochores are characterized by specific density and replication time (Costantini et al. 2006; Schmegner et al. 2007). Thus, the boundary represents a region of dramatic change in chromatin structure and function. Replication, which starts early in high-CG isochore, pauses for several hours at the transition to low-CG isochore (Schmegner et al. 2007). It was proposed that replication disturbance at the transition between these zones may explain “fragility” of certain regions (Debatisse et al. 2006). The presence of satellite-rich sequences with a potential to form unusual secondary structures may enhance this instability effect, as it was shown for rare and common fragile sites (Zlotorynski et al. 2003; Gericke 2006) and for human rRNA genes (Lebofsky and Bensimon 2005). Palindromic repeat mediated genomic plasticity has been demonstrated to promote a variety of translocations (Tapia-Paez et al. 2000, 2001; Kurahashi and Emanuel 2001; Kurahashi et al. 2003) and even cancer-associated somatic rearrangements (Barbouti et al. 2004). Replication asynchrony and delay also characterize polysomic regions (Kost-Alimova et al. 2004) and gene duplications (Gimelbrant and Chess 2006). Thus, incomplete replication within these regions of special organization may lead to breakage and chromosomal rearrangements under certain conditions.
Another instability mechanism within the TBSDs may be related to over-representation of LTR retrotransposons. They predominantly belong to several families of human endogenous retroviruses (HERVs). HERV elements are usually densely methylated in normal somatic cells, but if activated, they may contribute to chromosomal instability in cancer (for review, see Schulz et al. 2006). We found that HERVE elements flanked by SATR repeats at the TBSD boundaries are juxtaposed to the most flexible, frequently duplicated, or deleted parts of the TBSD (see “putative instability element” in Fig. 3). This “putative instability element” was formed at the same evolutionary time when the TBSDs spread. While the HERVE and SATR repeats are separated in the rhesus genome, they combine and spread together with TBSDs in chimpanzee and human genomes. Probably, the HERVE/SATR complex could function as a transposable element during recent primate evolution. This complex might participate in duplicative transposition of larger genomic material into new sites of the genome, as it was proposed for primate-specific retroposons of the Alu family (Bailey et al. 2003).
Selective value of the changes within TBSDs
Evolutionary and tumor-related instability features of the described break-prone regions, which include breakage and accumulation of SDs, suggests that these regions can be “recycled” in different species and in cancer cells. What is the selective value of these changes? We found that at least one group of functional genes was involved in the “recycling.” The FAM86 family has expanded together with the TBSDs in higher primates. The genes of this family are active in the human genome, as shown by mRNA alignments, but their function is not known. It may be noted that they contain the S-adenosyl-L-methionine-dependent methyltransferase superfamily domain. Methyl transfer from the ubiquitous donor S-adenosyl-L-methionine to nitrogen, oxygen, or carbon atoms occurs frequently in many organisms ranging from bacteria to plants and mammals. The reaction is catalyzed by methyltransferases and modifies DNA, RNA, proteins, and small molecules such as catechol, which participate in gene regulation and differentiation.
The selective growth advantage of the rearrangements in TBSD regions may not be dependent entirely on their gene content. Breakage in tumors often leads to the loss of large chromosomal fragments. Changes in gene expression that occur in the wake of such losses may convey growth advantage. Alternatively, it can change the copy number of specific genes. For example, the rearrangements within chr3:75 resulted in 3p12-pter losses in the majority of cell lines analyzed by us. The loss of distal 3p genes may be advantageous due to the well-documented presence of multiple tumor suppressor genes on 3p (Zabarovsky et al. 2002; Imreh et al. 2003).
Conclusion
Identification and analysis of chr 3 regions 3p12-p13 and 3q21.3-q22.1, involved in tumor-related breaks, showed that they have also played an important role both in the recent evolution of primates and generally in mammalian evolution. The main feature common to both regions was the presence of specific SDs, designated tumor break-prone segmental duplications (TBSDs). The chromosomal regions containing TBSD share common sequence features related to the regional instability:
They are located at the transition between more and less CG-rich areas at sites, which may represent ancestral terminal chromosomal segments.
They contain large segmental duplications. The gene clusters and the functional diversity created within contributes to speciation.
They remain structurally unstable both in evolution and in malignancy. Their instability may be explained by disturbed replication at the transition between different isochores, which is catalyzed by the presence of unusual structures like large SDs, satellite repeats, and by reactivation of retroviral elements during evolution and in cancer cells.
The plasticity in these sites is maintained during long evolutionary time. Among the explanations, it may be that the regional instability leads to chromosomal rearrangements that fuse areas that are even more dissimilar CG-content, and to increased ability to accept satellite repeats, transposable elements, and SDs. These changes would in turn perpetuate the instability.
