Abstract
The genus Populus consists of dioecious woody species with largely unknown genetic mechanisms for gender determination. We have discovered genetic and genomic features in the peritelomeric region of chromosome XIX that suggest this region of the Populus genome is in the process of developing characteristics of a sex chromosome. We have identified a gender-associated locus that consistently maps to this region. Furthermore, comparison of genetic maps across multiple Populus families reveals consistently distorted segregation within this region. We have intensively characterized this region using an F1 interspecific cross involving the female genotype that was used for genome sequencing. This region shows suppressed recombination and high divergence between the alternate haplotypes, as revealed by dense map-based genome assembly using microsatellite markers. The suppressed recombination, distorted segregation, and haplotype divergence were observed only for the maternal parent in this cross. Furthermore, the progeny of this cross showed a strongly male-biased sex ratio, in agreement with Haldane’s rule that postulates that the heterogametic sex is more likely to be absent, rare, or sterile in interspecific crosses. Together, these results support the role of chromosome XIX in sex determination and suggest that sex determination in Populus occurs through a ZW system in which the female is the heterogametic gender.
The genus Populus is composed of dioecious woody plants that arose during a period of global cooling in the late Miocene (5–10 million years ago [Mya]) (Schoell et al. 1994; Shevenell et al. 2004) and consists of six subgenera—Abaso, Leuce, Leucoides, Aigeiros, Turanga, and Tacamahaca (Eckenwalder 1996). Cytological studies reveal that all Populus exist in the diploid form with a haploid number of chromosomes equal to 19, although triploid and tetraploid individuals are commonly observed as well (Smith 1943; Bradshaw and Stettler 1993). The chromosomes of Populus are metacentric, small, and of uniform size (Blackburn and Harrison 1924; Meurman 1925; Erlanson and Hermann 1927; Nakajima 1937). Moreover, there have been no definite sex chromosomes found in any Populus species (Peto 1938; van Buijtenen and Einspahr 1959).
Mechanisms of sex determination in plants are extremely heterogeneous, ranging from XY systems common in many animal species to completely autosomal determination of gender (Ainsworth 2000; Ming et al. 2007). There is evidence that sex chromosomes originate from autosomes, and dioecy almost certainly evolves from ancestral hermaphrodites that lacked sex chromosomes (Muller 1914; Liu et al. 2004). Although Populus trees are normally dioecious, they show evidence of labile sex expression (Stettler 1971; Rowland et al. 2002). The observed sex ratios in certain genetic backgrounds of Salix viminalis, a close dioecious relative of Populus, suggest a multilocus epistatic model of gender determination (Alstrom-Rapaport et al. 1998). However, the genetic mechanism for gender determination in Populus remains largely unknown (McLetchie and Tuskan 1994).
There has been considerable interest recently in the evolution of sex chromosomes, and major insights have emerged from genomic investigations of sex chromosomes in model organisms in recent years, including several dioecious plants (for review, see Charlesworth et al. 2005; Ming and Moore 2007). One salient characteristic of all sex chromosomes described thus far is suppressed recombination, which is seen as essential to the evolution of separate sexes from hermaphroditic organisms when multiple loci are involved in gender determination (Nei 1969; Charlesworth et al. 2005). Furthermore, following suppression of recombination, the chromosomes will begin to diverge in the gender determination region, and one chromosome (the Y in heteromorphic males or the W in heteromorphic females) tends to degrade due to the accumulation of deleterious mutations and large-scale rearrangements and insertions (Charlesworth et al. 2005; Nicolas et al. 2005). Therefore, incipient sex chromosomes should show regions of suppressed recombination with high levels of haplotype divergence.
Due to its experimental tractability and abundant genetic and genomic resources, the genus Populus is widely used in basic scientific investigations geared toward perennial woody plant biology, biofuels production, carbon sequestration, and production of forest products (Tuskan et al. 2004a). Populus genomic resources have grown rapidly in recent years, culminating in the recent release of the whole-genome sequence database (Tuskan et al. 2006). As a sequenced dioecious plant, the genus Populus provides an excellent model and a rare opportunity to study chromosomal-scale evolution, including the transition of autosomes into sex chromosomes. However, to allow inferences about genome structure and chromosomal evolution, multiple genomes from closely related species must be examined. The main objectives of this article are to (1) elucidate the genetic mechanism for gender determination in Populus and (2) determine whether there is a sex chromosome in Populus. To achieve these goals, we established a comprehensive consensus genetic map and conducted extensive map comparisons among several pedigrees, whose parents involve alternate species and the sequenced genotype “Nisqually-1.”
