Abstract
Lemurs and the other strepsirrhine primates are of great interest to the primate genomics community due to their phylogenetic placement as the sister lineage to all other primates. Previous attempts to resolve the phylogeny of lemurs employed limited mitochondrial or small nuclear data sets, with many relationships poorly supported or entirely unresolved. We used genomic resources to develop 11 novel markers from nine chromosomes, representing ∼9 kb of nuclear sequence data. In combination with previously published nuclear and mitochondrial loci, this yields a data set of more than 16 kb and adds ∼275 kb of DNA sequence to current databases. Our phylogenetic analyses confirm hypotheses of lemuriform monophyly and provide robust resolution of the phylogenetic relationships among the five lemuriform families. We verify that the genus Daubentonia is the sister lineage to all other lemurs. The Cheirogaleidae and Lepilemuridae are sister taxa and together form the sister lineage to the Indriidae; this clade is the sister lineage to the Lemuridae. Divergence time estimates indicate that lemurs are an ancient group, with their initial diversification occurring around the Cretaceous-Tertiary boundary. Given the power of this data set to resolve branches in a notoriously problematic area of primate phylogeny, we anticipate that our phylogenomic toolkit will be of value to other studies of primate phylogeny and diversification. Moreover, the methods applied will be broadly applicable to other taxonomic groups where phylogenetic relationships have been notoriously difficult to resolve.
In recent years, the increasing amount of genomic sequence data available for major evolutionary lineages has been paralleled by an increase in the use of multilocus phylogenetic approaches to resolve difficult and previously intractable evolutionary relationships (Rokas et al. 2003; Murphy et al. 2004; James et al. 2006; Weisrock et al. 2006). Some of these studies have been based exclusively on publicly available databases, mining DNA sequence information to yield large data sets, a number of which contain hundreds of thousands of orthologous nucleotide positions (Bapteste et al. 2002; Rokas et al. 2003; Pollard et al. 2006; Cannarozzi et al. 2007; Nikolaev et al. 2007). Still others have used comparative genomic patterns of insertions and deletions to reconstruct relationships, owing in part to the nonhomoplastic and conserved nature of such events (Kriegs et al. 2006; Murphy et al. 2007). Ideally, phylogenetic studies will sample broadly across a wide range of independently segregating loci distributed throughout the genome. Such a strategy conforms to a fundamental assumption of phylogenetic analysis, that characters will be identically and independently distributed. Nonetheless, up to the present, the influx of multilocus data sets has been driven largely by studies using only a very limited number of loci, usually utilizing conserved PCR primers for orthologous nuclear loci that were previously developed in other taxonomic groups (e.g., Malcomber 2002; Townsend et al. 2004; Poux et al. 2005; Weisrock et al. 2005; Galewski et al. 2006; Harlin-Cognato and Honeycutt 2006; James et al. 2006; Noonan and Chippindale 2006; Zhang et al. 2006; Heckman et al. 2007; Li and Orti 2007).
Increasing the number of nuclear loci for phylogenetic analysis is more readily accomplished in some clades than others. For example, clades that contain model organisms with substantial genomic resources allow for the rapid assembly of large and broadly distributed phylogenomic loci (e.g., Rokas et al. 2003). This process is more problematic for nonmodel organisms. To date, the typical approach for increasing the number of amplifiable and informative nuclear sequence markers is to compare distantly related species with available genomic resources to develop conserved PCR primers for use in a more inclusive set of taxa (Palumbi and Baker 1994). This approach for nuclear marker generation has been successful for studies within diverse groups, such as placental mammals (Murphy et al. 2001) and fish (Li et al. 2007), and successful when subsequently applied to more exclusive terminal groups, for example, bats (Teeling et al. 2005) and cats (Johnson et al. 2006).
A major drawback to this approach is that increased evolutionary divergence between genomes used in primer development will likely result in an increased PCR failure rate, leading to incomplete data matrices in phylogenetic analyses. For example, Murphy et al. (2001) developed a set of 15 nuclear DNA primer pairs based on human–mouse genomic comparisons, for use in placental mammalian phylogenetics. Of the 66 taxa used in Murphy et al. (2001), an average of six taxa were missing from each data set, with some matrices missing sequence data from as many as 14 taxa. Notably, the phylogenetically-mysterious primate genus Tarsius (Yoder 2003) is represented in only 10 of the Murphy et al. (2001) nuclear data sets and contains only 55% of the total aligned nuclear sequence data. Given the importance of taxon sampling in phylogenetic reconstruction (Zwickl and Hillis 2002; Hedtke et al. 2006), minimizing missing data should be a central objective of phylogenomic primer design methods. The analysis of data sets that are compromised by weak sampling, whether it be taxonomic or character insufficiencies, often results in phylogenetic hypotheses that either are conflicting or are weakly resolved.
Previous character-based phylogenetic analyses of lemurs (Primates, suborder Lemuriformes) have been subject to weak taxonomic sampling and small data sets that are based on only one or a few genetic loci (Crovella et al. 1993, 1995; Adkins and Honeycutt 1994; Yoder 1994, 2003; DelPero et al. 1995, 2001; Dutrillaux and Rumpler 1995; Porter et al. 1995, 1997b; Stanger 1996; Stanger-Hall 1997; Stanger-Hall and Cunningham 1998; Yoder and Irwin 1999; Murphy et al. 2001; Pastorini et al. 2003; Poux et al. 2005). Despite these weaknesses, virtually all phylogenetic studies of lemurs and related primates agree on three points: (1) lemurs are monophyletic; (2) the lemuriform clade is most closely related to the African and Asian lorisiform clade, together forming the tooth-combed primate clade strepsirrhini; and (3) the strepsirrhine clade is sister to all other living primates. Moreover, the majority of these studies have found that the aye-aye, genus Daubentonia (family Daubentoniidae), is sister to a clade composed of all other lemurs, including the recently extinct giant lemurs (Karanth et al. 2005). Beyond this result, however, there has been complete disagreement among studies as to the relationships among the other four major evolutionary lineages, also classified as families. Virtually every conceivable phylogenetic resolution has been presented among the Cheirogaleidae, Indriidae, Lemuridae, and Lepilemuridae (containing the single genus Lepilemur) in studies where taxon sampling has been adequate to investigate their relationships.
The accurate resolution of lemuriform phylogeny is critical for reconstructing primate evolutionary history and, by extension, for providing a comparative framework for understanding human biology. Lemurs constitute more than 15% of extant primate species diversity, and their position in the primate evolutionary tree offers unique power for hypothesizing the timing and phylogenetic placement of primate-specific traits. For example, the current project to sequence the genome of the gray mouse lemur, Microcebus murinus (http://www.genome.gov/10002154), will allow genomic researchers to identify genetic traits that are diagnostic of the primates, distinct from those of all other mammals. Given the independent evolution of lemurs, a fully resolved lemur phylogeny can be used as the foundation for quantitative comparative analyses, thus allowing for more powerful tests of evolutionary hypotheses across all primates (Nunn and Barton 2001). Moreover, our study is timely in addressing the biological community’s current interest in both the methods (Telford 2007) and the organisms (Yoder 2007).
In order to develop this robust phylogeny, we have capitalized on available lemur genome sequence to create a molecular toolkit for use in phylogenetic comparisons across all strepsirrhine taxa targeted in our study. This phylogenomic toolkit includes PCR primer pairs designed across the genome and corresponding genomic sequences from all strepsirrhine primates in our study. Our toolkit resolves long-standing questions of lemur phylogeny and is used to calculate divergence time estimates for lemuriform evolutionary history. Importantly, this primer design approach should be useful for application to other diverse groups of species for which limited genomic sequence exists.