Methods
Cell lines
Ten carcinoma-derived cell lines were analyzed. Eight were derived from renal-cell carcinoma: KH39, A498, UOK115, UOK125, UOK147, TK-164 (Bear et al. 1987; Gnarra et al. 1994; Tomita et al. 1996), Caki-1 (ATCC catalog no. HTB46), and Caki-2 (ATCC catalog no. HTB47). U2020 was derived from small cell lung cancer (Heppell-Parton et al. 1999). HONE1 was a human nasopharyngeal carcinoma cell line (Glaser et al. 1989). All cell lines were cultured on IMDM medium with 10% fetal calf serum, 1% penicillin-streptomycin, and 1% glutamine. For fluorescence in situ hybridization (FISH) experiments, cells were treated with 0.1 μg/mL colcemid for 3–4 h to obtain metaphase chromosomes. After treatment with hypotonic solution, cells were fixed in methanol:acetic acid (3:1).
FISH
FISH was performed on metaphase spreads prepared from the described cell lines using chr 3-, chr 2-, and chr 12-specific painting probes labeled with FITC, Cy3, and biotin, respectively (Cambio). The biotin-labeled probe hybridization was detected with Cy5 conjugated streptavidin (Amersham Biosciences, GE Healthcare Worldwide).
A total of 179 BAC/PAC clones, which covered the chr 3 with a resolution of ∼1 Mb, were used for comparative analysis of carcinoma cell lines by mpFISH and arrayCGH in our earlier study (Darai-Ramqvist et al. 2006). Metaphase mpFISH data from 176 clones, which were proven to be on chr 3, were used in the present study to analyze distribution of breakpoints on chr 3 in 10 cell lines. To map the breakpoints within TBSDs and to analyze TBR1 in human, gorilla, and orangutan chromosomes, we used 11 clones that were inside and close to TBSDs (BACPAC Resources Center, Children's Hospital Oakland, CA). BAC/PAC DNA was isolated using QIAGEN columns (QIAGEN, Inc.) and labeled with nick-translation either with biotin-dUTP or digoxigenin-dUTP (BIO-Nick Translation Mix or Digoxigenin-Nick Translation Mix, Roche Molecular Biochemicals). FISH and analysis were performed as described (Darai-Ramqvist et al. 2006). Biotin-labeled probes were detected with Cy3-conjugated streptavidin (Amersham Biosciences, GE Healthcare Worldwide) and digoxigenin-labeled probes with FITC-conjugated anti-digoxigenin antibodies (Roche Molecular Biochemicals Mannheim).
Metaphase plates were analyzed for each painting and mpFISH probe using a fluorescence microscope (Leitz-DMRB, Leica) equipped with a Hamamatsu C 4800 cooled CCD camera (Hamamatsu) and Adobe Photoshop 7.0 (Adobe Systems).
Database analysis
UCSC Genome Bioinformatics Site (http://genome.ucsc.edu) was used for majority of searches. The March 2006 human reference sequence (NCBI Build 36.1), as well as other specie latest-reference sequences were analyzed using the tracks listed below in the UCSC Genome Browser (Kent et al. 2002). The May 2004 human assembly was used for analysis of retroposed sequences, including pseudogenes and some bacterial artificial chromosome (BAC) end sequences, which were missing from the March 2006 assembly. The results obtained in the May 2004 assembly were converted to positions corresponding to the March 2006 assembly.
BAC End Pairs Track and FISH Clones Track, helped us to identify positions of mpFISH probes and to order FISH probes for TBR1 analysis. To identify segmental duplications, Segmental Duplications Track, was analyzed. Repeat contents were determined using Repeating Elements by RepeatMasker Track. Conservation of chromosome segments in other sequenced species was identified using Net Traks. To obtain information about approximate location of bands seen on Giemsa-stained chromosomes, Chromosome Band Track was used. GC Percent Track was used to analyze the percentage of G (guanine) and C (cytosine) bases. Known Genes Track identified known protein-coding genes. The mRNA track showed alignments between human mRNAs in GenBank and the genome, produced using the BLAT program. The Superfamily track shows proteins having homologs with known structures or functions.
For the analysis of carcinoma-related breakpoint distribution along the human chromosomes, we used the Mitelman Database of Chromosome Aberrations in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman).
The Dot Plot output of PipMaker (http://pipmaker.bx.psu.edu/pipmaker) (Schwartz et al. 2000) program was used to identify and illustrate similar parts in two sequences of >100 bp with at least 70% nucleotide identity.