Results
Map construction, chromosome-level assembly, and map comparison
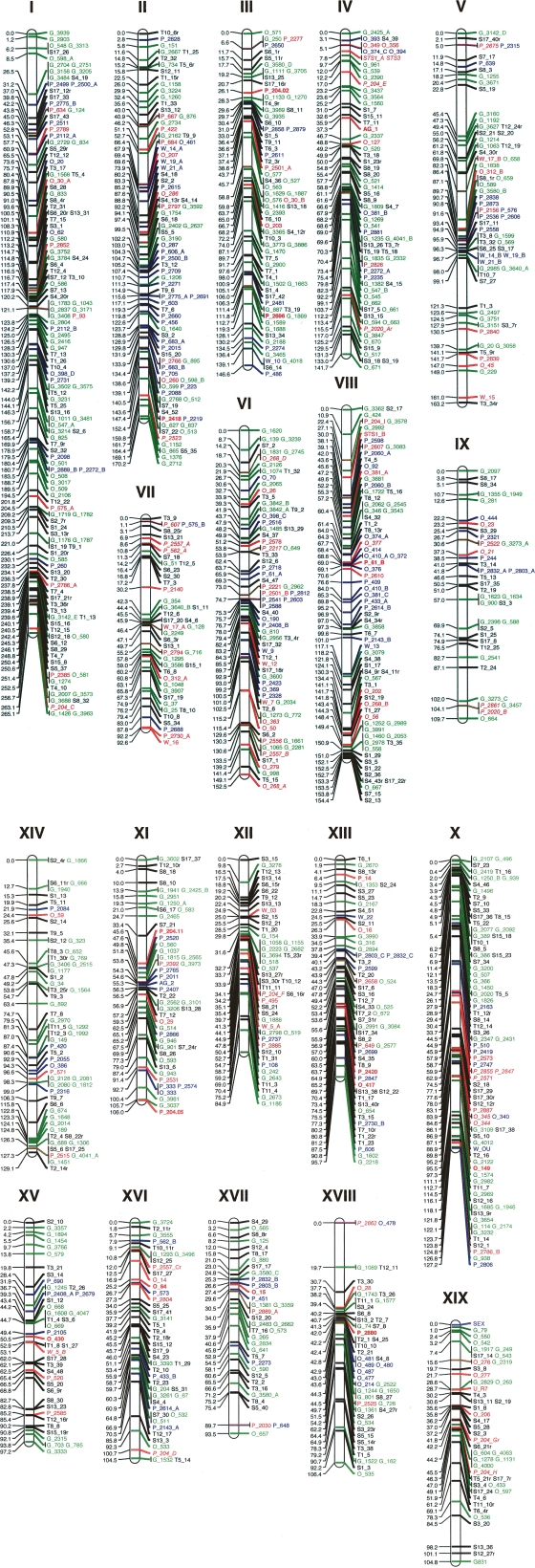
A comprehensive consensus genetic map containing 1138 markers was established for Family 13 and Family 331 (Fig. 1). The overall average genetic length between markers was 2.1 cM, and the average physical length between the mapped simple sequence repeats (SSRs) was <1 Mb. This map achieved inclusive coverage of the entire 480-Mb Populus genome with linkage groups corresponding to the 19 haploid chromosomes. Based on the mapped primer sequence tags, 162 large sequence scaffolds representing a physical distance of 385 Mb were mapped unambiguously along the 19 Populus chromosomes. The vast majority (91%) of the mapped microsatellite markers were colinear with the sequence assembly, indicating that both the genetic map and sequence scaffolds achieved high fidelity. There was a high degree of marker colinearity among the genetic maps built for alternate pedigrees (Supplemental Figs. 2, 3). However, there were some discrepancies in marker order observed among different genetic maps. Scaffold assembly was verified by referencing the marker order derived in Family 545, whose maternal genotype is Nisqually-1. For example, there is a marker, P_2861, in Family 545 on chromosome IX that mapped in divergent order on the consensus map. This marker initially caused a discrepancy in the scaffold assembly for chromosome IX; yet, its position on the Nisqually-1 genetic map was identical to that revealed in the sequence assembly. The alternate position of this marker suggests that there is a chromosomal rearrangement event on chromosome IX in the maternal parent of Family 13 relative to Nisqually-1.
Figure 1.
The consensus genetic map for Populus. (Black) AFLP markers; (red) the framework SSR markers in Family 13 and Family 331; (green) the alternative SSR markers genotyped in Family 13; (blue) the alternative SSR markers genotyped in Family 331; (SEX) the gender determination locus. The markers in bins have the same consensus position. The consensus map was established from genotypes of different pedigrees, and thus, the genetic distance does not reflect the recombination rate between markers but rather only marker orders on the consensus map. Note SSR primers were developed from different sequence data (for marker nomenclature, see text).
Consistent segregation distortion and mapping of the gender determination locus
Based on a genome-wide clustering test for SSR position by chromosome, the average density of SSR was relatively uniform across the Populus genome. However, SSR frequency was significantly higher on chromosomes II and X and significantly lower on chromosome XIX. Thirty-five SSR markers were mapped to the 44.5-cM (∼5.1-Mb) peritelomeric portion of chromosome XIX. In contrast, in the distal portion of chromosome XIX, only four SSR markers were mapped when 21 were expected by chance alone (based on a Poisson distribution with P[bij ≥ λij] = 0.0005). However, based on the Nisqually-1 assembly, SSRs are distributed relatively evenly across chromosome XIX; no substantial SSR clusters or deserts were observed (Supplemental Fig. 4). Together, these findings suggested that physical distance in the peritelomeric portion of chromosome XIX is larger than what the genetic distance indicates and that recombination is suppressed in this region.
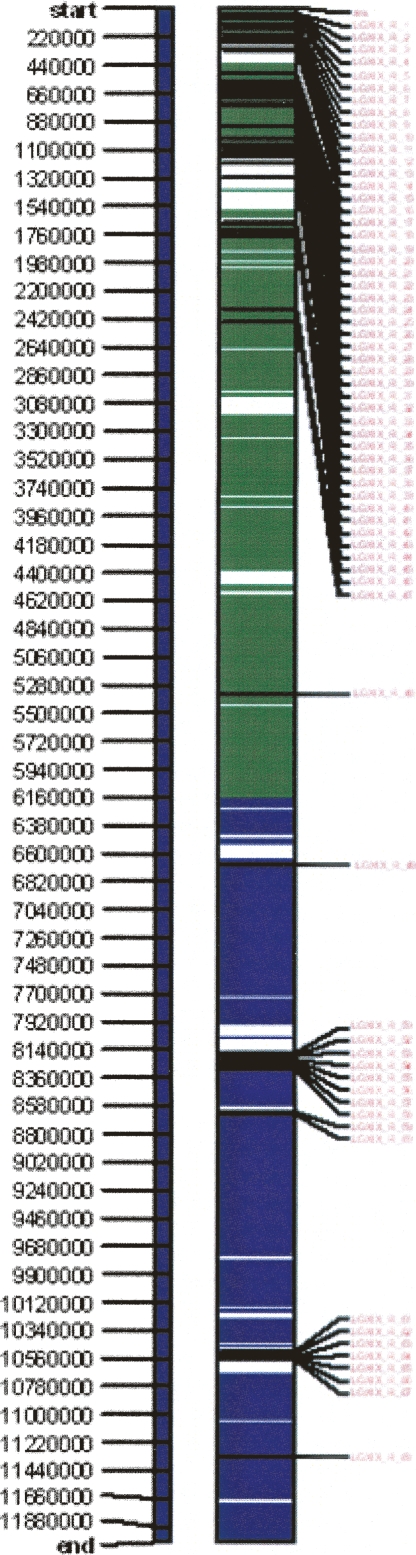
Map comparisons among the different mapping pedigrees demonstrated high marker colinearity on chromosome XIX (Supplemental Figs. 2, 3). However, a striking feature occurred consistently among the examined genomes, i.e., a region of segregation distortion extending over very large portions of chromosome XIX within and among the interspecific mapping pedigrees. Sequence annotation revealed that multiple members of a nucleotide-binding site/leucine rich repeat (NBS-LRR) gene family occur in the peritelomeric region of chromosome XIX. There are 192 members of this gene family in the mapped scaffolds of Nisqually-1, of which 47 are located in a 2.45-Mb segment (composed of 1.77 Mb of assembled reads and 0.68 Mb of captured gaps) occurring at the peritelomeric region on chromosome XIX (Fig. 2). This region contains significantly more NBS-LRR genes than expected by chance alone (P[bij ≥ λij] ≈ 0.0 based on genome-wide average NBS-LRR distribution within a random 2.45-Mb region).
