Abstract
A large number of cis-regulatory motifs involved in transcriptional control have been identified, but the regulatory context and biological processes in which many of them function are unknown. Here, we computationally identify the sets of human core promoters targeted by motifs, and systematically characterize their function by using a robust gene-set-based approach and diverse sources of biological data. We find that the target sets of most motifs contain both genes with similar function and genes that are coregulated in vivo, thereby suggesting both the biological process regulated by the motifs and the conditions in which this regulation may occur. Our analysis also identifies many motifs whose target sets are predicted to be regulated by a common microRNA, suggesting a connection between transcriptional and post-transcriptional control processes. Finally, we predict novel roles for uncharacterized motifs in the regulation of specific biological processes and certain types of human cancer, and experimentally validate four such predictions, suggesting regulatory roles for four uncharacterized motifs in cell cycle progression. Our analysis thus provides a concrete framework for uncovering the biological function of cis-regulatory motifs genome wide.
Coordinated control of gene expression is key in nearly all biological processes. The instructions for achieving this coordination are encoded in the DNA sequence by a regulatory network that, for each gene, specifies a small number of transcription factors responsible for controlling its expression. Each transcription factor recognizes short DNA-binding site motifs, typically 6–12 bp in length, and by binding these sites can induce or inhibit transcription of the nearby gene. Various experimental and computational approaches, most notably genome-scale chromatin immunoprecipitation analysis (ChIP-chip) and comparative genomic methods that exploit evolutionary conservation (Cliften et al. 2003; Kellis et al. 2003; Xie et al. 2005), have had much success in cataloging regulatory motifs for transcription factors (Wingender et al. 2001). However, the next task of identifying the biological functions regulated by these motifs remains an important challenge, and the function of many of the motifs is not known or is poorly characterized based on examination of a handful of target genes of the motif.
Here, we present a gene-set-based approach to systematically characterize the function of cis-regulatory motifs in human core promoters. Our approach consists of two main steps. First, we use a probabilistic approach to computationally identify the targets of each motif and motif combination. Next, we identify the biological function of the motifs and the conditions in which they are active by testing the overlap of their predicted targets with sets of genes known to have similar biological functions and analyze the behavior of their targets in large compendiums of gene expression profiles.
For the first step of identifying the targets of each motif, simply searching for motif occurrences in promoters results in many false positive predictions, since the motifs, being short sequences, appear by happenstance in many promoter regions. For this reason, approaches for identifying transcription-factor targets genome wide are based on discovering statistically significant clusters of motifs (Berman et al. 2002; Blanchette et al. 2006; Chang et al. 2006; Hallikas et al. 2006). We previously developed a hidden Markov model (HMM) for this task (Sinha et al. 2003), which computes the likelihood that a regulatory region was generated by the model, thereby removing the need for using ad-hoc thresholds and conservation filters for defining motifs. Through extensive experimental validation, our approach was shown to have much utility for identifying regulatory regions in fly (Schroeder et al. 2004). Here, we extend our HMM model into a computational pipeline for identifying targets of both individual motifs and motif combinations (pairs and triplets of nonredundant motifs), and apply it to human core promoters, thus deriving a “motif target map” of human (Fig. 1A). This map assigns a set of core promoters (and respective genes) as being the target set of each motif or motif combination considered.
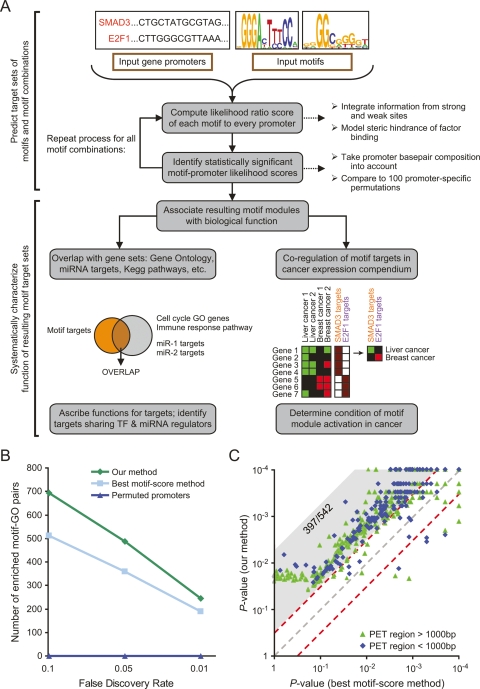
Figure 1.
Overview of our approach and comparison to other methods. (A) Flowchart of our motif functional characterization pipeline. (B) Comparison of the number of significant enrichments between gene sets from Gene Ontology (GO) (Ashburner et al. 2000) and motif modules defined using our approach, GO and an alternative approach based on best motif occurrences, or GO and our approach applied to permuted promoters. For each particular FDR threshold f, the comparison shows the number of GO-motif target set pairs with FDR < f. (C) Comparison of the ability of our method (Y-axis) and best motif occurrences method (X-axis) to predict TP53 binding to 542 sequences experimentally measured to be strongly bound to TP53 by genome-wide chromatin immunoprecipitation followed by sequencing of paired end ditags (PET) (Wei et al. 2006). For each experimentally identified PET region, shown is the predicted binding P-value of each method, determined by comparing the likelihood score of the original region to that obtained in 10,000 regions randomly selected from the human genome. PET regions are separated into two types by their length (green and blue). Of the 542 PET regions, 397 have a fivefold lower P-value in our method (gray shaded area above diagonal), compared with nine with a fivefold lower P-value in the score-based method.
Given these target sets, our second step attempts to characterize the biological function of each motif by testing the association of its target set with biologically meaningful gene sets. Motifs whose targets are significantly enriched with genes of similar biological function are probable candidates for regulating that biological function. Similarly, motifs whose targets are coordinately expressed in specific genome-wide expression microarrays are likely to regulate their targets under the biological conditions represented by those microarrays. A critical feature of our approach is that when characterizing the function of each motif, all of the analyses are done at the gene-set level by comparing the target set of the motif to biologically meaningful gene sets. Since significant enrichments can still be identified even if some members of the compared gene sets are incorrectly classified, such a gene-set-based approach will be robust to errors in predictions of individual motif–target interactions from the first step, and can thus be used for associating biological functions to motifs with high confidence.
Our analysis reconstructs validated roles for known motifs, suggests novel roles for uncharacterized motifs in the regulation of diverse biological processes, and identifies links between transcriptional and post-transcriptional control processes by the finding that targets of many motifs are also predicted to have a common microRNA regulator. By examining the behavior of motif targets in large compendiums of genome-wide expression profiles, we identify prominent roles for many uncharacterized motifs in the dysregulation of gene expression in certain types of human cancers, suggesting novel mechanisms of pathogenesis. Finally, we experimentally validate four such novel predictions generated by our approach, suggesting regulatory roles for four uncharacterized motifs in cell cycle progression.
Results
Constructing a motif target map of human core promoters
To construct a motif target map of human core promoters, we applied our computational pipeline (described below) to find targets of 432 motifs (Wingender et al. 2001; Xie et al. 2005) in 17,365 human core promoters, defined as the 500 bp upstream and 200 bp downstream from the annotated transcription start site. This choice of promoter size is motivated by the finding that evolutionarily conserved motifs show a strong bias toward these regions (Xie et al. 2005). The 432 motifs included 258 experimentally characterized motifs (Wingender et al. 2001) and 174 computationally predicted motifs (Xie et al. 2005).