Results
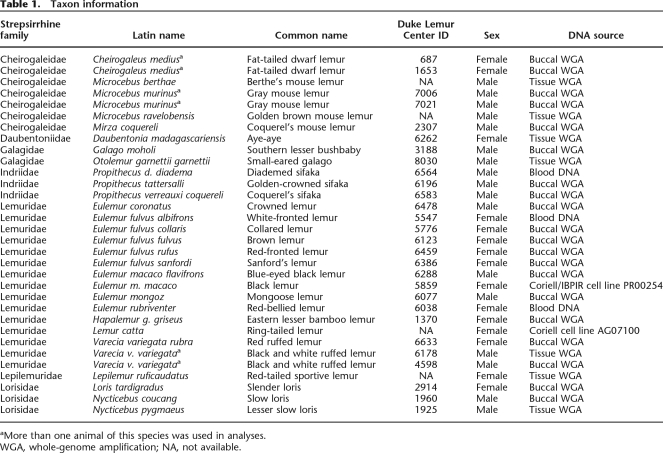
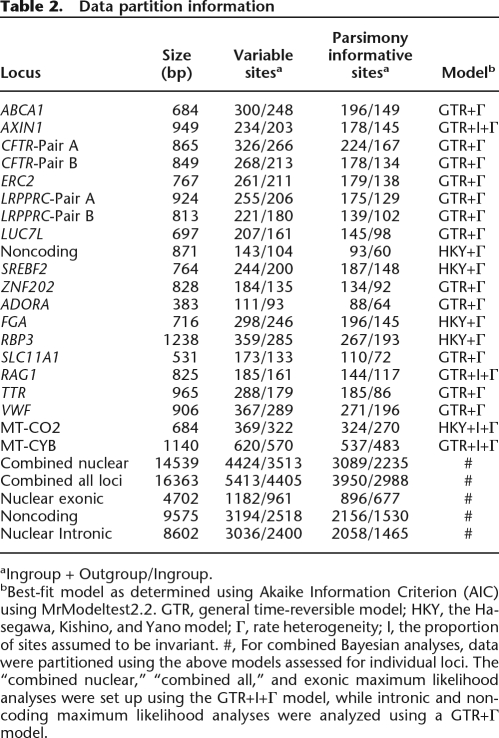
Our study includes 29 species that span strepsirrhine evolutionary diversity (Table 1). The 11 new loci total 9 kb and include exons, introns, and noncoding regions and provide a largely balanced representation of loci across the genome (Supplemental Table 1). An additional 7.3 kb of nuclear DNA and mitochondrial DNA (mtDNA) sequence data was assembled from GenBank or generated de novo with published nuclear PCR primers (Supplemental Table 1). These published sequences represent seven nuclear loci (both intronic and exonic) and two mtDNA genes (MT-CO2 and MT-CYB). Combined, these loci yield 16,363 bp of aligned data, containing 5413 variable and 3950 parsimony informative characters. When mtDNA sequence data are excluded, the combined nuclear character matrix contains 14,539 bp of aligned sequence data, of which 4424 characters are variable and 3089 are parsimony informative. For a full description of character variation by locus, see Table 2 and Supplemental Table 2.
Table 1.
Taxon information
aMore than one animal of this species was used in analyses.
WGA, whole-genome amplification; NA, not available.
Table 2.
Data partition information
aIngroup + Outgroup/Ingroup.
bBest-fit model as determined using Akaike Information Criterion (AIC) using MrModeltest2.2. GTR, general time-reversible model; HKY, the Hasegawa, Kishino, and Yano model; Γ, rate heterogeneity; I, the proportion of sites assumed to be invariant. #, For combined Bayesian analyses, data were partitioned using the above models assessed for individual loci. The “combined nuclear,” “combined all,” and exonic maximum likelihood analyses were set up using the GTR+I+Γ model, while intronic and noncoding maximum likelihood analyses were analyzed using a GTR+Γ model.
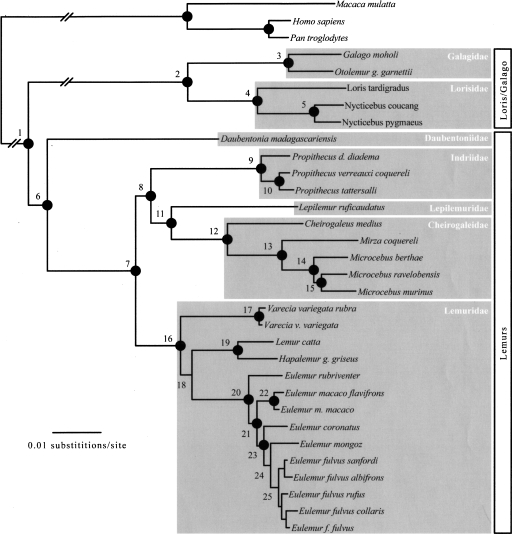
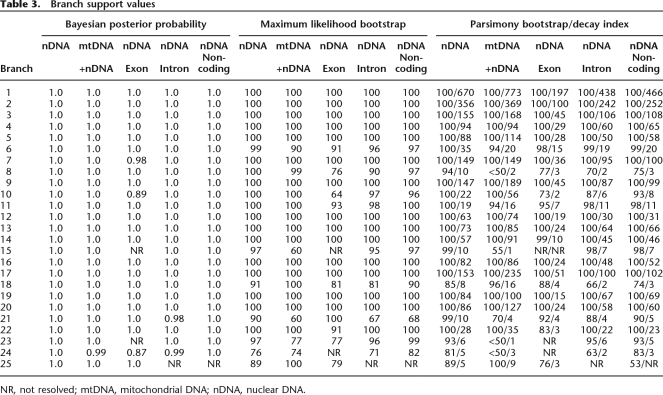
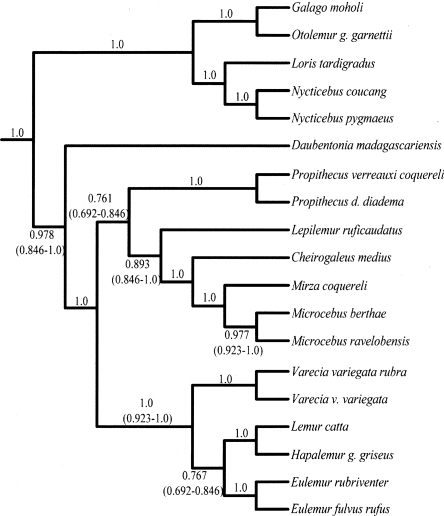
Phylogenetic analysis of the combined nuclear and nuclear plus mtDNA data sets yielded similar topologies across all optimality criteria. Branch support for several nodes diminished when the nuclear data were combined with the mtDNA data. Given the high degree of homoplasy seen in previous mtDNA-based reconstructions of lemur phylogeny (Yoder et al. 1996b), we focus on the topology favored by the combined nuclear data set as our best approximation of lemuriform phylogeny. The results of the Bayesian analysis are presented in Figure 1 and are consistent with maximum parsimony (MP) and maximum likelihood (ML) analyses. Branch support values for all combined analyses are provided in Table 3. The topology is well supported with MP and ML bootstrap values >90 and Bayesian posterior probabilities (PPs) equal to 1.0 for 22 of 25 numbered nodes. This result supports the hypothesis that lemurs are monophyletic, with the deepest split within the clade between the family Daubentoniidae and all remaining lemurs. The Cheirogaleidae and Lepilemuridae are supported as sister lineages. Moreover, the Indriidae (represented here by the three species contained within genus Propithecus) is resolved as the sister lineage to the Cheirogaleidae plus Lepilemuridae clade. As with most previous analyses, species of the family Lemuridae are resolved as a well-supported monophyletic group (Yoder and Irwin 1999; Pastorini et al. 2003; Roos et al. 2004).
Figure 1.
Lemur Bayesian consensus phylogram using 18 nuclear loci combined. This phylogram is based on the combined posterior distribution of four independent analyses of the combined and partitioned nuclear data sets. The topology is identical to those resolved using maximum parsimony (MP) and maximum likelihood (ML). Nodes with strong measures of branch support (posterior probabilities = 1.0, MP bootstrap >90%, ML >90%) are marked with filled black circles. Branch support values are indicated in Table 3. Open vertical boxes span the two infraorders of strepsirrhine primates. Shaded boxes encompass family-level taxonomic groups.
Table 3.
Branch support values
NR, not resolved; mtDNA, mitochondrial DNA; nDNA, nuclear DNA.
Tree topologies for most individual loci were not substantially different from the total evidence trees generated from the concatenated nuclear data. Even so, branch support in the single-locus trees was often weak, leaving some uncertainty about how informative the individual loci were (data not shown; for discussion, see Methods). Separate analyses of exonic, intronic, or noncoding partitions yielded decreased support for some branches relative to the total evidence trees, but the overall topologies were not substantially altered (see Supplemental Fig. 1A–C; Supplemental material). Similarly, analyses excluding single loci (“leave-one-out” analysis) from the concatenated set did not yield substantially different branch support from the overall total evidence topology (Supplemental material; Supplemental Table 3).
Concordance across data sets
A Bayesian analysis of concordance across 13 of our nuclear loci produced a primary concordance tree with a topology completely congruent with our total evidence trees (Fig. 2; Supplemental Table 4). Identical results were obtained when using α priors of 0.1, 1, and 10. A key result of these analyses was the high level of concordance across loci for all branches. All but two branches in the primary concordance tree have clade concordance factors (CCFs) >0.89, with 95% confidence intervals including a CCF of 1 (i.e., these nodes are supported by all 13 assessed loci). The two nodes with slightly lower CCFs are the placement of Propithecus (family Indriidae) in a clade with the Cheirogaleidae and Lepilemuridae (Fig. 1, node 8), and the placement of Eulemur as sister to the Lemur plus Hapalemur clade (Fig. 1, node 18). Our analyses still yield relatively high CCFs for these nodes (>0.761), in favor of the primary concordance tree. However, these analyses do identify three loci (ABCA1, CFTR-pair A, and RBP3) that appear to strongly contradict these latter two relationships, with joint posterior densities that favor the placement of the Indriidae as the sister lineage to the Lemuridae and favor the sister relationship of the genera Eulemur and Varecia (Supplemental Table 4). Further details regarding concordance among loci can be found in the Supplemental materials.