Figure 2.
Forty-seven NBS-LRR genes localize to a 2.45-Mb region at the peritelomeric end of chromosome XIX. The physical length is estimated by the assembled sequence scaffold lengths plus the averaged gap length among scaffolds. The green portion of the vertical bar indicates the distorted region; the blue portion indicates the nondistorted region; the white portion indicates sequence gaps; the horizontal black bars show the physical positions of NBS-LRR genes; and the red letters are the serial number of NBS-LRR genes based on their physical orders along the chromosome. The ruler to the left of the chromosomal bar indicates the physical length in base pairs. Only one NBS-LRR gene would be expected in the corresponding 2.45-Mb region based on a uniform average genome-wide distribution assumption.
Utilizing the consensus map and a 14-yr-old, flowering Populus deltoides × Populus nigra BC1 pedigree (Supplemental Fig. 1), we colocated SSR markers and a locus associated with gender determination to the peritelomeric region of chromosome XIX. This pedigree also showed a significantly skewed gender ratio in favor of male progeny (197 males vs. 115 females). Sequence annotations for the assembled scaffolds mapped on chromosome XIX revealed no obvious candidate genes for gender determination.
Recombination suppression revealed by fine-scale mapping
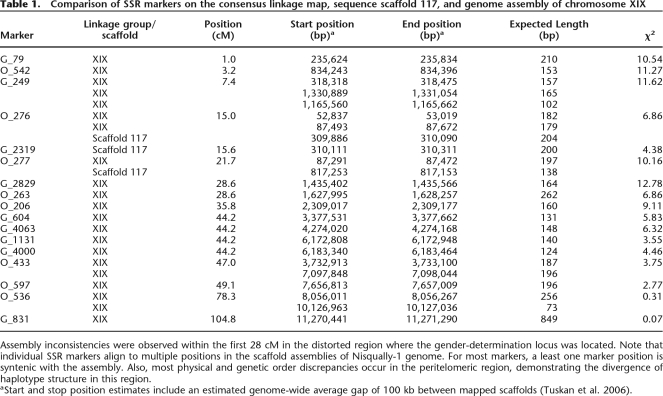
As mentioned above, consistent segregation distortion occurs across the peritelomeric region of chromosome XIX, a region that contains a higher than expected SSR frequency, an overabundance of NBS-LRR genes, and a gender-associated locus. From these observations, we hypothesized that chromosome XIX is an incipient sex chromosome. To resolve the issue of suppressed recombination and to lend support to the hypothesis that chromosome XIX is evolving into a sex chromosome, we created a new integrated assembly for scaffold 117, a single haplotype of chromosome XIX, which was not included in the original map-based assembly (Table 1; Fig. 3; Supplemental Fig. 5). By using Family 545, scaffold 117 was mapped to the peritelomeric region of chromosome XIX using 27 microsatellite markers. However, based on gene content comparisons, we detected substantial divergence between the alternate haplotypes in this region. There were 107 genes on scaffold 117, of which only 30 were shared with chromosome XIX. Strikingly, 26 of these are NBS-LRR genes.
Table 1.
Comparison of SSR markers on the consensus linkage map, sequence scaffold 117, and genome assembly of chromosome XIX
Assembly inconsistencies were observed within the first 28 cM in the distorted region where the gender-determination locus was located. Note that individual SSR markers align to multiple positions in the scaffold assemblies of Nisqually-1 genome. For most markers, a least one marker position is syntenic with the assembly. Also, most physical and genetic order discrepancies occur in the peritelomeric region, demonstrating the divergence of haplotype structure in this region.
aStart and stop position estimates include an estimated genome-wide average gap of 100 kb between mapped scaffolds (Tuskan et al. 2006).
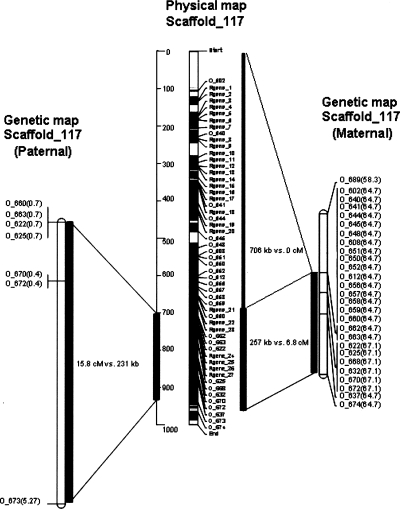
Figure 3.
Relationship between the physical (middle) and gender-specific genetic maps (left and right) of scaffold 117 in Family 545. This scaffold was positioned in the peritelomeric region of chromosome XIX. We detected recombination suppression in the upper 706-kb region of this scaffold containing an overabundant number of NBS-LRR genes. In contrast, the lower 257-kb region is rich with recombination hotspots in both the maternal and paternal parents. Moreover, genotyping revealed more heterozygous loci in the maternal genotype than in the paternal genotype on this scaffold. Values in parentheses are χ2 statistics for tests of segregation distortion. Alleles from the maternal parent showed strong and consistent segregation distortion, whereas alleles from the paternal parent generally conformed to Mendelian expectations (χ2 critical value = 3.84 for α ≤ 0.05).