Acknowledgments
This work was supported by grants from the Swedish Cancer Society, the Swedish Medical Research Council, the Cancer Society in Stockholm, the Cancer Research Institute/Concern Foundation NY-LA, USA, the Karolinska Hospital, and the Karolinska Institute.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7010208
References
- Bailey J.A., Eichler E.E., Eichler E.E. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Liu G., Eichler E.E., Liu G., Eichler E.E., Eichler E.E. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballarati L., Piccini I., Carbone L., Archidiacono N., Rollier A., Marozzi A., Meneveri R., Ginelli E., Piccini I., Carbone L., Archidiacono N., Rollier A., Marozzi A., Meneveri R., Ginelli E., Carbone L., Archidiacono N., Rollier A., Marozzi A., Meneveri R., Ginelli E., Archidiacono N., Rollier A., Marozzi A., Meneveri R., Ginelli E., Rollier A., Marozzi A., Meneveri R., Ginelli E., Marozzi A., Meneveri R., Ginelli E., Meneveri R., Ginelli E., Ginelli E. Human genome dispersal and evolution of 4q35 duplications and interspersed LSau repeats. Gene. 2002;296:21–27. doi: 10.1016/s0378-1119(02)00858-2. [DOI] [PubMed] [Google Scholar]
- Barbouti A., Stankiewicz P., Nusbaum C., Cuomo C., Cook A., Hoglund M., Johansson B., Hagemeijer A., Park S.S., Mitelman F., Stankiewicz P., Nusbaum C., Cuomo C., Cook A., Hoglund M., Johansson B., Hagemeijer A., Park S.S., Mitelman F., Nusbaum C., Cuomo C., Cook A., Hoglund M., Johansson B., Hagemeijer A., Park S.S., Mitelman F., Cuomo C., Cook A., Hoglund M., Johansson B., Hagemeijer A., Park S.S., Mitelman F., Cook A., Hoglund M., Johansson B., Hagemeijer A., Park S.S., Mitelman F., Hoglund M., Johansson B., Hagemeijer A., Park S.S., Mitelman F., Johansson B., Hagemeijer A., Park S.S., Mitelman F., Hagemeijer A., Park S.S., Mitelman F., Park S.S., Mitelman F., Mitelman F., et al. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am. J. Hum. Genet. 2004;74:1–10. doi: 10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear A., Clayman R.V., Elbers J., Limas C., Wang N., Stone K., Gebhard R., Prigge W., Palmer J., Clayman R.V., Elbers J., Limas C., Wang N., Stone K., Gebhard R., Prigge W., Palmer J., Elbers J., Limas C., Wang N., Stone K., Gebhard R., Prigge W., Palmer J., Limas C., Wang N., Stone K., Gebhard R., Prigge W., Palmer J., Wang N., Stone K., Gebhard R., Prigge W., Palmer J., Stone K., Gebhard R., Prigge W., Palmer J., Gebhard R., Prigge W., Palmer J., Prigge W., Palmer J., Palmer J. Characterization of two human cell lines (TK-10, TK-164) of renal cell cancer. Cancer Res. 1987;47:3856–3862. [PubMed] [Google Scholar]
- Becker N.A., Thorland E.C., Denison S.R., Phillips L.A., Smith D.I., Thorland E.C., Denison S.R., Phillips L.A., Smith D.I., Denison S.R., Phillips L.A., Smith D.I., Phillips L.A., Smith D.I., Smith D.I. Evidence that instability within the FRA3B region extends four megabases. Oncogene. 2002;21:8713–8722. doi: 10.1038/sj.onc.1205950. [DOI] [PubMed] [Google Scholar]
- Bodega B., Cardone M.F., Rocchi M., Meneveri R., Marozzi A., Ginelli E., Cardone M.F., Rocchi M., Meneveri R., Marozzi A., Ginelli E., Rocchi M., Meneveri R., Marozzi A., Ginelli E., Meneveri R., Marozzi A., Ginelli E., Marozzi A., Ginelli E., Ginelli E. The boundary of macaque rDNA is constituted by low-copy sequences conserved during evolution. Genomics. 2006;88:564–571. doi: 10.1016/j.ygeno.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Bourque G., Pevzner P.A., Tesler G., Pevzner P.A., Tesler G., Tesler G. Reconstructing the genomic architecture of ancestral mammals: Lessons from human, mouse, and rat genomes. Genome Res. 2004;14:507–516. doi: 10.1101/gr.1975204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini M., Clay O., Auletta F., Bernardi G., Clay O., Auletta F., Bernardi G., Auletta F., Bernardi G., Bernardi G. An isochore map of human chromosomes. Genome Res. 2006;16:536–541. doi: 10.1101/gr.4910606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courseaux A., Richard F., Grosgeorge J., Ortola C., Viale A., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Richard F., Grosgeorge J., Ortola C., Viale A., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Grosgeorge J., Ortola C., Viale A., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Ortola C., Viale A., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Viale A., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Dutrillaux B., Gaudray P., Nahon J.L., Gaudray P., Nahon J.L., Nahon J.L. Segmental duplications in euchromatic regions of human chromosome 5: A source of evolutionary instability and transcriptional innovation. Genome Res. 2003;13:369–381. doi: 10.1101/gr.490303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darai E., Kost-Alimova M., Kiss H., Kansoul H., Klein G., Imreh S., Kost-Alimova M., Kiss H., Kansoul H., Klein G., Imreh S., Kiss H., Kansoul H., Klein G., Imreh S., Kansoul H., Klein G., Imreh S., Klein G., Imreh S., Imreh S. Evolutionarily plastic regions at human 3p21.3 coincide with tumor breakpoints identified by the “elimination test.”