To identify targets of individual motifs, the pipeline uses our HMM-based model (Sinha et al. 2003) to compute a likelihood score for each input motif and promoter sequence, which captures the number and strengths of occurrences of the motif in the sequence. This computation integrates contributions from both weak and strong binding sites and does not use any arbitrary cutoffs on binding-site strength. To take the local basepair composition of each input sequence into account, we compare this likelihood score with that obtained in a fixed number of permutations of the tested sequence, and only consider as significant the likelihood scores that are higher than those obtained in all permutations. Such comparisons are particularly important in the human genome due to the large variability in basepair composition among promoters in general and among promoters of particular gene sets (Supplemental Fig. S1; Supplemental Table S1).
Next, we systematically identify motif combinations that are likely to act together on a set of target genes. Specifically, we iterate over every pair and three-way combination of constituent motifs and report it as a motif combination if it meets two criteria. First, the target set of the putative motif combination, as determined by intersecting the target set of its constituent motifs, must be significantly larger than that expected by chance. Second, the likelihood score of the motif combination has to be significantly higher than that obtained from each constituent motif alone. This latter requirement allows us to distinguish between true combinatorial interactions and apparent interactions due to redundant or similar motifs. Finally, by iterating this step of identifying motif combinations, we identify motif combinations of higher order (Fig. 1A).
Overall, we found target sets for all individual motifs and for 471 motif combinations that were deemed significant based on the above criteria. Each of these 903 target sets, henceforth called “motif modules”, thus represents a unique combination of enriched transcription-factor binding sites. As a negative control, we applied the same procedure to a promoter set generated by permuting the sequence of each promoter. We found only 41,015 predicted targets (for individual motifs) in these permutations, significantly less than the 281,012 targets found in the real promoter set. Moreover, the permuted promoter set did not produce any significant motif combinations, strongly validating the specificity of our pipeline.
As an independent validation of our method, we tested whether genes in the predicted target sets (motif modules) are significantly enriched in nonredundant biological categories from Gene Ontology (Ashburner et al. 2000). Indeed, among ∼4 × 105 (motif, GO category) pairs that were tested, 487 pairs, covering 160 different motifs and 164 different GO categories, significantly associated at a false discovery rate (FDR) threshold of 0.05. Moreover, no such statistical association was observed in the negative control of permuted promoters (Fig. 1B), strongly suggesting that the observed enrichments represent true biological associations. This result highlights the robustness of our gene-set-based approach for identifying the biological functions of motifs: although 41,015 false positive targets are predicted in permuted promoters, their resulting motif target map has no associations with known biological functions, in contrast to the large number of such associations found for the map constructed on real promoters. As another control, we also created permuted versions of the input position-specific scoring matrixes (PSSMs), and ran our analysis pipeline on these motifs. As with the control of permuting sequences, we find that the resulting motif map in this control has zero significant associations between GO categories and the target genes predicted for these permuted motifs (data not shown).
To evaluate the utility of integrating contributions from both weak and strong sites, and of not using arbitrary cutoffs on binding-site strength, we compared our probabilistic HMM-based method with an alternative method in which the best motif score within the tested promoter is taken as the score of the motif on each promoter, and the target set of each motif is taken as the top N scoring promoters. By setting the target set size of each motif, N, to be equal to that which our HMM model obtained for that motif, we obtain the most fair comparison and find that our method leads to significantly more (motif, GO category) associations than this alternative method (Fig. 1B).
As another validation, we tested the ability of our method to predict target genes of the tumor suppressor TP53 in 542 strongly bound regions that were recently identified using chromatin immunoprecipitation experiments (Wei et al. 2006). Our method assigned a significant score (P < 0.01 by comparison with scores assigned to randomly shuffled versions of each region) to 440 (81%) of these regions. Notably, for 397 (73%) regions, our method was significantly better than the above described highest motif occurrence score method at discriminating these sequences from their randomly shuffled versions. Only nine (2%) sequences scored higher using the highest motif occurrence score method (Fig. 1C). These improved results were largely attributable to our integration of weak binding sites, which we found to be particularly important in the longer TP53-bound regions obtained by the ChIP experiment (Wei et al. 2006) due to their higher probability of containing multiple low-affinity but functional sites (data not shown).
Identifying biological processes regulated by cis-regulatory motifs
Given our motif target map, which specifies the targets of motifs and motif combinations, we set out to identify the biological process regulated by each motif. To this end, we compiled a collection of 3519 gene sets from various sources, comprising groups of genes that share similar functions (Ashburner et al. 2000; Dahlquist et al. 2002; Kanehisa et al. 2002), possess similar pattern of expression (Segal et al. 2004; Su et al. 2004), or are predicted to be regulated by the same microRNAs (Krek et al. 2005; Lewis et al. 2005), and tested the overlap of these gene sets with our motif modules. If a set of coordinately functioning or coexpressed genes were to share a common upstream regulatory motif, such motifs may be important regulatory mechanisms for that biological process. We found 9940 such significant associations (FDR < 0.05) for 809 gene sets and 568 motifs (or combinations thereof), yielding what we call a “motif function map” (Fig. 2A). This map assigns zero or more gene sets to each motif (or combinations) in our collection, corresponding to the gene sets whose member genes are enriched in the target gene set of the motif. We next discuss several intriguing insights of the map, but note that the full map is available for further exploration from our website.
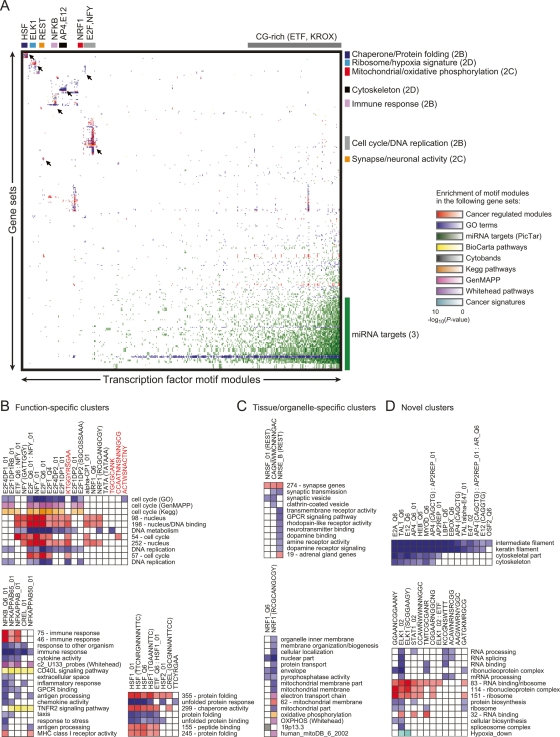
Figure 2.
Functional analysis of motif modules. (A) Global view of specific transcription-factor motifs that are enriched in gene sets (by hypergeometric P-value). Different types of gene sets are distinguished by color, and their intensity corresponds to the significance of the motif module enrichment. Unsupervised clustering was applied to the resulting matrix. Several clusters are shown in detail (indicated by arrows), with the figure location in parentheses. (B) Detailed image of motif module clusters with function-specific enrichment. Novel conserved motif modules that we experimentally validated are highlighted in red. Notations following some transcription-factor names (ex. Q6_01) are identifiers for variants of the motifs according to TRANSFAC. Stand-alone sequences or sequences in parentheses following a transcription-factor name represent consensus binding motifs from ref. (Xie et al. 2005) (key for combination of nucleotides: Y = C or T; R = A or G; W = A or T; S = C or G; K = T or G; M = C or A; N = unknown). Individual motifs of motif combinations are separated by a colon. (C) Detailed image of tissue/organelle-specific enrichments. (D) Detailed image of novel enrichments.