Fine-scale mapping for scaffold 117 within Family 545 indicated that there is no recombination within the upper 706-kb region of chromosome XIX (Fig. 3). In contrast, the lower portion (a 257-kb region) included (1) several recombination positions, (2) significant segregation distortion (χ2 ≥ 58.3) for alleles located on the maternal haplotypes, and (3) no distortion within the paternal haplotypes. Moreover, recombination suppression extends along the maternal haplotype into a region homologous to the paternal haplotype (i.e., 257 kb representing 6.8 cM in the maternal parent vs. 231 kb representing 15.8 cM in the paternal parent). Therefore, it appears that recombination suppression is confined to the peritelomeric portion of the chromosome XIX containing the gender-association locus.
Discussion
In a previous study, high conservation of SSR priming sequences was observed across Populus species (Tuskan et al. 2004b). In this study, near-complete marker colinearity was observed in pedigrees established from multiple species within the Tacamahaca and Aigeiros subgenera. The comparative mapping results demonstrate that the tested Populus species, and other members of the Salicaceae (Hanley et al. 2006), have maintained the basic genome structure formed following a whole-genome duplication event (Tuskan et al. 2006). Thus, our consensus map extends the sequence database of Populus trichocarpa Nisqually-1 to other Populus and Salix species, supplying a bridge between genetic and genomic information related to candidate genes.
A provocative characteristic of chromosome XIX is the location of a gender-determination locus, which we mapped to the peritelomeric region of chromosome XIX and which has recently been reported in a similar position in a P. nigra pedigree (Gaudet et al. 2007). Markussen et al. (2007) also reported mapping of a gender-determination locus in a Populus tremula × Populus tremuloides pedigree. However, they did not align this locus to chromosome XIX. We were able to align SSR from their map to our consensus map and the primer sequences to the Nisqually-1 genome and confirmed that this locus also maps to the peritelomeric chromosome XIX (Supplemental Fig. 6). Therefore, all three mapping studies validate the position of the gender-determination locus in Populus genome to the peritelomeric region of chromosome XIX.
Although the mapping studies revealed that there is a single locus that controls gender determination, recombination suppression would render all genes within this region as one locus. Thus the gender locus might encompass several genes underlying gender determination. Multigene-based gender determination can be used to explain labile sex expression in Populus. If there is more than one gene determining gender, recombination would impair sexual differentiation. When the recombination suppression is relaxed or translocation of sex-determination loci occurs, hermaphrodites will be generated. To maintain separate sexes, the genes determining maleness or femaleness would have to be closely linked on opposing haplotypes of a single chromosome, and this region would have to develop local mechanisms to prevent recombination (Ohno 1967; Nei 1969). The conclusions of Ohno (1967) and Nei (1969) are consistent with our findings for the female genetic maps and genome assemblies of chromosome XIX. Interestingly, incipient sex chromosomes have also been reported in papaya, and recombination was found to be severely suppressed in the vicinity of the sex determination locus (Liu et al. 2004). Furthermore, Bergero et al. (2007) report several linked sex-related genes in the dioecious plant Silene latifolia.
Multiple lines of evidence point to a ZW sex determination system in Populus, with the female being the heterogametic sex. First, the sequenced tree, Nisqually-1, is a female, and it showed highly divergent haplotypes in the sex determination region. Second, suppressed recombination in this region was only observed in the female parent. Finally, we have observed distorted segregation ratios in only the maternal genotypes of several mapping populations in Populus, including Family 545 and Family 13 (Yin et al. 2004).
The complex haplotype divergence observed in the sex determination region on chromosome XIX could be a contributing mechanism that reinforces recombination suppression (Rick 1969; Chetelat and Meglic 2000). It is also possible that gene content in this region plays a direct role in suppressing recombination, thus contributing to the evolution of a sex chromosome in Populus. There is a distinct overabundance of NBS-LRR genes in this region, and these may play a role as recombination suppressors. Previous work has demonstrated recombination rates that were 20 times lower than expected around MXC3, a locus conferring disease resistance in Populus (Stirling et al. 2001). Recombination suppression has also been observed around the Mla resistance cluster in barley (Wei et al. 1999), the Tm-2a virus resistance gene, the Mi nematode resistance gene in tomato (Ganal et al. 1989), the RPP5 gene conferring resistance to the oomycete pathogen in Arabidopsis (Yang and Hua 2004), and the Hessian fly resistance locus in wheat (Behura et al. 2004). Therefore, recombination suppression seems to be a relatively common feature of R-gene clusters. It is unclear if this is due to structural effects of the complex tandem repeats, if this is due to some direct action of the gene products themselves, or if there is a selective advantage to accumulating tandem clusters of these genes in regions of suppressed recombination.
Consistent segregation distortion was also observed within the peritelomeric region on chromosome XIX across pedigrees, indicating the existence of genomic segments with unequal transmission of the parental alleles to their progeny. This pattern was not due to interspecific differences or selective primer amplification. That is, segregation distortion and recombination suppression were observed for this region of chromosome XIX in Family 545, an F1 pedigree involving a pure P. trichocarpa mother and a pure P. deltoides father. This suggests that the causal mechanism is an inherent characteristic of the alternative haplotypes of chromosome XIX. The relationship between gender determination and segregation distortion, if any, is currently unclear. However, it is interesting to note that NBS-LRR genes have recently been shown to be involved in hybrid necrosis, which is emerging as a primary mechanism for the establishment and maintenance of barriers to gene flow among related species (Bomblies and Weigel 2007). Related mechanisms may create negative genetic interactions between different haplotypes in this region of suppressed recombination, thus contributing to the observed patterns of segregation distortion.