. Genomics. 2005;86:1–12. doi: 10.1016/j.ygeno.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Darai-Ramqvist E., de Stahl T.D., Sandlund A., Mantripragada K., Klein G., Dumanski J., Imreh S., Kost-Alimova M., de Stahl T.D., Sandlund A., Mantripragada K., Klein G., Dumanski J., Imreh S., Kost-Alimova M., Sandlund A., Mantripragada K., Klein G., Dumanski J., Imreh S., Kost-Alimova M., Mantripragada K., Klein G., Dumanski J., Imreh S., Kost-Alimova M., Klein G., Dumanski J., Imreh S., Kost-Alimova M., Dumanski J., Imreh S., Kost-Alimova M., Imreh S., Kost-Alimova M., Kost-Alimova M. Array-CGH and multipoint FISH to decode complex chromosomal rearrangements. BMC Genomics. 2006;7:330. doi: 10.1186/1471-2164-7-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M., El Achkar E., Dutrillaux B., El Achkar E., Dutrillaux B., Dutrillaux B. Common fragile sites nested at the interfaces of early and late-replicating chromosome bands: Cis acting components of the G2/M checkpoint? Cell Cycle. 2006;5:578–581. doi: 10.4161/cc.5.6.2574. [DOI] [PubMed] [Google Scholar]
- Dennehey B.K., Gutches D.G., McConkey E.H., Krauter K.S., Gutches D.G., McConkey E.H., Krauter K.S., McConkey E.H., Krauter K.S., Krauter K.S. Inversion, duplication, and changes in gene context are associated with human chromosome 18 evolution. Genomics. 2004;83:493–501. doi: 10.1016/j.ygeno.2003.08.017. [DOI] [PubMed] [Google Scholar]
- Eichler E.E. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- Gericke G.S. Chromosomal fragility, structural rearrangements and mobile element activity may reflect dynamic epigenetic mechanisms of importance in neurobehavioural genetics. Med. Hypotheses. 2006;66:276–285. doi: 10.1016/j.mehy.2005.06.032. [DOI] [PubMed] [Google Scholar]
- Gimelbrant A.A., Chess A., Chess A. An epigenetic state associated with areas of gene duplication. Genome Res. 2006;16:723–729. doi: 10.1101/gr.5023706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser R., Zhang H.Y., Yao K.T., Zhu H.C., Wang F.X., Li G.Y., Wen D.S., Li Y.P., Zhang H.Y., Yao K.T., Zhu H.C., Wang F.X., Li G.Y., Wen D.S., Li Y.P., Yao K.T., Zhu H.C., Wang F.X., Li G.Y., Wen D.S., Li Y.P., Zhu H.C., Wang F.X., Li G.Y., Wen D.S., Li Y.P., Wang F.X., Li G.Y., Wen D.S., Li Y.P., Li G.Y., Wen D.S., Li Y.P., Wen D.S., Li Y.P., Li Y.P. Two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein-Barr virus that were derived from nasopharyngeal carcinomas. Proc. Natl. Acad. Sci. 1989;86:9524–9528. doi: 10.1073/pnas.86.23.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnarra J.R., Tory K., Weng Y., Schmidt L., Wei M.H., Li H., Latif F., Liu S., Chen F., Duh F.M., Tory K., Weng Y., Schmidt L., Wei M.H., Li H., Latif F., Liu S., Chen F., Duh F.M., Weng Y., Schmidt L., Wei M.H., Li H., Latif F., Liu S., Chen F., Duh F.M., Schmidt L., Wei M.H., Li H., Latif F., Liu S., Chen F., Duh F.M., Wei M.H., Li H., Latif F., Liu S., Chen F., Duh F.M., Li H., Latif F., Liu S., Chen F., Duh F.M., Latif F., Liu S., Chen F., Duh F.M., Liu S., Chen F., Duh F.M., Chen F., Duh F.M., Duh F.M., et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- Goidts V., Armengol L., Schempp W., Conroy J., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Armengol L., Schempp W., Conroy J., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Schempp W., Conroy J., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Conroy J., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Estivill X., Enard W., Szamalek J.M., Enard W., Szamalek J.M., Szamalek J.M., et al. Identification of large-scale human-specific copy number differences by inter-species array comparative genomic hybridization. Hum. Genet. 2006;119:185–198. doi: 10.1007/s00439-005-0130-9. [DOI] [PubMed] [Google Scholar]