Figure 2.
Bayesian primary concordance tree. The Bayesian primary concordance tree is shown, resulting from analyses of 13 nuclear loci, using the program BUCKy (Larget 2006). Numbers on branches represent the average concordance factor. The 95% confidence intervals that are not exclusively 1.0 are included in parentheses. Results are identical across analyses using α priors of 0.1, 1, and 10. Loci not used in this analysis are listed in the Supplementary material.
Lemur divergence estimates
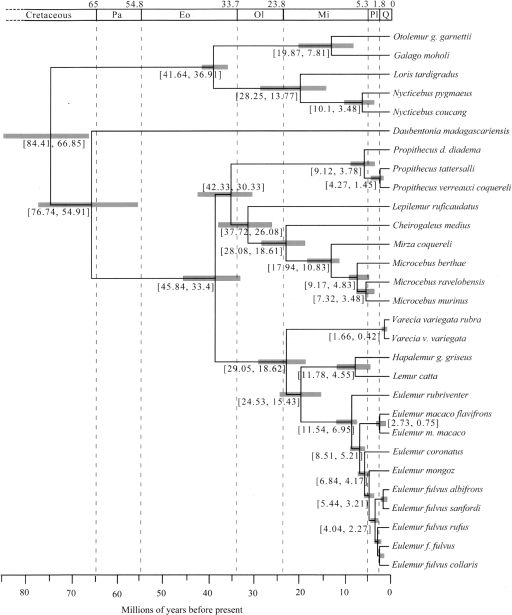
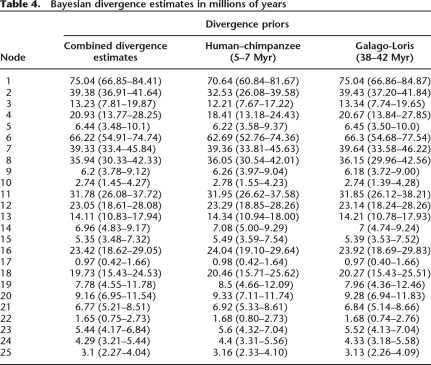
Divergence times were estimated using the combined nuclear data and two calibrations: the Human–Chimpanzee divergence at 5–7 million years ago (Mya) (Kumar et al. 2005), and the Galagidae–Lorisidae divergence at 38–42 Mya (Fig. 3; Table 4) (Seiffert et al. 2003). Divergence between the Lemuriformes and Lorisiformes is estimated at 75 Mya (with a 95% credibility interval [CI] of 66.9–84.4 Mya). The mean age estimate for the earliest lemuriform divergence between Daubentonia and all other lemurs is 66 Mya, with a CI (54.9–74.7 Mya) that overlaps the 65 Mya Cretaceous/Tertiary (K/T) boundary. These age estimates are congruent with previous estimates of strepsirrhine and lemuriform antiquity (Roos et al. 2004; Yoder and Yang 2004) (Supplemental Fig. 2). Similarly, these analyses agree with previous findings suggesting an Eocene/Oligocene timing for diversification of the majority of family-level lemur lineages (excluding Daubentoniidae).
Figure 3.
Divergence time estimates for the strepsirrhine lineages. An ultrametric tree with divergence time estimates resulting from the combined posterior distribution of four independent BEAST analyses (see Methods) of the combined nuclear data set. The results are based on prior date estimates of (1) the 6 Mya (5–7 Mya) split between Homo sapiens and Pan troglodytes (Kumar et al. 2005) and (2) the 40 Mya (38–42 Mya) split between the Lorisidae and Galagidae (Seiffert et al. 2003). Shaded gray boxes and numbers within brackets span the 95% highest posterior density of divergence time estimates. The scale bar is divided up by time (in millions of years before present) according to the Cretaceous, Tertiary, and Quaternary periods. The Tertiary period is shown according to epochs (Pa indicates Paleocene; Eo, Eocene; Ol, Oligocene; Mi, Miocene; Pl, Pliocene). Full details of time estimates from all BEAST analyses are presented in Table 4.
Table 4.
Bayesian divergence estimates in millions of years
Discussion
The power of our data set for resolving the evolutionary history of the lemuriformes demonstrates the utility of phylogenetic marker development utilizing genomic resources from a focal set of taxa. In our study, the use of BAC library sequence data from two relatively divergent species (Lemur and Microcebus) led to the development of PCR primer pairs with exceptionally high efficacy across strepsirrhine taxa. In contrast, the use of previously published mammalian primers yielded data sets featuring incomplete taxon sampling (see Supplemental Table 5), some of which are unable to contribute to the phylogenetic resolution of key lemuriform family-level lineages. Overall, the combination of 11 newly developed loci combined with seven previously published nuclear loci, provides the most comprehensive strepsirrhine phylogenetic analysis to date, with more than 400 kb compared among 29 strepsirrhine and three anthropoid primate taxa. The unequivocal support of lemur monophyly and the position of Daubentoniidae as the most basal lineage is not surprising given past studies (Yoder et al. 1996a, 2003; Porter et al. 1997a; Stanger-Hall and Cunningham 1998; Pastorini et al. 2003; Poux and Douzery 2004; Roos et al. 2004; Yoder and Yang 2004; Poux et al. 2005; DelPero et al. 2006). However, the fact that many of these past phylogenies did not sufficiently resolve particular family relationships (or did so in the absence of convincing branch support) (Yoder and Yang 2004; Poux et al. 2005) raised the issue of whether these data sets were merely too small or whether these phylogenetic relationships represented a hard polytomy (i.e., rapid speciation event into three or more lineages essentially simultaneously). Our phylogenetic resolution of the major lemur lineages with high levels of concordance across loci is strong evidence against a polytomous history among the major lemur lineages and instead argues for a deep bifurcating history. It also accentuates the point that difficult phylogenetic problems are best addressed via the application of large data sets drawn from across the breadth of the genome.
Implications of loci concordance
Both the Bayesian concordance and “leave-one-out” analyses indicate that most loci contribute phylogenetic signal to the overall topology and that most loci are in agreement (Fig. 2; Supplemental Tables 3, 4; Methods). The Bayesian concordance analyses did, however, uncover an interesting pattern in which three loci were consistent in yielding joint posterior densities that favor an alternative hypothesis that shifts the Indriidae to a sister position with the Lemuridae and places Eulemur and Varecia as sister lineages (Supplemental Table 4). The cause for this discordance may be related to the short evolutionary durations preceding nodes 8 and 18. As the durations between branching events decrease, there is an increased chance that gene trees will disagree due to an incomplete lineage sorting process (Hudson 1992). While this incongruence is more often thought to affect genealogical discordance at the population-species boundary, it has also been shown to have substantial effects at deeper levels of evolutionary divergence (Takahashi et al. 2001).
Short evolutionary durations between deep branching events are also expected to challenge phylogenetic resolution due to a limited set of synapomorphic substitutions and the loss of signal due to multiple substitutions. This challenge is well recognized in phylogenetics (Felsenstein 1978; Huelsenbeck 1998; Fishbein et al. 2001; Weisrock et al. 2005) and has been highlighted in previous attempts at resolving lemur phylogeny with mtDNA sequence data (Adkins and Honeycutt 1994; Yoder et al. 1996b) in which lemur monophyly is only recovered when using third position transversions in protein-coding genes. These previous results are the likely explanation for the decreased branch support in our analyses of the combined nuclear and mtDNA data. Furthermore, it should not be too surprising to find alternative phylogenetic hypotheses for some taxa in a small number of individual nuclear gene trees. Overall, the vast majority of loci used in this study resolve concordant relationships for nearly all branches in our tree, including the placement of the Indriidae, Cheirogaleidae, and Lepilemuridae into a clade. These consistent phylogenetic patterns across numerous independent loci are indicative of an evolutionary history that is recoverable, despite relatively short durations between deep branching events.