An additional piece of evidence from natural populations is the extensive occurrence of skewed sex ratios in Populus in favor of male trees (Grant and Mitton 1979; Rottenberg et al. 2000). There is some evidence that sex ratios are male-biased in more severe environments in Populus (Farmer 1964; Grant and Mitton 1979; Rottenberg et al. 2000), perhaps driven by differential resource usage by the genders (Casper and Charnov 1982; Bierzychudek and Eckhart 1988). However, male-biased sex ratios were also observed for our pedigree for which gender has been determined in Populus. This suggests that male-biased sex ratios may have a genetic basis in Populus. Haldane’s rule (1922) postulates that in hybrids, the heterogametic sex is more likely to be absent, rare, or sterile (Coyne and Orr 2004). This phenomenon is widespread in plants and animals and is probably due to the effects of deleterious recessive alleles that accumulate on sex chromosomes (Coyne and Orr 2004), coupled with the faster rate of evolution (and divergence) of alleles on sex chromosomes (Charlesworth et al. 1987). Perhaps the incipient heterogametic sex chromosomes in female Populus trees harbor deleterious polymorphisms in linkage disequilibrium with positively selected alleles, which result in reduced average fitness and skewed sex ratios. With the availability of the whole-genome sequence and the initiation of a number of efforts to characterize adaptive polymorphisms and gender determination in natural populations, answers to this and other intriguing questions about the evolutionary biology of this model genus are rapidly emerging.
Methods
Plant materials and mapping of the gender locus
Five mapping pedigrees were used in this study (Supplemental Fig. 1). Family 13 is an interspecific backcross involving female parent 52–225, an F1 hybrid derived from P. trichocarpa (T), 93–968 × P. deltoides (D) (ILL-101), back-crossed to a male parent (F109R6P15), a pure P. deltoides (Yin et al. 2004). Family 331 is an F2 (TD×TD) inbred interspecific hybrid family arising from sib-mating of female 53–246 and male 53–242 of F1 hybrid (T×D) Family 53, which is a cross between a female P. trichocarpa (93–968) and a male P. deltoides (ILL-129). Thus, Family 13 and Family 331 share the same grandmother, clone 93–968, and were used to construct the consensus map. A total of 171 progeny were genotyped in Family 13, while 310 progeny were genotyped in Family 331. Family BCN is a cross between a Populus angustifolia (clone 996) and a natural hybrid (clone WSU-6, a Populus fremontii × P. angustifolia). None of these pedigrees involved the sequenced individual, Nisqually-1. To compare the genetic map and genome sequence of Nisqually-1 directly, we created a genetic map for Family 545, an interspecific F1 pedigree obtained from a cross between P. trichocarpa (clone Nisqually-1) and P. deltoides (clone ILL-101). Among the parental species, P. fremontii, P. deltoides, and P. nigra belong to the Aigeiros subgenus, and P. angustifolia and P. trichocarpa belong to the Tacamahaca subgenus.
The Populus gender-determination locus was mapped in a cross between a female P. deltoides and a male hybrid derived from P. deltoides and P. nigra. The paternal parent was a natural hybrid, whereas the female parent is a pure P. deltoides tree originating from a natural population in Illinois. Among the 312 progeny investigated, 115 were females and 197 were males. A χ2 test was used to test for bias in the sex ratio.
Consensus map construction and genome assembly
Marker generation, genotyping, and nomenclature were as described in Tuskan et al. (2004b) and Yin et al. (2004). Four sources of SSR primers were used for map construction in this study: ORNL primers (O_), PMGC primers (P_), WPMS primers (W_) (Smulders et al. 2001), and GCPM primers (G_). The majority of the G_ SSR primers were designed based on end-sequences of BAC contigs used for Populus physical map construction (Kelleher et al. 2007). The microsatellite locations on scaffold 117 were identified using the Sputnik program (C. Abajian, University of Washington, Seattle). Additional primer information is available at http://www.ornl.gov/sci/ipgc/ssr_resource.htm. In total, we examined 640 SSR primer pairs, selected from 1230 pairs, and generated data for 701 loci for the consensus map construction.
Maternally informative testcross locus data in Family 13 and fully informative markers in Family 331 were linked using JoinMap 3.0 (Van Ooijen and Voorrips, 2001). We first analyzed each population data set separately and then applied the “Combine Groups for Map Integration” command to integrate linkage groups based on shared orthologs. When shared orthologs, defined as a common interval in both pedigrees, were detected, the combined value of the recombination rate was replaced by a weighted average value (Sewell et al. 1999; Stam 2004). The consensus position for the alternative SSR markers was derived by the following formula:
where Pi is the consensus position of the ith alternative SSR marker, Dfram is the size of the framework interval, Dalt is the size of the new interval after new alternative markers added in, Pu is the position of the upper boundary of the framework interval, and Liu is the distance between the ith alternative markers to the upper boundary of the new interval.
Scaffold assembly was conducted with the Populus genome assembly (06252004 version compiled using the JAZZ assembler (Aparicio et al. 2002) at the U.S. Department of Energy Joint Genome Institute [JGI]). SSR priming site sequences were used as queries, and then the genome sequence location was determined based on “best” hits against the sequence scaffolds using the BLASTN (Altschul et al. 1997). Primer hits were accepted only if priming sites were in the correct orientation and separated by the expected physical distance. Multiple SSR were used to map scaffolds to linkage groups in cases where multiple priming sites were present on alternate scaffolds.
Discovery of the genetic features of the incipient sex chromosome
Segregation distortion in the progeny was tested using a χ2 test. NBS-LRR genes were identified using the Advanced Search feature of the poplar genome portal (http://genome.jgi-psf.org/poplar), based on identification of genes with NBS, LRR, TIR, and CNL domains. A total of 528 genes were found, which were subsequently manually curated using the Gene Ontology tool. Genes containing just a TIR or CNL domain were excluded. NBS-LRR gene distribution was visualized by MapChart 2.2 (Voorrips 2006). Thereafter, NBS-LRR gene clusters were evaluated by the observed number in each cluster compared with the whole-genome average number under an assumed Poisson distribution. The average number of NBS-LRR genes in 1-Mb DNA sequence, λ, was calculated as
where mi is the number of NBS-LRR genes detected on chromosome or scaffold i; Li is the base pair length of chromosome or scaffold i. Gene content was analyzed based on gene annotation by Eugene, Grail, Genewise, and FgenesH predictors available at http://genome.jgi-psf.org/cgi-bin/browserLoad/474c6f8a3e06fd24124c70ab. The survey of recombination suppression was conducted using Family 545. The maternal genotype of this pedigree is Nisqually-1, the sequenced individual, so genetic and genomic comparisons were conducted on the same individual.
