Abstract
Consomic strains, also known as chromosome substitution strains, are powerful tools for assigning polygenes that control quantitative complex traits to specific chromosomes. Here, we report generation of a full set of mouse consomic strains, in which each chromosome of the common laboratory strain C57BL/6J (B6) is replaced by its counterpart from the inbred strain MSM/Ms, which is derived from Japanese wild mouse, Mus musculus molossinus. The genome sequence of MSM/Ms is divergent from that of B6, whose genome is predominantly derived from Western European wild mouse, Mus musculus domesticus. MSM/Ms exhibits a number of quantitative complex traits markedly different from those of B6. We systematically determined phenotypes of these inter-subspecific consomic strains, focusing on complex traits related to reproduction, growth, and energy metabolism. We successfully detected more than 200 statistically significant QTLs affecting 26 traits. Furthermore, phenotyping of the consomic strains revealed that the measured values for quantitative complex traits often far exceed the range between B6 host and MSM/Ms donor strains; this may result from segregation of alleles or nonadditive interactions among multiple genes derived from the two mouse subspecies (that is, epistasis). Taken together, the results suggest that the inter-subspecific consomic strains will be very useful for identification of latent genetic components underlying quantitative complex traits.
Quantitative complex traits observed at the whole-body level, such as body weight, blood pressure, and blood glucose levels, are controlled by the cooperative functions of multiple genes and by interactions between genetic and environmental factors. Laboratory inbred mouse strains have long served as prime model organisms for uncovering the genetic components of quantitative complex traits as well as for understanding the pathogenesis of human diseases (Paigen 2003a, b). Since the inception of mouse genetic research, numerous inbred strains with characteristic phenotypes have been established for analysis of various complex traits (Beck et al. 2000).
Consomic strains, also known as chromosome substitution strains (CSSs), have been proposed and constructed in many species for genetic dissection of a variety of traits (Seiger 1966; Rick 1969). In mouse, consomic strains were reported as powerful tools for dissection of genetic factors responsible for quantitative polygenic traits (Nadeau et al. 2000; Guénet and Bonhomme 2003). The first example was a consomic strain with chromosome 19 from MOLF/Ei (Mus musculus molossinus-derived inbred strain) in the 129/Sv-Ter/+ background, which was successfully used to map a gene that modifies testicular tumor development (Matin et al. 1999). We reported consomic strains with either X chromosome or chromosome 17 of the Japanese wild mouse-derived inbred strain MSM/Ms in the C57BL/6J (hereafter referred to as B6) background, which showed male sterility (Oka et al. 2004, 2007) and resistance to late-onset hearing loss (Nemoto et al. 2004), respectively.
The first full set of consomic strains was established using A/J as the chromosome donor and B6 as the host (background) strain (Singer et al. 2004). These consomic strains are now widely used for genetic dissection of complex traits, including anxiety (Singer et al. 2005), pubertal timing (Krewson et al. 2004), and cis- and trans-regulation of transcripts in the kidney and liver (Shockley and Churchill 2006). A full set of consomic strains, referred to as a consomic panel, provides a tool for identifying QTLs for a complex trait of interest that differs between donor and host strains. Major advantages of using consomic panels for genetic analysis are reported elsewhere (Nadeau et al. 2000; Guénet and Bonhomme 2003; Singer et al. 2004; Gregorová et al. 2008, this issue).
Here, we report construction of a new full set of mouse consomic strains, using inbred strain MSM/Ms as the chromosome donor and B6 as the host strain. It is well established that commonly used laboratory mouse strains, including B6, derive predominantly from Western European wild mice, Mus musculus domesticus (Yonekawa et al. 1980; Ferris et al. 1982; Bonhomme et al. 1984; Moriwaki 1994; Frazer et al. 2007; Yang et al. 2007). Because their genomes were derived from a relatively small gene pool (Wade et al. 2002), it is thought that genetic variation among them is limited (Guénet and Bonhomme 2003). The inbred strain MSM/Ms was established from the Japanese wild mouse, M. m. molossinus (Moriwaki 1994). As these two inbred strains originated from two different subspecies, the new strains are referred to collectively as inter-subspecific consomic strains. The time of divergence between the two subspecies is estimated at roughly one million years (Yonekawa et al. 1980; Moriwaki 1994). Therefore, we found a vast quantity of genomic differences (∼1% divergence) between B6 and MSM/Ms (Abe et al. 2004).
As a consequence of its genetic divergence from other strains, MSM/Ms has unique complex traits that are significantly different from those of B6 and other common laboratory strains (Moriwaki 1994; Shiroishi et al. 1994; Yonekawa et al. 1994; Yoshiki and Moriwaki 2006). For example, MSM/Ms has extremely low susceptibility for the development of a variety of tumor types (Moriwaki et al. 1999), significant resistance to age-dependent hearing loss (Nemoto et al. 2004), and unique behavioral traits such as very high locomotor activity (Koide et al. 2000). In addition, because this strain was selected as a subject of the Mouse Phenome Project, accumulated data about its phenotypes are now available online (Paigen and Eppig 2000). The MSM/Ms strain likely bears additional intriguing traits that are specific to M. m. molossinus and not found in other mouse subspecies. Thus, by using inter-subspecific consomic strains, we are able to expand the spectrum of target complex traits that can be analyzed, as the extent of overall phenotypic difference will undoubtedly be larger between members of inter-subspecific pairs than between members of intra-subspecific pairs. Furthermore, inter-subspecific genome difference provides numerous positional DNA markers, which is a significant benefit for fine mapping of QTLs. All these features of inter-subspecific consomic strains suggest their utility for dissection of a wide-range of quantitative complex traits.
Results
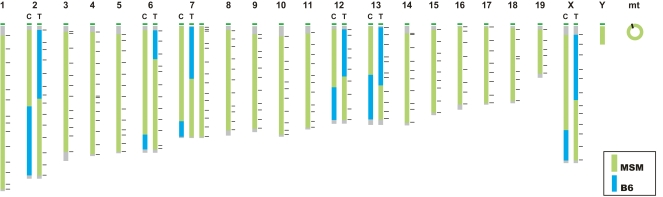
The complete consomic panel we established consists of 29 strains and includes consomic strains bearing the Y chromosome and mitochondrial DNA of MSM/Ms (Fig. 1). According to the consomic nomenclature, each strain was designated by B6-ChrNMSM, where N is the chromosome number transferred from MSM/Ms.