A biogeographical context for lemur divergence time estimates
This resolved lemur phylogeny provides an opportunity to explore basal primate origins, the historical biogeography of Madagascar (Yoder et al. 1996a), and primate speciation events (Horvath and Willard 2007) and should contribute to conservation efforts (Ryder 2005; Lehman 2006). Divergence age analyses calculated here and elsewhere (Yoder and Yang 2004; Poux et al. 2005) estimated that lemurs did not arrive in Madagascar until 50–80 Mya. Given that the time of lemuriform divergence clearly postdates the separation of Madagascar from other landmasses by many millions of years, it seems clear that lemurs arrived via oceanic dispersal (Kappeler 2000; Yoder et al. 1996a, 2003) though other biogeographic mechanisms have been proposed (McCall 1997; Arnason et al. 2000). Once the ancestral lemurs arrived on Madagascar, there appear to have been at least two prominent episodes of lineage diversification, at the family and species levels. Following the deepest divergence between Daubentonia and the common ancestor of all remaining lemurs, the four remaining family-level lineages diverged within a window of 16 million years (Myr), between the end of the Eocene and through the Oligocene (23.8–40 Mya) (see Fig. 3). Coincident with this timeframe, rainforests began emerging (Wells 2003; Yoder and Yang 2004) and many other mammals such as carnivores, tenrecs, and rodents may have colonized Madagascar (Poux et al. 2005). As a working hypothesis, one can imagine that the combination of the changing climate combined with the arrival of new predators and competitors would have altered the ecological landscape and thus served as an impetus for lineage diversification within the lemuriform clade.
The divergence time estimates for the origin of diversification in the genera Eulemur and Microcebus, two of the most diverse lemur genera, are both hypothesized to have emerged in the late Miocene (∼8 Mya). The past 8 Myr on Madagascar initially began with a wetter climate from monsoons (Wells 2003) and, if congruent with global patterns, would have experienced plant biome changes (Cerling et al. 1997), some of which may have been in response to Pleistocene glaciation cycles (Wilme et al. 2006). Again, though cause and effect are hypothetical, such environmental changes may have stimulated speciation within the Eulemur and Microcebus lineages.
The value of phylogenomic toolkits
Despite the large investment required to develop phylogenomic toolkits such as the one presented here, the analytical payoff can be enormous. Although our taxonomic focus was strepsirrhine phylogeny, the primer design approach used in this study can be applied to any organismal group, even those with limited amounts of available genomic sequence data. Our resulting lemur-based PCR primers proved to be applicable to divergent strepsirrhine taxa separated by as much as 84 Myr of evolutionary divergence (Fig. 3; Table 4). Consequently, toolkit design can span great divergences but will be more easily applied to closely related species. The development of conserved primer pairs that work in a diverse set of species is obviously geared toward phylogenetic analysis at and above the species level, but may also prove useful in analyses at the intraspecific level, either in genealogical-based analyses (Rosenberg and Nordborg 2002), or as the basis for single nucleotide polymorphism development (Brumfield et al. 2003). Consequently, intraspecific application of our toolkit may provide useful estimates of genetic diversity within populations and be useful for directing conservation strategies (Ryder 2005; Lehman 2006). Our hope is that these PCR primers will become a standard molecular resource utilized by lemur researchers with access to species not represented here, thus offering a means for filling in missing taxa and enhancing the overall lemur phylogeny. While we have extensive species sampling within some genera (e.g., Eulemur), other genera are missing entirely (Phaner, Allocebus, Indri, Avahi) and will be important to include in future studies to verify their placement with respect to those currently assessed. As these missing taxa are well established as being included within the major family-level lineages of focus in this study, we do not expect the tree topology to change substantially.
The ultimate goal of our approach was to design a method applicable to any clade-specific phylogenetic problem wherein at least one species offers genomic sequence. This strategy was tested in the evolutionarily diverse lemurs and was successful for resolving their evolutionary history. While we anticipate that our phylogenomic toolkit (the 11 newly designed primer pairs and corresponding strepsirrhine genomic sequence) will be of use to other primatologists for genomic comparisons and for resolution of the evolutionary history of their study taxa, its application extends much further. This primer design approach can be applied to any organism with available genomic sequence for phylogenetic analyses, diversity assessments, or other molecular evolutionary comparisons across diverse taxa. Primer design in other nonmodel organisms will facilitate the exploration of more complete comparative genomics, and will lead to a better understanding of the tree of life.
Methods
Taxon sampling
Sequence data were collected from 29 strepsirrhine taxa representing all five lemuriform families and both lorisiform families (Galagidae and Lorisidae) (Table 1). Sequence data from three outgroup taxa with complete genome sequence were also used (Homo sapiens, Pan troglodytes, Macaca mulatta). The majority of ingroup samples were obtained from the Duke Lemur Center (http://lemur.duke.edu/). Samples of Microcebus ravelobensis, Microcebus berthae, and Lepilemur ruficaudatus are from the personal collection of the authors (A.D. Yoder and P. Kappeler). The sample for Lemur catta (cell line AG07100) was obtained from Coriell Cell Repositories (http://locus.umdnj.edu/). The sample for Eulemur m. macaco (cell line PR00254) was obtained from Integrated Primate Biomaterials and Information Resource (http://ccr.coriell.org/Sections/Collections/IPBIR/?SsId=18).
Genomic DNA sample collection
Genomic DNA was isolated from buccal cells collected on brushes (PUREGENE kit, Qiagen), from blood, and from cell line material. Genomic DNA was isolated using the PUREGENE kit and the manufacturer’s recommended protocol (Qiagen). Approximately 10–100 ng genomic DNA (1/20 volume buccal or 1/80 volume blood DNA isolated) was used in whole-genome amplification (WGA) reactions (REPLI-g [Qiagen] or GenomiPhi [GE Healthcare]) according to the manufacturer’s recommendations. Each set of WGA reactions was set up with a negative control to ensure no cross-contamination of genomic DNA samples occurred during the WGA procedure.
Primer design
We chose two approaches to design PCR primer pairs to obtain loci conserved in a diverse set of strepsirrhine taxa. First, we reasoned that loci conserved between human and a nonprimate outgroup, such as the dog, would likely also be conserved in most primate taxa and could be used to develop loci for deep phylogenetic comparisons (i.e., presently the standard approach to primer design). Initial primer design using human and dog sequence targeted six genomic regions that were assessed for conservation in both species using the conservation track in the UCSC genome browser (http://genome.ucsc.edu/). By use of the conservation track alignments, 17 primer pairs were designed by hand to be 100% identical between human and dog, 20–24 bp in length and 45%–55% GC rich, and to terminate with a G or C nucleotide at the 3′ end. Two primers were designed with degenerate bases at two sites not identical between human and dog. No primers were designed within, or spanning, known repetitive elements.
Our second approach used lemur and human sequence comparisons since sequence conserved between one lemur species and human would likely be conserved among other strepsirrhine taxa. At the onset of this project, as is typically true for nonmodel organisms, no partially or fully sequenced genome existed for any strepsirrhine primate. Therefore, we relied upon publicly available bacterial artificial chromosome (BAC) sequences from two lemur species (the ringtailed lemur, L. catta, and the gray mouse lemur, M. murinus). We downloaded three ringtailed lemur BAC sequences (accession nos. AC123971, AC145463, and AC145484) from GenBank (http://www.ncbi.nlm.nih.gov/) and masked them for known primate repeat elements (RepeatMasker 2004, A. Smit; http://www.repeatmasker.org). The repeat masked BAC sequences were compared with the human genome (hg18, March 2006) by BLAST (blastall v2.2.10, http://www.ncbi.nlm.nih.gov/BLAST/), and 16 primer pairs were designed by hand using the above parameters.
The human and lemur sequence comparisons were much more successful than human and dog and therefore were used for further primer design with a semi-automated program, primer_ conservation.py. Using primer_conservation.py, 48 L. catta and 42 M. murinus, repeat masked lemur BACs were compared with the human genome by BLAST. Coordinates of 100% identity were submitted to Primer3 for primer design (v 0.3.0) (Rozen and Skaletsky 2000; source code available at http://fokker.wi.mit.edu/primer3/). Parameters for primer design were as follows: 600–1000 bp product size, 22–26 bp primer length, 45%–65% GC, 1-bp GC clamp at the 3′ end, and primer pairs that did not span, or contain, a known repeat element. The output primers were assessed in the human genome to ensure no duplicated regions were utilized for further analyses. By use of this program with human and lemur sequence comparisons, an additional 31 primer pairs were designed in 16 of the 90 newly downloaded BAC sequences. The remaining BAC sequences did not return primer pairs with our parameters and were not utilized further.
Combined, these approaches yielded a total of 64 primer pairs that were assessed for success in PCR assays on a panel of four taxa chosen for diverse placement in the phylogenetic tree (H. sapiens, Daubentonia madagascariensis, L. catta, and Otolemur g. garnettii). A successful primer pair was one that amplified a single band in PCR assays in all four taxa. Of the 17 human–dog primer pairs, only one (6%) was successful, while 14 of 16 (88%) human–lemur hand-designed primer pairs and 15 of 31 (48%) human–lemur program-designed primer pairs were successful in the four-taxon panel. The human–dog genomic regions chosen for primer design were different from the human–lemur regions and could have contributed to the lower level of primer pair success.