- Heppell-Parton A.C., Nacheva E., Carter N.P., Rabbitts P.H., Nacheva E., Carter N.P., Rabbitts P.H., Carter N.P., Rabbitts P.H., Rabbitts P.H. A combined approach of conventional and molecular cytogenetics for detailed karyotypic analysis of the small cell lung carcinoma cell line U2020. Cancer Genet. Cytogenet. 1999;108:110–119. doi: 10.1016/s0165-4608(98)00130-7. [DOI] [PubMed] [Google Scholar]
- Imreh S., Wang Y., Panda C.K., Babonits M., Axelson H., Silva S., Szeles A., Wiener F., Klein G., Wang Y., Panda C.K., Babonits M., Axelson H., Silva S., Szeles A., Wiener F., Klein G., Panda C.K., Babonits M., Axelson H., Silva S., Szeles A., Wiener F., Klein G., Babonits M., Axelson H., Silva S., Szeles A., Wiener F., Klein G., Axelson H., Silva S., Szeles A., Wiener F., Klein G., Silva S., Szeles A., Wiener F., Klein G., Szeles A., Wiener F., Klein G., Wiener F., Klein G., Klein G. Hypersomy of chromosome 15 with retrovirally rearranged c-myc, loss of germline c-myc and IgK/c-myc juxtaposition in a macrophage-monocytic tumour line. Eur. J. Cancer. 1994;7:994–1002. doi: 10.1016/0959-8049(94)90131-7. [DOI] [PubMed] [Google Scholar]
- Imreh S., Klein G., Zabarovsky E.R., Klein G., Zabarovsky E.R., Zabarovsky E.R. Search for unknown tumor-antagonizing genes. Genes Chromosomes Cancer. 2003;38:307–321. doi: 10.1002/gcc.10271. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodnyuk I.D., Kost-Alimova M., Yang Y., Kiss H., Fedorova L., Klein G., Imreh S., Kost-Alimova M., Yang Y., Kiss H., Fedorova L., Klein G., Imreh S., Yang Y., Kiss H., Fedorova L., Klein G., Imreh S., Kiss H., Fedorova L., Klein G., Imreh S., Fedorova L., Klein G., Imreh S., Klein G., Imreh S., Imreh S. The microcell hybrid-based “elimination test” identifies a 1-Mb putative tumor-suppressor region at 3p22.2-p22.1 centromeric to the homozygous deletion region detected in lung cancer. Genes Chromosomes Cancer. 2002;34:341–344. doi: 10.1002/gcc.10068. [DOI] [PubMed] [Google Scholar]
- Kiss H., Darai E., Kiss C., Kost-Alimova M., Klein G., Dumanski J.P., Imreh S., Darai E., Kiss C., Kost-Alimova M., Klein G., Dumanski J.P., Imreh S., Kiss C., Kost-Alimova M., Klein G., Dumanski J.P., Imreh S., Kost-Alimova M., Klein G., Dumanski J.P., Imreh S., Klein G., Dumanski J.P., Imreh S., Dumanski J.P., Imreh S., Imreh S. Comparative human/murine sequence analysis of the common eliminated region 1 from human 3p21.3. Mamm. Genome. 2002;13:646–655. doi: 10.1007/s00335-002-3037-y. [DOI] [PubMed] [Google Scholar]
- Kost-Alimova M., Imreh S., Imreh S. Modeling non-random deletions in cancer. Semin. Cancer Biol. 2007;17:19–30. doi: 10.1016/j.semcancer.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Kost-Alimova M., Kiss H., Fedorova L., Yang Y., Dumanski J.P., Klein G., Imreh S., Kiss H., Fedorova L., Yang Y., Dumanski J.P., Klein G., Imreh S., Fedorova L., Yang Y., Dumanski J.P., Klein G., Imreh S., Yang Y., Dumanski J.P., Klein G., Imreh S., Dumanski J.P., Klein G., Imreh S., Klein G., Imreh S., Imreh S. Coincidence of synteny breakpoints with malignancy-related deletions on human chromosome 3. Proc. Natl. Acad. Sci. 2003;100:6622–6627. doi: 10.1073/pnas.0430971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost-Alimova M., Fedorova L., Yang Y., Klein G., Imreh S., Fedorova L., Yang Y., Klein G., Imreh S., Yang Y., Klein G., Imreh S., Klein G., Imreh S., Imreh S. Microcell-mediated chromosome transfer provides evidence that polysomy promotes structural instability in tumor cell chromosomes through asynchronous replication and breakage within late-replicating regions. Genes Chromosomes Cancer. 2004;40:316–324. doi: 10.1002/gcc.20054. [DOI] [PubMed] [Google Scholar]
- Kost-Alimova M., Darai-Ramqvist E., Yau W.L., Sandlund A., Fedorova L., Yang Y., Kholodnyuk I., Cheng Y., Li Lung M., Stanbridge E., Darai-Ramqvist E., Yau W.L., Sandlund A., Fedorova L., Yang Y., Kholodnyuk I., Cheng Y., Li Lung M., Stanbridge E., Yau W.L., Sandlund A., Fedorova L., Yang Y., Kholodnyuk I., Cheng Y., Li Lung M., Stanbridge E., Sandlund A., Fedorova L., Yang Y., Kholodnyuk I., Cheng Y., Li Lung M., Stanbridge E., Fedorova L., Yang Y., Kholodnyuk I., Cheng Y., Li Lung M., Stanbridge E., Yang Y., Kholodnyuk I., Cheng Y., Li Lung M., Stanbridge E., Kholodnyuk I., Cheng Y., Li Lung M., Stanbridge E., Cheng Y., Li Lung M., Stanbridge E., Li Lung M., Stanbridge E., Stanbridge E., et al. Mandatory chromosomal segment balance in aneuploid tumor cells. BMC Cancer. 2007;7:21. doi: 10.1186/1471-2407-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Emanuel B.S., Emanuel B.S. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Shaikh T., Takata M., Toda T., Emanuel B.S., Shaikh T., Takata M., Toda T., Emanuel B.S., Takata M., Toda T., Emanuel B.S., Toda T., Emanuel B.S., Emanuel B.S. The constitutional t(17;22): Another translocation mediated by palindromic AT-rich repeats. Am. J. Hum. Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R., Bensimon A., Bensimon A. DNA replication origin plasticity and perturbed fork progression in human inverted repeats. Mol. Cell. Biol. 2005;25:6789–6797. doi: 10.1128/MCB.25.15.6789-6797.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke D.P., Archidiacono N., Misceo D., Cardone M.F., Deschamps S., Roe B., Rocchi M., Eichler E.E., Archidiacono N., Misceo D., Cardone M.F., Deschamps S., Roe B., Rocchi M., Eichler E.E., Misceo D., Cardone M.F., Deschamps S., Roe B., Rocchi M., Eichler E.E., Cardone M.F., Deschamps S., Roe B., Rocchi M., Eichler E.E., Deschamps S., Roe B., Rocchi M., Eichler E.E., Roe B., Rocchi M., Eichler E.E., Rocchi M., Eichler E.E., Eichler E.E. Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster. Genome Biol. 2003;4:R50. doi: 10.1186/gb-2003-4-8-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloni-Ehrig A.M. Renal cancer: Cytogenetic and molecular genetic aspects. Am. J. Med. Genet. 2002;115:164–172. doi: 10.1002/ajmg.10697. [DOI] [PubMed] [Google Scholar]
- Muller S., Stanyon R., Finelli P., Archidiacono N., Wienberg J., Stanyon R., Finelli P., Archidiacono N., Wienberg J., Finelli P., Archidiacono N., Wienberg J., Archidiacono N., Wienberg J., Wienberg J. Molecular cytogenetic dissection of human chromosomes 3 and 21 evolution. Proc. Natl. Acad. Sci. 2000;97:206–211. doi: 10.1073/pnas.97.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Finelli P., Neusser M., Wienberg J., Finelli P., Neusser M., Wienberg J., Neusser M., Wienberg J., Wienberg J. The evolutionary history of human chromosome 7. Genomics. 2004;84:458–467. doi: 10.1016/j.ygeno.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Agarwala R., Schaffer A.A., Stephens R., Smith C., Crumpler N.J., David V.A., O'Brien S.J., Agarwala R., Schaffer A.A., Stephens R., Smith C., Crumpler N.J., David V.A., O'Brien S.J., Schaffer A.A., Stephens R., Smith C., Crumpler N.J., David V.A., O'Brien S.J., Stephens R., Smith C., Crumpler N.J., David V.A., O'Brien S.J., Smith C., Crumpler N.J., David V.A., O'Brien S.J., Crumpler N.J., David V.A., O'Brien S.J., David V.A., O'Brien S.J., O'Brien S.J. A rhesus macaque radiation hybrid map and comparative analysis with the human genome. Genomics. 2005a;86:383–395. doi: 10.1016/j.ygeno.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Chowdhary B.P., Galibert F., Gatzke L., Galibert F., Gatzke L., Gatzke L., et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005b;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Nahon J.L. Birth of 'human-specific' genes during primate evolution. Genetica. 2003;118:193–208. doi: 10.1023/a:1024157714736. [DOI] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Fiegler H., Shapero M.H., Carson A.R., Chen W., Shapero M.H., Carson A.R., Chen W., Carson A.R., Chen W., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonte R.V., Eichler E.E., Eichler E.E. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 2002;3:65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- Schmegner C., Hameister H., Vogel W., Assum G., Hameister H., Vogel W., Assum G., Vogel W., Assum G., Assum G. Isochores and replication time zones: A perfect match. Cytogenet. Genome Res. 2007;116:167–172. doi: 10.1159/000098182. [DOI] [PubMed] [Google Scholar]
- Schulz W.A., Steinhoff C., Florl A.R., Steinhoff C., Florl A.R., Florl A.R. Methylation of endogenous human retroelements in health and disease. Curr. Top. Microbiol. Immunol. 2006;310:211–250. doi: 10.1007/3-540-31181-5_11. [DOI] [PubMed] [Google Scholar]
- Schwartz S., Zhang Z., Frazer K.A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W., Zhang Z., Frazer K.A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W., Frazer K.A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W., Bouck J., Gibbs R., Hardison R., Miller W., Gibbs R., Hardison R., Miller W., Hardison R., Miller W., Miller W. PipMaker–a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M., Zlotorynski E., Kerem B., Zlotorynski E., Kerem B., Kerem B. The molecular basis of common and rare fragile sites. Cancer Lett. 2006;232:13–26. doi: 10.1016/j.canlet.2005.07.039. [DOI] [PubMed] [Google Scholar]