Figure 1.
Genomes of consomic mouse strains. Centromeres are shown at the top. (Green) MSM/Ms-derived chromosomes; (blue) B6 chromosomes. The genetic background and mitochondria DNA were confirmed to be from the B6 genome by genotyping with the markers listed in Supplemental Table S1.
During the course of production of the consomic strains, we found it difficult for a small set of chromosomes to transfer the entire MSM/Ms chromosomes as a whole into the B6 background, mainly due to male sterility and partial reduction of viability of the homozygous lines. We overcame this problem by meiotic recombination-based dividing of the chromosome to be substituted into two pieces, a centromeric and a telomeric region, with an overlapping segment in the middle of the chromosome. Strains carrying the partial chromosomes were maintained as homozygous lines and are referred to as sub-consomic strains. Two sub-consomic strains were established for chromosomes 2, 6, 7, 12, 13, and X (Fig. 1). For these sub-consomic strains, we appended C (centromere) or T (telomere) following the chromosome number N, e.g., B6-Chr2CMSM and B6-Chr2TMSM. The microsatellite markers used to locate the MSM/Ms and B6 chromosomal breakpoints in these sub-consomic strains are shown in Supplemental Table S2.
After the strains were established as homozygous lines, we genotyped the progeny of each line once again in order to confirm that the genetic background had been replaced by B6 markers located on the whole genome at intervals of less than 20 Mbp (Fig. 1; Supplemental Table S1). The result indicated that all markers had been fully replaced by the B6 type, except for chromosome 13. In two sub-consomic strains, B6-Chr13CMSM and B6-Chr13TMSM, a small region between D13Mit311 and D13Mit9, which is in an ∼18 Mb segment that overlaps in these two strains, was not replaced by the MSM/Ms genome, as the two homozygous lines are inviable for undetermined reasons (data not shown). We obtained the homozygous line of B6-Chr10MSM by natural mating and in vitro fertilization using heterozygous animals. However, we found that it was very hard to maintain and expand a colony of this strain for experimental use, mostly due to reproduction problems. Currently, we are trying to establish sub-consomic strains by splitting chromosome 10 into two pieces (B6-Chr10CMSM and B6-Chr10TMSM; data not shown) as was done for other chromosomes.
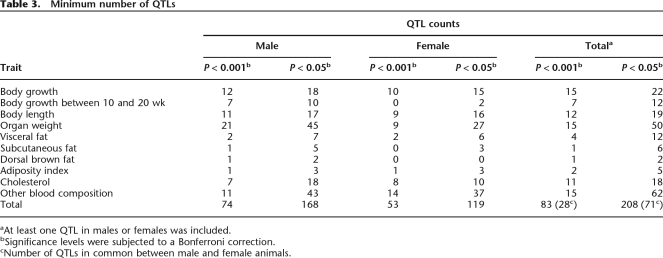
We performed systematic phenotyping of the B6-ChrNMSM consomic strains as well as the chromosome donor and host strains, MSM/Ms and B6. Our measurements were focused on complex traits related to reproduction, growth, and energy metabolism. For each phenotyping, significance levels were established to account for multiple hypothesis testing in comparisons of the consomic strains and the host B6 strain. The level of significance was adjusted to 0.001 (highly significant) or 0.05 (significant) using the Bonferroni correction for individual traits. We estimated the minimum number of QTLs for separate groups of traits that are related to one another, circumventing the need to count identical QTLs twice or more (see Methods). The minimum estimated number of highly significant QTLs detected in this study is 74 in males and 53 in females (Table 3). A total of 208 significant QTLs that affect 26 traits were detected. Many QTLs were detected only in males or females, with fewer than half (34.1%) of significant QTLs commonly observed in both males and females.
Table 3.
Minimum number of QTLs
aAt least one QTL in males or females was included.
bSignificance levels were subjected to a Bonferroni correction.
cNumber of QTLs in common between male and female animals.
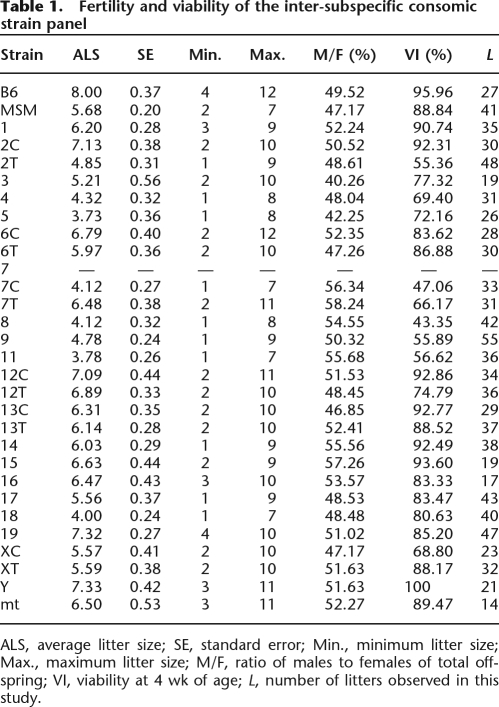
The result of phenotyping of reproduction-related traits is summarized in Table 1. Average litter size shows large variation among the consomic strains, ranging from 3.73 for B6-Chr5MSM to 7.33 for B6-ChrYMSM. No strain had a litter size exceeding that of B6. Notably, reductions in both litter size and viability are observed in consomic strains bearing chromosome 7C, 8, or 9. The number of pups that were weaned at 4 wk of age significantly decreased to ∼50% of that of the B6 strain in consomic strains bearing chromosome 7C or 8 and to 70% in chromosome 4, 7T, 9, or 11.
Table 1.
Fertility and viability of the inter-subspecific consomic strain panel
ALS, average litter size; SE, standard error; Min., minimum litter size; Max., maximum litter size; M/F, ratio of males to females of total offspring; VI, viability at 4 wk of age; L, number of litters observed in this study.