Next, the 29 successful human–lemur–based primers were assessed in PCR assays using an expanded panel of 10 primates (H. sapiens, Cheirogaleus medius, D. madagascariensis, Eulemur f. fulvus, L. catta, M. murinus, Nycticebus coucang, O. g. garnettii, Propithecus tattersalli, and Varecia v. variegata). This resulted in 16 of 29 (55%) primer pairs that successfully amplified a product in all 10 primate taxa. The primer pairs with the largest, most robust amplicons (using L. catta accession nos. AC145533, AC146284, and AC139880 in addition to M. murinus accession nos. AC145757 and AC186699) were selected, and 11 primer pairs successfully amplified all 32 primates in our panel (see Supplemental Table 5). Primer pairs that were not successful in the panel of four or 10 taxa often failed to amplify in multiple taxa, though no consistent pattern emerged.
PCR and sequencing
Standard PCR assays were conducted using ∼50–150 ng DNA as template and Platinum Taq High Fidelity (Invitrogen) enzyme (for details, see Supplemental material). PCR products were directly sequenced using both forward and reverse PCR primers and BigDye Terminator v3.1 (Applied Biosystems) (for details, see Supplemental material). PCR products failing to produce high-quality sequence by direct sequencing methods were ligated using the pGEM-T Easy Vector System II (Promega) with ∼20–40 ng PCR product as insert (for details, see Supplemental Material). Sequence quality was assessed using either phred/phrap/consed (Gordon et al. 1998) or Sequencher (Gene Codes Corporation). Base calls with a phred quality score of less than 30 were scored as Ns. Accession numbers corresponding to each sequence are deposited in GenBank (Supplemental Table 6). Outgroup sequences (human, chimpanzee, macaque) were obtained using PSL map (http://hgdownload.cse.ucsc.edu/downloads.html) based on the human coordinates.
Phylogenetic analyses
All sequences were globally aligned using MUSCLE (http://www.drive5.com/muscle/; Edgar 2004a, b), with comparisons to ClustalW (Wilbur and Lipman 1983; Myers and Miller 1988) for manual editing. Regions of ambiguous alignment were excluded from analyses. Parsimony analyses were performed on each individual locus data set as well as on a combined data set with all 18 nuclear loci, including and excluding previously published mtDNA sequence data. Parsimony analyses were performed using the program PAUP* v4.0 (Swofford 2002). Heuristic searches were performed with 100 random-addition replicates and TBR branch swapping. All parsimony analyses gave equal weight to all character changes. To assess support for branches in parsimony trees, bootstrap percentages (BPs) were calculated using 1000 pseudoreplicates with 10 random additions per replicate. Decay indices were calculated using constraint trees generated in TreeRot v2 (Sorenson 1999) and analyzed in PAUP*.
Maximum likelihood and Bayesian analyses were performed using the best-fit model selected using the Akaike Information Criterion (AIC) in the program MrModeltest v2.2 (Nylander 2004). ML analyses were performed in the program GARLI v0.9.5.1 (www.bio.utexas.edu/faculty/antisense/garli/Garli.html; Zwickl 2006). Three replicate analyses were performed using random starting trees. Default conditions were used for the genetic algorithm search. Branch support for ML trees was generated using bootstrapping with 100 pseudoreplicates. Bootstrap analyses were performed using default conditions except that runs were set to terminate after 5000 generations without an improvement in topology. Combined GARLI analyses were unpartitioned using the general time-reversible (GTR) model with rate heterogeneity (Γ) and a proportion of sites assumed to be invariant (I). All Bayesian analyses (Huelsenbeck et al. 2001; Ronquist and Huelsenbeck 2003) were run with MrBayes v3.1.2 and used four Markov chains with the temperature profile at the default setting of 0.2. Flat dirichlet priors were used for GTR substitution-rate parameters and for all base-frequency parameters. A flat Beta prior was used in estimating the transition/transversion substitution-rate parameter. Uniform priors were used for the Γ and I parameters. Unconstrained, uniform priors were used for topology and branch-length estimation. A molecular clock was not enforced. Combined analyses were performed in a partitioned framework, allowing for independent parameter estimation at each locus. Partitioned analyses involving mtDNA data treated MT-CO2 and MT-CYB as separate partitions. All Bayesian analyses were run for 1 million generations and replicated four times. Individual runs were compared with each other in the program Tracer v1.3 (Drummond et al. 2003) to assess convergence. Tree and parameter samplings from the latter 500,000 generations of each run were pooled and used as an estimate of the posterior distribution. A Bayesian concordance analysis was performed to assess concordance in gene trees across loci following the method of Ane et al. (2007). Full details of this method are given in the Supplementary materials.
Divergence time estimation
A rate-smoothed tree with divergence times was estimated from our 18 locus nuclear data set using a Bayesian relaxed clock method in BEAST v1.4 (Drummond et al. 2006). BEAST analyses used a relaxed lognormal model of lineage rate variation, a GTR+I+Γ model of nucleotide substitution, and a Yule prior for branching rates. No fossil-based time calibrations are currently available for the Lemuriformes (Simons et al. 1995). However, two calibrations are available that are relevant to our data set: (1) The split between slow lorises (Loris and Nycticebus) and galagos (Galago and Otolemur) has been dated at 38–42 Mya (Seiffert et al. 2003), and we used this date as a normal-distribution prior on this node with a mean of 40 Mya and a SD of 1.2 Myr. (2) The split between H. sapiens and P. troglodytes has been estimated through fossil and molecular evidence to have occurred at 5–7 Mya (Kumar et al. 2005), and we use this date as a normal-distribution prior with a mean of 6 Mya and a SD of 0.6 Myr. The result of each of these divergence time priors was assessed individually and in combination. In all BEAST analyses, we performed four replicate analyses of 10,000,000 generations each. The sampling distributions of each run were visualized using the program Tracer v1.3 (Drummond et al. 2003) to ensure the achievement of a stable posterior distribution in all parameter estimates, and then combined using the program LogCombiner v1.4. A consensus chronogram with node height distributions was then generated using TreeAnnotater v1.4. Because we only have two priors to calibrate our divergence calculations, our results may suffer from underestimates of nodal divergence times (Yoder and Yang 2004). Until basal terrestrial Tertiary fossil lemurs are discovered (if any even exist), these molecular calculations will serve as our best estimate of divergence dates for the Malagasy lemurs.