- Shaw C.J., Lupski J.R., Lupski J.R. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum. Mol. Genet. 2004;13:R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- She X., Liu G., Ventura M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Liu G., Ventura M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Ventura M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Zhao S., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Rocchi M., Green E.D., Archidiacano N., Green E.D., Archidiacano N., Archidiacano N., et al. A preliminary comparative analysis of primate segmental duplications shows elevated substitution rates and a great-ape expansion of intrachromosomal duplications. Genome Res. 2006;16:576–583. doi: 10.1101/gr.4949406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J.R., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P., Shaw C.J., Dapper J.D., Wakui K., Shaffer L.G., Withers M., Elizondo L., Park S.S., Lupski J.R., Shaw C.J., Dapper J.D., Wakui K., Shaffer L.G., Withers M., Elizondo L., Park S.S., Lupski J.R., Dapper J.D., Wakui K., Shaffer L.G., Withers M., Elizondo L., Park S.S., Lupski J.R., Wakui K., Shaffer L.G., Withers M., Elizondo L., Park S.S., Lupski J.R., Shaffer L.G., Withers M., Elizondo L., Park S.S., Lupski J.R., Withers M., Elizondo L., Park S.S., Lupski J.R., Elizondo L., Park S.S., Lupski J.R., Park S.S., Lupski J.R., Lupski J.R. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am. J. Hum. Genet. 2003;72:1101–1116. doi: 10.1086/374385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P., Shaw C.J., Withers M., Inoue K., Lupski J.R., Shaw C.J., Withers M., Inoue K., Lupski J.R., Withers M., Inoue K., Lupski J.R., Inoue K., Lupski J.R., Lupski J.R. Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res. 2004;14:2209–2220. doi: 10.1101/gr.2746604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Paez I., O'Brien K.P., Kost-Alimova M., Sahlen S., Kedra D., Bruder C.E., Andersson B., Roe B.A., Hu P., Imreh S., O'Brien K.P., Kost-Alimova M., Sahlen S., Kedra D., Bruder C.E., Andersson B., Roe B.A., Hu P., Imreh S., Kost-Alimova M., Sahlen S., Kedra D., Bruder C.E., Andersson B., Roe B.A., Hu P., Imreh S., Sahlen S., Kedra D., Bruder C.E., Andersson B., Roe B.A., Hu P., Imreh S., Kedra D., Bruder C.E., Andersson B., Roe B.A., Hu P., Imreh S., Bruder C.E., Andersson B., Roe B.A., Hu P., Imreh S., Andersson B., Roe B.A., Hu P., Imreh S., Roe B.A., Hu P., Imreh S., Hu P., Imreh S., Imreh S., et al. Fine mapping of the constitutional translocation t(11;22)(q23;q11) Hum. Genet. 2000;106:506–516. doi: 10.1007/s004390000287. [DOI] [PubMed] [Google Scholar]
- Tapia-Paez I., Kost-Alimova M., Hu P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Kost-Alimova M., Hu P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Hu P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Fedorova L., Imreh S., Dumanski J.P., Imreh S., Dumanski J.P., Dumanski J.P. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum. Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- Tomita Y., Bilim V., Kawasaki T., Takahashi K., Okan I., Magnusson K.P., Wiman K.G., Bilim V., Kawasaki T., Takahashi K., Okan I., Magnusson K.P., Wiman K.G., Kawasaki T., Takahashi K., Okan I., Magnusson K.P., Wiman K.G., Takahashi K., Okan I., Magnusson K.P., Wiman K.G., Okan I., Magnusson K.P., Wiman K.G., Magnusson K.P., Wiman K.G., Wiman K.G. Frequent expression of Bcl-2 in renal-cell carcinomas carrying wild-type p53. Int. J. Cancer. 1996;66:322–325. doi: 10.1002/(SICI)1097-0215(19960503)66:3<322::AID-IJC9>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Tsend-Ayush E., Grutzner F., Yue Y., Grossmann B., Hansel U., Sudbrak R., Haaf T., Grutzner F., Yue Y., Grossmann B., Hansel U., Sudbrak R., Haaf T., Yue Y., Grossmann B., Hansel U., Sudbrak R., Haaf T., Grossmann B., Hansel U., Sudbrak R., Haaf T., Hansel U., Sudbrak R., Haaf T., Sudbrak R., Haaf T., Haaf T. Plasticity of human chromosome 3 during primate evolution. Genomics. 2004;83:193–202. doi: 10.1016/j.ygeno.2003.08.012. [DOI] [PubMed] [Google Scholar]