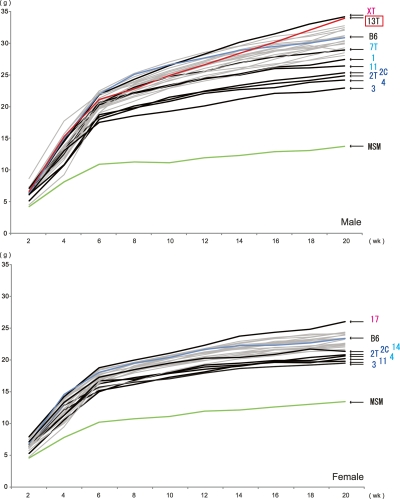
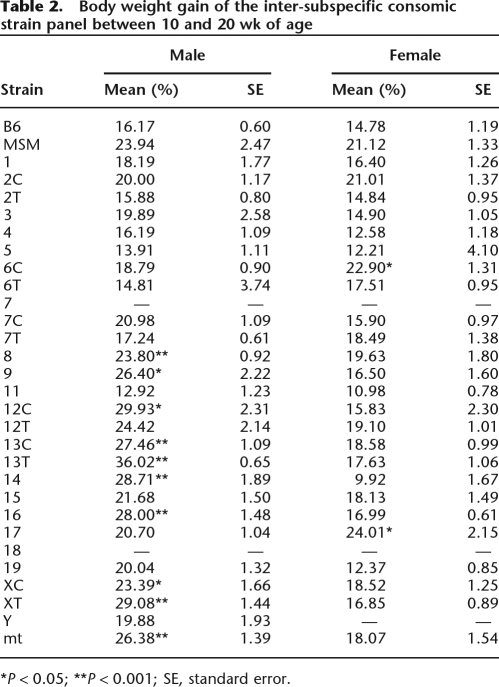
The MSM/Ms strain has an average body weight (g) far less than B6, in both males and females and at different ages (Supplemental Table S3). Many consomic strains carrying different MSM/Ms autosomes and mitochondrial DNA have reduced body weight. Among them, the most significant reductions of body weight (P < 0.001) are observed in males from consomic strains bearing MSM/Ms chromosome 1, 2, 3, 4, or 11 (Figs. 2, 3). In contrast, males from consomic strains bearing MSM/Ms chromosome 5, 17, or XT gain body weight beyond what is observed for males of the B6 strain. In general, highly significant QTLs are more frequently detected in males than in females. This is partly due to gender differences in the body weight of donor MSM/Ms and host B6 strains; correspondingly, the range of measured values is significantly larger in males than in females (Fig. 3). The effect of most QTLs on body weight depends on the age of the mice (Figs. 2, 3). Consomic strains bearing MSM/Ms chromosome 15 or mitochondrial DNA have reduced body weight only at earlier stages, from 2 to 6 wk of age, and not at later stages. The reverse is true for body weight increase in animals of consomic strains bearing MSM/Ms chromosomes 17 or XT. Another remarkable example is consomic strain B6-Chr13TMSM. In this strain, males but not females have significantly lower body weight than B6 mice until 6 wk of age, but thereafter, body weight increases dramatically, such that these mice exhibit their highest rate of growth (36.02%) between 10 and 20 wk of age (Fig. 3; Table 2). Females of the B6-Chr17MSM strain also have a fast growth rate (24.01%; Table 2).
Figure 2.
Summary of comprehensive phenotyping of the consomic panel. The upper half of each box indicates the result obtained in males, and the lower half, the result in females. Red and magenta boxes indicate values larger than those of the host B6 strain. Highly significant values (P < 0.001) are shown in red, and all other statistically significant values (P < 0.05) are shown in magenta. Dark blue and light blue boxes indicate values lower than those of B6. Highly significant values (P < 0.001) are shown in dark blue, and all other statistically significant values (P < 0.05) are shown in light blue.
Figure 3.
Growth of consomic strain mice. Every 2 wk, from 2 to 20 wk of age, the body weight of mice from each consomic strain (B6 and MSM/Ms) was measured, and the mean value of each strain was plotted. The bold line denotes a growth curve that deviates significantly from that of B6. The names of consomic strains with statistically significant differences in body weight at 20 wk of age are shown, with color-coding as described for Figure 2. The red line denotes the growth curve of B6-Chr13TMSM, for which the highest rate of growth was between 10 and 20 wk of age.
Table 2.
Body weight gain of the inter-subspecific consomic strain panel between 10 and 20 wk of age
*P < 0.05; **P < 0.001; SE, standard error.
Total body lengths of MSM/Ms are ∼20%–30% shorter than those of B6 at 20 wk of age (Supplemental Table S3). Reflecting the shorter body length of MSM/Ms, the transfer of several individual MSM/Ms chromosomes into the B6 background causes reduction in body length (Fig. 2).
The weight of the kidneys and heart relative to lean body weight is heavier in MSM/Ms than in B6 strain mice, in both males and females. As shown in Figure 2, many consomic strains show increased weight of the spleen, kidney, and heart in comparison with B6, indicating an additive effect of the MSM/Ms chromosomes on weight gain of these three organs. By contrast, several consomic strains show reduced testis and liver weights, although the relative weights of these organs in MSM/Ms are indistinguishable from those of the B6 strain (Fig. 2).
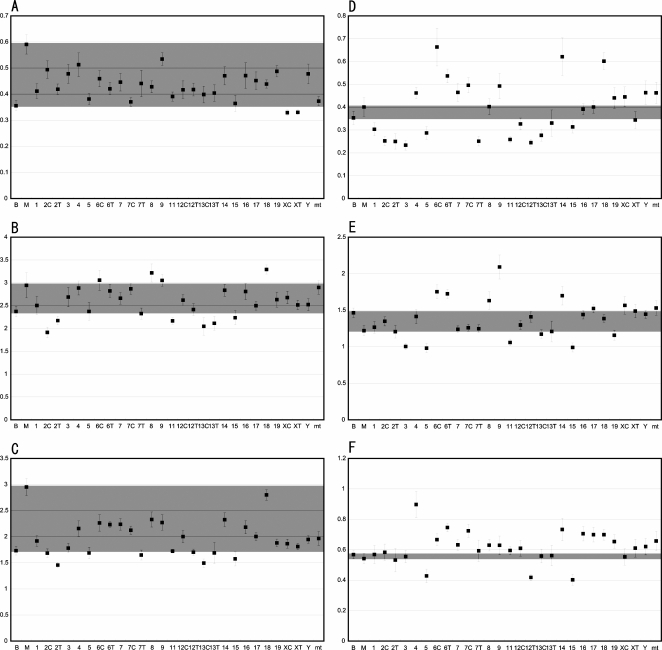
We also carried out extensive phenotyping of fat pad weight. We measured epididymal, perirenal, and mesenteric fat pads for visceral adipose accumulation and inguinal and dorsal fat pads for subcutaneous adipose accumulation. We also measured scapular brown fat, which is an indicator of ability to dissipate stored energy as heat. As shown in Figures 2 and 4, many consomic strains exhibit increased or decreased fat pad in comparison with B6. We detect a few highly significant and many significant QTLs affecting the visceral fat pad on many chromosomes. In particular, chromosomes 6C and 9 have a highly significant QTL(s) that affects the epididymal and/or mesenteric fat pad specifically in females. The MSM/Ms allele(s) at this QTL(s) leads to significant gain in the mesenteric fat pad. Likewise, several MSM/Ms chromosomes lead to a gain in subcutaneous fat pads relative to B6. For example, consomic males bearing MSM/Ms chromosome 18 show a significant gain in the inguinal fat pad. Consomic strains bearing MSM/Ms chromosome 9 or 19 show gains of the brown fat pad specifically in males. The difference in the fat pad content between MSM/Ms and B6 is smaller in visceral white adipose tissues than in subcutaneous white adipose tissues or brown adipose tissue (Fig. 4). It is notable that, for several consomic strains, the values for the visceral but not the subcutaneous fat pad of several consomic strains lie far outside the range of values between MSM/Ms and B6 strains (Fig. 4D–F).