Acknowledgments
We thank Karen Hayden, Darin London, Stacey Smith, and Olivier Fedrigo for computational assistance and advice, and Lisa Bukovnik for technical assistance. J.E.H. was supported in part by a fellowship from the Center for Evolutionary Genomics in the Institute for Genome Sciences & Policy at Duke University. This research was conducted under Duke Lemur Center research project BS-4-06-1 and Institutional Animal Care and Use Committee (IACUC) project A094-06-03. This paper is Duke Lemur publication no. 1127.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. EU057196–EU057514 and EU342218–EU342345.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7265208
References
- Adkins R.M., Honeycutt R.L., Honeycutt R.L. Evolution of the primate cytochrome c oxidase subunit II gene. J. Mol. Evol. 1994;38:215–231. doi: 10.1007/BF00176084. [DOI] [PubMed] [Google Scholar]
- Ane C., Larget B., Baum D.A., Smith S.D., Rokas A., Larget B., Baum D.A., Smith S.D., Rokas A., Baum D.A., Smith S.D., Rokas A., Smith S.D., Rokas A., Rokas A. Bayesian estimation of concordance among gene trees. Mol. Biol. Evol. 2007;24:412–426. doi: 10.1093/molbev/msl170. [DOI] [PubMed] [Google Scholar]
- Arnason U., Gullberg A., Burguete A.S., Janke A., Gullberg A., Burguete A.S., Janke A., Burguete A.S., Janke A., Janke A. Molecular estimates of primate divergences and new hypotheses for primate dispersal and the origin of modern humans. Hereditas. 2000;133:217–228. doi: 10.1111/j.1601-5223.2000.00217.x. [DOI] [PubMed] [Google Scholar]
- Bapteste E., Brinkmann H., Lee J.A., Moore D.V., Sensen C.W., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Brinkmann H., Lee J.A., Moore D.V., Sensen C.W., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Lee J.A., Moore D.V., Sensen C.W., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Moore D.V., Sensen C.W., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Sensen C.W., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Durufle L., Gaasterland T., Lopez P., Muller M., Gaasterland T., Lopez P., Muller M., Lopez P., Muller M., Muller M., et al. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc. Natl. Acad. Sci. 2002;99:1414–1419. doi: 10.1073/pnas.032662799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumfield R.T., Beerli P., Nickerson D.A., Edwards S.V., Beerli P., Nickerson D.A., Edwards S.V., Nickerson D.A., Edwards S.V., Edwards S.V. The utility of single nucleotide polymorphisms in inferences of population history. Trends Ecol. Evol. 2003;18:249–256. [Google Scholar]
- Cannarozzi G., Schneider A., Gonnet G., Schneider A., Gonnet G., Gonnet G. A phylogenomic study of human, dog, and mouse. PLoS Comput. Biol. 2007;3:e2. doi: 10.1371/journal.pcbi.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerling T.E., Harris J.M., MacFadden B.J., Leakey M.G., Quade J., Eisenmann V., Ehleringer J.R., Harris J.M., MacFadden B.J., Leakey M.G., Quade J., Eisenmann V., Ehleringer J.R., MacFadden B.J., Leakey M.G., Quade J., Eisenmann V., Ehleringer J.R., Leakey M.G., Quade J., Eisenmann V., Ehleringer J.R., Quade J., Eisenmann V., Ehleringer J.R., Eisenmann V., Ehleringer J.R., Ehleringer J.R. Global vegetation change through the Miocene/Pliocene boundary. Nature. 1997;389:153–158. [Google Scholar]
- Crovella S., Montagnon D., Rumpler Y., Montagnon D., Rumpler Y., Rumpler Y. Highly repeated DNA analysis and systematics of the Lemuridae, a family of Malagasy prosimians. Primates. 1993;34:61–69. [Google Scholar]
- Crovella S., Montagnon D., Rumpler Y., Montagnon D., Rumpler Y., Rumpler Y. Highly repeated DNA sequences and systematics of malagasy primates. Hum. Evol. 1995;10:35–44. [Google Scholar]
- DelPero M., Crovella S., Cervella P., Ardito G., Rumpler Y., Crovella S., Cervella P., Ardito G., Rumpler Y., Cervella P., Ardito G., Rumpler Y., Ardito G., Rumpler Y., Rumpler Y. Phylogenetic relationships among Malagasy lemurs as revealed by mitochondrial DNA sequence analysis. Primates. 1995;36:431–440. [Google Scholar]
- DelPero M., Masters J.C., Cervella P., Crovella S., Ardito G., Rumpler Y., Masters J.C., Cervella P., Crovella S., Ardito G., Rumpler Y., Cervella P., Crovella S., Ardito G., Rumpler Y., Crovella S., Ardito G., Rumpler Y., Ardito G., Rumpler Y., Rumpler Y. Phylogenetic relationships among the Malagasy lemuriforms (Primates: Strepsirrhini) as indicated by mitochondrial sequence data from the 12S rRNA gene. Zool. J. Linn. Soc. 2001;133:83–103. [Google Scholar]
- DelPero M., Pozzi L., Masters J.C., Pozzi L., Masters J.C., Masters J.C. A composite molecular phylogeny of living lemuroid primates. Folia Primatol. 2006;77:434–445. doi: 10.1159/000095390. [DOI] [PubMed] [Google Scholar]
- Drummond A., Pybus O.G., Rambaut A., Pybus O.G., Rambaut A., Rambaut A. Inference of viral evolutionary rates from molecular sequences. Adv. Parasitol. 2003;54:331–358. doi: 10.1016/s0065-308x(03)54008-8. [DOI] [PubMed] [Google Scholar]
- Drummond A.J., Ho S.Y., Phillips M.J., Rambaut A., Ho S.Y., Phillips M.J., Rambaut A., Phillips M.J., Rambaut A., Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutrillaux B., Rumpler Y., Rumpler Y. Phylogenetic relations among Prosimii with special reference to Lemuriformes and Malagasy nocturnals. In: Alterman L., et al., editors. Creatures of the dark. The nocturnal prosimians. Plenum Press; New York: 1995. pp. 141–150. [Google Scholar]
- Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004a;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004b;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 1978;27:401–410. [Google Scholar]
- Fishbein M., Hibsch-Jetter C., Soltis D.E., Hufford L., Hibsch-Jetter C., Soltis D.E., Hufford L., Soltis D.E., Hufford L., Hufford L. Phylogeny of saxifragales (angiosperms, eudicots): Analysis of a rapid, ancient radiation. Syst. Biol. 2001;50:817–847. doi: 10.1080/106351501753462821. [DOI] [PubMed] [Google Scholar]
- Galewski T., Tilak M.K., Sanchez S., Chevret P., Paradis E., Douzery E.J., Tilak M.K., Sanchez S., Chevret P., Paradis E., Douzery E.J., Sanchez S., Chevret P., Paradis E., Douzery E.J., Chevret P., Paradis E., Douzery E.J., Paradis E., Douzery E.J., Douzery E.J. The evolutionary radiation of Arvicolinae rodents (voles and lemmings): Relative contribution of nuclear and mitochondrial DNA phylogenies. BMC Evol. Biol. 2006;6:80. doi: 10.1186/1471-2148-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Abajian C., Green P., Abajian C., Green P., Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Harlin-Cognato A.D., Honeycutt R.L., Honeycutt R.L. Multi-locus phylogeny of dolphins in the subfamily Lissodelphininae: Character synergy improves phylogenetic resolution. BMC Evol. Biol. 2006;6:87. doi: 10.1186/1471-2148-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman K.L., Mariani C.L., Rasoloarison R., Yoder A.D., Mariani C.L., Rasoloarison R., Yoder A.D., Rasoloarison R., Yoder A.D., Yoder A.D. Multiple nuclear loci reveal patterns of incomplete lineage sorting and complex species history within Western mouse lemurs (Microcebus) Mol. Phylogenet. Evol. 2007;43:353–367. doi: 10.1016/j.ympev.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Hedtke S.M., Townsend T.M., Hillis D.M., Townsend T.M., Hillis D.M., Hillis D.M. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst. Biol. 2006;55:522–529. doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- Horvath J.E., Willard H.F., Willard H.F. Primate comparative genomics: Lemur biology and evolution. Trends Genet. 2007;23:173–182. doi: 10.1016/j.tig.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Hudson R.R. Gene trees, species trees and the segregation of ancestral alleles. Genetics. 1992;131:509–513. doi: 10.1093/genetics/131.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J.P. Systematic bias in phylogenetic analysis: Is the Strepsiptera problem solved? Syst. Biol. 1998;47:519–537. [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F., Nielsen R., Bollback J.P., Ronquist F., Nielsen R., Bollback J.P., Nielsen R., Bollback J.P., Bollback J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- James T.Y., Kauff F., Schoch C.L., Matheny P.B., Hofstetter V., Cox C.J., Celio G., Gueidan C., Fraker E., Miadlikowska J., Kauff F., Schoch C.L., Matheny P.B., Hofstetter V., Cox C.J., Celio G., Gueidan C., Fraker E., Miadlikowska J., Schoch C.L., Matheny P.B., Hofstetter V., Cox C.J., Celio G., Gueidan C., Fraker E., Miadlikowska J., Matheny P.B., Hofstetter V., Cox C.J., Celio G., Gueidan C., Fraker E., Miadlikowska J., Hofstetter V., Cox C.J., Celio G., Gueidan C., Fraker E., Miadlikowska J., Cox C.J., Celio G., Gueidan C., Fraker E., Miadlikowska J., Celio G., Gueidan C., Fraker E., Miadlikowska J., Gueidan C., Fraker E., Miadlikowska J., Fraker E., Miadlikowska J., Miadlikowska J., et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Johnson W.E., Eizirik E., Pecon-Slattery J., Murphy W.J., Antunes A., Teeling E., O’Brien S.J., Eizirik E., Pecon-Slattery J., Murphy W.J., Antunes A., Teeling E., O’Brien S.J., Pecon-Slattery J., Murphy W.J., Antunes A., Teeling E., O’Brien S.J., Murphy W.J., Antunes A., Teeling E., O’Brien S.J., Antunes A., Teeling E., O’Brien S.J., Teeling E., O’Brien S.J., O’Brien S.J. The late Miocene radiation of modern Felidae: A genetic assessment. Science. 2006;311:73–77. doi: 10.1126/science.1122277. [DOI] [PubMed] [Google Scholar]