- van den Berg A., Buys C.H., Buys C.H. Involvement of multiple loci on chromosome 3 in renal cell cancer development. Genes Chromosomes Cancer. 1997;19:59–76. doi: 10.1002/(sici)1098-2264(199706)19:2<59::aid-gcc1>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- Ventura M., Weigl S., Carbone L., Cardone M.F., Misceo D., Teti M., D'Addabbo P., Wandall A., Bjorck E., de Jong P.J., Weigl S., Carbone L., Cardone M.F., Misceo D., Teti M., D'Addabbo P., Wandall A., Bjorck E., de Jong P.J., Carbone L., Cardone M.F., Misceo D., Teti M., D'Addabbo P., Wandall A., Bjorck E., de Jong P.J., Cardone M.F., Misceo D., Teti M., D'Addabbo P., Wandall A., Bjorck E., de Jong P.J., Misceo D., Teti M., D'Addabbo P., Wandall A., Bjorck E., de Jong P.J., Teti M., D'Addabbo P., Wandall A., Bjorck E., de Jong P.J., D'Addabbo P., Wandall A., Bjorck E., de Jong P.J., Wandall A., Bjorck E., de Jong P.J., Bjorck E., de Jong P.J., de Jong P.J., et al. Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–1703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., Antonacci F., Cardone M.F., Stanyon R., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Antonacci F., Cardone M.F., Stanyon R., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Cardone M.F., Stanyon R., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Stanyon R., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Eichler E.E., Archidiacono N., Rocchi M., Archidiacono N., Rocchi M., Rocchi M. Evolutionary formation of new centromeres in macaque. Science. 2007;316:243–246. doi: 10.1126/science.1140615. [DOI] [PubMed] [Google Scholar]
- Yang Y., Kiss H., Kost-Alimova M., Kedra D., Fransson I., Seroussi E., Li J., Szeles A., Kholodnyuk I., Imreh M.P., Kiss H., Kost-Alimova M., Kedra D., Fransson I., Seroussi E., Li J., Szeles A., Kholodnyuk I., Imreh M.P., Kost-Alimova M., Kedra D., Fransson I., Seroussi E., Li J., Szeles A., Kholodnyuk I., Imreh M.P., Kedra D., Fransson I., Seroussi E., Li J., Szeles A., Kholodnyuk I., Imreh M.P., Fransson I., Seroussi E., Li J., Szeles A., Kholodnyuk I., Imreh M.P., Seroussi E., Li J., Szeles A., Kholodnyuk I., Imreh M.P., Li J., Szeles A., Kholodnyuk I., Imreh M.P., Szeles A., Kholodnyuk I., Imreh M.P., Kholodnyuk I., Imreh M.P., Imreh M.P., et al. A 1-Mb PAC contig spanning the common eliminated region 1 (CER1) in microcell hybrid-derived SCID tumors. Genomics. 1999;62:147–155. doi: 10.1006/geno.1999.5952. [DOI] [PubMed] [Google Scholar]
- Yue Y., Haaf T., Haaf T. 7E olfactory receptor gene clusters and evolutionary chromosome rearrangements. Cytogenet. Genome Res. 2006;112:6–10. doi: 10.1159/000087507. [DOI] [PubMed] [Google Scholar]
- Yue Y., Grossmann B., Ferguson-Smith M., Yang F., Haaf T., Grossmann B., Ferguson-Smith M., Yang F., Haaf T., Ferguson-Smith M., Yang F., Haaf T., Yang F., Haaf T., Haaf T. Comparative cytogenetics of human chromosome 3q21.3 reveals a hot spot for ectopic recombination in hominoid evolution. Genomics. 2005;85:36–47. doi: 10.1016/j.ygeno.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Yue Y., Tsend-Ayush E., Grutzner F., Grossmann B., Haaf T., Tsend-Ayush E., Grutzner F., Grossmann B., Haaf T., Grutzner F., Grossmann B., Haaf T., Grossmann B., Haaf T., Haaf T. Segmental duplication associated with evolutionary instability of human chromosome 3p25.1. Cytogenet. Genome Res. 2006;112:202–207. doi: 10.1159/000089871. [DOI] [PubMed] [Google Scholar]
- Zabarovsky E.R., Lerman M.I., Minna J.D., Lerman M.I., Minna J.D., Minna J.D. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002;21:6915–6935. doi: 10.1038/sj.onc.1205835. [DOI] [PubMed] [Google Scholar]
- Zhao S., Shetty J., Hou L., Delcher A., Zhu B., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Shetty J., Hou L., Delcher A., Zhu B., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Hou L., Delcher A., Zhu B., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Delcher A., Zhu B., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Zhu B., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Nierman W.C., Strausberg R.L., Fraser C.M., Strausberg R.L., Fraser C.M., Fraser C.M. Human, mouse, and rat genome large-scale rearrangements: Stability versus speciation. Genome Res. 2004;14:1851–1860. doi: 10.1101/gr.2663304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotorynski E., Rahat A., Skaug J., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Rahat A., Skaug J., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Skaug J., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Levi A., Scherer S.W., Margalit H., Kerem B., Scherer S.W., Margalit H., Kerem B., Margalit H., Kerem B., Kerem B. Molecular basis for expression of common and rare fragile sites. Mol. Cell. Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]