The motif function map identified the known master regulators of specific biological functions, including E2F for cell proliferation (Giacinti and Giordano 2006), NF-κB for immune response (Hayden and Ghosh 2004), and the heat-shock factor HSF for the unfolded protein response (Morimoto et al. 1997) (Fig. 2B). The enrichments also predicted the known role of several tissue and organelle-specific transcription factors, such as REST (also known as NRSF) for neuronal genes (Coulson 2005) and NRF1 (nuclear respiratory factor 1) for mitochondria-related genes (Goffart and Wiesner 2003) (Fig. 2C). The motif enrichments also suggested novel regulators for various gene sets. For example, cytoskeletal genes were enriched for the motif module MYOD, a known regulator of myogenesis genes (Bergstrom et al. 2002), but these genes were also enriched for the factors TCF3 (E12/E47) and AP4 (TFAP4), neither of which have known cytoskeleton regulation roles (Fig. 2D). As another example, ELK1, a known downstream effector of multiple MAPK pathways (Yordy and Muise-Helmericks 2000), is predicted to regulate RNA processing and translation (Fig. 2D). Finally, we identified 204 functional enrichments for targets of uncharacterized motifs, suggesting specific regulatory contexts and biological functions for 55 of 105 uncharacterized motifs (e.g., see Fig. 2B, discussed in detail below). Overall, the comparison of motif modules with gene sets from various sources predicts roles for hundreds of motifs and motif combinations in the regulation of diverse biological processes, a large number of which represent novel regulatory interaction hypotheses. These predictions reconstruct validated regulatory relationships from the literature and do not identify any significant associations in permuted promoters, thus increasing the likelihood that many of the predicted associations are biologically meaningful.
A link between transcriptional regulation and microRNA regulation
An intriguing finding from the above comparisons of motif modules and biological gene sets (Fig. 2A) is that 334 motif modules have significant overlaps with sets of microRNA targets (Krek et al. 2005; Lewis et al. 2005). MicroRNAs bind the 3′ untranslated regions of target mRNAs and either degrade or block the translation of the target mRNA (Lim et al. 2005). Since microRNAs are predicted to target ∼20% of human genes (Xie et al. 2005), the fact that their target genes often are motif targets is not surprising. However, our above finding goes beyond a generic overlap between motif targets and microRNA targets, and identifies a large number (6865) of specific motif and microRNA pairs, in which the motif targets are significantly enriched with targets for its paired microRNA. This correspondence thus suggests a strong connection between transcriptional and post-transcriptional control mechanisms, whereby many sets of genes that share a common transcription-factor regulator also share a common post-transcriptional microRNA regulator.
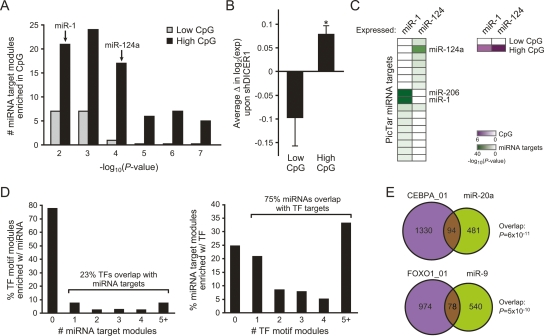
Notably, the strongest such overlaps are for motifs whose consensus sequence is enriched in CG dinucleotides (e.g., ETF, KROX), raising the possibility that the CG-richness of the motifs, rather than their specific sequences, underly this association. Promoters of many genes contain clustered regions of CG-dinucleotides (CpG islands), and a recent computational study found a natural partition of human promoters into high and low levels of CG-dinucleotides (Saxonov et al. 2006). Indeed, separate from the comparison with our motif modules, we found that microRNA targets from two independent prediction algorithms (Krek et al. 2005; Lewis et al. 2005) are strongly enriched for genes with high CG-dinucleotides, beyond the number of such genes that would be expected by chance given the large number of genes with high CG-dinucleotides (Fig. 3A; data not shown). Analysis of experimental data also confirmed this intriguing enrichment of microRNA targets in genes with high CG-dinucleotides in their promoters. First, depletion of DICER1 (Schmitter et al. 2006), required for microRNA biogenesis, led to coordinate genome-wide induction of genes with high CpG promoters (Fig. 3B). Second, overexpression of miR-1 or miR-124 (Lim et al. 2005) coordinately repressed their respective predicted microRNA target genes, most of which had high CG-dinucleotides in their promoters (Fig. 3C). Thus, both gain and loss of function of microRNAs preferentially target genes with high CpG promoters, as predicted by the association of microRNA targets with our motif modules.
Figure 3.
Extensive coupling between mechanisms of transcriptional and post-transcriptional control. (A) Shown is the number of gene sets of predicted microRNA targets (Krek et al. 2005; Lewis et al. 2005) that are significantly enriched for High or Low CpG promoter genes (Saxonov et al. 2006) (P < 0.05). The P-value of enrichment of the microRNAs analyzed in C is shown. (B) Concomitant change in genes expression in High vs. Low CpG promoter genes. Average expression (± SE) is shown. (*) P = 0.001, Student’s t-test. (C) Gene repression by microRNA overexpression (Lim et al. 2005). (Left) Coordinate repression of predicted microRNA target genes. (Right) Coordinate repression of high CpG promoter genes. Each column is a microRNA overexpression experiment; each row is a gene set of microRNA targets or CpG promoters. The degree of coordinate gene regulation over expectation by chance alone is quantified by the color scale. The enrichment of miR-206 targets in miR-1 overexpression is due to the 98% overlap of their predicted targets by PicTar (Krek et al. 2005). (D) A common modularity between motif modules and microRNA targets. We compared singleton motif modules with fewer than 1000 member genes against predicted microRNA targets (PicTar). (Left) Percentage of motif modules whose targets are enriched in the targets of k different microRNAs, for k = 1, 2, 3, 4, >5. (Right) Percentage of microRNA target modules whose targets are enriched in the targets of k different motif modules, for k = 1, 2, 3, 4, >5. (E) CEBPA_01 targets significantly overlap with miR-20a targets (P = 6 × 10−11), and FOXO1_01 targets significantly overlap with miR-9 targets (P = 5 × 10−10).
To test whether the connection between motif modules and microRNA targets extends beyond genes with high CG-dinucleotides in their promoters, we first excluded motifs that are enriched in CG-dinucleotides. To this end, we only extracted single cis-motifs with fewer than 1000 targets, as CG-related motifs almost always contained large numbers of targets (data not shown). Next, we tested the enrichment of these extracted motifs in microRNA targets. Indeed, we found 601 significant associations, to the extent that 23% of the motifs coregulate target genes with one or more microRNA, and conversely, 75% of the microRNAs coregulate targets with at least one motif (Fig. 3D). For example, target genes of the CEBPA transcription factor significantly overlap with the targets of miR-20a (P < 10−10), and FOXO1 targets significantly overlap with miR-9 targets (P < 10−9; Fig. 3E). Taken together, our results suggest that there is a high correspondence between the transcriptional and post-transcriptional networks, whereby many sets of genes share both their transcription factor and microRNA regulators.