- Kappeler P.M. Lemur origins: Rafting by groups of hibernators? Folia Primatol. 2000;71:422–425. doi: 10.1159/000052741. [DOI] [PubMed] [Google Scholar]
- Karanth K.P., Delefosse T., Rakotosamimanana B., Parsons T.J., Yoder A.D., Delefosse T., Rakotosamimanana B., Parsons T.J., Yoder A.D., Rakotosamimanana B., Parsons T.J., Yoder A.D., Parsons T.J., Yoder A.D., Yoder A.D. Ancient DNA from giant extinct lemurs confirms single origin of Malagasy primates. Proc. Natl. Acad. Sci. 2005;102:5090–5095. doi: 10.1073/pnas.0408354102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs J.O., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J., Kiefmann M., Jordan U., Brosius J., Schmitz J., Jordan U., Brosius J., Schmitz J., Brosius J., Schmitz J., Schmitz J. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Filipski A., Swarna V., Walker A., Hedges S.B., Filipski A., Swarna V., Walker A., Hedges S.B., Swarna V., Walker A., Hedges S.B., Walker A., Hedges S.B., Hedges S.B. Placing confidence limits on the molecular age of the human-chimpanzee divergence. Proc. Natl. Acad. Sci. 2005;102:18842–18847. doi: 10.1073/pnas.0509585102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larget B. 2006. Bayesian untangling of Concordance Knots (BUCKy) [Google Scholar]
- Lehman S.M. Conservation biology of Malagasy strepsirhines: A phylogenetic approach. Am. J. Phys. Anthropol. 2006;130:238–253. doi: 10.1002/ajpa.20239. [DOI] [PubMed] [Google Scholar]
- Li C., Orti G., Orti G. Molecular phylogeny of Clupeiformes (Actinopterygii) inferred from nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2007;44:386–398. doi: 10.1016/j.ympev.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Li C., Orti G., Zhang G., Lu G., Orti G., Zhang G., Lu G., Zhang G., Lu G., Lu G. A practical approach to phylogenomics: The phylogeny of ray-finned fish (Actinopterygii) as a case study. BMC Evol. Biol. 2007;7:44. doi: 10.1186/1471-2148-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber S.T. Phylogeny of Gaertnera Lam. (Rubiaceae) based on multiple DNA markers: Evidence of a rapid radiation in a widespread, morphologically diverse genus. Evolution Int. J. Org. Evolution. 2002;56:42–57. doi: 10.1111/j.0014-3820.2002.tb00848.x. [DOI] [PubMed] [Google Scholar]
- McCall R.A. Implications of recent geological investigations of the Mozambique Channel for the mammalian colonization of Madagascar. Proc. Biol. Sci. 1997;264:663–665. doi: 10.1098/rspb.1997.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W.J., Eizirik E., Johnson W.E., Zhang Y.P., Ryder O.A., O’Brien S.J., Eizirik E., Johnson W.E., Zhang Y.P., Ryder O.A., O’Brien S.J., Johnson W.E., Zhang Y.P., Ryder O.A., O’Brien S.J., Zhang Y.P., Ryder O.A., O’Brien S.J., Ryder O.A., O’Brien S.J., O’Brien S.J. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Pevzner P.A., O’Brien S.J., Pevzner P.A., O’Brien S.J., O’Brien S.J. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Pringle T.H., Crider T.A., Springer M.S., Miller W., Pringle T.H., Crider T.A., Springer M.S., Miller W., Crider T.A., Springer M.S., Miller W., Springer M.S., Miller W., Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E.W., Miller W., Miller W. Optimal alignments in linear space. Comput. Appl. Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Nikolaev S., Montoya-Burgos J.I., Margulies E.H., Rougemont J., Nyffeler B., Antonarakis S.E., Montoya-Burgos J.I., Margulies E.H., Rougemont J., Nyffeler B., Antonarakis S.E., Margulies E.H., Rougemont J., Nyffeler B., Antonarakis S.E., Rougemont J., Nyffeler B., Antonarakis S.E., Nyffeler B., Antonarakis S.E., Antonarakis S.E. Early history of mammals is elucidated with the ENCODE multiple species sequencing data. PLoS Genet. 2007;3:e2. doi: 10.1371/journal.pgen.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan B.P., Chippindale P.T., Chippindale P.T. Vicariant origin of malagasy reptiles supports late cretaceous antarctic land bridge. Am. Nat. 2006;168:730–741. doi: 10.1086/509052. [DOI] [PubMed] [Google Scholar]
- Nunn C.L., Barton R.A., Barton R.A. Comparative methods for studying primate adaptation and allometry. Evol. Anthropol. 2001;10:81–98. [Google Scholar]
- Nylander J.A.A. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre; Uppsala University, Uppsala, Sweden: 2004. [Google Scholar]
- Palumbi S.R., Baker C.S., Baker C.S. Contrasting population structure from nuclear intron sequences and mtDNA of humpback whales. Mol. Biol. Evol. 1994;11:426–435. doi: 10.1093/oxfordjournals.molbev.a040115. [DOI] [PubMed] [Google Scholar]
- Pastorini J., Thalmann U., Martin R.D., Thalmann U., Martin R.D., Martin R.D. A molecular approach to comparative phylogeography of extant Malagasy lemurs. Proc. Natl. Acad. Sci. 2003;100:5879–5884. doi: 10.1073/pnas.1031673100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard D., Iyer V.N., Moses A.M., Eisen M.B., Iyer V.N., Moses A.M., Eisen M.B., Moses A.M., Eisen M.B., Eisen M.B. Widespread discordance of gene trees with species tree in Drosophila: Evidence for incomplete lineage sorting. PLoS Genet. 2006;2:e173. doi: 10.1371/journal.pgen.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter C.A., Sampaio I., Schneider H., Schneider M.P., Czelusniak J., Goodman M., Sampaio I., Schneider H., Schneider M.P., Czelusniak J., Goodman M., Schneider H., Schneider M.P., Czelusniak J., Goodman M., Schneider M.P., Czelusniak J., Goodman M., Czelusniak J., Goodman M., Goodman M. Evidence on primate phylogeny from epsilon-globin gene sequences and flanking regions. J. Mol. Evol. 1995;40:30–55. doi: 10.1007/BF00166594. [DOI] [PubMed] [Google Scholar]
- Porter C.A., Czelusniak J., Schneider H., Schneider M.P., Sampaio I., Goodman M., Czelusniak J., Schneider H., Schneider M.P., Sampaio I., Goodman M., Schneider H., Schneider M.P., Sampaio I., Goodman M., Schneider M.P., Sampaio I., Goodman M., Sampaio I., Goodman M., Goodman M. Sequences of the primate epsilon-globin gene: Implications for systematics of the marmosets and other New World primates. Gene. 1997a;205:59–71. doi: 10.1016/s0378-1119(97)00473-3. [DOI] [PubMed] [Google Scholar]
- Porter C.A., Page S.L., Czelusniak J., Schneider H., Schneider M.P.C., Sampio I., Goodman M., Page S.L., Czelusniak J., Schneider H., Schneider M.P.C., Sampio I., Goodman M., Czelusniak J., Schneider H., Schneider M.P.C., Sampio I., Goodman M., Schneider H., Schneider M.P.C., Sampio I., Goodman M., Schneider M.P.C., Sampio I., Goodman M., Sampio I., Goodman M., Goodman M. Phylogeny and evolution of selected primates as determined by sequences of the ε-globin locus and 5′ flanking regions. Int. J. Primatol. 1997b;18:261–295. [Google Scholar]
- Poux C., Douzery E.J., Douzery E.J. Primate phylogeny, evolutionary rate variations, and divergence times: A contribution from the nuclear gene IRBP. Am. J. Phys. Anthropol. 2004;124:1–16. doi: 10.1002/ajpa.10322. [DOI] [PubMed] [Google Scholar]
- Poux C., Madsen O., Marquard E., Vieites D.R., de Jong W.W., Vences M., Madsen O., Marquard E., Vieites D.R., de Jong W.W., Vences M., Marquard E., Vieites D.R., de Jong W.W., Vences M., Vieites D.R., de Jong W.W., Vences M., de Jong W.W., Vences M., Vences M. Asynchronous colonization of Madagascar by the four endemic clades of primates, tenrecs, carnivores, and rodents as inferred from nuclear genes. Syst. Biol. 2005;54:719–730. doi: 10.1080/10635150500234534. [DOI] [PubMed] [Google Scholar]