Figure 4.
Distribution of measured values for fat pad deposits from the consomic strains as compared with the range observed for parental strains. The fat pad deposits of males were measured at 10 wk of age. Error bars show standard errors of the mean. Gray shading refers to the range of values between the MSM/Ms and B6 strains. (A) Dorsal brown adipose tissue; (B) dorsal white adipose tissue; (C) inguinal white adipose tissue; (D) perirenal white adipose tissue; (E) epididymal white adipose tissue; (F) mesenteric white adipose tissue. Measured values were used to determine the percentage of fat pad weight as compared to total fasted body weight.
We measured a total of 16 clinical biochemical parameters in serum, including lipid metabolism-related components, enzyme activities, and other protein and mineral components that are good indicators of liver and kidney function. The results of the measurements are summarized in Figure 2 and Supplemental Table S3. Almost all MSM/Ms chromosomes, as well as the mitochondrial DNA, affect blood chemical constituents for at least one parameter, with the exception of chromosome 6T, which has no observable effect on any parameter we tested. Notably, several MSM/Ms chromosomes have decreased HDL cholesterol, non-HDL cholesterol, and total cholesterol levels relative to B6, despite the fact that females from the donor MSM/Ms strain have rather higher levels of HDL and total cholesterol. Likewise, multiple MSM/Ms chromosomes significantly increase alkaline phosphatase (ALP) activity in either males or females, or both, even though MSM/Ms has a value comparable to B6.
Discussion
As expected from phenotypic differences between MSM/Ms and B6 (Moriwaki 1994; Yoshiki and Moriwaki 2006), we successfully detected a large number of QTLs for a variety of complex traits. It is not easy to directly compare efficacy of QTL detection in this study with a previously reported study using B6-A/J consomic panel (Singer et al. 2004) because only a small fraction of complex traits were measured in both of the studies and the protocols used for measurement were different. However, for certain traits, the inter-subspecific consomic panel yielded a higher number of QTLs. For instance, seven QTLs affecting serum cholesterol level in males were detected on different chromosomes using the B6-A/J consomic panel (Singer et al. 2004), whereas in this study, QTLs affecting total cholesterol levels in males were detected on at least nine chromosomes and mitochondrial DNA, and a total of 14 chromosomes appear to affect cholesterol-related measurements in both males and females (Fig. 2). Thus, taken together with the detection of over 200 QTLs in this study, these results indicate that phenotyping of an inter-subspecific consomic panel is a powerful way to dissect genetic determinants underlying quantitative complex traits.
Recently, Forejt and colleagues constructed another set of inter-subspecific consomic strains, using M. m. musculus-derived PWD/Ph strain as the donor and B6 as the host strain (Gregorová et al. 2008). Phenotyping of these consomic strains also detected a number of QTLs. The phenotype data from the two different sets of inter-subspecific consomic strains indicated their usefulness for genetic dissection of many complex traits.
MSM/Ms shows significantly lower body weight and shorter length than those of B6, as are characteristics of the Japanese wild mouse, M. m. molossinus. This study showed that many different MSM/Ms chromosomes significantly reduce body size (Fig. 2). The results indicate that polygenes controlling body weight and length of MSM/Ms have an additive effect when combined with the B6 genomic background. The biological processes that determine body and organ size are complex and regulated by different processes, including cell proliferation and growth, cell localization, and cell death (Conlon and Raff 1999). The mechanisms of cell proliferation in the context of body and organ size determinants are still not well understood, but one pathway known to act in growth control of mammals is the phosphoinositide 3-kinase (PI3K) signaling cascade activated by growth factors such as IGF1 (Conlon and Raff 1999; Shioi et al. 2000; Sutter et al. 2007). In our study, we detected various QTLs affecting body and organ size (Fig. 2). However, these QTLs are not linked to genes involved in the PI3K/IGF1 signaling pathway. Thus, it is likely that as-yet unidentified genes also regulate mouse body and organ size and that these are responsible for the size difference between MSM/Ms and B6.
Many QTLs affecting obese or lean phenotypes have been detected by genetic linkage analysis using various combinations of laboratory mouse strains (Corva and Medrano 2001; Rankinen et al. 2006). We anticipated that MSM/Ms would have significantly different metabolic parameters than the B6 strain, because of large genomic divergence and differences in the two strains’ histories. To capitalize on this difference, we carried out extensive phenotyping of the B6-ChrNMSM consomic panel mice in terms of energy metabolism-related traits with the goal of learning more about metabolic control. Accumulation of fat pads is important for overall body weight gain and serves as a good landmark of thrifty phenotypes, such as energy deposition. Our extensive measurements of fat pads in B6-ChrNMSM consomic panel mice revealed that many MSM/Ms chromosomes have effects on fat pad deposition (Figs. 2, 4). Notably, several chromosomes tend to increase subcutaneous and brown fat pad depositions. The same chromosomes also increase visceral fat pad deposition, whereas other chromosomes decrease visceral fat pad deposition. Because it is well known that visceral but not subcutaneous fat is a risk factor for cardiovascular disease in humans, MSM/Ms likely has a genetic system that directs excess energy to be stored safely as subcutaneous fat pads. Of particular interest are other consomic strains that have shown age- and sex-dependent obesity phenotypes after sexual maturation. For example, the increase in weight of animals of the B6-Chr13TMSM strain between 10 and 20 wk of age (36.02% increase) is higher than that observed for any other consomic strain (Fig. 3; Table 2). In humans, obesity-related genes are located on human chromosome 5q (Rankinen et al. 2006). That human region is counterpart to the distal segment of mouse chromosome 13, which corresponds to the substituted segment (D13Mit9-D13Mit196) of B6-Chr13TMSM. Thus, this consomic strain provides a useful tool to ask whether these genes might be causative for obesity in humans.