Diverse roles for cis-regulatory motifs in human cancers
Next, we asked whether we can identify the roles that our motif modules may have in driving gene expression programs in cancer. Previous approaches to mine cis-regulatory motifs in cancer gene expression identified only a small number of enriched motifs, mostly those associated with cell proliferation (Rhodes et al. 2004, 2005). Since human cancers demonstrate large-scale and systematic variations in gene expression (Segal et al. 2004), it is clear that the associations identified thus far account for only a small fraction of the involved motifs. To systematically identify roles for our motifs in human cancer, we used a compendium of 1975 expression profiles representing 22 distinct human cancers (Segal et al. 2004), and applied a gene-set-based method (Segal et al. 2004) to identify motif modules that are coordinately induced or repressed in each sample of the compendium. As a next step, we asked whether the arrays in which the targets of each motif are coexpressed are further enriched for particular clinical annotations. Indeed, we found 751 motifs and motif combinations (FDR < 0.05) whose targets had similar expression in at least one cancer annotation, resulting in a higher-order compendium of activated and deactivated cis-regulatory motifs in clinical outcomes in cancer (Fig. 4A).
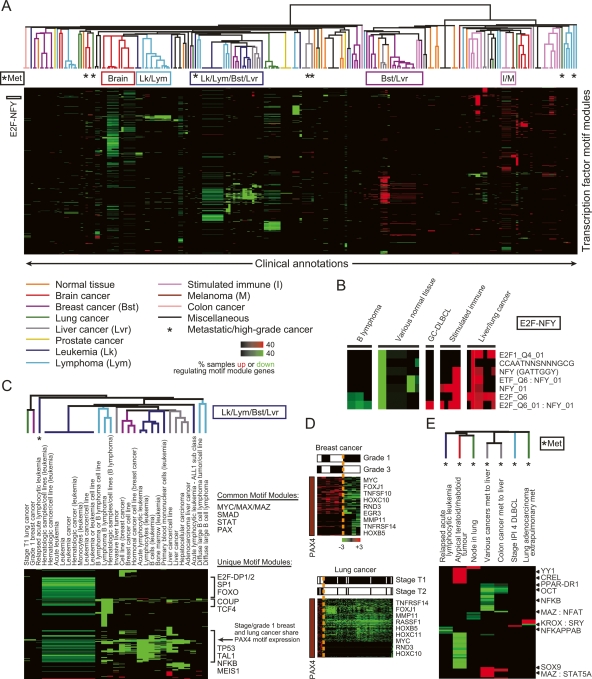
Figure 4.
Global view of cis-regulation in cancer using motif modules. (A) Clinical annotations organized by their associated activated and deactivated motif modules. We used the “module map” method (Segal et al. 2004) on a compendium of 1975 arrays spanning 22 different tumor types. We found arrays in which the genes of each motif module were coordinately induced or repressed (P < 0.05). We then tested the enrichment of these coregulated arrays of each motif module in clinical annotations of the arrays (Segal et al. 2004) (P < 0.05, corrected for multiple hypothesis testing using FDR), and applied unsupervised hierarchical clustering to group together clinical annotations that show enrichments for the same motif modules. Intensity of each enrichment corresponds to the percentage of microarray samples that have motif module target genes significantly induced (up) or repressed (down). The colors of the branch arms represent specific groups of clinical annotations (the specific tissues listed correspond to the tissue origin of the cancer). (*) Groups of metastatic/high-grade cancers. Locations of clusters analyzed in detail are shown at the top and left of the figure. (B) Clinical annotation enrichment of the array signature of E2F and NFY-containing motif modules (E2F-NFY). (C) Motif modules enriched in the leukemia/lymphoma/breast/liver (Lk/Lym/Bst/Lvr) cluster. (D) Shown are PAX4 motif module genes noted in C that are significantly induced or repressed in the indicated grade/stage of breast/lung cancer, respectively. PAX4 target gene induction is enriched in the higher grade/stage tumors (P < 0.05, χ2 test). (E) Motif modules enriched in metastatic/high-grade tumors (*Met) of different histologic origins.
The association between motif modules and expression patterns provided several insights into the transcriptional regulation of cancer. First, it confirmed known roles for several regulators. For example, consistent with a previous study (Rhodes et al. 2005), E2F and NFY motif modules, alone and in combination, were induced in many types of cancers and solid tumors, yet repressed in their normal tissue counterparts, supporting a role for these factors in regulating cell proliferation of these tumors (Fig. 4B; Supplemental Fig. S2). Consistent with a role in regulating the motif modules, E2F1 (along with additional E2F genes) and NFYA gene expression levels were highly correlated with expression levels of their predicted motif modules (E2F1: R = 0.61, P < 10−37; NFYA: R = 0.33, P < 10−12) (data not shown). E2F modules (and correspondingly multiple E2F genes) also showed reduced expression in B-cell lymphomas, consistent with the previous observation that E2F1 is weakly expressed in this type of cancer (Moller et al. 2000). Second, the compendium identified several factors that had widespread roles in cancer, including breast, liver, lung, leukemia, lymphoma, and brain samples (Fig. 4C; Supplemental Figs. S3–S6). For example, we found that activity of the PAX4 motif module could distinguish lower grade tumors of both breast and lung from higher grade: higher grade tumors had increased expression of PAX4 target genes, including MYC, MMP11, and several HOX genes (Fig. 4D). Third, we predicted novel roles for 92 uncharacterized motifs, alone or in combination with a known motif, in the regulation of gene expression in cancer. In total, 991 significant enrichments were identified in the overlap between targets of uncharacterized motifs and genes coordinately induced or repressed in cancers of distinct clinical behaviors, suggesting potentially widespread roles of uncharacterized regulatory motifs in the biology of cancer. Finally, the compendium identified a property of advanced cancers that was shared across different tumor types. We found that primary tumors of the same histologic origin tended to have similar patterns of activated and repressed motif modules, while metastatic tumors are characterized by motif modules that are often distinct from those of primary tumors of the same histologic origin (Fig. 4A,E). While it is possible that the difference in surrounding stromal cells may contribute to the different motif modules observed in metastatic tumor samples, histological analysis of most of the samples used in our study confirmed the purity of the tumor tissue, and thus the contribution of surrounding tissue in these samples is likely minimal. These results suggest that distinct transcriptional pathways are sequentially altered during cancer progression. By examining the behavior of motif targets in genome-wide expression profiles from human cancer, we identify roles for many motifs and paint a rich and mechanistically-revealing portrait of human cancers that provides multiple research directions for hypothesis-driven experiments.