Acknowledgments
We thank the anonymous reviewers for their help in formulating the final manuscript. Special thanks goes to J.C. Tuskan for his assistance in the final stage of the manuscript. Oak Ridge National Laboratory (ORNL) is managed by UT-Battelle, LLC for the U.S. Department of Energy under contract no. DE-AC05-00OR22725. Funding for this research was provided by the Laboratory Directed Research and Development Program of ORNL and the U.S. Department of Energy, Office of Science, Biological and Environmental Research Carbon Sequestration Program. Funding was also provided by the National Science Foundation FIBR (DEB-0425908), Genome Canada and the Province of BC (Treenomix I project), and the Educational Department of China (NCET-04-0516).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7076308
References
- Ainsworth C. Boys and girls come out to play: The molecular biology of dioecious plants. Ann. Bot. 2000;86:211–221. [Google Scholar]
- Alstrom-Rapaport C., Lascoux M., Wang Y.C., Roberts G., Tuskan G.A., Lascoux M., Wang Y.C., Roberts G., Tuskan G.A., Wang Y.C., Roberts G., Tuskan G.A., Roberts G., Tuskan G.A., Tuskan G.A. Identification of a RAPD marker linked to sex determination in the basket willow (Salix viminalis L.) J. Hered. 1998;89:44–49. [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S., Chapman J., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smith A., Chapman J., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smith A., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smith A., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smith A., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smith A., Dehal P., Christoffels A., Rash S., Hoon S., Smith A., Christoffels A., Rash S., Hoon S., Smith A., Rash S., Hoon S., Smith A., Hoon S., Smith A., Smith A., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Behura S.K., Valicente F.H., Rider S.D., Shun-Chen M., Jackson S., Stuart J.J., Valicente F.H., Rider S.D., Shun-Chen M., Jackson S., Stuart J.J., Rider S.D., Shun-Chen M., Jackson S., Stuart J.J., Shun-Chen M., Jackson S., Stuart J.J., Jackson S., Stuart J.J., Stuart J.J. A physically anchored genetic map and linkage to avirulence reveals recombination suppression over the proximal region of Hessian fly chromosome A2. Genetics. 2004;167:343–355. doi: 10.1534/genetics.167.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R., Forrest A., Kamau E., Charlesworth D., Forrest A., Kamau E., Charlesworth D., Kamau E., Charlesworth D., Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: Evidence from new sex-linked genes. Genetics. 2007;175:1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierzychudek P., Eckhart V., Eckhart V. Spatial segregation of the sexes of dioecious plants. Am. Nat. 1988;132:34–43. [Google Scholar]
- Blackburn K.B., Harrison J.W.H., Harrison J.W.H. A preliminary account of the chromosomes and chromosome behavior in the Salicaceae. Ann. Bot. (Lond.) 1924;38:361–378. [Google Scholar]
- Bomblies K., Weigel D., Weigel D. Hybrid necrosis: Autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 2007;8:382–393. doi: 10.1038/nrg2082. [DOI] [PubMed] [Google Scholar]
- Bradshaw H.D., Stettler R.F., Stettler R.F. Molecular genetics of growth and development in Populus. I. Triploidy in hybrid poplars. Theor. Appl. Genet. 1993;86:301–307. doi: 10.1007/BF00222092. [DOI] [PubMed] [Google Scholar]
- Casper B.B., Charnov E.L., Charnov E.L. Sex allocation in heterostylous plants. J. Theor. Biol. 1982;96:143–149. [Google Scholar]
- Charlesworth D., Charlesworth B., Charlesworth B. The effect of investment in attractive structures on allocation to male and female functions in plants. Evolution. 1987;41:948–968. doi: 10.1111/j.1558-5646.1987.tb05869.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G., Charlesworth B., Marais G., Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Chetelat R.T., Meglic V., Meglic V. Molecular mapping of chromosome segments introgressed from Solanum lycopersicoides into cultivated tomato (Lycopersicon esculentum) Theor. Appl. Genet. 2000;100:232–241. [Google Scholar]
- Coyne J.A., Orr H.A., Orr H.A. Speciation. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- Eckenwalder J.E. Systematics and evolution of Populus. In: Stettler R.F., et al., editors. Biology of Populus and its implications for management and conservation. NRC Research Press; Ottawa: 1996. pp. 7–32. [Google Scholar]
- Erlanson E.W., Hermann F.J., Hermann F.J. The morphology and cytology of perfect flowers in Populus tremuloides. Michx. Papers Mich. Acad. Sci. 1927;8:97–110. [Google Scholar]
- Farmer R.E. Sex ratio and sex-related characteristics in eastern cottonwood. Silvae Genet. 1964;13:116–118. [Google Scholar]
- Ganal M.W., Young N.D., Tanksley S.D., Young N.D., Tanksley S.D., Tanksley S.D. Pulsed field gel electrophoresis and physical mapping of large DNA fragments in the Tm-2a region of chromosome 9 in tomato. Mol. Gen. Genet. 1989;215:395–400. [Google Scholar]