In this study, genetic determinants for some phenotypes of MSM/Ms are simply dissected into multiple chromosomes on the B6 background (Fig. 2). However, measured values in many consomic strains often lay outside the range between the parental strains MSM/Ms and B6. The prominent examples are several fat pad depositions in adipose tissues (Fig. 4) and clinical biochemical constituents, such as serum cholesterol level and ALP activity (Fig. 2). This result may be due to segregation of MSM/Ms alleles by chromosome substitution. It is possible that MSM/Ms has alleles on different chromosomes that have opposite effects on measured values of the quantitative traits. In that case, substitution of the chromosomes concerned would elicit visible effects of the MSM/Ms alleles on the relevant consomic strains. Alternatively, it is also possible that chromosome substitution either gives rise to non-additive interactions or disrupts conventional genetic interactions between multiple genes located on the substituted chromosome and other chromosomes. In these cases, chromosome substitution would elicit epistatic effects. We previously reported one prominent example of such epistasis in the consomic strain B6-ChrXMSM. In this strain, genetic incompatibility between the MSM/Ms X chromosome and the B6 autosomes including chr 1 and chr 11 causes male sterility (Oka et al. 2004, 2007). At present, it is not clear whether it is segregation of alleles with positive and negative effects in MSM/Ms, or epistasis, that gives rise to the measured values lying outside the range between the parental strains for some quantitative complex traits. However, it is clear that use of inter-subspecific consomic strains provides a unique opportunity to uncover latent genetic components and genetic interactions that are otherwise very difficult to recognize.
Methods
Construction of inter-subspecific consomic strains
MSM/Ms is maintained at National Institute of Genetics (NIG), Mishima. Inbreeding of MSM/Ms reached 80 generations in February 2007. B6 was purchased from CLEA Japan (Tokyo, Japan). To construct a full set of consomic strains, B6-ChrNMSM, F1 females were back-crossed to B6 males for 7–16 generations. Females heterozygous for a non-recombinant chromosome were selected by genotyping and used in each back-cross. All mice were housed in a SPF facility (12-h light and dark cycles) with free access to food and water. The feed contained 346.8 kcal/100 g metabolic energy and contained 24.9% crude protein, 4.6% crude fat, 3.7% crude fiber, and 6.7% crude ash (CE-2 diet, CREA Japan). B6-Chr5MSM animals were fed a CA-1 diet (from the same supplier), which contained 347.4 kcal/100 g metabolic energy and contained 26.8% crude protein, 5.0% crude fat, 3.4% crude fiber, and 7.6% crude ash. The room temperature (°C) was 23 ± 2, and relative humidity (%) was 50 ± 10. All animals were weaned at 4 wk of age. Thereafter, males were housed individually and females were grouped (<5) in plastic cages. All animal experiments were approved by the Animal Care and Use Committee of NIG.
Genotyping
Genotyping was performed with a panel of 233 microsatellite markers located at intervals of less than 20 Mbp (Fig. 1; Supplemental Table S1). Genotyping of the mitochondrial genome was performed by direct sequencing of PCR-amplified products using the complete sequence of the M. m. molossinus mitochondria genome as a reference (Akimoto et al. 2005). Progenitor animals of each consomic panel were used in a final quality check by genotyping with 217 markers to detect unwanted MSM/Ms-derived chromosomal segments remaining in each established strain. A list of the markers and primers is presented in Supplemental Table S1.
Reproduction and growth
Animals were mated at 6 wk of age or later, and pregnancy and delivery were monitored until 90 d from the first day of mating. We recorded the gender and any abnormalities of litter mates. For phenotyping of body growth-related traits, pup number was adjusted to less than eight per cage by discarding extra pups within a few days of birth. The average sex ratio was calculated until weaning. Survival and growth defects of pups were monitored until weaning. For analysis of reproductive performance, we measured fertility or viability of the 28 consomic strains as well as B6 and MSM/Ms strains, using data from 942 litters in total. Body weight was measured every 2 wk from 2 to 20 wk of age. Organ weight of the consomic strains was measured at 10 wk of age. Organ and lean weights were also determined, and organ weight is shown as a percentage of total lean weight. In total, 752 animals (387 males and 365 females) were used for measuring body growth, and 613 animals (300 males and 313 females) were used for anatomical and blood chemical measurements. The raw phenotypic data and the number of animals used for measurements are summarized in Supplemental Table S3.
Measurement of fat pad weight
Mice 10 wk of age were fasted from 6 am to 10 am (±30 min) prior to blood sampling. Non-fasting and fasting body weight and body and tail lengths were measured. Subsequently, tissue and fat pad measurements were done immediately thereafter. The process proceeded from 10 am to 12 pm. Heart, liver, spleen, kidney, and testis were dissected and weighed. The carcasses were weighed to determine lean weight measurements. The tissue weights were measured and indicated as a percentage of total lean weight. To measure fat pad weight, dorsal and inguinal deposits of subcutaneous white adipose tissues; epididymal, gonadal, and mesenteric deposits for visceral white adipose tissues; and dorsal brown adipose deposits (inter- and subscapular) of brown adipose tissue were dissected and then weighed individually. Adipose tissue weights were measured and indicated as a percentage of total fasting body weight.