Experimental validation of regulatory roles for four uncharacterized motifs in cell cycle progression
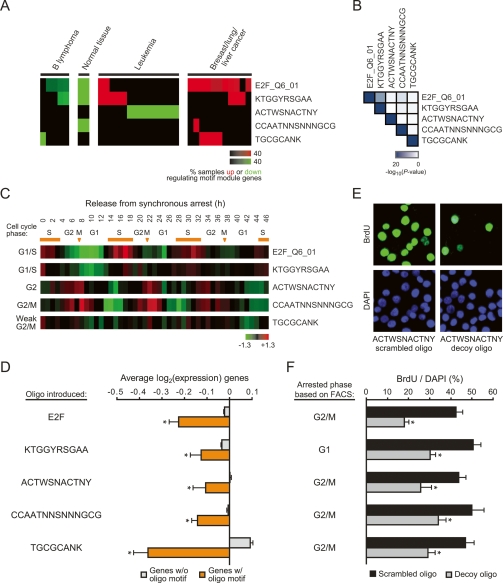
As an example of novel hypotheses suggested by our analysis, we found four evolutionarily conserved but uncharacterized motifs (Xie et al. 2005) whose targets were enriched in cell cycle genes (Fig. 2B, highlighted in red) and induced in at least four types of human cancers (Fig. 5A), suggesting a role for these motifs in cell proliferation. The target genes associated with each of these four motifs had little overlap with each other (Fig. 5B), further suggesting that these motifs regulate distinct sets of genes during cell cycle progression. Indeed, these motif modules were periodically induced at distinct stages of the cell cycle (Whitfield et al. 2002): the KTGGYRSAGAA motif module, whose consensus sequence is similar to that of the canonical cell cycle motif E2F, is induced during the G1/S phase (similar to E2F), while the ACTWSNACTNY motif module is induced during the G2 phase, and the CCAATNNSNNNGCG motif module is induced during the G2/M phase (Fig. 5C). The motif module of TGCGCANK showed weak association with the G2/M phase.
Figure 5.
Uncharacterized motifs regulate cell cycle progression. (A) Clinical annotation enrichment of the array signature of E2F_Q6_01 and the four uncharacterized motif modules. (B) Significance of enrichment of the indicated motif module target genes with all other motif targets is shown. (C) Shown is the average expression of the indicated motif module genes found in a set of periodically expressed cell cycle genes in HeLa cells (Whitfield et al. 2002). The stage of cell cycle that the pattern of expression most resembles is indicated on the left. Orange bars signify S phase; arrows signify mitoses. (D) Decoy oligos corresponding to the uncharacterized motif sequences inhibit target gene expression in HeLa cells. Shown is the average log2 gene expression (± SE) of genes predicted to have the indicated motif (orange) or those that lack the motif (gray). (*) P < 0.05 compared with genes without motif, Student’s t-test. (E,F) Decoy oligos inhibit cell cycle progression in HeLa cells. (E) Example of BrdU immunofluorescence staining and DAPI counterstain after introduction of scrambled oligos or decoy oligos for the ACTWSNACTNY motif. (F) Average percentage (± SE) of BrdU-positive cells after transfection of the indicated scrambled or decoy oligos. The cell cycle phase in which the cells arrested (as determined by FACS) is indicated on the left. (*) P < 0.01 compared with the respective scrambled oligo, Student’s t-test.
Given this multitude of evidence, we set out to experimentally validate this hypothesis. We transfected double-stranded oligonucleotide decoys corresponding to each of the four uncharacterized motifs into HeLa cells. As previously illustrated for many motif decoys (Cutroneo and Ehrlich 2006), decoy oligodeoxynucleotides can bind and sequester cognate TFs, thereby revealing the physiologic functions of the endogenous motifs. We performed microarray experiments following decoy addition to globally characterize the response at the molecular level. Notably, by analyzing these genome-wide expression profiles, we found that genes predicted to contain each motif, as determined by our motif module map, are significantly repressed as compared with genes that are predicted to lack the motif (Fig. 5D), suggesting that these motifs act as transcriptional activators of our predicted targets. Moreover, we measured DNA synthesis following decoy addition and found that each of these four decoys inhibited cell cycle progression (Fig. 5E,F), with efficacies approaching that of the E2F motif (Morishita et al. 1995). In contrast, scrambled oligonucleotides preserving the nucleotide content of each of the four tested motifs had no effect. We also performed FACS analysis, and found that for all four uncharacterized motifs, cell cycle arrest occurred in the same phase as predicted above by target gene induction during cell cycle progression (Fig. 5F). One exception was with the E2F motif decoy, where cells arrested in G2/M phase of the cell cycle despite the induction of E2F target genes during the G1/S phase. However, E2F has also been shown to regulate the expression of its targets during G2/M phase in addition to G1/S (Polager et al. 2002; Ren et al. 2002; Zhu et al. 2005), suggesting that these G2/M target genes may be more sensitive to E2F function in our motif decoy experiments. Together, these results thus provide strong experimental evidence that supports our prediction of a regulatory role in cell cycle progression for these four uncharacterized motifs and further confirms the ability of the motif module map to predict novel motif targets.
Discussion
We presented a gene-set-based approach for characterizing the biological function of cis-regulatory motifs and their condition of activation consisting of two main steps. In the first step, we take a set of motifs as input and use a probabilistic approach to identify their target promoters, and in the second step, we use gene-set statistical tools to compare the targets of each motif and motif combination with biologically meaningful gene sets and large compendiums of gene expression data to characterize their function and condition of activation. A key advantage of our approach is the robustness gained by considering the entire set of targets of each motif when characterizing its biological function, rather than considering its targets individually. This robustness is evident in the large number of significant overlaps between our sets of motif targets and functional gene sets, an overlap that is not seen when predicting motif targets from permuted promoter sequences.
Our approach compliments recent work (Pennacchio et al. 2007) that combined gene expression data and transcription-factor binding site analysis for the purpose of identifying tissue-specific enhancers in human. However, there are key differences between the two approaches. First, the goal of Pennacchio and colleagues was to predict tissue-specific enhancers, whereas our goal was to provide functional analysis of motif modules, including characterizing their biological functions, overlap with miRNA regulation, and relation to human disease states such as cancer. Second, they predict TF targets at the single binding-site level and use sequence conservation as one of the filters. In contrast, our method computes the overall score of a promoter and does not restrict itself only to promoters that can be aligned in multiple genomes.
We applied our approach to characterize the function of motifs in human core promoters and identified candidate biological functions for a large number of motifs, including putative functions for over 50 uncharacterized motifs. Our approach generated novel regulatory hypotheses for directed experimentation, and we experimentally validated roles for four uncharacterized regulatory motifs in cell cycle progression. When comparing our motif targets with predicted targets of microRNAs (Krek et al. 2005; Lewis et al. 2005), we found a strong correspondence between the transcriptional and post-transcriptional regulatory networks, whereby many sets of genes share both their transcription factor and microRNA regulators. While each of these networks is known to exhibit a modular organization, the higher level of organization that we find between these networks has not, to our knowledge, been reported previously. This finding suggests that modularity is an important design principle of biological interactions: sets of genes that are coregulated at one level remain as indivisible regulatory units in other levels of regulation. Moreover, we find that genes with high CpG promoters are generally targeted by microRNA genes. Since genes with high CpG promoters tend to be broadly expressed (Saxonov et al. 2006), it may be that their targeting by microRNAs allows fine-tuning their expression levels in specific tissues.
Despite its successes, our approach has several limitations that need to be addressed. First, additional experiments are needed to derive more accurate transcription-factor binding-site descriptions, as some motifs are derived from a small number of experimentally determined sites. Second, we only examined 700 bp surrounding the annotated transcription start sites, as these regions are highly enriched in conserved motifs (Xie et al. 2005). Clearly, other more distal enhancer regions are known to have important regulatory roles, but including them will require more sensitivity in the detection due to the additional noise they bring. Finally, for microRNA targets we relied on computationally derived databases, only a fraction of which have been experimentally verified. However, the concordance of our findings using two different microRNA prediction algorithms and our use of gene set level statistical analysis, suggests that the robust overlap between the transcriptional and post-transcriptional networks will still hold in improved future versions of microRNA target predictions.