- Rokas A., Williams B.L., King N., Carroll S.B., Williams B.L., King N., Carroll S.B., King N., Carroll S.B., Carroll S.B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Roos C., Schmitz J., Zischler H., Schmitz J., Zischler H., Zischler H. Primate jumping genes elucidate strepsirrhine phylogeny. Proc. Natl. Acad. Sci. 2004;101:10650–10654. doi: 10.1073/pnas.0403852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N.A., Nordborg M., Nordborg M. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat. Rev. Genet. 2002;3:380–390. doi: 10.1038/nrg795. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S., Misener S., Misener S., editors. Bioinformatics methods and protocols: Methods in molecular biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Ryder O.A. Conservation genomics: Applying whole genome studies to species conservation efforts. Cytogenet. Genome Res. 2005;108:6–15. doi: 10.1159/000080796. [DOI] [PubMed] [Google Scholar]
- Seiffert E.R., Simons E.L., Attia Y., Simons E.L., Attia Y., Attia Y. Fossil evidence for an ancient divergence of lorises and galagos. Nature. 2003;422:421–424. doi: 10.1038/nature01489. [DOI] [PubMed] [Google Scholar]
- Simons E.L., Burney D.A., Chatrath P.S., Godfrey L.R., Jungers W.L., Rakotosamimanana B., Burney D.A., Chatrath P.S., Godfrey L.R., Jungers W.L., Rakotosamimanana B., Chatrath P.S., Godfrey L.R., Jungers W.L., Rakotosamimanana B., Godfrey L.R., Jungers W.L., Rakotosamimanana B., Jungers W.L., Rakotosamimanana B., Rakotosamimanana B. AMS 14C dates for extinct lemurs from caves in the Ankarana Massif, Northern Madagascar. Quaternary Res. 1995;43:249–254. [Google Scholar]
- Sorenson M.D. TreeRot, version 2. Boston University; Boston: 1999. [Google Scholar]
- Stanger K.F. The phylogenetic relationships of the extant malagasy lemurs (Lemuriformes) based on DNA sequence data. Am. Zool. 1996;36:134A. [Google Scholar]
- Stanger-Hall K.F. Phylogenetic affinities among the extant Malagasy lemurs (Lemuriformes) based on morphology and behavior. J. Mamm. Evol. 1997;4:163–194. [Google Scholar]
- Stanger-Hall K., Cunningham C.W., Cunningham C.W. Support for a monophyletic lemuriformes: overcoming incongruence between data partitions. Mol. Biol. Evol. 1998;15:1572–1577. doi: 10.1093/oxfordjournals.molbev.a025885. [DOI] [PubMed] [Google Scholar]
- Swofford D.L. PAUP*. Phylogenetic analysis using parsimony (and other methods). Version 4. Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- Takahashi K., Terai Y., Nishida M., Okada N., Terai Y., Nishida M., Okada N., Nishida M., Okada N., Okada N. Phylogenetic relationships and ancient incomplete lineage sorting among cichlid fishes in Lake Tanganyika as revealed by analysis of the insertion of retroposons. Mol. Biol. Evol. 2001;18:2057–2066. doi: 10.1093/oxfordjournals.molbev.a003747. [DOI] [PubMed] [Google Scholar]
- Teeling E.C., Springer M.S., Madsen O., Bates P., O’Brien S.J., Murphy W.J., Springer M.S., Madsen O., Bates P., O’Brien S.J., Murphy W.J., Madsen O., Bates P., O’Brien S.J., Murphy W.J., Bates P., O’Brien S.J., Murphy W.J., O’Brien S.J., Murphy W.J., Murphy W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Telford M.J. Phylogenomics. Curr. Biol. 2007;17:R945–R946. doi: 10.1016/j.cub.2007.09.023. [DOI] [PubMed] [Google Scholar]
- Townsend T., Larson A., Louis E., Macey J.R., Larson A., Louis E., Macey J.R., Louis E., Macey J.R., Macey J.R. Molecular phylogenetics of squamata: The position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst. Biol. 2004;53:735–757. doi: 10.1080/10635150490522340. [DOI] [PubMed] [Google Scholar]
- Weisrock D.W., Harmon L.J., Larson A., Harmon L.J., Larson A., Larson A. Resolving deep phylogenetic relationships in salamanders: Analyses of mitochondrial and nuclear genomic data. Syst. Biol. 2005;54:758–777. doi: 10.1080/10635150500234641. [DOI] [PubMed] [Google Scholar]
- Weisrock D.W., Shaffer H.B., Storz B.L., Storz S.R., Voss S.R., Shaffer H.B., Storz B.L., Storz S.R., Voss S.R., Storz B.L., Storz S.R., Voss S.R., Storz S.R., Voss S.R., Voss S.R. Multiple nuclear gene sequences identify phylogenetic species boundaries in the rapidly radiating clade of Mexican ambystomatid salamanders. Mol. Ecol. 2006;15:2489–2503. doi: 10.1111/j.1365-294X.2006.02961.x. [DOI] [PubMed] [Google Scholar]
- Wells N.A. Some hypotheses on the Mesozoic and Cenozoic paleoenvironmental history of Madagascar. In: Goodman S.M., Benstead J.P., Benstead J.P., editors. The natural history of Madagascar. University of Chicago Press; Chicago: 2003. pp. 16–74. [Google Scholar]
- Wilbur W.J., Lipman D.J., Lipman D.J. Rapid similarity searches of nucleic acid and protein data banks. Proc. Natl. Acad. Sci. 1983;80:726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilme L., Goodman S.M., Ganzhorn J.U., Goodman S.M., Ganzhorn J.U., Ganzhorn J.U. Biogeographic evolution of Madagascar’s microendemic biota. Science. 2006;312:1063–1065. doi: 10.1126/science.1122806. [DOI] [PubMed] [Google Scholar]
- Yoder A.D. Relative position of the Cheirogaleidae in strepsirrhine phylogeny: A comparison of morphological and molecular methods and results. Am. J. Phys. Anthropol. 1994;94:25–46. doi: 10.1002/ajpa.1330940104. [DOI] [PubMed] [Google Scholar]
- Yoder A.D. Phylogeny of the lemurs. In: Goodman S.M., Benstead J., Benstead J., editors. The natural history of Madagascar. University of Chicago Press; Chicago: 2003. pp. 1242–1247. [Google Scholar]
- Yoder A.D. Lemurs. Curr. Biol. 2007;17:R866–R868. doi: 10.1016/j.cub.2007.07.050. [DOI] [PubMed] [Google Scholar]
- Yoder A.D., Irwin J.A., Irwin J.A. Phylogeny of the Lemuridae: Effects of character and taxon sampling on resolution of species relationships within Eulemur. Cladistics. 1999;15:351–361. doi: 10.1111/j.1096-0031.1999.tb00271.x. [DOI] [PubMed] [Google Scholar]
- Yoder A.D., Yang Z., Yang Z. Divergence dates for Malagasy lemurs estimated from multiple gene loci: Geological and evolutionary context. Mol. Ecol. 2004;13:757–773. doi: 10.1046/j.1365-294x.2004.02106.x. [DOI] [PubMed] [Google Scholar]
- Yoder A.D., Cartmill M., Ruvolo M., Smith K., Vilgalys R., Cartmill M., Ruvolo M., Smith K., Vilgalys R., Ruvolo M., Smith K., Vilgalys R., Smith K., Vilgalys R., Vilgalys R. Ancient single origin for Malagasy primates. Proc. Natl. Acad. Sci. 1996a;93:5122–5126. doi: 10.1073/pnas.93.10.5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder A.D., Vilgalys R., Ruvolo M., Vilgalys R., Ruvolo M., Ruvolo M. Molecular evolutionary dynamics of cytochrome b in strepsirrhine primates: The phylogenetic significance of third-position transversions. Mol. Biol. Evol. 1996b;13:1339–1350. doi: 10.1093/oxfordjournals.molbev.a025580. [DOI] [PubMed] [Google Scholar]
- Yoder A.D., Burns M.M., Zehr S., Delefosse T., Veron G., Goodman S.M., Flynn J.J., Burns M.M., Zehr S., Delefosse T., Veron G., Goodman S.M., Flynn J.J., Zehr S., Delefosse T., Veron G., Goodman S.M., Flynn J.J., Delefosse T., Veron G., Goodman S.M., Flynn J.J., Veron G., Goodman S.M., Flynn J.J., Goodman S.M., Flynn J.J., Flynn J.J. Single origin of Malagasy Carnivora from an African ancestor. Nature. 2003;421:734–737. doi: 10.1038/nature01303. [DOI] [PubMed] [Google Scholar]
- Zhang N., O’Donnell K., Sutton D.A., Nalim F.A., Summerbell R.C., Padhye A.A., Geiser D.M., O’Donnell K., Sutton D.A., Nalim F.A., Summerbell R.C., Padhye A.A., Geiser D.M., Sutton D.A., Nalim F.A., Summerbell R.C., Padhye A.A., Geiser D.M., Nalim F.A., Summerbell R.C., Padhye A.A., Geiser D.M., Summerbell R.C., Padhye A.A., Geiser D.M., Padhye A.A., Geiser D.M., Geiser D.M. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J. Clin. Microbiol. 2006;44:2186–2190. doi: 10.1128/JCM.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl D.J. Ph.D. dissertation, The University of Texas at Austin; Austin: 2006. “Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion.”. [Google Scholar]
- Zwickl D.J., Hillis D.M., Hillis D.M. Increased taxon sampling greatly reduces phylogenetic error. Syst. Biol. 2002;51:588–598. doi: 10.1080/10635150290102339. [DOI] [PubMed] [Google Scholar]