Measurement of clinical biochemical constituents
In total, we measured 16 parameters in blood samples from the 27 consomic strains, B6 and MSM/Ms strains at 10 wk of age. Concentrations of blood constituents, inorganic phosphate (IP), high-density lipoprotein (HDL) cholesterol, total cholesterol (TCHOL), triglyceride (TG), alanine aminotransferase (ALT), lactate dehydrogenase (LDH), amylase (AMY), and albumin (ALB) were measured immediately after blood sampling using an automated dry chemistry analyzer (Drychem, Fuji-Film Medical). Concentrations of alkaline phosphatase (ALP), aspartate aminotransferase (AST), blood urea nitrogen (BUN), total bilirubin (TBIL), total protein (TP), calcium, and potassium were measured with a VetScan blood chemistry analyzer (Abaxis) using whole blood samples. Low-density lipoprotein (LDLC) cholesterol was calculated by subtraction of HDLC from TCHOL. We excluded data from abnormal individuals, i.e., those exhibiting an eye-opening defect, facial and body contortion, growth defects with hydrocephalus, or abnormal kidney shape (data not shown).
Statistics and estimation of the minimum number of QTLs for each phenotype tested
Statistical significance was determined in a t-test between each consomic strain and the chromosome host strain B6. Significance levels were subjected to a Bonferroni correction to account for multiple-hypotheses testing. A measured value was considered significant if the corrected t-test P value was less than 0.05 and highly significant if the P-value was less than 0.001. Because of the multiple-hypothesis correction, the expected rate of false positives is only 0.05–0.001 per trait (or trait/gender combination) across the entire consomic panel. To prevent overestimation in counting QTLs, we integrated some related traits into a single group and counted number of QTLs for the group. For example, we measured epididymal, perirenal, and mesenteric fat pad deposits separately (Fig. 2). The same consomic strain showed fat pad weights statistically different from B6 for more than one type of fat pad. Although we cannot exclude the possibility that two different QTLs independently regulate the different fat pads, we estimate that the number of QTLs in one consomic strain is only one for a group of visceral fat deposits. Likewise, we estimated the total number of QTLs in the same way for subcutaneous fat pad deposit traits, blood cholesterol traits, and body growth-related traits. For other traits, the number of QTLs was estimated independently.
Resource distribution
The B6-ChrNMSM strains are available through The Genetic Strains Research Center of National Institute of Genetics (http://www.shigen.nig.ac.jp/mouse/strain/) and will be soon available from RIKEN BioResource Center (http://www.brc.riken.jp/lab/animal/en/).
Acknowledgments
We thank all past and present members of the Mammalian Genetics Laboratory of NIG and the Laboratory Animal Science of The Tokyo Metropolitan Institute of Medical Science (Rinshoken) who contributed to the development and maintenance of the strains that made this project possible. We also thank S. Kuriki of The Institute of Statistical Mathematics for advice on statistical analyses, and J. Nadeau for critically reading the manuscript and providing useful comments. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas Comparative Genomics from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and supported in part by the Bio-diversity Research Project of the Transdisciplinary Research Integration Center, Research Organization of Information and Systems.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7175308
References
- Abe K., Noguchi H., Tagawa K., Yuzuriha M., Toyoda A., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Noguchi H., Tagawa K., Yuzuriha M., Toyoda A., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Tagawa K., Yuzuriha M., Toyoda A., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Yuzuriha M., Toyoda A., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Toyoda A., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Ezawa K., Saitou N., Hattori M., Sakaki Y., Saitou N., Hattori M., Sakaki Y., Hattori M., Sakaki Y., Sakaki Y., et al. Contribution of Asian mouse subspecies Mus musculus molossinus to genomic constitution of strain C57BL/6J, as defined by BAC-end sequence–SNP analysis. Genome Res. 2004;14:2439–2447. doi: 10.1101/gr.2899304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto M., Niikura M., Ichikawa M., Yonekawa H., Nakada K., Honma Y., Hayashi J., Niikura M., Ichikawa M., Yonekawa H., Nakada K., Honma Y., Hayashi J., Ichikawa M., Yonekawa H., Nakada K., Honma Y., Hayashi J., Yonekawa H., Nakada K., Honma Y., Hayashi J., Nakada K., Honma Y., Hayashi J., Honma Y., Hayashi J., Hayashi J. Nuclear DNA but not mtDNA controls tumor phenotypes in mouse cells. Biochem. Biophys. Res. Commun. 2005;327:1028–1035. doi: 10.1016/j.bbrc.2004.12.105. [DOI] [PubMed] [Google Scholar]
- Beck J.A., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Eppig J.T., Festing M.F., Fisher E.M., Festing M.F., Fisher E.M., Fisher E.M. Genealogies of mouse inbred strains. Nat. Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Bonhomme F., Catalan J., Britten-Davidian J., Chapman V.M., Moriwaki K., Nevo E., Thaler L., Catalan J., Britten-Davidian J., Chapman V.M., Moriwaki K., Nevo E., Thaler L., Britten-Davidian J., Chapman V.M., Moriwaki K., Nevo E., Thaler L., Chapman V.M., Moriwaki K., Nevo E., Thaler L., Moriwaki K., Nevo E., Thaler L., Nevo E., Thaler L., Thaler L. Biochemical diversity and evolution in the genus Mus. Biochem. Genet. 1984;22:275–303. doi: 10.1007/BF00484229. [DOI] [PubMed] [Google Scholar]
- Conlon I., Raff M., Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Corva P.M., Medrano J.F., Medrano J.F. Quantitative trait loci (QTL) mapping for growth traits in the mouse: A review. Genet. Sel. Evol. 2001;33:105–132. doi: 10.1186/1297-9686-33-2-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris S.D., Sage R.D., Wilson A.C., Sage R.D., Wilson A.C., Wilson A.C. Evidence from mtDNA sequences that common laboratory strains of inbred mice are descended from a single female. Nature. 1982;295:163–165. doi: 10.1038/295163a0. [DOI] [PubMed] [Google Scholar]