- Gaudet M., Jorge V., Paolucci I., Beritognolo I., Scarascia-Mugnozza G., Sabatti M., Jorge V., Paolucci I., Beritognolo I., Scarascia-Mugnozza G., Sabatti M., Paolucci I., Beritognolo I., Scarascia-Mugnozza G., Sabatti M., Beritognolo I., Scarascia-Mugnozza G., Sabatti M., Scarascia-Mugnozza G., Sabatti M., Sabatti M. Genetic linkage maps of Populus nigra L. including AFLPs, SSRs, SNPs, and sex trait. Tree Genet. Genomes. 2007;4:25–36. [Google Scholar]
- Grant M.C., Mitton J.B., Mitton J.B. Elevational gradients in adult sex-ratios and sexual-differentiation in vegetative growth-rates of Populus tremuloides. Evolution Int. J. Org. Evolution. 1979;33:914–918. doi: 10.1111/j.1558-5646.1979.tb04744.x. [DOI] [PubMed] [Google Scholar]
- Haldane J.B.S. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 1922;12:101–109. [Google Scholar]
- Hanley S.J., Mallott M.D., Karp A., Mallott M.D., Karp A., Karp A. Alignment of a Salix linkage map to the Populus genomic sequence reveals macrosynteny between willow and poplar genomes. Tree Genet. Genomes. 2006;3:35–48. [Google Scholar]
- Kelleher C.T., Chiu R., Shin H., Bosdet I.E., Krzywinski M.I., Fjell C.D., Wilkin J., Yin T.M., DiFazio S.P., Ali J., Chiu R., Shin H., Bosdet I.E., Krzywinski M.I., Fjell C.D., Wilkin J., Yin T.M., DiFazio S.P., Ali J., Shin H., Bosdet I.E., Krzywinski M.I., Fjell C.D., Wilkin J., Yin T.M., DiFazio S.P., Ali J., Bosdet I.E., Krzywinski M.I., Fjell C.D., Wilkin J., Yin T.M., DiFazio S.P., Ali J., Krzywinski M.I., Fjell C.D., Wilkin J., Yin T.M., DiFazio S.P., Ali J., Fjell C.D., Wilkin J., Yin T.M., DiFazio S.P., Ali J., Wilkin J., Yin T.M., DiFazio S.P., Ali J., Yin T.M., DiFazio S.P., Ali J., DiFazio S.P., Ali J., Ali J., et al. A physical map of the highly heterozygous Populus genome: integration with the genome sequence and genetic map and analysis of haplotype variation. Plant J. 2007;50:1063–1078. doi: 10.1111/j.1365-313X.2007.03112.x. [DOI] [PubMed] [Google Scholar]
- Liu Z., Moore P.H., Ma H., Ackerman C.M., Ragiba M., Yu Q., Pearl H.M., Kim M.S., Charlton J.W., Stiles J.I., Moore P.H., Ma H., Ackerman C.M., Ragiba M., Yu Q., Pearl H.M., Kim M.S., Charlton J.W., Stiles J.I., Ma H., Ackerman C.M., Ragiba M., Yu Q., Pearl H.M., Kim M.S., Charlton J.W., Stiles J.I., Ackerman C.M., Ragiba M., Yu Q., Pearl H.M., Kim M.S., Charlton J.W., Stiles J.I., Ragiba M., Yu Q., Pearl H.M., Kim M.S., Charlton J.W., Stiles J.I., Yu Q., Pearl H.M., Kim M.S., Charlton J.W., Stiles J.I., Pearl H.M., Kim M.S., Charlton J.W., Stiles J.I., Kim M.S., Charlton J.W., Stiles J.I., Charlton J.W., Stiles J.I., Stiles J.I., et al. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature. 2004;427:348–352. doi: 10.1038/nature02228. [DOI] [PubMed] [Google Scholar]
- Markussen T., Pakull B., Fladung M., Pakull B., Fladung M., Fladung M. Positioning of sex-correlated markers for Populus in an AFLP- and SSR-marker based genetic map of Populus tremula × tremuloides. Silvae Genet. 2007;56:180–184. [Google Scholar]
- McLetchie D.N., Tuskan G.A., Tuskan G.A. Gender determination in Populus. Norw. J. Agric. Sci. 1994;18:57–66. [Google Scholar]
- Meurman O. The chromosome behavior of some dioecious plants and their relatives with special reference to the sex chromosomes. Comm. Biol. Soc. Sci. Fennica. 1925;2:1–104. [Google Scholar]
- Ming R., Moore P.H., Moore P.H. Genomics of sex chromosomes. Curr. Opin. Plant Biol. 2007;10:123–130. doi: 10.1016/j.pbi.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Ming R., Wang J., Moore P.H., Paterson A.H., Wang J., Moore P.H., Paterson A.H., Moore P.H., Paterson A.H., Paterson A.H. Sex chromosomes in flowering plants. Am. J. Bot. 2007;94:141–150. doi: 10.3732/ajb.94.2.141. [DOI] [PubMed] [Google Scholar]
- Muller H.J. A gene for the fourth chromosome of Drosophila. J. Exp. Zool. 1914;17:325–336. [Google Scholar]
- Nakajima G. Cytological studies in some dioecious plants. Cytologia Fuji Jub. 1937:282–292. [Google Scholar]
- Nei M. Linkage modification and sex difference in recombination. Genetics. 1969;63:681–699. doi: 10.1093/genetics/63.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M., Marais G., Hykelova V., Janousek B., Laporte V., Vyskot B., Mouchiroud D., Negrutiu I., Charlesworth D., Moneger F., Marais G., Hykelova V., Janousek B., Laporte V., Vyskot B., Mouchiroud D., Negrutiu I., Charlesworth D., Moneger F., Hykelova V., Janousek B., Laporte V., Vyskot B., Mouchiroud D., Negrutiu I., Charlesworth D., Moneger F., Janousek B., Laporte V., Vyskot B., Mouchiroud D., Negrutiu I., Charlesworth D., Moneger F., Laporte V., Vyskot B., Mouchiroud D., Negrutiu I., Charlesworth D., Moneger F., Vyskot B., Mouchiroud D., Negrutiu I., Charlesworth D., Moneger F., Mouchiroud D., Negrutiu I., Charlesworth D., Moneger F., Negrutiu I., Charlesworth D., Moneger F., Charlesworth D., Moneger F., Moneger F.2005A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants PLoS Biol. 3e4 . 10.1371/journal.pbio.0030004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Sex chromosomes and sex-linked genes. Springer-Verlag; New York: 1967. [Google Scholar]
- Peto F.H. Cytology of poplar species and natural hybrids. Can. J. For. Res. 1938;16:445–455. [Google Scholar]