Overall, our approach provides a functional characterization of cis-regulatory motifs in human core promoters, identifying both the biological process targeted by the motifs and the conditions in which this regulation occurs. The results of our analysis as well as our probabilistic method are publicly available on a data-mining website, which researchers can use to identify transcription factors that are likely to bind to their own sequences of interest or predict specific motifs that drive observed patterns of gene expression. Thus, our method and tools provide valuable resources for guiding the identification of cis-regulatory mechanisms that control a wide range of biological processes.
Methods
Sequences and motifs
We downloaded the genome sequence and locations of transcription start sites from the UCSC genome browser (Karolchik et al. 2003), and used these to define each promoter as 500 bp upstream and 200 bp downstream from each transcription start site. We downloaded DNA-binding sites from TRANSFAC (Wingender et al. 2001) and Xie et al. (2005) and represented each binding site as a position-specific scoring matrix (PSSM).
Identifying motif targets
For all input motifs, we identified the set of genes whose promoters significantly contain the particular motif. To this end, for each motif and a particular gene promoter, we used our probabilistic model (Sinha et al. 2003) to assign a likelihood score that represents the probability that the promoter sequence contains the motif, normalized against the background probability inferred from the local nucleotide composition of the same sequence. (We use the “HMM0” model described in Sinha et al. 2003.) The probabilistic model treats the PSSMs as prescribing binding free energies and takes frequency and affinities of sites, as well as competition among PSSMs into account when computing the score of a particular promoter. Each promoter was scored for every motif separately, and considered as being a “target” of the motif if the score had an empirical P-value < 0.01, estimated from 100 mononucleotide-preserving permutations of that promoter sequence.
Identifying motif combinations
Each combination Mk of k motifs was combined with each single motif M1 to form a candidate k+1-motif combination Mk+1 that was accepted only if three conditions were met: (1) The target set size for Mk+1 was significantly large compared with that expected at random from intersecting the target sets of Mk and M1 (hypergeometric test, FDR [Benjamini and Hochberg 1995] < 0.05). (2) Mk+1 had a higher statistical significance, as per the above test, than that of Mk. (3) The likelihood score of Mk+1 on a promoter (using our probabilistic model; Sinha et al. 2003) improved (on average, across all promoters) upon the likelihood scores of Mk and M1. To enforce this, we computed the (log) likelihood score Fk, F1, and Fk+1, for Mk, M1, and Mk+1, respectively, on each promoter, and took the ratio of the observed increase in score (min[Fk+1 − Fk, Fk+1 − F1]) to the approximate increase in score expected if the two motifs are nonredundant (min[Fk, F1]). We required the average of this ratio to be >0.8, to ensure that M1 is not redundant with Mk. This step thus allows us to identify the coordinately acting motif combinations and exclude the combinations of redundant motifs that may result simply from the high similarity between the input motifs.
Comparison with alternative methods of scoring promoters
We implemented an alternative strategy for computing the target set of genes for any motif. It relies on the log likelihood ratio (LLR) score of a substring s, defined as LLR(s) = log[Pr(s | W)/Pr(s | Wb)], where Pr(s | W) is the probability of sampling s from PSSM W, and Pr(s | Wb) is the background sampling probability of s (local background, as used in our score). The highest LLR score over all substrings in a promoter is assigned as the score of the gene. Genes are sorted by their score, and a threshold is applied to choose the top N genes, where N is the size of the target set that our HMM model obtained for that motif. To compare our probabilistic scoring scheme with this alternative strategy, we considered each Gene Ontology (GO) (Ashburner et al. 2000) category with 10–1000 genes (the upper bound on the gene sets was done to remove nonspecific GO categories) and formed a “GO gene set” corresponding to each category. We tested each motif’s target set for enrichment for each of the resulting 999 GO gene sets (hypergeometric test), and counted the number of significant associations.
Controls for motif target map
In the first control, the sequence of each promoter was randomly permuted. In the second control, each input motif (PSSM) was permuted—columns first, followed by entries in each column—so as to obtain a random PSSM with the same Information Content (specificity) as the original. This was repeated 10 times for each input PSSM, and the random motif most distinct from input PSSMs was chosen, resulting in a compendium of 432 random motifs.
Identifying gene sets enriched in motif modules
Motif modules were analyzed for their enrichment in the following gene sets: Gene Ontology terms (Ashburner et al. 2000) (1665 categories; several broad GO terms were manually removed and then a nonredundant set of categories was obtained by removing categories with a correlation of ≥0.9 between their membership vector and that of another category); gene modules coregulated in cancer (Segal et al. 2004) (456 modules); predicted microRNA targets (Krek et al. 2005) (178 modules); BioCarta pathways (http://www.biocarta.com) (289 modules); cytobands (Karolchik et al. 2003) (624 modules); KEGG pathways (Kanehisa et al. 2002) (104 modules); GenMAPP (Dahlquist et al. 2002) (52 modules); Whitehead pathways (123 modules); and cancer prognostic gene expression signatures (Adler et al. 2006) (28 modules). Significant enrichment of motif modules (P < 0.05; corrected for multiple hypotheses using FDR) (Benjamini and Hochberg 1995) was determined using the “gene module map method” implemented in Genomica (Segal et al. 2004).
MicroRNA analyses
Global gene expression of DICER1 knockdown in 293 cells (Schmitter et al. 2006) was downloaded from GEO, converted to log2 space, and median centered by gene. Data from shDICER1 clone 2b2 had consistent DICER1 knockdown after 2 d, which was used for subsequent analysis. Replicates of control cells were averaged and subtracted from all samples (zero-transformation). Paired Student’s t-tests were used to identify genes that were significantly induced or repressed upon DICER1 knockdown (P < 0.05). These genes were split into Low CpG or High CpG groups and averaged: Low CpG genes have normalized CpG score < 0.35 as defined by Saxonov et al. (2006); High CpG genes have normalized CpG score > 0.35. Genes that were repressed following miR-1 or miR-124 overexpression are as defined (Lim et al. 2005). Gene module map was used to determine the significance of enrichment of the repressed genes with microRNA targets (Krek et al. 2005; Lewis et al. 2005) and High and Low CpG groups as defined above (P < 0.01, FDR = 0.01) (Benjamini and Hochberg 1995). To identify microRNA target modules that are enriched for TF target modules independent of CpG content, we isolated single motif modules that had <1000 targets (346 modules) and used gene module map to identify significant enrichments (P < 0.05, FDR = 0.05) (Benjamini and Hochberg 1995) with microRNA targets.
Identifying expression patterns of motif modules in cancer
The cancer compendium of 1975 microarrays and assortment into clinical annotations is as described (Segal et al. 2004). Enrichment of motif modules was first calculated for each microarray experiment (P < 0.05) using the “motif module map method” implemented in Genomica (Segal et al. 2004). The resulting expression signatures of each motif module, consisting of the set of arrays in which it is significantly up- or down-regulated, was subsequently tested for its enrichment in clinical annotations (P < 0.05, FDR = 0.05) (Benjamini and Hochberg 1995). For the detailed clusters in Figure 4, C and E, and Supplemental Figures S3–S6, all motif modules represented in one or more of the listed clinical annotations were isolated and reclustered; since the same cluster algorithm was used, the associated cluster trees are the same as in Figure 4A, but without any gaps in the clustergram.