- Frazer K.A., Eskin E., Kang H.M., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Eskin E., Kang H.M., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Kang H.M., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Montgomery J., Morenzoni M.M., Nilsen G.B., Morenzoni M.M., Nilsen G.B., Nilsen G.B., et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Gregorová S., Divina P., Storchova R., Trachtulec Z., Fotopulosova V., Swenson K.L., Donahue L.R., Paigen B., Forejt J., Divina P., Storchova R., Trachtulec Z., Fotopulosova V., Swenson K.L., Donahue L.R., Paigen B., Forejt J., Storchova R., Trachtulec Z., Fotopulosova V., Swenson K.L., Donahue L.R., Paigen B., Forejt J., Trachtulec Z., Fotopulosova V., Swenson K.L., Donahue L.R., Paigen B., Forejt J., Fotopulosova V., Swenson K.L., Donahue L.R., Paigen B., Forejt J., Swenson K.L., Donahue L.R., Paigen B., Forejt J., Donahue L.R., Paigen B., Forejt J., Paigen B., Forejt J., Forejt J. Mouse consomic strains: Exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res. 2008 doi: 10.1101/gr.7160508. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénet J.L., Bonhomme F., Bonhomme F. Wild mice: An ever-increasing contribution to a popular mammalian model. Trends Genet. 2003;19:24–31. doi: 10.1016/s0168-9525(02)00007-0. [DOI] [PubMed] [Google Scholar]
- Koide T., Moriwaki K., Ikeda K., Niki H., Shiroishi T., Moriwaki K., Ikeda K., Niki H., Shiroishi T., Ikeda K., Niki H., Shiroishi T., Niki H., Shiroishi T., Shiroishi T. Multi-phenotype behavioral characterization of inbred strains derived from wild stocks of Mus musculus. Mamm. Genome. 2000;11:664–670. doi: 10.1007/s003350010129. [DOI] [PubMed] [Google Scholar]
- Krewson T.D., Supelak P.J., Hill A.E., Singer J.B., Lander E.S., Nadeau J.H., Palmert M.R., Supelak P.J., Hill A.E., Singer J.B., Lander E.S., Nadeau J.H., Palmert M.R., Hill A.E., Singer J.B., Lander E.S., Nadeau J.H., Palmert M.R., Singer J.B., Lander E.S., Nadeau J.H., Palmert M.R., Lander E.S., Nadeau J.H., Palmert M.R., Nadeau J.H., Palmert M.R., Palmert M.R. Chromosomes 6 and 13 harbor genes that regulate pubertal timing in mouse chromosome substitution strains. Endocrinology. 2004;145:4447–4451. doi: 10.1210/en.2004-0543. [DOI] [PubMed] [Google Scholar]
- Matin A., Collin G.B., Asada Y., Varnum D., Nadeau J.H., Collin G.B., Asada Y., Varnum D., Nadeau J.H., Asada Y., Varnum D., Nadeau J.H., Varnum D., Nadeau J.H., Nadeau J.H. Susceptibility to testicular germ-cell tumours in a 129.MOLF-Chr 19 chromosome substitution strain. Nat. Genet. 1999;23:237–240. doi: 10.1038/13874. [DOI] [PubMed] [Google Scholar]
- Moriwaki K. Wild mouse from geneticist’s viewpoint. In: Moriwaki K., et al., editors. Genetics in wild mice: Its application to biomedical research. Japan Scientific Press/Karger; Tokyo: 1994. pp. xiii–xxiv. [Google Scholar]
- Moriwaki K., Miyashita N., Yamaguchi Y., Shiroishi T., Miyashita N., Yamaguchi Y., Shiroishi T., Yamaguchi Y., Shiroishi T., Shiroishi T. Multiple genes governing biological functions in the genetic background of laboratory mice and Asian wild mice. In: Hiai H., Hino O., Hino O., editors. Progress in Experimental Tumor Research. Animal Models of Cancer Predisposition Syndromes. Vol. 35. Karger; Basel: 1999. pp. 1–12. [DOI] [PubMed] [Google Scholar]
- Nadeau J.H., Singer J.B., Matin A., Lander E.S., Singer J.B., Matin A., Lander E.S., Matin A., Lander E.S., Lander E.S. Analysing complex genetic traits with chromosome substitution strains. Nat. Genet. 2000;24:221–225. doi: 10.1038/73427. [DOI] [PubMed] [Google Scholar]
- Nemoto M., Morita Y., Mishima Y., Takahashi S., Nomura T., Ushiki T., Shiroishi T., Kikkawa Y., Yonekawa H., Kominami R., Morita Y., Mishima Y., Takahashi S., Nomura T., Ushiki T., Shiroishi T., Kikkawa Y., Yonekawa H., Kominami R., Mishima Y., Takahashi S., Nomura T., Ushiki T., Shiroishi T., Kikkawa Y., Yonekawa H., Kominami R., Takahashi S., Nomura T., Ushiki T., Shiroishi T., Kikkawa Y., Yonekawa H., Kominami R., Nomura T., Ushiki T., Shiroishi T., Kikkawa Y., Yonekawa H., Kominami R., Ushiki T., Shiroishi T., Kikkawa Y., Yonekawa H., Kominami R., Shiroishi T., Kikkawa Y., Yonekawa H., Kominami R., Kikkawa Y., Yonekawa H., Kominami R., Yonekawa H., Kominami R., Kominami R. Ahl3, a third locus on mouse chromosome 17 affecting age-related hearing loss. Biochem. Biophys. Res. Commun. 2004;324:1283–1288. doi: 10.1016/j.bbrc.2004.09.186. [DOI] [PubMed] [Google Scholar]
- Oka A., Mita A., Sakurai-Yamatani N., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Mita A., Sakurai-Yamatani N., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Sakurai-Yamatani N., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Toshimori K., Moriwaki K., Shiroishi T., Moriwaki K., Shiroishi T., Shiroishi T. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics. 2004;166:913–924. doi: 10.1534/genetics.166.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Aoto T., Totsuka Y., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Aoto T., Totsuka Y., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Totsuka Y., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Yamamoto H., Kuriki S., Takagi N., Kuriki S., Takagi N., Takagi N., et al. Disruption of genetic interaction between two autosomal regions and the X chromosome causes reproductive isolation between mouse strains derived from different subspecies. Genetics. 2007;175:185–197. doi: 10.1534/genetics.106.062976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K. One hundred years of mouse genetics: An intellectual history. I. The classical period (1902–1980) Genetics. 2003a;163:1–7. doi: 10.1093/genetics/163.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K. One hundred years of mouse genetics: An intellectual history. II. The molecular revolution (1981–2002) Genetics. 2003b;163:1227–1235. doi: 10.1093/genetics/163.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K., Eppig J.T., Eppig J.T. A mouse phenome project. Mamm. Genome. 2000;11:715–717. doi: 10.1007/s003350010152. [DOI] [PubMed] [Google Scholar]