- Rick C.M. Controlled introgression of chromosomes of Solanum pennellii into Lycopersicon esculentum: Segregation and recombination. Genetics. 1969;62:753–768. doi: 10.1093/genetics/62.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg A., Nevo E., Zohary D., Nevo E., Zohary D., Zohary D. Genetic variability in sexually dimorphic and monomorphic populations of Populus euphratica (Salicaceae) Can. J. For. Res. 2000;30:482–486. [Google Scholar]
- Rowland D.L., Garner R., Jespersen M., Garner R., Jespersen M., Jespersen M. A rare occurrence of seed formation on male branches of the dioecious tree, Populus deltoides. Am. Midl. Nat. 2002;147:185–187. [Google Scholar]
- Schoell M., Schouten S., Damste J.S.S., de Leeuw J.W., Summons R.E., Schouten S., Damste J.S.S., de Leeuw J.W., Summons R.E., Damste J.S.S., de Leeuw J.W., Summons R.E., de Leeuw J.W., Summons R.E., Summons R.E. A molecular organic carbon isotope record of Miocene climate changes. Science. 1994;263:1122–1125. doi: 10.1126/science.263.5150.1122. [DOI] [PubMed] [Google Scholar]
- Sewell M.M., Sherman B.K., Neale D.B., Sherman B.K., Neale D.B., Neale D.B. A consensus map for loblolly pine (Pinus taeda L.). I. Construction and integration of individual linkage maps from two outbred three-generation pedigrees. Genetics. 1999;151:321–330. doi: 10.1093/genetics/151.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevenell A.E., Kennett J.P., Lea D.W., Kennett J.P., Lea D.W., Lea D.W. Middle Miocene southern ocean cooling and Antarctic cryosphere expansion. Science. 2004;305:1766–1770. doi: 10.1126/science.1100061. [DOI] [PubMed] [Google Scholar]
- Smith E.C. A study of cytology and speciation in the genus Populus L. J. Arnold Arbor. 1943;24:275–305. [Google Scholar]
- Smulders M.J.M., Van Der Schoot J., Arens P., Vosman B., Van Der Schoot J., Arens P., Vosman B., Arens P., Vosman B., Vosman B. Trinucleotide repeat microsatellite markers for black poplar (Populus nigra L.) Mol. Ecol. Notes. 2001;1:188–190. [Google Scholar]
- Stam P. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 2004;3:739–744. [Google Scholar]
- Stettler R. Variation in sex expression of black cottonwood and related hybrids. Silvae Genet. 1971;20:42–46. [Google Scholar]
- Stirling B., Newcombe G., Vrebalov J., Bosdet I., Bradshaw H.D., Newcombe G., Vrebalov J., Bosdet I., Bradshaw H.D., Vrebalov J., Bosdet I., Bradshaw H.D., Bosdet I., Bradshaw H.D., Bradshaw H.D. Suppressed recombination around the MXC3 locus, a major NBS-LRR gene for resistance to poplar leaf rust. Theor. Appl. Genet. 2001;103:1129–1137. [Google Scholar]
- Tuskan G.A., DiFazio S.P., Teichmann T., DiFazio S.P., Teichmann T., Teichmann T. Poplar genomics is getting popular: The impact of the poplar genome project on tree research. Plant Biol. 2004a;6:2–4. doi: 10.1055/s-2003-44715. [DOI] [PubMed] [Google Scholar]
- Tuskan G.A., Gunter L.E., Yang Z.M., Yin T.M., Sewell M.M., DiFazio S.P., Gunter L.E., Yang Z.M., Yin T.M., Sewell M.M., DiFazio S.P., Yang Z.M., Yin T.M., Sewell M.M., DiFazio S.P., Yin T.M., Sewell M.M., DiFazio S.P., Sewell M.M., DiFazio S.P., DiFazio S.P. Characterization of microsatellites revealed by genomic sequencing of Populus trichocarpa. Can. J. For. Res. 2004b;34:85–93. [Google Scholar]
- Tuskan G.A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Putnam N., Ralph S., Rombauts S., Salamov A., Ralph S., Rombauts S., Salamov A., Rombauts S., Salamov A., Salamov A., et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- van Buijtenen J.P., Einspahr D.W., Einspahr D.W. Note on the presence of sex chromosomes in Populus tremuloides. Bot. Gaz. 1959;121:60–61. [Google Scholar]
- Van Ooijen J.W., Voorrips R.E., Voorrips R.E. JoinMap 3.0, Software for the calculation of genetic linkage maps. Plant Research International; Wageningen, The Netherlands: 2001. [Google Scholar]
- Voorrips R.E. MapChart 2.2: Software for the graphical presentation of linkage maps and QTLs. Plant Research International; Wageningen, The Netherlands: 2006. [Google Scholar]
- Wei F., Gobelman-Werner K., Morroll S.M., Kurth J., Mao L., Wing R., Leister D., Schulze-Lefert P., Wise R.P., Gobelman-Werner K., Morroll S.M., Kurth J., Mao L., Wing R., Leister D., Schulze-Lefert P., Wise R.P., Morroll S.M., Kurth J., Mao L., Wing R., Leister D., Schulze-Lefert P., Wise R.P., Kurth J., Mao L., Wing R., Leister D., Schulze-Lefert P., Wise R.P., Mao L., Wing R., Leister D., Schulze-Lefert P., Wise R.P., Wing R., Leister D., Schulze-Lefert P., Wise R.P., Leister D., Schulze-Lefert P., Wise R.P., Schulze-Lefert P., Wise R.P., Wise R.P. The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics. 1999;153:1929–1948. doi: 10.1093/genetics/153.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Hua J., Hua J. A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell. 2004;16:1060–1071. doi: 10.1105/tpc.020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T.M., DiFazio S.P., Gunter L.E., Riemenschneider D., Tuskan G.A., DiFazio S.P., Gunter L.E., Riemenschneider D., Tuskan G.A., Gunter L.E., Riemenschneider D., Tuskan G.A., Riemenschneider D., Tuskan G.A., Tuskan G.A. Large-scale heterospecific segregation distortion in Populus revealed by a dense genetic map. Theor. Appl. Genet. 2004;109:451–463. doi: 10.1007/s00122-004-1653-5. [DOI] [PubMed] [Google Scholar]