Analysis of uncharacterized motif modules
Gene module map was used to identify the P-value of significance for the enrichment of targets of E2F_Q6_01 and the four uncharacterized motifs (Fig. 2B, highlighted in red) with each other (Fig. 3A). Gene expression time course following release from double thymidine block in HeLa cells was as described (Whitfield et al. 2002); genes defined as being periodically expressed were isolated, and the average expression of all represented motif module genes is shown. These expression patterns were clustered with “ideal expression profiles” (Whitfield et al. 2002) of well-characterized genes from each phase of the cell cycle to determine the stage in which the uncharacterized motif target genes are periodically expressed.
Oligo sequences and cell cycle analyses
Single-stranded oligos listed below (and the corresponding reverse compliment) were synthesized and annealed; underlined regions correspond to the consensus motif sequence. E2F decoy sequence was as described (Morishita et al. 1995). Motif module map scores for all target genes for each of the uncharacterized motifs were ranked, and the highest scoring sequence was used for the decoy oligos:
E2F: CTAGATTTCCCGCGGATC (decoy); CTAGACTCTGCTCG GATC (scrambled)
KTGGYRSGAA: CTAGATTCCCGCCAAGGATC (decoy); CTAGA CAGCTACTCCGGATC (scrambled)
TGCGCANK: CTAGACATGCGCAGGATC (decoy); CTAGATCA CAGGCGGATC (scrambled)
CCAATNNSNNNGCG: CTAGACGCCCTCCGATTGGGGATC (decoy); CTAGATGCACGCTCGGTCCGGATC (scrambled)
ACTWSNACTNY: CTAGAGGAGTTGTAGTGGATC (decoy); CTA GAGATAGTGTGTGGGATC (scrambled)
HeLa cells were propagated in DMEM (Invitrogen) plus 10% FBS and transfected with 0.5 μM of double-stranded DNA. Cell proliferation was monitored by measuring the incorporation of the thymidine nucleotide analog 5-bromo-2′-deoxyuridine (BrdU) (Sigma) into DNA as described (Sage et al. 2003). Briefly, 10 μM BrdU was added to the medium for 1.5 h prior to immunofluorescent staining with anti-BrdU antibody (Becton Dickinson) and Alexa Fluor-conjugated secondary antibody (Molecular Probes). The percentage of BrdU-positive cells among >250 DAPI-positive cells in multiple high-power fields was determined. Fluorescence activated cell sorting (FACS) combined with propidium iodide staining to determine the specific stage of cell cycle arrest was analyzed as described (Whitfield et al. 2002).
Microarray profile of uncharacterized motif modules
Total RNA was extracted with TRIzol (Invitrogen) from HeLa cells 2 d after transfection of the indicated decoy oligo or scrambled oligo in duplicate. RNA was amplified using the Ambion Amino Allyl MessageAmp II aRNA kit. For each motif, decoy oligo transfected samples (labeled with Cy5) and the corresponding scrambled oligo transfected samples (labeled with Cy3) were competitively hybridized to HEEBO microarrays as described (http://www.microarray.org/sfgf/heebo.do). Genes selected for analysis had a fluorescent hybridization signal at least 1.5-fold over local background in either Cy5 or Cy3 channel and had technically adequate data in at least 70% of experiments. For each array, genes that were induced or repressed >1 standard deviation from the mean were isolated, and then motif module map was used to identify genes predicted to contain the given motif in the promoter. Log2 expression values from duplicate arrays were averaged, and then values for all genes with or without the predicted motif were averaged.
Additional methods and URLs
For our data, model, genome-wide motif predictions, full enrichment analyses, and tools for predicting motifs in your own sequences, see http://genie.weizmann.ac.il/pubs/motifs07. Our results can be viewed in Genomica (http://Genomica.weizmann.ac.il). Full microarray data are available for download at Stanford Microarray Database (http://smd.stanford.edu/).
Acknowledgments
We thank C.Z. Chen, N. Kaplan, and D. Pe’er for useful comments on the manuscript. A.S.A was supported by NCI, DHHS grant CA09302, and the California Breast Cancer Research Program. H.Y.C. was supported by the American Cancer Society and is the Kenneth G. and Elaine A. Langone Scholar of the Damon Runyon Cancer Research Foundation. E.S. was supported by a European ENFIN grant and by NIH grant R01 CA119176-01 and is the incumbent of the Soretta and Henry Shapiro career development chair.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6828808
References
- Adler A.S., Lin M., Horlings H., Nuyten D.S., de van Vijver M.J., Chang H.Y., Lin M., Horlings H., Nuyten D.S., de van Vijver M.J., Chang H.Y., Horlings H., Nuyten D.S., de van Vijver M.J., Chang H.Y., Nuyten D.S., de van Vijver M.J., Chang H.Y., de van Vijver M.J., Chang H.Y., Chang H.Y. Genetic regulators of large-scale transcriptional signatures in cancer. Nat. Genet. 2006;38:421–430. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Bergstrom D.A., Penn B.H., Strand A., Perry R.L., Rudnicki M.A., Tapscott S.J., Penn B.H., Strand A., Perry R.L., Rudnicki M.A., Tapscott S.J., Strand A., Perry R.L., Rudnicki M.A., Tapscott S.J., Perry R.L., Rudnicki M.A., Tapscott S.J., Rudnicki M.A., Tapscott S.J., Tapscott S.J. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Berman B.P., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Levine M., Rubin G.M., Eisen M.B., Rubin G.M., Eisen M.B., Eisen M.B. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophilagenome. Proc. Natl. Acad. Sci. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Deblois G., Giguere V., Ferretti V., Bergeron D., Giguere V., Ferretti V., Bergeron D., Ferretti V., Bergeron D., Bergeron D., et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.W., Nagarajan R., Magee J.A., Milbrandt J., Stormo G.D., Nagarajan R., Magee J.A., Milbrandt J., Stormo G.D., Magee J.A., Milbrandt J., Stormo G.D., Milbrandt J., Stormo G.D., Stormo G.D. A systematic model to predict transcriptional regulatory mechanisms based on over-representation of transcription factor binding profiles. Genome Res. 2006;16:405–413. doi: 10.1101/gr.4303406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Majors J., Waterston R., Cohen B.A., Johnston M., Waterston R., Cohen B.A., Johnston M., Cohen B.A., Johnston M., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Coulson J.M. Transcriptional regulation: Cancer, neurons and the REST. Curr. Biol. 2005;15:R665–R668. doi: 10.1016/j.cub.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Cutroneo K.R., Ehrlich H., Ehrlich H. Silencing or knocking out eukaryotic gene expression by oligodeoxynucleotide decoys. Crit. Rev. Eukaryot. Gene Expr. 2006;16:23–30. doi: 10.1615/critreveukargeneexpr.v16.i1.20. [DOI] [PubMed] [Google Scholar]
- Dahlquist K.D., Salomonis N., Vranizan K., Lawlor S.C., Conklin B.R., Salomonis N., Vranizan K., Lawlor S.C., Conklin B.R., Vranizan K., Lawlor S.C., Conklin B.R., Lawlor S.C., Conklin B.R., Conklin B.R. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- Giacinti C., Giordano A., Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- Goffart S., Wiesner R.J., Wiesner R.J. Regulation and co-ordination of nuclear gene expression during mitochondrial biogenesis. Exp. Physiol. 2003;88:33–40. doi: 10.1113/eph8802500. [DOI] [PubMed] [Google Scholar]