- Rankinen T., Zuberi A., Chagnon Y.C., Weisnagel S.J., Argyropoulos G., Walts B., Pérusse L., Bouchard C., Zuberi A., Chagnon Y.C., Weisnagel S.J., Argyropoulos G., Walts B., Pérusse L., Bouchard C., Chagnon Y.C., Weisnagel S.J., Argyropoulos G., Walts B., Pérusse L., Bouchard C., Weisnagel S.J., Argyropoulos G., Walts B., Pérusse L., Bouchard C., Argyropoulos G., Walts B., Pérusse L., Bouchard C., Walts B., Pérusse L., Bouchard C., Pérusse L., Bouchard C., Bouchard C. The human obesity gene map: The 2005 update. Obesity. 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Rick C.M. Controlled introgression of chromosomes of Solanum pennellii into Lycopersicon esculentum: Segregation and recombination. Genetics. 1969;62:753–768. doi: 10.1093/genetics/62.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiger M.B. The effects of chromosome substitution on male body weight of Drosophila melanogaster. Genetics. 1966;53:237–248. doi: 10.1093/genetics/53.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioi T., Kang P.M., Douglas P.S., Hampe J., Yballe C.M., Lawitts J., Cantley L.C., Izumo S., Kang P.M., Douglas P.S., Hampe J., Yballe C.M., Lawitts J., Cantley L.C., Izumo S., Douglas P.S., Hampe J., Yballe C.M., Lawitts J., Cantley L.C., Izumo S., Hampe J., Yballe C.M., Lawitts J., Cantley L.C., Izumo S., Yballe C.M., Lawitts J., Cantley L.C., Izumo S., Lawitts J., Cantley L.C., Izumo S., Cantley L.C., Izumo S., Izumo S. The conserved phosphoinositide 3-kinase pathway determines heart size in mice. EMBO J. 2000;19:2537–2548. doi: 10.1093/emboj/19.11.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroishi T., Sagai T., Moriwaki K., Sagai T., Moriwaki K., Moriwaki K. Recombinational hotspot in the H2 haplotypes derived from the Asian wild mouse. In: Moriwaki K., et al., editors. Genetics in wild mice: Its application to biomedical research. Japan Scientific Press/Karger; Tokyo: 1994. pp. 141–151. [Google Scholar]
- Shockley K.R., Churchill G.A., Churchill G.A. Gene expression analysis of mouse chromosome substitution strains. Mamm. Genome. 2006;17:598–614. doi: 10.1007/s00335-005-0176-y. [DOI] [PubMed] [Google Scholar]
- Singer J.B., Hill A.E., Burrage L.C., Olszens K.R., Song J., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Hill A.E., Burrage L.C., Olszens K.R., Song J., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Burrage L.C., Olszens K.R., Song J., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Olszens K.R., Song J., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Song J., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Conti D.V., Witte J.S., Lander E.S., Witte J.S., Lander E.S., Lander E.S., et al. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- Singer J.B., Hill A.E., Nadeau J., Lander E.S., Hill A.E., Nadeau J., Lander E.S., Nadeau J., Lander E.S., Lander E.S. Mapping quantitative trait loci for anxiety in chromosome substitution strains of mice. Genetics. 2005;169:855–862. doi: 10.1534/genetics.104.031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter N.B., Bustamante C.D., Chase K., Gray M.M., Zhao K., Zhu L., Padhukasahasram B., Karlins E., Davis S., Jones P.G., Bustamante C.D., Chase K., Gray M.M., Zhao K., Zhu L., Padhukasahasram B., Karlins E., Davis S., Jones P.G., Chase K., Gray M.M., Zhao K., Zhu L., Padhukasahasram B., Karlins E., Davis S., Jones P.G., Gray M.M., Zhao K., Zhu L., Padhukasahasram B., Karlins E., Davis S., Jones P.G., Zhao K., Zhu L., Padhukasahasram B., Karlins E., Davis S., Jones P.G., Zhu L., Padhukasahasram B., Karlins E., Davis S., Jones P.G., Padhukasahasram B., Karlins E., Davis S., Jones P.G., Karlins E., Davis S., Jones P.G., Davis S., Jones P.G., Jones P.G., et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade C.M., Kulbokas E.J., Kirby A.W., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J., Kulbokas E.J., Kirby A.W., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J., Kirby A.W., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J., Lander E.S., Lindblad-Toh K., Daly M.J., Lindblad-Toh K., Daly M.J., Daly M.J. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- Yang H., Bell T.A., Churchill G.A., de Pardo-Manuel Villena F., Bell T.A., Churchill G.A., de Pardo-Manuel Villena F., Churchill G.A., de Pardo-Manuel Villena F., de Pardo-Manuel Villena F. On the subspecific origin of the laboratory mouse. Nat. Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- Yonekawa H., Moriwaki K., Gotoh O., Watanabe J., Hayashi J.-I., Miyashita N., Petras M.L., Tagashira Y., Moriwaki K., Gotoh O., Watanabe J., Hayashi J.-I., Miyashita N., Petras M.L., Tagashira Y., Gotoh O., Watanabe J., Hayashi J.-I., Miyashita N., Petras M.L., Tagashira Y., Watanabe J., Hayashi J.-I., Miyashita N., Petras M.L., Tagashira Y., Hayashi J.-I., Miyashita N., Petras M.L., Tagashira Y., Miyashita N., Petras M.L., Tagashira Y., Petras M.L., Tagashira Y., Tagashira Y. Relationship between laboratory mice and the subspecies Mus musculus domesticus based on restriction endonuclease cleavage sites. Jpn. J. Genet. 1980;55:289–296. [Google Scholar]
- Yonekawa H., Takahama S., Gotoh O., Miyashita N., Moriwaki K., Takahama S., Gotoh O., Miyashita N., Moriwaki K., Gotoh O., Miyashita N., Moriwaki K., Miyashita N., Moriwaki K., Moriwaki K. Genetic diversity and geographic distribution of Mus musculus subspecies based on the polymorphism of mitochondria DNA. In: Moriwaki K., et al., editors. Genetics in wild mice: Its application to biomedical research. Japan Scientific Press/Karger; Tokyo: 1994. pp. 25–40. [Google Scholar]
- Yoshiki A., Moriwaki K., Moriwaki K. Mouse phenome research: Implications of genetic background. ILAR J. 2006;47:94–102. doi: 10.1093/ilar.47.2.94. [DOI] [PubMed] [Google Scholar]