- Hallikas O., Palin K., Sinjushina N., Rautiainen R., Partanen J., Ukkonen E., Taipale J., Palin K., Sinjushina N., Rautiainen R., Partanen J., Ukkonen E., Taipale J., Sinjushina N., Rautiainen R., Partanen J., Ukkonen E., Taipale J., Rautiainen R., Partanen J., Ukkonen E., Taipale J., Partanen J., Ukkonen E., Taipale J., Ukkonen E., Taipale J., Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Hayden M.S., Ghosh S., Ghosh S. Signaling to NF-κB. Genes & Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Nakaya A., Goto S., Kawashima S., Nakaya A., Kawashima S., Nakaya A., Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Schwartz M., Sugnet C.W., Thomas D.J., Sugnet C.W., Thomas D.J., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S., Patterson N., Endrizzi M., Birren B., Lander E.S., Endrizzi M., Birren B., Lander E.S., Birren B., Lander E.S., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., da Piedade I., Gunsalus K.C., Stoffel M., Gunsalus K.C., Stoffel M., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Bartel D.P., Linsley P.S., Johnson J.M., Linsley P.S., Johnson J.M., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Moller M.B., Kania P.W., Ino Y., Gerdes A.M., Nielsen O., Louis D.N., Skjodt K., Pedersen N.T., Kania P.W., Ino Y., Gerdes A.M., Nielsen O., Louis D.N., Skjodt K., Pedersen N.T., Ino Y., Gerdes A.M., Nielsen O., Louis D.N., Skjodt K., Pedersen N.T., Gerdes A.M., Nielsen O., Louis D.N., Skjodt K., Pedersen N.T., Nielsen O., Louis D.N., Skjodt K., Pedersen N.T., Louis D.N., Skjodt K., Pedersen N.T., Skjodt K., Pedersen N.T., Pedersen N.T. Frequent disruption of the RB1 pathway in diffuse large B cell lymphoma: Prognostic significance of E2F-1 and p16INK4A. Leukemia. 2000;14:898–904. doi: 10.1038/sj.leu.2401761. [DOI] [PubMed] [Google Scholar]
- Morimoto R.I., Kline M.P., Bimston D.N., Cotto J.J., Kline M.P., Bimston D.N., Cotto J.J., Bimston D.N., Cotto J.J., Cotto J.J. The heat-shock response: Regulation and function of heat-shock proteins and molecular chaperones. Essays Biochem. 1997;32:17–29. [PubMed] [Google Scholar]
- Morishita R., Gibbons G.H., Horiuchi M., Ellison K.E., Nakama M., Zhang L., Kaneda Y., Ogihara T., Dzau V.J., Gibbons G.H., Horiuchi M., Ellison K.E., Nakama M., Zhang L., Kaneda Y., Ogihara T., Dzau V.J., Horiuchi M., Ellison K.E., Nakama M., Zhang L., Kaneda Y., Ogihara T., Dzau V.J., Ellison K.E., Nakama M., Zhang L., Kaneda Y., Ogihara T., Dzau V.J., Nakama M., Zhang L., Kaneda Y., Ogihara T., Dzau V.J., Zhang L., Kaneda Y., Ogihara T., Dzau V.J., Kaneda Y., Ogihara T., Dzau V.J., Ogihara T., Dzau V.J., Dzau V.J. A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc. Natl. Acad. Sci. 1995;92:5855–5859. doi: 10.1073/pnas.92.13.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A., Loots G.G., Nobrega M.A., Ovcharenko I., Loots G.G., Nobrega M.A., Ovcharenko I., Nobrega M.A., Ovcharenko I., Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S., Kalma Y., Berkovich E., Ginsberg D., Kalma Y., Berkovich E., Ginsberg D., Berkovich E., Ginsberg D., Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002;21:437–446. doi: 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Terragni J., Young R.A., Dynlacht B.D., Young R.A., Dynlacht B.D., Dynlacht B.D. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes & Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M., Barrette T., Pandey A., Chinnaiyan A.M., Pandey A., Chinnaiyan A.M., Chinnaiyan A.M. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl. Acad. Sci. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Barrette T.R., Ghosh D., Chinnaiyan A.M., Kalyana-Sundaram S., Mahavisno V., Barrette T.R., Ghosh D., Chinnaiyan A.M., Mahavisno V., Barrette T.R., Ghosh D., Chinnaiyan A.M., Barrette T.R., Ghosh D., Chinnaiyan A.M., Ghosh D., Chinnaiyan A.M., Chinnaiyan A.M. Mining for regulatory programs in the cancer transcriptome. Nat. Genet. 2005;37:579–583. doi: 10.1038/ng1578. [DOI] [PubMed] [Google Scholar]
- Sage J., Miller A.L., Perez-Mancera P.A., Wysocki J.M., Jacks T., Miller A.L., Perez-Mancera P.A., Wysocki J.M., Jacks T., Perez-Mancera P.A., Wysocki J.M., Jacks T., Wysocki J.M., Jacks T., Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- Saxonov S., Berg P., Brutlag D.L., Berg P., Brutlag D.L., Brutlag D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitter D., Filkowski J., Sewer A., Pillai R.S., Oakeley E.J., Zavolan M., Svoboda P., Filipowicz W., Filkowski J., Sewer A., Pillai R.S., Oakeley E.J., Zavolan M., Svoboda P., Filipowicz W., Sewer A., Pillai R.S., Oakeley E.J., Zavolan M., Svoboda P., Filipowicz W., Pillai R.S., Oakeley E.J., Zavolan M., Svoboda P., Filipowicz W., Oakeley E.J., Zavolan M., Svoboda P., Filipowicz W., Zavolan M., Svoboda P., Filipowicz W., Svoboda P., Filipowicz W., Filipowicz W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M.D., Pearce M., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Pearce M., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Rajewsky N., Siggia E.D., Gaul U., Siggia E.D., Gaul U., Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:e271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., Friedman N., Koller D., Regev A., Friedman N., Koller D., Regev A., Koller D., Regev A., Regev A. A module map showing conditional activity of expression modules in cancer. Nat. Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- Sinha S., van Nimwegen E., Siggia E.D., van Nimwegen E., Siggia E.D., Siggia E.D. A probabilistic method to detect regulatory modules. Bioinformatics. 2003;19:i292–i301. doi: 10.1093/bioinformatics/btg1040. [DOI] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Zhang J., Soden R., Hayakawa M., Kreiman G., Soden R., Hayakawa M., Kreiman G., Hayakawa M., Kreiman G., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Shahab A., Yong H.C., Fu Y., Weng Z., Yong H.C., Fu Y., Weng Z., Fu Y., Weng Z., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Whitfield M.L., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Perou C.M., Hurt M.M., Brown P.O., Hurt M.M., Brown P.O., Brown P.O., et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E., Chen X., Fricke E., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Chen X., Fricke E., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Fricke E., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Krull M., Matys V., Michael H., Ohnhauser R., Matys V., Michael H., Ohnhauser R., Michael H., Ohnhauser R., Ohnhauser R., et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lander E.S., Kellis M., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordy J.S., Muise-Helmericks R.C., Muise-Helmericks R.C. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]
- Zhu W., Giangrande P.H., Nevins J.R., Giangrande P.H., Nevins J.R., Nevins J.R. Temporal control of cell cycle gene expression mediated by E2F transcription factors. Cell Cycle. 2005;4:633–636. doi: 10.4161/cc.4.5.1650. [DOI] [PubMed] [Google Scholar]